Abstract
Allosteric regulation is a ubiquitous phenomenon exploited in biological processes to control cells in a myriad of ways. It is also of emerging interest in the design of functional proteins and therapeutics. Even though allostery was proposed over 50 years ago and has been studied intensively from a structural perspective, many key details of allosteric mechanisms remain mysterious. Over the last decade significant attention has been paid to the “dynamic component” of allostery, as opposed to the analysis of rigid structures. Nuclear magnetic resonance spectroscopy and its ability to detect conformationally dynamic processes at atomic resolution have played an important role in expanding our understanding of allosteric mechanisms and opening up new questions. This article focuses on work that highlights how protein dynamics can factor into allosteric processes in distinct ways. Two cases are contrasted. The first considers the “traditionally allosteric” protein CheY, which undergoes a conformational change as a key element of its allostery. The second considers the more rarely observed “dynamic allostery” in a PDZ domain, in which allosteric behavior arises from changes in internal structural dynamics. Interestingly, the dynamic processes in these two contrasting examples occur on different timescales. In the case of the PDZ domain, subsequent experimental and computational work is reviewed to reveal a more complete picture of this interesting case of allostery.
Keywords: Allostery, Protein dynamics, NMR, CheY, PSD-95, PDZ domain, Dynamic allostery
Introduction
In recent years, our appreciation of the ways in which proteins achieve allosteric effects have expanded significantly. Many new insights have arisen from the realization that dynamics are likely to play a central role in mediating allosteric control (Lee 2013; Manley and Loria 2012; Motlagh et al. 2014; Tsai et al. 2009; Tzeng and Kalodimos 2011; Zhuravlev and Papoian 2010). This is obviously true for cases of allostery in the absence of conformational change (Tsai et al. 2008), but a dynamics-based perspective has also infused insight into mechanistic possibilities for “traditional” allostery, in which the conformational change is viewed as the critical event responsible for a change of activity away from the effector site. To highlight different allosteric mechanisms that employ dynamics as an essential feature, two systems under study in my laboratory will be reviewed here. The first, the bacterial flagellar motor regulatory protein CheY, is a model for traditional allostery because it utilizes a conformational change. While the conformational change is ultimately responsible for CheY’s increased affinity for its target protein, CheY undergoes intrinsic dynamic switching on the microsecond–millisecond (μs–ms) timescale that enables the allosteric structural transition. The second system, the third PDZ domain from postsynaptic density-95 (“PDZ3” from PSD-95), does not appear to undergo any significant conformational changes, and the allosteric effect is mediated by changes in side-chain dynamics on the picosecond–nanosecond (ps–ns) timescale. For both of these proteins and for countless other allosteric systems, nuclear magnetic resonance (NMR) spin relaxation is an indispensable tool for detecting and quantifying molecular motion with residue-level resolution. Assessment of motion at this detailed level, along with accurate timescale information, will be needed for a breakdown of how allostery actually happens and what drives allosteric events. This review therefore has the dual purpose of underscoring the diversity in how proteins achieve allostery and emphasizing that NMR spectroscopy is a critical tool for identification and further characterization of allosteric mechanisms.
Allostery and dynamics in CheY
A major goal of structural studies of allosteric mechanisms is to use NMR and the detailed information it can provide to quantitatively characterize dynamic conformational switching events in allosteric proteins. This, however, is challenging as the great majority of well-known allosteric proteins are large and multimeric. As a result there are very few such studies with “large” proteins (by NMR standards, >25 kDa) since high-quality relaxation data are needed for quantitative analysis of switching rate constants and structural features that can yield insights into allosteric processes. Nevertheless, there are a number of very good NMR relaxation studies on larger proteins, many of which are allosteric (Lipchock and Loria 2010; Shi and Kay 2014; Tzeng and Kalodimos 2012; Venditti et al. 2015). Kern, Wemmer, and colleagues recognized that “response regulator” (RR) proteins could be useful for characterizing the dynamic aspects of allostery through NMR relaxation (Volkman et al. 2001). The so-called receiver domains of RRs undergo a conformational change upon phosphorylation of an aspartyl side chain, allowing a distal surface of the receiver domain to bind with greater affinity to its targets, which are variable depending on the RR (Bourret 2010). Even though receiver domains are essentially monomeric, most researchers consider this state to be true allostery: a change in activity distant from the site of phosphorylation. While the Kern lab pioneered studies of dynamics in the RR receiver domain NtrC (nitrogen regulatory protein C) using NMR combined with mutational studies, the rate at which NtrC switches is rather fast for straightforward characterization of switching motions using Carr–Purcell–Meiboom–Gill (CPMG) relaxation dispersion methodology (Gardino et al. 2009), which is briefly overviewed in Fig. 1. By contrast, the RR CheY, which consists only of a receiver domain, has the same general allosteric features as NtrC but switches intrinsically at a slower rate (McDonald et al. 2012) such that CPMG relaxation dispersion has greater potential. Thus, CheY appears to be a bona fide allosteric protein that operates via conformational switching between low- and high-affinity states, and yet it is small enough (approx. 16 kDa) for detailed study by NMR relaxation dispersion methods. I now discuss findings on CheY dynamics as they pertain to allostery.
