Abstract
Allostery is fundamentally thermodynamic in nature. Long-range communication in proteins may be mediated not only by changes in the mean conformation with enthalpic contribution but also by changes in dynamic fluctuations with entropic contribution. The important role of protein motions in mediating allosteric interactions has been established by NMR spectroscopy. By using CAP as a model system, we have shown how changes in protein structure and internal dynamics can allosterically regulate protein function and activity. The results indicate that changes in conformational entropy can give rise to binding enhancement, binding inhibition, or have no effect in the expected affinity, depending on the magnitude and sign of enthalpy–entropy compensation. Moreover, allosteric interactions can be regulated by the modulation a low-populated conformation states that serve as on-pathway intermediates for ligand binding. Taken together, the interplay between fast internal motions, which are intimately related to conformational entropy, and slow internal motions, which are related to poorly populated conformational states, can regulate protein activity in a way that cannot be predicted on the basis of the protein’s ground-state structure.
Keywords: NMR spectroscopy, Catabolite activator protein, Cyclic nucleotide-binding
The catabolite activator protein (CAP), a gene regulatory protein, is a textbook example of how the binding of small molecules controls protein activity through allosteric interactions. CAP is a 47-kDa homodimer comprising 209 residues per subunit. The protein is very well-folded and stable with a melting temperature of ∼63 °C and dimerization equilibrium constant of ∼1010 M−1. Each subunit is organized in two distinct regions: (i) an N-terminal domain (residues 1–135) that contains the cyclic nucleotide-binding (CNB) domain and a long a-helix (C-helix) that mediates dimerization, and (ii) a DNA-binding domain (DBD; residues 139–209), located at the C-terminus, that adopts the classical DNA-binding helix-turn-helix (HTH) motif. A short hinge region (residues 136–138) connects the N- and C-domains and is essential for mediating the allosteric response. The DNA-binding domain (DBD; residues 139–209) in cAMP-free CAP (apo-CAP) adopts an orientation that is incompatible with DNA binding (Popovych et al. 2009). cAMP elicits an allosteric transition that switches CAP from the “off” state, which binds DNA weakly and nonspecifically, to the “on” state, which binds DNA strongly and specifically. In the cAMP-bound state, CAP binds to DNA sites located in or adjacent to target promoters resulting in modulation of interactions of RNA polymerase with target promoters (Lawson et al. 2004).
Structural basis for allosteric control
How does cAMP switch CAP from the inactive to the active conformation? The primary mechanism of allosteric control in CAP is clear and simple (Popovych et al. 2009): cAMP binding to the cAMP-binding domain (CBD; residues 1–135) of CAP induces a coil-to-helix transition that extends the C-helix, and the intersubunit C-helix/C′-helix coiled coil, by three turns of helix. This coil-to-helix transition results in rotation of the DBDs of the CAP dimer by ≈60° and translation of the DBDs of the CAP dimer by ≈7 Å (distance of intersubunit F-helices is 41 Å in apo-CAP and 34 Å, matching the distance between successive DNA major grooves in CAP-cAMP2). This rotation and translation places the F-helices (“recognition helices”) of the DBDs of the CAP dimer in the correct orientation and correct position to interact with successive DNA major grooves.
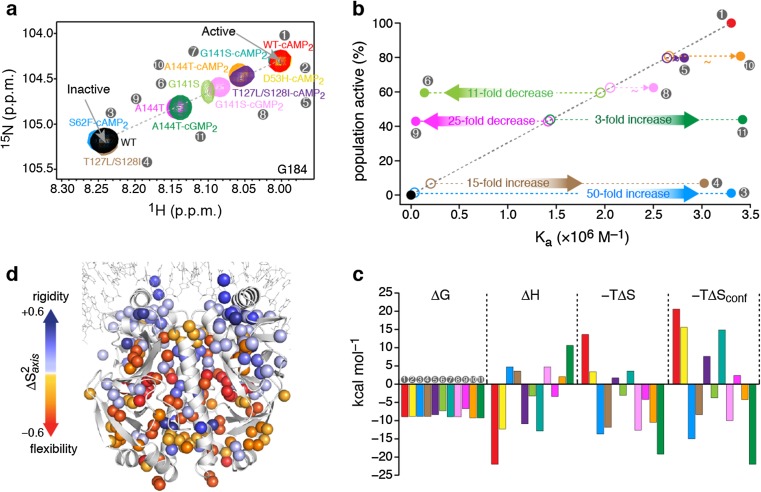
We explored the mechanisms underpinning allostery in CAP by engineering mutations at sites remote from the DNA-binding interface, but which nevertheless modulate DNA binding (Tzeng and Kalodimos 2009, 2011, 2012). To dissect the contribution of structure and internal dynamics to binding energetics, we studied a series of allosteric mutants of CAP in the unliganded, cNMP-liganded (cAMP- and cGMP-liganded), and DNA-liganded states (Tzeng et al. 2012). NMR chemical-shift analysis of all of the CAP variants in the unliganded and nucleotide-liganded states showed that the DBD resonances fall on the line that connects the resonances of the corresponding apo form of wild-type CAP (apo WT-CAP; the inactive DBD state) and cAMP-bound wild-type CAP (WT-CAP-cAMP2; the active DBD state) (Fig. 1a). The relative populations in the two DBD states can be determined from the chemical shift of the DBD resonances because the average chemical shift, δ, will be a weighted fraction of the population in the two states.
