Abstract
Vibrational spectroscopic mapping (point-by-point measurement) and imaging of biological samples (cells and tissues) covering Fourier-transform infrared (FTIR) and Raman spectroscopies has opened up many exciting new avenues to explore biochemical architecture and processes within healthy and diseased cells and tissues, including medical diagnostics and drug design.
Keywords: Biological tissues, Cells, Imaging, Mapping, Vibrational spectroscopy
Introduction
Vibrational spectroscopy, mapping and imaging
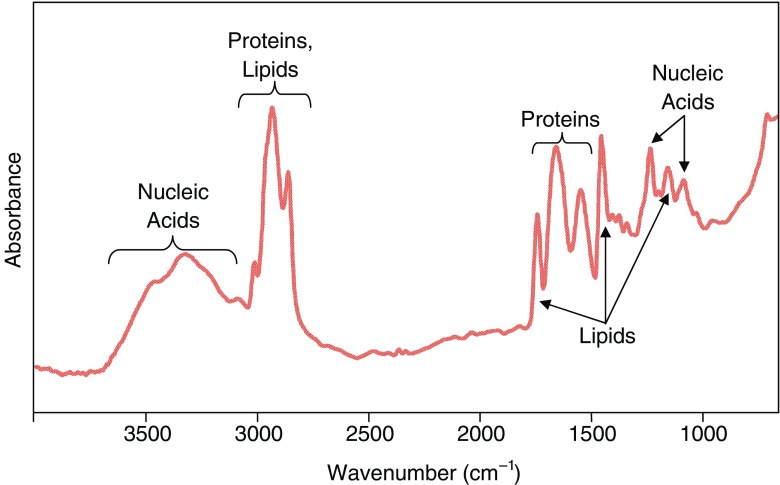
Vibrational spectroscopies measure signals due to molecular vibrations from chemical bonds exposed to infrared (IR) or, in the case of Raman spectroscopy, near infrared (NIR), visible or ultraviolet (UV) radiation. Signal intensities vary with the concentration and the nature of the functional groups in the molecule (primary structure) and its conformation (secondary structure). The latter two factors also dictate the energy of the vibrational spectroscopic bands. The mid-IR (MIR) energy (wavenumbers, cm−1) range contains bands due to vibrations of the major classes of biological molecules, such as carbohydrates, organophosphates, phospholipids and proteins, and provides considerable information about the structures and relative concentrations of these molecules within a biological sample (Miller and Dumas 2006). Typical spectra from breast tissues (Tam 2006) illustrate the positions of these characteristic bands (Fig. 1). False color images and/or maps of the content of biological molecules are generated by integrating the intensity or area of one or more of these bands, thereby illustrating the distribution of these biochemicals.
Fig. 1.
Typical Fourier-transform infrared (FTIR) spectrum from a sample of breast tissue. The spectrum illustrates the positions of bands from the major biochemical components of the tissue
The use of microprobe vibrational spectroscopic analysis of cells and tissues has undergone a rapid expansion in recent years because of the large amount of biochemical information that can be mined from changes in vibrational spectra across and between biological samples. This has been assisted by the availability of a raft of new techniques that are designed to obtain specific information. The differentiation of biological samples relies on subtle but multiple changes in the ratios and wavenumbers of bands that are objectively compared with chemometrics to remove sampling variability, rather than on absolute values that are subject to experimental conditions (Geladi 2003; Geladi et al. 2004; Levin and Bhargava 2005). Thus, while functional group maps provide useful information, the more objective discrimination of organelles or tissue type (and changes within these with treatments or disease states) requires the use of chemometric analyses.
A number of recent reviews (Bailo and Deckert 2008; Bhargava 2007; Boskey and Mendelsohn 2005; Chan et al. 2008; Gierlinger and Schwanninger 2007; Levin and Bhargava 2005; Moreira et al. 2008; Movasaghi et al. 2007; Petibois and Guidi 2008; Petter et al. 2009; Srinivasan and Bhargava 2007; Swain and Stevens 2007) have highlighted various aspects of vibrational spectroscopic mapping and the imaging of cells and tissues, as well as the use of complementary microscopic imaging/mapping (Aitken et al. 2009; Eichert et al. 2007; Miller et al. 2007). This review will discuss the techniques used in obtaining vibrational spectroscopic images and maps of cells and tissues, and the types of biochemical information that are available through the use of these techniques.
Vibrational spectroscopies used in the imaging and mapping of cells and tissues
Mapping versus imaging
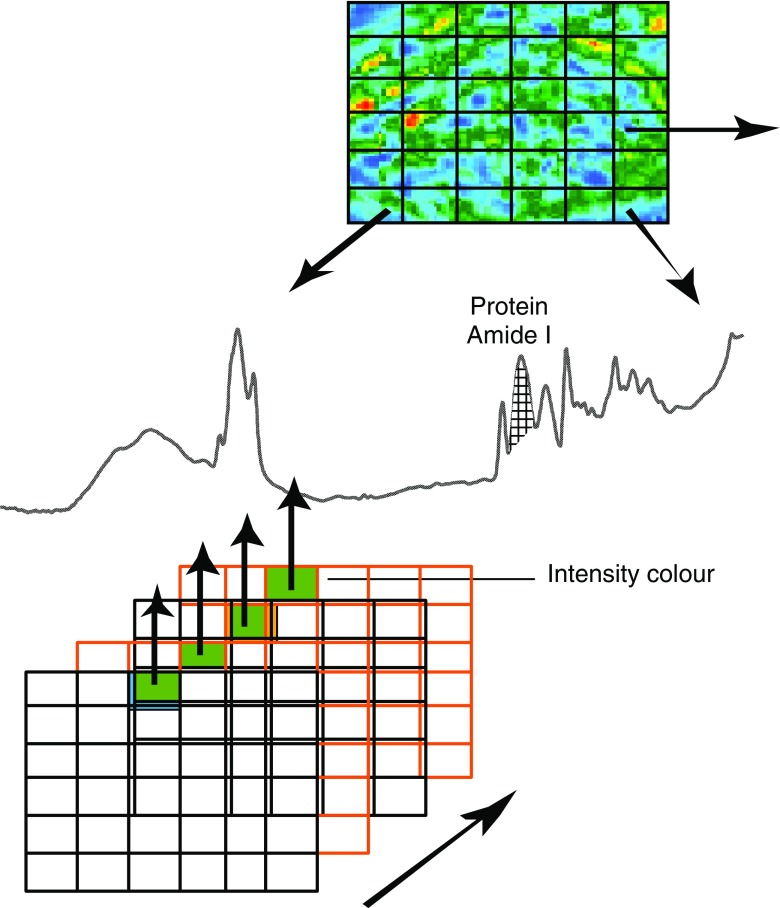
In mapping experiments, point-by-point spectra are measuring sequentially from a defined region in the sample as it is scanned, whereas in imaging experiments, multiple spectra are recorded simultaneously at different points using a focal plane array (FPA) detector in Fourier-transform IR (FTIR) spectroscopy (Fig. 2). Modern FTIR instruments have the capabilities for imaging relatively large areas (macro-imaging, 2 × 2 mm2), which can then be used to find areas of interest for high-spatial-resolution (5–10 μm) imaging of areas of around 170 × 170 μm2. Even larger areas can be imaged by producing a mosaic that stitches together a number of areas.
