Abstract
Molecular motors are enzymes that convert chemical potential energy into controlled kinetic energy for mechanical work inside cells. Understanding the biophysics of these motors is essential for appreciating life as well as apprehending diseases that arise from motor malfunction. This review focuses on kinesin motor enzymology with special emphasis on the literature that reports the chemistry, structure and physics of several different kinesin superfamily members.
Electronic supplementary material
The online version of this article (doi:10.1007/s12551-014-0150-6) contains supplementary material, which is available to authorized users.
Keywords: Molecular motors, Kinesin motor enzymology, Kinesin superfamily, Chemical models, Enzymes
Introduction
Molecular motors are enzymes that convert chemical potential energy into controlled kinetic energy for mechanical work (i.e., motion or movement) inside cells. These biological motors come in a variety of flavors, including cytoskeletal motors, rotary motors, nucleic acid motors, protein synthesis and translocation machinery, and dynamic biopolymers. Each motor is designed to perform various tasks inside cells, such as intracellular transport, signal transduction, oxidative phosphorylation, cell motility, nucleic acid translocation, nucleic acid synthesis, endocytosis, protein synthesis, protein translocation across membranes, and cell division. Understanding the chemistry, structure, and physics of these biological motors is essential for appreciating life, as well as for apprehending diseases that arise from malfunction of these motors.
Since the discovery of the first force-generating, microtubule-dependent ATPase that drives fast axonal transport in neurons (Brady 1985; Vale et al. 1985), a wealth of research has focused on characterizing the structure and function of various members of the kinesin superfamily. These motors convert the free energy of nucleoside 5′-triphosphate hydrolysis (typically ATP) into directed mechanical motion during their interaction with a microtubule track. Most kinesins contain: (1) a catalytic domain (also known as a motor or head domain) responsible for nucleotide catalysis, microtubule binding, and force production, (2) a central α-helical stalk domain involved in coiled–coil interactions to organize the motor into higher order oligomers (e.g., dimers, tetramers), and (3) a tail domain responsible for various subfamily specific functions [e.g., light chain binding domain for kinesin-1 (Hackney 2007), DNA binding for kinesin-10 (Vanneste et al. 2011), ATP-independent microtubule binding domain for kinesin-8 (Weaver et al. 2011) and kinesin-14 (Karabay and Walker 1999)]. Several reviews have been written that summarize the various features of the kinesin superfamily (Hirokawa and Takemura 2004; Marx et al. 2009; Sack et al. 1999; Vale and Fletterick 1997).
Studying the biophysics of molecular motors has been the focus of many laboratories for decades. The fundamental understanding of enzyme biochemistry, structure, and (particularly for motors) mechanics has been investigated, yet networking the results from these scientific disciplines into a coherent model is often a difficult task. This review focuses on kinesin enzymology with special emphasis on the literature that reports the chemistry, structure and physics of several different kinesin molecular motors. In the first section, I summarize the biochemistry of kinesins, with a focus on the types of kinetic and thermodynamic experiments that researchers have used to study these motors. In this section I also discuss how the interpretation of these results lead to a chemical model for how kinesins catalyze nucleotide hydrolysis and how their partner filaments (microtubules) stimulate their ATPase cycle. In the second section, I provide a detailed summary of the current knowledge of kinesin structure, highlighting the conserved and divergent structural characteristics among the kinesin superfamily, as well as recent structures that have provided much insight into the function of molecular motors. In the third section, I describe the physics of kinesin motors, focusing on the movements and mechanics that are driven by the biochemical reactions in kinesin enzymes for force generation. A dominant feature of this section is the connection of biochemical free energies with the work performed by the motors. In the fourth and final section, several conclusions and future directions for the study of molecular motors are presented.
Chemistry
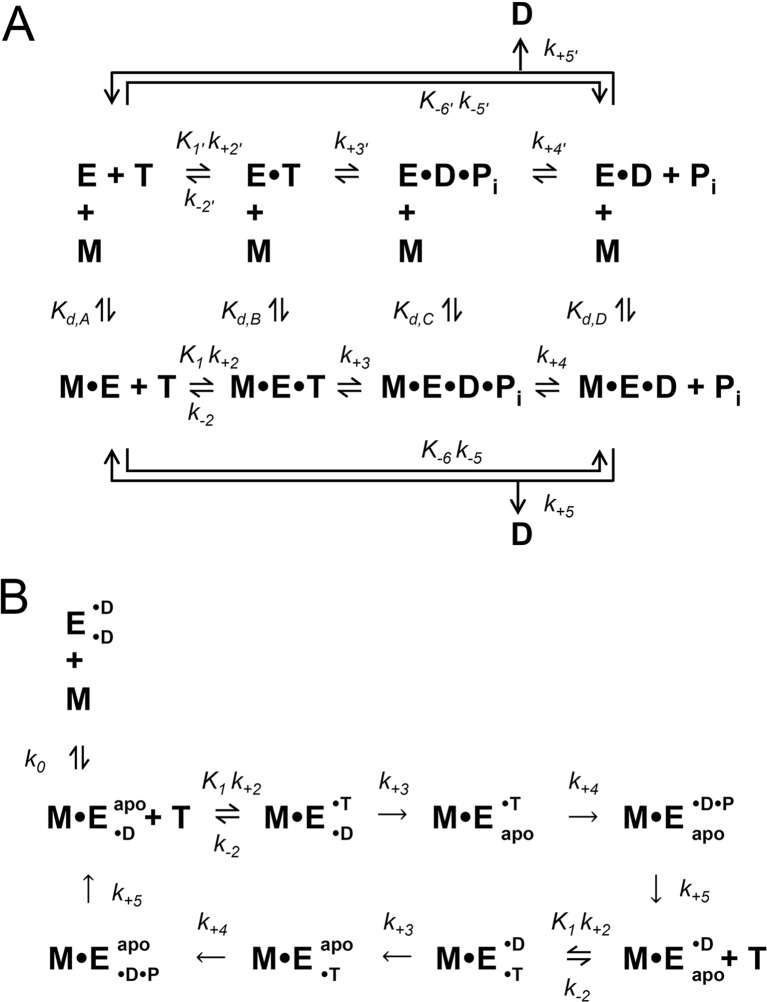
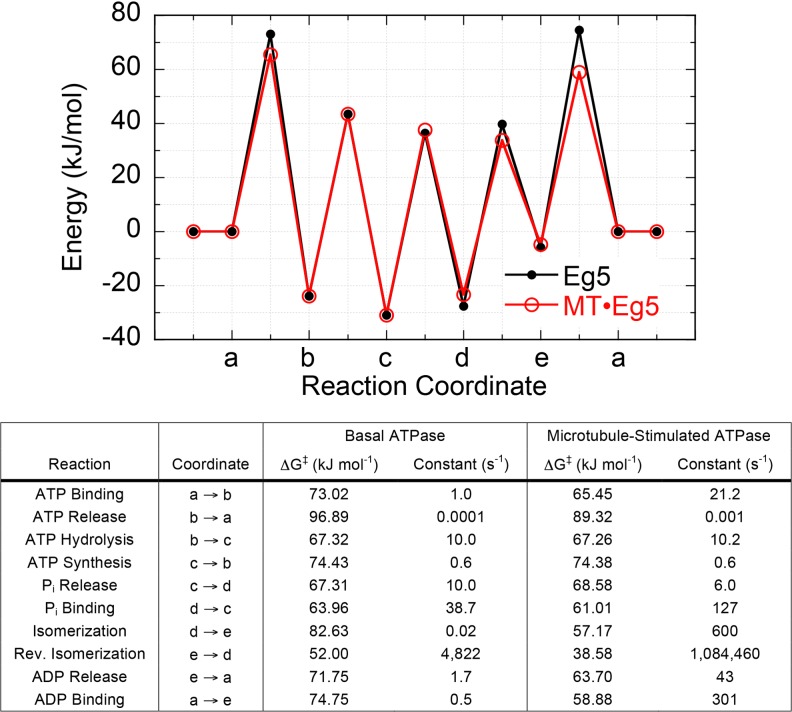
Although the kinesin superfamily consists of 14–17 different subfamilies with several individual members per subfamily (Dagenbach and Endow 2004; Lawrence et al. 2004; Miki et al. 2005; Wickstead et al. 2010), only a select few have been studied extensively using a combination of biophysical, biochemical, and structural methodologies. Based on a series of elegant kinetic, thermodynamic, and structural studies, ATPase mechanisms of truncated, recombinant monomeric (Fig. 1a) and dimeric (Fig. 1b) kinesin motors have been defined (note: the minimal mechanisms presented here are simplified in order to highlight the dominant steps in the cycle and for the purpose of comparing many different members of the superfamily; Tables 1 and 2). For monomeric kinesins, the minimal mechanism consists of four consecutive biochemical reactions [ATP binding, ATP hydrolysis, inorganic phosphate (H2PO4 -; Pi) release, and ADP release]. These four reactions can occur in either the microtubule-bound or unbound state. Depending on the family member, the motor affinity for the microtubule varies with the nucleotide state at the kinesin active site (e.g., kinesin-1 binds the microtubule tightly in the nucleotide-free/apo/rigor, ATP-like, and ADP⋅Pi-like states, but binds the microtubule weakly in the ADP state). Therefore, the detachment/dissociation and attachment/association rate constants define the four equilibrium constants at each nucleotide state in the cycle (K d,A = nucleotide-free, K d,B = ATP-like, K d,C = ADP⋅Pi-like, K d,D = ADP). The ATP-like and ADP⋅Pi-like states are typically achieved with nucleotide and Pi analogs, respectively (e.g., AMPPNP, AMPPCP, ATPγS as non-hydrolyzable or slowly hydrolyzable ATP analogs, and ADP with BeFx, AlFx, VO4 -, SO4 - as ADP⋅Pi analogs). These nucleotide states have revealed structural details for a variety of different ATPases, including kinesins (Table 3), myosins (Coureux et al. 2004; Gulick et al. 1997; Menetrey et al. 2008; Smith and Rayment 1996), helicases (Bernstein et al. 2003; Halbach et al. 2012; Luo et al. 2008; Schutz et al. 2010; Xiol et al. 2014), AAA+ motors (Schmidt et al. 2012; Stinson et al. 2013; Yamada et al. 2001), and RecA (Datta et al. 2003; Rajan and Bell 2004).
Fig. 1.
Minimal ATPase mechanisms for monomeric and dimeric kinesins. Monomeric (a) and dimeric (b) kinesin ATPase mechanisms are shown. E kinesin, M microtubule, T ATP substrate, D ADP product, P i inorganic phosphate product, apo nucleotide-free/rigor. For dimeric kinesins that have two inter-molecularly regulated active sites, each site is depicted as either a superscript or subscript (for example: E• ADP• ADP indicates tha3 both sites of the dimeric kinesin (E) are bound to ADP)
Table 1.
Comparison of rate and equilibrium constants for truncated monomeric kinesins in published studiesa
| Constant (units) | Reaction | Kinesin-1 | Kinesin-5 | Kinesin-7 | Kinesin-10 | Kinesin-13 | Kinesin-14 | ||
|---|---|---|---|---|---|---|---|---|---|
| Hs/BtKHC1-3 | DmKHC4,5 | HsEg56-8 | XlCENP-E9 | DmNOD10,11 | MCAK12 | DmNCD13 | ScKar314,15 | ||
| K 1′ k +2′ (μΜ-1 s-1) | ATP Binding | 300-400 | ND | 0.08 ± 0.01 | ND | 13.8 ± 0.6 | ND | ND | ND |
| k +2′ (s-1) | Isomerization | 170 ± 30 | ND | 0.5-1.2 | ND | 1800 ± 500 | ND | ND | ND |
| k -2′ (s-1) | ATP release | <1 | ND | <0.01 | ND | 5 ± 1 | ND | ND | ND |
| k +3′ (s-1) | Hydrolysis | 7 | ND | >10 | ND | 0.016 ± 0.002 | ND | ND | ND |
| k +4′ (s-1) | Pi release | >0.1 | ND | >10 | ND | ND | ND | ND | ND |
| k +5′ (s-1) | ADP release | 0.003 | 0.025 | 0.02 ± 0.001 | ND | 0.23 ± 0.004 | ND | 0.003 ± 0.0006 | 0.006 ± 0.0004 |
| K -6′ k -5′ (μΜ-1 s-1) | ADP binding | 333 | ND | 0.07 ± 0.01 | ND | 19.7 ± 0.4 | ND | ND | ND |
| k -5′ (s-1) | Isomerization | >300 | ND | 0.6-1.0 | ND | 1400 ± 220 | ND | ND | ND |
| K 1 k +2 (μΜ-1 s-1) | ATP binding | 19 ± 3 | 20 ± 5 | 3.4 ± 0.3 | 4.9 ± 0.7 | 5.1 ± 0.2 | ND | 1.1 ± 0.1 | 1.2 ± 0.2 |
| k +2 (s-1) | isomerization | >800 | 565 ± 50 | 21.2 ± 1.3 | 28.7 ± 3.9 | 900 ± 120 | ND | >400 | 500 ± 75 |
| k -2 (s-1) | ATP release | 105 ± 11 | 113 ± 60 | <1 | 123 ± 51 | 0.02 ± 0.004 | ND | 7 ± 7 | 50 |
| k +3 (s-1) | Hydrolysis | ND | >300 | 10.2 ± 0.7 | ND | ND | ND | 11.0 ± 2.3 | 70 ± 25 |
| k +4 (s-1) | Pi release | 92 ± 1 | ND | 6.0 ± 0.1 | 11.4 ± 0.9 | ND | ND | ND | ND |
| k +5 (s-1) | ADP release | 300 | 303 ± 23 | 43.3 ± 0.2 | 193 ± 35 | 0.60 ± 0.01 | ND | 3.9 ± 0.4 | 0.4 ± 0.01 |
| K -6 k -5 (μΜ-1 s-1) | ADP binding | 3.3 | 11 ± 1 | 2.8 ± 0.5 | 1.4 ± 0.4 | 10.6 ± 0.4 | ND | 0.6 ± 0.04 | 1.1 ± 0.01 |
| k -5 (s-1) | Isomerization | >300 | ND | >300 | ND | 760 ± 120 | ND | >100 | >100 |
| k cat (s-1 site-1) | Basal ATPase | 0.005 ± 0.0004 | ND | 0.02 ± 0.003 | ND | 0.001 ± 0.0001 | 0.03 | 0.004 | 0.008 |
| K m,ATP (μΜ) | ND | ND | 0.17 ± 0.03 | ND | 14.1 ± 2.6 | ND | ND | ND | |
| k cat / K m,ATP | ND | ND | 0.12 ± 0.01 | ND | 5 x 10-5 | ND | ND | ND | |
| k cat (s-1 site-1) | MT-ATPase | 60 ± 7 | 81 ± 2 | 5.5 ± 0.3 | 21.0 ± 0.7 | 2.5 ± 0.1 | 0.49 ± 0.06 | 2.1 ± 0.1 | 0.49 ± 0.02 |
| K m,ATP (μΜ) | ND | 91 ± 10 | 9.5 ± 0.4 | ND | 40.9 ± 9.3 | 34 ± 6 | 28 ± 4 | 12.2 ± 2.8 | |
| k cat / K m,ATP | ND | 0.9 | 0.6 | ND | 0.06 | 0.014 | 0.075 | 0.04 | |
| K 0.5,MT (μΜ) | 3.4 ± 1.5 | 0.2 ± 0.01 | 0.71 ± 0.60 | 1.9 ± 0.2 | 38.5 ± 3.3 | 0.01 ± 0.01 | 9.6 ± 1.6 | 6.0 ± 0.7 | |
| k cat / K 0.5,MT | 18 | 405 | 8 | 11 | 0.07 | 49 | 0.22 | 0.08 | |
| K d,A (μΜ) | Apo | 0.4 ± 0.2 | ND | 0.03 – 0.05 | ND | 0.07 ± 0.02 | 0.7 ± 0.1 | 0.2 ± 0.06 | 0.7 ± 0.1 |
| K d,B (μΜ) | AMPPNP | 1 ± 0.1 | ND | 0.03 – 0.05 | ND | 9.4 ± 0.3 | ND | ND | ND |
| K d,C (μΜ) | ADP•Pi | 7 ± 0.6 | ND | ND | ND | 8.0 ± 0.7 | ND | ND | ND |
| K d,D (μΜ) | ADP | 22 ± 2 | ND | ND | ND | 11.6 ± 0.8 | ND | ND | ND |
See Fig. 1 for basal (i.e. no microtubules) and microtubule-simulated ATPase mechanisms
ND Not determined
a1Rosenfeld et al. (2001), 2Sadhu and Taylor (1992), 3Cochran et al. (2011), 4Moyer et al. (1998), 5Jiang et al. (1997), 6Cochran et al. (2004), 7Cochran and Gilbert (2005), 8Cochran et al. (2006), 9Rosenfeld et al. (2009), 10Cochran et al. (2009), 11Sontag et al. (unpublished), 12Hertzer et al. (2006), 13Mackey and Gilbert (2000), 14Song and Endow (1998), 15Mackey and Gilbert (2003)
Table 2.
Comparison of rate and equilibrium constants for truncated dimeric kinesins in published studiesa
| Constant (units) | Reaction | Kinesin-1 | Kinesin-5 | Kinesin-7 | Kinesin-14 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| HsKHC1-4 | DmKHC5-8 | RnKHC9,10 | HsEg511-14 | HsCENP-E15 | XlCENP-E16,17 | DmNCD18-23 | ScKar3Cik124,25 | ScKar3Vik126,27 | ||
| k 0 (s-1) | ADP release from head 1 | ND | 306 ± 25 | >1000 | 28 ± 0.5 | 0.9 ± 0.01 | ND | 17.8 ± 1.6 | 110 ± 8 | 14.4 ± 0.3 |
| K +1 k +2 (μΜ-1 s-1) | ATP Binding | 2.6 ± 0.3 | 2.0 ± 0.8 | 1.7 | 1.2 ± 0.1 | 1.4 ± 0.1 | 1.7 ± 0.3 | 2.3 ± 0.1 | 8.4 ± 0.02 | 18.6 ± 0.03 |
| k +2 (s-1) | Head isomerization | 457 ± 56 | >200 | 210 ± 25 | 50 ± 3 | 47.5 ± 0.8 | 13.7 ± 2.7 | >200 | 69 ± 1.4 | 54.9 ± 1.0 |
| k -2 (s-1) | ATP dissociation | 55–135 | 71 ± 9 | 18 | 1.4 ± 0.6 | 1.6 ± 0.7 | 101 ± 13 | 14 ± 5 | 16.6 ± 1.0 | 7.4 ± 0.9 |
| k +3 (s-1) | ADP release from head 2 | 170 ± 17 | 300 ± 100 | >1000 | 0.62 ± 0.04 | 19.4 ± 0.6 | 56.8 ± 3.5 | 1.4 ± 0.02 | N/A | N/A |
| k +4 (s-1) | Hydrolysis | 79 ± 4 | 100 ± 30 | 500 | 5–10 | 24.7 ± 0.5 | ND | 35 ± 5 | 26 ± 0.8 | 25.9 ± 1.4 |
| k +5 (s-1) | Pi Release / Dissociation | 48–55 | 50 ± 8 | 80 | 6.6 ± 3.0 | 1.4 ± 0.01 | 4.7 ± 0.4 | 12.2 ± 0.3 | 11.5 ± 0.3 | 6.2 ± 0.1 |
| k cat (s-1 site-1) | MT-ATPase | 20 ± 2 | 20 ± 2 | 40.3 ± 1.1 | 0.5 ± 0.02 | 0.9 ± 0.02 | 12.1 ± 0.8 | 2.1 ± 0.02 | 5.0 ± 0.1 | 6.7 ± 0.1 |
| K m,ATP (μΜ) | 23 ± 9 | 61 ± 8 | 53.6 ± 4.8 | 7.9 ± 2.4 | 18.3 ± 2.5 | 35 ± 5 | 23.1 ± 1.5 | 65.2 ± 4.1 | 27.1 ± 2.9 | |
| k cat / K m,ATP | 0.87 | 0.33 | 0.72 | 0.06 | 0.05 | 0.35 | 0.09 | 0.08 | 0.25 | |
| K 0.5,MT (μΜ) | 0.3 ± 0.1 | 0.9 ± 0.3 | ND | 1.8 ± 0.2 | 1.5 ± 0.2 | 0.10 ± 0.04 | 18.3 ± 5.1 | 0.7 ± 0.05 | 0.4 ± 0.02 | |
| k cat / K 0.5,MT | 66.7 | 22.2 | ND | 0.3 | 0.6 | 121 | 0.1 | 7.1 | 16.8 | |
| K 0 (μΜ) | No added nucleotide | ND | ND | ND | 1.0 ± 0.1a | 0.9 ± 0.1 | ND | 0.22 ± 0.05 | 0.6 ± 0.1 | 0.13 ± 0.02 |
| V max (μm/min) | MT gliding velocity | 42.2 ± 4.4 | 51.2 ± 1.5 | 39–48 | 2.8 ± 0.1 | 0.66 ± 0.003 | 20.5 ± 0.6 | 10.2 ± 0.7 | 2.7 ± 0.1 | 5.9 ± 0.1 |
| MT end directionality | Plus | Plus | Plus | Plus | Plus | Plus | Minus | Minus | Minus | |
| Stall force (pN) | 6 ± 1 | 7 ± 1 | ND | 5 ± 1 | ND | 6 ± 1 | 1 ± 0.5c | ND | ND | |
ND Not determined; N/A Not applicable
aPublished studies: 1Rosenfeld et al. (2002), 2Rosenfeld et al. (2003), 3Clancy et al. (2011), 4Schnitzer et al. (2000), 5Gilbert et al. (1998), 6Coy et al. (1999), 7Moyer et al. (1998), 8Higuchi et al. (2004), 9Auerbach and Johnson (2005a), 10Mazumdar and Cross (1998), 11Krzysiak et al. (2006), 12Kikkawa and Hirokawa (2006), 13Krzysiak et al. (2008), 14 Valentine et al. (2006), 15Sardar and Gilbert (2012), 16Yardimci et al. (2008), 17Rosenfeld et al. (2009), 18Chandra et al. (1993), 19Foster et al. (1998), 20Foster and Gilbert (2000), 21Foster et al. (2001), 22Furuta et al. (2013), 23Heuston et al. (2010), 24Chen et al. (2011), 25Sproul et al. (2005), 26Allingham et al. (2007), 27Chen et al. (2012)
b K Hill value is the product of the K d values for cooperative sites in the Eg5 dimer (Krzysiak et al. 2006)
cStall force determined for 2–4 NCD motors linked via a rigid DNA scaffold with an ~6 or ~22-nm spacing (Furuta et al. 2013)
Table 3.
