Abstract
Wherever nanoparticles (NPs) come in contact with a living organism, physical and chemical interactions take place between the surfaces of the NPs and biomatter, in particular proteins. When NP are exposed to biological fluids, an adsorption layer of proteins, a “protein corona” forms around the NPs. Consequently, living systems interact with the protein-coated NP rather than with a bare NP. To anticipate biological responses to NPs, we thus require comprehensive knowledge of the interactions at the bio–nano interface. In recent years, a wide variety of biophysical techniques have been employed to elucidate mechanistic aspects of NP–protein interactions. In this brief review, we present the latest findings regarding the composition of the protein corona as it forms on NPs in the blood stream. We also discuss molecular aspects of this adsorption layer and its time evolution. The current state of knowledge is summarized, and issues that still need to be addressed to further advance our understanding of NP–protein interactions are identified.
Keywords: Nanoparticles, Protein corona, Nanoparticle–protein interactions, Nanoparticle imaging, Nanoparticle spectroscopy, Nanoparticle toxicity
Introduction
The use of nanoparticles (NPs) and nanomaterials in general in both scientific (Ye-Qin et al. 2008) and commercial applications is continuously expanding (Aitken et al. 2006; Anselmann 2001; OECD 2008; Roco 2008). As a consequence, there is an unavoidable increasing release and accumulation of NPs in the environment (Handy et al. 2008a; Kreyling et al. 2004; Owen and Depledge 2005; Wiesner et al. 2006), leading to an unintentional but nevertheless growing human exposure. A profound risk assessment of nanotechnology is overdue (Abbas et al. 2010; Barnard 2006; Keller 2007; Maynard et al. 2006; Nel et al. 2006; Service 2006), particularly so as the current level of our understanding of interactions of nanomaterials with biomatter is still incomplete (Abbas et al. 2010; Barnard 2006; Keller 2007; Maynard et al. 2006; Nel et al. 2006; Service 2006), largely due to a lack of mechanistic knowledge (Abbas et al. 2010; Handy et al. 2008b). Even in the case of intended therapeutic applications of NPs, knowledge of the risk potential is still insufficient (Linse et al. 2007; Lunov et al. 2011, 2010).
In recent years, the interaction of NPs with biological systems has become one of the most intriguing areas of basic and applied research at the interface between the physical and life sciences (Fillafer et al. 2009; Watari et al. 2009). As NPs are of similar size as typical cellular components and proteins, they may evade the natural defenses of biological organisms, utilize the endocytosis machinery for intruding cells and, thereby, lead to permanent cell damage (Asha Rani et al. 2008; Lunov et al. 2010). While many studies have been performed using cell biology or toxicology approaches, recent work has shed light on the molecular aspects of the biological action of NPs. The application of sophisticated techniques of molecular biophysics may facilitate the elucidation of the biomolecular interactions involved and, in the long run, reveal the fundamental factors governing the biological effects of NPs.
Wherever NPs come in contact with biological systems, physical and chemical interactions take place between the surfaces of the NPs and biological components (e.g., proteins, membranes, phospholipids, endocytic vesicles, organelles, DNA, etc.)—the so-called “bio–nano interface.” It is well established that, upon the exposure of an organism to NPs, proteins from body fluids bind to NP surfaces (Cedervall et al. 2007a, b; De Paoli Lacerda et al. 2010; Lundqvist et al. 2008; Lynch 2007; Röcker et al. 2009), resulting in living systems actually interacting with protein-coated NPs (Cedervall et al. 2007b; Klein 2007). This so-called “protein corona” that forms around the NPs (Cedervall et al. 2007b; Klein 2007; Röcker et al. 2009; Treuel et al. 2010) largely defines the biological identity of the NP, and the efficiency of this interaction can be a decisive factor driving the biological response of an organism to NP exposure (Leszczynski 2010). Nel and co-authors (Nel et al. 2009) have presented an in-depth discussion of the basic physical interactions happening at the bio–nano interface. Table 1 summarizes the relevant forces responsible for the interfacial interactions between nanomaterials and biosystems.
Table 1.
Main forces governing the interfacial interaction between nanomaterials and biological systems (adapted from Nel et al. 2009)
| Force | Origin and Nature | Range (nm) | Possible impact on the interface |
|---|---|---|---|
| Hydrodynamic interactions | Convective drag, shear, lift, and Brownian diffusion are often influenced at nanoscale separations between interacting interfaces | 102–106 | Increases the collision frequencies between NPs and other surfaces responsible for transport |
| Electrostatic interactions | Charged interfaces attract counter-ions and repel co-ions (Coulomb forces) giving rise to the formation of an electrostatic double layer | 1–100 | Overlapping double layers are generally repulsive for equally charged objects but can be attractive for oppositely charged objects |
| Electrodynamic interactions | Van der Waals (vdW) interactions | 1–100 | Attractive in aqueous media, substantially smaller in biological media |
| Solvent interactions | Lyophilic materials interact favorably with solvent molecules | 1–10 | Lyophilic materials are thermodynamically stable in the solvent and do not aggregate |
| Lyophobic materials interact unfavorably with solvent molecules | Lyophobic materials are spontaneously expelled from the bulk of the solvent and forced to minimize the contact surface. | ||
| Steric interactions | Polymeric species adsorbed onto NPs give rise to repulsive interactions with other interfaces | 1–100 | Generally increase stability of individual NPs but can interfere in cellular uptake, especially when surface polymers are highly water-soluble |
| Polymer bridging interactions | Polymeric species adsorbed to inorganic NPs containing charged functional groups can be attracted by oppositely charged moieties on a substrate surface. | 1–100 | Generally promote aggregation, particularly when charge functionality is carboxylic acid and dispersed in aqueous media containing calcium ions. |
NPs, Nanoparticles
Understanding the formation and persistence of the protein corona is a complex task and of great importance for the elucidation, interpretation, and assessment of the biological effects of NPs. The formation process is essentially a competition of proteins and other biomolecules for binding to the NP surface. The notion of “hard” and “soft” protein coronae has been introduced (Cedervall et al. 2007b), and it is believed that the “soft” corona forms on short time scales (seconds to minutes) and evolves to a “hard” corona over incubation times of the order of hours (Monopoli et al. 2011).
An important aspect of protein adsorption onto NP surfaces is that structural changes of the protein may occur, giving rise to altered protein conformations (Aubin-Tam and Hamad-Schifferli 2005; Medintz et al. 2004; Roach et al. 2006; Zhou et al. 1997). Associated with this can be the loss of function (Rodriguez et al. 2005; Vertegel et al. 2004). Structural changes in the protein upon adsorption onto the NP surface may well lead to the exposure of novel “cryptic” peptide epitopes (Klein 2007; Lynch et al. 2006), altered function, and/or avidity effects (Cedervall et al. 2007b; Linse et al. 2007; Lundqvist et al. 2008). When a protein containing cryptic epitopes is denatured on a particle surface, the exposure of new antigenic sites may elicit an immune response (Baron et al. 1999; Brandes et al. 2006), which, if launched against a self-protein, could promote autoimmune diseases (Nel et al. 2009). However, the driving forces and mechanistic details of protein unfolding at NP surfaces remain still elusive (Des Rieux et al. 2006; Liu et al. 2009; Nel et al. 2009; Yan et al. 2009). Fortunately, there are many experimental techniques available for studying the structure of proteins in solution and in NP–protein aggregates (Havel 1996), including circular dichroism (Greenfield 1999; Kelly et al. 2005; Shang et al. 2007), Fourier transform infrared spectroscopy (Chittur 1998; Wang et al. 2012; Zhang and Yan 2005), Raman spectroscopy (RS)/surface-enhanced RS (SERS) (Schlücker 2008; Shao et al. 2009), and fluorescence spectroscopy (Mátyus et al. 2006; Royer 2006), size-exclusion chromatography (Carpenter et al. 2010; Mori and Barth 1999; Printz and Friess 2012; Vogt et al. 2008; Wiedenmann et al. 2004, 2002), isothermal titration calorimetry (ITC) (Baier et al. 2011; Nienhaus 2005), and surface plasmon resonance (SPR) (Cedervall et al. 2007b; Cheng et al. 2011).