Fig. 2.
Focal-plane array FTIR imaging technique showing multiple grids. Each grid represents an image of the absorbances of the sample at a particular wavenumber. The spectrum of a point on the sample is represented by the line of a particular pixel through all the grids. The IR images can be constructed based on the intensity, area or relative diagnostic band ratios, resulting in a functional group map showing the distribution of specific band(s) of interest on the sample
Infrared and Raman spectroscopies
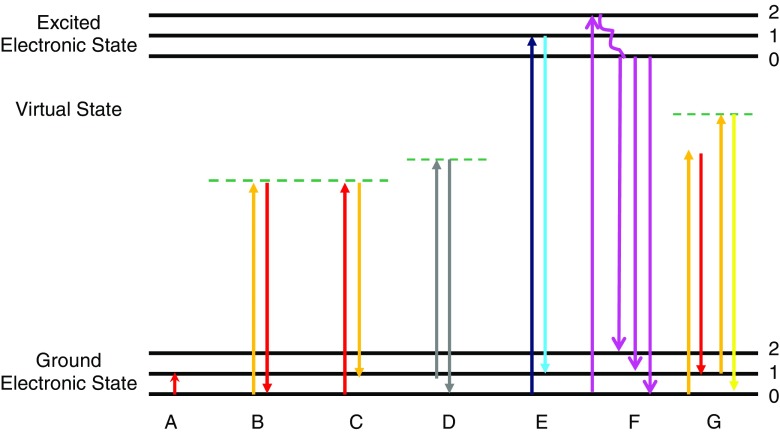
The basic phenomena involved in IR and Raman spectroscopies are outlined in Fig. 3. In IR spectroscopy, the sample is irradiated with polychromatic light, and a photon of light is absorbed when the frequency (energy) of the absorbed light matches the energy required for a particular bond to vibrate within the sample. In order for a vibration to be IR active, the molecular dipole moment must change during the vibration. The energy of mid-IR light provides a molecule with sufficient energy to vibrate but not enough energy to result in ionization or to break bonds and, consequently, there is no photodamage to the sample; local heating has been found to occur when using a synchrotron source, but it was reported to be too small (0.5°C) to be a significant problem (Martin et al. 2001). This enables other mapping/imaging spectroscopies to be performed on the same sample after IR imaging or mapping (Aitken et al. 2009). Signal (band) intensities vary with the concentration and the nature of functional groups in the molecule (primary structure) and with its conformation (secondary structure). The latter two factors also dictate the energy of the vibrational spectroscopic bands.
Fig. 3.
Schematic representations of infrared absorption (A), Rayleigh scattering (B), Stokes Raman scattering (C), anti-Stokes Raman scattering (D), resonance Raman scattering (E), fluorescence (F), coherent anti-Stokes Raman spectroscopy (CARS) (G). The numbers 0–2 represent different vibrational levels (νvib) within each electronic state
In Raman spectroscopy, the sample is irradiated with monochromatic light, i.e. UV, visible or NIR excitation, and the photons are either inelastically or elastically scattered. The inelastically scattered light, known as Raman scatter, has lost (Stokes) or gained (anti-Stokes) energy during this interaction, and the emitted photon contains information about the molecular structure of the sample. The elastically scattered light has the same energy as the incident laser light and is called Rayleigh scatter. Raman scattering is a very low probability process (except under specialist conditions discussed in subsequent paragraphs) and relies on lasers to produce enough photons to observe the weak signals. Under ambient conditions, the Boltzmann distribution of vibrational states has most molecules in their ground vibrational states. The Raman-scattered photons from the ground vibrational state have a lower energy than the incident photons, with energy differences that correspond to those of vibrational modes (Stokes scattering). Anti-Stokes Raman scattering occurs from vibrationally excited states that are thermally populated according to a Boltzmann distribution and lead to scattered photons that return the energy to the ground vibrational state. Because the thermal population of vibrational excited states is low under ambient conditions, anti-Stokes Raman scattering results in much weaker bands than does Stokes scattering. Hence, Stokes scattering is used in most mapping experiments.
Biological samples typically autofluoresce when irradiated with the higher energy (UV and visible) excitation wavelengths often used to collect Raman spectra. The fluorescence is generated from the many fluorophores that are contained within the sample. The high-energy incident radiation not only produces Raman scattered photons but also excites electronic states that can result in a strong fluorescence signal. This can lead to a strong and broad background that can swamp the Raman bands of the biological sample (Carter and Edward 2001); this process is illustrated in Fig. 3f. In order to avoid or reduce the likelihood of inducing fluorescence, which is a considerably more efficient process than Raman scattering, lower energy (i.e. longer wavelength) laser excitations are typically used, i.e., 785 or 1064 nm.