X-ray crystallographic structural summary for kinesin motors
| Family | Name | PDB ID | Notes | Nucleotide | Ligand(s) | Resolution (Å) | Reference |
|---|---|---|---|---|---|---|---|
| Kinesin-1 | HsKHC | 1bg2 | MgADP | Acetate | 1.8 | Kull et al. 1996 | |
| HsKHC | 1mkj | MgADP | SO4 | 2.7 | Sindelar et al. 2002 | ||
| RnKHC | 2kin | MgADP | SO4 | 2.0 | Sack et al. 1997 | ||
| NcKHC | 1goj | MgADP | 2.3 | Song et al. 2001 | |||
| RnKHC | 3kin | Dimer | ADP | 3.1 | Kozielski et al. 1997 | ||
| DmKHC | 2y5w | Dimer | MgADP | 2.7 | Kaan et al. 2011 | ||
| DmKHC | 2y65 | Dimer; Tail | MgADP | 2.2 | Kaan et al. 2011 | ||
| HsKHC | 4hna | MgADP⋅AlFx | Tubulin | 3.2 | Gigant et al. 2013 | ||
| Kinesin-2 | GiKIN2A | 2vvg | MgADP | 1.6 | Hoeng et al. 2008 | ||
| Kinesin-3 | MmKIF1A | 2zfi | MgADP | 1.6 | Nitta et al. 2008 | ||
| MmKIF1A | 1i6i | Collision | MgAMPPCP | 2.0 | Kikkawa et al. 2001 | ||
| MmKIF1A | 1vfv | Isomerization | MgAMPPNP | 1.9 | Nitta et al. 2004 | ||
| MmKIF1A | 1vfx | MgADP⋅AlFx | 2.6 | Nitta et al. 2004 | |||
| MmKIF1A | 1vfz | MgADP⋅VO4 | 2.2 | Nitta et al. 2004 | |||
| MmKIF1A | 2zfm | ADP | 2.3 | Nitta et al. 2008 | |||
| NcKIN3 | 2owm | MgADP | 3.3 | Nitta et al. 2008 | |||
| HsKIF3B | 3b6u | MgADP | 1.8 | Zhu et al. (SGC) | |||
| HsKIF3C | 3b6v | MgADP | 2.7 | Tempel et al. (SGC) | |||
| HsKIF13B | 3gbj | MgADP | 2.1 | Tong et al. (SGC) | |||
| MmKIF14 | 4ozq | β-sheet twist | ADP | 2.7 | Arora et al. 2014 | ||
| Kinesin-4 | MmKIF4A | 3zfd | MgAMPPNP | 1.7 | Chang et al. 2013 | ||
| HsKIF7 | 4a14 | MgADP | 1.6 | Klejnot and Kozielski 2012 | |||
| HsKIF7 | 2xt3 | MgADP | 1.9 | Klejnot and Kozielski 2012 | |||
| Kinesin-5 | HsEg5/KSP | 1ii6 | MgADP | NO3 | 2.1 | Turner et al. 2001 | |
| HsEg5/KSP | 3hqd | Isomerization | MgAMPPNP | PO4 | 2.2 | Parke et al. 2010 | |
| HsEg5/KSP | 1q0b | L5 Inhibitor | MgADP | Monastrol | 1.9 | Yan et al. 2004 | |
| HsEg5/KSP | 4ap0 | L5 Inhibitor | MgADP | Ispinesib | 2.6 | Talapatra et al. 2012 | |
| HsEg5/KSP | 4as7 | L5 Inhibitor | CoADP | SB743921 | 2.4 | Talapatra et al. 2013 | |
| HsEg5/KSP | 3zcw | α4 Inhibitor | MgADP | 4A2 | 1.7 | Ulaganathan et al. 2013 | |
| Kinesin-7 | HsCENP-E | 1t5c | MgADP | NO3 | 2.5 | Garcia-Saez et al. 2004 | |
| Kinesin-8 | HsKIF18A | 3lre | MgADP | 2.2 | Peters et al. 2010 | ||
| Kinesin-9 | HsKIF9 | 3nwn | MgADP | 2.0 | Zhu et al. (SGC) | ||
| Kinesin-10 | HsKIF22 | 3bfn | MgADP | 2.3 | Zhu et al. (SGC) | ||
| DmNOD | 3 dc4 | MgADP | 1.9 | Cochran et al. 2009 | |||
| DmNOD | 3dcb | MgAMPPNP | 2.5 | Cochran et al. 2009 | |||
| DmNOD | 3pxn | MnADP | 2.6 | Cochran et al. 2011 | |||
| Kinesin-12 | HsKIF15 | 4bn2 | MgADP | 2.7 | Klejnot et al. 2014 | ||
| Kinesin-13 | MmKIF2C | 1v8j | MgADP | 3.2 | Ogawa et al. 2004 | ||
| MmKIF2C | 1v8k | MgAMPPNP | 2.3 | Ogawa et al. 2004 | |||
| PfKINI | 1ry6 | SO4 | 1.6 | Shipley et al. 2004 | |||
| HsKIF2A | 2gry | MgADP | 2.4 | Wang et al. (SGC) | |||
| HsKIF2C | 2heh | MgADP | 2.2 | Wang et al. (SGC) | |||
| Kinesin-14 | DmNCD | 2ncd | Dimer | ADP | SO4 | 2.5 | Sablin et al. 1998 |
| DmNCD | 1cz7 | Dimer | MgADP | 2.9 | Kozielski et al. 1998 | ||
| DmNCD | 1n6m | Dimer | MgADP | 2.5 | Yun et al. 2003 | ||
| DmNCD | 3l1c | Dimer; T436S | MgADP | 2.8 | Heuston et al. 2010 | ||
| DmNCD | 3u06 | Dimer; G347D | MgADP | 2.4 | Liu et al. 2012b | ||
| ScKAR3 | 3kar | MgADP | 2.3 | Gulick et al. 1998 | |||
| ScKAR3 | 1f9t | WT | MgADP | 1.5 | Yun et al. 2001 | ||
| ScKAR3 | 1f9u | N650K | MgADP | 1.7 | Yun et al. 2001 | ||
| ScKAR3 | 1f9v | R598A | MgADP | 1.3 | Yun et al. 2001 | ||
| ScKAR3 | 1f9w | E631A | MgADP | 2.5 | Yun et al. 2001 | ||
| ScKAR3/VIK1 | 4etp | MgADP | 2.3 | Rank et al. 2012 | |||
| CgKAR3 | 4gkr | MgADP | 2.7 | Duan et al. 2012b | |||
| CaKAR3 | 4h1g | MBP fusion | MgADP | 2.2 | Delorme et al. 2012 | ||
| AgKAR3 | 3t0q | MgADP | 2.4 | Duan et al. 2012a | |||
| StKCBP | 1sdm | MgADP | 2.3 | Vinogradova et al. 2004 | |||
| StKCBP | 3cob | Extended α4 | MgADP | 2.2 | Vinogradova et al. 2008 | ||
| StKCBP | 3h4s | Regulator KIC | MgADP | 2.4 | Vinogradova et al. 2009 | ||
| AtKCBP | 4frz | MgADP | 2.4 | Vinogradova et al. 2013 | |||
| HsKIFC1 | 2rep | MgADP | 2.6 | Zhu et al. (SGC) | |||
| HsKIFC3 | 2 h58 | MgADP | 1.9 | Wang et al. (SGC) | |||
| ScVIK1 | 2o0a | Nonmotor | N/A | 1.6 | Allingham et al. 2007 |
SGC Structural Genomics Consortium Toronto; N/A Not applicable
For dimeric kinesins, the ATPase mechanism is more complicated due to the intramolecular communication between the two heads of the dimer (Fig. 2). This intramolecular communication (aka gating) is both chemical and mechanical in nature and functions to ensure one head is tightly bound to the filament at any given point in the cycle for processive motility (Block 2007). Historically, kinesins have been purified with ADP bound to the active site (Muller et al. 1999); and upon interaction with the microtubule, the first head (head 1) tightly binds the filament and releases its bound ADP very rapidly, while the second head (head 2) remains tightly bound to ADP and weakly bound to the microtubule (Ma and Taylor 1997). Upon ATP binding to head 1, a conformational change in the motor domain induces the neck linker to dock (Asenjo et al. 2006; Khalil et al. 2008; Rice et al. 1999; Rosenfeld et al. 2001; Skiniotis et al. 2003; Sugata et al. 2004; Sugata et al. 2009) and thus bias the diffusional search by head 2 toward its next microtubule binding site in the direction of the plus-end. Head 2 will tightly bind the microtubule and rapidly release its ADP, leading to a “strained” complex with two heads tightly bound to the microtubule, thus resulting in significant head–head tension (Hariharan and Hancock 2009; Hyeon and Onuchic 2007). This intramolecular strain is thought to decrease the affinity of the front head (head 2) for ATP until the rear head (head 1) hydrolyzes ATP and detaches from the microtubule (Guydosh and Block 2006; Klumpp et al. 2004b; Rosenfeld et al. 2003), as well as to trigger ATP hydrolysis and accelerate detachment of the rear head (Crevel et al. 2004; Farrell et al. 2002; Schief et al. 2004; Yildiz et al. 2008). After ATP hydrolysis and Pi release from the rear head (head 1), this head detaches and effectively brings the motor to the beginning of the cycle, with the exception that the heads have swapped position (i.e., head 1 is now tightly bound to ADP and weakly bound to the microtubule, and head 2 is tightly bound to the microtubule where it awaits ATP binding). This finely tuned mechanochemical cycle maintains head–head coordination in order to keep each head’s ATPase cycle out of phase so that the motor is able to take hundreds of steps (for kinesin-1, kinesin-7 and kinesin-8), five to ten steps (for kinesin-5), or thousands of steps (for kinesin-3) along the microtubules without completely detaching and diffusing away from the track (this topic is discussed in more detail in the “Physics” section of this review).
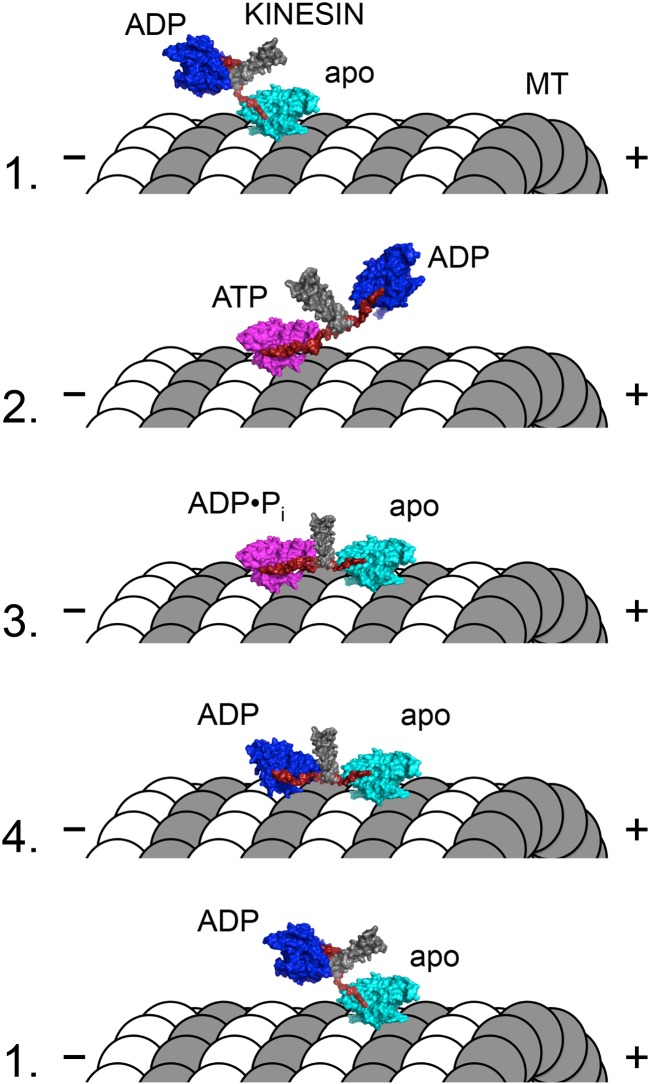
Fig. 2.
Model for the kinesin-1 “hand-over-hand” walking mechanism. Molecular representations of the dimeric kinesin-1 motor and microtubule (MT) are shown in each of four states (labeled numbers to the left of figure). The coiled–coil stalk (gray) and neck linker (red) are highlighted in each state. Coloring of the kinesin motor core represents three structural states: dark blue strongly ADP-bound, weakly microtubule-bound, neck linker undocked; cyan weakly nucleotide-bound, strongly microtubule-bound, neck linker undocked; magenta strongly ATP- or ADP•Pi-bound, strongly microtubule-bound, neck linker docked. Dimeric kinesin-1 motors are purified with ADP tightly bound to both active sites. The first head to strongly interact with the microtubule (head 1) will rapidly release its bound ADP to reach the nucleotide-free/apo/rigor state (state 1). Since head 2 remains unattached to the microtubule, head 1 is able to bind ATP, which docks the neck linker of head 1, biasing head 2 forward (i.e., toward the MT plus-end) to the next binding site on the same MT protofilament (state 2). ATP hydrolysis on head 1 is concomitant with tight microtubule-binding by head 2 to rapidly release its ADP product (state 3). Upon Pi release from head 1, intramolecular strain coupled to conformational changes in the ADP state of head 1 lead to weakening of head 1’s interaction with the MT (state 3). After detachment of head 1 from the MT, the kinesin effectively reaches state 1 with the exception that the heads trade places (i.e., head 2 is nucleotide-free/apo/rigor while head 1 is ADP-bound and detached from the MT). The transition from state 3 to state 4 leads to a release of the intramolecular tension that is thought to provide a mechanical gating of ATP binding to the front head
In the absence of microtubules, most purified kinesin motors are unstable and will denature when the nucleotide is removed from its active site (Pechatnikova and Taylor 1997; Sadhu and Taylor 1992). Past studies with monomeric kinesin-5 (Eg5) have demonstrated that the motor domain is stable and active when purified away from the bound ADP (Cochran and Gilbert 2005). Using kinetic (Cochran and Gilbert 2005; Cochran et al. 2006; Cochran et al. 2004; Rosenfeld et al. 2005) and thermodynamic (Zhao et al. 2010) methodologies, the energy landscape has been defined in the absence and presence of microtubules (note: a more thorough discussion of this energy landscape is found in the “Physics” section of this review). The microtubule modulates the energy landscape in three major ways: (1) by accelerating the rate of ATP binding, (2) by weakening ADP binding, and (3) by accelerating the rate of ADP release. Thus, the microtubule acts as an ATPase-activating protein as well as an ADP-exchange factor, analogous to GTPase-activating proteins (GAPs) and guanine nucleotide exchange factors (GEFs) for small G proteins (Bos et al. 2007).
Steady-state ATPase
The kinetic study of kinesin family members has been made possible through the purification of truncated constructs from a recombinant source (usually Escherichia coli or baculovirus). After preparation, the protein concentration must be established using available methods (e.g. Bradford assay or ultraviolet light absorbance at 280 nm). The first step in understanding the kinetic properties of the motor is to determine the microtubule-activated Michaelis–Menten steady-state ATPase activity of kinesin, which simply requires measuring the products of ATP hydrolysis (ADP or Pi) produced as a function of time [note: see De La Cruz and Ostap (2009); Gilbert and Mackey (2000) for methods to monitor hydrolysis product formation]. The aim of such experiments is to characterize the steady-state kinetic parameters (k cat, K m,ATP, K 0.5,MT) using a set of reciprocal experiments. In the first experiment, the microtubule concentration is kept constant and in excess (ideally 10-fold higher than the K 0.5,MT) and the ATP concentration is varied. The initial ATPase velocity (i.e., v o, slope of a plot of product formed over time) is then plotted against ATP concentration and the data fit to the Michaelis–Menten equation (Michaelis and Menten 1913; Michaelis et al. 2011):
| 1 |
where [E]o is the total concentration of kinesin [which ideally is 100- to 1,000-fold lower than the lowest ATP concentration ([ATP]) in the experiment], k cat is the maximum rate of ATP turnover per enzyme active site at infinite [ATP], and K m,ATP is the Michaelis constant, which is defined as the [ATP] which yields ½ [E]o k cat.
In the second experiment, the ATP concentration is kept constant and in excess (ideally 10-fold higher than K m,ATP) and the microtubule concentration is varied. The initial ATPase velocity is plotted against microtubule concentration and the data fit to the following quadratic equation since (1) [E]o is rarely 100- to 1,000-fold lower than the lowest microtubule concentration in the experiment, (2) the microtubules are not consumed in the reaction (whereas ATP does get consumed as a substrate), and (3) microtubules typically bind very tightly to kinesin (Morrison 1969). Thus,
| 2 |
where [E]o is the total concentration of kinesin {typically 2- to 10-fold lower than the lowest microtubule concentration ([MT])]}, k cat is the maximum rate of ATP turnover per enzyme active site at infinite [MT], and K 0.5,MT is the [MT] that yields ½ [E]o k cat.
It has been observed that unpolymerized tubulin heterodimers stimulate the ATPase activity of various kinesin motors (Alonso et al. 2007; Arora et al. 2014; Friel and Howard 2011; Gigant et al. 2013; Hertzer et al. 2006). Tubulin-stimulated ATPase activity in kinesin can be assayed in a similar manner as described above [i.e., with [ATP] held constant and in excess (10-fold higher than K m,ATP) while the concentration of unpolymerized tubulin is varied]. One must be thorough and also perform an ATP-concentration dependence test with excess tubulin heterodimer (ideally 10-fold higher than the K 0.5,tub) to test if the substrate binding affinity is different when kinesin is bound to tubulin heterodimers rather than polymerized microtubules. An important consideration for this analysis would be to test that the tubulin is not polymerizing under the experimental conditions. Sedimentation and light microscopy using fluorescently labeled tubulin are two commonly used experimental strategies to provide evidence that tubulin remains soluble and unpolymerized. Also, recent studies have demonstrated a designed ankyrin repeat protein (DARPin) that blocks longitudinal interaction between tubulin dimers along the protofilament axis, thus preventing microtubule polymer formation (Gigant et al. 2013; Pecqueur et al. 2012). This biochemical reagent can be useful to ensure non-polymerizable tubulin conditions.
Steady-state ATPase assays have also been useful for assessing the binding affinity of allosteric inhibitors of multiple different kinesin motors (Catarinella et al. 2009; Cochran et al. 2005; DeBonis et al. 2003; Maliga et al. 2002; Skoufias et al. 2006; Tcherniuk et al. 2010; Wood et al. 2010). Typically, these steady-state assays are designed by varying the inhibitor concentration in the presence of excess ATP and microtubule concentrations. However, with this added reagent to the ATPase reaction, one can determine the ATP affinity (vary [ATP] while keeping microtubule and inhibitor concentrations constant) and microtubule affinity (vary microtubule concentration while keeping [ATP] and inhibitor concentration constant) in the presence of the inhibitor (Cochran et al. 2005). Results from these experiments are important for deciphering the mechanism of kinesin inhibition and establish the foundation for the design of subsequent kinetic and thermodynamic experiments.
Together, these steady-state assays provide information on important properties of the kinesin enzyme, including the apparent binding affinities for ATP (K m,ATP) and microtubules (K 0.5,MT), as well as the quantitative measure of the maximum rate of ATPase activity (k cat). However, since the Michaelis–Menten theory does not adequately model even the minimal ATPase mechanism for kinesin (Fig. 1), one must be careful in the interpretation of the meaning of these steady-state kinetic parameters. Also, the determination of the k cat requires an accurate measure of the active enzyme population, which can be a very challenging task for a protein enzymologist. Monomeric kinesins have been shown to have a range of microtubule-stimulated k cat values, i.e., 0.5–80 s-1, K m,ATP values of between 9 and 90 μM, and K 0.5,MT values of between 0.01 and 10 μM (Table 1). The enzymatic efficiency (k cat/K m,ATP) and the biochemical “processivity” [k bi = k cat/K 0.5,MT; (Hackney 1995a; Hackney 1995b)] of the motor can be derived from the steady-state constants, which range from 0.04 to 0.9 μM-1 s-1 and from 0.07 to 405 μM-1 s-1, respectively. For dimeric kinesin motors (Table 2), k cat values range from 0.5 to 40 s-1, K m,ATP values range from 8 to 65 μM, and K 0.5,MT values range from 0.1 to 20 μM, with the enzymatic efficiency and biochemical processivity ranging from 0.05 to 0.9 μM-1 s-1 and from 0.1 to 121 μM-1 s-1, respectively. Despite the relatively minimal information attained through these assays, this information is essential for the design of more complicated pre-steady-state experiments to measure intrinsic rate constants for ATP binding/release, microtubule association/dissociation, Pi release, and ADP binding/release, as well as for the design of parameters for cosedimentation experiments.
Microtubule⋅kinesin cosedimentation
Cosedimentation experiments have historically been performed to quantify the concentration of kinesin that pellets (or sediments) with microtubule polymers after ultra-high speed centrifugation. Since this methodology provides the effective dissociation constant (K d) of an ensemble of kinesin states, various nucleotides (or nucleotide analogs) can be added in excess to the reaction to assess how different nucleotide states can influence microtubule binding affinity. For this assay, the kinesin concentration is typically kept constant, and microtubules are titrated into the reaction where they are allowed to equilibrate prior to centrifugation at ultra-high speed. Samples of the supernatant and pellet are prepared for analysis by SDS-PAGE. The fraction (f) of total kinesin [E]o bound to microtubules [E⋅MT] is plotted as a function of microtubule concentration, and the data are fit to a quadratic equation (Eq. 6):
| 3 |
| 4 |
| 5 |
| 6 |
To ascertain mechanistic information for the kinesin affinity for microtubules at various nucleotide states (nucleotide-free/apo/rigor: apyrase or alkaline phosphatase treatment; ATP-like: AMPPNP, AMPPCP, ATPγS; ADP⋅Pi-like: ADP + BeFx, AlFx, VO4 -, SO4 -; ADP; steady-state ATP hydrolysis), care must be taken to ensure that the “equilibrium population” being studied demonstrates a valid biochemical state that will lead to sound interpretation. For example, if you want to characterize the fraction of kinesin motors bound to microtubules under steady-state ATP hydrolysis conditions, the concentrations of reagents and timing for incubation prior to sedimentation must be correct to monitor the binding under “steady-state” ATP turnover conditions. Often an ATP-regeneration system [pyruvate kinase/phospho(enol)pyruvate or creatine kinase/phosphocreatine] will assist in keeping ATP concentrations “constant,” but may interfere with densitometry analysis of Coomassie-stained SDS-polyacrylamide gels. Overall, this cosedimentation methodology is very powerful in elucidating the mechanochemistry of a kinesin motor and is a relatively simple experiment to perform in a biochemistry laboratory.