Another issue that requires further attention is if and how different NPs are degraded in a biological environment. Degradation may not only remove the protein corona on the NP, it may also modify or remove the original surface functionality, and it may ultimately lead to an exposure of the NP core and even its complete dissolution. These processes can induce toxic effects through the release of molecular species or metal ions into the biological environment. In many cases (e.g., heavy metal ions, silver ions, etc.), the adverse effects of metal ions are well known, and the toxicity of degrading the NPs will generally be a combination of ionic/molecular toxicity and toxicity aspects related to the particulate nature of the material.
The physical and chemical characteristics of NPs determine their interactions with the surrounding medium by promoting the adsorption of ions, proteins, natural organic materials, and detergents, particle dissolution, or even by allowing the free surface energy to be minimized by surface restructuring (Gilbert et al. 2004; Min et al. 2008; Nel et al. 2009). The effect of degradation on these parameters should always be considered whenever the biological impact of the NPs is assessed. These characteristics can also be changed upon protein adsorption and so may influence the internalization by biological systems (Kreyling et al. 2004; Oberdörster et al. 2005b). In addition, even though the as-synthesized NPs may be rather insoluble in aqueous solvents, interactions with proteins may dramatically change this behavior. Detailed knowledge of the mechanistic steps involved in these processes is still scarce (Geiser et al. 2005; Rothen-Rutishauser et al. 2005).
The overall protein concentrations in typical body fluids (e.g., blood, lung, gut) and intracellular environments can be as high as 0.35 g mL-1 (Klein 2007). These fluids may contain more than 3,000 different proteins (Jeong et al. 2009) at widely differing concentrations. While highly abundant proteins will likely dominate the protein corona at early times after exposure, proteins with a lower abundance but higher affinities might prevail on longer time scales (Cedervall et al. 2007b; Nel et al. 2009).
In this brief review, we present recent findings on the composition of the protein corona in human blood plasma. We also focus on its time evolution and the mechanistic aspects of its formation. Finally, the current level of understanding is discussed, and remaining issues as well as open questions are identified.
Composition of the protein corona
To understand the mechanistic aspects of protein corona formation, knowledge of the composition of the protein corona and the factors governing its formation within the physiological context is required. NPs intended for biomedical applications are frequently administered by intravenous injection and thus exposed to blood. In a recent work, Tenzer and co-workers (Tenzer et al. 2011) determined the detailed composition of the strongly bound, “hard” protein corona upon exposure of the NP to blood plasma. To achieve this, they exposed monodisperse amorphous silica nanoparticles (SiNPs) to blood plasma and subsequently quantitatively analyzed the proteins associated with the SiNPs using liquid chromatography mass spectrometry (MS), one- and two-dimensional gel electrophoresis, and immunoblotting the composition of the protein corona. To observe a possible effect of NP size, they used SiNPs with three different diameters (20, 30, and 100 nm).
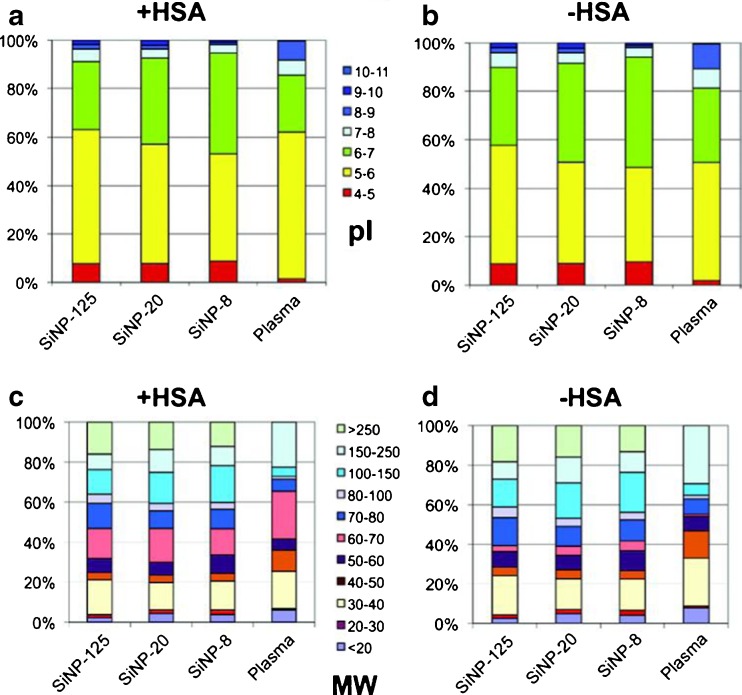
Figure 1 shows the SiNP-specific protein signatures identified by quantitative MS with a classification of proteins by their calculated isoelectric point. Negatively charged proteins (pI < 7) preferentially bound to the SiNPs at pH 7.3, i.e., the physiological pH of blood plasma (Fig. 1a, b). Moreover, proteins with high molecular mass were significantly enriched on the NP surfaces, whereas proteins with a low molecular mass were less abundant than in the plasma (Fig. 1c, d).
Fig. 1.
Composition of the protein coronae on silica nanoparticles (SiNPs) identified by quantitative mass spectrometry. Proteins were classified by their isoelectric points (pI) as calculated from the sequence information (a, b) as well as their molecular mass, or weight (MW) (c, d). a, b At pH 7.3 (pH measured in plasma) SiNPs preferentially bound negatively charged proteins (pI < 7). Proteins with pI < 5 were enhanced in the protein corona, independent of particle size, as compared with plasma. c, d A distinct, protein size-dependent particle binding pattern was absent. Proteins with low molecular mass were less abundant in the corona than in the plasma, whereas large proteins which were almost undetectable in plasma were significantly enriched in the corona. The observed patterns remained unchanged when the most abundant plasma protein, human serum albumin (HSA; b, d), was omitted from the data analysis. (Figure reproduced with kind permission from Tenzer et al. 2011)
Further analysis revealed that the binding of 37% of the identified corona proteins was significantly affected by particle size. Even a change in particle size from 20 to 30 nm was observed to significantly influence the composition of the NP corona. There were proteins with enhanced binding to larger (100 nm) SiNPs (e.g., prothrombin or the actin regulatory protein gelsolin), but also proteins with higher affinities for smaller (20 nm) particles (e.g., clusterin). However, the adsorption of many proteins, including immunoglobulin (IgG) or actin showed no dependence on NP size. These results study clearly show that a single physical parameter, i.e., the particle size, can affect the composition of the protein corona around chemically identical particles and underline the complexity of protein corona formation.