Fourier-transform IR spectroscopic mapping and imaging
All instruments used for IR spectroscopic mapping and imaging rely on Fourier-transform techniques, whereby the intensities of all signals from vibrational modes are produced simultaneously from a fast Fourier-transform conversion of an interferogram (as described below) into a spectrum. The spectrum is a plot of the intensity of signal versus the energy of the vibration (Griffiths and de Haseth 2007). An interferometer consists of a beamsplitter and two mirrors, one fixed and one moving. The beamsplitter divides the incoming polychromatic beam into two component beams, with one beam directed to the fixed mirror and the other to the moving mirror that moves a very short distance from the beamsplitter (the larger the distance, the higher the spectral resolution). The reflected beams recombine at the beamsplitter and the two beams interfere either constructively or destructively, depending on the distance the mirror has traveled, to produce the interferogram. In order to obtain the rapid acquisition and high sensitivity required for the mapping and imaging of biological samples, the distance that the moving mirror transverses is kept to a minimum, which means that the wavenumber resolution is usually around 4 cm−1.
The conventional source in benchtop instruments is a globar source, which produces an intense IR radiation over the entire mid-IR range. A synchrotron source relies on collecting the IR radiation component of synchrotron radiation that is emitted when the trajectory of an intense beam of electrons traveling close to the speed of light is changed by a bending magnet in the synchrotron ring (Aitken et al. 2009; Carr et al. 1995; Miller et al. 2007). Both sources produce a comparable number of IR photons per unit of time, but radiation from the globar source is widely dispersed, while the synchrotron source is highly focused. This means that the brightness of the IR radiation on the sample is at least two orders of magnitude higher at high spatial resolution (Carr et al. 1995; Miller et al. 2007), which has profound effects on the ways in which the measurements are performed with these different sources. For both techniques, the spatial resolution of maps and images are diffraction limited (typically 2–10 μm over the mid-IR range), with the spatial resolution becoming poorer with a decrease in the wavenumbers of the vibrational bands.
The mapping of biological samples with a benchtop globar source is slow because of the relatively weak IR intensity resulting from the use of apertures in the microscope to physically restrict the IR absorption to the sample area of interest (Levin and Bhargava 2005). The long time required for such measurements means that the spatial resolution is normally nowhere near the diffraction limit. As such, most benchtop measurements on biological samples require FPA detection (Fig. 2) whereby a large number of pixels are measured simultaneously, and many spectra are co-added to provide both high spatial resolution and good signal-to-noise ratios. This leads to images with spatial resolution that is close to the diffraction limit on biological samples, although FPA detection causes some complications associated with blurring of the image at this resolution. While such FPA detection has been of enormous benefit for the rapid imaging of micro- and macro-samples with benchtop instruments, its use with synchrotron–radiation sources is more problematic (Miller and Dumas 2006). This arises from the small synchrotron IR spot size, which means that only the inner detectors of the array have sufficient responses to be used in imaging. As discussed previously (Aitken et al. 2009), specialist array detectors have been designed that achieve an effective geometric pixel size of 0.54 × 0.54 μm2, compared to the 6.25 × 6.25 μm2 pixels that are the best case scenario for a globar. Such oversampling of the IR bandwidth results in an improvement in the deconvolution algorithms and, hence, the quality of the diffraction-limited images (Nasse et al. 2007), leading to rapid measurements of high-resolution maps of cells and tissues. Compressive imaging can also increase the speed of acquisition, thereby enabling sub-diffraction-limited spatial resolution (Gallet et al. 2008). A germanium attenuated total reflectance (ATR) sample accessory, when combined with a FPA detector, also achieves rapid sub-diffraction-limited spatial resolution of a few microns even with a benchtop instrument (Kazarian 2007; Ricci et al. 2007). A combination of these hardware and software developments coupled to a synchrotron source offers the possibility of rapid mapping with a spatial resolution of approximately 1 µm or less, which can be used to monitor changes in live single cells under different stimuli or drug treatment regimes. An important distinction needs to be made between the pixel size (best possible optical resolution) and spatial resolution, which may be limited by longer wavelengths in the spectrum and optical effects to yield a poorer measured lateral resolution (Bhargava 2007).
An emerging technique for obtaining maps with a spatial resolution of well below the diffraction limit is photothermal micro-spectroscopy (PTMS), alternatively known as PTTF (photothermal temperature fluctuation) (Hammiche et al. 1999; Pollock and Smith 2002). This technique combines FTIR and atomic force microscopy (AFM) using scanning thermal microscopy. The sub-micron temperature sensor is used to obtain IR spectra from sample regions that are well below the IR diffraction limit by determining the thermal response to IR radiation of different wavenumbers. Such a technique could ultimately lead to spatial resolutions of 20–30 nm, and the measurements are unaffected by IR opacity of the sample, which can be an important issue for other FTIR microscopies. The technique has now been applied to a variety of important biological problems, such as discrimination of human stem cells (Grude et al. 2007, 2009), monitoring of cell cycles (Hammiche et al. 2005) and the differentiation of exfoliative cervical cytology (Hammiche et al. 2007).
Raman spectroscopic microprobes
Fourier-transform-Raman spectroscopy uses the same optical and processing principles as FT-IR spectroscopy, but the wavelengths of radiation that are suitable for this optical detection and sampling mode are limited to the NIR. Thus, the advantages of fast acquisition using FT techniques are generally restricted to 1064-nm excitation using a Nd:YAG laser (Parker 1994). For biological samples, this has the disadvantages that the Raman effect diminishes rapidly with longer excitation wavelengths and the diffraction-limited spatial resolution is much worse than for UV/visual light (Vis) excitation wavelengths. Conversely, apart from the advantage of being able to use FT techniques with 1064-nm excitation, the lack of fluorescence, which often accompanies visible irradiation with biological samples and swamps the Raman spectrum, is an additional advantage. These advantages combined often outweigh the disadvantages and have resulted in the extensive use of FT-Raman studies on biological samples (Gierlinger and Schwanninger 2007; Moreira et al. 2008; Movasaghi et al. 2007; Parker 1994).
