Abstract
Single-molecule fluorescence microscopy experiments on RNA molecules brought to light the highly complex dynamics of key biological processes, including RNA folding, catalysis of ribozymes, ligand sensing of riboswitches and aptamers, and protein synthesis in the ribosome. By using highly advanced biophysical spectroscopy techniques in combination with sophisticated biochemical synthesis approaches, molecular dynamics of individual RNA molecules can be observed in real time and under physiological conditions in unprecedented detail that cannot be achieved with bulk experiments. Here, we review recent advances in RNA folding and functional studies of RNA and RNA-protein complexes addressed by using single-molecule Förster (fluorescence) resonance energy transfer (smFRET) technique.
Keywords: RNA, RNA folding, RNA-protein complexes, Single-molecule Förster resonance energy transfer technique
Introduction
Ribonucleic acid (RNA) is a key biopolymer on which all life forms on Earth are based. Remarkably, precursors of modern life have been envisioned that existed without proteins and deoxyribonucleic acid (DNA), relying almost exclusively on the unique properties of RNA. In addition to encoding information, RNA molecules can perform complex biological functions, based on their ability to assume sophisticated three-dimensional architectures that may form ligand binding pockets and catalytic sites. RNA’s paucity of sequence information, encoded by only four different building blocks, gives rise to the RNA folding problem: A given RNA sequence can typically assume a multitude of states of similar thermodynamic stability, which are stabilized by strong π-stacking and Watson-Crick base-pairing interactions. These states are separated by large energy barriers, so that RNA molecules possess a highly rugged free energy landscape (Thirumalai and Hyeon 2005). As a result, metastable structures and folding intermediates are often fairly long-lived and, in particular, large RNAs frequently show kinetic folding traps and misfolding. RNA’s conformational rearrangements are often large and thus ideal for observation with single molecule techniques, and typical interconversion times in the millisecond to second range are particularly well suited for single molecule Förster (fluorescence) resonance energy transfer (smFRET) methods. Another beneficial property of RNA is its predictability with respect to solubility and surface adhesion, which allows the development of generic procedures of immobilizing individual RNA molecules. Its largely invariant chemical properties greatly facilitate chemical manipulation of RNA, e.g., site-directed labeling with two fluorophores or attachment of biotin. Consequently, we have witnessed an explosion of the field in the past few years, which we will try to outline here. Much of this work has already been summarized in a number of excellent reviews on single-molecule studies of RNA (Al-Hashimi and Walter 2008; Blanchard 2009; Bokinsky and Zhuang 2005; Ditzler et al. 2007; Marshall et al. 2008a; Munro et al. 2008; Zhuang 2005; Zhuang and Rief 2003). Here, we focus on smFRET studies of plain RNA and RNA-protein complexes, also touching on the exciting field of polymerases at the end. Results of other biophysical techniques with single-molecule sensitivity will not be reviewed in depth but just briefly referred to in conjunction with smFRET results.
Hierarchy of RNA folding
RNA folding is strongly dependent on the concentration of cations in the surrounding solvent, in particular magnesium ions (Brion and Westhof 1997). Because the backbone phosphates carry one negative charge per nucleotide, the resulting strong electrostatic repulsion does not permit folding at low concentrations of shielding cations. Thus, RNA is essentially unfolded in pure water, in the U state. Starting in this state, a progressive increase of salt concentration leads to an increased shielding of the RNA charges by the cations in solution. First, structural elements form, thus “collapsing” the previously random coil polymer. The resulting “collapsed” state contains essentially elements of the secondary structure, i.e., mostly Watson-Crick helices. Magnesium ions play an import role: because of their high charge-to-volume ratio, they provide much more effective shielding and, therefore, induce polymer collapse at significantly lower concentrations than do monovalent cations. Very frequently, more than one collapsed state is found; such I states may be considered early intermediates in the progression towards a native state. As the salt concentration is further increased, formation of structure also includes tertiary interactions. Here, the role of magnesium ions is even more important, because the tertiary structure often contains specific binding pockets, to which monovalent ions do not have sufficient affinity. Thus, monovalent ions can collapse RNA and enable formation of the secondary structure, but magnesium ions are essential for the formation of a spatially well-defined, native N state. Physiological concentrations of magnesium ions range around 1 mM, but many in vitro studies of RNA folding and activity are performed at higher concentrations for a variety of reasons, including (1) compensation for the stabilization of native RNA structure by proteins or other factors in the cell (Qu et al. 2008), (2) accessibility of kinetic or thermodynamic parameters from titration with magnesium ions (Kobitski et al. 2008), (3) investigation of minimal constructs that retain activity only at higher concentrations (Zhuang et al. 2002), or (4) investigations of entirely artificial RNAs, for which the term ‘physiological’ is not relevant (Kobitski et al. 2007).
Titration with counterions, as outlined above, clearly reveals a pronounced hierarchy of RNA folding, with relatively stable secondary structures forming as autonomous units before tertiary contacts are being established. This experimental approach is thermodynamic rather than kinetic, preferably sampling states near the minimum of the free energy landscape. Kinetic data on folding/unfolding or ligand binding are, however, accessible from single-molecule studies even under equilibrium conditions, without the requirement of synchronization to any perturbing event.
In order to obtain statistically significant information, large ensembles of molecules need to be investigated. However, even registration of several thousand events may not be sufficient to identify rarely populated intermediate states. One way to enhance such hidden states is to perform experiments under non-equilibrium conditions. Relaxation methods employing rapid jumps in ion concentration have provided access to folding intermediates further away from the known energy minima and to particular aspects of molecular memory (Liu et al. 2007; Qu et al. 2008). Microfluidic devices provide very convenient ways to create controllable perturbations to the systems under investigation (Dittrich et al. 2004; Kauffmann et al. 2001; Lipman et al. 2003; Pfeil et al. 2009).
Techniques and technical developments in smFRET
Dye labeling and photophysics
Radiationless Förster resonance energy transfer from an excited donor fluorophore to an acceptor fluorophore depends on the inverse sixth power of the inter-dye distance. Because fluorescence emission can be detected with excellent time resolution, smFRET is an attractive method for the observation of distance variations between two dye-labeled domains of a given RNA, or RNA-protein complex (Bokinsky and Zhuang 2005; Hengesbach et al. 2008). In principle, measurement of the donor fluorescence alone contains sufficient information (e.g., fluorescence decay rates) to detect donor–acceptor distance changes. Indeed, certain applications have used a quencher instead of a fluorescent acceptor dye. In single-molecule experiments, however, monitoring the fluorescence of both dyes of a FRET pair is usually preferred because it allows one to unambiguously assign anticorrelated intensity changes to FRET efficiency, E FRET, variations and to calculate this quantity ratiometrically,
| 1 |
from the fluorescence intensities registered from the acceptor, I A, and the donor, I D. Both intensities have to be corrected for background and cross-talk between the two detection channels, and the parameter γ accounts for differences in the detection efficiencies of the two channels and the quantum yields of the dyes. While it is often assumed that the inter-dye distance is the major parameter determining E FRET, other effects, including dye photochemistry and photophysics (blinking), changes in the refractive index or variations of the dyes’ orientations and rotational freedom may also affect the dipolar coupling of the two dye molecules and, therefore, the resulting E FRET values. Interactions of the dyes with the host biomolecule may prevent the complete averaging of the dipole orientation during the fluorescence lifetime (Kobitski et al. 2007; Kuzmenkina et al. 2006; Schuler et al. 2005). A change in FRET efficiency due to dye stacking on dsRNA/DNA was also noticed (Iqbal et al. 2008a, b). Multi-parameter smFRET measurements, which not only register emission intensity but also fluorescence polarization/anisotropy and decay times, have been introduced to enable more reliable distance measurements based on FRET coupling (Widengren et al. 2006; Wozniak et al. 2005). Still, observations of dynamic events by changes in FRET coupling are generally preferred in the field; measurements of absolute distances are typically approached with great caution.
Dye-labeled RNA should be thoroughly validated to exclude artifacts as a consequence of the presence of the dyes. Besides anisotropy measurements that measure interactions between the dye and the host molecule, functional tests must assure that the pertinent biochemical function of the biomacromolecule is not perturbed by the dyes (Walter 2003). If the molecule is no longer functional after labeling, the dyes must be “walked” to a different attachment site, and the functional test reiterated. Dye attachment on proteins is frequently achieved by alkylating accessible cysteine residues with iodoacetyl-derivatives of fluorescent dyes. For RNA, dyes can be positioned at the 5’-end during transcription (Xie et al. 2004). Postsynthetic modification schemes of both the 5’-end and the 3’-end of RNA (Chan et al. 1999) are known but rarely applied in smFRET studies. Ligation techniques permit the incorporation of RNAs with 5’- or 3’-labels, or functional groups for labeling introduced during solid phase phosphoramidite synthesis (Bokinsky et al. 2003; de Silva and Walter 2009; Ha et al. 1999; Hengesbach et al. 2008; Zhuang et al. 2002). In numerous cases, dyes have been introduced into large RNAs through hybridization to DNA or RNA oligos carrying the actual dye label (Dorywalska et al. 2005; Smith et al. 2008). This approach is convenient and straightforward especially in large RNAs, where entire domains can be engineered without significant loss of biochemical activity. For smaller RNAs, structural engineering in combination with end labeling is more likely to be inadequate (Voigts-Hoffmann et al. 2007). Here, internal modifications can be postsynthetically introduced into synthetic RNA oligos equipped with amino or thiol groups (de Silva and Walter 2009). These fragments may then be joined by ligation (Hengesbach et al. 2008; Walter 2003).
Despite impressive advances in the development of bright and stable fluorophores for single-molecule measurements (Kapanidis and Weiss 2002), the dyes are still the Achilles’ heel of single-molecule FRET experiments. First of all, dye photobleaching limits the observation time, and thus is a major problem in fluorescence experiments. In addition, phenomena such as quenching, blinking, emission spectral jumps, or anisotropy changes can easily lead to erroneous results. While typical first generation smFRET dyes like fluorescein and TMR have mostly yielded to the Cy3-Cy5 pair, these widely used carbocyanine dyes exhibit extensive blinking due to cis-trans isomerization and triplet formation (Bates et al. 2005; Heilemann et al. 2005). However, the use of oxygen scavenging systems in combination with triplet quenchers can enormously improve dye performance and substantially lengthen the observation times (Vogelsang et al. 2008). A better understanding of the underlying dye photophysics in recent years (Aitken et al. 2008; Eggeling et al. 2006; Rasnik et al. 2006; Widengren et al. 2007) has helped in the rational development of fluorophores with higher photostability. Transient interaction of fluorophores with the local environment may also cause transitions to non-emissive states or states having a shifted emission spectrum. The latter case can be especially troublesome because part of the emission can bleed through to another detection channel, and such transient changes of the donor–acceptor intensity ratio can be mistaken as FRET efficiency changes due to inter-dye distance variations (W.A. Eaton, personal communication).