Fig. 1.
a Overlaid 1H–15N heteronuclear single quantum coherence (HSQC) spectra of the CAP variants, showing the resonance of the G184 amino acid, a residue located in the DNA recognition helix. The chemical shift of apo WT-CAP (labeled WT) indicates the inactive conformation of the DBD, whereas the chemical shift of WT-CAP-cAMP2 indicates the active conformation of the DBD. Each CAP variant is denoted by a number (gray circles) and color, which are the same in each figure panel. b The relative population of each CAP variant in the active DBD state as a function of the variant’s theoretical (open circles) and experimentally determined (filled circles) affinity for DNA. c Thermodynamic components of the binding of the CAP variants to DNA. d Effect of DNA binding on the methyl order parameters, S 2 axis, of CAP-T127L/S128I. Figure reproduced from Tzeng and Kalodimos 2012
Allosteric regulation by slow dynamics
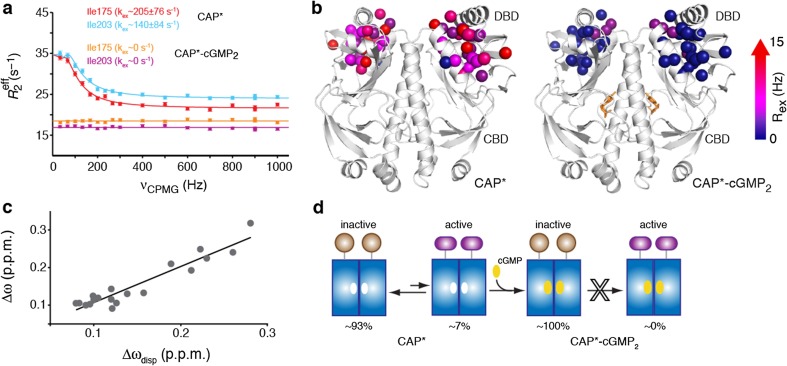
We performed a series of relaxation dispersion experiments (Bouvignies et al. 2010; Hansen et al. 2008a, b; Korzhnev et al. 2004; Loria et al. 1999) suited for detecting conformational states that are poorly populated and thus only transiently visited. The results indicated that two variants—CAP-T127L/S128I and CAP-S62F-cAMP2—underwent an exchange between conformational states on the microsecond-to-millisecond timescale (Tzeng and Kalodimos 2009, 2012) (Fig. 2a, b). Data fitting was indicative of a two site exchange process for both variants, with the population of the excited state being ∼7 % for CAP-T127L/S128I and ∼2 % for CAP-S62F-cAMP2. The differences in chemical-shift (∆ω) values between the major and the minor conformations of CAP-T127L/S128I and CAP-S62F-cAMP2, as determined by relaxation dispersion measurements (∆ωdisp), correlates with the absolute 15 N or 13C ∆ω values of the DBD residues measured between apo WT-CAP (inactive) and WT-CAP-cAMP2 (active) (Fig. 2c). Thus, the data provide strong evidence that the excited state that the DBD transiently populates in CAP-T127L/S128I and CAPS62F-cAMP2 closely resembles the active DBD conformation.