The composition of the protein corona around silica NPs and, for comparison, sulfonated polystyrene NPs, has also been studied by Monopoli and co-workers at different plasma concentrations (Monopoli et al. 2011). By using differential centrifugal sedimentation, dynamic light scattering, and zeta-potential measurements, these researchers analyzed protein corona formation both in situ, i.e., in the presence of plasma, and after spinning down, separating, and washing the NP–protein complexes. They also semi-quantitatively determined the hard corona composition using one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrospray liquid chromatography-tandem MS. They reported that the structures of the NP protein complexes from in situ studies were almost identical to those found in vitro after isolation from excess plasma, indicating a high biological relevance of the in vitro findings. Interestingly, an essentially complete NP surface coverage with proteins was inferred even at very low plasma concentrations. The authors claimed that protein binding was essentially irreversible on the time scale of their experiments, although a progressive displacement of proteins with a lower affinity in favor of those with a higher affinity was also suggested. Apparently, a detailed physicochemical picture of protein adsorption is direly needed to further our understanding of the temporal aspects of protein corona formation, i.e., which processes are irreversible and which are in dynamic equilibrium on the relevant time scales.
The time evolution of the protein corona
To address the intriguing problem of the time dependence of protein corona formation, Lundqvist and co-workers (Lundqvist et al. 2011) studied the evolution of the protein corona following the transfer of silica, polystyrene, and carboxyl-modified polystyrene NPs from plasma to cytosolic fluid. They reported a significant evolution of the corona on time scales of seconds. However, not all proteins were exchanged, and they concluded that the final corona contains a “fingerprint” of its history, an effect they suggested could be employed to trace biological transport of NPs.
The “hardening” of the protein corona, i.e., the development from a reversible to an irreversible adsorption layer, has also been investigated by Casals and co-workers (Casals et al. 2011). These researchers monitored the tendency of a variety of inorganic NPs (Au, Ag, Fe3O4, CoO, and CeO2) to adsorb proteins from serum-containing cell culture medium. Based on their measurements of surface charge, hydrodynamic diameter and the red shift of SPR spectra, they reported the formation of an “irreversible coating” or “hard” corona on Au and Ag NPs. Moreover, they probed a number of biological and toxicological implications of protein corona formation by exposing cells of the human monocytic cell line THP-1 to CoO nanoparticles, observing that the production of reactive oxygen species was decreased if the nanoparticles were pre-incubated with serum for 48 h. They also studied the time evolution of the protein corona around Au NPs with diameters of between 4 and 40 nm (Casals et al. 2010) and reported a “hardening” of the protein corona over time, accompanied by an increase of its thickness and a decrease of the net surface charge. An interesting aspect of this work was an increase in NP size following the mixing of NPs coated with a hard corona with bovine serum albumin-specific antibodies, whereas such an effect was absent upon the addition of a control antibody. This finding clearly shows the presence of albumin on the NP surface that is still capable of presenting a native epitope. Apparently, protein structural changes upon absorption were limited so that an antibody was still able to recognize its epitope.
Overall, the present state of knowledge suggests that further biophysical studies are needed with a higher time resolution to reveal the mechanistic details of the initial formation of the protein corona and its evolution over time. Importantly, changes in the structure of the proteins that occur upon adsorption to the NP surface have to be elucidated, including their time dependence. This knowledge is also important to assess the biological responses to NP exposure.
Mechanistic aspects of protein corona formation
Both the causes and effects of protein corona formation can only be thoroughly understood within a physicochemical framework on the molecular level. Experimental studies revealing the complex composition and time evolution of the protein corona in biological media are often descriptive and fall short of elucidating the mechanistic aspects responsible for the observed effects. Thermodynamic and kinetic studies of protein adsorption to NPs are required to understand the forces driving the process. To this end, a number of studies have been performed recently that shed light on the mechanisms responsible for forming and shaping the protein corona.
Shang and co-workers (Shang et al. 2009) studied the structure, thermodynamic and kinetic stability, and activity of cytochrome c (cyt c) on silica nanoparticles and provide evidence that the structure, function, and stability of cyt c adsorbed onto silica NPs strongly depend on NP size.
Cedervall et al. (2007b) introduced ITC as a suitable methodology for studying the affinity and stoichiometry of protein binding to a series of copolymer NPs if varying size and hydrophobicity. These researchers reported a strong dependence of protein adsorption on particle surface characteristics and size, but also noticed a strong protein specificity. They found higher association and dissociation rates for albumin and fibrinogen than for apolipoprotein A-I (ApoA-I) and various other plasma proteins. For albumin, they found shorter residence times on the more hydrophobic particles than on the more hydrophilic ones. A higher degree of surface coverage was observed on hydrophobic particles at equilibrium.
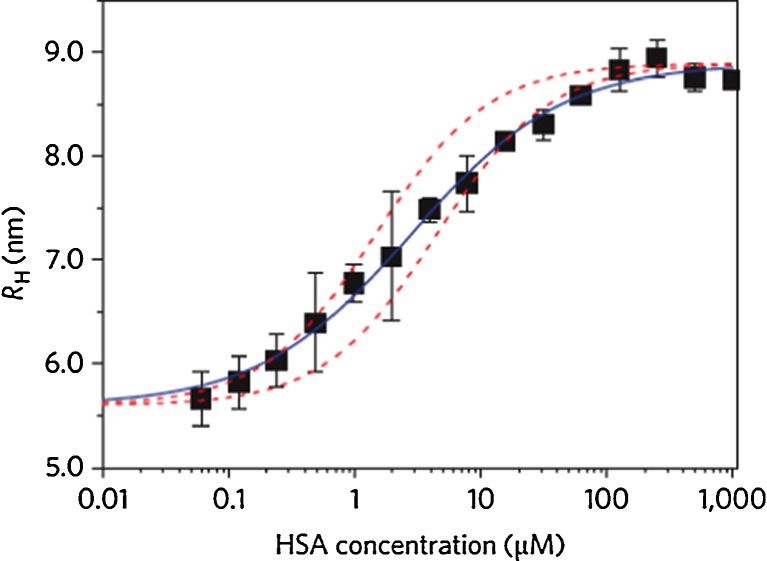
Nienhaus and co-workers quantitatively analyzed the adsorption of human serum albumin (HSA) onto small (diameter 10–20 nm) polymer-coated, fluorescence-labeled FePt and CdSe/ZnS NPs displaying carboxyl functions on the surface (Röcker et al. 2009). Using fluorescence correlation spectroscopy (FCS), they measured the thickness of the HSA corona formed around these NPs with sub-nanometer precision. Their result, 3.3 nm, confirmed that a single protein monolayer formed around the NP (Fig. 2). Apparent binding affinities associated with the NP–protein interaction were determined based on the response of the corona thickness to varying HSA concentration; the dissociation coefficients were found to be in the micromolar concentration range. Interestingly, HSA binding occurred with an anti-cooperative binding curve. Determination of these apparent binding affinities (transition midpoints) is very useful since these coefficients allow the strengths of the interactions between proteins and NPs to be quantified and data from different experimental techniques to be compared. However, a word of caution is in order here. These parameters are not proper equilibrium coefficients or affinities because the adsorption layer—or at least a significant fraction of it—is persistent and thus not appropriately described by a binding equilibrium. The fact that the protein concentration dependence of corona formation (Fig. 2) resembles an equilibrium binding curve when, in fact, it is not an equilibrium process is presently still a conundrum.
Fig. 2.
Formation of a HSA corona on polymer-coated iron–platinum (FePt) NPs as measured by fluorescence correlation spectroscopy (FCS). The (average) hydrodynamic radii (R H) of the NPs is plotted as a function of HSA concentration. The data points are averages from three independent sets of measurements. Blue solid line Fit of an anti-cooperative binding model to the data, red dashed lines Langmuir binding isotherms fitted to the first and last 20% of the transition to show the anti-cooperative behavior. (Figure reproduced with kind permission from Röcker et al. 2009)
The Nienhaus group recently extended their FCS studies to various other important plasma proteins. Transferrin was shown to bind to negatively charged FePt NPs with an affinity of approximately 26 μM (Jiang et al. 2010). The transferrin monolayer had a thickness of 7 nm. As for HSA, this value can be related to the molecular dimensions of the protein molecule as inferred from the crystal structure. By using confocal laser scanning fluorescence microscopy, the uptake of bare FePt NPs by live human cancer (HeLa) cells was compared to that of FePt NPs carrying a transferrin or HSA layer. The protein corona led to a significant reduction of the amount of NPs on the cell surface and in the cellular interior with respect to bare particles.