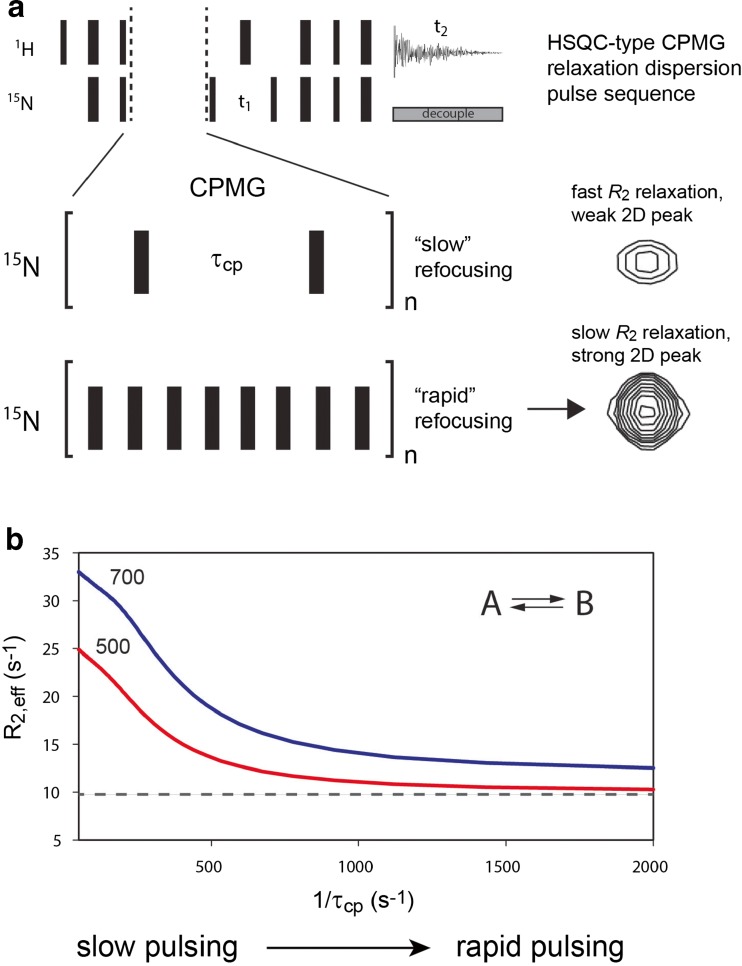
Fig. 1.
Schematic overview of Carr–Purcell–Meiboom–Gill (CPMG) relaxation dispersion. a Relation of pulse sequence to two-dimensional (2D) peak intensity. CPMG relaxation dispersion is collected in a 2D heteronuclear single quantum correlation (HSQC) format, with 90° and 180° pulses shown as thin and thick black bars, respectively. Multiple 2D spectra are acquired with different values of τcp and n such that the total CPMG time (n times the duration within the backets shown in Figure 1a) is constant. For residues undergoing chemical shift exchange (e.g., conformational exchange) on the microsecond to millisecond (μs–ms) timescale, peak intensities diminish as τcp increases because of an increase in line broadening due to chemical exchange. Rapid 180° pulsing suppresses 15N transverse relaxation (R 2,effective), leading to stronger peak intensities. b Theoretical CPMG relaxation dispersion plot, in which R 2,eff is plotted as a function of 1/τcp for a two-state chemical shift exchange. R 2,eff can be calculated directly from intensities of “A” state peaks (Mulder et al. 2001). Data are shown at 500 and 700 MHz. Curves were calculated for population of A and B states of 95 and 5 %, respectively, an exchange rate value (k ex) of 700 s−1 (sum of forward and reverse rate constants), and a difference in 15N chemical shift between states A and B (Δω) of 2.0 ppm. Dashed line Expected dispersion curve at 500 MHz in the absence of exchange
An interesting reason for studying CheY switching dynamics derives from earlier crystallography studies. Although numerous published studies have been critical to defining the primary inactive and active states and the conformational change model (Lee et al. 2001; Simonovic and Volz 2001; Volz and Matsumura 1991), a study by Dyer and Dahlquist focused on CheY conformations that fall in-between the fully inactive and active conformations (Dyer and Dahlquist 2006). The presence of these intermediate conformations suggests that CheY’s allosteric conformational change may involve more than two states. Complementing this, molecular dynamics simulations on CheY’s conformational change trajectory showed that different key regions that undergo significant movement during the inactive–active transition can move independently (Ma and Cui 2007).
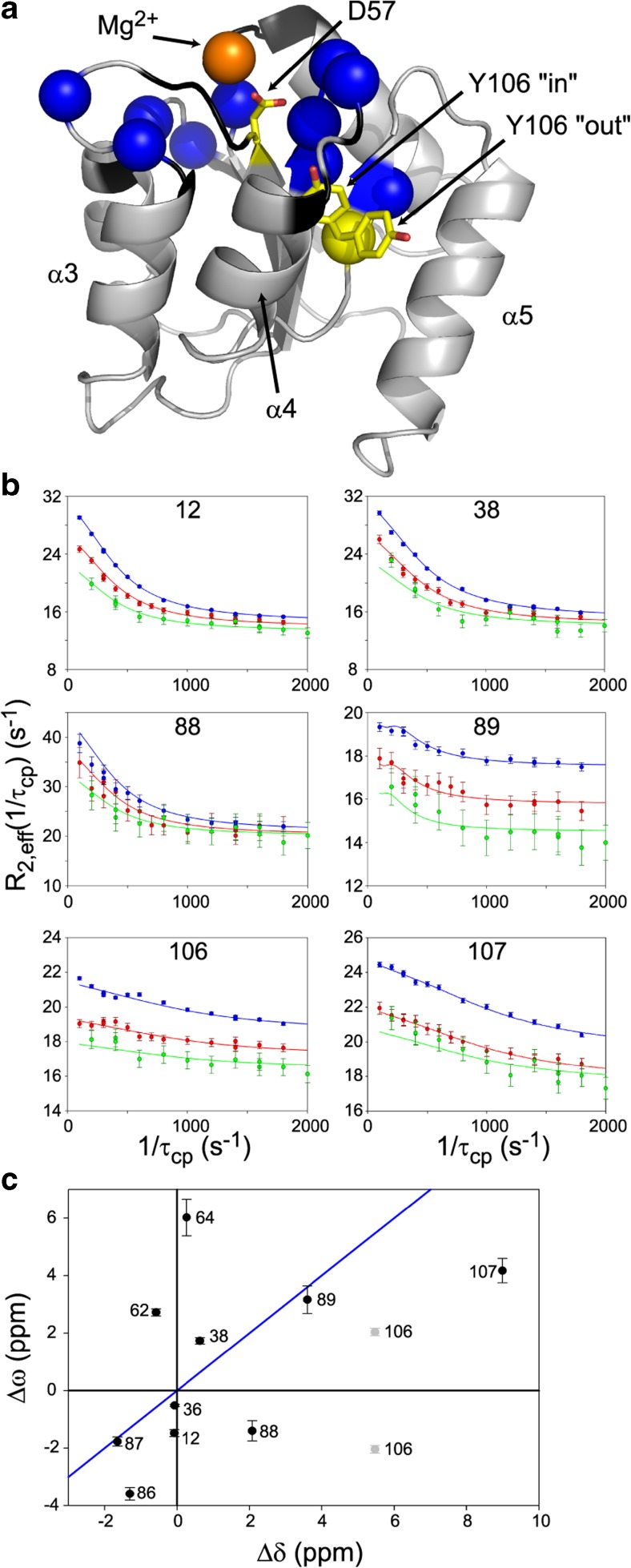
The attractiveness of NMR relaxation studies on CheY is that such studies have the potential to experimentally integrate the structural questions raised by the observed intermediate states with the dynamics and timescales associated with the conformational change(s) as observed in the molecular dynamics simulations. Our research group thus undertook CPMG relaxation dispersion studies of unphosphorylated CheY since this state appears to sample the active conformation at a low population (Schuster et al. 2001; Volz and Matsumura 1991), analogous to NtrC (Volkman et al. 2001). 15N CPMG relaxation dispersion allows protein motions on the timescale of approximately 200 μs to 10 ms (i.e., rate constants in the range 100–5000 s−1) to be characterized (see Fig. 1). Applied to CheY, these experiments revealed motion on this timescale at locations distributed throughout regions known to undergo conformational change, and little μs–ms motion was detected elsewhere (Fig. 2a). Assuming two-state conformational switching, analysis of CPMG relaxation dispersion under favorable conditions allows determination of the overall rate constant for switching (k ex = k fwd + k rev), of the equilibrium populations (pmaj and pmin), and of the chemical shift difference between the major and minor states (Δω). For the CheY residues along the allosteric pathway that exhibited pronounced dispersion curves (Fig. 2), two-state fitting yielded these parameters for each residue. The key finding from this analysis was that the values of k ex (and also pmaj/pmin) varied considerably among the residues, from approximately 300 to 3000 s−1. This result suggested that intrinsic conformational switching of CheY is not two-state and concerted, as would be expected from a Monod–Wyman–Changeux (MWC) model of allostery and which also has been the prevailing conceptual framework for receiver domain activation/allostery and many other allosteric systems. Two additional factors are consistent with the non-uniform k ex values that argue against simple two-state switching: (1) fitted Δω values did not agree with the known difference in 15N chemical shifts (Δδ) based on assigning the shifts for the inactive and active states of CheY (Fig. 2c); (2) the relaxation dispersion data could not be satisfactorily fitted when residues were grouped together (i.e., having shared k ex and pmaj/pmin values). Based on these results, CheY switching appears to be more complex. Unfortunately, insufficient data were collected to fit rigorously to three-state models of switching. Further analysis of chemical shifts of various CheY states led to the proposal that even though switching is not concerted, multiple sub-regions may switch as two-state concerted elements; this was described as “segmental switching” (McDonald et al. 2012).