Microscopy apparatus
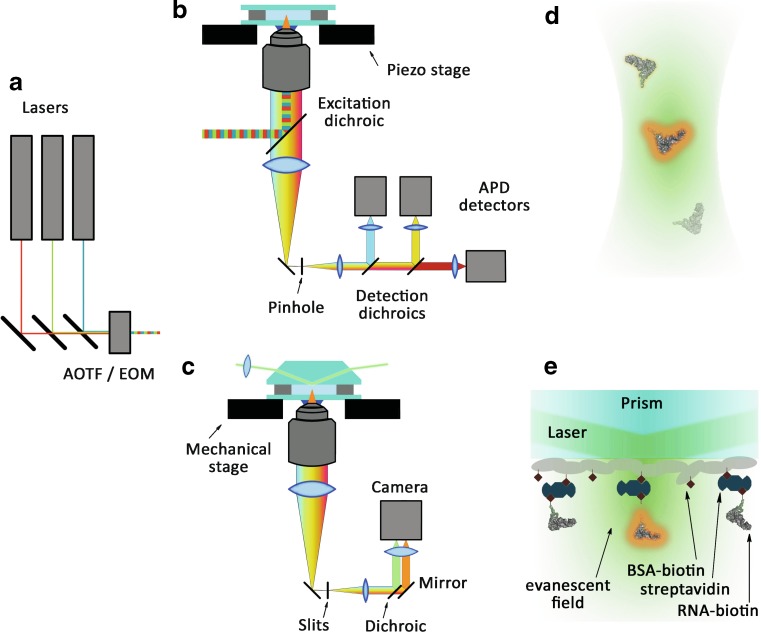
Over the last decade, single-molecule fluorescence techniques have been substantially advanced and adapted to probe individual molecules under various conditions. Although commercial systems have become available from different manufacturers, it is quite straightforward to assemble a home-built microscope with single-molecule sensitivity from off-the-shelf components (Rieger et al. 2005; Roy et al. 2008; Walter et al. 2008). Advanced single-molecule microscopy setups feature alternating excitation with several laser lines and multi-channel detection in three or more channels (Fig. 1) (Hohng et al. 2004a; Kapanidis et al. 2005; Lee et al. 2007a; Muller et al. 2005). Moreover, instead of monitoring only the fluorescence intensity, the simultaneous measurement of anisotropy, lifetime and emission spectrum in real time allows one to get unprecedented control of the photophysical properties of the probes under investigation (Tinnefeld and Sauer 2005; Widengren et al. 2006).
Fig. 1.
Experimental configurations of single-molecule fluorescence microscopy. a Lasers serve as excitation sources for fluorescent labels. Typical excitation lines are 488/492 nm for fluorescein, Cy2 and Alexa 488, 514/532 nm for TMR and Cy3, and 633/635 nm for Cy5. Acousto-optical tunable filter (AOTF) or electro-optical modulator (EOM) allow for fast switching between different laser lines in alternating excitation (ALEX) experiments. b Stage scanning confocal fluorescence microscopy. An excitation dichroic mirror reflects the excitation light into the sample and transmits the emitted fluorescence to the detectors. A pinhole in the focal plane of the tube lens reduces the detection volume to ~1 fl. Detection dichroics separate the emission among several, highly sensitive avalanche photo diode (APD) detectors. Images are acquired by a consecutive shift of the sample in a pixel-by-pixel manner by using a piezoelectric stage. c Prism-type total internal reflection fluorescence microscope (TIRFM). Focused excitation light reflected at the quartz-water interface at a large incident angle (Θc > 68°) produces an evanescent field penetrating ~100 nm in the aqeueous solution. For two-channel detection, donor and acceptor emission is separated by detection dichroic mirrors and imaged side-by-side on a CCD chip. d Confocal microscopy on molecules diffusing freely in solution. Single-molecule conditions are obtained by diluting the sample to concentrations of <1 nM. e TIRFM experiments with molecules immobilized on a surface via biotin-streptavidin coupling. A layer of bovine serum albumin sparsely decorated with biotin serves to anchor streptavidin and, thereon, biotinylated RNA molecules
The two main approaches to single-molecule fluorescence detection are confocal microscopy and wide-field microscopy using the total internal reflection effect (Haustein and Schwille 2007; Moerner and Fromm 2003; Tinnefeld and Sauer 2005). In both cases, sensitivity on the single-molecule level is achieved by minimizing the excitation and detection volumes, either by introducing a confocal pinhole in the detection path (in confocal microscopy, Fig. 1b) or by exciting molecules in a ~100-nm layer close to a surface by using an evanescent field (total internal reflection fluorescence microscopy, TIRF; Fig. 1c and 1e). Both methods are complementary, and the choice of one over the other is in general governed by the time scale of the processes under investigation. In high-sensitivity confocal microscopy, avalanche photodiode detectors are employed, offering a time resolution better than microseconds. However, images have to be acquired in a time-consuming pixel-by-pixel manner. TIRF microscopy relies on high-sensitivity, charge-coupled device (CCD) cameras, which permit collection of entire images containing many molecules the fluorescence of which can be recorded in parallel. However, the time resolution is limited by the frame rate of the CCD camera to typically 5–100 ms. In TIRF microscopy, molecules have to be immobilized on a surface, whereas experiments may be performed either on immobilized or freely diffusing molecules in confocal microscopy. In the latter case, the maximum time of signal detection from each individual molecule is governed by their diffusional transit through the confocal volume (typically 100–1000 µs) (Fig. 1d), whereas in the case of immobilized molecules, the time of observation is only limited by the unavoidable photobleaching of the dye, which may permit measurements over many minutes (Vogelsang et al. 2008).
Surface immobilization
Immobilization procedure ought to be carried out with special care. While specific attachment is easily achieved with a variety of straightforward approaches, including the widely used biotin-(strept)avidin coupling (Fig. 1e), unspecific adsorption to the surface has to be strictly minimized to at most a few percent. Also, the interaction of the immobilized biomolecule with the underlying surface needs to be as low as possible. Otherwise, static heterogeneity of the observed properties due to the environment may result and, moreover, intramolecular dynamics may also be strongly affected by such interactions. A number of protein and polymer surface coatings as well as vesicle encapsulation have been examined with respect to their homogeneity and interaction with tethered or caged biomolecules (Amirgoulova et al. 2004; Groll et al. 2004, Christensen and Stamou 2007; Heyes et al. 2004, 2007; Koopmans et al. 2008; Okumus et al. 2004; Rasnik et al. 2005). While many such surfaces affect protein folding and unfolding to a large extent, RNA/DNA molecules seem to be less sensitive to interactions with protein coatings such as BSA and streptavidin, which could be due to the repulsion between negatively charged nucleic acids and proteins at neutral pH (Ha et al. 1999; Kobitski et al. 2007; Tan et al. 2003; Xie et al. 2004; Zhuang et al. 2000, 2002). Modified PEG surfaces have also been employed for the immobilization of nucleic acids (Elenko et al. 2009; Qu et al. 2008; Rasnik et al. 2004), and it has further been suggested that lipid vesicles offer a native-like environment for biomolecules (Boukobza et al. 2001; Lee et al. 2005; Okumus et al. 2004; Rhoades et al. 2003). Unfortunately, the lipid membrane acts as a barrier for solvent, so that in situ solvent exchange becomes impossible, and the lipid bilayer may also become unstable under harsh solvent conditions. Recently, an approach was developed that introduces pores into vesicles, which affords biomolecule immobilization in a most gentle way and, in addition, allows one to increase the effective concentration of molecules with low binding affinity and to trigger the reaction between them via a change of external buffer conditions (Cisse et al. 2007).
FRET studies of RNA molecules
Dynamics of small natural RNAs
Minimal constructs
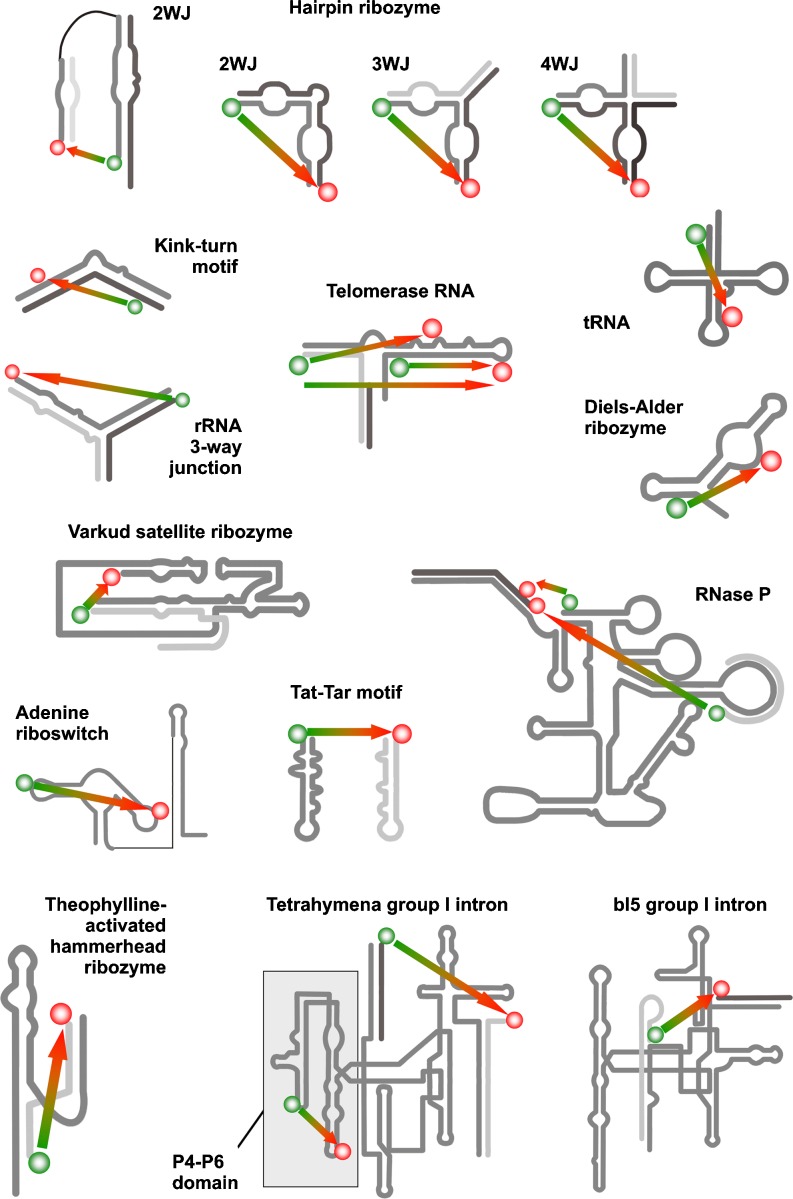
Figure 2 gives an overview of RNA molecules investigated by smFRET. Among the first RNAs to be investigated by smFRET was a three-way junction derived from ribosomal RNA (rRNA) (Ha et al. 1999). This junction, which also provides a binding site for the ribosomal S15 protein, was assembled by hybridization of three separate RNA oligomers carrying fluorescein, Cy3, or biotin for anchoring on the surface (Fig. 2). This work introduced many smFRET features that are now standard in this field. It confirmed the expectation that the average behavior of many individual RNA molecules should closely resemble bulk behavior. Two conformations, denoted as open and closed according to the arrangement of the arms of its Y-shape, were identified, and their interconversion was observed in response to their interaction with Mg2+ ions or the cognate S15 protein. Thus, it was not only the first demonstration of a structural rearrangement in RNA by smFRET but also pioneering work regarding RNA−protein interactions. This model system was further investigated by an application of fluorescence correlation spectroscopy to immobilized biomolecules, from which the transition rates of the conformational changes were measured as functions of Mg2+ and Na+ ion concentrations (Kim et al. 2002).