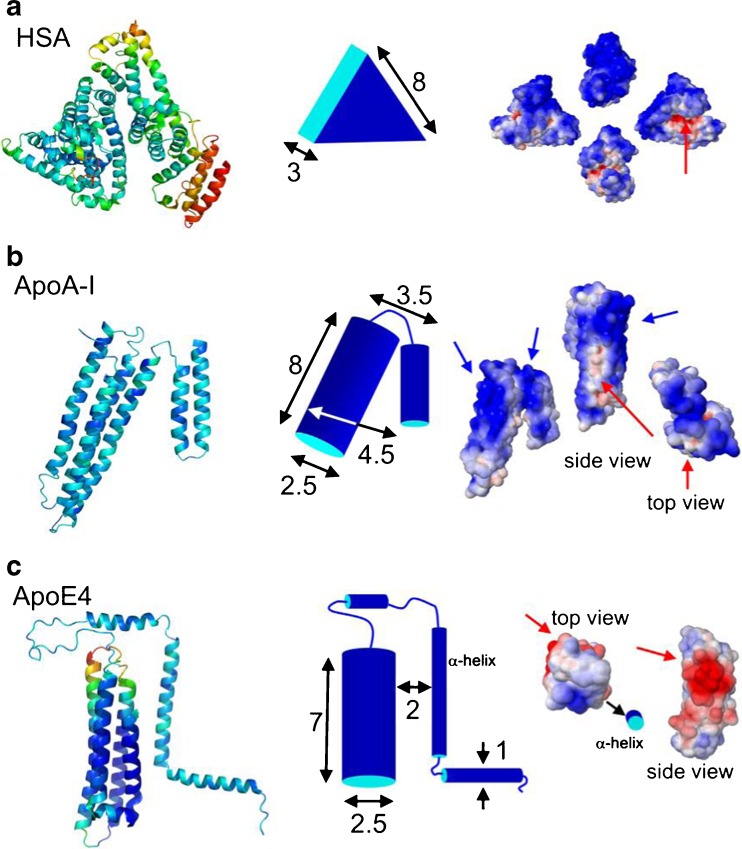
In a subsequent study, Maffre et al. (2011) employed a refined version of FCS, i.e., dual-focus (2f)FCS, to measure the adsorption of two human blood serum lipoproteins (ApoA-I and ApoE4) and, for reference, HSA, onto negatively charged FePt NPs carrying carboxyl groups on their surface. The formation of a protein corona was observed for all three proteins. The apparent binding affinities were observed to spread over almost four orders of magnitude, ranging from 0.021 μM for ApoE4 to >9.9 μM for HSA and to 140 μM for ApoA-I. An intriguing novel aspect of this work was the attempt to explain both the binding affinities and the monolayer thicknesses based on the known structural properties of the proteins (Fig. 3). The absolute increase in hydrodynamic radius due to corona formation was found to be well correlated with the molecular shapes of the proteins known from X-ray crystallography and solution experiments. The electrostatic properties of the proteins were computed by structure-based calculations of the surface potentials, and the ensuing analysis showed that the binding affinities were not governed by the net charge of the protein. Instead, they appear to be related to the presence of positively charged patches on the protein surface. From the locations of these patches, it was possible to qualitatively deduce the specific orientations of the corona proteins for optimal electrostatic interaction between the positively charged patches on the proteins and the negatively charged, carboxylated surfaces of the NPs. Binding in these orientations led to a protein corona with a thickness compatible with the observed increase in NP size. The intriguing revelation of a close structure–affinity relation by this work opens up a promising perspective for a better understanding of NP–protein interactions at the molecular level and, ultimately, even to a predictability of biological responses.
Fig. 3.
Structural depictions of HSA (a), apolipoprotein A-I (ApoA-I; b), and ApoE4 (c). Left column Cartoon representations of HSA [protein data bank accession (pdb) code 1AO6], apoE4 (pdb code 1GS9), and apoA-I (pdb code 2A01). For apoE4, only the molecular structure of the 22-K domain (4-helix bundle) is known. Center column Simplified structures of the proteins including approximate dimensions (in nm). Right column Space-filling models colored according to their surface electrostatics at pH 7.4 (blue negative potential, red positive potential; range −5 to +5 kT/e; calculated online using the website http://kryptonite.nbcr.net/pdb2pqr/. (Figure reproduced with kind permission from Maffre et al. 2011)
The physical and chemical nature of the NP surface controls not only the type of biomolecules interacting with it but also the strength of this interaction. Consequently, the effect of the chemical surface composition of the NPs on protein corona formation is an important issue. With a view toward biotechnology applications, it is of paramount interest to identify surface properties that can enhance or diminish the ability of NP surfaces to interact with proteins and other biomolecules. These properties mediate NP access to cells because it is the protein corona through which the cells initially interact with the NPs.
Treuel et al. (2010) used circular dichroism (CD) spectroscopy to study the thermodynamic and structural aspects of NP–protein interactions. A fundamental aspect of this work has been the elucidation of the influence of the surface composition of NPs on their interaction with proteins. These researchers determined dissociation constants for the interaction of serum albumin with citrate-coated Ag and Au NPs as well as polyvinylpyrrolidone-coated Ag NPs and polystyrene NPs (Treuel et al. 2010). Their results show apparent binding affinities stretching over three orders of magnitude, depending on the surface composition of the NPs, and underline the importance of the original surface functionality of the NPs, which determines the interaction with the adsorbed proteins. Moreover, their study emphasizes that the persistence of the surface functionality is an important parameter affecting protein corona formation in the physiological context. Only a stable molecular surface functionality on the NP surface under physiological conditions will be able to play a significant role in shaping the in vivo protein corona.
Shang et al. (2011a) recently measured protein adsorption onto ultra-small (diameter 1.4 ± 0.3 nm) Au nanoclusters (NCs) (Shang et al. 2011b, c) coated with dihydrolipoic acid (DHLA). They showed that protein binding resulted in a substantial increase in the fluorescence intensity of the NCs. Utilizing this effect, they measured the apparent binding affinities of four different proteins (HSA, transferrin, lysozyme, ApoE4) to their Au NCs (Fig. 4). All proteins were observed to bind with roughly micromolar apparent affinities (K D[HSA] = 0.9 μM, K D[transferrin] = 0.7 μM, K D[lysozyme] = 3.0 μM, K D[ApoE4] = 2.7 μM). An interesting aspect of their results is the observation of Hill coefficients n > 1 for their binding curves. However, this parameter may not necessarily reflect cooperativity because the experimental readout, namely, the fluorescence intensity, may not be proportional to the fraction of protein ligands bound to the NPs.