Care must be taken in the design and interpretation of cosedimentation results. Typically, these experiments rely on Coomassie-stained SDS-polyacrylamide gel and densitometry analysis, which sets the lower limit for the concentration of kinesin used in the experiment. Under most nucleotide conditions, kinesin binds tightly to microtubules (usually a 10- to 100-fold lower K d than the kinesin concentration) and renders an inaccurate measurement of the binding constant. Alternative SDS-gel staining methods (e.g., Sypro Ruby; Life Technologies, Carlsbad, CA) have been utilized to perform the cosedimentation experiments at lower kinesin concentrations (Allingham et al. 2007), however the error in the standard curve using low quantities of protein (5–20 ng) renders experimental reproducibility difficult, ultimately leading to large standard deviations in observed binding constants (±10–30 %). It is possible that emerging technologies, such as microscale thermophoresis (Seidel et al. 2013) or isothermal titration calorimetry (Devred et al. 2010; Tsvetkov et al. 2013), will provide a means to accurately determine the kinesin–microtubule binding constant under various nucleotide conditions.
ATP binding
The kinetic mechanism of substrate binding to an enzyme is very important in enzymology. This is especially true for nucleotide binding to kinesin motors since conformational changes during MgATP binding are central to harnessing of the free energy of ATP hydrolysis for force production (Rice et al. 1999). Depending on how the observed rate of the reaction changes as a function of ATP concentration, the data can reveal the presence of intermediate steps after (or before) ATP binding that limit the rate of this reaction. Several methodologies are available for measuring the presteady-state ATP binding kinetics for kinesins, including fluorescent nucleotide binding experiments (mant-ATP), radiolabeled pulse-chase methodologies, FRET methods, and intrinsic protein fluorescence experiments (in the absence of microtubules). All kinesins studied to date have demonstrated (at least) a two-step rapid equilibrium binding mechanism in the presence or absence of microtubules. This binding mechanism can follow two similar (yet distinct) pathways:
| 7 |
| 8 |
In the first mechanism (Eq. 7), the ATP substrate collides with the kinesin motor (E) and weakly interacts. Then, upon a reversible isomerization (i.e., change in conformational state) of the E⋅S complex, the motor tightens its interaction with ATP to form the E*⋅S intermediate. In the second mechanism (Eq. 8), the motor undergoes a reversible isomerization prior to binding the ATP substrate, which is followed by tight ATP binding. The difference between these two mechanisms centers on the timing of the reversible isomerization step (i.e., a conformation change after or before ATP binding, respectively). Either mechanism satisfies the curvature in the concentration dependence of the observed reaction rate, which is indicative of (at least) a two-step mechanism approaching a maximum rate that is limited by a first-order isomerization of the E⋅S complex (or E prior to binding ATP).
Monomeric kinesin motors have shown a wide range of ATP binding affinities in the absence and presence of microtubules (K +1′ k +2′ = 0.08–400 μM-1 s-1 and 1–20 μM-1 s-1, respectively; Table 1) as well as a range of isomerization rates (k +2′ = 0.5–1800 s-1 and 20–900 s-1, respectively; Table 1). Dimeric kinesins have also demonstrated a wide range of ATP binding kinetics when bound to microtubules (K +1 k +2 = 1–20 μM-1 s-1 and k +2 = 10–500 s-1; Table 2). These kinetic constants indicate that kinesin motors have diverse ways of interacting with ATP substrate and that the structural interpretation for these kinetics may not be as straightforward as some have suggested (Chang et al. 2013).
ATP hydrolysis
After ATP binds tightly to the kinesin core, the motor can adopt the necessary active site configuration that promotes rapid hydrolysis of the γ-phosphate from ATP. In certain kinesins [e.g., monomeric Drosophila kinesin-10 in the absence and presence of microtubules (Cochran et al. 2009); monomeric human kinesin-13 in the absence of microtubules (Friel and Howard 2011)] and microtubule-stimulated dimeric human kinesin-5 (Krzysiak and Gilbert 2006)], this conformational change in the core represents the slow, rate-limiting step in the pathway. For other well-characterized kinesins, this reaction is relatively fast during the ATPase cycle (Tables 1 and 2), so the time spent in the ATP-bound state is transient. Subsequent to the characterization of ATP binding kinetics, rapid acid-quench-flow methodologies are used to measure the kinetics of the ATP hydrolysis reaction (Gilbert and Mackey 2000; Johnson 1992). However, since the kinetics of ATP binding precede the hydrolysis reaction, care must be taken in the interpretation of observed kinetics determined by acid-quench experiments. Normally kinetic simulation and global data fitting software are utilized to fit presteady-state data to the minimal mechanism based on numerical integration of the rate equations [e.g. DynaFit (Kuzmic 1996), KINSIM/FITSIM (Barshop et al. 1983; Zimmerle and Frieden 1989), or Global Kinetic Explorer (Johnson et al. 2009)].
ATP hydrolysis is a thermodynamically favorable reaction (ΔG° = –35.6 kJ mol-1; see Appendix A), yet, when uncatalyzed, is a very slow reaction (ΔG‡ = 115 kJ mol-1, k uncat = 3.8 × 10-8 s-1, t 0.5 = 211 days; Stockbridge and Wolfenden 2009). ATPases catalyze this reaction by lowering the energy of the transition state relative to the reactants through either a “dissociative” mechanism, an “associative” mechanism, or a “concerted” mechanism between these two extremes (Admiraal and Herschlag 1995; Khan and Kirby 1970; Thatcher and Kluger 1989). A substantial amount of data supports a largely “dissociative” mechanism for phosphoanhydride hydrolysis in which bond cleavage precedes nucleophilic attack by the lytic water molecule, thus leading to a metaphosphate-like transition state (Bunton and Chaimovi 1965; Halkides and Frey 1991; Lightcap and Frey 1992; Miller and Ukena 1969; Miller and Westheim 1966a; Miller and Westheim 1966b). Recent studies using combined quantum–mechanical/molecular–mechanical metadynamics simulations support the “dissociative” model of ATP hydrolysis by kinesin whereby two water molecules participate in proton transfer to ultimately the Pi product (McGrath et al. 2013). These two water molecules (one “lytic” water molecule located proximal to the γ-phosphate and a second “relay” water molecule positioned between the lytic water molecule and the switch-2 glutamic acid that forms a salt bridge with the switch-1 arginine; for more detail, see the “Structure” section of this review) establish a network of hydrogen bonds in the hydrolysis-competent conformation of motor ATPases (Dittrich et al. 2003; Dittrich et al. 2004; Grigorenko et al. 2011; Grigorenko et al. 2007; Hayashi et al. 2012; Onishi et al. 2004; Parke et al. 2010; Schwarzl et al. 2006).
This “hydrolysis-competent” conformational state of the motor must be a very transient (i.e., short-lived) structural intermediate that rapidly isomerizes to a state that is not conducive for ATP synthesis for monomeric kinesins in the absence and presence of microtubules, yet the trailing head in a dimeric kinesin does show propensity for ATP synthesis (Hackney 2005). Despite having an abundant quantity of data available on the ATP hydrolysis reaction and various conformational states of motor ATPases, it remains unknown how sequential conformational changes in hydrolases catalyze nucleotide bond cleavage.
Pi release
After ATP hydrolysis produces its reaction products (ADP and Pi), Pi product release occurs first, followed by ADP release. For most kinesins, Pi release leads to an allosteric change in the microtubule-binding region that weakens the motor’s affinity for the microtubule (see below). Therefore, Pi release is one of the pivotal reactions in the kinesin ATPase cycle, yet arguably the most difficult to experimentally study since it is a transient step buried deep within the mechanism. Although Pi release is typically a rapid reaction in the basal ATPase cycle, monomeric kinesin-1, kinesin-5, and kinesin-7 motors demonstrate rate-limiting Pi release for their microtubule-stimulated ATPase cycles [(Cochran and Gilbert 2005; Cochran et al. 2006; Cochran et al. 2004; Cochran et al. 2011; Moyer et al. 1998; Rosenfeld et al. 2001; Rosenfeld et al. 2009); Table 1].
Experimental strategies have been developed to directly measure the rapid kinetics of Pi release from kinesin in the absence and presence of microtubules using a fluorescence-coupled assay (Brune et al. 1994; White et al. 1997). This strategy relies on the very rapid (i.e., near diffusion-limited reaction) and tight (K d,Pi ~ 100 nM) binding of Pi to a modified phosphate-binding protein with the MDCC fluorophore (MDCC-PBP; Phosphate Sensor® from Life Technologies). This reagent can be rapidly mixed in a kinesin-catalyzed ATPase reaction using a stopped-flow fluorimeter, and fluorescence can be monitored over time to measure the kinetics of Pi release. After ATP hydrolysis, Pi is released from the kinesin active site, followed by rapid and tight Pi binding to MDCC-PBP, thus triggering a five- to sixfold fluorescence enhancement of the MDCC-PBP⋅Pi complex. Since this assay is very sensitive to Pi in solution, a “Pi mop” consisting of 7-methylguanosine and purine nucleoside phosphorylase must be added to effectively eliminate background Pi before the ATPase reaction is initiated (Cochran and Gilbert 2005). Depending on the intrinsic rate constant for Pi release in relation to the other steps in the ATPase cycle, various outcomes are possible for the observed rate of reaction (e.g., linear kinetics if Pi release is the rate-limiting step or burst kinetics if a step after Pi release is rate-limiting). Also, the presence or absence of an initial exponential lag phase can provide insight into the rate of a given reaction prior to Pi release, which can be compared with observed rates of ATP binding or ATP hydrolysis reactions. Kinetic simulation and global data fitting software ought to be utilized to model the intrinsic rate constant for Pi release since both the ATP binding and ATP hydrolysis steps precede this reaction (note: Pi rebinding to kinesin can be ignored due to the Pi binding to MDCC-PBP rendering the reaction kinetically irreversible).
MgADP release
The final reaction that completes the kinesin ATPase cycle is MgADP release to reach the nucleotide-free/apo/rigor state. For the basal ATPase cycle, this step represents the rate-limiting reaction for most (but not all) kinesin motors (Table 1). The observed presteady-state rate of ADP release in the absence of microtubules is typically similar to the measured basal ATPase activity, strengthening the argument for ADP release being rate-limiting in the cycle (Table 1). However, in the presence of microtubules, the rate of ADP release is dramatically accelerated for most kinesins (3–5 orders of magnitude). For kinesin-14 motors, the rate of MgADP release remains the rate-limiting step in the mechanism, but for other kinesins, the rate of this reaction no longer limits steady-state ATPase activity (Tables 1 and 2).
The mechanism of MgADP release has been the subject of multiple investigations since the discovery of kinesin, yet it remains a poorly understood step in the kinesin ATPase cycle. Several techniques are available to measure the presteady-state rate constants for MgADP binding and release, including mant-ADP fluorescence (both direct excitation/emission and FRET from protein tryptophans and tyrosines), intrinsic tryptophan fluorescence (in the absence of microtubules), and radiolabeled ADP chase experiments using a rapid ATP regeneration system. One can also measure the kinesin or microtubule⋅kinesin complex binding affinity for ADP using mant-ATP binding competition assays (De La Cruz and Ostap 2009; Zhao et al. 2010). Each method has advantages and limitations that must be taken into consideration when determining the mechanism of ADP product release. If nucleotide-free kinesin can be attained, the kinetics of MgADP binding can be measured in the absence of microtubules (Cochran et al. 2009; Zhao et al. 2010). Most kinesins are stable in the nucleotide-free state when bound tightly to microtubules. Therefore, presteady-state MgADP binding kinetics can be measured for the microtubule⋅kinesin complex (Table 1). The mechanism of MgADP binding ought to provide insight into the pathway for ADP release from kinesins since the intrinsic rate constants that govern the minimal mechanism can be derived from the fit of the ADP concentration dependence of the observed reaction rate (Johnson 1992).
All kinesins studied to date have demonstrated (at least) a two-step rapid equilibrium binding mechanism for MgADP binding in the presence or absence of microtubules. As with MgATP binding, this binding mechanism can follow two similar (yet distinct) pathways:
| 9 |
| 10 |
The difference between these two mechanisms centers on the timing of the reversible isomerization step (i.e., a conformation change after or before ADP binding, respectively). Although the reversible isomerization of the kinesin motor domain has been demonstrated using enzyme kinetics, the details of the structural changes that drive this isomerization remain mysterious.
Monomeric kinesin motors have shown a wide range of ADP binding affinities in the absence and presence of microtubules (K -6′ k -5′ = 0.07–330 μM-1 s-1 and 0.6–11 μM-1 s-1, respectively; Table 1), as well as a range of isomerization rates (k -5′ = 0.6–1400 s-1 and 100–760 s-1, respectively; Table 1). As with MgATP binding, the variability of rate constants governing MgADP binding suggests that the conformational changes occurring in each motor extend beyond the conserved active site motifs. What remains an unexplainable phenomenon is the symmetry of the rate constants that limit the isomerization reactions for MgATP and MgADP binding. For example, these rates were measured at 0.5–1 s-1 for monomeric kinesin-5 in the absence of microtubules using multiple different methodologies (Cochran and Gilbert 2005), suggesting the rate-determining structural change is dictated by a reaction that does not require the presence of the nucleotide γ-phosphate.
Microtubule detachment and binding
How kinesin motors convert the energy of ATP hydrolysis into directed mechanical work largely relies on allosteric changes in the region of the motor that interacts with the microtubule in order to modulate binding affinity. A wealth of data supports the notion that the kinesin nucleotide state modulates the microtubule binding affinity and vice versa. Thus, not only measuring the equilibrium binding constants under different nucleotide conditions (as described above), but also measuring the kinetics of microtubule binding (aka microtubule association) and microtubule detachment (aka microtubule dissociation) is important for deciphering the kinesin ATPase mechanism.
These presteady-state experiments have been adopted from similar methodologies from the actomyosin (Finlayson et al. 1969; Finlayson and Taylor 1969) and axonemal dynein systems (Porter and Johnson 1983). Solution turbidity (λ = 340–350 nm at 180° to the incident beam) and light scattering (λ = 420 nm at 90° to the incident beam) provide optical signals for kinesin binding to or detaching from microtubules, which can be monitored over time in a rapid mixing stopped-flow fluorimeter. This experimental design depends on: (1) the turbidity/light scattering signal from the microtubule⋅kinesin complex is greater than the sum of turbidity/light scattering signals from the individual components; (2) the formation of or dissociation of the microtubule⋅kinesin complex follows simple exponential kinetics; (3) the initial and final equilibrium states can be characterized using other biophysical methodologies, such as cosedimentation or analytical ultracentrifugation, to confirm either complex formation or dissociation (Finlayson et al. 1969). For measuring the kinetics of microtubule⋅kinesin complex formation, kinesin motors under various nucleotide conditions are rapidly mixed with increasing concentrations of taxol-stabilized microtubules, and the observed rate of the change of turbidity/light scattering is plotted as a function of microtubule concentration. The fit of the data provides the apparent second-order rate constant for kinesin association with microtubules. For measuring the kinetics of microtubule⋅kinesin complex dissociation, the microtubule⋅kinesin complex is rapidly mixed with increasing concentrations of MgATP + extra salt (typical ionic strength 100–200 mM). The MgATP triggers the kinesin to detach from the microtubule during the ATPase cycle, whereas the increased ionic strength promotes weak microtubule⋅kinesin interaction, thus shifting the kinesin population towards its detached state. The observed rate of turbidity/light scattering is plotted as a function of MgATP concentration, and the hyperbolic fit provides the maximal rate of kinesin dissocation and the relative binding affinity of kinesin for MgATP.
Structure
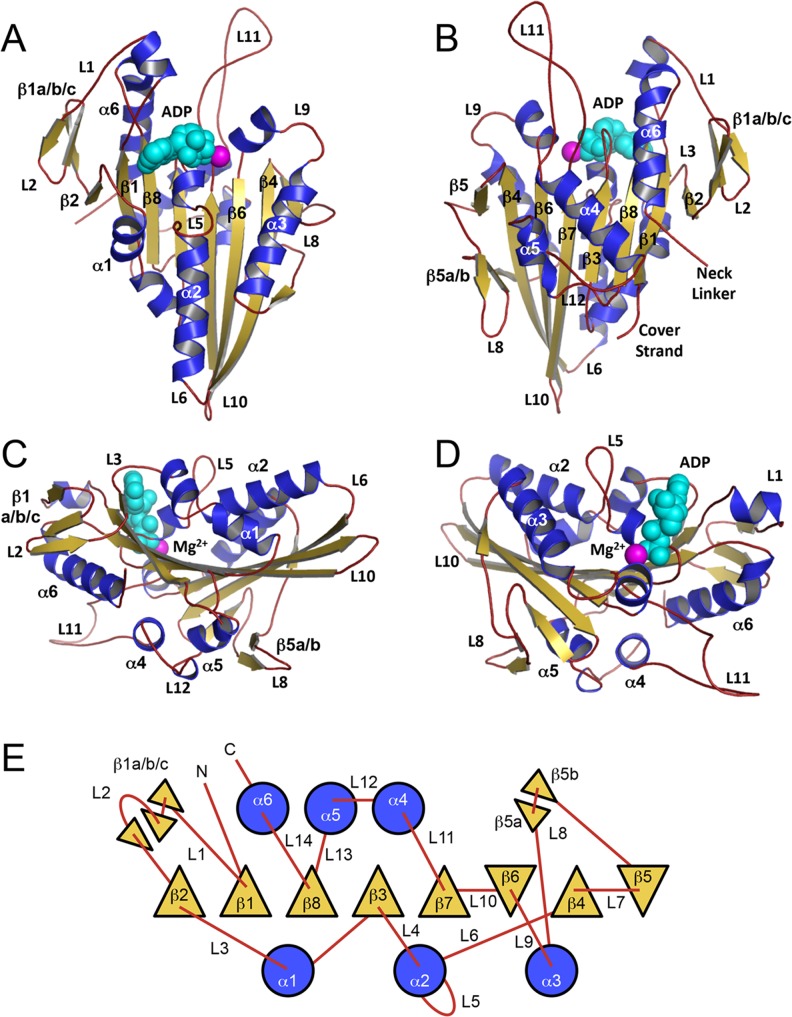
The very first glimpse into how the kinesin motor is built came from determination of the high-resolution structure of the kinesin-1 motor domain based on by X-ray crystallography [Fig. 3; (Kull et al. 1996)], which showed that the kinesin-1 motor has many similarities with the related myosin motor domain (Rayment et al. 1993). Since then, over 100 kinesin structures from multiple families have been deposited into the Protein Data Bank (PDB; Table 3). and detailed reviews of these structures have been published (Kull 2000; Kull and Endow 2002; Marx et al. 2009; Marx et al. 2005; Woehlke 2001).
Fig. 3.
Kinesin structure and topology. The monomeric human kinesin-1 structure (1BG2; Kull et al. 1996) is depicted in four distinct orientations: a front, b back, c left side, d right side. Secondary structure elements (α-helices, β-strands, and loops) are colored blue, gold, and red, respectively. Mg2+ and ADP are shown as space-fill model and are colored magenta and cyan, respectively. e Topology diagram of the kinesin motor domain shows α-helices (circles; shaded blue; up/down direction indicated by loop attachment), β-strands (triangles; shaded gold; up/down to depict orientation), and loops (red lines) as rendered from (Kull et al. 1998)
The kinesin motor domain contains: (1) the motor core consisting of an αβα fold with three α-helices on each side of an eight-stranded β-sheet; (2) a family-specific neck-linker sequence immediately preceding or following the core in its primary sequence; (3) a motif called the cover strand at the opposite terminus relative to the neck linker (e.g., kinesin-5 has its neck linker at the C-terminus and the cover strand has its neck-linker at the N-terminus relative to the core). The neck linker is central for energy transduction in kinesin motors (Rice et al. 1999; Schief and Howard 2001) and is also important for communicating strain between the two motor domains of dimeric kinesins to chemically and physically gate the mechanochemical cycle (Guydosh and Block 2006; Hwang and Lang 2009; Hyeon and Onuchic 2007; Jaud et al. 2006; Miyazono et al. 2010; Rosenfeld et al. 2003; Shastry and Hancock 2010; Yildiz et al. 2008). Although the neck linker and the cover strand are at opposite ends of the core in the primary sequence, they are in close tertiary-structural proximity due to the physical location for where the N- and C-termini enter and exit the core, respectively (Fig. 3). The cover strand interacts with the neck linker to form the cover-neck bundle, which is thought to participate in force generation (Hwang et al. 2008; Khalil et al. 2008).
The core contains several key structural motifs present in all kinesin motors. First, located on opposite sides of the eight-stranded β-sheet are the nucleotide binding pocket and the microtubule-binding interface (β5-L8 lobe, L11, and the “Sw2 cluster” comprised of α4-L12-α5; Sosa et al. 1997; Woehlke et al. 1997). Second, conserved motifs called the phosphate binding loop (P-loop: Walker A motif consensus GxxxxGKT/S located at L4), switch-1 (Sw1: NxxSSR located at the β6-L9 junction), and switch-2 (Sw2: Walker B motif, DxxGxE located at the β7-L11 junction) undergo conformational changes during the ATPase cycle in response to the presence (e.g., ATP-bound state) or absence (e.g., ADP-bound or nucleotide-free/apo/rigor states) of the γ-phosphate. These structural changes are reciprocally related to–and thus modulate–the conformations of the neck linker/cover strand, and vice versa (Block 2007; Hwang et al. 2008; Khalil et al. 2008; Rice et al. 1999; Zhao et al. 2010). Third, a conserved motif called N-4 (consensus: RxxP) establishes interaction with the purine nucleotide base.