Fig. 2.
Overview of RNA structures investigated by smFRET. The schematic secondary structures are shown; labeling sites with donor dye (green) and acceptor dye (red) are indicated as dots of the respective color. Green-red arrows indicate the distances monitored by smFRET
Along structurally similar lines, the kink-turn motif (labeled with Alexa488 and Cy5; Fig. 2) was recently studied on the confocal microscope in solution in the context of a two-stem motif derived from U4 snRNA, a constituent of the major spliceosome (Wozniak et al. 2005). It was observed that the kink-turn depends on the presence of two subsequent purine-purine basepairs followed by a three-nucleotide bulge, and offers a binding site for the 15.5 K protein. Binding of the protein was found to increase bending of the two stems: the interhelical angle was decreased from 78° in the absence to 48° in the presence of protein.
tRNA
Among the first RNAs to be investigated by single molecule fluorescence was tRNAPhe from E. coli carrying a single TMR label. By measurement of fluorescence decay times of immobilized molecules, at least two conformational states were identified (Jia et al. 1997). A more detailed analysis of several alternative conformations was performed on human mitochondrial tRNALys , which was labeled intramolecularly (Fig. 2) using the Cy3/Cy5 FRET pair and biotin at the 3’-end (Voigts-Hoffmann et al. 2007). Three conformations were distinguished in this unusual tRNA based on the observed FRET levels, corresponding to an unfolded state, an extended hairpin conformation, and the classical cloverleaf. Interestingly, equilibria between these states could be shifted by introducing a single methyl group in a modified nucleotide and by varying the concentration of magnesium ions. The latter approach enabled a detailed picture of the energy landscape to be obtained (Kobitski et al. 2008). As observed before with metal ions and small molecule ligands, a natural covalent modification was thus identified as another entity capable of shifting the conformational equilibrium towards the functional structure.
Folding and catalysis of natural ribozymes
The insight that RNA holds the potential for catalysis has greatly changed our outlook on the central dogma of life and even on the entire process of evolution. Despite many open questions, the idea of a prebiotic RNA world, in which RNA acts as a catalyst as well as an information storage device, offers an attractive hypothesis to explain the origins of life without DNA and proteins (Gesteland and Atkins 1993). In vitro selection experiments have identified a variety of reactions catalyzed by RNA (Gold et al. 1995; Wilson and Szostak 1999), but RNA hydrolysis is still dominating both artificial and natural ribozymes. Although ultimate proof may remain elusive, comparison with protein RNases suggests that RNAs must build larger structures that are still catalytically less efficient than protein enzymes. Such lower activity may, in part, be due to alternative structures and folding traps, the numbers of which increase with the size of the RNA. Clearly, the most fascinating and complex folding trajectories have been observed by smFRET of large catalytic RNAs.
Hairpin ribozyme
The hairpin ribozyme is one of the most intensely studied RNA molecules (Bokinsky et al. 2003; Ditzler et al. 2008; Hohng et al. 2004b; Liu et al. 2007; Nahas et al. 2004; Rueda et al. 2004; Tan et al. 2003; Zhuang et al. 2002). Three forms have been explored, a construct retaining a four-way junction (4WJ) from the natural ribozyme, a structurally reduced three-way junction construct (Pljevaljcic et al. 2004), and finally a minimalist construct with a two-way junction (2WJ; Fig. 2). The latter construct, which had already been investigated by smFRET early on (Zhuang et al. 2002), has since served as a minimalist paradigm for studying RNA folding and function, especially by using smFRET. Altogether six states, at least three distinct folding pathways, and a multitude of rate coefficients have been determined (Bokinsky et al. 2003; Ditzler et al. 2008; Liu et al. 2007; Rueda et al. 2004; Zhuang 2002, 2005). In a first step, the substrate strand binds to the ribozyme main body by hybridization, and then undergoes a pivotal “docking” step, during which two internal loops on adjacent arms associate in a strongly Mg2+-ion-dependent manner to form the catalytic pocket. From this state, catalytic cleavage produces another docked state, still binding the hybridized product. These two docked states are in equilibrium, as ligation activity has also been observed. Before product release, the catalytic pocket has to be dissolved in an undocking step. Many of the more multistep folding pathways have been identified in the 2WJ construct by Mg2+-ion pulse-chase experiments, in analogy to the nonequilibrium Mg2+-ion relaxation experiments of Qu et al. (2008) termed single-molecule kinetic fingerprinting. Evidence for a pronounced ruggedness of the free energy landscape of the 2WJ hairpin has recently been presented (Ditzler et al. 2008). Different stable folding intermediates have been preparatively isolated based on their differential migration behavior in non-denaturing gel electrophoresis. Analysis of these isolates by smFRET then showed that they had retained certain kinetic folding characteristics throughout the isolation procedure. The authors report the retention of molecular heterogeneity even after exposure to very harsh denaturing conditions commonly believed to dissolve all features of secondary and tertiary structure (Ditzler et al. 2008). The 2WJ construct is catalytically active, but overall cleavage is slow due to several cycles of internal cleavage and the reverse reaction, i.e., ligation, before product release. For an activity comparable to the natural 4WJ construct, 2WJ requires a two to three orders of magnitude higher concentration of magnesium ions. The natural ribozyme strongly relies on the four-way junction to facilitate formation of the active site from its other structural elements (Hohng et al. 2004b; Tan et al. 2003). Investigation of the natural form by smFRET revealed fluctuations among three distinct states before cleavage: the folded state and two additional, rapidly interconverting states that strongly depended on the junction, and at least one of which is missing in the 2WJ construct (Tan et al. 2003). Further investigations brought a breakthrough in ribozyme research, as they succeeded in assignment of the relevant intermediates to E FRET states and real-time observation of cleavage and ligation (Nahas et al. 2004). The kinetic data suggest that, in vivo, the RNA may also undergo several cycles of internal cleavage and ligation, but the ultimate release of the cleaved product is greatly enhanced compared to the 2WJ construct. It was further suggested that the active site is stably formed by docking of two internal loops, but substrate cleavage accelerates undocking by two orders of magnitude, thus favoring the subsequent substrate release step (Nahas et al. 2004). This discrepancy between the two-way and four-way ribozymes has been interpreted as an allosteric effect of the junction on the docked structure of the loop domains (Zhuang 2005).
Tetrahymena ribozyme
Discovery of the catalytic properties of the Tetrahymena ribozyme (Kruger et al. 1982), a group I self-splicing intron, has resulted in the Nobel price being awarded to Tom Cech. Consequently, a vast amount of biochemical (Cech 1995) and structural data (Cate et al. 1996; Guo et al. 2004) have made this complex structure a paradigm of RNA folding, its importance even surpassing that of the hairpin ribozyme. Early investigations had already identified several folding pathways and intermediates (Silverman et al. 2000), before the first groundbreaking smFRET investigations took place on the full length RNA (Fig. 2). These studies identified kinetic traps in two previously known slow and one newly identified fast pathways, with folding rate coefficients of 1 s, 1 min, and 0.01 h, respectively (Zhuang et al. 2000). In an extension of this work, folding was investigated starting from several distinct “collapsed” states. Remarkably, these states could be selectively populated by controlled variation of the Na+ ion concentration in the absence of divalent cations. Formation of tertiary structure was then initiated selectively by addition of magnesium ions (Russell et al. 2002). Bartley et al. (2003) have used smFRET to apply the concept of Φ-value analysis to RNA folding. This concept, developed for protein folding (Fersht et al. 1992), uses site-directed mutagenesis (or modification) in combination with measurements of the folding to determine at which point on the reaction pathway interactions with the modified group are made. In the course of this work, the docking step of the P1 helix to the core of the ribozyme was investigated. The absence of effects on the docking constant, both from atomic modifications and global perturbations, strongly suggested that the transition state for docking occurs early and does not closely resemble the docked state (Bartley et al. 2003). The P4–P6 domain is a Tetrahymena ribozyme fragment of about 160 nucleotides, which has gained importance as one of the first complex RNA structures (apart from tRNA) to be solved by X-ray crystallography (Cate et al. 1996). Despite not being catalytically active, it has since served as a model system in its own right. Lee et al. (2007c) have used this system to establish a new, time-apertured photon correlation method for resolving the transition time between two states of RNA folding. Application to the P4–P6 domain yielded a folding transition time of 240 µs. Cooperativity of tertiary contact formation in the folding of the P4–P6 domain has been investigated by Sattin et al. (2008).
A related RNA, the bl5 group I intron (Fig. 2) has been investigated in conjunction with a cognate auxiliary protein CBP2 (Bokinsky et al. 2006). Two binding states of the protein were observed, a non-specific state and one in which the protein specifically recognizes features of the native RNA fold. Interestingly, both binding states appear to contribute to the overall chaperoning effect, with which the protein paves a way through the rugged free energy landscape to accelerate RNA folding into the native state.