Fig. 4.
a Fluorescence emission spectra of HSA (curve A) and Au nanoclusters (NCs) in the presence of different concentrations of HSA (curves B–G), taken with excitation at 550 nm. b Photographs of dihydrolipoic acid–Au NCs in the presence of different concentrations of HSA [0 (A), 0.3 (B), 0.6 (C), 1.0 (D), 1.5 μM (E)], under a UV light source with wavelength 365 nm. c–f Fluorescence intensity of Au NCs, plotted as a function of the protein concentration in the solution: c HSA, d transferrin, e lysozyme, f ApoE4. Data points are averages of three independent series of measurements. Gray lines Fits to the data points using the adapted Hill equation. (Figure reproduced with kind permission from Shang et al. 2011a)
Current situation and outlook
Overall, the current state of knowledge indicates an urgent need for further biophysical studies of protein adsorption to engineered NPs on the molecular level. Experiments providing higher time resolution may help to unravel the mechanistic details of the initial formation of the protein corona and its evolution over time. The evolution from a weakly bound, or “soft”, to a “hard” protein corona in particular needs a mechanistic explanation. Also, the effects of the protein adsorption process on the protein structure and its possible time dependence has to be elucidated in more detail to better understand the biological effects of NP exposure.
In the future, the development of the protein corona under complex biological conditions, with proteins exchanging with a multitude of competing proteins, needs to be addressed. Large gaps still exist in our understanding of the fundamental physicochemical aspects of corona formation; however, even larger gaps exist in applying this knowledge to a realistic biological situation. Overall, the consequences of the protein corona and, hence, its properties on the biological behavior of NPs are still elusive and still poorly understood (Casals et al. 2010; Chithrani and Chan 2007; Ehrenberg et al. 2009; Oberdörster 2010). The surface properties of NPs can influence their cytotoxicity as well as their intracellular fate (Nativo et al. 2008), and the dependence of the cytotoxicity of NPs on size (Pan et al. 2007) and surface coating (Goodman et al. 2004; Harris et al. 2001; Niidome et al. 2006) has also been described in the literature (Abbas et al. 2010; Maynard et al. 2006; Oberdörster et al. 2005a; Poland et al. 2008). It has also been observed that NPs can translocate from the tissue where they have been absorbed to other target tissues, further adding to the complexity (Abbas et al. 2010; Oberdörster et al. 2005a; Semmler-Behnke et al. 2007).
The first examples of correlations between protein corona formation and biological responses have been shown. For example, immunoglobulin binding has been shown to lead to particle opsonization, thereby promoting receptor-mediated phagocytosis (Owens and Peppas 2006). Decreased protein absorption on injected polyethylene glycol-coated particles was found to lead to longer circulation times in the vascular system and altered bio-distribution (Kah et al. 2009; Owens and Peppas 2006). It has also been proposed that, because the proteins constituting the corona may be transported with the NPs across membranes, these proteins appear in biological compartments where they would normally not be present (Klein 2007).
Current findings underline the dependence of the composition of the protein corona in biological fluids on surface properties of the NPs. However, it is likely that some of the original surface properties may be modified in the biological environment, thereby illustrating the importance of the persistence of NP surface functionalities under biological conditions: only stable surface coatings can shape the formation of the “hard” corona by resisting removal from the surface in the early stages of corona formation.
While the surface properties of the NP and thereby also the protein corona will affect the uptake yield and delivery to certain cells and compartments, many particles will be decomposed on longer time scales. In fact, the biological action of internalized NPs may also result from the decomposition of its core, which is largely independent of surface functionality. Various studies of Ag NPs with and without polymer surface coatings have revealed that the formation of the protein corona strongly depends on the surface coatings around these NPs (Treuel et al. 2010), while their severe cytotoxicity arises from the release of Ag ions (Greulich et al. 2009; Kittler et al. 2010a, b).
While recent years have witnessed considerable progress toward a better understanding of the interactions at the bio–nano interface, there are still many open questions that need to be answered before the biological responses to NP exposure can be predicted. This knowledge, however, is necessary for a beneficial and safe use of nanomaterials in science and technology.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) through the Center for Functional Nanostructures (CFN) and the Priority Program SPP1313.
Conflict of interest
None.
Contributor Information
Lennart Treuel, Email: lennart.treuel@kit.edu.
Gerd Ulrich Nienhaus, Email: uli@uiuc.edu.
References
- Abbas K, Cydzik I, Del Torchio R, Farina M, Forti E, Gibson N, Holzwarth U, Simonelli F, Kreyling W. Radiolabelling of TiO2 nanoparticles for radiotracer studies. J Nanopart Res. 2010;12:2435–2443. doi: 10.1007/s11051-009-9806-8. [DOI] [Google Scholar]
- Aitken RJ, Chaudhry MQ, Boxall ABA, Hull M. Manufacture and use of nanomaterials: current status in the UK and global trends. Occup Med. 2006;56:300–306. doi: 10.1093/occmed/kql051. [DOI] [PubMed] [Google Scholar]
- Anselmann R. Nanoparticles and nanolayers in commercial applications. J Nanopart Res. 2001;3:329–336. doi: 10.1023/A:1017529712314. [DOI] [Google Scholar]
- Asha Rani PV, Low Kah Mun G, Hande MP, Valiyaveettil S. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano. 2008;3(2):279–290. doi: 10.1021/nn800596w. [DOI] [PubMed] [Google Scholar]
- Aubin-Tam M-E, Hamad-Schifferli K. Gold nanoparticle–cytochrome c complexes: the effect of nanoparticle ligand charge on protein structure. Langmuir. 2005;21:12080–12084. doi: 10.1021/la052102e. [DOI] [PubMed] [Google Scholar]
- Baier G, Costa C, Zeller A, Baumann D, Sayer C, Araujo PHH, Mailänder V, Musyanovych A, Landfester K. BSA adsorption on differently charged polystyrene nanoparticles using isothermal titration calorimetry and the influence on cellular uptake. Macromol Biosci. 2011;11:628–638. doi: 10.1002/mabi.201000395. [DOI] [PubMed] [Google Scholar]
- Barnard AS. Nanohazards: knowledge is our first defence. Nat Mater. 2006;5(4):245–248. doi: 10.1038/nmat1615. [DOI] [PubMed] [Google Scholar]
- Baron MH, Revault M, Servagent-Noinville S, Abadie J, Qui-Quampoix HJ. Chymotrypsin adsorption on montmorillonite: enzymatic activity and kinetic FTIR structural analysis. J Coll Interf Sci. 1999;214:319–332. doi: 10.1006/jcis.1999.6189. [DOI] [PubMed] [Google Scholar]