Fig. 2.
15N CPMG relaxation dispersion data from unphosphorylated CheY in the presence of 10 mM Mg2+. a Structure of unphosphorylated CheY using 3CHY with the side-chain orientation of D57 and Mg2+ location from 2CHE. Residues with non-zero R ex values at 10 mM Mg2+ are displayed as blue spheres. Black indicates residues that are too broad to be measured. Highlighted in yellow are Y106 and the site of phosphorylation, D57. For Y106, the “in” rotamer indicates active and “out” indicates inactive CheY. b Raw CPMG dispersion curves are displayed for six example residues at 700 MHz (blue), 600 MHz (red), and 500 MHz (green). c Δω values from local fits of relaxation dispersion are plotted against Δδ using the difference between unphosphorylated CheY and BeFx-bound CheY both with 10 mM Mg2+. Δδ was determined using , where N is the 15N chemical shift and to get the final Δδ value we made an additional adjustment. Since unphosphorylated CheY is not completely inactive and BeFx-bound CheY is not completely active, we used Δδ = 1.05Δδ ′. The blue line has a slope of 1. Figure taken from McDonald et al. (2012)
In summary, the NMR-based work in McDonald et al. (2012) showed that intrinsic allosteric conformational switching is a complex dynamic event (or collection of smaller events) that can be quantitatively characterized at the residue level by NMR CPMG relaxation dispersion. These measurements indicate that the allosteric switching does not conform to MWC-type concerted, two-state switching but rather undergoes a more complex switching mechanism that may be segmental in nature, characterized by a variety of clustered residues moving with different time constants. Characterizing the structural and temporal aspects of conformational change mechanisms will provide us with a deeper understanding of allostery and may allow for allostery to be rationally designed into proteins or for intermediate states to be exploited in drug/ligand design.
Dynamic allostery in PDZ3 from PSD-95
PDZ domains and PDZ3 from PSD-95
In 1999, Ranganathan and coworkers published a provocative paper showing that from a multiple sequence alignment of a protein family, residue covariation could be used to track evolutionarily conserved “pathways” that connect structurally distant sites (Lockless and Ranganathan 1999). This approach is now known as statistical coupling analysis (SCA) and, remarkably, in that initial study the statistical couplings generally correlated with thermodynamic couplings between residue pairs, as measured from double mutant cycles. This study and the original SCA methodology have been subject to continuous debate, but this was an important study in terms of the idea of a “communication pathway”, and it therefore provided stimulation for both experimental and computational research, including our work on PDZ domains. The protein in which the thermodynamic coupling measurements were made was the third PDZ domain (“PDZ3”) from the synaptic scaffolding protein postsynaptic density-95 (PSD-95). PDZ3 is also the first PDZ domain to have its structure determined by X-ray crystallography (Doyle et al. 1996; Morais Cabral et al. 1996) and, therefore, it has probably become the most studied PDZ domain out of the approximately 270 PDZ domains in the human genome. In particular, it has been the subject of numerous computational studies focusing on the transmission of dynamics through protein structure.
PDZ domains are globular proteins composed of approximately 80–90 residus that fold up into a curved β-sheet formed from five to six strands and, typically, two α-helices (Ernst et al. 2014; Ye and Zhang 2013). They are often found in tandem arrays in signaling proteins operating near cell membranes. Their primary function is binding C-terminal tails of other proteins, in some cases membrane receptors which are thereby tethered via PDZ scaffolding. PDZ domains bind their C-terminal ligands through specific recognition of four to six—but in some cases as many as nine—residues (Birrane et al. 2003; Erlendsson et al. 2014; Feng et al. 2008). C-terminal ligands bind at a shallow groove of the PDZ domain between β-strand 2 and α-helix 2, in a backbone hydrogen bonding pattern that extends the antiparallel sheet by one strand and introduces hydrophobic interactions between PDZ and the ligand.