RNase P
RNase P is a ribonucleoprotein (RNP) containing a single protein subunit in bacteria and several more in eukaryotes, in addition to a single RNA subunit. The main task of the holoenzyme in vivo is the hydrolytic processing of polycistronic transcripts, thereby releasing 5’-phosphate-containing tRNAs or tRNA precursors. The discovery of an intrinsic activity of bacterial RNase P RNA subunit (Guerrier-Takada et al. 1983) has resulted in the Nobel prize being awarded to Sid Altman (Altman 2007) and made this particular RNA the subject of intense research efforts. Recently, an RNA of 255 nucleotides corresponding to the catalytic core of RNase P has been investigated, with special attention devoted to early intermediates in the folding pathway (Qu et al. 2008; Smith et al. 2008; Xie et al. 2004). Here, either fluorescein-Cy3 or Cy3-Cy5 dye pairs were incorporated together with biotin by end labeling and by hybridization of labeled DNA to the extended RNA molecule (Fig. 2). As many as seven folding states were discovered based on distinguishable E FRET levels, connectivity of states, and conversion rates. Among these conformations, some are infrequently populated and have previously escaped detection in equilibrium measurements, as they primarily sample states near the minimum of the free energy landscape. By observation of nonequilibrium relaxation events, Qu et al. (2008) were able to analyze individual molecules in hitherto inaccessible regions of the free energy landscape.
VS ribozyme
The Varkud satellite (VS) RNA is an abundant transcript from DNA found in mitochondria of a number of natural isolates of Neurospora. The VS transcript contains the ~170-nt VS ribozyme that undergoes self cleavage as a result of its nucleolytic activity.
Using single-molecule FRET, Pereira et al. (2008) have addressed alternative global folds, their rates of interconversion, and how they relate to the cleavage activity. Cy3, Cy5, and biotin were attached to the 5’-end and the ends of the loop in the interior of RNA strand that is removed during self-cleavage (Fig. 2). FRET traces were recorded on a TIRF microscope with prism-type excitation. Three E FRET states were identified and the analysis of the dynamics showed that two transitions largely dominated the ensemble, in addition to some dynamic heterogeneity which is commonly noticed in single molecule studies on various other RNAs. It was suggested that a slow local conformational change, invisible to the smFRET experiment, must take place prior to ribozyme self-cleavage (Pereira et al. 2008).
Structural dynamics and small molecule binding in aptamers, allosteric ribozymes, riboswitches and artificial ribozymes
Aptamers are small synthetic RNAs capable of binding specific ligands, which may be large biomolecules, e.g., proteins, but also small molecules such as lysine, arginine, thiamine, and biologically active purines such as cyclic di-GMP, GTP, adenosine, and theophylline (Montange and Batey 2008; Sudarsan et al. 2008). Their small molecule binding properties largely distinguish them from antibodies, with which they are often compared. Like antibodies, most aptamers have been obtained by a selection process. This selection is typically conducted in vitro through a process called SELEX (systematic evolution of ligands by exponential enrichment) (Famulok and Mayer 1999; Wilson and Szostak 1999), which has also yielded numerous artificial ribozymes including the Diels-Alderase discussed below. In a recent publication (Elenko et al. 2009), the binding of a small molecule ligand, namely TMR-labeled GTP, to an in vitro selected aptamer was observed in a TIRF setting. This particular aptamer is known for its unusually strong binding: a KD of ~0.3–0.5 nM for the labeled GTP ligand was measured both in bulk studies and from on and off times measured by smFRET. The aptamer displayed great uniformity in its small molecule binding kinetics, which is in notable contrast to the heterogeneity typically observed in the folding kinetics of many natural ribozymes. By combination of both RNA types, engineered ‘‘aptazymes’’ are produced. To this end, in vitro selected aptamers are fused with ribozymes to create allosteric biocatalysts, which can be employed as biosensing components and artificial gene regulatory switches through ligand-induced conformational rearrangement and activation. These molecules are promising systems for application in smFRET experiments. A case in point is an aptazyme built from a theophylline aptamer with micromolar ligand affinity and the hammerhead ribozyme (Fig. 2) (Breaker 2002; de Silva and Walter 2009; Soukup and Breaker 1999; Soukup et al. 2000). Several FRET states with anti-correlated donor- and acceptor-fluorescence were observed in a dynamic equilibrium, in which the catalytically active state was frequently accessed in the theophylline-bound and, less frequently, in the ligand-free state. The latter finding is in agreement with the observed residual hammerhead ribozyme activity in the absence of theophylline (de Silva and Walter 2009). Such leakage, or in other terms, suboptimal allosteric transmission of structural rearrangements from the aptamer to the catalytic site, is still the major problem in aptazyme design and application. Ironically, the fact that nature has optimized solutions to this problem in the form of so-called riboswitches has escaped detection until recently (Blount and Breaker 2006; Grundy and Henkin 2006; Montange and Batey 2008). Thus, the principles of aptamers as well as of aptazymes have historically been detected and investigated on synthetic sequences, and only later it was discovered that certain organisms exploit the same principles for the regulation of gene expression. Such riboswitches are generally composed of two modular domains: the aptamer domain and the expression platform. In analogy to artificial aptazymes, the latter may be a ribozyme, as is the case of the GlmS riboswitch (Klein et al. 2007; Klein and Ferre-D’Amare 2006). Alternatively, the expression platform may contain Shine Dalgarno sequences or transcription terminator structures, which may be masked or accessible, depending on the binding state of the regulatory small molecule (Montange and Batey 2008). One particular natural riboswitch, which senses adenine in vivo and in vitro, is a rare type of an ‘‘on’’ switch, which disfavors the formation of a transcription terminator hairpin structure upon ligand binding, and thus activates gene expression (Johansen et al. 2003; Nygaard and Saxild 2005). Investigations on internally Cy3/Cy5 labeled riboswitch RNA (Fig. 2) by smFRET (Lemay et al. 2006) showed that folding and unfolding rates exhibited great heterogeneity, which was partially remedied by addition of the adenine ligand. Such restriction of folding pathways by binding of small molecules is most interesting because it is thought that co-transcriptional ligand binding and RNA folding plays a decisive role in this system as well as in other natural riboswitches. Interestingly, the investigations also uncovered the existence of an intermediate state in the folding process of a pivotal loop–loop interaction structure. Formation of the intermediate state was promoted by adenine, suggesting that the ligand actively participates in loop–loop interaction, and thus in the overall folding process of the riboswitch aptamer domain (Lemay et al. 2006). Notably, much additional insight into the mechanics of the adenine riboswitch has recently been obtained by single molecule force spectroscopy (Greenleaf et al. 2008).
Diels-Alderase ribozyme
The Diels-Alderase ribozyme is among the smallest RNAs whose intramolecular folding has been investigated by smFRET. This unusual catalytically active RNA is among the few to act in trans, thus not only being a self-modifying RNA, but one that can also perform multiple turnovers. As its ultimate outstanding feature, it is, as yet, the only known ribozyme to process two small organic molecules: anthracene and maleimide, neither of which contain any RNA-related structures (Seelig and Jaschke 1999; Seelig et al. 2000). These molecules undergo a Diels-Alder cycloaddition, which is strongly accelerated by this non-natural ribozyme. Introduction of the Cy3/Cy5 pair at various internal positions of the 49mer RNA led to one particular combination of attachment points (Fig. 2) that allowed the direct observation of three distinguishable folding intermediates; indications for the presence of additional states were also noticed. A large heterogeneity of folding rates was revealed despite the small size of the RNA (Kobitski et al. 2007). However, the magnesium dependence of the average rate coefficients followed a ‘chevron’ shape, as was also observed for large ribozymes (Fang et al. 1999).
RNA-protein complexes
Ribosomal protein synthesis
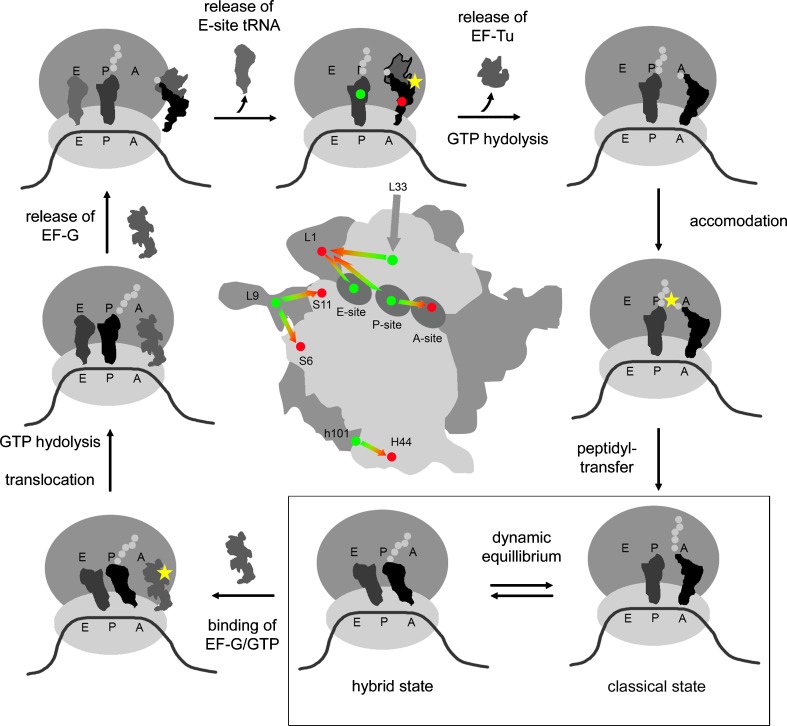
Peptide synthesis, i.e., the formation of an amide bond from an amine and an activated carboxylic acid, is the only known example of a reaction outside phosphodiester chemistry that is catalyzed by a natural RNA. In a process called translation, the ribosome utilizes aminoacylated tRNAs as adaptor molecules that decode the genetic code, contained in a nucleotide sequence on the mRNA, to translate it chemically into a polypeptide sequence. The ribosome is an immensely complex structure, containing dozens of proteins in addition to 3 or 4 rRNA units, but it has become clear that its active site contains no protein: the ribosome is a ribozyme (Cech 2000). Primed especially by the recently solved ribosome structures (Ramakrishnan 2002), research on ribosomal protein biosynthesis has greatly benefited from advances in single molecule techniques (Blanchard 2009; Marshall et al. 2008a; Munro et al. 2008). Out of the plethora of known and postulated states of bacterial peptide synthesis, some central aspects are depicted in Fig. 3, starting with the docking of the ternary complex of aminoacylated tRNA, GTP, and EF-Tu, followed by tRNA selection, accommodation, and peptide transfer to reach the so-called pre-translocation state, which, after EF-G binding, translocation, and GTP cleavage and EF-G ejection, reenters the cycle.