For other excitation wavelengths, dispersive measurement modes are used, which include normal Raman and resonance- and/or surface-enhanced Raman spectroscopies. For resonance enhancement, the absorbing photon is of sufficient energy to excite a chromophore into an electronic excited state, which results in a markedly increased probability that the scattered photon differs in energy by an amount that corresponds to the energy of a molecular vibration. This can increase the intensity of the Raman effect by up to six orders of magnitude and can, for instance, be used to selectively monitor heme proteins in biological samples using visible excitation into the Soret or Q bands (Wood et al. 2005) or selective enhancement of nucleic acids and certain aromatic amino acid residues in proteins by UV excitation (Neugebauer et al. 2007). This advantage, however, can often be outweighed by fluorescence and photodamage caused by such irradiation; thus, NIR excitation is also often used in dispersive Raman measurements with biological samples, albeit using shorter wavelength excitation than is used in FT-Raman spectroscopy, in order to enhance the Raman effect. The large variety of measurement modes available in Raman spectroscopy requires a judicious choice of excitation wavelength, and often more than one excitation wavelength is used to obtain complementary information.
For surface-enhanced Raman spectroscopy (SERS), roughened metal surfaces or nanoparticles of Cu, Ag or Au (and other metals) are used as substrates to obtain these enhancements (Smith 2008). Increases of many orders of magnitude in the Raman intensities can be achieved, and the surface can, for instance, be an electrode, which can enable studies of the response of the biomolecules to electrode potential (Bernad et al. 2007) or a nanoparticle for solution studies (Smith 2008). When combined with the resonance Raman effect to produce surface-enhanced resonance Raman scattering (SERRS), this Raman microscopic technique becomes an extremely sensitive probe (Smith 2008). It also has the advantage that the metal quenches fluorescence, so that a much larger range of excitation wavelengths can be used without the signal from the biological sample being swamped by fluorescence. The disadvantages of the technique are that surface enhancement is highest for the material directly attached to the metal surface and that the surface can affect the morphology (Smith 2008). This can result in incomplete information on the biochemical content of the whole sample or in spectroscopic data from samples that are further removed from a physiologically relevant environment.
The ability of Raman spectroscopy to use optical fiber probes also allows correlated simultaneous microscopies on a sample. For example, Raman microprobe fibre optics have been combined with many other techniques (Aitken et al. 2009; Williams et al. 2003) including AFM (Biju et al. 2007; Eronen et al. 2009), microdiffraction (Davies et al. 2009) and scanning electron microscopy (SEM), which is interfaced with the Raman spectrometer via a structural chemical analyzer allowing elemental and chemical information to be obtained from the same area of the same sample (Williams et al. 2003).
Certain combined microscopies can also include Raman enhancement techniques within the probe used for other microscopies, for example, the use of silicon nanowires by a vapor–liquid–solid (VLS) mechanism catalyzed by gold, which resulted in gold caps (droplets) of approximately 20–500 nm in diameter (Becker et al. 2008). These gold droplets can give rise to a large SERS effect that results in complementary high-resolution AFM and Raman mapping of small biological entities, such as bacterial cells or organelles within larger cells.
The SERS effect can also image organs in live animals by the use of particles consisting of a gold core, a Raman-active molecular layer and a silica coating as biotags to attach molecules that target the organ of interest within a living organism. The strong resonance-enhanced Raman signals from single-walled carbon nanotubes (SWNTs) also allow highly sensitive in vivo detection. As such, both SWNT nanoparticles and silica-coated biotags have been used to image small animals noninvasively using a 785-nm laser (Keren et al. 2008; Schipper et al. 2008).
The emergence of sensitive fast-scanning technologies, such as the Streamline Plus, have resulted in data acquisition that is orders of magnitude faster than was previously available (Aitken et al. 2009). This technology can be used to scan large areas of tissue, such as investigating tooth decay in a whole tooth cross-section (9 × 16 mm2), which was imaged in 58 min at a spatial resolution of 42 µm. Alternatively, small samples, such as single cells, can be scanned rapidly and at higher spatial resolution (Aitken et al. 2009). The combination of Streamline technology with SERRS using Ag nanoparticles incorporated into cells that also contain fluorophores allows rapid correlative fluorescence and Raman images/maps (Stokes et al. 2009). As discussed previously, the SERRS is not very sensitive to the presence of the fluorophores, unlike normal or resonance Raman spectroscopies. Thus, the greatly enhanced SERRS signal and the rapid scanning that is achievable with the Streamline capability enables low laser powers to be used so as to minimize cell damage and maximize the rapid collection of detailed biochemical information.
Coherent anti-Stokes Raman scattering (CARS) microscopy is the only technique relevant to biological mapping that monitors anti-Stokes Raman scattering, albeit a special type of anti-Stokes spectroscopy. The CARS technique uses multiple photons to produce a signal in which the emitted waves are coherent with one another to result in Raman spectra that are orders of magnitude stronger than spontaneous Raman emission (Fig. 3). As discussed previously (Aitken et al. 2009), it has been an important new development in microprobe mapping because it is sufficiently fast and non-destructive to enable live-cell mapping of metabolic processes and of the time-dependence of drug interactions with cells (Botvinick and Shah 2007; Djaker et al. 2007), In addition, improved sampling techniques have the potential to enable background-free CARS microprobe studies to follow a single molecule distribution in live-cell imaging in biological samples (Gachet et al. 2008; Serge et al. 2007). While the CARS technique has opened up exciting possibilities in live-cell imaging, the optical limitations of this multi-photon spectroscopy means it suffers from being able to only monitor one or two narrow vibrational spectral regions at the one time. Hence, it does not provide the more extensive biochemical information that is obtained from full spectral data at each point on the sample using other Raman and FTIR mapping and imaging methods.