In hindsight, the early X-ray structures of PDZ3 were unusual for PDZ domains. Although not always shown in the figures, PDZ3 contains a third α-helix (residues 394–399) that packs against the conserved, core PDZ structure. In addition, these early PDZ3 structures contained additional C-terminal residues that formed a β-hairpin, although those residues derive from a cloning artifact and are, in fact, not part of PSD-95. Because α3 is located away from the observed binding site in these structures, this “extradomain structure” did not appear to serve any particular function, although since then it has been shown that this helix influences PDZ3 function. Interestingly, in recent years the presence of a third helix has been discovered in a modest fraction of other PDZ domains (Wang et al. 2010).
Dynamic allostery in PDZ3, conferred by the α3 helix
The role of the α3-helix in PSD-95 PDZ3 was first probed by truncation of residues 396–402 (Petit et al. 2009). These residues are the seven C-terminal residues of the canonical PDZ3 construct studied by Ranganathan (Lockless and Ranganathan 1999), with residue 402 being the last wild-type residue in the constructs used for crystallographic structures (Doyle et al. 1996). This truncated PDZ3, referred to here as PDZ3303–395, was found to bind to the canonical C-terminal peptide ligand from CRIPT (the 9-mer Ac-TKNYKQTSV) with a 21-fold lower affinity (Kd = 26 μM) than PDZ3303–402 (Kd = 1.2 μM) (Petit et al. 2009). Although there is currently no structure of PDZ3303–395 bound to peptide ligand, peptide binding to PDZ3 containing α3 results in only negligible changes in PDZ3 structure (Doyle et al. 1996). More recent experiments, published by the Martinez laboratory and unpublished from the author’s laboratory, have shown PDZ3303–395 to be stably folded at 25 °C and physiological pH ranges (Murciano-Calles et al. 2014). Also, comparison of Nuclear Overhauser effect spectroscopy (NOESY) data from PDZ3303–395 and PDZ3303–402 indicate that the NOE patterns within the PDZ3 core domain are essentially unchanged (unpublished data), consistent with these two PDZ3 cores having indistinguishable structural properties in solution at 25 °C. Given that the time-averaged structures of PDZ3303–395 and PDZ3303–402 appear to be very similar—either in the absence or presence of the C-terminal ligand—and α3 that does not make direct contact with the C-terminal ligand, how can the 21-fold difference in binding affinity be explained? Stated differently, what is the basis for allostery if α3 is considered to be an effector?
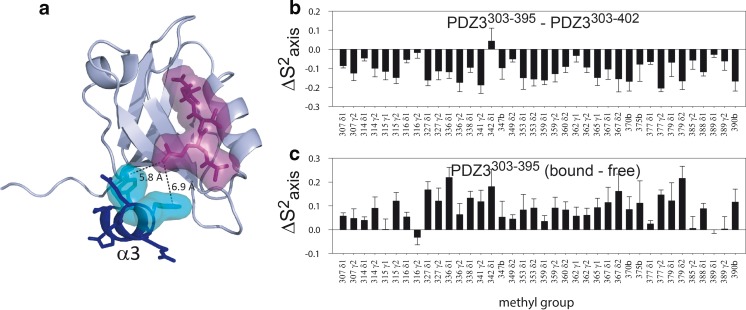
To address this question, other facets of peptide binding were studied (Petit et al. 2009). Isothermal titration calorimetry (ITC) was used to determine Kd (equilibrium dissociation constant) values, but this method also allows determination of ΔH and ΔS for the binding event. Table 1 shows these thermodynamic parameters for both forms of PDZ3. While both PDZ3 forms bind to the CRIPT peptide in an enthalpically driven manner, ΔH for the binding peptide is essentially unchanged upon deletion of the α3-helix. Rather, it is a larger –TΔS value that is responsible for the higher Kd for PDZ3303–395. Because of scant evidence for a structural explanation for the difference in binding affinities between the +α3 and –α3 PDZ3 forms, dynamics measurements were made on the free and bound forms of PDZ3303–402 and PDZ3303–395 using NMR spin relaxation. Relaxation studies focused on the ps–ns timescale because PDZ3 constructs show very little motion on the slower μs–ms timescales. Backbone dynamics were characterized using 15N R 1, R 2, and heteronuclear NOE measurements, which were interpreted within the Lipari–Szabo “model-free” formalism as so-called “order parameters,” or S 2. The order parameter is essentially a parameter of rigidity that varies on a scale of 0 to 1 and which corresponds to an individual bond vector. For NMR relaxation it encompasses reorientational motions of the bond vector (e.g., NH) occurring on the ps scale (and faster) and up to just below the characteristic time for molecular tumbling, which in the case of PDZ3 is approximately 7 ns. Thus, S 2 provides a measure of the amplitude of reorientational motion on the ps–ns timescale. Backbone NH order parameters were determined for all four states of PDZ3. While there are clearly some differences in S 2 that indicate some change in backbone dynamics, overall the differences are rather small, as is often seen for backbone dynamics (Clarkson and Lee 2004; Lee et al. 2000). Ps–ns side-chain dynamics were characterized using 2H-labeled methyl relaxation (Igumenova et al. 2006; Muhandiram et al. 1995; Petit and Lee 2014), which affords order parameters of the methyl symmetry axis (collinear with C–CH3 bond vector), S 2 axis (Igumenova et al. 2006). In contrast to the backbone, there is significantly more variability in the methyl S 2 axis parameters as a function of the ± α3 and ± CRIPT peptide. Considering the unbound forms, removal of α3 has the effect of increasing ps–ns side-chain dynamics, as indicated by the approximately ~0.1 lower S 2 axis values of PDZ3303–395 compared to PDZ3303–402 for nearly all methyl groups observed (Fig. 3b). Upon binding CRIPT 9-mer peptide, S 2 axis values in PDZ3303–402 show both positive and negative changes locally, with little overall change in side-chain dynamics (Petit et al. 2009). However, CRIPT binding to PDZ3303–395 leads to a large overall increase in S 2 axis values to a level comparable with those of PDZ3303–402 (Fig. 3c). Thus, unlike “normal” PDZ3303–402, PDZ3303–395 undergoes significant rigidification upon binding the peptide ligand, and there is an expected entropic penalty for doing so because of the loss of conformational entropy associated with an increase in order parameters. We refer the reader to a recent review by Wand for in-depth discussion of the relationship between order parameters and conformational entropy (Wand 2013).