Fig. 3.
Overview of the cycle of ribosomal protein biosynthesis and smFRET investigation of ribosomal dynamics during translation. Sites of labeling with donor (green) and acceptor (red) dyes are indicated as dots of the respective color. Yellow stars symbolize conversion of chemical energy contained in GTP or aminoacyl-tRNA. The investigated dynamic equilibrium of classical and hybrid states of tRNA on the ribosome is boxed. A top view of the ribosome in the center shows the positioning of dyes on the large (dark grey) and small (light grey) ribosomal subunits. Green-red arrows indicate the distances monitored by smFRET. The grey arrow points to the position of the Cy3 label on L33, which is obscured by the small subunit
Single-molecule studies on the ribosome have recently been expertly reviewed (Marshall et al. 2008a; Blanchard 2009; Munro et al. 2008). As with tRNA, initial single-molecule fluorescence studies relied on a single TMR dye to yield information through decay times (Sytnik et al. 1999). An important prerequisite in the field was the development of immobilization methods for ribosomes that retained their activity in accommodation, translocation, and peptide-bond formation (Blanchard et al. 2004a, b). By application of smFRET, accommodation of the tRNA-elongation factor Tu-GTP ternary complex, and a subsequent multi-step movement of the tRNA on the ribosome, were observed in real time. By application of various antibiotics and non-hydrolyzable GTP analogs, various intermediates were observed, starting from an initial selection step, and including kinetic proofreading steps following GTP hydrolysis. Such studies also elucidated kinetic details of the action of antibiotics including tetracyclines, pyromycin, and thiostrepton (Gonzalez et al. 2007). The classic mechanistic description of ribosome function involves tRNA binding with both the 3’-end and the anticodon to the corresponding parts of the A- and the P-sites on each ribosomal subunit, respectively (Marshall et al. 2008a). smFRET investigations have also observed the so-called hybrid conformation, in which the tRNA is anchored to the small subunit P-site with its anticodon, while the deacylated 3’-end is already in the E-site of the large subunit (Lee et al. 2007b; Munro et al. 2007).
The unique capability of single molecule methods to detect and characterize subpopulations in structurally heterogeneous samples had led to the discovery of numerous unexpected aspects of ribosomes in early cryo-electron microscopy studies. Although cryo-electron microscopy cannot directly reveal movements by itself, certain of the characterized ribosome subpopulations showed features pointing to dynamic aspects of protein biosynthesis (Frank 1997; Frank et al. 2007; Julian et al. 2008). The observation of a relative torsion between the ribosomal subunits led to the postulate of a ratchet mechanism (Frank and Agrawal 2000), whose rotational movement might have a coordinating function for the various different dynamic aspects of ribosomal peptide synthesis. The ratchet motion has recently been investigated by three different teams (Cornish et al. 2008; Fei et al. 2008; Marshall et al. 2008b), who employed smFRET to look at a possible synchronization of the ratchet movement with a conformational equilibrium of classical and hybrid tRNA states on the ribosome (Lee et al. 2007b; Munro et al. 2007), as illustrated in the box in Fig. 3.
Because protein synthesis is driven by the conversion of chemical energy, e.g., during hydrolysis of the energy-rich aminoacyl-tRNA bond or during GTPase activity of EF-Tu and EF-G, these events have obviously received special attention in single-molecule investigations of ribosome dynamics. All three of the above groups reported a direct observation of the ratchet movement, but do so at different sites of the ribosome. As shown in Fig. 3, the teams of Noller and Ha attached Cy3 and Cy5 dyes to proteins S6 and L9 near the L1-stalk on the large ribosomal subunit (Cornish et al. 2008). A spontaneous, reversible relative motion of both dyes was observed, which, after extensive characterization, allowed a correlation with the tRNA equilibrium between classical and hybrid states (Fig. 3). The observed dynamics was characterized as thermally driven, thus not directly dependent on GTP hydrolysis or peptidyl transfer. A similar correlation of the dynamics between the L1-stalk itself and a tRNA oscillating between the classical and hybrid states was reported by the group of Gonzalez, Jr., who placed Cy3 and Cy5 dyes directly on the L1-stalk and tRNA (Fig. 3) (Fei et al. 2008). Finally, the teams of Puglisi and Chu positioned their Cy3 and Cy5 FRET dyes at a large distance to mRNA, tRNA and the peptidyl transfer active site (Fig. 3) (Marshall et al. 2008b). These sites were considered as more rigid and thus more representative for global movements of the ribosomal subunits as opposed to local flexibility near the L1-stalk. In contrast to the previous findings, these researchers did not report a thermally driven equilibrium of the ratchet movement at this part of the ribosome. Instead, association of small and large subunits was observed in real time, followed by a relative distortion of the subunits, which could be correlated with the arrival of aminoacylated tRNA on the ribosome in a three-color experiment also involving the Cy2 dye. The inhibition of the first part of the ratchet movement by chloramphenicol, but not by puromycin, suggested that this movement is associated with formation of the new peptide bond. The reverse ratchet movement proved to be dependent on GTP hydrolysis by EF-G. Subject to differences in the experimental setups, including ionic strengths, syntheses of dye-labeled ribosomes, or different methods for the assembly of translationally active complexes on the ribosome, the apparently contradictory data may be consolidated in a model, in which both parts of the global ratchet movement coincide with biochemical events in protein biosynthesis and are possibly driven by the corresponding conversion of chemical energy. This model would comprise the commonly known flexible L1-stalk region, where thermally driven local equillibria of local structure would be associated with the different tRNA states on the ribosome. Indeed, the movement of the L1 stalk relative to the body of the large subunit has recently been observed by labeling the L33 protein in addition to the L1 stalk. These studies have revealed at least three distinct states, whose population is indeed linked to the pre- versus post-translocation states of tRNAs on the ribosomes (Cornish et al. 2009). Since not all data seem to fit with the above model, it appears that the debate remains open. Recently, fascinating new insights into the mechanisms of protein biosynthesis have also come from force spectroscopy of individual active ribosomes (Uemura et al. 2007; Wen et al. 2008).
Replication
Telomerase
Telomerase is an RNP containing an RNA and a telomerase reverse transcriptase (TERT) protein subunit. The polymerase activity restores chromosome ends by adding telomeric DNA to the termini of linear chromosomes. As a reverse transcriptase, it uses parts of the RNA subunit as a template for DNA synthesis. The real-time assembly of individual telomerase RNP complexes, which in Tetrahymena thermophila is assisted by the p65 protein, was investigated in vitro by smFRET using RNA labeled at various positions with the Cy3/Cy5 FRET pair (Fig. 2) (Stone et al. 2007). A sequential addition of p65 first and then the TERT protein was evidenced, accompanied by substantial reorientation of the RNA domains. This process corresponds to a hierarchical assembly mechanism for telomerase RNP, in which the protein subunits mould a specific RNA tertiary structure in a stepwise fashion.
HIV Reverse Transcription
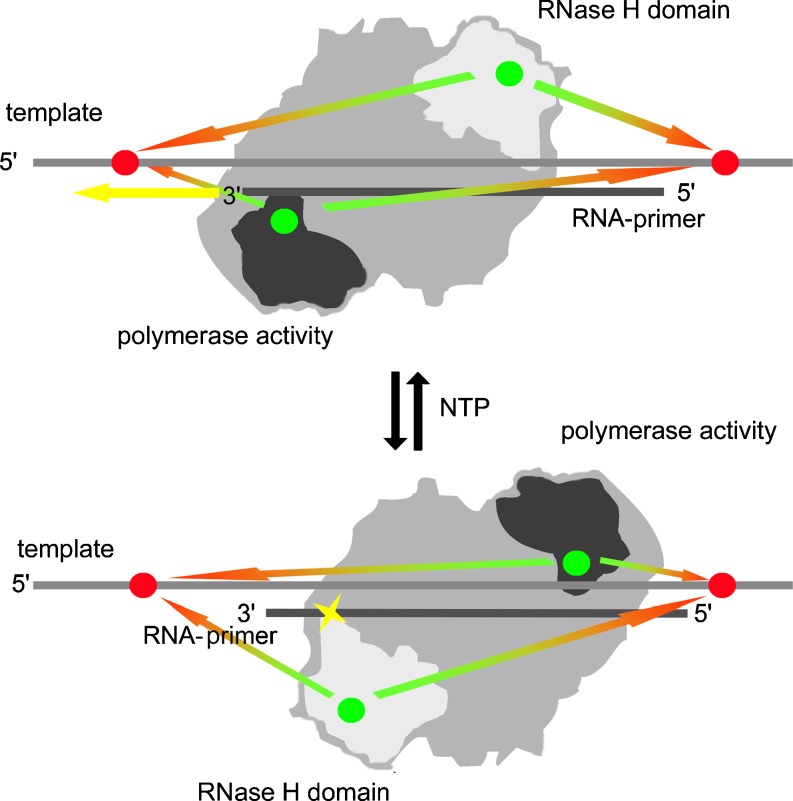
A particular highlight of recent smFRET science is the investigation of differential activities on HIV reverse transcriptase (RT) (Abbondanzieri et al. 2008; Liu et al. 2008). This enzyme, which is central to the highly complex conversion of viral RNA to double-stranded DNA, possesses two catalytic centers, combining polymerase activity with RNase H activity. It synthesizes a DNA strand using RNA or cDNA as a template and, in parallel, the RNase H function leads to degradation of the very RNA that has just been used as a template and is, therefore, still hybridized to the newly synthesized DNA. Among the many poorly understood characteristics of HIV replication, polypurine tracks (PPT) feature prominently (Abbondanzieri et al. 2008). These purine-rich stretches of nucleotides escape degradation by RNase H, and may thus serve as primers for second strand DNA synthesis. To investigate the dynamic behavior of HIV-RT in different complexes with nucleic acids by smFRET, Cy3 labels were attached to either the polymerase or the RNase H domain of the HIV-RT protein, and the corresponding acceptor dye Cy5 was placed near the 3’- or the 5’-end of the template strand, respectively (Fig. 4). Two binding modes, one for degradation and one for polymerization, could be identified in this way (Abbondanzieri et al. 2008). Remarkably, real time interconversion between the two modes was observed, which occurred without intermittent dissociation from the template (Fig. 4). Of particular interest was that the degradation mode was predominant with normal RNA primers on a DNA template. By contrast, the polymerization mode was favored with DNA or PPT as primers, which was even more preferred in the presence of dNTPs (Abbondanzieri et al. 2008). In further studies, the sliding motion of the RT protein along a RNA–DNA hybrid could be observed. On the RNA-3’-end, the protein was in the degradation mode, but switched to the polymerization mode upon reaching the 3’-end of the DNA primer (Liu et al. 2008). This sliding motion is in agreement with the observation of RNase H activity at several different locations on the RNA–DNA hybrid, and repeated sliding away from the polymerization site at the DNA 3’-end might explain the known low processivity of HIV-RT (Abbondanzieri et al. 2008). The presence of Nevapirin, a non-nucleoside RT-inhibitor, was observed to accelerate the interconversion between the two binding modes, yielding perspectives for the elucidation of the hithero ill-understood mechanism of Nevapirin (Abbondanzieri et al. 2008; Liu et al. 2008) .