- Brandes N, Welzel PB, Werner C, Kroh LW. Adsorption-induced conformational changes of proteins onto ceramic particles: differential scanning calorimetry and FTIR analysis. J Coll Interf Sci. 2006;299:56–69. doi: 10.1016/j.jcis.2006.01.065. [DOI] [PubMed] [Google Scholar]
- Carpenter JF, Randolph TW, Jiskoot W, Crommelin DJA, Middaugh CR, Winter G. Potential inaccurate quantitation and sizing of protein aggregates by size exclusion chromatography: essential need to use orthogonal methods to assure the quality of therapeutic protein products. J Pharmaceut Sci. 2010;99(5):2200–2208. doi: 10.1002/jps.21989. [DOI] [PubMed] [Google Scholar]
- Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V. Time evolution of the nanoparticle protein corona. ACS Nano. 2010;4(7):3623–3632. doi: 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]
- Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes VF. Hardening of the nanoparticle–protein corona in metal (Au, Ag) and oxide (Fe3O4, CoO, and CeO2) nanoparticles. Small. 2011;7(24):3479–3486. doi: 10.1002/smll.201101511. [DOI] [PubMed] [Google Scholar]
- Cedervall T, Lynch I, Foy M, Berggård T, Donnelly SC, Cagney G, Linse S, Dawson KA. Detailed identification of plasma proteins adsorbed on copolymer nanoparticles. Angew Chem Int Ed. 2007;46:5754–5756. doi: 10.1002/anie.200700465. [DOI] [PubMed] [Google Scholar]
- Cedervall T, Lynch I, Lindman S, Berggård T, Thulin E, Nilsson H, Dawson KA, Linse S. Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc Natl Acad Sci USA. 2007;104(7):2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Wang M, Borghs G, Chen H. Gold nanoparticle dimers for plasmon sensing. Langmuir. 2011;27:7884–7891. doi: 10.1021/la200840m. [DOI] [PubMed] [Google Scholar]
- Chithrani BD, Chan WCW. Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett. 2007;7(6):1542–1550. doi: 10.1021/nl070363y. [DOI] [PubMed] [Google Scholar]
- Chittur KK. FTIR/ATR for protein adsorption to biomaterial surfaces. Biomaterials. 1998;19(4–5):357–369. doi: 10.1016/S0142-9612(97)00223-8. [DOI] [PubMed] [Google Scholar]
- De Paoli Lacerda SH, Park JJ, Meuse C, Pristinski D, Becker ML, Karim A, Douglas JF. Interaction of gold nanoparticles with common human blood proteins. ACS Nano. 2010;4(1):365–379. doi: 10.1021/nn9011187. [DOI] [PubMed] [Google Scholar]
- Des Rieux A, Fievez V, Garinot M, Schneider Y-J, Préat V. Nanoparticles as potential oral delivery systems of proteins and vaccines: a mechanistic approach. J Control Release. 2006;116:1–27. doi: 10.1016/j.jconrel.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Ehrenberg MS, Friedman AE, Finkelstein JN, Oberdörster G, McGrath JL. The influence of protein adsorption on nanoparticle association with cultured endothelial cells. Biomaterials. 2009;30(4):603–610. doi: 10.1016/j.biomaterials.2008.09.050. [DOI] [PubMed] [Google Scholar]
- Fillafer C, Friedl DS, Ilyes AK, Wirth M, Gabor F. Bionanoprobes to study particle-cell interactions. J Nanosci Nanotechnol. 2009;9:3239–3245. doi: 10.1166/jnn.2009.226. [DOI] [PubMed] [Google Scholar]
- Geiser M, Rothen-Rutishauser B, Kapp N, Schürch S, Kreyling W, Schulz H, Semmler M, Im Hof V, Heyder J, Gehr P. Ultrafine particles cross cellular membranes by nonphagocytic mechanisms in lungs and in cultured cells. Environ Health Perspect. 2005;113(11):1555–1560. doi: 10.1289/ehp.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert B, Huang F, Zhang H, Waychunas GA, Banfield JF. Nanoparticles: strained and stiff. Science. 2004;305(5684):651–654. doi: 10.1126/science.1098454. [DOI] [PubMed] [Google Scholar]
- Goodman CM, McCusker CD, Yilmaz T, Rotello VM. Toxicity of gold nanoparticles functionalized with cationic and anionic side chains. Bioconjug Chem. 2004;15:897–900. doi: 10.1021/bc049951i. [DOI] [PubMed] [Google Scholar]
- Greenfield NJ. Applications of circular dichroism in protein and peptide analysis. Trends Anal Chem. 1999;18(4):236–244. doi: 10.1016/S0165-9936(98)00112-5. [DOI] [Google Scholar]
- Greulich C, Kittler S, Epple M, Muhr G, Köller M. Studies on the biocompatibility and the interaction of silver nanoparticles with human mesenchymal stem cells (hMSCs) Langenbecks Arch Surg. 2009;394:495–502. doi: 10.1007/s00423-009-0472-1. [DOI] [PubMed] [Google Scholar]
- Handy RD, Henry TB, Scrown TM, Johnston BD, Tyler CR. Manufactured nanoparticles: their uptake and effects on fish—a mechanistic analysis. Ecotoxicology. 2008;17(5):396–409. doi: 10.1007/s10646-008-0205-1. [DOI] [PubMed] [Google Scholar]
- Handy RD, von der Kammer F, Lead JR, Hassellöv M, Owen R, Crane M. The ecotoxicology and chemistry of manufactured nanoparticles. Ecotoxicology. 2008;17:287–314. doi: 10.1007/s10646-008-0199-8. [DOI] [PubMed] [Google Scholar]
- Harris JM, Martin NE, Modi M. Pegylation: a novel process for modifying pharmacokinetics. Clin Pharmacokinet. 2001;40:539–551. doi: 10.2165/00003088-200140070-00005. [DOI] [PubMed] [Google Scholar]
- Havel HA. Spectroscopic methods for determining protein structure in solution. New York: Wiley-VCH; 1996. [Google Scholar]
- Jeong SK, Kwon MS, Lee EY, Lee HJ, Cho SY, Kim H, Yoo JS, Omenn GS, Aebersold R, Hanash S, Paik YK. BiomarkerDigger: a versatile disease proteome database and analysis platform for the identification of plasma cancer biomarkers. Proteomics. 2009;9(14):3729–3740. doi: 10.1002/pmic.200800593. [DOI] [PubMed] [Google Scholar]
- Jiang X, Weise S, Hafner M, Röcker C, Zhang F, Parak WJ, Nienhaus GU. Quantitative analysis of the protein corona on FePt nanoparticles formed by transferrin binding. J R Soc Interface. 2010;7:S5–S13. doi: 10.1098/rsif.2009.0272.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kah JC, Wong KY, Neoh KG, Song JH, Fu JW, Mhaisalkar S, Olivo M, Sheppard CJ. Critical parameters in the pegylation of gold nanoshells for biomedical applications: an in vitro macrophage study. J Drug Target. 2009;17:181–193. doi: 10.1080/10611860802582442. [DOI] [PubMed] [Google Scholar]
- Keller KH. Nanotechnology and society. J Nanopart Res. 2007;9:5–10. doi: 10.1007/s11051-006-9193-3. [DOI] [Google Scholar]
- Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochim Biophys Acta. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Kittler S, Greulich C, Diendorf J, Köller M, Epple M. The toxicity of silver nanoparticles increases during storage due to slow dissolution under release of silver ions. Chem Mater. 2010;22(16):4548–4554. doi: 10.1021/cm100023p. [DOI] [Google Scholar]
- Kittler S, Greulich C, Gebauer JS, Diendorf J, Treuel L, Ruiz L, Gonzalez-Calbet JM, Vallet-Regi M, Zellner R, Köller M, Epple M. The influence of proteins on the dispersability and cell-biological activity of silver nanoparticles. J Mater Chem. 2010;20(3):512–518. doi: 10.1039/b914875b. [DOI] [Google Scholar]