Table 1.
Thermodynamic parametersc for PDZ3 constructs binding to CRIPT peptides (9-mer or 7-mer), determined by isothermal titration calorimetry
| PDZ3 constructs | Kd (μM) | ΔH (kcal/mol) | −TΔS (kcal/mol) |
|---|---|---|---|
| PDZ3303–402 (binding 9-mer)a | 1.2 (0.1) | −9.70 (0.12) | 1.66 (0.06) |
| PDZ3303–395 (binding 9-mer)a | 25.7 (3.6) | −10.3 (0.7) | 4.00 (0.59) |
| PDZ3303–402 (binding 7-mer)b | 3.60 (0.28) | −8.57 (0.31) | 1.16 (0.26) |
| pY-PDZ3303–402 (binding 7-mer)b | 14.0 (0.5) | −8.80 (0.29) | 2.18 (0.31) |
Standard errors are given in parenthesis
aPetit et al. (2009). CRIPT peptide used was Ac-TKNYKQTSV-COOH
bZhang et al. (2011). CRIPT peptide used was Ac-NYKQTSV-COOH. Note that in Zhang et al. (2011) the –TΔS terms in Table 1 were reported with incorrect signs. That error is corrected here
cKd is the equilibrium dissociation constant; Δ H is the change in enthalpy upon binding, and TΔ S is the temperature (in Kelvin) times the change in entropy upon binding
Fig. 3.
Dynamic allostery in PDZ3 from PSD-95. a Crystal structure of PDZ3 (including wild-type residues up to position 402) bound to C-terminal peptide from CRIPT, PDB ID 1BE9. α3 is shown as a dark blue ribbon, with the closest side chains highlighted in cyan. The CRIPT peptide is highlighted in magenta, and the distances of closest approach of α3 are shown. b, c Differences in side-chain methyl order parameters (ΔS 2 axis) are shown for removal of α3 from PDZ3303–402 (b) and binding of 9-mer CRIPT peptide to PDZ3303–395
The structural, thermodynamic, and dynamics data on PDZ3 suggest an explanation for how the α3 helix can regulate peptide binding. The most obvious mechanism of an allosteric conformational change in the PDZ core appears not to be supported by the experimental data (see above). Given the lack of a clear structural explanation, it is noteworthy that there is an approximately 2.3 kcal/mol difference in TΔS for binding peptide to PDZ3303–402 and PDZ3303–395 and a negligible difference in ΔH for the two forms (Table 1). Because the entropy change is less favorable (−TΔS is more positive) for PDZ3303–395, this represents an entropy penalty that is qualitatively well explained by the loss of side-chain entropy, observed as described above, upon binding peptide. Based on these findings, PDZ3 from PSD-95 serves as an example of “dynamic allostery”—whereby allostery is achieved through the modulation of conformational entropy, and in this particular example, via side-chain dynamics. In this model of allostery, the physical changes are not merely located in discrete structural regions, but rather they are delocalized throughout the PDZ domain. This type of allosteric mechanism is precisely that postulated by Cooper some 30 years ago (Cooper and Dryden 1984). It is not easily visualized with static models but rather must be envisioned with the basic understanding that, in principle, entropy can contribute to binding affinity just as it can with enthalpy (Motlagh et al. 2014). Examples from the Wand and Kalodimos laboratories are particularly striking (Frederick et al. 2007; Tzeng and Kalodimos 2012). In terms of relating this back to the original SCA study on PDZ3, these findings offer some insight into how a conserved energetic pathway might exist throughout the PDZ family, as well as into how side-chain dynamics may play a role since dynamics can influence peptide binding affinity. However, it is important to realize that this work does not provide a direct assessment of the original SCA study since individual couplings were not measured.
Is PDZ3 allostery of biological significance or just proof of principle?
Undoubtedly obvious to many readers up to this point is that the “allostery” under consideration in PDZ3 has been entirely artificial because the α3 “effector” has only been removed through recombinant DNA methods. Not only is the α3 sequence always present in PDZ3 (and PSD-95) in the cell, but it would presumably always be folded and packed into the PDZ3 domain—indeed, it is conceivably best considered as an inseparable part of the PDZ3 structure in vivo. Even if this were the case, from a biophysical perspective this serves as a useful proof of principle for dynamic allostery. However, mass spectrometry studies have shown that Tyr-397, within α3, is phosphorylated in brain tissue (Ballif et al. 2008). This has raised the question of whether somehow α3 can be reversibly perturbed and hence serve as a bona fide effector or allosteric signal transduction site of action. Phosphorylation of Tyr-397 was studied biochemically and biophysically to address this question (Zhang et al. 2011). Because PDZ303–402 only has two tyrosine residues, it was straightforward to phosphorylate Tyr-397 and purify this specific phosphorylated form of PDZ3 for ITC and NMR studies. Even though in this study the C-terminal CRIPT peptide ligand was a 7-mer (as opposed to a 9-mer), the same behavior was observed (Table 1); namely, phosphorylation reduced the binding affinity of PDZ3, and the effect was observed to be almost entirely in the entropy term. This result showed that peptide ligand binding affinity can be regulated by phosphorylation at a site not known to participate directly in ligand binding.