Applications to biological systems
Because of the rapid expansion of biological applications of vibrational spectroscopic mapping and imaging techniques, it is not possible to provide an extensive review of recent research within the space limitations of this article, but an overview of the diversity of applications in medicine and biology, including medical diagnostics, studies of physiological and diseases processes and treatments of diseases and plant biology, can be found in a number of recent reviews (Bailo and Deckert 2008; Bhargava 2007; Boskey and Mendelsohn 2005; Chan et al. 2008; Gierlinger and Schwanninger 2007; Levin and Bhargava 2005; Moreira et al. 2008; Movasaghi et al. 2007; Petibois and Guidi 2008; Petter et al. 2009; Srinivasan and Bhargava 2007; Swain and Stevens 2007). As outlined in these reviews and the current review, the increase in the speed of acquisition, sensitivity, discrimination and spatial resolution of the emerging techniques of vibrational spectroscopic microscopies has greatly expanded the horizons of what is possible in terms of an explosion of new knowledge that is being generated. In the near future, this should lead to these techniques rivaling complementary fluorescence microscopy and electron microscopy as mainstay techniques for interrogation of the architecture and dynamics of biological systems at a molecular and cellular level.
Infrared imaging of a cribriform ductal carcinoma in situ
As an example of an application of imaging, we describe studies on a ductal carcinoma in situ (DCIS), which is a pre-invasive cancer localized within the breast ducts (Leonard and Swain 2004). As epithelial cells proliferate, the architecture of the duct or the terminal ductal-lobular unit (TDLU) of the breast alters (Burstein et al. 2004; Skinner 2003). The major difference between a low-grade DCIS and a normal or benign duct is the presence of tumor cells arranged in a cribriform architecture within the duct lumen (Burstein et al. 2004; Skinner 2003).
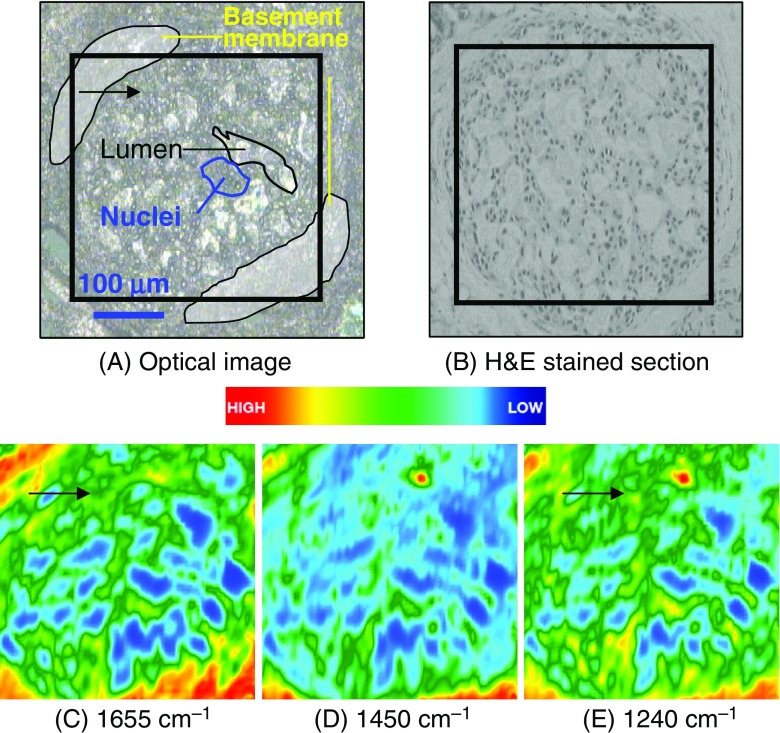
The optical images of an unstained cribriform DCIS (low-grade) deparaffinized formalin-fixed, paraffin-embedded (FFPE) section and an adjacent hematoxylin and eosin (H&E)-stained section are presented in Figs. 4a and 4b, respectively (Tam 2006). A selection of nuclei and duct openings are highlighted, and the box indicates the area imaged (350 × 350 μm2). The functional group maps were plotted using Cytospec (Lasch and Naumann 2006) and illustrate the protein (amide I), lipid and nucleic acid contents obtained from integration of bands centered at 1655, 1450 and 1240 cm−1, respectively (Figs. 4a–c). These maps can be used together with the unstained and H&E-stained sections to identify regions with a high density of protein and nucleic acids that can be correlated to regions of high cell proliferation. A normal breast duct should not contain any cells; any spectral evidence of biological components within the duct is, therefore, indicative of abnormalities.
Fig. 4.
Images of a 5-μm thick deparaffinized ormalin-fixed, paraffin-embedded ductal carcinoma in situ tissue section. Optical images of unstained (a) and adjacent hematoxylin and eosin-stained (b) sections. The area imaged using FTIR spectroscopy is marked by the black square. IR maps of proteins (c), lipids (d) and nucleic acids (e) are shown. Experimental conditions: 64 × 64 array (4096 spectra); 256 scans; 4 cm−1;×15 magnification; image area: 350 × 350 μm2
The protein (amide I) functional group map (Fig. 4c) shows that a network-shaped feature of medium intensity (green) is found within the center of the duct and is surrounded by another feature displaying a medium-to-high intensity (red) on the peripheral region. When compared with the H&E-stained section, the peripheral region is attributed to the basement membrane, while the network-shape feature is due to the tumor cells within the duct lumen arranged in a DCIS cribriform architecture.