Fig. 4.
Two orientations of HIV reverse transcriptase on a hybrid duplex comprised of a DNA template and an RNA primer. The upper panel shows the enzyme in its polymerization mode, characterized by the spatial proximity of the respective catalytic center to the 3’-end of the primer. The direction of polymerization is indicated by a yellow arrow. This conformation exists in a dynamic equilibrium with the opposite orientation, where the catalytic center of the RNase H activity is in spatial proximity to the RNA. The equilibrium is shifted to polymerization activity in the presence of NTPs. Sites of labeling with donor (green) and acceptor (red) dyes are indicated as dots of the respective color. Green-red arrows indicate the distances monitored by smFRET
Another step in the reverse transcription of the HIV-1 genome, the so-called minus-strand transfer was also investigated by smFRET (Zeng et al. 2007). In minus-strand transfer, the transactivation response region (TAR) RNA of the genome is annealed to the complementary "TAR DNA" generated during minus-strand strong-stop DNA synthesis. The smFRET investigations focused on the annealing of TAR DNA to an in vitro model consisting of cTAR DNA/TAR RNA. The results demonstrate that the HIV-1 nucleocapsid protein induces reversible annealing at various stages along the reaction path of the annealing reaction. Certain steps of the annealing sequence appear to be inhibited by argininamide (Landes et al. 2007).
Outlook
In recent years, smFRET measurements have yielded unprecedented insights into the conformational dynamics in the folding and function of RNA molecules. Based on the different FRET efficiency levels and their temporal behavior (Qu et al. 2008; Xie et al. 2004), multiple discrete conformations have been identified, structurally assigned and interpreted in functional terms. Even for rather small RNA molecules, a multitude of intermediate states has been observed in single molecule folding studies. Such investigations provide direct experimental access to the highly structured, hierarchical energy landscape governing conformational states and their interconversion dynamics.
Despite the impressive successes of smFRET-based experiments, there is still ample opportunity for further optimization. A sophisticated statistical analysis of the time traces of detected photon events will allow one to extract the relevant information more efficiently and to make proper assignments of conformational states (Gopich and Szabo 2009; Kalinin et al. 2007; McKinney et al. 2006; Noe and Fischer 2008). Conventional analysis distinguishes different conformations only on the basis of spectroscopic observables. Novel time-trace analysis tools based on Hidden Markov Models (Horenko et al. 2007) detect relevant conformations of the molecular complexes by observing the temporal sequence of the measured signal, even if the spectroscopic observables themselves overlap. While this technique has already been tested successfully in one-dimensional FRET measurements (McKinney et al. 2006), a multi-dimensional extension promises to yield an extremely detailed conformational analysis.
Another area in which further progress will be highly appreciated is the labeling of the RNA molecules with a FRET pair of dyes. Typically, dyes are attached with fairly long, flexible linkers, which introduce considerable spatial uncertainties. However, many biological processes involve minor rearrangements of the RNA molecules that may be hidden in FRET measurements due to the flexible dye linker. To visualize small-scale structural movements, changes in transition dipole orientation rather than inter-dye distance could be exploited by novel derivatization schemes involving direct dye attachment to the nucleotide bases (Ehrenschwender et al. 2008). Indeed, a noticeable dependence of the FRET efficiency was observed (Iqbal et al. 2008a) as a function of the relative orientation of Cy3 and Cy5 dyes stacked on the opposite ends of DNA duplex. Biophysical studies of RNA structure, dynamics and function on the single molecule level are still at the beginning, and exciting research problems in this field will likely keep us occupied for years to come.
Acknowledgements
The authors acknowledge funding by the Volkswagen Foundation and the Deutsche Forschungsgemeinschaft (DFG). We thank Martin Hengesbach for technical assistance.
References
- Abbondanzieri EA, Bokinsky G, Rausch JW, Zhang JX, Le Grice SF, Zhuang X. Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature. 2008;453:184–189. doi: 10.1038/nature06941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hashimi HM, Walter NG. RNA dynamics: it is about time. Curr Opin Struct Biol. 2008;18:321–329. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman S. A view of RNase P. Mol Biosyst. 2007;3:604–607. doi: 10.1039/b707850c. [DOI] [PubMed] [Google Scholar]
- Amirgoulova E, Groll J, Heyes CD, Ameringer T, Röcker C, Möller M, Nienhaus GU. Biofunctionalized polymer surfaces exhibiting minimal interaction towards immobilized proteins. Chem Phys Chem. 2004;5:552–555. doi: 10.1002/cphc.200400024. [DOI] [PubMed] [Google Scholar]
- Bartley LE, Zhuang X, Das R, Chu S, Herschlag D. Exploration of the transition state for tertiary structure formation between an RNA helix and a large structured RNA. J Mol Biol. 2003;328:1011–1026. doi: 10.1016/s0022-2836(03)00272-9. [DOI] [PubMed] [Google Scholar]
- Bates M, Blosser TR, Zhuang X. Short-range spectroscopic ruler based on a single-molecule optical switch. Phys Rev Lett. 2005;94:108101. doi: 10.1103/PhysRevLett.94.108101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard SC. Single-molecule observations of ribosome function. Curr Opin Struct Biol. 2009;19:103–109. doi: 10.1016/j.sbi.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- Bokinsky G, Zhuang X. Single-molecule RNA folding. Acc Chem Res. 2005;38:566–573. doi: 10.1021/ar040142o. [DOI] [PubMed] [Google Scholar]
- Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X. Single-molecule transition-state analysis of RNA folding. Proc Natl Acad Sci USA. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokinsky G, Nivon LG, Liu S, Chai G, Hong M, Weeks KM, Zhuang X. Two distinct binding modes of a protein cofactor with its target RNA. J Mol Biol. 2006;361:771–784. doi: 10.1016/j.jmb.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukobza E, Sonnenfeld A, Haran G. Immobilization in surface-tethered lipid vesicles as a new tool for single biomolecule spectroscopy. J Phys Chem B. 2001;105:12165–12170. [Google Scholar]
- Breaker RR. Engineered allosteric ribozymes as biosensor components. Curr Opin Biotechnol. 2002;13:31–39. doi: 10.1016/s0958-1669(02)00281-1. [DOI] [PubMed] [Google Scholar]
- Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu Rev Biophys Biomol Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- Cech T. Group I introns: new molecular mechanisms for mRNA repair. Biotechnology (NY) 1995;13:323–326. doi: 10.1038/nbt0495-323. [DOI] [PubMed] [Google Scholar]
- Cech TR. Structural biology. The ribosome is a ribozyme. Science. 2000;289:878–879. doi: 10.1126/science.289.5481.878. [DOI] [PubMed] [Google Scholar]
- Chan B, Weidemaier K, Yip WT, Barbara PF, Musier-Forsyth K. Intra-tRNA distance measurements for nucleocapsid proteindependent tRNA unwinding during priming of HIV reverse transcription. Proc Natl Acad Sci USA. 1999;96:459–464. doi: 10.1073/pnas.96.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SM, Stamou D. Surface-based lipid vesicle reactor systems: fabrication and applications. Soft Matter. 2007;3:828–836. doi: 10.1039/b702849k. [DOI] [PubMed] [Google Scholar]
- Cisse I, Okumus B, Joo C, Ha T. Fueling protein DNA interactions inside porous nanocontainers. Proc Natl Acad Sci USA. 2007;104:12646–12650. doi: 10.1073/pnas.0610673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci USA. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva C, Walter NG. Leakage and slow allostery limit performance of single drug-sensing aptazyme molecules based on the hammerhead ribozyme. RNA. 2009;15:76–84. doi: 10.1261/rna.1346609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich PS, Muller B, Schwille P. Studying reaction kinetics by simultaneous FRET and cross-correlation analysis in a miniaturized continuous flow reactor. Phys Chem Chem Phys. 2004;6:4416–4420. [Google Scholar]
- Ditzler MA, Aleman EA, Rueda D, Walter NG. Focus on function: single molecule RNA enzymology. Biopolymers. 2007;87:302–316. doi: 10.1002/bip.20819. [DOI] [PubMed] [Google Scholar]
- Ditzler MA, Rueda D, Mo J, Hakansson K, Walter NG. A rugged free energy landscape separates multiple functional RNA folds throughout denaturation. Nucleic Acids Res. 2008;36:7088–7099. doi: 10.1093/nar/gkn871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorywalska M, Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 2005;33:182–189. doi: 10.1093/nar/gki151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggeling C, Widengren J, Brand L, Schaffer J, Felekyan S, Seidel CA. Analysis of photobleaching in single-molecule multicolor excitation and Forster resonance energy transfer measurements. J Phys Chem A. 2006;110:2979–2995. doi: 10.1021/jp054581w. [DOI] [PubMed] [Google Scholar]
- Ehrenschwender T, Wanninger-Weiss C, Wagenknecht HA. BODIPY-modified uridines as potential fluorescent probes for nucleic acids that are recognized by DNA-polymerases. Nucleic Acids Symp Ser. 2008;52:349–350. doi: 10.1093/nass/nrn176. [DOI] [PubMed] [Google Scholar]
- Elenko MP, Szostak JW, van Oijen AM. Single-Molecule Imaging of an in Vitro-Evolved RNA Aptamer Reveals Homogeneous Ligand Binding Kinetics. J Am Chem Soc. 2009;131:9866–9867. doi: 10.1021/ja901880v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famulok M, Mayer G. Aptamers as tools in molecular biology and immunology. Curr Top Microbiol Immunol. 1999;243:123–136. doi: 10.1007/978-3-642-60142-2_7. [DOI] [PubMed] [Google Scholar]