To see how phosphorylation imparts weaker binding to PDZ3, NMR was used to characterize phosphorylated PDZ3303–402 (pY397-PDZ3) for comparison to the unphosphorylated PDZ3303–402 and PDZ3303–395 states (Zhang et al. 2011). The most striking finding was that the pattern of chemical shift perturbations upon phosphorylation bears a striking resemblance to that for removal of the α3 helix. Closer inspection of tow dimensional (2D) HSQC overlays showed that many peaks for PDZ3303–402, PDZ3303–395, and pY397-PDZ3 fell into a linear pattern, with the pY397-PDZ3 falling in the middle position. Such behavior is a clear signature of an equilibrium between two states in rapid exchange on the NMR timescale. In fact, the end states are well-characterized: they are the highly structured PDZ3 domain with α3 stably packed and the structured but more dynamic (at the side-chain level) PDZ3 domain lacking α3. With the pY397-PDZ3 peak falling roughly in the middle of these end peaks, phosphorylation at Tyr-397 can be considered to induce the undocking of α3 from the PDZ3 core, with re-docking occurring with roughly equal probability on a sub-millisecond timescale. The α3-undocked state therefore mimics the dynamic PDZ3303–395 form and likewise has a lower binding affinity. The effect is dampened in the Kd values because approximately one-half of the population is presumably in the high-affinity form (Table 1). In summary, the characterization of Tyr-397-phosphorylated PDZ3303–402 suggests that truncation of the α3-helix (i.e., PDZ303–395) serves as a mimic of the α3-undocked, phosphorylated PDZ3 domain which appears to exist in vivo. This suggests yet further that the dynamic allostery observed using PDZ3303–395 may have biological significance, although precisely what this biological significance is is currently not known. It may, however, be the subject for future studies.
Computational studies of PDZ3 suggest a role for transient interactions
Since the reporting of α3 and its role in dynamic allostery in PDZ3 (Petit et al. 2009), there have been a number of studies that examined the allosteric basis in more detail. Here, I first discuss the computational studies on PDZ3, then the experimental studies. In one of the first computational studies of PDZ3 and the role of α3 in regulating CRIPT binding, the free and bound states of PDZ3306–402 and PDZ3306–395 were simulated using molecular dynamics (Mostarda et al. 2012). From these simulations, the α3 salt bridges that are evident in the original PDZ3 structures (1BFE, 1BE9), R399–E334 and E401-K355, appear to have a role in stabilizing PDZ3 core elements that become more flexible upon truncation of α3. In the truncated PDZ3306–395, the backbone atoms of the β2–β3 loop, the carboxylate binding loop (318–323), and residues 342–357 increase in flexibility on a timescale of approximately 100 ns. In addition, two acidic residues in the β2–β3 loop make transient interactions with CRIPT peptide lysine residues at the −4 and −7 positions, residues that are not observed in the crystal structures due to flexibility. Intermolecular contact between some combinations of these residues is observed 44 % of the time in these simulations. Based on these observations, the authors proposed that the α3 helix influences C-terminal ligand binding by stabilizing the β2–β3 loop which makes transient contacts with the ligand and by indirectly stabilizing the carboxylate binding loop. Consistent with the notion of allostery, the simulations revealed no direct interactions between α3 and the CRIPT 9-mer peptide. Also consistent with a direct role of dynamics in allostery, these authors observed increased dynamics in PDZ3 upon truncation of α3. However, this study looked only at backbone dynamics—not side-chain dynamics. Interestingly, the enhanced backbone flexibility in PDZ3306–395 was only observed on the slower timescale of approximately100 ns and longer. One possibility is that the conformational entropy that is quenched upon ligand binding is present on slow timescales for backbone atoms and on faster timescales for side-chain atoms. Further study will be required to resolve these questions.
In a separate interesting computational study, Steiner and Caflisch (2012) focused on dynamic conformational switching in PDZ3303–402. In that study, the authors observed that PDZ3303–402 samples three major conformational “basins” during a total simulation time of 650 ns, thus showing that the native state contains these three basins. The conformational basins (“A,” “B,” and “C”) differ by the relative orientation of the α2 helix and the β3 strand, which together form the shallow binding groove for PDZ ligands. Upon binding CRIPT peptide, PDZ3 remains strictly in the “A” basin. Thus, PDZ3303–402 undergoes conformational selection to some degree. Even though the complex was made using a 5-mer of CRIPT peptide, the peptide remains bound to PDZ3, unlike in the previously described study of Mostarda et al. (2012). Especially interesting in this study is that the salt bridges (e.g., R399–E334, R399–E331) correlate strongly with transitions between the three basins.
Experimental studies of PDZ3 probing ligand residues −4 to −9
In 2010, Martinez and coworkers solved the crystal structure of PDZ3 without the additional C-terminal (after position 402, see above) cloning artifact sequences (Camara-Artigas et al. 2010). Two crystal forms were obtained that yielded similar findings. Interestingly, crystallized PDZ3 underwent succinimide ring formation at D332 in the important β2–β3 loop, although the lifetime of the succinimide group is only approximately 1 h in solution (Murciano-Calles et al. 2014). With the artificial structure from the C-terminal residues removed, α3 relaxes, changing the positioning of some of the side chains. As a result, α3 and the β2–β3 loop move approximately 3 Å closer, relative to the original PDZ3 structures. These new structures raise the question once again of whether a simpler structural explanation exists for how α3 affects ligand binding. For example, a new interaction which had not been seen previously is that between the side chain of E401 and the side chain of E334 in the β2–β3 loop. It is not clear if the newly observed structural interactions are due to the use of a natural PDZ3 construct, succinimide group formation (and β2–β3 rigidification), or both. Furthermore, because this construct has not been crystallized with peptide ligand, potentially different interactions between PDZ3 and ligand remain unknown.
In an interesting study by Jemth and coworkers, NMR evidence was presented that implicates CRIPT residues that are N-terminal to position −4 in making interactions with PDZ3 (Chi et al. 2012). This could potentially provide a direct mechanism for at least part of the loss of binding strength upon truncation of α3 if those N-terminal residues interact favorably with α3. The primary finding was the observation of weak NOEs to a ring proton on tyrosine at the −5 position of CRIPT; the NOEs were hypothesized (but not confirmed) to originate from PDZ3 protons, which would be consistent with the patterns of chemical shift perturbations observed in the peptide (9-mer) upon addition of PDZ3 (Chi et al. 2012). A few caveats should be mentioned for the hypothesis that direct interactions between N-terminal CRIPT residues and α3 residues contribute to the “high” affinity between PDZ3303–402 and CRIPT. First, in contrast to the original study by Petit et al. (2009) which was carried out at 25 °C, these NMR studies were performed at 5–10 °C since raising the temperature decreased the quality of the spectra. Second, it is not clear exactly what PDZ3 construct was used in the Jemth study (Chi et al. 2012). Prior studies from the same laboratory used a F337W mutation (which is located at the interface with α3) and a PDZ3 construct that terminates at E401. Prior studies also use an N-terminal poly-histidine tag, which could conceivably change interactions with α3-bound peptide, or the exposed interface upon α3 deletion, relative to the natural PDZ3 N-terminal/linker residues. Third, the originating protons to the NOEs observed to Y(−5) when bound to PDZ3 were not assigned, and the authors state that these could be from either PDZ3 or CRIPT side-chains. Nevertheless, these studies should help to gain a more complete understanding of PDZ3-ligand interactions.