Fig. 2.
a Relaxation dispersion profiles of 13C side-chain methyls of representative CAP-T127L/S128I (CAP*) DBD residues in the apo and cGMP-bound form. b Enhanced R 2 relaxation rate (R ex) values of CAP* and CAP*–cGMP2. c Correlation between the 13CH3 Δω and Δωdisp chemical shifts of selected DBD residues. d CAP* interconverts between a ground state, which adopts the inactive conformation and is 93 % populated, and an excited state, which adopts the active conformation and is only ∼7 % populated. cGMP binding to CAP* results in the suppression of the active conformation through an allosteric mechanism. Figure reproduced from Tzeng and Kalodimos 2013
Because the affinity of the active DBD conformation for DNA (for example, in CAP-cAMP2) is many orders of magnitude higher than that of the inactive DBD conformation (for example in apo-CAP), DNA will preferentially bind to the active DBD conformation of CAP-T127L/S128I and CAP-S62F-cAMP2, despite being so poorly populated. Thus, the data indicate that DNA binding to CAP-T127L/S128I and CAP-S62F-cAMP2 proceeds with a population-shift mechanism. The lowly populated conformational state serves as an on-pathway intermediate for DNA binding and depletion of this intermediate is thus expected to result in DNA binding inhibition. Binding of cGMP to CAP-T127L/S128I seems to have no effect on the conformation of the DBD, and the chemical shifts of the DBD residues of CAP-T127L/S128I in apo and cGMP-bound states are essentially identical (Tzeng and Kalodimos 2012). However, relaxation dispersion experiments of CAP-T127L/S128I–cGMP2 complex showed profiles that are characteristic of the absence of any alternative conformational states that form on the ms–μs time scale (the detection limit of the relaxation dispersion experiments is ∼0.5 %). The data suggested that cGMP binding suppresses the active conformation and then results in allosteric inhibition of DNA binding (Tzeng and Kalodimos 2013) (Fig. 2c, d). Collectively, the allosteric interaction can be effected by stabilizing or destabilizing a low-populated conformation state that serves as an on-pathway intermediate for ligand binding, without altering the protein’s ground-state structure. Therefore, protein function cannot be simply predicted on the basis of the ground state of protein structures.
Allosteric regulation by fast dynamics
In general, the affinity of a protein for a ligand is directly proportional to the concentration of the active binding species. Eleven CAP variants that differentially populate the active DBD state were characterized. The population of the active DBD state by the CAP variants is expected to correlate with the affinity of the CAP variants for DNA, with higher populations of the active state giving rise to stronger binding (Fig. 1b, open circles). However, the experimentally measured affinities showed no correlation with the population of the active DBD state (Fig. 1b, filled circles). CAP-S62F-cAMP2 (∼2 % active DBD state) and CAP-T127L/S128I (∼7 % active DBD state) were expected to bind with a 50- and 15-fold lower affinity than WT-CAP-cAMP2 (∼100 % active DBD state); however, all three of these CAP variants bound to DNA with the same affinity. Similarly, CAP-A144T (∼50 % active) was expected to bind to DNA with only a twofold lower affinity than WT-CAP-cAMP2; however, it bound with a 50-fold lower affinity. Even for CAP variants that populate the active and inactive states to the same extent (for example, CAP-A144T and CAP-A144T-cGMP2), a 75-fold difference in binding affinity for DNA was measured experimentally. Clearly, factors in addition to the structure have a major role in modulating the affinity of the protein–DNA interactions.
To explore the thermodynamic basis for binding of CAP variants to DNA, we used isothermal titration calorimetry (ITC) to measure the association free energy (ΔG), binding stoichiometry, affinity, and its enthalpic (ΔH) and entropic (ΔS) components. Interestingly, the various CAP proteins bound to DNA using alternative thermodynamic strategies, with some interacting with favorable enthalpy (ΔH) and others with favorable entropy (ΔS) (Fig. 1c). To gain more insight into the origin of this large variation in binding entropy, we sought to determine the role of fast (picosecond to nanosecond; one picosecond is 10−12 s) protein motions in the binding process by measuring changes in the order parameter S 2. The order parameter is a measure of the amplitude of internal motions on the ps–ns timescale and may vary from S 2 = 1, for a bond vector having no internal motion, to S 2 = 0, for a bond vector rapidly sampling multiple orientations (Li and Bruschweiler 2009; Yang and Kay 1996). We determined the changes in the S 2 of the side-chain methyl groups (S 2 axis) of CAP that were elicited by DNA binding (Tugarinov et al. 2007). S 2 values are indicative of the amplitude of spatial fluctuations experienced by a bond vector and thus can be related to conformational entropy (Marlow et al. 2010). The results showed that DNA binding to each of the CAP variants resulted in a notable redistribution of the amplitude of motions throughout the entire protein in a distinct manner (Tzeng and Kalodimos 2009, 2012) (Fig. 1d). DNA binding to WT-CAP-cAMP2 results in widespread increase in S 2, indicating a global rigidification of the protein and thereby giving rise to a large and unfavorable change in conformational entropy. By contrast, DNA binding to either CAP-S62F-cAMP2 or CAP-T127L/S128I only causes residues of DBD to decrease their motions, as evidenced by the corresponding increase in their S 2 values, but resulted in the majority of the residues remote from the DNA-binding interface becoming more flexible, thereby giving rise to a favorable conformational entropy change. Notably, the large changes in amplitudes of motion might have been expected only at regions close to the binding interface, but such changes extend much farther away, involving methyl groups more than 50 Å from the interface. In the absence of such a significant change in conformational entropy, the two CAP variants might have bound poorly to DNA, and the results indicated that large net changes in conformational entropy can significantly increase the stability of the complex.