- Fang XW, Pan T, Sosnick TR. Mg2+-dependent folding of a large ribozyme without kinetic traps. Nat Struct Biol. 1999;6:1091–1095. doi: 10.1038/70016. [DOI] [PubMed] [Google Scholar]
- Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Fersht AR, Matouschek A, Serrano L. The folding of an enzyme. I. Theory of protein engineering analysis of stability and pathway of protein folding. J Mol Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- Frank J. The ribosome at higher resolution - the donut takes shape. Curr Opin Struct Biol. 1997;7:266–272. doi: 10.1016/s0959-440x(97)80035-8. [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proc Natl Acad Sci USA. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland RF, Atkins JF (1993) The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
- Gold L, Polisky B, Uhlenbeck O, Yarus M. Diversity of oligonucleotide functions. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- Gonzalez RL, Jr, Chu S, Puglisi JD. Thiostrepton inhibition of tRNA delivery to the ribosome. RNA. 2007;13:2091–2097. doi: 10.1261/rna.499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopich IV, Szabo A (2009) Decoding the Pattern of Photon Colors in Single-Molecule FRET. J Phys Chem B (in press) [DOI] [PMC free article] [PubMed]
- Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll J, Amirgoulova EV, Ameringer T, Heyes CD, Röcker C, Nienhaus GU, Möller M. Biofunctionalized, ultrathin coatings of cross-linked star-shaped poly(ethylene oxide) allow reversible folding of immobilized proteins. J Am Chem Soc. 2004;126:4234–4239. doi: 10.1021/ja0318028. [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM. From ribosome to riboswitch: control of gene expression in bacteria by RNA structural rearrangements. Crit Rev Biochem Mol Biol. 2006;41:329–338. doi: 10.1080/10409230600914294. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Guo F, Gooding AR, Cech TR. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol Cell. 2004;16:351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Ha T, Zhuang X, Kim HD, Orr JW, Williamson JR, Chu S. Ligand-induced conformational changes observed in single RNA molecules. Proc Natl Acad Sci USA. 1999;96:9077–9082. doi: 10.1073/pnas.96.16.9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haustein E, Schwille P. Trends in fluorescence imaging and related techniques to unravel biological information. HFSP J. 2007;1:169–180. doi: 10.2976/1.2778852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilemann M, Margeat E, Kasper R, Sauer M, Tinnefeld P. Carbocyanine dyes as efficient reversible single-molecule optical switch. J Am Chem Soc. 2005;127:3801–3806. doi: 10.1021/ja044686x. [DOI] [PubMed] [Google Scholar]
- Hengesbach M, Kobitski A, Voigts-Hoffmann F, Frauer C, Nienhaus GU, Helm M (2008) RNA intramolecular dynamics by single-molecule FRET. Curr Protoc Nucleic Acid Chem Chapter 11:Unit 11 12 [DOI] [PubMed]
- Heyes CD, Kobitski AY, Amirgoulova EV, Nienhaus GU. Biocompatible surfaces for specific tethering of individual protein molecules. J Phys Chem B. 2004;108:13387–13394. [Google Scholar]
- Heyes CD, Groll J, Moller M, Nienhaus GU. Synthesis, patterning and applications of star-shaped poly(ethylene glycol) biofunctionalized surfaces. Mol Biosyst. 2007;3:419–430. doi: 10.1039/b700055n. [DOI] [PubMed] [Google Scholar]
- Hohng S, Joo C, Ha T. Single-molecule three-color FRET. Biophys J. 2004;87:1328–1337. doi: 10.1529/biophysj.104.043935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohng S, Wilson TJ, Tan E, Clegg RM, Lilley DM, Ha T. Conformational flexibility of four-way junctions in RNA. J Mol Biol. 2004;336:69–79. doi: 10.1016/j.jmb.2003.12.014. [DOI] [PubMed] [Google Scholar]
- Horenko I, Dittmer E, Fischer A, Christof S. Automated model reduction for complex systems exhibiting metastability. Multiscale Model Simul. 2007;5:802–827. [Google Scholar]
- Iqbal A, Arslan S, Okumus B, Wilson TJ, Giraud G, Norman DG, Ha T, Lilley DM. Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proc Natl Acad Sci USA. 2008;105:11176–11181. doi: 10.1073/pnas.0801707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal A, Wang L, Thompson KC, Lilley DM, Norman DG. The structure of cyanine 5 terminally attached to double-stranded DNA: implications for FRET studies. Biochemistry. 2008;47:7857–7862. doi: 10.1021/bi800773f. [DOI] [PubMed] [Google Scholar]
- Jia Y, Sytnik A, Li L, Vladimirov S, Cooperman BS, Hochstrasser RM. Nonexponential kinetics of a single tRNAPhe molecule under physiological conditions. Proc Natl Acad Sci USA. 1997;94:7932–7936. doi: 10.1073/pnas.94.15.7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen LE, Nygaard P, Lassen C, Agerso Y, Saxild HH. Definition of a second Bacillus subtilis pur regulon comprising the pur and xpt-pbuX operons plus pbuG, nupG (yxjA), and pbuE (ydhL) J Bacteriol. 2003;185:5200–5209. doi: 10.1128/JB.185.17.5200-5209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian P, Konevega AL, Scheres SH, Lazaro M, Gil D, Wintermeyer W, Rodnina MV, Valle M. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci USA. 2008;105:16924–16927. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinin S, Felekyan S, Antonik M, Seidel CA. Probability distribution analysis of single-molecule fluorescence anisotropy and resonance energy transfer. J Phys Chem B. 2007;111:10253–10262. doi: 10.1021/jp072293p. [DOI] [PubMed] [Google Scholar]
- Kapanidis A, Weiss S. Fluorescent probes and bioconjugation chemistries for single-molecule fluorescence analysis of biomolecules. J Chem Phys. 2002;117:10953–10964. [Google Scholar]
- Kapanidis AN, Laurence TA, Lee NK, Margeat E, Kong X, Weiss S. Alternating-laser excitation of single molecules. Acc Chem Res. 2005;38:523–533. doi: 10.1021/ar0401348. [DOI] [PubMed] [Google Scholar]
- Kauffmann E, Darnton NC, Austin RH, Batt C, Gerwert K. Lifetimes of intermediates in the beta -sheet to alpha -helix transition of beta -lactoglobulin by using a diffusional IR mixer. Proc Natl Acad Sci USA. 2001;98:6646–6649. doi: 10.1073/pnas.101122898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HD, Nienhaus GU, Ha T, Orr JW, Williamson JR, Chu S. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc Natl Acad Sci USA. 2002;99:4284–4289. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Ferre-D’Amare AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- Klein DJ, Been MD, Ferre-D’Amare AR. Essential role of an active-site guanine in glmS ribozyme catalysis. J Am Chem Soc. 2007;129:14858–14859. doi: 10.1021/ja0768441. [DOI] [PubMed] [Google Scholar]
- Kobitski AY, Nierth A, Helm M, Jaschke A, Nienhaus GU. Mg2+-dependent folding of a Diels-Alderase ribozyme probed by single-molecule FRET analysis. Nucleic Acids Res. 2007;35:2047–2059. doi: 10.1093/nar/gkm072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobitski AY, Hengesbach M, Helm M, Nienhaus GU. Sculpting an RNA conformational energy landscape by a methyl group modification–a single-molecule FRET study. Angew Chem Int Ed Engl. 2008;47:4326–4230. doi: 10.1002/anie.200705675. [DOI] [PubMed] [Google Scholar]
- Koopmans WJ, Schmidt T, van Noort J. Nucleosome immobilization strategies for single-pair FRET microscopy. ChemPhysChem. 2008;9:2002–2009. doi: 10.1002/cphc.200800370. [DOI] [PubMed] [Google Scholar]
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Kuzmenkina EV, Heyes CD, Nienhaus GU. Single-molecule FRET study of denaturant induced unfolding of RNase H. J Mol Biol. 2006;357:313–324. doi: 10.1016/j.jmb.2005.12.061. [DOI] [PubMed] [Google Scholar]
- Landes CF, Zeng Y, Liu HW, Musier-Forsyth K, Barbara PF. Single-molecule study of the inhibition of HIV-1 transactivation response region DNA/DNA annealing by argininamide. J Am Chem Soc. 2007;129:10181–10188. doi: 10.1021/ja071491r. [DOI] [PubMed] [Google Scholar]
- Lee JY, Okumus B, Kim DS, Ha T. Extreme conformational diversity in human telomeric DNA. Proc Natl Acad Sci USA. 2005;102:18938–18943. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NK, Kapanidis AN, Koh HR, Korlann Y, Ho SO, Kim Y, Gassman N, Kim SK, Weiss S. Three-color alternating-laser excitation of single molecules: monitoring multiple interactions and distances. Biophys J. 2007;92:303–312. doi: 10.1529/biophysj.106.093211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Blanchard SC, Kim HD, Puglisi JD, Chu S. The role of fluctuations in tRNA selection by the ribosome. Proc Natl Acad Sci USA. 2007;104:13661–13665. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Lapidus LJ, Zhao W, Travers KJ, Herschlag D, Chu S. Measuring the folding transition time of single RNA molecules. Biophys J. 2007;92:3275–3283. doi: 10.1529/biophysj.106.094623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem Biol. 2006;13:857–368. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Lipman EA, Schuler B, Bakajin O, Eaton WA. Single-molecule measurement of protein folding kinetics. Science. 2003;301:1233–1235. doi: 10.1126/science.1085399. [DOI] [PubMed] [Google Scholar]
- Liu S, Bokinsky G, Walter NG, Zhuang X. Dissecting the multistep reaction pathway of an RNA enzyme by single-molecule kinetic "fingerprinting". Proc Natl Acad Sci USA. 2007;104:12634–12639. doi: 10.1073/pnas.0610597104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Abbondanzieri EA, Rausch JW, Le Grice SF, Zhuang X. Slide into action: dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science. 2008;322:1092–1097. doi: 10.1126/science.1163108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RA, Aitken CE, Dorywalska M, Puglisi JD. Translation at the single-molecule level. Annu Rev Biochem. 2008;77:177–203. doi: 10.1146/annurev.biochem.77.070606.101431. [DOI] [PubMed] [Google Scholar]
- Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci USA. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerner WE, Fromm D. Methods of single-molecule fluorescence spectroscopy and microscopy. Rev Sci Instrum. 2003;74:3597–3619. [Google Scholar]
- Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- Muller BK, Zaychikov E, Brauchle C, Lamb DC. Pulsed interleaved excitation. Biophys J. 2005;89:3508–3522. doi: 10.1529/biophysj.105.064766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Vaiana A, Sanbonmatsu KY, Blanchard SC. A new view of protein synthesis: mapping the free energy landscape of the ribosome using single-molecule FRET. Biopolymers. 2008;89:565–577. doi: 10.1002/bip.20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahas MK, Wilson TJ, Hohng S, Jarvie K, Lilley DM, Ha T. Observation of internal cleavage and ligation reactions of a ribozyme. Nat Struct Mol Biol. 2004;11:1107–1113. doi: 10.1038/nsmb842. [DOI] [PubMed] [Google Scholar]
- Noe F, Fischer S. Transition networks for modeling the kinetics of conformational change in macromolecules. Curr Opin Struct Biol. 2008;18:154–162. doi: 10.1016/j.sbi.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Nygaard P, Saxild HH. The purine efflux pump PbuE in Bacillus subtilis modulates expression of the PurR and G-box (XptR) regulons by adjusting the purine base pool size. J Bacteriol. 2005;187:791–794. doi: 10.1128/JB.187.2.791-794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumus B, Wilson TJ, Lilley DM, Ha T. Vesicle encapsulation studies reveal that single molecule ribozyme heterogeneities are intrinsic. Biophys J. 2004;87:2798–2806. doi: 10.1529/biophysj.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira MJ, Nikolova EN, Hiley SL, Jaikaran D, Collins RA, Walter NG. Single VS ribozyme molecules reveal dynamic and hierarchical folding toward catalysis. J Mol Biol. 2008;382:496–509. doi: 10.1016/j.jmb.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeil SH, Wickersham CE, Hoffmann A, Lipman EA. A microfluidic mixing system for single-molecule measurements. Rev Sci Instrum. 2009;80:5105. doi: 10.1063/1.3125643. [DOI] [PubMed] [Google Scholar]
- Pljevaljcic G, Millar DP, Deniz AA. Freely diffusing single hairpin ribozymes provide insights into the role of secondary structure and partially folded states in RNA folding. Biophys J. 2004;87:457–467. doi: 10.1529/biophysj.103.036087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Smith GJ, Lee KT, Sosnick TR, Pan T, Scherer NF. Single-molecule nonequilibrium periodic Mg2+-concentration jump experiments reveal details of the early folding pathways of a large RNA. Proc Natl Acad Sci U S A. 2008;105:6602–6607. doi: 10.1073/pnas.0801436105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- Rasnik I, Myong S, Cheng W, Lohman TM, Ha T. DNA-binding orientation and domain conformation of the E. coli rep helicase monomer bound to a partial duplex junction: single-molecule studies of fluorescently labeled enzymes. J Mol Biol. 2004;336:395–408. doi: 10.1016/j.jmb.2003.12.031. [DOI] [PubMed] [Google Scholar]
- Rasnik I, McKinney SA, Ha T. Surfaces and orientations: much to FRET about? Acc Chem Res. 2005;38:542–548. doi: 10.1021/ar040138c. [DOI] [PubMed] [Google Scholar]
- Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- Rhoades E, Gussakovsky E, Haran G. Watching proteins fold one molecule at a time. Proc Natl Acad Sci USA. 2003;100:3197–3202. doi: 10.1073/pnas.2628068100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger R, Rocker C, Nienhaus GU. Fluctuation correlation spectroscopy for the advanced physics laboratory. Am J Phys. 2005;73:1129–1134. [Google Scholar]
- Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda D, Bokinsky G, Rhodes MM, Rust MJ, Zhuang X, Walter NG. Single-molecule enzymology of RNA: essential functional groups impact catalysis from a distance. Proc Natl Acad Sci USA. 2004;101:10066–10071. doi: 10.1073/pnas.0403575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R, Zhuang X, Babcock HP, Millett IS, Doniach S, Chu S, Herschlag D. Exploring the folding landscape of a structured RNA. Proc Natl Acad Sci USA. 2002;99:155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattin BD, Zhao W, Travers K, Chu S, Herschlag D. Direct measurement of tertiary contact cooperativity in RNA folding. J Am Chem Soc. 2008;130:6085–6087. doi: 10.1021/ja800919q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler B, Lipman EA, Steinbach PJ, Kumke M, Eaton WA. Polyproline and the "spectroscopic ruler" revisited with single-molecule fluorescence. Proc Natl Acad Sci USA. 2005;102:2754–2759. doi: 10.1073/pnas.0408164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig B, Jaschke A. A small catalytic RNA motif with Diels-Alderase activity. Chem Biol. 1999;6:167–176. doi: 10.1016/S1074-5521(99)89008-5. [DOI] [PubMed] [Google Scholar]
- Seelig B, Keiper S, Stuhlmann F, Jaschke A. Enantioselective ribozyme catalysis of a bimolecular cycloaddition reaction. Angew Chem Int Ed Engl. 2000;39:4576–4579. [PubMed] [Google Scholar]
- Silverman SK, Deras ML, Woodson SA, Scaringe SA, Cech TR. Multiple folding pathways for the P4–P6 RNA domain. Biochemistry. 2000;39:12465–12475. doi: 10.1021/bi000828y. [DOI] [PubMed] [Google Scholar]
- Smith GJ, Lee KT, Qu X, Xie Z, Pesic J, Sosnick TR, Pan T, Scherer NF. A large collapsed-state RNA can exhibit simple exponential single-molecule dynamics. J Mol Biol. 2008;378:943–953. doi: 10.1016/j.jmb.2008.01.078. [DOI] [PubMed] [Google Scholar]
- Soukup GA, Breaker RR. Engineering precision RNA molecular switches. Proc Natl Acad Sci USA. 1999;96:3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Emilsson GA, Breaker RR. Altering molecular recognition of RNA aptamers by allosteric selection. J Mol Biol. 2000;298:623–632. doi: 10.1006/jmbi.2000.3704. [DOI] [PubMed] [Google Scholar]
- Stone MD, Mihalusova M, O’Connor CM, Prathapam R, Collins K, Zhuang X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–461. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sytnik A, Vladimirov S, Jia Y, Li L, Cooperman BS, Hochstrasser RM. Peptidyl transferase center activity observed in single ribosomes. J Mol Biol. 1999;285:49–54. doi: 10.1006/jmbi.1998.2312. [DOI] [PubMed] [Google Scholar]
- Tan E, Wilson TJ, Nahas MK, Clegg RM, Lilley DM, Ha T. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc Natl Acad Sci USA. 2003;100:9308–9313. doi: 10.1073/pnas.1233536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumalai D, Hyeon C. RNA and protein folding: common themes and variations. Biochemistry. 2005;44:4957–4970. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- Tinnefeld P, Sauer M. Branching out of single-molecule fluorescence spectroscopy: challenges for chemistry and influence on biology. Angew Chem Int Ed Engl. 2005;44:2642–2671. doi: 10.1002/anie.200300647. [DOI] [PubMed] [Google Scholar]
- Uemura S, Dorywalska M, Lee TH, Kim HD, Puglisi JD, Chu S. Peptide bond formation destabilizes Shine-Dalgarno interaction on the ribosome. Nature. 2007;446:454–457. doi: 10.1038/nature05625. [DOI] [PubMed] [Google Scholar]
- Vogelsang J, Kasper R, Steinhauer C, Person B, Heilemann M, Sauer M, Tinnefeld P. A reducing and oxidizing system minimizes photobleaching and blinking of fluorescent dyes. Angew Chem Int Ed Engl. 2008;47:5465–5469. doi: 10.1002/anie.200801518. [DOI] [PubMed] [Google Scholar]
- Voigts-Hoffmann F, Hengesbach M, Kobitski AY, van Aerschot A, Herdewijn P, Nienhaus GU, Helm M. A methyl group controls conformational equilibrium in human mitochondrial tRNA(Lys) J Am Chem Soc. 2007;129:13382–11383. doi: 10.1021/ja075520+. [DOI] [PubMed] [Google Scholar]
- Walter NG (2003) Probing RNA structural dynamics and function by fluorescence resonance energy transfer (FRET). Curr Protoc Nucleic Acid Chem Chapter 11:Unit 11 10 [DOI] [PubMed]
- Walter NG, Huang CY, Manzo AJ, Sobhy MA. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat Methods. 2008;5:475–489. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, Bustamante C, Tinoco I. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widengren J, Kudryavtsev V, Antonik M, Berger S, Gerken M, Seidel CA. Single-molecule detection and identification of multiple species by multiparameter fluorescence detection. Anal Chem. 2006;78:2039–2050. doi: 10.1021/ac0522759. [DOI] [PubMed] [Google Scholar]
- Widengren J, Chmyrov A, Eggeling C, Lofdahl PA, Seidel CA. Strategies to improve photostabilities in ultrasensitive fluorescence spectroscopy. J Phys Chem A. 2007;111:429–440. doi: 10.1021/jp0646325. [DOI] [PubMed] [Google Scholar]
- Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- Wozniak AK, Nottrott S, Kuhn-Holsken E, Schroder GF, Grubmuller H, Luhrmann R, Seidel CA, Oesterhelt F. Detecting protein-induced folding of the U4 snRNA kink-turn by single-molecule multiparameter FRET measurements. RNA. 2005;11:1545–1554. doi: 10.1261/rna.2950605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Srividya N, Sosnick TR, Pan T, Scherer NF. Single-molecule studies highlight conformational heterogeneity in the early folding steps of a large ribozyme. Proc Natl Acad Sci USA. 2004;101:534–539. doi: 10.1073/pnas.2636333100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Liu HW, Landes CF, Kim YJ, Ma X, Zhu Y, Musier-Forsyth K, Barbara PF. Probing nucleation, reverse annealing, and chaperone function along the reaction path of HIV-1 single-strand transfer. Proc Natl Acad Sci USA. 2007;104:12651–12656. doi: 10.1073/pnas.0700350104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X. Single-molecule RNA science. Annu Rev Biophys Biomol Struct. 2005;34:399–414. doi: 10.1146/annurev.biophys.34.040204.144641. [DOI] [PubMed] [Google Scholar]
- Zhuang X, Rief M. Single-molecule folding. Curr Opin Struct Biol. 2003;13:88–97. doi: 10.1016/s0959-440x(03)00011-3. [DOI] [PubMed] [Google Scholar]
- Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- Zhuang X, Kim H, Pereira MJ, Babcock HP, Walter NG, Chu S. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]