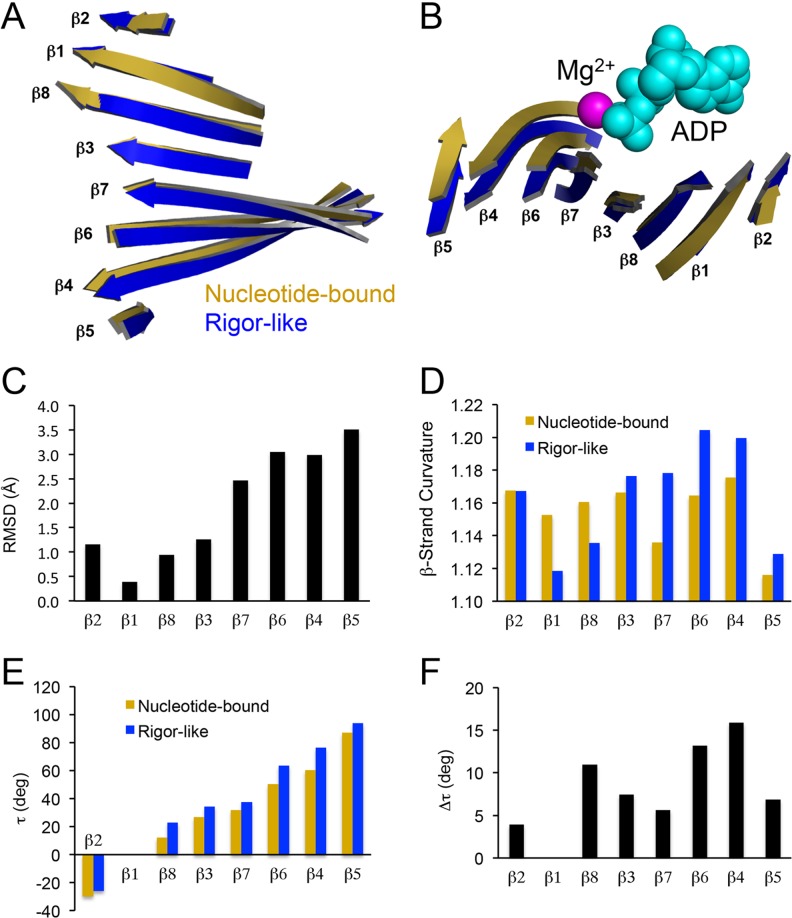
Central to the kinesin motor domain is an eight-stranded β-sheet (topology β2-β1-β8-β3-β7-β6-β4-β5; Fig. 3e) which serves as the core element that structurally connects the nucleotide-binding pocket and the microtubule-binding region. This central β-sheet has shown slightly variable degrees of right-handed twisting, which is thought to be important for the geometry and function of the motor domain (Kull and Endow 2013). Since most kinesins have been crystallized with bound nucleotide (Table 3), the degree of twisting has been very similar. However, recent crystal structures have demonstrated significant increases in right-handed twist or “over-twisting” of the kinesin β-sheet (Fig. 4; (Arora et al. 2014; Gigant et al. 2013), similar to the “over-twisting” seen in myosin’s β-sheet as it transitions from the nucleotide-bound state to the nucleotide-free state (Cecchini et al. 2008; Coureux et al. 2003; Reubold et al. 2003) [note: the “nucleotide-free” state has historically been called the “rigor” state since myosin binds very tightly to actin filaments in this nucleotide state]. The degree of “over-twisting” (τavg = 9.1 ± 4.3°; range 3.9–13.2°; Fig. 4f) observed in kinesins transitioning between the nucleotide-bound to nucleotide-free rigor states was comparable to that seen in myosin-V (τavg = 6.7 ± 4.8°; range 1–14°; Cecchini et al. 2008), which supports the hypothesis for common allosteric communication in cytoskeletal motors via the twisting of the central β-sheet. Indeed, theoretical studies on the origin of β-sheet twisting have demonstrated that the conformational energy associated with perturbing the structure of a β-sheet (through “twisting”, “bulging,” and “bending”) provide physical “reservoirs” of conformational free energy in globular proteins (Shamovsky et al. 2000a; Shamovsky et al. 2000b). Examination of the number of hydrogen bonds formed between residues of the central β-sheet reveals that five more hydrogen bonds are formed in the “under-twisted” nucleotide-bound state compared to the “over-twisted” rigor-like state, which amounts to 10–50 kJ mol-1 bond energy. This “stored” bond energy may provide the biophysical basis whereby kinesin motors can accelerate their rates of product release during their ATPase cycles (for more details, see the “Physics” section of this review). Interestingly, Hsp70 ATPases have also demonstrated nucleotide-dependent conformational changes in their central β-sheet (Zhuravleva et al. 2012; Zhuravleva and Gierasch 2011), suggesting this means of allosteric communication in proteins may be quite prevalent.
Fig. 4.
Structural changes in the eight-stranded central β-sheet of kinesin. Side (a) and end (b) views of the kinesin β-sheet after a nucleotide-bound structure (1BG2; Kull et al. 1996) was aligned with a rigor-like structure (4OZQ; Arora et al. 2014) using the main chain atoms of the P-loop. Mg2+ and ADP are shown as space-fill model and are colored magenta and cyan, respectively. c The root-mean-square deviation (RMSD) of complementary residues is plotted for each individual β-strand. d Individual β-strand curvature based on the sum of sequential Cα distances for each β-strand residue (i = initial residue, i + x = final residue) divided by the total distance from initial to final plotted for nucleotide-bound and rigor-like structures. e β-strand torsion angles (τ) relative to β1 are plotted to quantify the degree of twisting within the entire β-sheet. Torsion angles were calculated using the following equation for each individual β-strand: , where are the unit vectors for each respective β-strand (Cecchini et al. 2008). f The change in each individual β-strand torsion angle (Δτ) as the kinesin central β-sheet transitions from nucleotide-bound (1BG2) to rigor-like (4OZQ) states
Structural differences among kinesin members result in differences in the intrinsic rate and equilibrium constants that govern the ATPase cycle. Although the detailed structural mechanism of the nucleotide- and microtubule-driven conformational changes remains largely unknown, a current model exists for processive kinesins and is largely based on studies of kinesin-1. The nucleotide state at the active site is thought to trigger a conformational change that is transmitted to the adjacent microtubule-binding regions of the core. Communication between the active site and the microtubule binding region is achieved through two prominent structural pathways: (1) Sw1 to α3 to the β5-L8 lobe (Cochran et al. 2009; Ogawa et al. 2004) and (2) Sw2 to L11 to helix α4 to the remainder of the “Sw2 cluster” (Rice et al. 1999; Sindelar and Downing 2007; Vale and Milligan 2000). The Sw2 cluster also controls the orientation of the neck linker in alternate conformations that result in directed force production and motility stabilization (Gigant et al. 2013; Kikkawa et al. 2001). Cryo-electron microscopy (cyro-EM) studies have provided key insights into the conformational changes that kinesin motor domains undergo during their ATPase cycle, and recent advances in image processing algorithms have provided moderate/high resolution structures of these complexes (Table 4).
Table 4.
Summary for kinesin⋅microtubule interactions via cryo-electron microscopy methodologies
| Family | Name | PDB ID | Nucleotide | Notes | Resolution (Å) | Reference |
|---|---|---|---|---|---|---|
| Kinesin-1 | HsKHC | 2p4n | Free/Rigor | 9.0 | Sindelar and Downing 2007 | |
| RnKHC | 4atx | Free/Rigor | Dcxa; Pseudo model | 8.2 | Liu et al. 2012b | |
| Kinesin-3 | MmKIF1A | 2hxf | MgAMPPNP | 10.0 | Kikkawa and Hirokawa 2006 | |
| MmKIF1A | 2hxh | MgADP | 11.0 | Kikkawa and Hirokawa 2006 | ||
| MmKIF1A | 1ai0 | MgAMPPCP | 15.0 | Kikkawa et al. 2001 | ||
| Kinesin-5 | HsEg5/KSP | 4aqw | Free/Rigor | 9.5 | Goulet et al. 2012 | |
| HsEg5/KSP | 4aqv | MgAMPPNP | 9.7 | Goulet et al. 2012 | ||
| DmKLP61F | 2wbe | MgAMPPNP | KLP61F homology model | 9.4 | Bodey et al. 2009 | |
| Kinesin-10 | DmNOD | 3dco | Free/Rigor | 11.0 | Cochran et al. 2009 | |
| Kinesin-13 | DmKLP10A | 3j2o | Unknown | 10.8 | Asenjo et al. 2013 | |
| DmKIF2C | 3edl | MgAMPPNP | 28 | Tan et al. 2008 |
aDcx, Microtubule-associated protein called doublecortin
Family-specific motifs modulate the structural communication of various kinesins during their mechanochemical cycles. For example, L5 plays an important role during the kinesin ATPase cycle and has been the subject of several studies due to its location in the motor domain (it interrupts the long α2 helix immediately downstream of the P-loop; Fig. 3), variability in length across the superfamily (6–21 residues), and forming part of the binding pocket for various kinesin-5/Eg5 inhibitors (Brier et al. 2004; Kaan et al. 2010; Maliga et al. 2006; Talapatra et al. 2012; Yan et al. 2004). L5 wast proposed to first undergo conformational changes upon nucleotide binding during the kinesin-5 ATPase cycle (Cochran et al. 2005) and has since been found to function as a “conformational latch” to structurally and kinetically regulate its stepping behavior (Behnke-Parks et al. 2011; Parke et al. 2010; Waitzman et al. 2011). Specific interactions of the “microtubule-sensing latch” L7 with Sw1 and Sw2 have been implicated in the mechanism of MgADP product release from the active site of the kinesin-3 motor KIF1A (Nitta et al. 2008). For kinesin-13 motors, microtubule depolymerization activity relies on an extended L2 that contains the sequence “KVD” (also called the “KVD finger”), which has been shown to be necessary for microtubule depolymerization activity of these motors (Ogawa et al. 2004). This extended L2 is located within the conserved β-lobe near α6 (Fig. 3); however, the actual function of this β-lobe has remained elusive. The authors of a recent molecular dynamics simulation study aimed at investigating the role of this β-lobe in the mechanical stepping behavior in kinesin-1 suggested that it functions to amplify movements in the neck linker for processive motility (Geng et al. 2014).
Most kinesins walk along the microtubule lattice in a directional manner, either moving towards the plus-end or minus-end of the microtubule (Wade and Kozielski 2000). Traditionally, kinesins which have their motor domain located at the N-terminus of the heavy chain (i.e., N-class kinesins; e.g., kinesin-1) have shown plus-end directed motility, while kinesins with their motor domain located at the C-terminus (i.e., C-class kinesins; e.g., kinesin-14) have demonstrated minus-end directed motility. It was originally thought that directionality was caused by inherent differences in the structure of the motor domains of N-class and C-class kinesins (Henningsen and Schliwa 1997). However, crystal structures of the motor domains of kinesin-1 and kinesin-14 show very few structural differences (Kull et al. 1996; Sablin et al. 1996), while dimeric structures obtained through cryo-EM show significant differences in the orientation of the two motor domains relative to one another, with both having their unattached heads pointing in the direction they move along the microtubule (Arnal et al. 1996; Hirose et al. 1996; Kozielski et al. 1999; Sablin et al. 1998). Furthermore, dimeric chimeras with the motor core from kinesin-14 and stalk and neck regions from kinesin-1 show plus-end directed movement, despite the fact that the motor domains are from a minus-end directed motor (Henningsen and Schliwa 1997). Likewise, chimeras with the kinesin-1 motor core and kinesin-14 neck and stalk regions show minus-end motility (Endow and Waligora 1998). Taken together, these results suggest that directionality is determined primarily by structures outside the conserved motor core (Case et al. 1997).
The neck linker, which connects the motor core to the stalk, has been shown to reorient in response to the nucleotide state of the motor domain (Rice et al. 1999) and is thought to be responsible for positioning the detached motor head in the dimers towards either the plus- or minus-end of the microtubule, as seen in the dimeric structures of kinesin-1 and kinesin-14 (Rice et al. 1999). Additionally, mutant studies of kinesin-14 chimeras have also indicated that interactions between the neck region and the stalk are critical for maintaining minus-end directed movement. Mutant motors that lack critical core-interacting residues result in the slowing of kinesin-14 walking towards the plus-end of microtubules (Sablin et al. 1998). Disrupting the interaction between the neck linker and the motor domain in kinesin-1 also significantly impairs motility (Case et al. 2000), suggesting that interactions of the motor core with both the neck linker and neck region are responsible for determining directional movement. Lastly, the finding that kinesin-14 moves towards the plus-end of microtubules when interactions between the motor core and neck are interrupted suggests that the catalytic motor core inherently has an slight plus-ended directionality (Sablin et al. 1998), which in plus-ended motors is amplified by their neck linker and neck interactions, and in minus-ended motors is over-ridden by stronger interactions resulting in minus-directed movement.
Recently, the kinesin-5’s isolated from Saccharomyces cerevisiae (Cin8p and Kip1p; Fridman et al. 2013; Gerson-Gurwitz et al. 2011; Roostalu et al. 2011; Thiede et al. 2012) and from Schizosaccharomyces pombe [Cut7p; (Edamatsu 2014)] have been shown to be able to switch directionality between slow plus-end movement to rapid minus-end movement. In vitro, the switch to plus-end directionality for both Cin8 and Kip1 can be induced by motor coupling when they bind two antiparallel microtubules or by decreasing the ionic strength. At high (physiological) ionic strength, or when on single microtubules, they act as rapid minus-end directed motors, while at low ionic strength, or when cross-linking two antiparallel microtubules, they act as slow plus-end directed motors. This ability for single kinesin motors to change directionality based on environmental conditions provides an opportunity to better understand the underlying factors that regulate directional movement of all members of the kinesin superfamily.
Kinesin motors are allosteric enzymes by nature. Therefore, amino acid residue substitutions in the motor domain can yield expected results, yet often produce unexpected or unpredicted pleiotropic effects (Table 5). Many different substitutions have been discovered in kinesin motors that have been implicated in diseases such as cancer (Rath and Kozielski 2012) or motor neuron degeneration [e.g., hereditary spastic paraplegia; (Ebbing et al. 2008; Fuger et al. 2012)]. As a general trend, but one not yet considered to be universal across all kinesin members, most tested substitutions lead to a reduced turnover number (k cat), altered binding affinity for microtubules, and slower in vitro microtubule gliding velocity (Table 5). Substitutions in the conserved active site motifs lead to drastically reduced (if not completely abolished) ATPase activity, whereas substitutions in adjacent loops involved in the structural communication pathways between the active site and the microtubule binding region lead to disruption of allosteric regulation of the motor. Recently, the conserved switch-1 serine residue that physically interacts with the divalent metal ion in the active site was substituted with a cysteine; this rendered several kinesin family members inactive with Mg2+ but swapping metals with Mn2+ led to the restoration of activity (Cochran et al. 2011). Interestingly, single substitutions in the neck of kinesin-14 leads to bidirectional motility (Endow and Higuchi 2000), which should shed light on how the yeast kinesin-5 motors (Cin8p, Kip1p, and Cut7p) show directionality switching behavior (Fridman et al. 2013; Gerson-Gurwitz et al. 2011; Roostalu et al. 2011; Thiede et al. 2012). Despite the numerous residue substitutions that have been characterized, researchers still have only scratched the surface for using site-specific mutagenesis techniques to probe kinesin structure and function.
Table 5.