Furthermore, 50 % of the CAP-A144T molecules populate the active conformation and the addition of cGMP to CAP-A144T seems to have no effect on the DBD (Tzeng and Kalodimos 2012) (Fig. 1b). Because both CAP-A144T and CAP-A144T-cGMP2 equally populate the active species, they are expected to bind to DNA with similar affinities. However, CAP-A144T binds to DNA with a 75-fold lower affinity than does CAP-A144T-cGMP2. Thermodynamic analysis showed that the basis of this affinity difference is exclusively of an entropic nature, with the difference in entropy, Δ(−TΔS), for DNA binding to CAP-A144T and CAP-A144T-cGMP2 amounting to ∼15 kcal mol−1 (Fig. 1c). The difference in the conformational entropy of binding between the two complexes, Δ(−TΔSconf), is ∼25 kcal mol−1. It is of particular interest that although the two CAP variants populate the active DBD state to the same extent, they have remarkably different affinities for DNA because of the distinct responses of their conformational entropy to DNA binding. Similar behavior is seen in the case of CAP-G141S and CAP-G141S-cGMP2, with the two variants populating the active species to an almost equal extent, but with CAP-G141S binding to DNA with an order of magnitude weaker affinity, driven by the large difference in conformational entropy. Thus, our data show that unfavorable changes in conformational entropy can markedly suppress a protein’s DNA binding affinity, giving rise to a binding inhibition that cannot be rationalized on the basis of protein structural data.
For some of the CAP variants, the experimentally determined affinity for DNA was similar (within twofold) to the theoretical affinity (expected on the basis of the population of the active DBD state). This is the case for CAP-D53H-cAMP2, CAP-T12L/S128I-cAMP2, CAP-G141S-cAMP2, and CAP-A144T-cAMP2. However, the thermodynamic strategies used by these variants to interact with DNA are distinct, with the binding being either enthalpically or entropically driven and accompanied by large changes in conformational entropy. The difference in the entropy of binding of these complexes spans a range of ∼35 kcal mol−1. Dynamic analysis demonstrated that the response of each of the CAP variants to DNA binding is distinct, with the change in conformational entropy spanning a range of ∼40 kcal mol−1. However, and despite the markedly different dynamic changes, there seems to be a strong enthalpy–entropy compensation that results in little or no effect on the affinity for DNA (Kalodimos 2012; Tzeng and Kalodimos 2012).
Conclusions
Recent results on the catabolite activator protein (CAP) have revised our view about how allosteric interactions can be modulated. In particular, NMR studies of the binding of cAMP and DNA to CAP have established that (i) allostery can be mediated through changes in protein motions, in the absence of detectable changes in the mean structure of the protein; (ii) changes in conformational entropy can give rise to binding enhancement or binding inhibition; and (iii) allosteric interactions can be effected by stabilizing or destabilizing low-populated conformation states that serve as on-pathway intermediates for ligand binding, without altering the protein’s ground-state structure.
Compliance with Ethical Standards
Funding
This work was supported by the US National Science Foundation grant MCB1121896 to C.G.K.
Conflict of interest
S.-R. Tzeng and C.G. Kalodimos declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
This article is part of a Special Issue on 'The Role of Protein Dynamics in Allosteric Effects' edited by Gordon Roberts.
Contributor Information
Shiou-Ru Tzeng, Email: srtzeng@ntu.edu.tw.