Lipids are usually present in the breast in the form of adipose tissue and within the basement membrane (Dukor 2002; Jackson and Mantsch 2002). The presence of lipids in the duct (green in Fig. 4d) is due to the lipoprotein from the tumor cell membranes. The medium-to-high intensity of lipids on the periphery correlates with the protein map (Fig. 4c), which is attributed to the basement membrane. Confirmation that this region is that of basement membrane is made by comparison of the image with the H&E-stained section.
The unstained and H&E-stained cribriform DCIS sections clearly show there are tumor cells contained within the duct. The presence of nuclei was deduced from the intensities of the bands due to nucleic acids in the IR map. Fig. 4d reveals a medium intensity of bands due to nucleic acids within the duct lumen, which are indicative of cellular proliferation. The periphery, again, displays a medium-to-high intensity of bands due to nucleic acids, which have the histopathological characteristics of the basement membrane of the duct containing myoepithelial and cuboidal epithelial cells.
Collagen is present in the intra- and interlobular stromal tissues and has a number of IR absorptions that overlap with the bands due to the anti-symmetric and symmetric stretching vibrations of the phosphate modes of nucleic acids within the 1300–1000 cm−1 spectral region (Ci et al. 1999). Therefore, the medium-to-high intensity of nucleic acid bands, located on the periphery of the image, is likely to be associated with both the basement membrane and stromal tissues. Similarly, the high concentration also indicates areas of stroma and basement membrane that physically overlap. However, the difference between stroma and basement membranes is not obvious from the functional group maps.
A normal breast duct is composed of a bi-layer of myoepithelial and cuboidal epithelial cells that are surrounded by basement membranes (McKee 2002). The ducts and lobules are supported by the intra- and interlobular stromal tissues (McKee 2002). The arrows in Figs. 4c and e show the distribution of protein and nucleic acids that indicate a sieve-like architecture of cellular components present within the duct lumen, which is highly correlated with the adjacent H&E section (Fig. 4b) (McKee 2002). Although functional group maps provide information on the distribution of bands attributed to proteins, nucleic acids and lipids over the mid-IR region, direct tissue classification cannot be undertaken without more in-depth multivariate statistical analyses, as discussed in the section Vibrational spectroscopy, mapping and imaging.
Conclusions
The use of vibrational spectroscopic microprobes for the mapping and imaging of cells and tissues is undergoing a rapid expansion in the range of techniques, sampling procedures and applications that span from fundamental studies to clinical applications. The research results obtained from these rapidly evolving techniques are providing many new insights into biochemical architectures and processes, and are having a significant impact on the development of new treatments and diagnostics.
Acknowledgments
The breast cancer research was supported by a grant from The University of Sydney, Cancer Research Fund, the Australian Synchrotron Research Program, which is funded by the Commonwealth of Australia under the Major National Research Facilities Program for research conducted at NSSRC, and the Australian Synchrotron. The authors also thank Dr. Brian Reedy for the use of the FTIR imaging instrument at the University of Technology, Sydney. We are grateful to the ARC for funding of some of the research reported herein through RIEF and LIEF grants and ARC Discovery grants to PAL, including Australian Professorial Fellowships. We also thank Carolyn Mountford from the Institute for Magnetic Resonance Research, The University of Sydney, and Peter Russell from the Department of Anatomical Pathology, Royal Prince Alfred Hospital, for provision of the breast cancer samples used to obtain the spectra in Figs. 1, 2 and 4.
References
- Aitken JB, Carter EA, Eastgate H, et al (2009) Biomedical applications of X-Ray absorption and vibrational spectroscopic microscopies in obtaining structural information from complex systems. Radiat Phys Chem (in press). doi:10.1016/jradphyschem.2009.03.068
- Bailo E, Deckert V. Tip-enhanced Raman scattering. Chem Soc Rev. 2008;37:921–930. doi: 10.1039/b705967c. [DOI] [PubMed] [Google Scholar]
- Becker M, Sivakov V, Goesele U, et al. Nanowires enabling signal-enhanced nanoscale Raman spectroscopy. Small. 2008;4:398–404. doi: 10.1002/smll.200701007. [DOI] [PubMed] [Google Scholar]
- Bernad S, Leygue N, Korri-Youssoufi H, et al. Kinetics of the electron transfer reaction of Cytochrome c552 adsorbed on biomimetic electrode studied by time-resolved surface-enhanced resonance Raman spectroscopy and electrochemistry. Eur Biophys J. 2007;36:1039–1048. doi: 10.1007/s00249-007-0173-z. [DOI] [PubMed] [Google Scholar]
- Bhargava R. Towards a practical Fourier transform infrared chemical imaging protocol for cancer histopathology. Anal Bioanal Chem. 2007;389:1155–1169. doi: 10.1007/s00216-007-1511-9. [DOI] [PubMed] [Google Scholar]
- Biju V, Pan D, Gorby YA, et al. Combined spectroscopic and topographic characterization of nanoscale domains and their distributions of a redox protein on bacterial cell surfaces. Langmuir. 2007;23:1333–1338. doi: 10.1021/la061343z. [DOI] [PubMed] [Google Scholar]