Residue Some of the WT sequence is in bold. Should the bold font not be explained in a footnote? substitutions in kinesin motor domain with reported effects relative to the respective WT construct
| Family | Name | Substitution | WT Sequence | Structure | Effects | Reference |
|---|---|---|---|---|---|---|
| Kinesin-1 | HsKHC | R14A | MCRFR | β1/N-4 | R14A/R14A homodimer; 35-fold tighter K m; 18-fold slower k cat; 18-fold slower gliding V obs | Kaseda et al. 2003 |
| R14A | MCRFR | β1/N-4 | WT/R14A heterodimer; 2-fold tighter K m; 9-fold slower k cat; 8-fold slower gliding V obs; similar stall force | Kaseda et al. 2003 | ||
| D140A | YLDKI | L7 | 3.5-fold tighter K m; 1.7-fold slower k cat; 1.8-fold slower gliding V obs | Woehlke et al. 1997 | ||
| K141A | LDKIR | L7 | 1.8-fold weaker K m; 1.1-fold faster k cat; 1.1-fold slower gliding V obs | Woehlke et al. 1997 | ||
| K159A | EDKNR | L8 | 2.3-fold weaker K m; similar k cat; 1.3-fold slower gliding V obs | Woehlke et al. 1997 | ||
| E170A | CTERF | L8 | 2.6-fold tighter K m; 1.2-fold slower k cat; 1.6-fold slower gliding V obs | Woehlke et al. 1997 | ||
| S202C | HSSRS | L9/Sw1 | 10-fold slower MgATPase; similar MnATPase; 100-fold slower MgATP gliding V obs; 10-fold slower MnATP gliding V obs | Cochran et al. 2011 | ||
| E236A | GSEKV | L11/Sw2 | No MT gliding; >1000-fold slower k cat; similar ATP binding kinetics; increased ADP release kinetics; can take one ATP-dependent step | Rice et al. 1999 | ||
| K240A | VSKTG | L11 | 4.2-fold weaker K m; similar k cat; 1.6-fold slower gliding V obs | Woehlke et al. 1997 | ||
| L248A | AVLDE | L11 | 3.1-fold weaker K m; 1.3-fold slower k cat; 1.1-fold faster gliding V obs | Woehlke et al. 1997 | ||
| E250A | LDEAK | L11 | 1.9-fold tighter K m; 1.3-fold slower k cat; 1.8-fold slower gliding V obs | Woehlke et al. 1997 | ||
| K252A | EAKNI | α4 ext. | 4.4-fold weaker K m; 2.7-fold slower k cat; 1.1-fold faster gliding V obs | Woehlke et al. 1997 | ||
| K256A | INKSL | α4 | 2.9-fold weaker K m; 1.4-fold slower k cat; similar gliding V obs | Woehlke et al. 1997 | ||
| E270A | LAEGS | L12 | 2.1-fold tighter K m; 1.1-fold faster k cat; 1.1-fold slower gliding V obs | Woehlke et al. 1997 | ||
| Y274A | STYVP | L12 | 3.2-fold weaker K m; 1.3-fold slower k cat; 1.2-fold slower gliding V obs | Woehlke et al. 1997 | ||
| R278A | PYRDS | L12 | 15.5-fold weaker K m; 1.5-fold slower k cat; 1.3-fold slower gliding V obs | Woehlke et al. 1997 | ||
| D279A | YRDSK | L12 | 2.3-fold tighter K m; 1.3-fold slower k cat; 1.2-fold slower gliding V obs | Woehlke et al. 1997 | ||
| K281A | DSKMT | L12/α5 | 2.4-fold weaker K m; 1.1-fold slower k cat; 1.2-fold slower gliding V obs | Woehlke et al. 1997 | ||
| R284A | MTRIL | α5 | 2.2-fold weaker K m; 2.3-fold slower k cat; 1.1-fold slower gliding V obs | Woehlke et al. 1997 | ||
| E311A | ESETK | α6 | 3.1-fold tighter K m; 2.9-fold slower k cat; 3.2-fold slower gliding V obs | Woehlke et al. 1997 | ||
| W340F | EQWKK | Neck | Similar k cat; 4-fold tighter K 0.5,MT; Cys-light K413-C333 construct | Rosenfeld et al. 2001 | ||
| DmKHC | T94S | GQTSS | L4/P-loop | 3.6-fold faster basal ADP release; 1.8-fold faster MT-stimulated ADP release; 3.3-fold slower MT gliding V obs; similar stall force | Higuchi et al. 2004 | |
| E164A | VHEDK | β5a/L8 | 4.2-fold slower k cat; 4-fold faster ATP hydrolysis; no observed MT dissociation; 2–3-fold slower ADP release | Klumpp et al. 2004a; Klumpp et al. 2004b; Klumpp et al. 2003 | ||
| E164K | VHEDK | β5a/L8 | 3-fold slower k cat; 2-fold faster ATP hydrolysis; 3–4-fold slower ADP release | Klumpp et al. 2004a; Klumpp et al. 2004b | ||
| E164N | VHEDK | β5a/L8 | 2.6-fold slower k cat; 3-fold faster MT dissociation; 3-fold slower MT association; 2–3-fold slower ADP release | Klumpp et al. 2004a | ||
| E164D | VHEDK | β5a/L8 | 1.5-fold slower k cat; 4-fold faster MT dissociation | Klumpp et al. 2004a | ||
| E164Q | VHEDK | β5a/L8 | 1.4-fold slower k cat; 3-fold faster MT dissociation; 2-fold slower MT association; 2-fold slower ADP release from both heads | Klumpp et al. 2004a | ||
| D165A | HEDKN | β5a/L8 | 1.7-fold slower k cat; 1.5-fold slower ADP release | Klumpp et al. 2004a | ||
| R210A | SSRSH | L9/Sw1 | >500-fold slower ATP hydrolysis; 350-fold slower k cat; 12–25-fold slower MT association; 5–6-fold slower ADP release from both heads | Farrell et al. 2002 | ||
| R210K | SSRSH | L9/Sw1 | >800-fold slower ATP hydrolysis; 350-fold slower k cat; 1.3-fold slower MT association; processive motility by WT/R210K heterodimer | Klumpp et al. 2003; Thoresen and Gelles 2008 | ||
| T291M | KLTRI | α5 | 3.8-fold slower MT gliding V obs; 2.5-fold weaker K m; 4-fold weaker K 0.5,MT; 2.5-fold faster ATP hydrolysis; 3-fold faster MT dissociation | Brendza et al. 1999; Brendza et al. 2000 | ||
| RnKHC | E200A | MNEHS | L9/Sw1 | 1.4-fold slower k cat; 5–10-fold slower MT-stimulated ADP release | Auerbach and Johnson 2005b | |
| E200D | MNEHS | L9/Sw1 | 1.7-fold slower k cat; 2-fold faster mantATP binding; 4.3-fold slower MT-stimulated mantADP release | Auerbach and Johnson 2005b | ||
| E237A | GSEKV | L11/Sw2 | >400-fold slower k cat; >500-fold faster basal ADP release; 3–4-fold slower MT-stimulated ADP release | Auerbach and Johnson 2005b | ||
| E237D | GSEKV | L11/Sw2 | 9.1-fold slower k cat; 2.7-fold slower ATP binding; 4.4-fold slower MT-stimulated ADP release | Auerbach and Johnson 2005b | ||
| N256K | NINKS | α4 ext. | 133-fold slower k cat; 1.3-fold faster ATP binding; 400–500-fold slower MT-stimulated ADP release | Auerbach and Johnson 2005b | ||
| MmKIF5C | V240Q/S241K | EKVSKT | L11 | Double substitution; similar k cat; ~2-fold weaker K 0.5,MT | Chang et al. 2013 | |
| S258G | NKSLS | α4 | 2-fold faster k cat; 10-fold weaker K 0.5,MT | Chang et al. 2013 | ||
| GDDKKG | AEGTK | L12 | Chimeric construct; 2-fold faster k cat; 1.6-fold tighter K 0.5,MT; 3.4-fold increase in k cat/K 0.5,MT | Chang et al. 2013 | ||
| Kinesin-3 | MmKIF1A | E148A | YMEIY | β4 | 37-fold slower k cat; 2.9-fold slower basal ATPase | Nitta et al. 2008 |
| Y150F | EIYCE | β4/L7 | 5.3-fold slower k cat; 2.1-fold slower basal ATPase | Nitta et al. 2008 | ||
| E152A | YCERV | L7 | 11.5-fold slower k cat; 8.1-fold tighter K 0.5,MT; >7-fold faster MT-stimulated ADP release; 1.5-fold slower basal ATPase | Nitta et al. 2008 | ||
| R153A | CERVR | L7/β5 | 2.4-fold slower k cat; similar basal ATPase | Nitta et al. 2008 | ||
| K161A/R167A R169A/K183A |
L8 | Quadruple substitution; 6-fold weaker K d,MT with AMPPNP; 3.9-fold weaker K d,MT with ADP | Nitta et al. 2004 | |||
| R203A | KARTV | α3 | 15-fold slower k cat; 3.3-fold slower basal ATPase | Nitta et al. 2008 | ||
| R216A | SSRSH | L9/Sw1 | 240-fold slower k cat; 8.3-fold slower basal ATPase | Nitta et al. 2008 | ||
| D248A | LVDLA | β7/Sw2 | 1670-fold slower k cat; 3.1-fold slower basal ATPase | Nitta et al. 2008 | ||
| K261A/R264A/ K266A |
L11 | Triple substitution; 4.8-fold weaker K d,MT with AMPPNP; 1.8-fold weaker K d,MT with ADP | Nitta et al. 2004 | |||
| E267A | LKEGA | α4 ext. | 22-fold slower k cat; 5.6-fold slower basal ATPase | Nitta et al. 2008 | ||
| GTKT | MDSGPNKN(K)6TD | L12 | Chimeric construct; 3.5-fold weaker K d,MT with ADP; 3.8-fold weaker K d,MT with ATP; 7.8-fold weaker K d,MT with ADP + VO4 | Nitta et al. 2004 | ||
| Kinesin-4 | MmKIF4A | Q248V/K249S | ERQKKT | L11 | Double substitution; Slightly increased k cat and K 0.5,MT; similar k cat/K 0.5,MT | Chang et al. 2013 |
| G267S | NRGLL | α4 | Slightly decreased k cat and K 0.5,MT; similar k cat/K 0.5,MT | Chang et al. 2013 | ||
| AEGTK | GDDKKG | L12 | Chimeric construct; 2-fold decreased k cat; similar K 0.5,MT; 2.3-fold decrease in k cat/K 0.5,MT | Chang et al. 2013 | ||
| Kinesin-5 | HsEg5 | P121A | RSPNE | L5 | 2-fold slower neck-linker isomerization; 4.3-fold slower ADP binding; 5.1-fold slower ADP release | Behnke-Parks et al. 2011 |
| P131A | EDPLA | L5 | 50-fold slower ATP binding; 5-fold slower neck-linker isomerization; 7.7-fold slower ADP release; 3.5-fold slower MT dissociation | Behnke-Parks et al. 2011 | ||
| S233C | YSSRS | L9/Sw1 | 5–6-fold slower MgATPase; similar MnATPase; ATP hydrolysis slowed for MgATPase; similar ATP binding and ADP release kinetics | Cochran et al. 2011 | ||
| R234K | SSRSH | L9/Sw1 | 125-fold slower ATP hydrolysis; 10-fold faster MT association; similar AMPPNP binding affinity | Zhao et al. 2010 | ||
| SC/RK | YSSRSH | L9/Sw1 | Double substitution; similar ATP binding; complete loss of ATP hydrolysis | Krzysiak et al. 2008; Zhao et al. 2010 | ||
| Kinesin-10 | DmNOD | S203C | NSSRS | L9/Sw1 | ~20-fold slower basal MgATPase; similar basal MnATPase | Cochran et al. 2011 |
| Kinesin-13 | MmKIF2C | K293A | KLKVD | L2 | ~50 % MT depolymerization activity | Ogawa et al. 2004 |
| V294A | LKVDL | L2 | ~55 % MT depolymerization activity | Ogawa et al. 2004 | ||
| D295A | KVDLT | L2 | ~65 % MT depolymerization activity | Ogawa et al. 2004 | ||
| K293A/V294A/ D295A |
L2 | Triple substitution; ~35 % MT depolymerization activity | Ogawa et al. 2004 | |||
| G489A | LAGNE | L11/Sw2 | Defective in tubulin detachment; very slow ATP hydrolysis | Wagenbach et al. 2008 | ||
| E491A | GNERR | L11/Sw2 | Defective in tubulin detachment; 20-fold tighter K d,tubulin; catalytically depolymerizes MTs in ADP | Wagenbach et al. 2008 | ||
| PfKINI | K40A/V41A/ D42A |
L2 | Triple substitution; 10-fold reduction in MT depolymerization; ~10-fold slower MT-stimulated k cat; similar tubulin-stimulated k cat | Shipley et al. 2004 | ||
| R242A | SERGA | L11 | ~2-fold reduction in MT depolymerization; ~2-fold slower MT-stimulated k cat; ~3-fold slower tubulin-stimulated k cat | Shipley et al. 2004 | ||
| D245A | GADTV | L11 | Similar MT depolymerization; ~1.5-fold faster MT-stimulated k cat; ~1.2-fold faster tubulin-stimulated k cat | Shipley et al. 2004 | ||
| K268A/E269A/ C270A |
α4 | Triple substitution; no MT depolymerization; ~20-fold slower MT-stimulated k cat; ~4-fold slower tubulin-stimulated k cat | Shipley et al. 2004 | |||
| R272A | CIRAM | α4 | ~2-fold reduction in MT depolymerization; ~3-fold slower MT-stimulated k cat; ~3-fold slower tubulin-stimulated k cat | Shipley et al. 2004 | ||
| Kinesin-14 | DmNCD | R335A/K336A | MERKEL | Neck | Double substitution; 2.0-fold slower MT gliding V obs | Sablin et al. 1998 |
| H339A/N340A | ELHNTV | Neck | Double substitution; 1.3-fold slower MT gliding V obs | Sablin et al. 1998 | ||
| N340K | LHNTV | Neck | Bidirectional MT gliding; similar MT gliding velocities in either direction; unstable neck position with AMPPNP | Endow and Higuchi 2000; Endres et al. 2006 | ||
| N340K/K640N | Double substitution; bidirectional MT gliding | Endow and Higuchi 2000 | ||||
| ESGAKQ GEKGES |
RKELHN
TVMDLR |
Neck | WT (aa 335-346); 50-fold slower MT gliding V obs | Sablin et al. 1998 | ||
| G347D | LRGNI | Neck | Dimer X-ray structure (PDB ID: 3u06); 1.5-fold slower basal ADP release and ATPase; no MT gliding observed; 6-fold weaker K 0.5,MT | Liu et al. 2012b | ||
| Q420A/S421A | LIQSAL | α1 | Double substitution; 2.3-fold slower MT gliding V obs | Sablin et al. 1998 | ||
| Y426A | DGYNI | L3 | 1.8-fold slower MT gliding V obs | Sablin et al. 1998 | ||
| Q420A/S421A/Y426A | α1/L3 | Triple substitution; 10-fold slower MT gliding V obs | Sablin et al. 1998 | |||
| T436S | GQTGS | L4/P-loop | Dimer X-ray structure (PDB ID: 3l1c); 10-fold faster basal ADP release; similar MT-stimulated ADP release; 2-fold faster k cat; 1.5-fold slower MT gliding V obs | Heuston et al. 2010 | ||
| Y485E | EIYNE | β4/L7 | 3.8-fold faster basal ADP release; 1.5-fold weaker K d,MT; similar MT-stimulated ATPase; similar MT gliding V obs | Liu et al. 2012a | ||
| Y485N | EIYNE | β4/L7 | 10-fold faster basal ADP release and ATPase; 1.9-fold weaker K d,MT; 2-fold faster MT-stimulated ATPase; 1.3-fold faster MT gliding V obs | Liu et al. 2012a | ||
| Y485K | EIYNE | β4/L7 | 26-fold faster basal ADP release; 3-fold weaker K d,MT; 2-fold faster k cat; 10-fold tighter K 0.5,MT; 2-fold faster MT gliding V obs | Liu et al. 2012a | ||
| S551C | RSSRS | L9/Sw1 | ~6-fold slower basal MgATPase; similar basal MnATPase | Cochran et al. 2011 | ||
| R552A | SSRSH | L9/Sw1 | 4-fold weaker K d,MT (no nucleotide); 70-fold weaker K d,MT (AMPPNP); >9-fold weaker K d,MT (ADP); 2-fold slower k cat; 3-fold weaker K 0.5,MT; 5-fold slower MT gliding V obs | Kocik et al. 2009 | ||
| V556F | HAVTK | β6 | 3-fold slower k cat and weaker K 0.5,MT; 2-fold weaker K d,MT; 3-fold weaker K d,MT in apyrase; 4-fold tighter K d,MT in AMPPNP | Moore et al. 1996 | ||
| E567A/K568A | HAEKQE | L10 | Double substitution; similar MT gliding V obs | Sablin et al. 1998 | ||
| E585A | GSESK | L11/Sw2 | 3-fold weaker K d,MT (no nucleotide); 5–7-fold weaker K d,MT; 7-fold slower k cat; 28-fold slower MT gliding V obs | Kocik et al. 2009 | ||
| E585D | GSESK | L11/Sw2 | 3-fold weaker K d,MT (no nucleotide); 5–7-fold weaker K d,MT; 7-fold slower k cat; 28-fold slower MT gliding V obs | Kocik et al. 2009 | ||
| N600K | NINRS | α4 ext. | 8-fold weaker K d,MT (no nucleotide); 17-fold weaker K d,MT (AMPPNP); 2-fold slower k cat and weaker K 0.5,MT; 1.4-fold slower MT gliding V obs | Kocik et al. 2009; Song and Endow 1998 | ||
| G636A/G637A | SLGGNS | L13 | Double substitution; 1.3-fold slower MT gliding V obs | Sablin et al. 1998 | ||
| K640A | NSKTL | L13/β8 | 3-fold slower MT gliding V obs | Sablin et al. 1998 | ||
| K640N | NSKTL | L13/β8 | Bidirectional MT gliding | Endow and Higuchi 2000 | ||
| ScKAR3 | R598A | SSRSH | L9/Sw1 | X-ray structure (PDB ID: 1f9v); similar basal ATPase; complete loss of MT-stimulated ATPase | Yun et al. 2001 | |
| E631A | GSERR | L11/Sw2 | X-ray structure (PDB ID: 1f9w); similar basal ATPase; complete loss of MT-stimulated ATPase | Yun et al. 2001 | ||
| N650K | NINKS | α4 ext. | X-ray structure (PDB ID: 1f9u); similar basal ATPase; complete loss of MT-stimulated ATPase; complete loss of MT gliding; mant-ADP release not stimulated by MT binding | Song and Endow 1998; Yun et al. 2001 |
The bold font indicates the amino acid position that was substituted
Physics
Kinesin-1 is a processive translocase that can take hundreds of steps along the microtubule while consuming 1 ATP molecule per 8-nm (approx.) step (Carter and Cross 2005; Coy et al. 1999; Hua et al. 1997; Iwatani et al. 1999; Ray et al. 1993; Schnitzer and Block 1997; Schnitzer et al. 2000; Svoboda et al. 1993; Vale et al. 1996). Using the law of mass action and standard thermodynamic equations, the average available free energy of ATP hydrolysis under typical in vitro steady-state ATP turnover conditions equals –74.9 kJ mol-1 or –124 pN nm (see Appendix A for details). The stall force for kinesin-1 has been measured at 5–7 pN using optical trapping methodologies (Schnitzer and Block 1997; Svoboda and Block 1994; Visscher et al. 1999); therefore, with an average step size at 8.3 nm (Yildiz et al. 2004), kinesin-1 is 33–47 % efficient (e.g. 5 pN × 8.3 nm = 42 pN nm = 33 % efficiency) in its use of ATP hydrolysis-free energy for moving along the microtubule.
The stall force in other dimeric kinesin motors has been determined: kinesin-2 (KLP11/KLP20 heterodimer) at ~4 pN (Brunnbauer et al. 2010), kinesin-3 (Unc104/KIF1A) at ~6 pN (Tomishige et al. 2002), kinesin-5 (Eg5/KSP) at ~5 pN (Valentine et al. 2006), kinesin-7 (CENP-E) at ~6 pN (Yardimci et al. 2008), kinesin-8 (KIF18A/Kip3) at ~1 pN (Jannasch et al. 2013), and kinesin-14 (Ncd) at ~1 pN for two to four motors linked via a rigid DNA scaffold with ~6- or ~22-nm spacing (Furuta et al. 2013). The motility velocities for these different kinesin family members were typically much slower than that for kinesin-1, and their run lengths were much shorter. Therefore, kinesin motors appear to have a wide range of efficiencies (namely, 9–64 %) for converting the energy of ATP hydrolysis to mechanical motion, as well as for using this energy to carry out different functions in cellular processes.
Processivity is a characteristic of most motor proteins that is defined by the distance traveled along its track before it detaches and diffuses away. Several classes of kinesin motors are quite processive, traveling from several microns through hundreds of ATP hydrolysis cycles before dissociating from their microtubule track. These families include kinesin-1 (Howard et al. 1989), kinesin-2 (Muthukrishnan et al. 2009), kinesin-3 (Soppina et al. 2014), kinesin-5 (Valentine et al. 2006), kinesin-7 (Yardimci et al. 2008), and kinesin-8 (Varga et al. 2006). Early experiments designed to measure the movement of single dimeric kinesin motors tethered to a bead provided the first direct evidence of kinesin processivity (Svoboda et al. 1993). Presteady-state kinetic analyses of the dimeric kinesin-1 ATP hydrolysis cycle subsequently provided additional evidence for chemical processivity (Gilbert et al. 1995; Hackney 1995a). Single fluorescently labeled kinesin-1 motors were shown to demonstrate processive motility in solution (Vale et al. 1996), again supporting this important biophysical characteristic for transport kinesins. The molecular basis for kinesin processivity is chiefly thought to stem from an asymmetric hand-over-hand mechanism that keeps the catalytic cycles in each head “out-of-phase” with its partner, thus allowing one head to remain tightly bound to the microtubule at any given point along a processive run (Toprak et al. 2009). It still remains a mystery why certain kinesin members take a few steps during a processive run (5–10 steps for kinesin-5; Valentine et al. 2006), while other kinesin members can take thoursands of steps during a processive run (e.g., kinesin-3; Soppina et al. 2014; Soppina and Verhey 2014). It has been postulated that increased positive charge in the neck region and/or the C-terminal region functions to enhance kinesin processivity (Grissom et al. 2009; Kim et al. 2008; Mayr et al. 2011; Rosenfeld et al. 2009; Stumpff et al. 2011; Su et al. 2011; Thorn et al. 2000; Varga et al. 2009; Weaver et al. 2011). Another prevailing mechanism for modulating kinesin processivity centers on the length of the neck linker, which is thought to affect mechanochemical gating between partner heads during a processive run (Clancy et al. 2011; Duselder et al. 2012; Miyazono et al. 2010; Shastry and Hancock 2010; Shastry and Hancock 2011; Yildiz et al. 2008). However, recent work from the Verhey laboratory on kinesin-3 motors clearly shows that these so-called “superprocessive” motors utilize an unknown molecular mechanism to enable this level of processivity (Soppina et al. 2014; Soppina and Verhey 2014). It is also important to note that not all kinesins are processive (e.g., kinesin-10/Kid and kinesin-14/Ncd are non-processive mitotic motors), and that in fact some kinesins are not transport motors at all (e.g., kinesin-10/NOD is a non-motile microtubule plus-end tracker and kinesin-13/MCAK is a potent microtubule depolymerase). This variation highlights the diversity of ways that the energy of ATP hydrolysis can be converted into mechanical motion for a variety of biophysical tasks.
Cytoskeletal molecular motors such as kinesin, myosin, and dynein have demonstrated the ability to generate force tangential to the direction of movement along their tracks, thus producing “torque” in the motor–filament interaction (Nishizaka et al. 1993; Vale and Toyoshima 1988; Walker et al. 1990). Kinesin-1 has been shown to processively step along a single protofilament of a microtubule consisting of 13–16 protofilaments (Mori et al. 2007; Ray et al. 1993; Ray et al. 1995; Schaap et al. 2011), whereas kinesin-2 and kinesin-5 have demonstrated an ability to step along adjacent protofilaments of the microtubule, thus causing the motors to spiral around the microtubule (Brunnbauer et al. 2012; Pan et al. 2010; Yajima et al. 2008). It was initially thought that the degree of processivity was inversely proportional to torque generation (i.e., that increasing torque generation decreases processivity; Yajima et al. 2008); however, the combination of results from several studies strongly disfavor this hypothesis (Brunnbauer et al. 2012; Brunnbauer et al. 2010; Muthukrishnan et al. 2009; Yildiz et al. 2008). Indeed, recent results suggest that the stability of the neck (not that of the neck linker with the motor core) is responsible for conferring the side-stepping behavior (Brunnbauer et al. 2012). Understanding how conformational changes in the kinesin dimer’s motor domains and the directionally biased selection of microtubule binding sites are biophysically linked during asymmetric hand-over-hand stepping remains a subject for future investigation.
Analysis of the energy landscape of kinesin-5 using isothermal titration calorimetry (ITC) (Fig. 5) revealed an the observed free energy of ATP binding and hydrolysis (ΔG1 + ΔG2) of –31 kJ mol-1 (–51 pN nm), which represents an energy efficiency of 43 % based on the conditions used in the ITC experiment (ΔGrxn = –72.4 kJ mol-1 or –120 pN nm; Zhao et al. 2010). This observed free energy is comparable to the range of work performed by dimeric kinesin-5 (–42 to 49 pN nm; Valentine et al. 2006), suggesting that the observed thermodynamics of a single kinesin head provides valuable insight into dissecting the structure and function of the motor. One outstanding question: where is the remainder of free energy made available from the hydrolysis of ATP (i.e., approximately –41.4 kJ mol-1 missing free energy)? This remaining chemical free energy that is not converted to mechanical work could get rapidly exchanged (within picoseconds) as entropy production (i.e., dissipated “heat”) in the surrounding physiological milieu (Ge and Qian 2013; Henry et al. 1986). If this hypothesis were true, the excess heat should be “observed” in the ITC experiment since the instrument physically measures the heat associated with a biochemical reaction. Yet this heat was not observed (Zhao et al. 2010), therefore refuting the “dissipated heat” hypothesis. Another hypothesis centers on the “deposition” or “storage” of the energy into the motor as conformational potential energy, which is used by the motor to promote ADP product release from the active site at a much faster rate than theoretically possible (see below). To put the quantity of missing energy into perspective, this represents a net gain of two to six hydrogen bonds (or some other combination of non-covalent interactions within the motor). It is worth speculating that the source of this conformational potential energy resides within the “under-twisted” central β-sheet seen in nucleotide-bound kinesin structures (Fig. 4). Nevertheless, the accounting of the free energy from the reaction remains a mystery.
Fig. 5.
Kinesin energy landscape in the absence and presence of microtubules. The energy landscapes for human monomeric kinesin-5 (Eg5) in the absence (black) and presence (red) of microtubules are shown. The thermodynamically and kinetically defined intermediate states are defined in the table. The magnitude of energy for each transition state was calculated from the Arrhenius/Eyring/Kramer equation using previously determined kinetic rate constants for the monomeric kinesin-5 ATPase mechanism (Table 1). Adapted and updated from Zhao et al. (2010)
Kinesin motors typically bind Pi very weakly and ADP very tightly (Hackney 1988). Consequently, the release of these products is thermodynamically unfavorable–but necessary–to get the enzyme to the beginning of the next cycle. The calculated free energy of Pi binding to kinesin-5 (-ΔG3) was –3.3 kJ mol-1, which equates to a binding affinity (K d,Pi) of 260 mM. The observed ADP binding free energy (-ΔG4 + -ΔG5) was –27.6 kJ mol-1, which equates to a binding affinity (K d,ADP) of 7–8 μM. As the rates of ATP and ADP binding to kinesin-5 have been determined in the absence of microtubules (k +2 and k -5 ≈ 1 s-1, respectively; Cochran and Gilbert 2005), the Arrhenius/Eyring/Kramer equation can be used to calculate the free energy of these transition states (ΔG‡ 1 for ATP binding and ΔG‡ 5 for ADP binding = +73 kJ mol-1). The transition state free energy for ATP hydrolysis (ΔG‡ 2) and Pi release (ΔG‡ 3) are modeled using the minimum intrinsic rate constants for both steps at 10 s-1 [see Cochran and Gilbert (2005) for details on how this minimum rate was determined].