- Klein J. Probing the interactions of proteins and nanoparticles. Proc Natl Acad Sci USA. 2007;104(7):2029–2030. doi: 10.1073/pnas.0611610104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreyling WG, Semmler M, Möller W. Dosimetry and toxicology of ultrafine particles. J Aerosol Med. 2004;17(2):140–152. doi: 10.1089/0894268041457147. [DOI] [PubMed] [Google Scholar]
- Leszczynski J. Bionanoscience: nano meets bio at the interface. Nat Nanotechnol. 2010;5(9):633–634. doi: 10.1038/nnano.2010.182. [DOI] [PubMed] [Google Scholar]
- Linse S, Cabaleiro-Lago C, Xue W-F, Lynch I, Lindman S, Thulin E, Radford SE, Dawson KA. Nucleation of protein fibrillation by nanoparticles. Proc Natl Acad Sci USA. 2007;104(21):8691–8696. doi: 10.1073/pnas.0701250104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Xu K, Wang H, Tan PKJ, Fan W, Venkatraman SS, Li L, Yang Y-Y. Self-assembled cationic peptide nanoparticles as an efficient antimicrobial agent. Nat Nanotechnol. 2009;4:457–463. doi: 10.1038/nnano.2009.153. [DOI] [PubMed] [Google Scholar]
- Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc Natl Acad Sci USA. 2008;105(38):14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundqvist M, Stigler J, Cedervall T, Berggård T, Flanagan MB, Lynch I, Elia G, Dawson K. The evolution of the protein corona around nanoparticles: a test study. ACS Nano. 2011;5(9):7503–7509. doi: 10.1021/nn202458g. [DOI] [PubMed] [Google Scholar]
- Lunov O, Syrovets T, Röcker C, Tron K, Nienhaus GU, Rasche V, Mailänder V, Landfester K, Simmet T. Lysosomal degradation of the carboxydextran shell of coated superparamagnetic iron oxide nanoparticles and the fate of professional phagocytes. Biomaterials. 2010;31(34):9015–9022. doi: 10.1016/j.biomaterials.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Lunov O, Zablotskii V, Syrovets T, Röcker C, Tron K, Nienhaus GU, Simmet T. Modeling receptor-mediated endocytosis of polymer-functionalized iron oxide nanoparticles by human macrophages. Biomaterials. 2011;32(2):547–555. doi: 10.1016/j.biomaterials.2010.08.111. [DOI] [PubMed] [Google Scholar]
- Lynch I. Are there generic mechanisms governing interactions between nanoparticles and cells? Random epitope mapping for the outer layer of the protein-material interface. Physica A. 2007;373:511–520. doi: 10.1016/j.physa.2006.06.008. [DOI] [Google Scholar]
- Lynch I, Dawson KA, Linse S. Detecting cryptic epitopes created by nanoparticles. Sci STKE. 2006;2006(327):14. doi: 10.1126/stke.3272006pe14. [DOI] [PubMed] [Google Scholar]
- Maffre P, Nienhaus K, Amin F, Parak WJ, Nienhaus GU. Characterization of protein adsorption onto FePt nanoparticles using dual-focus fluorescence correlation spectroscopy. Beilstein J Nanotechnol. 2011;2:374–383. doi: 10.3762/bjnano.2.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mátyus L, Szöllösi J, Jenei A. Steady-state fluorescence quenching applications for studying protein structure and dynamics. J Photochem Photobiol B: Biol. 2006;83:223–236. doi: 10.1016/j.jphotobiol.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Maynard AD, Aitken RJ, Butz T, Colvin V, Donaldsen K, Oberdörster G, Philbert MA, Ryan J, Seaton A, Stone V, Tinkle SS, Tran L, Walker NJ, Warheit DB. Safe handling of nanotechnology. Nature. 2006;444:267–269. doi: 10.1038/444267a. [DOI] [PubMed] [Google Scholar]
- Medintz IL, Konnert JH, Clapp AR, Stanish I, Twing ME, Mattoussi H, Mauro JM, Deschamps JR. A fluorescence resonance energy transfer-derived structure of a quantum dot-protein bioconjugate nanoassembly. Proc Natl Acad Sci USA. 2004;101(26):9612–9617. doi: 10.1073/pnas.0403343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min Y, Akbulut M, Kristiansen K, Golan Y, Israelachvili J. The role of interparticle and external forces in nanoparticle assembly. Nat Mater. 2008;7(7):527–538. doi: 10.1038/nmat2206. [DOI] [PubMed] [Google Scholar]
- Monopoli MP, Walczyk D, Campbell A, Elia G, Lynch I, Bombelli FB, Dawson KA. Physical-chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles. J Am Chem Soc. 2011;133(8):2525–2534. doi: 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- Mori S, Barth HG. Size exclusion chromatography. Berlin: Springer; 1999. [Google Scholar]
- Nativo P, Prior IA, Brust M. Uptake and intracellular fate of surface-modified gold nanoparticles. ACS Nano. 2008;2:1639–1644. doi: 10.1021/nn800330a. [DOI] [PubMed] [Google Scholar]
- Nel A, Xia T, Mädler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311:622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- Nel AE, Mädler L, Velegol D, Xia T, Hoek EM, Somasundaran P, Klaessig F, Castranova V, Thompson M. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8(7):543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- Nienhaus GU, editor. Protein-ligand interactions: methods and applications. New York: Humana Press; 2005. [Google Scholar]
- Niidome T, Yamagata M, Okamoto Y, Akiyama Y, Takahashi H, Kawano T, Katayama Y, Niidome Y. PEG-modified gold nanorods with a stealth character for in vivo applications. J Control Release. 2006;114:343–347. doi: 10.1016/j.jconrel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Oberdörster G. Safety assessment for nanotechnology and nanomedicine: concepts of nanotoxicology. J Intern Med. 2010;267(1):89–105. doi: 10.1111/j.1365-2796.2009.02187.x. [DOI] [PubMed] [Google Scholar]
- Oberdörster G, Maynard A, Donaldson K, Castranova V, Fitzpatrick J, Ausman K, Carter J, Karn B, Kreyling W, Lai D, Olin S, Monteiro-Riviere N, Warheit D, Yang H. Principles for characterizing the potential human health effects from exposure to nanomaterials: elements of a screening strategy. Part Fibre Toxicol. 2005;2(8):1–35. doi: 10.1186/1743-8977-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdörster G, Oberdörster E, Oberdörster J. Nanotoxicology: an emerging discipline evolving from studies of ultrafine particles. Environ Health Perspect. 2005;113(7):823–839. doi: 10.1289/ehp.7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization for Economic Co-operation and Development (OECD) (2008) Current developments/activities on the safety of manufactured nanomaterials. OECD environment, health and safety publication series on the safety of manufactured nanomaterials. OECD, Paris
- Owen R, Depledge M. Nanotechnology in the environment: risks and rewards. Mar Pollut Bull. 2005;50:609–612. doi: 10.1016/j.marpolbul.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Owens DE, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Pan Y, Neuss S, Leifert A, Fischler M, Wen F, Simon U. Size-dependent cytotoxicity of gold nanoparticles. Small. 2007;3:1941–1949. doi: 10.1002/smll.200700378. [DOI] [PubMed] [Google Scholar]
- Poland CA, Duffin R, Kinloch I, Maynard A, Wallace WAH, Seaton A, Stone V, Brown S, Macnee W, Donaldson K. Carbon nanotubes introduced into the abdominal cavity of mice show asbestoslike pathogenicity in a pilot study. Nature. 2008;3:423–428. doi: 10.1038/nnano.2008.111. [DOI] [PubMed] [Google Scholar]