A recurring theme arising from these recent experimental and computational studies is the existence of transient interactions that contribute to binding affinity. Although transient interactions are more difficult to detect than stable interactions, further progress towards understanding the unusual ligand affinity effects observed in the PSD-95–PDZ3 system are likely to involve better characterization of these transient interactions, including how these may change with temperature. More broadly, it will be interesting to see future examples of conformational entropy entering into the thermodynamics of protein–protein and protein–ligand interactions. It is noted that the atypical α3 element is conserved in other PDZ3 domains for the PSD-95 homologs SAP97, SAP102, PSD-93, and the Drosophila protein Dlg. In NHERF1, the second PDZ domain has a more extensive extra-domain structure consisting of a helix-turn-helix motif, which confers approximately a tenfold increase in ligand binding affinity, similar to PDZ3 (Bhattacharya et al. 2010). Strikingly, a comprehensive study by Zhang and coworkers indicates that approximately 30 % of all PDZ domains possess extra-domain structures (Wang et al. 2010). Further characterization of the role of these extra-domain structures—in PDZ and non-PDZ proteins alike—as well as of dynamic allostery will be of great interest to protein biophysics in the future.
Acknowledgments
Compliance with ethical standards
ᅟ
Funding
This work was supported by National Institutes of Health grant GM083059.
Conflict of interest
Andrew L. Lee declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on ‘The Role of Protein Dynamics in Allosteric Effects’ edited by Gordon Roberts.
References
- Ballif BA, Carey GR, Sunyaev SR, Gygi SP. Large-scale identification and evolution indexing of tyrosine phosphorylation sites from murine brain. J Proteome Res. 2008;7:311–318. doi: 10.1021/pr0701254. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Dai Z, Li J, Baxter S, Callaway DJ, Cowburn D, Bu Z (2010) A conformational switch in the sodium/hydrogen exchange regulatory factor 1(NHERF1) controls autoinhibition and complex formation. J Biol Chem 85(13):9981–9994. doi: 10.1074/jbc.M109.074005 [DOI] [PMC free article] [PubMed]
- Birrane G, Chung J, Ladias JA. Novel mode of ligand recognition by the Erbin PDZ domain. J Biol Chem. 2003;278:1399–1402. doi: 10.1074/jbc.C200571200. [DOI] [PubMed] [Google Scholar]
- Bourret RB. Receiver domain structure and function in response regulator proteins. Curr Opin Microbiol. 2010;13:142–149. doi: 10.1016/j.mib.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camara-Artigas A, Murciano-Calles J, Gavira JA, Cobos ES, Martinez JC. Novel conformational aspects of the third PDZ domain of the neuronal post-synaptic density-95 protein revealed from two 1.4 A X-ray structures. J Struct Biol. 2010;170:565–569. doi: 10.1016/j.jsb.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Chi CN, Haq SR, Rinaldo, Jemth P (2012) Interactions outside the boundaries of the canonical binding groove of a PDZ domain influence ligand binding. Biochemistry 51:8971–8979. doi:10.1021/bi300792h [DOI] [PubMed]
- Clarkson MW, Lee AL. Long-range dynamic effects of point mutations propagate through side chains in the serine protease inhibitor eglin c. Biochemistry. 2004;43:12448–12458. doi: 10.1021/bi0494424. [DOI] [PubMed] [Google Scholar]
- Cooper A, Dryden DT. Allostery without conformational change. A plausible model. Eur Biophys J. 1984;11:103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/S0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- Dyer CM, Dahlquist FW. Switched or not?: The structure of unphosphorylated CheY bound to the N terminus of FliM. J Bacteriol. 2006;188:7354–7363. doi: 10.1128/JB.00637-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlendsson S, Rathje M, Heidarsson PO, Poulsen FM, Madsen KL, Teilum K, Gether U. Protein interacting with C-kinase 1 (PICK1) binding promiscuity relies on unconventional PSD-95/discs-large/ZO-1 homology (PDZ) binding modes for nonclass II PDZ ligands. J Biol Chem. 2014;289:25327–25340. doi: 10.1074/jbc.M114.548743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst A, Appleton BA, Ivarsson Y, Zhang Y, Gfeller D, Wiesmann C, Sidhu SS. A structural portrait of the PDZ domain family. J Mol Biol. 2014;426:3509–3519. doi: 10.1016/j.jmb.2014.08.012. [DOI] [PubMed] [Google Scholar]
- Feng W, Wu H, Chan LN, Zhang M. Par-3-mediated junctional localization of the lipid phosphatase PTEN is required for cell polarity establishment. J Biol Chem. 2008;283:23440–23449. doi: 10.1074/jbc.M802482200. [DOI] [PubMed] [Google Scholar]
- Frederick KK, Marlow MS, Valentine KG, Wand AJ. Conformational entropy in molecular recognition by proteins. Nature. 2007;448:325–329. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardino AK, Villali J, Kivenson A, Kern D (2009) Transient non-native hydrogen bonds promote activation of a signaling protein. Cell 139:1109–1118. doi:10.1016/j.cell.2009.11.022 [DOI] [PMC free article] [PubMed]
- Igumenova TI, Frederick KK, Wand AJ. Characterization of the fast dynamics of protein amino acid side chains using NMR relaxation in solution. Chem Rev. 2006;106:1672–1699. doi: 10.1021/cr040422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AL. Protein dynamics and allostery. In: Roberts GCK, editor. Encyclopedia of biophysics. New York: Springer; 2013. [Google Scholar]
- Lee AL, Kinnear SA, Wand AJ. Redistribution and loss of side chain entropy upon formation of a calmodulin-peptide complex. Nat Struct Biol. 2000;7:72–77. doi: 10.1038/71280. [DOI] [PubMed] [Google Scholar]
- Lee SY, Cho HS, Pelton JG, Yan D, Berry EA, Wemmer DE. Crystal structure of activated CheY. Comparison with other activated receiver domains. J Biol Chem. 2001;276:16425–16431. doi: 10.1074/jbc.M101002200. [DOI] [PubMed] [Google Scholar]