Once kinesin-5 releases the bound Pi product and reaches the ADP-bound state (state “d” in Fig. 5), a very large thermodynamically unfavorable transition must occur for ADP to be released from the active site (i.e., to reach state “a” in Fig. 5). The height of this energy barrier is approximately +101 kJ mol-1, which means that if the mechanism of ADP release happened in a single step (where the motor utilizes thermal energy from the surrounding environment), then the rate of the reaction (determined using Arrhenius/Eyring/Kramers rate theory) should be 7.2 × 10-6 s-1, which is approximately 2,900-fold slower than the observed rate of ADP release from kinesin-5 (k +5’ = 0.02 s-1; Cochran and Gilbert 2005; Cochran et al. 2004; Zhao et al. 2010). Therefore, another intermediate state along the pathway must exist (state “e” in Fig. 5), and this state must somehow avoid microscopic reversibility (i.e., when the enzyme is in state “e”, the probability of returning to state “d” rather than proceeding to state “a” is 671,000:1 under the fundamental principles of microscopic reversibility) and thus act as a molecular thermal ratchet. It has been postulated that the release of Mg2+ and ADP from the kinesin⋅MgADP intermediate (state “d”) occurs sequentially along the ATPase pathway. For kinesin-1, this takes the form of slow, rate-limiting Mg2+ release followed by rapid ADP release (Hackney and Stock 2008). For kinesin-5, it appears that a similar mechanism of MgADP release is utilized, but the intrinsic rate constants which govern these reactions lead to relatively rapid Mg2+ release followed by slow, rate-limiting ADP release (unpublished data). The existence of isomerization state “e” on the reaction path, as presented in Fig. 5, is supported since the concentration of Mg2+ modulates the observed rate of ADP product release in kinesin-5 (unpublished data), as well as in other kinesins (Hackney and Stock 2008; Nitta et al. 2008). Also, kinesin-5 shows very weak ADP binding in the absence of Mg2+ (unpublished data), which is consistent with the notion that the free energy of state “e” is linked to the occupancy of the divalent ion in the active site. Future biophysical investigations surrounding these reactions in the kinesin ATPase cycle will be central to gaining an understanding of force generation and mechanochemical gating in kinesin motors.
Where is the mechanical motion produced in the kinesin ATPase cycle? Even for kinesin-1, this question has been answered differently and the issue remains controversial (Block 2007). Nevertheless, this is a very important and outstanding question for other kinesin families, particularly for the mitotic kinesins, since inhibition of force production would provide a plausible therapeutic strategy. Mechanical output requires communication between the two motor domains in a dimeric kinesin-1 molecule; yet just how this communication occurs is still unclear. Most theories suggest that communication occurs through a “gating” mechanism where a mechanistic step in one motor domain is blocked until a certain step is taken in the other motor domain (Block 2007). This gating mechanism could be chemical in nature, such that ATP binding to the one head is prohibited until the other head dissociates from, or attaches to, the microtubule (Block et al. 2003; Guydosh and Block 2006; Klumpp et al. 2004b; Rosenfeld et al. 2003), or mechanical in nature, such that a conformational change in one head pulls the other head off the microtubule (Hancock and Howard 1999; Spudich 2006; Yildiz et al. 2008). Of course, it is likely that the actual mechanism of communication utilizes both types of gating, as they are not mutually exclusive.
Conclusions
As we near the 30th anniversary of the discovery of kinesin motors, we have uncovered many interesting chemical, structural, and physical insights into several members of the superfamily. Yet a series of important outstanding questions remain unanswered:
What are the biophysical characteristics of “unconventional” kinesins? When you compare the number of kinesin motors that have been studied to date and the number of kinesins that exist in nature (1,624 putative members; Wickstead et al. 2010), it is clear that much work is ahead of us. Several studies of various “conventional” kinesins (not to be confused with conventional kinesin, kinesin-1) have provided a biochemical/structural/mechanical model with variations between the different families studied (e.g., similarities and differences between kinesin-1, kinesin-5, kinesin-13, and kinesin-14 families). However, it remains to be seen how long this model will persist. Indeed, with the characterization of “unconventional” kinesins, researchers are led back to their favorite kinesin and design a series of experiments to test various aspects of the already well-characterized family members. As just one example, when we solved the crystal structure of one of the most distant “unconventional” kinesins in the superfamily (Drosophila melanogaster NOD), we identified two proline residues in L5 that physically interacted with helix α3 (Cochran et al. 2009). Another research group proceeded to mutate one of these conserved L5 proline residues in human kinesin-5 to demonstrate that L5 acts a conformational latch in kinesins (Behnke-Parks et al. 2011).
How do different kinesins utilize distinct nucleoside triphosphorylase mechanisms to either promote MT motility or to regulate MT polymerization dynamics for force generation? Differences in the kinesin superfamily show that some kinesins walk along microtubules (e.g., kinesin-1, kinesin-5, and kinesin-7), some take a few steps along microtubules in order to establish tension in the mitotic spindle (e.g., kinesin-14), while some kinesins are not motile, but instead use this energy to destabilize microtubule polymers (e.g., kinesin-13) or promote microtubule polymerization (e.g., kinesin-10). How are these vastly different outputs generated from a common motor core? Structures of these motor cores are known, but it is still unclear how subtle differences give rise to these different functions.
How can the same kinesin walk in different directions on the microtubule? At the turn of the century, the kinesin community would have overwhelmingly agreed that regions outside of the kinesin motor core determine the direction of movement (for review, see Wade and Kozielski 2000). The experiments that generated chimeric versions of kinesin-1 (plus-end directed) and kinesin-14 (minus-end directed) demonstrated that the linker sequences immediately adjacent (~2–3 residues) to the motor core (β1–α6) dictate directionality along the microtubule. However, recent studies with native yeast kinesin-5 motors (i.e., not chimeric constructs) show directional switching that is dependent on motor–microtubule density, ionic strength, and/or parallel versus antiparallel microtubule orientation (Fridman et al. 2013; Gerson-Gurwitz et al. 2011; Roostalu et al. 2011; Thiede et al. 2012). It will be interesting to develop a better understanding of the molecular mechanism that dictates this biophysical conundrum.
Why does the β-sheet “twist” during the ADP–ATP exchange process? Study of the twisting of the central β-sheet during the ADP–ATP exchange steps of the cycle is an emerging trend in kinesin biophysics. How does “twisting” of the β-sheet in molecular motors facilitate mechanochemistry and force production? Does the twisting provide a mechanism for storing conformational potential energy in order for kinesins to rapidly release ADP product and start a new cycle with ATP binding? Does the “over-twisting” and “under-twisting” of the β-sheet contribute to the thermodynamics and mechanics of force generation in kinesins? These questions will be important to answer in the near future, yet the appropriate experimental design will be essential to narrow the interpretation of data in supporting or refuting these hypotheses.
How do specific macromolecular interactions between kinesin and microtubules (or soluble tubulin for some kinesins) stimulate enzymatic activity? Although we have very solid experimental support for microtubule stimulation of the kinesin ATPase cycle, we still lack a detailed structural model that answers this question. Cryo-EM structures and X-ray crystal structures of kinesin–tubulin complexes have advanced our understanding of the molecular interface, although much work remains to complete our understanding of the molecular dynamics of this interaction. Given the large number of atoms that comprise the kinesin–tubulin complex, technological advances in computational power will be necessary to simulate this interaction in order to generate detailed structural hypotheses that can be tested in the laboratory.
How are kinesin, myosins, and G proteins related? Since the structures of myosin (Rayment et al. 1993) and kinesin (Kull et al. 1996) were initially determined, many structural and functional similarities have been observed to be shared between the motor proteins and small G proteins. These similarities include common nucleotide binding motifs (mentioned earlier), common active site chemistry, and common secondary structure topology in the catalytic domain (Kull et al. 1998). G proteins show exclusive preference for using GTP hydrolysis for its cellular function (GTP binds ~107-fold tighter than ATP; Goody et al. 1992), while kinesins have historically been thought to use ATP hydrolysis (ATP binds ~102-fold tighter than GTP; Gilbert and Johnson 1993). Nevertheless, given the vast wealth of independent studies on molecular motors and small G proteins, there remain many unanswered questions on the conservation of base specificity and nucleotide hydrolysis mechanisms for these hydrolases. Given the allosteric nature of kinesin enzymes and the relatively “loose” pocket for the nucleotide base, can certain kinesin family members use either GTP or ATP for energy transduction? If so, what are the unique features in the catalytic domain that provide a structural basis to explain these differences in the catalytic mechanism? Future research will provide insight into the conserved and non-conserved mechanisms between molecular motors and molecular switch proteins.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOCX 127 kb)
Acknowledgments
I would like to thank Joseph Eskew for his help with Appendix A, and Kayla Bell, Benjamin Walker, and Jeff Ewer for helpful discussions. I am grateful for comments on the manuscript provided by Claire Walczak and F. Jon Kull.
Compliance with Ethical Standards
ᅟ
Conflict of interest
Jarod C Cochran declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
Abbreviations
- AlFx
Aluminum fluoride
- AMPPCP
Adenosine 5′-(β,γ-methylene)triphosphate
- AMPPNP
Adenosine 5′-(β,γ-imido)triphosphate
- ATP
Adenosine 5′-triphosphate
- ATPγS
Adenosine 5′-(γ-thio)triphosphate
- BeFx
Beryllium fluoride
- E
Kinesin
- FRET
Förster resonance energy transfer
- mant-ATP
2'-(or-3')-O-(N-Methylanthraniloyl) adenosine 5'-triphosphate
- MDCC-PBP
7-Diethylamino-3-((((2-maleimidyl)ethyl)amino)carbonyl) coumarin)-labeled phosphate binding protein
- MT
Microtubule
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SO4-
Sulfate
- VO4-
Vanadate
References
- Admiraal SJ, Herschlag D. Mapping the transition state for ATP hydrolysis: implications for enzymatic catalysis. Chem Biol. 1995;2:729–739. doi: 10.1016/1074-5521(95)90101-9. [DOI] [PubMed] [Google Scholar]
- Allingham JS, Sproul LR, Rayment I, Gilbert SP. Vik1 modulates microtubule-Kar3 interactions through a motor domain that lacks an active site. Cell. 2007;128:1161–1172. doi: 10.1016/j.cell.2006.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso MC, Drummond DR, Kain S, Hoeng J, Amos L, Cross RA. An ATP gate controls tubulin binding by the tethered head of kinesin-1. Science. 2007;316:120–123. doi: 10.1126/science.1136985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnal I, Metoz F, DeBonis S, Wade RH. Three-dimensional structure of functional motor proteins on microtubules. Curr Biol. 1996;6:1265–1270. doi: 10.1016/s0960-9822(02)70712-4. [DOI] [PubMed] [Google Scholar]
- Asenjo AB, Chatterjee C, Tan D, et al (2013) Structural model for tubulin recognition and deformation by kinesin-13 microtubule depolymerases. Cell Rep 3:759–768. doi:10.1016/j.celrep.2013.01.030 [DOI] [PubMed]
- Arora K, Talje L, Asenjo AB, et al. KIF14 binds tightly to microtubules and adopts a rigor-like conformation. J Mol Biol. 2014 doi: 10.1016/j.jmb.2014.05.030. [DOI] [PubMed] [Google Scholar]
- Asenjo AB, Weinberg Y, Sosa H. Nucleotide binding and hydrolysis induces a disorder-order transition in the kinesin neck-linker region. Nat Struct Mol Biol. 2006;13:648–654. doi: 10.1038/nsmb1109. [DOI] [PubMed] [Google Scholar]
- Auerbach SD, Johnson KA. Alternating site ATPase pathway of rat conventional kinesin. J Biol Chem. 2005;280:37048–37060. doi: 10.1074/jbc.M502984200. [DOI] [PubMed] [Google Scholar]
- Auerbach SD, Johnson KA. Kinetic effects of kinesin switch I and switch II mutations. J Biol Chem. 2005;280:37061–37068. doi: 10.1074/jbc.M502985200. [DOI] [PubMed] [Google Scholar]
- Barshop BA, Wrenn RF, Frieden C. Analysis of numerical methods for computer simulation of kinetic processes: development of KINSIM–a flexible, portable system. Anal Biochem. 1983;130:134–145. doi: 10.1016/0003-2697(83)90660-7. [DOI] [PubMed] [Google Scholar]
- Behnke-Parks WM, Vendome J, Honig B, Maliga Z, Moores C, Rosenfeld SS. Loop L5 acts as a conformational latch in the mitotic kinesin Eg5. J Biol Chem. 2011;286:5242–5253. doi: 10.1074/jbc.M110.192930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein DA, Zittel MC, Keck JL. High-resolution structure of the E.coli RecQ helicase catalytic core. EMBO J. 2003;22:4910–4921. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block SM. Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys J. 2007;92:2986–2995. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block SM, Asbury CL, Shaevitz JW, Lang MJ. Probing the kinesin reaction cycle with a 2D optical force clamp. Proc Natl Acad Sci USA. 2003;100:2351–2356. doi: 10.1073/pnas.0436709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodey AJ, Kikkawa M, Moores CA. 9-Angstrom structure of a microtubule-bound mitotic motor. J Mol Biol. 2009;388:218–224. doi: 10.1016/j.jmb.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Brady ST. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature. 1985;317:73–75. doi: 10.1038/317073a0. [DOI] [PubMed] [Google Scholar]
- Brendza KM, Rose DJ, Gilbert SP, Saxton WM. Lethal kinesin mutations reveal amino acids important for ATPase activation and structural coupling. J Biol Chem. 1999;274:31506–31514. doi: 10.1074/jbc.274.44.31506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza KM, Sontag CA, Saxton WM, Gilbert SP. A kinesin mutation that uncouples motor domains and desensitizes the gamma-phosphate sensor. J Biol Chem. 2000;275:22187–22195. doi: 10.1074/jbc.M001124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brier S, Lemaire D, Debonis S, Forest E, Kozielski F. Identification of the protein binding region of S-trityl-L-cysteine, a new potent inhibitor of the mitotic kinesin Eg5. Biochemistry. 2004;43:13072–13082. doi: 10.1021/bi049264e. [DOI] [PubMed] [Google Scholar]
- Brune M, Hunter JL, Corrie JE, Webb MR. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry. 1994;33:8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- Brunnbauer M, Dombi R, Ho TH, Schliwa M, Rief M, Okten Z. Torque generation of kinesin motors is governed by the stability of the neck domain. Mol Cell. 2012;46:147–158. doi: 10.1016/j.molcel.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Brunnbauer M, Mueller-Planitz F, Kosem S, et al. Regulation of a heterodimeric kinesin-2 through an unprocessive motor domain that is turned processive by its partner. Proc Natl Acad Sci USA. 2010;107:10460–10465. doi: 10.1073/pnas.1005177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunton CA, Chaimovi H. Acid-catalyzed hydrolysis of pyrophosphoric acid. Inorgan Chem. 1965;4:1763–1766. [Google Scholar]
- Carter NJ, Cross RA. Mechanics of the kinesin step. Nature. 2005;435:308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- Case RB, Pierce DW, Hom-Booher N, Hart CL, Vale RD. The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell. 1997;90:959–966. doi: 10.1016/s0092-8674(00)80360-8. [DOI] [PubMed] [Google Scholar]
- Case RB, Rice S, Hart CL, Ly B, Vale RD. Role of the kinesin neck linker and catalytic core in microtubule-based motility. Curr Biol. 2000;10:157–160. doi: 10.1016/s0960-9822(00)00316-x. [DOI] [PubMed] [Google Scholar]
- Catarinella M, Gruner T, Strittmatter T, Marx A, Mayer TU. BTB-1: a small molecule inhibitor of the mitotic motor protein Kif18A. Angew Chem Int Ed Engl. 2009;48:9072–9076. doi: 10.1002/anie.200904510. [DOI] [PubMed] [Google Scholar]
- Cecchini M, Houdusse A, Karplus M. Allosteric communication in myosin V: from small conformational changes to large directed movements. PLoS Comput Biol. 2008;4:e1000129. doi: 10.1371/journal.pcbi.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra R, Salmon ED, Erickson HP, Lockhart A, Endow SA. Structural and functional domains of the Drosophila ncd microtubule motor protein. J Biol Chem. 1993;268:9005–9013. [PubMed] [Google Scholar]
- Chang Q, Nitta R, Inoue S, Hirokawa N. Structural basis for the ATP-induced isomerization of kinesin. J Mol Biol. 2013;425:1869–1880. doi: 10.1016/j.jmb.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Chen CJ, Porche K, Rayment I, Gilbert SP. The ATPase pathway that drives the kinesin-14 Kar3Vik1 powerstroke. J Biol Chem. 2012;287:36673–36682. doi: 10.1074/jbc.M112.395590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Rayment I, Gilbert SP. Kinesin Kar3Cik1 ATPase pathway for microtubule cross-linking. J Biol Chem. 2011;286:29261–29272. doi: 10.1074/jbc.M111.255554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy BE, Behnke-Parks WM, Andreasson JO, Rosenfeld SS, Block SM. A universal pathway for kinesin stepping. Nat Struct Mol Biol. 2011;18:1020–1027. doi: 10.1038/nsmb.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JC, Gatial JE, 3rd, Kapoor TM, Gilbert SP. Monastrol inhibition of the mitotic kinesin Eg5. J Biol Chem. 2005;280:12658–12667. doi: 10.1074/jbc.M413140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JC, Gilbert SP. ATPase mechanism of Eg5 in the absence of microtubules: insight into microtubule activation and allosteric inhibition by monastrol. Biochemistry. 2005;44:16633–16648. doi: 10.1021/bi051724w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JC, Krzysiak TC, Gilbert SP. Pathway of ATP hydrolysis by monomeric kinesin Eg5. Biochemistry. 2006;45:12334–12344. doi: 10.1021/bi0608562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JC, Sindelar CV, Mulko NK, Collins KA, Kong SE, Hawley RS, Kull FJ. ATPase cycle of the nonmotile kinesin NOD allows microtubule end tracking and drives chromosome movement. Cell. 2009;136:110–122. doi: 10.1016/j.cell.2008.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JC, Sontag CA, Maliga Z, Kapoor TM, Correia JJ, Gilbert SP. Mechanistic analysis of the mitotic kinesin Eg5. J Biol Chem. 2004;279:38861–38870. doi: 10.1074/jbc.M404203200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JC, Zhao YC, Wilcox DE, Kull FJ. A metal switch for controlling the activity of molecular motor proteins. Nat Struct Mol Biol. 2011;19:122–127. doi: 10.1038/nsmb.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coureux PD, Sweeney HL, Houdusse A. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J. 2004;23:4527–4537. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coureux PD, Wells AL, Menetrey J, Yengo CM, Morris CA, Sweeney HL, Houdusse A. A structural state of the myosin V motor without bound nucleotide. Nature. 2003;425:419–423. doi: 10.1038/nature01927. [DOI] [PubMed] [Google Scholar]
- Coy DL, Wagenbach M, Howard J. Kinesin takes one 8-nm step for each ATP that it hydrolyzes. J Biol Chem. 1999;274:3667–3671. doi: 10.1074/jbc.274.6.3667. [DOI] [PubMed] [Google Scholar]
- Crevel IM, Nyitrai M, Alonso MC, Weiss S, Geeves MA, Cross RA. What kinesin does at roadblocks: the coordination mechanism for molecular walking. EMBO J. 2004;23:23–32. doi: 10.1038/sj.emboj.7600042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagenbach EM, Endow SA. A new kinesin tree. J Cell Sci. 2004;117:3–7. doi: 10.1242/jcs.00875. [DOI] [PubMed] [Google Scholar]
- Datta S, Krishna R, Ganesh N, Chandra NR, Muniyappa K, Vijayan M. Crystal structures of Mycobacterium smegmatis RecA and its nucleotide complexes. J Bacteriol. 2003;185:4280–4284. doi: 10.1128/JB.185.14.4280-4284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz EM, Ostap EM. Kinetic and equilibrium analysis of the myosin ATPase. Methods Enzymol. 2009;455:157–192. doi: 10.1016/S0076-6879(08)04206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBonis S, Simorre JP, Crevel I, et al. Interaction of the mitotic inhibitor monastrol with human kinesin Eg5. Biochemistry. 2003;42:338–349. doi: 10.1021/bi026716j. [DOI] [PubMed] [Google Scholar]
- Delorme C, Joshi M, Allingham JS. Crystal structure of the Candida albicans Kar3 kinesin motor domain fused to maltose-binding protein. Biochem Biophys Res Commun. 2012;428:427–432. doi: 10.1016/j.bbrc.2012.10.101. [DOI] [PubMed] [Google Scholar]