- Printz M, Friess W. Simultaneous detection and analysis of protein aggregation and protein unfolding by size exclusion chromatography with post column addition of the fluorescent dye BisANS. J Pharmaceut Sci. 2012;101(2):826–837. doi: 10.1002/jps.22808. [DOI] [PubMed] [Google Scholar]
- Roach P, Farrar D, Perry CC. Surface tailoring for controlled protein adsorption: effect of topography at the nanometer scale and chemistry. J Am Chem Soc. 2006;128(12):3939–3945. doi: 10.1021/ja056278e. [DOI] [PubMed] [Google Scholar]
- Röcker C, Pötzl M, Zhang F, Parak WJ, Nienhaus GU. A quantitative fluoresence study of protein monolayer formation on colloidal nanoparticles. Nat Nanotechnol. 2009;4(9):577–580. doi: 10.1038/nnano.2009.195. [DOI] [PubMed] [Google Scholar]
- Roco MC. The journal of nanoparticle research at 10 years. J Nanopart Res. 2008;10(1):1–2. doi: 10.1007/s11051-008-9528-3. [DOI] [Google Scholar]
- Rodriguez CE, Fukuto JM, Taguchi K, Froines J, Cho AK. The interactions of 9,10-phenantrenequinone with glyceraldehyde-3-phosphatedehydrogenase (GAPDH), a potential site for toxic actions. Chem Biol Interact. 2005;155(1):97–110. doi: 10.1016/j.cbi.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Rothen-Rutishauser B, Kiama S, Gehr P. A three-dimensional cellular model of the human respiratory tract to study the interaction with particles. Am J Respir Cell Mol Biol. 2005;32(4):281–289. doi: 10.1165/rcmb.2004-0187OC. [DOI] [PubMed] [Google Scholar]
- Royer CA. Probing protein folding and conformational transitions with fluorescence. Chem Rev. 2006;106:1769–1784. doi: 10.1021/cr0404390. [DOI] [PubMed] [Google Scholar]
- Schlücker S. Gezielte Proteinlokalisierung. Biophotonik. 2008;3:18–20. [Google Scholar]
- Semmler-Behnke M, Takenaka S, Fertsch S, Wenk A, Seitz J, Mayer P, Oberdoerster G, Kreyling WG. Efficient elimination of inhaled nanoparticles from the alveolar region: evidence for interstitial uptake and subsequent reentrainment onto airways epithelium. Environ Health Perspect. 2007;115(5):728–733. doi: 10.1289/ehp.9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service RF. Priorities needed for nano-risk research and development. Science. 2006;314:45. doi: 10.1126/science.314.5796.45. [DOI] [PubMed] [Google Scholar]
- Shang L, Wang Y, Jiang J, Dong S. pH-dependent protein conformational changes in albumin—gold nanoparticle bioconjugates: a spectroscopic study. Langmuir. 2007;23:2714–2721. doi: 10.1021/la062064e. [DOI] [PubMed] [Google Scholar]
- Shang W, Nuffer JH, Muñiz-Papandrea VA, Colón W, Siegel RW, Dordick JS. cytochrome c on silica nanoparticles: influence of nanoparticle size on protein structure, stability, and activity. Small. 2009;5(4):470–476. doi: 10.1002/smll.200800995. [DOI] [PubMed] [Google Scholar]
- Shang L, Brandholt S, Stockmar F, Trouillet V, Bruns M, Nienhaus GU (2011a) Effect of protein adsorption on the fluorescence of ultrasmall gold nanoclusters. Small. doi:10.1002/smll.201101353 [DOI] [PubMed]
- Shang L, Doerlich RM, Brandholt S, Schneider R, Trouillet V, Bruns M, Gerthsen D, Nienhaus GU. Facile preparation of water-soluble fluorescent gold nanoclusters for cellular imaging applications. Nanoscale. 2011;3(5):2009–2014. doi: 10.1039/c0nr00947d. [DOI] [PubMed] [Google Scholar]
- Shang L, Dong S, Nienhaus GU. Ultra-small fluorescent metal nanoclusters: synthesis and biological applications. Nano Today. 2011;6(4):401–418. doi: 10.1016/j.nantod.2011.06.004. [DOI] [Google Scholar]
- Shao M, Lu L, Wang H, Luo S, Duo Duo Ma D. Microfabrication of a new sensor based on silver and silicon nanomaterials, and its application to the enrichment and detection of bovine serum albumin via surface-enhanced Raman scattering. Microchim Acta. 2009;164:157–160. doi: 10.1007/s00604-008-0051-0. [DOI] [Google Scholar]
- Tenzer S, Docter D, Rosfa S, Wlodarski A, Kuharev J, Rekik A, Knauer SK, Bantz C, Nawroth T, Bier C, Sirirattanapan J, Mann W, Treuel L, Zellner R, Maskos M, Schild H, Stauber RH. Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: a comprehensive quantitative proteomic analysis. ACS Nano. 2011;5(9):7155–7167. doi: 10.1021/nn201950e. [DOI] [PubMed] [Google Scholar]
- Treuel L, Malissek M, Gebauer JS, Zellner R. The influence of surface composition of nanoparticles on their interactions with serum albumin. Chem Phys Chem. 2010;11(14):3093–3099. doi: 10.1002/cphc.201000174. [DOI] [PubMed] [Google Scholar]
- Vertegel AA, Siegel RW, Dordic JS. Silica nanoparticle size influences the structure and enzymatic activity of adsorbed lysozyme. Langmuir. 2004;20(16):6800–6807. doi: 10.1021/la0497200. [DOI] [PubMed] [Google Scholar]
- Vogt A, D’Angelo C, Oswald F, Denzel A, Mazel CH, Matz MV, Ivanchenko S, Nienhaus GU, Wiedenmann J. A green fluorescent protein with photoswitchable emission from the deep sea. PLoS One. 2008;3(11):1–8. doi: 10.1371/journal.pone.0003766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Bai J, Jiang X, Nienhaus GU (2012) Cellular uptake of nanoparticles by membrane penetration: a study combining confocal microscopy with FTIR spectroelectrochemistry. ACS Nano. doi:10.1021/nn203892h [DOI] [PubMed]
- Watari F, Takashi N, Yokoyama A, Uo M, Akasaka M, Sato Y, Abe S, Totsuka Y, Tohji K. Material nanosizing effect on living organism: non-specific, biointeractive, physical size effects. J R Soc Interf. 2009;6:371–388. doi: 10.1098/rsif.2008.0488.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenmann J, Schenk A, Röcker C, Girod A, Spindler KD, Nienhaus GU. A far-red fluorescent protein with fast maturation and reduced oligomerization tendency from Entacmaea quadricolor (Anthozoa, Actinaria) Prod Natl Acad Sci USA. 2002;99:11646–11651. doi: 10.1073/pnas.182157199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenmann J, Ivanchenko S, Oswald F, Nienhaus GU. Identification of GFP-like proteins in nonbioluminescent, azooxanthellate anthozoa opens new perspectives for bioprospecting. Mar Biotechnol. 2004;6:270–277. doi: 10.1007/s10126-004-3006-4. [DOI] [PubMed] [Google Scholar]
- Wiesner MR, Lowry GV, Alvarez P, Dionysiou D, Biswas P. Assessing the risks of manufactured nanomaterials. Environ Sci Technol. 2006;40:4336–4345. doi: 10.1021/es062726m. [DOI] [PubMed] [Google Scholar]
- Yan M, Du J, Gu Z, Liang M, Hu Y, Zhang W, Priceman S, Wu L, Hong Zhou Z, Liu H, Segura T, Tang Y, Lu Y. A novel intracellular protein delivery platform based on single-protein nanocapsules. Nat Nanotechnol. 2009;5:48–53. doi: 10.1038/nnano.2009.341. [DOI] [PubMed] [Google Scholar]
- Ye-Qin Z, Wang Y-F, Jiang X-D. The application of nanoparticles in biochips. Recent Patents Biotechnol. 2008;2(1):55–59. doi: 10.2174/187220808783330938. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yan YB. Probing conformational changes of proteins by quantitative second-derivative infrared spectroscopy. Anal Biochem. 2005;340:89–98. doi: 10.1016/j.ab.2005.01.053. [DOI] [PubMed] [Google Scholar]
- Zhou HS, Aoki S, Honma I, Hirasawa M, Nagamune T, Komiyama H (1997) Conformational change of protein cytochrome b-562 adsorbed on colloidal gold particles; absorption band shift. Chem Commun. 605–606