- Boskey A, Mendelsohn R. Infrared analysis of bone in health and disease. J Biomed Opt. 2005;10(9):031102. doi: 10.1117/1.1922927. [DOI] [PubMed] [Google Scholar]
- Botvinick EL, Shah JV (2007) Laser-based measurements in cell biology. In: Botvinick EL, Shah JV (eds) Laser manipulation of cells and tissues, Elsevier, Amsterdam, pp 81–109 [DOI] [PubMed]
- Burstein H, Polyak K, Wong J, et al. Medical progress: ductal carcinoma in-situ of the breast. N Engl J Med. 2004;350:1430–1441. doi: 10.1056/NEJMra031301. [DOI] [PubMed] [Google Scholar]
- Carr GL, Hanfland M, Williams GP. Midinfrared beamline at the national synchrotron light source port U2B. Rev Sci Instrum. 1995;66:1643–1645. doi: 10.1063/1.1145870. [DOI] [Google Scholar]
- Carter EA, Edward HGM. Biological applications of Raman spectroscopy. In: Gremlich H-U, Yan B, editors. Infrared and Raman spectroscopy of biological materials. New York: Marcel Dekker; 2001. pp. 421–475. [Google Scholar]
- Chan J, Fore S, Wachsman-Hogiu S, et al. Raman spectroscopy and microscopy of individual cells and cellular components. Laser Photon Rev. 2008;2:325–349. doi: 10.1002/lpor.200810012. [DOI] [Google Scholar]
- Ci Y, Gao T, Feng J, et al. Fourier transform infrared spectroscopic characterization of human breast tissue: implications for breast cancer diagnosis. Appl Spectrosc. 1999;53:312–315. doi: 10.1366/0003702991946703. [DOI] [Google Scholar]
- Davies RJ, Burghammer M, Riekel C. A combined microRaman and microdiffraction set-up at the European Synchrotron Radiation Facility ID13 beamline. J Synchrotron Radiat. 2009;16:22–29. doi: 10.1107/S0909049508034663. [DOI] [PubMed] [Google Scholar]
- Djaker N, Gachet D, Sandeau N, Lenne P-F, Rigneault H. Refractive effects in coherent anti-Stokes Raman scattering microscopy. Appl Optics. 2007;45:7005–7011. doi: 10.1364/ao.45.007005. [DOI] [PubMed] [Google Scholar]
- Dukor R. Vibrational spectroscopy in the detection of cancer. In: Chalmers J, Griffiths P, editors. Handbook of vibrational spectroscopy. Chichester: John Wiley & Sons; 2002. pp. 3335–3361. [Google Scholar]
- Eichert D, Gregoratti L, Kaulich B, et al. Imaging with spectroscopic micro-analysis using synchrotron radiation. Anal Bioanal Chem. 2007;389:1121–1132. doi: 10.1007/s00216-007-1532-4. [DOI] [PubMed] [Google Scholar]
- Eronen P, Osterberg M, Jaaskelainen AS. Effect of alkaline treatment on cellulose supramolecular structure studied with combined confocal Raman spectroscopy and atomic force microscopy. Cellulose. 2009;16:167–178. doi: 10.1007/s10570-008-9259-8. [DOI] [Google Scholar]
- Gachet D, Billard F, Rigneault H. Focused field symmetries for background-free coherent anti-Stokes Raman spectroscopy. Phys Rev A. 2008;77:061802®)061801–061804 061802. doi: 10.1103/PhysRevA.77.061802. [DOI] [Google Scholar]
- Gallet J, Riley M, Hao Z, Martin MC. Increasing FTIR spectromicroscopy speed and resolution through compressive imaging. IR Physics Technol. 2008;51:420–422. [Google Scholar]
- Geladi P. Chemometrics in spectroscopy. Part 1. Classical chemometrics. Spectrochim Acta B. 2003;58:767–782. doi: 10.1016/S0584-8547(03)00037-5. [DOI] [Google Scholar]
- Geladi P, Sethson B, Nystrom J, et al. Chemometrics in spectroscopy. Part 2. Examples. Spectrochim Acta B. 2004;59:1347–1357. [Google Scholar]
- Gierlinger N, Schwanninger M. The potential of Raman microscopy and Raman imaging in plant research. Spectr-Int J. 2007;21:69–89. [Google Scholar]
- Griffiths PR, de Haseth JA. Fourier transform infrared spectrometry. New York: Wiley; 2007. [DOI] [PubMed] [Google Scholar]
- Grude O, Hammiche A, Pollock H, et al. Near-field photothermal microspectroscopy for adult stem-cell identification and characterization. J Microsc-Oxf. 2007;228:366–372. doi: 10.1111/j.1365-2818.2007.01853.x. [DOI] [PubMed] [Google Scholar]
- Grude O, Nakamura T, Hammiche A, et al. Discrimination of human stem cells by photothermal microspectroscopy. Vib Spectrosc. 2009;49:22–27. doi: 10.1016/j.vibspec.2008.04.008. [DOI] [Google Scholar]
- Hammiche A, Pollock HM, Reading M, et al. Photothermal FT-IR spectroscopy: A step towards FT-IR microscopy at a resolution better than the diffraction limit. Appl Spectrosc. 1999;53:810–815. doi: 10.1366/0003702991947379. [DOI] [Google Scholar]
- Hammiche A, German MJ, Hewitt R, et al. Monitoring cell cycle distributions in MCF-7 cells using near-field photothermal microspectroscopy. Biophys J. 2005;88:3699–3706. doi: 10.1529/biophysj.104.053926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammiche A, Walsh MJ, Pollock HM, et al. Non-contact micro-cantilevers detect photothermally induced vibrations that can segregate different categories of exfoliative cervical cytology. J Biochem Biophys Methods. 2007;70:675–677. doi: 10.1016/j.jbbm.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Jackson M, Mantsch HH. Pathology by infrared and Raman spectroscopy. In: Chalmers J, Griffiths P, editors. Handbook of vibrational spectroscopy. Chichester: John Wiley & Sons; 2002. pp. 3227–3245. [Google Scholar]
- Kazarian SG. Enhancing high-throughput technology and microfluidics with FTIR spectroscopic imaging. Anal Bioanal Chem. 2007;388:529–532. doi: 10.1007/s00216-007-1193-3. [DOI] [PubMed] [Google Scholar]