- Devred F, Barbier P, Lafitte D, Landrieu I, Lippens G, Peyrot V. Microtubule and MAPs: thermodynamics of complex formation by AUC, ITC, fluorescence, and NMR. Methods Cell Biol. 2010;95:449–480. doi: 10.1016/S0091-679X(10)95023-1. [DOI] [PubMed] [Google Scholar]
- Dittrich M, Hayashi S, Schulten K. On the mechanism of ATP hydrolysis in F1-ATPase. Biophys J. 2003;85:2253–2266. doi: 10.1016/s0006-3495(03)74650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich M, Hayashi S, Schulten K. ATP hydrolysis in the betaTP and betaDP catalytic sites of F1-ATPase. Biophys J. 2004;87:2954–2967. doi: 10.1529/biophysj.104.046128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D, Hnatchuk DJ, Brenner J, Davis D, Allingham JS. Crystal structure of the Kar3-like kinesin motor domain from the filamentous fungus Ashbya gossypii. Proteins. 2012;80:1016–1027. doi: 10.1002/prot.24004. [DOI] [PubMed] [Google Scholar]
- Duan D, Jia Z, Joshi M, et al. Neck rotation and neck mimic docking in the noncatalytic Kar3-associated protein Vik1. J Biol Chem. 2012;287:40292–40301. doi: 10.1074/jbc.M112.416529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duselder A, Thiede C, Schmidt CF, Lakamper S. Neck-linker length dependence of processive Kinesin-5 motility. J Mol Biol. 2012;423:159–168. doi: 10.1016/j.jmb.2012.06.043. [DOI] [PubMed] [Google Scholar]
- Ebbing B, Mann K, Starosta A, Jaud J, Schols L, Schule R, Woehlke G. Effect of spastic paraplegia mutations in KIF5A kinesin on transport activity. Hum Mol Genet. 2008;17:1245–1252. doi: 10.1093/hmg/ddn014. [DOI] [PubMed] [Google Scholar]
- Edamatsu M. Bidirectional motility of the fission yeast kinesin-5, Cut7. Biochem Biophys Res Commun. 2014;446:231–234. doi: 10.1016/j.bbrc.2014.02.106. [DOI] [PubMed] [Google Scholar]
- Endow SA, Higuchi H. A mutant of the motor protein kinesin that moves in both directions on microtubules. Nature. 2000;406:913–916. doi: 10.1038/35022617. [DOI] [PubMed] [Google Scholar]
- Endow SA, Waligora KW. Determinants of kinesin motor polarity. Science. 1998;281:1200–1202. doi: 10.1126/science.281.5380.1200. [DOI] [PubMed] [Google Scholar]
- Endres NF, Yoshioka C, Milligan RA, Vale RD. A lever-arm rotation drives motility of the minus-end-directed kinesin Ncd. Nature. 2006;439:875–878. doi: 10.1038/nature04320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell CM, Mackey AT, Klumpp LM, Gilbert SP. The role of ATP hydrolysis for kinesin processivity. J Biol Chem. 2002;277:17079–17087. doi: 10.1074/jbc.M108793200. [DOI] [PubMed] [Google Scholar]
- Finlayson B, Lymn RW, Taylor EW. Studies on the kinetics of formation and dissociation of the actomyosin complex. Biochemistry. 1969;8:811–819. doi: 10.1021/bi00831a008. [DOI] [PubMed] [Google Scholar]
- Finlayson B, Taylor EW. Hydrolysis of nucleoside triphosphates by myosin during the transient state. Biochemistry. 1969;8:802–810. doi: 10.1021/bi00831a007. [DOI] [PubMed] [Google Scholar]
- Foster KA, Correia JJ, Gilbert SP. Equilibrium binding studies of non-claret disjunctional protein (Ncd) reveal cooperative interactions between the motor domains. J Biol Chem. 1998;273:35307–35318. doi: 10.1074/jbc.273.52.35307. [DOI] [PubMed] [Google Scholar]
- Foster KA, Gilbert SP. Kinetic studies of dimeric Ncd: evidence that Ncd is not processive. Biochemistry. 2000;39:1784–1791. doi: 10.1021/bi991500b. [DOI] [PubMed] [Google Scholar]
- Foster KA, Mackey AT, Gilbert SP. A mechanistic model for Ncd directionality. J Biol Chem. 2001;276:19259–19266. doi: 10.1074/jbc.M008347200. [DOI] [PubMed] [Google Scholar]
- Fridman V, Gerson-Gurwitz A, Shapira O, Movshovich N, Lakamper S, Schmidt CF, Gheber L. Kinesin-5 Kip1 is a bi-directional motor that stabilizes microtubules and tracks their plus-ends in vivo. J Cell Sci. 2013 doi: 10.1242/jcs.125153. [DOI] [PubMed] [Google Scholar]
- Friel CT, Howard J. The kinesin-13 MCAK has an unconventional ATPase cycle adapted for microtubule depolymerization. EMBO J. 2011;30:3928–3939. doi: 10.1038/emboj.2011.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuger P, Sreekumar V, Schule R, et al. Spastic paraplegia mutation N256S in the neuronal microtubule motor KIF5A disrupts axonal transport in a Drosophila HSP model. PLoS Genet. 2012;8:e1003066. doi: 10.1371/journal.pgen.1003066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta K, Furuta A, Toyoshima YY, Amino M, Oiwa K, Kojima H. Measuring collective transport by defined numbers of processive and nonprocessive kinesin motors. Proc Natl Acad Sci USA. 2013;110:501–506. doi: 10.1073/pnas.1201390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Saez I, Yen T, Wade RH, Kozielski F. Crystal structure of the motor domain of the human kinetochore protein CENP-E. J Mol Biol. 2004;340:1107–1116. doi: 10.1016/j.jmb.2004.05.053. [DOI] [PubMed] [Google Scholar]
- Ge H, Qian H. Dissipation, generalized free energy, and a self-consistent nonequilibrium thermodynamics of chemically driven open subsystems. Phys Rev E Stat Nonlin Soft Matter Phys. 2013;87:062125. doi: 10.1103/PhysRevE.87.062125. [DOI] [PubMed] [Google Scholar]
- Geng YZ, Liu SX, Ji Q, Yan S. Mechanical amplification mechanism of kinesin's beta-domain. Arch Biochem Biophys. 2014;543:10–14. doi: 10.1016/j.abb.2013.12.017. [DOI] [PubMed] [Google Scholar]
- Gerson-Gurwitz A, Thiede C, Movshovich N, et al. Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. EMBO J. 2011;30:4942–4954. doi: 10.1038/emboj.2011.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigant B, Wang W, Dreier B, et al. Structure of a kinesin-tubulin complex and implications for kinesin motility. Nat Struct Mol Biol. 2013;20:1001–1007. doi: 10.1038/nsmb.2624. [DOI] [PubMed] [Google Scholar]
- Gilbert SP, Johnson KA. Expression, purification, and characterization of the Drosophila kinesin motor domain produced in Escherichia coli. Biochemistry. 1993;32:4677–4684. doi: 10.1021/bi00068a028. [DOI] [PubMed] [Google Scholar]
- Gilbert SP, Mackey AT. Kinetics: a tool to study molecular motors. Methods. 2000;22:337–354. doi: 10.1006/meth.2000.1086. [DOI] [PubMed] [Google Scholar]
- Gilbert SP, Moyer ML, Johnson KA. Alternating site mechanism of the kinesin ATPase. Biochemistry. 1998;37:792–799. doi: 10.1021/bi971117b. [DOI] [PubMed] [Google Scholar]
- Gilbert SP, Webb MR, Brune M, Johnson KA. Pathway of processive ATP hydrolysis by kinesin. Nature. 1995;373:671–676. doi: 10.1038/373671a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goody RS, Pai EF, Schlichting I, Rensland H, Scheidig A, Franken S, Wittinghofer A. Studies on the structure and mechanism of H-ras p21. Philos Trans R Soc Lond B Biol Sci. 1992;336:3–10. doi: 10.1098/rstb.1992.0037. [DOI] [PubMed] [Google Scholar]
- Goulet A, Behnke-Parks WM, Sindelar CV, Major J, Rosenfeld SS, Moores CA. The structural basis of force generation by the mitotic motor Kinesin-5. J Biol Chem. 2012;287:44654–44666. doi: 10.1074/jbc.M112.404228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorenko BL, Kaliman IA, Nemukhin AV. Minimum energy reaction profiles for ATP hydrolysis in myosin. J Mol Graph Model. 2011;31:1–4. doi: 10.1016/j.jmgm.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Grigorenko BL, Rogov AV, Topol IA, Burt SK, Martinez HM, Nemukhin AV. Mechanism of the myosin catalyzed hydrolysis of ATP as rationalized by molecular modeling. Proc Natl Acad Sci U S A. 2007;104:7057–7061. doi: 10.1073/pnas.0701727104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissom PM, Fiedler T, Grishchuk EL, Nicastro D, West RR, McIntosh JR. Kinesin-8 from fission yeast: a heterodimeric, plus-end-directed motor that can couple microtubule depolymerization to cargo movement. Mol Biol Cell. 2009;20:963–972. doi: 10.1091/mbc.E08-09-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick AM, Bauer CB, Thoden JB, Rayment I. X-ray structures of the MgADP, MgATPgammaS, and MgAMPPNP complexes of the Dictyostelium discoideum myosin motor domain. Biochemistry. 1997;36:11619–11628. doi: 10.1021/bi9712596. [DOI] [PubMed] [Google Scholar]
- Gulick AM, Song H, Endow SA, Rayment I. X-ray crystal structure of the yeast Kar3 motor domain complexed with Mg.ADP to 2.3 A resolution. Biochemistry. 1998;37:1769–1776. doi: 10.1021/bi972504o. [DOI] [PubMed] [Google Scholar]
- Guydosh NR, Block SM. Backsteps induced by nucleotide analogs suggest the front head of kinesin is gated by strain. Proc Natl Acad Sci USA. 2006;103:8054–8059. doi: 10.1073/pnas.0600931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD. Kinesin ATPase: rate-limiting ADP release. Proc Natl Acad Sci USA. 1988;85:6314–6318. doi: 10.1073/pnas.85.17.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD. Highly processive microtubule-stimulated ATP hydrolysis by dimeric kinesin head domains. Nature. 1995;377:448–450. doi: 10.1038/377448a0. [DOI] [PubMed] [Google Scholar]
- Hackney DD. Implications of diffusion-controlled limit for processivity of dimeric kinesin head domains. Biophys J. 1995;68:267S–269S. [PMC free article] [PubMed] [Google Scholar]
- Hackney DD. The tethered motor domain of a kinesin-microtubule complex catalyzes reversible synthesis of bound ATP. Proc Natl Acad Sci USA. 2005;102:18338–18343. doi: 10.1073/pnas.0505288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD. Jump-starting kinesin. J Cell Biol. 2007;176:7–9. doi: 10.1083/jcb.200611082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD, Stock MF. Kinesin tail domains and Mg2+ directly inhibit release of ADP from head domains in the absence of microtubules. Biochemistry. 2008;47:7770–7778. doi: 10.1021/bi8006687. [DOI] [PubMed] [Google Scholar]
- Halbach F, Rode M, Conti E. The crystal structure of S. cerevisiae Ski2, a DExH helicase associated with the cytoplasmic functions of the exosome. RNA. 2012;18:124–134. doi: 10.1261/rna.029553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkides CJ, Frey PA. The mechanism of the hydrolysis of mu-monothiopyrophosphate. J Am Chem Soc. 1991;113:9843–9848. [Google Scholar]
- Hancock WO, Howard J. Kinesin's processivity results from mechanical and chemical coordination between the ATP hydrolysis cycles of the two motor domains. Proc Natl Acad Sci USA. 1999;96:13147–13152. doi: 10.1073/pnas.96.23.13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan V, Hancock WO. Insights into the mechanical properties of the kinesin neck linker domain from sequence analysis and molecular dynamics simulations. Cell Mol Bioeng. 2009;2:177–189. doi: 10.1007/s12195-009-0059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Ueno H, Shaikh AR, et al. Molecular mechanism of ATP hydrolysis in F1-ATPase revealed by molecular simulations and single-molecule observations. J Am Chem Soc. 2012;134:8447–8454. doi: 10.1021/ja211027m. [DOI] [PubMed] [Google Scholar]
- Henningsen U, Schliwa M. Reversal in the direction of movement of a molecular motor. Nature. 1997;389:93–96. doi: 10.1038/38022. [DOI] [PubMed] [Google Scholar]
- Henry ER, Eaton WA, Hochstrasser RM. Molecular dynamics simulations of cooling in laser-excited heme proteins. Proc Natl Acad Sci U S A. 1986;83:8982–8986. doi: 10.1073/pnas.83.23.8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzer KM, Ems-McClung SC, Kline-Smith SL, Lipkin TG, Gilbert SP, Walczak CE. Full-length dimeric MCAK is a more efficient microtubule depolymerase than minimal domain monomeric MCAK. Mol Biol Cell. 2006;17:700–710. doi: 10.1091/mbc.E05-08-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuston E, Bronner CE, Kull FJ, Endow SA. A kinesin motor in a force-producing conformation. BMC Struct Biol. 2010;10:19. doi: 10.1186/1472-6807-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi H, Bronner CE, Park HW, Endow SA. Rapid double 8-nm steps by a kinesin mutant. EMBO J. 2004;23:2993–2999. doi: 10.1038/sj.emboj.7600306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R. Kinesin superfamily proteins and their various functions and dynamics. Exp Cell Res. 2004;301:50–59. doi: 10.1016/j.yexcr.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Hirose K, Lockhart A, Cross RA, Amos LA. Three-dimensional cryoelectron microscopy of dimeric kinesin and ncd motor domains on microtubules. Proc Natl Acad Sci U S A. 1996;93:9539–9544. doi: 10.1073/pnas.93.18.9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeng JC, Dawson SC, House SA, et al (2008) High-resolution crystal structure and in vivo function of a kinesin-2 homologue in Giardia intestinalis. Mol Biol Cell 19:3124–3137. doi:10.1091/mbc.E07-11-1156 [DOI] [PMC free article] [PubMed]
- Howard J, Hudspeth AJ, Vale RD. Movement of microtubules by single kinesin molecules. Nature. 1989;342:154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- Hua W, Young EC, Fleming ML, Gelles J. Coupling of kinesin steps to ATP hydrolysis. Nature. 1997;388:390–393. doi: 10.1038/41118. [DOI] [PubMed] [Google Scholar]
- Hwang W, Lang MJ. Mechanical design of translocating motor proteins. Cell Biochem Biophys. 2009;54:11–22. doi: 10.1007/s12013-009-9049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W, Lang MJ, Karplus M. Force generation in kinesin hinges on cover-neck bundle formation. Structure. 2008;16:62–71. doi: 10.1016/j.str.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Hyeon C, Onuchic JN. Internal strain regulates the nucleotide binding site of the kinesin leading head. Proc Natl Acad Sci USA. 2007;104:2175–2180. doi: 10.1073/pnas.0610939104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani S, Iwane AH, Higuchi H, Ishii Y, Yanagida T. Mechanical and chemical properties of cysteine-modified kinesin molecules. Biochemistry. 1999;38:10318–10323. doi: 10.1021/bi9904095. [DOI] [PubMed] [Google Scholar]
- Jannasch A, Bormuth V, Storch M, Howard J, Schaffer E. Kinesin-8 is a low-force motor protein with a weakly bound slip state. Biophys J. 2013;104:2456–2464. doi: 10.1016/j.bpj.2013.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaud J, Bathe F, Schliwa M, Rief M, Woehlke G. Flexibility of the neck domain enhances Kinesin-1 motility under load. Biophys J. 2006;91:1407–1412. doi: 10.1529/biophysj.105.076265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Stock MF, Li X, Hackney DD. Influence of the kinesin neck domain on dimerization and ATPase kinetics. J Biol Chem. 1997;272:7626–7632. doi: 10.1074/jbc.272.12.7626. [DOI] [PubMed] [Google Scholar]
- Johnson KA. Transient-state kinetic analysis of enzyme reaction pathways. In: The Enzymes, vol 20. New York: Academic Press; 1992. pp. 1–61. [Google Scholar]
- Johnson KA, Simpson ZB, Blom T. Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal Biochem. 2009;387:20–29. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- Kaan HY, Hackney DD, Kozielski F. The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science. 2011;333:883–885. doi: 10.1126/science.1204824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaan HY, Ulaganathan V, Rath O, Prokopcova H, Dallinger D, Kappe CO, Kozielski F. Structural basis for inhibition of Eg5 by dihydropyrimidines: stereoselectivity of antimitotic inhibitors enastron, dimethylenastron and fluorastrol. J Med Chem. 2010;53:5676–5683. doi: 10.1021/jm100421n. [DOI] [PubMed] [Google Scholar]
- Karabay A, Walker RA. Identification of microtubule binding sites in the Ncd tail domain. Biochemistry. 1999;38:1838–1849. doi: 10.1021/bi981850i. [DOI] [PubMed] [Google Scholar]
- Kaseda K, Higuchi H, Hirose K. Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat Cell Biol. 2003;5:1079–1082. doi: 10.1038/ncb1067. [DOI] [PubMed] [Google Scholar]
- Khalil AS, Appleyard DC, Labno AK, et al. Kinesin's cover-neck bundle folds forward to generate force. Proc Natl Acad Sci U S A. 2008;105:19247–19252. doi: 10.1073/pnas.0805147105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SA, Kirby AJ (1970) Reactivity of phosphate esters - multiple structure reactivity correlations for reactions of triesters with nucleophiles. J Chem Soc B:1172–1182, doi:10.1039/j29700001172
- Kikkawa M, Hirokawa N. High-resolution cryo-EM maps show the nucleotide binding pocket of KIF1A in open and closed conformations. EMBO J. 2006;25:4187–4194. doi: 10.1038/sj.emboj.7601299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa M, Sablin EP, Okada Y, Yajima H, Fletterick RJ, Hirokawa N. Switch-based mechanism of kinesin motors. Nature. 2001;411:439–445. doi: 10.1038/35078000. [DOI] [PubMed] [Google Scholar]
- Kim Y, Heuser JE, Waterman CM, Cleveland DW. CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J Cell Biol. 2008;181:411–419. doi: 10.1083/jcb.200802189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klejnot M, Falnikar A, Ulaganathan V, Cross RA, Baas PW, Kozielski F. The crystal structure and biochemical characterization of Kif15: a bifunctional molecular motor involved in bipolar spindle formation and neuronal development. Acta Crystallogr D Biol Crystallogr. 2014;70:123–133. doi: 10.1107/S1399004713028721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klejnot M, Kozielski F. Structural insights into human Kif7, a kinesin involved in Hedgehog signalling. Acta Crystallogr D Biol Crystallogr. 2012;68:154–159. doi: 10.1107/S0907444911053042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp LM, Brendza KM, Gatial JE, 3rd, Hoenger A, Saxton WM, Gilbert SP. Microtubule-kinesin interface mutants reveal a site critical for communication. Biochemistry. 2004;43:2792–2803. doi: 10.1021/bi035830e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp LM, Hoenger A, Gilbert SP. Kinesin's second step. Proc Natl Acad Sci U S A. 2004;101:3444–3449. doi: 10.1073/pnas.0307691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp LM, Mackey AT, Farrell CM, Rosenberg JM, Gilbert SP. A kinesin switch I arginine to lysine mutation rescues microtubule function. J Biol Chem. 2003;278:39059–39067. doi: 10.1074/jbc.M304250200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocik E, Skowronek KJ, Kasprzak AA. Interactions between subunits in heterodimeric Ncd molecules. J Biol Chem. 2009;284:35735–35745. doi: 10.1074/jbc.M109.024240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozielski F, Arnal I, Wade RH. A model of the microtubule-kinesin complex based on electron cryomicroscopy and X-ray crystallography. Curr Biol. 1998;8:191–198. doi: 10.1016/s0960-9822(98)70083-1. [DOI] [PubMed] [Google Scholar]
- Kozielski F, De Bonis S, Burmeister WP, Cohen-Addad C, Wade RH. The crystal structure of the minus-end-directed microtubule motor protein ncd reveals variable dimer conformations. Structure. 1999;7:1407–1416. doi: 10.1016/s0969-2126(00)80030-1. [DOI] [PubMed] [Google Scholar]
- Kozielski F, Sack S, Marx A, et al. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 1997;91:985–994. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- Krzysiak TC, Gilbert SP. Dimeric Eg5 maintains processivity through alternating-site catalysis with rate-limiting ATP hydrolysis. J Biol Chem. 2006;281:39444–39454. doi: 10.1074/jbc.M608056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzysiak TC, Grabe M, Gilbert SP. Getting in sync with dimeric Eg5. Initiation and regulation of the processive run. J Biol Chem. 2008;283:2078–2087. doi: 10.1074/jbc.M708354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzysiak TC, Wendt T, Sproul LR, Tittmann P, Gross H, Gilbert SP, Hoenger A. A structural model for monastrol inhibition of dimeric kinesin Eg5. EMBO J. 2006;25:2263–2273. doi: 10.1038/sj.emboj.7601108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kull FJ. Motor proteins of the kinesin superfamily: structure and mechanism. Essays Biochem. 2000;35:61–73. doi: 10.1042/bse0350061. [DOI] [PubMed] [Google Scholar]
- Kull FJ, Endow SA. Kinesin: switch I & II and the motor mechanism. J Cell Sci. 2002;115:15–23. doi: 10.1242/jcs.115.1.15. [DOI] [PubMed] [Google Scholar]
- Kull FJ, Endow SA. Force generation by kinesin and myosin cytoskeletal motor proteins. J Cell Sci. 2013;126:9–19. doi: 10.1242/jcs.103911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996;380:550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kull FJ, Vale RD, Fletterick RJ. The case for a common ancestor: kinesin and myosin motor proteins and G proteins. J Muscle Res Cell Motil. 1998;19:877–886. doi: 10.1023/a:1005489907021. [DOI] [PubMed] [Google Scholar]
- Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightcap ES, Frey PA. Discrete Monomeric Metaphosphate Anion as an Intermediate in the Hydrolysis of Mu-Monothiopyrophosphate. J Am Chem Soc. 1992;114:9750–9755. [Google Scholar]
- Liu HL, Hallen MA, Endow SA. Altered nucleotide-microtubule coupling and increased mechanical output by a kinesin mutant. PLoS One. 2012;7:e47148. doi: 10.1371/journal.pone.0047148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HL, Pemble Iv CW, Endow SA. Neck-motor interactions trigger rotation of the kinesin stalk. Sci Rep. 2012;2:236. doi: 10.1038/srep00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Xu T, Watson RP, et al. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008;27:3209–3219. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YZ, Taylor EW. Interacting head mechanism of microtubule-kinesin ATPase. J Biol Chem. 1997;272:724–730. doi: 10.1074/jbc.272.2.724. [DOI] [PubMed] [Google Scholar]