- Lipchock JM, Loria JP. Nanometer propagation of millisecond motions in V-type allostery. Structure. 2010;18:1596–1607. doi: 10.1016/j.str.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- Ma L, Cui Q. Activation mechanism of a signaling protein at atomic resolution from advanced computations. J Am Chem Soc. 2007;129:10261–10268. doi: 10.1021/ja073059f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley G, Loria JP. NMR insights into protein allostery. Arch Biochem Biophys. 2012;519:223–231. doi: 10.1016/j.abb.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald LR, Boyer JA, Lee AL. Segmental motions, not a two-state concerted switch, underlie allostery in CheY. Structure. 2012;20:1363–1373. doi: 10.1016/j.str.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais Cabral JH, Petosa C, Sutcliffe MJ, Liddington RC (1996) Crystal structure of a PDZ domain. Nature 382:649–652 [DOI] [PubMed]
- Mostarda S, Gfeller D, Rao F. Beyond the binding site: the role of the beta(2)-beta(3) loop and extra-domain structures in PDZ domains. PLoS Comput Biol. 2012;8:e1002429. doi: 10.1371/journal.pcbi.1002429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhandiram DR, Yamazaki T, Sykes BD, Kay LE. Measurement of 2H T1 and T1r relaxation times in uniformly 13C-labeled and fractionally 2H-labeled proteins in solution. J Am Chem Soc. 1995;117:11536–11544. doi: 10.1021/ja00151a018. [DOI] [Google Scholar]
- Mulder FA, Skrynnikov NR, Hon B, Dahlquist FW, Kay LE. Measurement of slow (micros-ms) time scale dynamics in protein side chains by (15)N relaxation dispersion NMR spectroscopy: application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J Am Chem Soc. 2001;123:967–975. doi: 10.1021/ja003447g. [DOI] [PubMed] [Google Scholar]
- Murciano-Calles J, Corbi-Verge C, Candel AM, Luque I, Martinez JC. Post-translational modifications modulate ligand recognition by the third PDZ domain of the MAGUK protein PSD-95. PLoS ONE. 2014;9:e90030. doi: 10.1371/journal.pone.0090030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit CM, Lee AL. Monitoring side-chain dynamics of proteins using (2)H relaxation. Methods Mol Biol. 2014;1084:3–27. doi: 10.1007/978-1-62703-658-0_1. [DOI] [PubMed] [Google Scholar]
- Petit CM, Zhang J, Sapienza PJ, Fuentes EJ, Lee AL. Hidden dynamic allostery in a PDZ domain. Proc Natl Acad Sci U S A. 2009;106:18249–18254. doi: 10.1073/pnas.0904492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Silversmith RE, Bourret RB. Conformational coupling in the chemotaxis response regulator CheY. Proc Natl Acad Sci USA. 2001;98:6003–6008. doi: 10.1073/pnas.101571298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Kay LE. Tracing an allosteric pathway regulating the activity of the HslV protease. Proc Natl Acad Sci U S A. 2014;111:2140–2145. doi: 10.1073/pnas.1318476111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonovic M, Volz K. A distinct meta-active conformation in the 1.1-A resolution structure of wild-type ApoCheY. J Biol Chem. 2001;276:28637–28640. doi: 10.1074/jbc.C100295200. [DOI] [PubMed] [Google Scholar]
- Steiner S, Caflisch A. Peptide binding to the PDZ3 domain by conformational selection. Proteins. 2012;80:2562–2572. doi: 10.1002/prot.24137. [DOI] [PubMed] [Google Scholar]
- Tsai CJ, del Sol A, Nussinov R. Allostery: absence of a change in shape does not imply that allostery is not at play. J Mol Biol. 2008;378:1–11. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Del Sol A, Nussinov R. Protein allostery, signal transmission and dynamics: a classification scheme of allosteric mechanisms. Mol BioSyst. 2009;5:207–216. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng SR, Kalodimos CG. Protein dynamics and allostery: an NMR view. Curr Opin Struct Biol. 2011;21:62–67. doi: 10.1016/j.sbi.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Tzeng SR, Kalodimos CG. Protein activity regulation by conformational entropy. Nature. 2012;488:236–240. doi: 10.1038/nature11271. [DOI] [PubMed] [Google Scholar]
- Venditti V, Tugarinov V, Schwieters CD, Grishaev A, Clore GM. Large interdomain rearrangement triggered by suppression of micro- to millisecond dynamics in bacterial Enzyme I. Nat Commun. 2015;6:5960. doi: 10.1038/ncomms6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- Volz K, Matsumura P. Crystal structure of Escherichia coli CheY refined at 1.7-A resolution. J Biol Chem. 1991;266:15511–15519. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- Wand AJ. The dark energy of proteins comes to light: conformational entropy and its role in protein function revealed by NMR relaxation. Curr Opin Struct Biol. 2013;23:75–81. doi: 10.1016/j.sbi.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CK, Pan L, Chen J, Zhang M. Extensions of PDZ domains as important structural and functional elements. Protein Cell. 2010;1:737–751. doi: 10.1007/s13238-010-0099-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Zhang M. Structures and target recognition modes of PDZ domains: recurring themes and emerging pictures. Biochem J. 2013;455:1–14. doi: 10.1042/BJ20130783. [DOI] [PubMed] [Google Scholar]
- Zhang J, Petit CM, King DS, Lee AL. Phosphorylation of a PDZ domain extension modulates binding affinity and interdomain interactions in postsynaptic density-95 (PSD-95) protein, a membrane-associated guanylate kinase (MAGUK) J Biol Chem. 2011;286:41776–41785. doi: 10.1074/jbc.M111.272583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuravlev PI, Papoian GA. Protein functional landscapes, dynamics, allostery: a tortuous path towards a universal theoretical framework. Q Rev Biophys. 2010;43:295–332. doi: 10.1017/S0033583510000119. [DOI] [PubMed] [Google Scholar]