- Keren S, Zavaleta C, Cheng Z, et al. Noninvasive molecular imaging of small living subjects using Raman spectroscopy. Proc Natl Acad Sci USA. 2008;105:5844–5849. doi: 10.1073/pnas.0710575105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasch P, Naumann D. Spatial resolution in infrared microspectroscopic imaging of tissues. Biochim Biophys Acta Biomembranes. 2006;1758:814–1729. doi: 10.1016/j.bbamem.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Leonard G, Swain S. Ductal carcinoma in-situ, complexities and challenges. J Natl Cancer Inst. 2004;96:906–920. doi: 10.1093/jnci/djh164. [DOI] [PubMed] [Google Scholar]
- Levin IW, Bhargava R. Fourier transform infrared vibrational spectroscopic imaging: Integrating microscopy and molecular recognition. Annu Rev Phys Chem. 2005;56:429–474. doi: 10.1146/annurev.physchem.56.092503.141205. [DOI] [PubMed] [Google Scholar]
- Martin MC, Tsvetkova NM, Crowe JH, et al. Negligible sample heating from synchrotron infrared beam. Appl Spectrosc. 2001;55:111–113. doi: 10.1366/0003702011951551. [DOI] [Google Scholar]
- McKee G. Cytopathology of the breast. Boston: Oxford University Press; 2002. [Google Scholar]
- Miller LM, Dumas P. Chemical imaging of biological tissues with synchrotron infrared light. Biochim Biophys Acta. 2006;1758:846–857. doi: 10.1016/j.bbamem.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Miller LM, Wang Q, Smith RJ, et al. A new sample substrate for imaging and correlating organic and trace metal composition in biological cells and tissues. Anal Bioanal Chem. 2007;387:1705–1715. doi: 10.1007/s00216-006-0879-2. [DOI] [PubMed] [Google Scholar]
- Moreira LM, Silveira L, Santos FV, et al. Raman spectroscopy: A powerful technique for biochemical analysis and diagnosis. Spectroscopy. 2008;22:1–19. [Google Scholar]
- Movasaghi Z, Rehman S, Rehman IU. Raman spectroscopy of biological tissues. Appl Spectrosc Rev. 2007;42:493–541. doi: 10.1080/05704920701551530. [DOI] [Google Scholar]
- Nasse MJ, Reininger R, Kubala T, et al. Synchrotron infrared microspectroscopy imaging using a multi-element detector (IRMSI-MED) for diffraction-limited chemical imaging. Nucl Instr Methods A. 2007;582:107–110. doi: 10.1016/j.nima.2007.08.073. [DOI] [Google Scholar]
- Neugebauer U, Schmid U, Baumann K, et al. Towards a detailed understanding of bacterial metabolism—spectroscopic characterization of Staphylococcus epidermidis. ChemPhysChem. 2007;8:124–137. doi: 10.1002/cphc.200600507. [DOI] [PubMed] [Google Scholar]
- Parker SF. A review of the theory of Fourier-transform Raman-spectroscopy. Spectrochim Acta [A] 1994;50:1841–1856. [Google Scholar]
- Petibois C, Guidi MC. Bioimaging of cells and tissues using accelerator-based sources. Anal Bioanal Chem. 2008;391:1599–1608. doi: 10.1007/s00216-008-2157-y. [DOI] [PubMed] [Google Scholar]
- Petter CH, Heigl N, Rainer M, et al. Development and application of Fourier-transform infrared chemical imaging of tumour in human tissue. Curr Med Chem. 2009;16:318–326. doi: 10.2174/092986709787002664. [DOI] [PubMed] [Google Scholar]
- Pollock H, Smith DA. The use of near-field probes for vibrational spectroscopy and photothermal imaging. In: Chalmers JM, Griffiths PR, editors. Handbook of vibrational spectroscopy. Chichester: John Wiley & Sons; 2002. pp. 1472–1492. [Google Scholar]
- Ricci C, Bloxham S, Kazarian SG. ATR-FTIR imaging of albumen photographic prints. J Cult Herit. 2007;8:387–395. doi: 10.1016/j.culher.2007.07.002. [DOI] [Google Scholar]
- Schipper ML, Nakayama-Ratchford N, Davis CR, et al. A pilot toxicology study of single-walled carbon nanotubes in a small sample of mice. Nat Nanotechnol. 2008;3:216–221. doi: 10.1038/nnano.2008.68. [DOI] [PubMed] [Google Scholar]
- Serge A, Bertaux N, Rigneault H, Marguet D (2007) Improved single molecule detection and tracing algorithms for the generation of a dynamic map of membrane diffusion in living cells. Biophys J Suppl S: 91A Supplement: Suppl. S
- Skinner KA. The clinical management of ductal carcinoma in-situ, lobular carcinoma in-situ and atypical hyperplasia of the breast. Sydney: National Breast Cancer Centre; 2003. [Google Scholar]
- Smith WE. Practical understanding and use of surface enhanced Raman scattering/surface enhanced resonance Raman scattering in chemical and biological analysis. Chem Soc Rev. 2008;37:955–964. doi: 10.1039/b708841h. [DOI] [PubMed] [Google Scholar]
- Srinivasan G, Bhargava R. Fourier transform-infrared spectroscopic imaging: The emerging evolution from a microscopy tool to a cancer imaging modality. Spectroscopy. 2007;22:30–43. [Google Scholar]
- Stokes RJ, McKenzie F, McFarlane E, et al. Rapid cell mapping using nanoparticles and SERRS. Analyst (Lond) 2009;134:170–175. doi: 10.1039/b815117b. [DOI] [PubMed] [Google Scholar]
- Swain RJ, Stevens MM. Raman microspectroscopy for non-invasive biochemical analysis of single cells. Biochem Soc Trans. 2007;35:544–549. doi: 10.1042/BST0350544. [DOI] [PubMed] [Google Scholar]
- Tam KK (2006) A non-destructive approach for breast cancer diagnosis and pathological strategy using infrared and Raman spectroscopy. PhD thesis. The University of Sydney, Sydney
- Williams K, Bennett R, Brooker A, et al. New methods in Raman spectroscopy—combining other microscopes. Microsc Microanal. 2003;9:1094–1095. [Google Scholar]
- Wood BR, Hammer L, Davis L, et al. Raman microspectroscopy and imaging provides insights into heme aggregation and denaturation within human erythrocytes. J Biomed Opt. 2005;10:014005. doi: 10.1117/1.1854678. [DOI] [PubMed] [Google Scholar]