Charalampos G. Kalodimos, Email: babis@rutgers.edu
References
- Bouvignies G, Korzhnev DM, Neudecker P, Hansen DF, Cordes MH, Kay LE. A simple method for measuring signs of (1)H (N) chemical shift differences between ground and excited protein states. J Biomol NMR. 2010;47:135–141. doi: 10.1007/s10858-010-9418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DF, Vallurupalli P, Kay LE. An improved 15N relaxation dispersion experiment for the measurement of millisecond time-scale dynamics in proteins. J Phys Chem B. 2008;112:5898–5904. doi: 10.1021/jp074793o. [DOI] [PubMed] [Google Scholar]
- Hansen DF, Vallurupalli P, Kay LE. Using relaxation dispersion NMR spectroscopy to determine structures of excited, invisible protein states. J Biomol NMR. 2008;41:113–120. doi: 10.1007/s10858-008-9251-5. [DOI] [PubMed] [Google Scholar]
- Kalodimos CG. Protein function and allostery: a dynamic relationship. Ann N Y Acad Sci. 2012;1260:81–86. doi: 10.1111/j.1749-6632.2011.06319.x. [DOI] [PubMed] [Google Scholar]
- Korzhnev DM, Kloiber K, Kanelis V, Tugarinov V, Kay LE. Probing slow dynamics in high molecular weight proteins by methyl-TROSY NMR spectroscopy: application to a 723-residue enzyme. J Am Chem Soc. 2004;126:3964–3973. doi: 10.1021/ja039587i. [DOI] [PubMed] [Google Scholar]
- Lawson CL, Swigon D, Murakami KS, Darst SA, Berman HM, Ebright RH. Catabolite activator protein: DNA binding and transcription activation. Curr Opin Struct Biol. 2004;14:10–20. doi: 10.1016/j.sbi.2004.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DW, Bruschweiler R. A dictionary for protein side-chain entropies from NMR order parameters. J Am Chem Soc. 2009;131:7226–7227. doi: 10.1021/ja902477s. [DOI] [PubMed] [Google Scholar]
- Loria JP, Rance M, Palmer AG., 3rd A TROSY CPMG sequence for characterizing chemical exchange in large proteins. J Biomol NMR. 1999;15:151–155. doi: 10.1023/A:1008355631073. [DOI] [PubMed] [Google Scholar]
- Marlow MS, Dogan J, Frederick KK, Valentine KG, Wand AJ. The role of conformational entropy in molecular recognition by calmodulin. Nat Chem Biol. 2010;6:352–358. doi: 10.1038/nchembio.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovych N, Tzeng SR, Tonelli M, Ebright RH, Kalodimos CG. Structural basis for cAMP-mediated allosteric control of the catabolite activator protein. Proc Natl Acad Sci U S A. 2009;106:6927–6932. doi: 10.1073/pnas.0900595106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugarinov V, Sprangers R, Kay LE. Probing side-chain dynamics in the proteasome by relaxation violated coherence transfer NMR spectroscopy. J Am Chem Soc. 2007;129:1743–1750. doi: 10.1021/ja067827z. [DOI] [PubMed] [Google Scholar]
- Tzeng SR, Kalodimos CG. Dynamic activation of an allosteric regulatory protein. Nature. 2009;462:368–372. doi: 10.1038/nature08560. [DOI] [PubMed] [Google Scholar]
- Tzeng SR, Kalodimos CG. Protein dynamics and allostery: an NMR view. Curr Opin Struct Biol. 2011;21:62–67. doi: 10.1016/j.sbi.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Tzeng SR, Kalodimos CG. Protein activity regulation by conformational entropy. Nature. 2012;488:236–240. doi: 10.1038/nature11271. [DOI] [PubMed] [Google Scholar]
- Tzeng SR, Kalodimos CG. Allosteric inhibition through suppression of transient conformational states. Nat Chem Biol. 2013;9:462–465. doi: 10.1038/nchembio.1250. [DOI] [PubMed] [Google Scholar]
- Tzeng SR, Pai MT, Kalodimos CG. NMR studies of large protein systems. Methods Mol Biol. 2012;831:133–140. doi: 10.1007/978-1-61779-480-3_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Kay LE. Contributions to conformational entropy arising from bond vector fluctuations measured from NMR-derived order parameters: application to protein folding. J Mol Biol. 1996;263:369–382. doi: 10.1006/jmbi.1996.0581. [DOI] [PubMed] [Google Scholar]