- Mackey AT, Gilbert SP. Moving a microtubule may require two heads: a kinetic investigation of monomeric Ncd. Biochemistry. 2000;39:1346–1355. doi: 10.1021/bi991918+. [DOI] [PubMed] [Google Scholar]
- Mackey AT, Gilbert SP. The ATPase cross-bridge cycle of the Kar3 motor domain. Implications for single head motility. J Biol Chem. 2003;278:3527–3535. doi: 10.1074/jbc.M206219200. [DOI] [PubMed] [Google Scholar]
- Maliga Z, Kapoor TM, Mitchison TJ. Evidence that monastrol is an allosteric inhibitor of the mitotic kinesin Eg5. Chem Biol. 2002;9:989–996. doi: 10.1016/s1074-5521(02)00212-0. [DOI] [PubMed] [Google Scholar]
- Maliga Z, Xing J, Cheung H, Juszczak LJ, Friedman JM, Rosenfeld SS. A pathway of structural changes produced by monastrol binding to Eg5. J Biol Chem. 2006;281:7977–7982. doi: 10.1074/jbc.M511955200. [DOI] [PubMed] [Google Scholar]
- Marx A, Hoenger A, Mandelkow E. Structures of kinesin motor proteins. Cell Motil Cytoskeleton. 2009;66:958–966. doi: 10.1002/cm.20392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx A, Muller J, Mandelkow E. The structure of microtubule motor proteins. Adv Protein Chem. 2005;71:299–344. doi: 10.1016/S0065-3233(04)71008-6. [DOI] [PubMed] [Google Scholar]
- Mayr MI, Storch M, Howard J, Mayer TU. A non-motor microtubule binding site is essential for the high processivity and mitotic function of kinesin-8 Kif18A. PLoS One. 2011;6:e27471. doi: 10.1371/journal.pone.0027471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar M, Cross RA. Engineering a lever into the kinesin neck. J Biol Chem. 1998;273:29352–29359. doi: 10.1074/jbc.273.45.29352. [DOI] [PubMed] [Google Scholar]
- McGrath MJ, Kuo IF, Hayashi S, Takada S. Adenosine triphosphate hydrolysis mechanism in kinesin studied by combined quantum-mechanical/molecular-mechanical metadynamics simulations. J Am Chem Soc. 2013;135:8908–8919. doi: 10.1021/ja401540g. [DOI] [PubMed] [Google Scholar]
- Menetrey J, Llinas P, Cicolari J, et al. The post-rigor structure of myosin VI and implications for the recovery stroke. EMBO J. 2008;27:244–252. doi: 10.1038/sj.emboj.7601937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis L, Menten ML. Die kinetik der invertinwirkung. Biochem Z. 1913;49:333–369. [Google Scholar]
- Michaelis L, Menten ML, Johnson KA, Goody RS. The original Michaelis constant: translation of the 1913 Michaelis-Menten paper. Biochemistry. 2011;50:8264–8269. doi: 10.1021/bi201284u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Miller DL, Ukena T. P1, P1-Diethyl Pyrophosphate. J Am Chem Soc. 1969;91:3050–3053. [Google Scholar]
- Miller DL, Westheim FH. Enzymic Hydrolysis of Gamma-Phenylpropyl Di- and Triphosphates. J Am Chem Soc. 1966;88:1511–1513. doi: 10.1021/ja00959a035. [DOI] [PubMed] [Google Scholar]
- Miller DL, Westheim FH. Hydrolysis of Gamma-Phenylpropyl Di- and Triphosphates. J Am Chem Soc. 1966;88:1507–1511. doi: 10.1021/ja00959a034. [DOI] [PubMed] [Google Scholar]
- Miyazono Y, Hayashi M, Karagiannis P, Harada Y, Tadakuma H. Strain through the neck linker ensures processive runs: a DNA-kinesin hybrid nanomachine study. EMBO J. 2010;29:93–106. doi: 10.1038/emboj.2009.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JD, Song H, Endow SA. A point mutation in the microtubule binding region of the Ncd motor protein reduces motor velocity. EMBO J. 1996;15:3306–3314. [PMC free article] [PubMed] [Google Scholar]
- Mori T, Vale RD, Tomishige M. How kinesin waits between steps. Nature. 2007;450:750–754. doi: 10.1038/nature06346. [DOI] [PubMed] [Google Scholar]
- Morrison JF. Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors. Biochim Biophys Acta. 1969;185:269–286. doi: 10.1016/0005-2744(69)90420-3. [DOI] [PubMed] [Google Scholar]
- Moyer ML, Gilbert SP, Johnson KA. Pathway of ATP hydrolysis by monomeric and dimeric kinesin. Biochemistry. 1998;37:800–813. doi: 10.1021/bi9711184. [DOI] [PubMed] [Google Scholar]
- Muller J, Marx A, Sack S, Song YH, Mandelkow E. The structure of the nucleotide-binding site of kinesin. Biol Chem. 1999;380:981–992. doi: 10.1515/BC.1999.122. [DOI] [PubMed] [Google Scholar]
- Muthukrishnan G, Zhang Y, Shastry S, Hancock WO. The processivity of kinesin-2 motors suggests diminished front-head gating. Curr Biol. 2009;19:442–447. doi: 10.1016/j.cub.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizaka T, Yagi T, Tanaka Y, Ishiwata S. Right-handed rotation of an actin filament in an in vitro motile system. Nature. 1993;361:269–271. doi: 10.1038/361269a0. [DOI] [PubMed] [Google Scholar]
- Nitta R, Kikkawa M, Okada Y, Hirokawa N. KIF1A alternately uses two loops to bind microtubules. Science. 2004;305:678–683. doi: 10.1126/science.1096621. [DOI] [PubMed] [Google Scholar]
- Nitta R, Okada Y, Hirokawa N. Structural model for strain-dependent microtubule activation of Mg-ADP release from kinesin. Nat Struct Mol Biol. 2008;15:1067–1075. doi: 10.1038/nsmb.1487. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Nitta R, Okada Y, Hirokawa N. A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops. Cell. 2004;116:591–602. doi: 10.1016/s0092-8674(04)00129-1. [DOI] [PubMed] [Google Scholar]
- Onishi H, Mochizuki N, Morales MF. On the myosin catalysis of ATP hydrolysis. Biochemistry. 2004;43:3757–3763. doi: 10.1021/bi040002m. [DOI] [PubMed] [Google Scholar]
- Pan X, Acar S, Scholey JM. Torque generation by one of the motor subunits of heterotrimeric kinesin-2. Biochem Biophys Res Commun. 2010;401:53–57. doi: 10.1016/j.bbrc.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parke CL, Wojcik EJ, Kim S, Worthylake DK. ATP hydrolysis in Eg5 kinesin involves a catalytic two-water mechanism. J Biol Chem. 2010;285:5859–5867. doi: 10.1074/jbc.M109.071233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechatnikova E, Taylor EW. Kinetic mechanism of monomeric non-claret disjunctional protein (Ncd) ATPase. J Biol Chem. 1997;272:30735–30740. doi: 10.1074/jbc.272.49.30735. [DOI] [PubMed] [Google Scholar]
- Pecqueur L, Duellberg C, Dreier B, et al. A designed ankyrin repeat protein selected to bind to tubulin caps the microtubule plus end. Proc Natl Acad Sci USA. 2012;109:12011–12016. doi: 10.1073/pnas.1204129109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C, Brejc K, Belmont L, et al. Insight into the molecular mechanism of the multitasking kinesin-8 motor. EMBO J. 2010;29:3437–3447. doi: 10.1038/emboj.2010.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Johnson KA. Transient state kinetic analysis of the ATP-induced dissociation of the dynein-microtubule complex. J Biol Chem. 1983;258:6582–6587. [PubMed] [Google Scholar]
- Rajan R, Bell CE. Crystal structure of RecA from Deinococcus radiodurans: insights into the structural basis of extreme radioresistance. J Mol Biol. 2004;344:951–963. doi: 10.1016/j.jmb.2004.09.087. [DOI] [PubMed] [Google Scholar]
- Rank KC, Chen CJ, Cope J, Porche K, Hoenger A, Gilbert SP, Rayment I. Kar3Vik1, a member of the Kinesin-14 superfamily, shows a novel kinesin microtubule binding pattern. J Cell Biol. 2012;197:957–970. doi: 10.1083/jcb.201201132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath O, Kozielski F. Kinesins and cancer. Nat Rev Cancer. 2012;12:527–539. doi: 10.1038/nrc3310. [DOI] [PubMed] [Google Scholar]
- Ray S, Meyhofer E, Milligan RA, Howard J. Kinesin follows the microtubule's protofilament axis. J Cell Biol. 1993;121:1083–1093. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Wolf SG, Howard J, Downing KH. Kinesin does not support the motility of zinc-macrotubes. Cell Motil Cytoskeleton. 1995;30:146–152. doi: 10.1002/cm.970300206. [DOI] [PubMed] [Google Scholar]
- Rayment I, Rypniewski WR, Schmidt-Base K, et al. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Reubold TF, Eschenburg S, Becker A, Kull FJ, Manstein DJ. A structural model for actin-induced nucleotide release in myosin. Nat Struct Biol. 2003;10:826–830. doi: 10.1038/nsb987. [DOI] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- Roostalu J, Hentrich C, Bieling P, Telley IA, Schiebel E, Surrey T. Directional switching of the kinesin Cin8 through motor coupling. Science. 2011;332:94–99. doi: 10.1126/science.1199945. [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, Fordyce PM, Jefferson GM, King PH, Block SM. Stepping and stretching. How kinesin uses internal strain to walk processively. J Biol Chem. 2003;278:18550–18556. doi: 10.1074/jbc.M300849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld SS, Jefferson GM, King PH. ATP reorients the neck linker of kinesin in two sequential steps. J Biol Chem. 2001;276:40167–40174. doi: 10.1074/jbc.M103899200. [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, van Duffelen M, Behnke-Parks WM, Beadle C, Corrreia J, Xing J. The ATPase cycle of the mitotic motor CENP-E. J Biol Chem. 2009;284:32858–32868. doi: 10.1074/jbc.M109.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld SS, Xing J, Jefferson GM, Cheung HC, King PH. Measuring kinesin's first step. J Biol Chem. 2002;277:36731–36739. doi: 10.1074/jbc.M205261200. [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, Xing J, Jefferson GM, King PH. Docking and rolling, a model of how the mitotic motor Eg5 works. J Biol Chem. 2005;280:35684–35695. doi: 10.1074/jbc.M506561200. [DOI] [PubMed] [Google Scholar]
- Sablin EP, Case RB, Dai SC, Hart CL, Ruby A, Vale RD, Fletterick RJ. Direction determination in the minus-end-directed kinesin motor ncd. Nature. 1998;395:813–816. doi: 10.1038/27463. [DOI] [PubMed] [Google Scholar]
- Sablin EP, Kull FJ, Cooke R, Vale RD, Fletterick RJ. Crystal structure of the motor domain of the kinesin-related motor ncd. Nature. 1996;380:555–559. doi: 10.1038/380555a0. [DOI] [PubMed] [Google Scholar]
- Sack S, Kull FJ, Mandelkow E. Motor proteins of the kinesin family. Structures, variations, and nucleotide binding sites. Eur J Biochem. 1999;262:1–11. doi: 10.1046/j.1432-1327.1999.00341.x. [DOI] [PubMed] [Google Scholar]
- Sack S, Muller J, Marx A, Thormahlen M, Mandelkow EM, Brady ST, Mandelkow E. X-ray structure of motor and neck domains from rat brain kinesin. Biochemistry. 1997;36:16155–16165. doi: 10.1021/bi9722498. [DOI] [PubMed] [Google Scholar]
- Sadhu A, Taylor EW. A kinetic study of the kinesin ATPase. J Biol Chem. 1992;267:11352–11359. [PubMed] [Google Scholar]
- Sardar HS, Gilbert SP. Microtubule capture by mitotic kinesin CENP-E. J Biol Chem. 2012;287(30):24894–904. doi: 10.1074/jbc.M112.376830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap IA, Carrasco C, de Pablo PJ, Schmidt CF. Kinesin walks the line: single motors observed by atomic force microscopy. Biophys J. 2011;100:2450–2456. doi: 10.1016/j.bpj.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schief WR, Clark RH, Crevenna AH, Howard J. Inhibition of kinesin motility by ADP and phosphate supports a hand-over-hand mechanism. Proc Natl Acad Sci USA. 2004;101:1183–1188. doi: 10.1073/pnas.0304369101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schief WR, Howard J. Conformational changes during kinesin motility. Curr Opin Cell Biol. 2001;13:19–28. doi: 10.1016/s0955-0674(00)00169-1. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Gleave ES, Carter AP. Insights into dynein motor domain function from a 3.3-A crystal structure. Nat Struct Mol Biol. 2012;19(492–497):S491. doi: 10.1038/nsmb.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzer MJ, Block SM. Kinesin hydrolyses one ATP per 8-nm step. Nature. 1997;388:386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- Schnitzer MJ, Visscher K, Block SM. Force production by single kinesin motors. Nat Cell Biol. 2000;2:718–723. doi: 10.1038/35036345. [DOI] [PubMed] [Google Scholar]
- Schutz P, Karlberg T, van den Berg S, et al. (2010) Comparative structural analysis of human DEAD-box RNA helicases. PLoS One 5. doi:10.1371/journal.pone.0012791 [DOI] [PMC free article] [PubMed]
- Schwarzl SM, Smith JC, Fischer S. Insights into the chemomechanical coupling of the myosin motor from simulation of its ATP hydrolysis mechanism. Biochemistry. 2006;45:5830–5847. doi: 10.1021/bi052433q. [DOI] [PubMed] [Google Scholar]
- Seidel SA, Dijkman PM, Lea WA, et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods. 2013;59:301–315. doi: 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamovsky IL, Ross GM, Riopelle RJ. Ab initio studies on the origin of beta-sheet twisting. Abstr Pap Am Chem Soc. 2000;219:U599–U599. [Google Scholar]
- Shamovsky IL, Ross GM, Riopelle RJ. Theoretical studies on the origin of beta-sheet twisting. J Phys Chem B. 2000;104:11296–11307. [Google Scholar]
- Shastry S, Hancock WO. Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Curr Biol. 2010;20:939–943. doi: 10.1016/j.cub.2010.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastry S, Hancock WO. Interhead tension determines processivity across diverse N-terminal kinesins. Proc Natl Acad Sci U S A. 2011;108:16253–16258. doi: 10.1073/pnas.1102628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikama K. Standard free energy maps for the hydrolysis of ATP as a function of pH, pMg and pCa. Arch Biochem Biophys. 1971;147:311–317. doi: 10.1016/0003-9861(71)90338-9. [DOI] [PubMed] [Google Scholar]
- Shipley K, Hekmat-Nejad M, Turner J, et al. Structure of a kinesin microtubule depolymerization machine. EMBO J. 2004;23:1422–1432. doi: 10.1038/sj.emboj.7600165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindelar CV, Budny MJ, Rice S, Naber N, Fletterick R, Cooke R. Two conformations in the human kinesin power stroke defined by X-ray crystallography and EPR spectroscopy. Nat Struct Biol. 2002;9:844–848. doi: 10.1038/nsb852. [DOI] [PubMed] [Google Scholar]
- Sindelar CV, Downing KH. The beginning of kinesin's force-generating cycle visualized at 9-A resolution. J Cell Biol. 2007;177:377–385. doi: 10.1083/jcb.200612090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiniotis G, Surrey T, Altmann S, Gross H, Song YH, Mandelkow E, Hoenger A. Nucleotide-induced conformations in the neck region of dimeric kinesin. EMBO J. 2003;22:1518–1528. doi: 10.1093/emboj/cdg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias DA, DeBonis S, Saoudi Y, et al (2006) S-trityl-L-cysteine is a reversible, tight binding inhibitor of the human kinesin Eg5 that specifically blocks mitotic progression. J Biol Chem 281:17559–17569. doi:10.1074/jbc.M511735200 [DOI] [PubMed]
- Smith CA, Rayment I. X-ray structure of the magnesium(II).ADP.vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 A resolution. Biochemistry. 1996;35:5404–5417. doi: 10.1021/bi952633+. [DOI] [PubMed] [Google Scholar]
- Song H, Endow SA. Decoupling of nucleotide- and microtubule-binding sites in a kinesin mutant. Nature. 1998;396:587–590. doi: 10.1038/25153. [DOI] [PubMed] [Google Scholar]
- Song YH, Marx A, Muller J, et al (2001) Structure of a fast kinesin: implications for ATPase mechanism and interactions with microtubules. EMBO J 20:6213–6225. doi:10.1093/emboj/20.22.6213 [DOI] [PMC free article] [PubMed]
- Soppina V, Norris SR, Dizaji AS, Kortus M, Veatch S, Peckham M, Verhey KJ. Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proc Natl Acad Sci USA. 2014;111:5562–5567. doi: 10.1073/pnas.1400759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppina V, Verhey KJ. The family-specific K-loop influences the microtubule on-rate but not the superprocessivity of kinesin-3 motors. Mol Biol Cell. 2014;25:2161–2170. doi: 10.1091/mbc.E14-01-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa H, Dias DP, Hoenger A, et al (1997) A model for the microtubule-Ncd motor protein complex obtained by cryo-electron microscopy and image analysis. Cell 90:217–224 [DOI] [PubMed]
- Sproul LR, Anderson DJ, Mackey AT, Saunders WS, Gilbert SP. Cik1 targets the minus-end kinesin depolymerase kar3 to microtubule plus ends. Curr Biol. 2005;15:1420–1427. doi: 10.1016/j.cub.2005.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA. Molecular motors take tension in stride. Cell. 2006;126:242–244. doi: 10.1016/j.cell.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Stinson BM, Nager AR, Glynn SE, Schmitz KR, Baker TA, Sauer RT. Nucleotide binding and conformational switching in the hexameric ring of a AAA+ machine. Cell. 2013;153:628–639. doi: 10.1016/j.cell.2013.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockbridge RB, Wolfenden R. The intrinsic reactivity of ATP and the catalytic proficiencies of kinases acting on glucose, N-acetylgalactosamine, and homoserine: a thermodynamic analysis. J Biol Chem. 2009;284:22747–22757. doi: 10.1074/jbc.M109.017806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Du Y, English CA, et al (2011) A tethering mechanism controls the processivity and kinetochore-microtubule plus-end enrichment of the kinesin-8 Kif18A. Mol Cell 43:764–775. doi:10.1016/j.molcel.2011.07.022 [DOI] [PMC free article] [PubMed]
- Su X, Qiu W, Gupta ML, Jr, Pereira-Leal JB, Reck-Peterson SL, Pellman D. Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol Cell. 2011;43:751–763. doi: 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugata K, Nakamura M, Ueki S, Fajer PG, Arata T. ESR reveals the mobility of the neck linker in dimeric kinesin. Biochem Biophys Res Commun. 2004;314:447–451. doi: 10.1016/j.bbrc.2003.12.093. [DOI] [PubMed] [Google Scholar]
- Sugata K, Song L, Nakamura M, Ueki S, Fajer PG, Arata T. Nucleotide-induced flexibility change in neck linkers of dimeric kinesin as detected by distance Measurements using spin-labeling EPR. J Mol Biol. 2009;386:626–636. doi: 10.1016/j.jmb.2008.12.079. [DOI] [PubMed] [Google Scholar]
- Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- Talapatra SK, Anthony NG, Mackay SP, Kozielski F. Mitotic kinesin Eg5 overcomes inhibition to the phase I/II clinical candidate SB743921 by an allosteric resistance mechanism. J Med Chem. 2013;56:6317–6329. doi: 10.1021/jm4006274. [DOI] [PubMed] [Google Scholar]
- Talapatra SK, Schuttelkopf AW, Kozielski F. The structure of the ternary Eg5-ADP-ispinesib complex. Acta Crystallogr D Biol Crystallogr. 2012;68:1311–1319. doi: 10.1107/S0907444912027965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D, Rice WJ, Sosa H. Structure of the kinesin13-microtubule ring complex. Structure. 2008;16:1732–1739. doi: 10.1016/j.str.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherniuk S, Skoufias DA, Labriere C, Rath O, Gueritte F, Guillou C, Kozielski F. Relocation of Aurora B and survivin from centromeres to the central spindle impaired by a kinesin-specific MKLP-2 inhibitor. Angew Chem Int Ed Engl. 2010;49:8228–8231. doi: 10.1002/anie.201003254. [DOI] [PubMed] [Google Scholar]
- Thatcher GRJ, Kluger R. Mechanism and catalysis of nucleophilic-substitution in phosphate-esters. Adv Phys Org Chem. 1989;25:99–265. [Google Scholar]
- Thiede C, Fridman V, Gerson-Gurwitz A, Gheber L, Schmidt CF. Regulation of bi-directional movement of single kinesin-5 Cin8 molecules. Bioarchitecture. 2012;2:70–74. doi: 10.4161/bioa.20395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoresen T, Gelles J. Processive movement by a kinesin heterodimer with an inactivating mutation in one head. Biochemistry. 2008;47:9514–9521. doi: 10.1021/bi800747e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn KS, Ubersax JA, Vale RD. Engineering the processive run length of the kinesin motor. J Cell Biol. 2000;151:1093–1100. doi: 10.1083/jcb.151.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomishige M, Klopfenstein DR, Vale RD. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 2002;297:2263–2267. doi: 10.1126/science.1073386. [DOI] [PubMed] [Google Scholar]
- Toprak E, Yildiz A, Hoffman MT, Rosenfeld SS, Selvin PR. Why kinesin is so processive. Proc Natl Acad Sci USA. 2009;106:12717–12722. doi: 10.1073/pnas.0808396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov PO, Barbier P, Breuzard G, Peyrot V, Devred F. Microtubule-associated proteins and tubulin interaction by isothermal titration calorimetry. Methods Cell Biol. 2013;115:283–302. doi: 10.1016/B978-0-12-407757-7.00018-9. [DOI] [PubMed] [Google Scholar]
- Turner J, Anderson R, Guo J, Beraud C, Fletterick R, Sakowicz R. Crystal structure of the mitotic spindle kinesin Eg5 reveals a novel conformation of the neck-linker. J Biol Chem. 2001;276:25496–25502. doi: 10.1074/jbc.M100395200. [DOI] [PubMed] [Google Scholar]
- Ulaganathan V, Talapatra SK, Rath O, Pannifer A, Hackney DD, Kozielski F. Structural insights into a unique inhibitor binding pocket in kinesin spindle protein. J Am Chem Soc. 2013;135:2263–2272. doi: 10.1021/ja310377d. [DOI] [PubMed] [Google Scholar]
- Vale RD, Fletterick RJ. The design plan of kinesin motors. Annu Rev Cell Dev Biol. 1997;13:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- Vale RD, Funatsu T, Pierce DW, Romberg L, Harada Y, Yanagida T. Direct observation of single kinesin molecules moving along microtubules. Nature. 1996;380:451–453. doi: 10.1038/380451a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Milligan RA. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Toyoshima YY. Rotation and translocation of microtubules in vitro induced by dyneins from Tetrahymena cilia. Cell. 1988;52:459–469. doi: 10.1016/s0092-8674(88)80038-2. [DOI] [PubMed] [Google Scholar]
- Valentine MT, Fordyce PM, Krzysiak TC, Gilbert SP, Block SM. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat Cell Biol. 2006;8:470–476. doi: 10.1038/ncb1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste D, Ferreira V, Vernos I. Chromokinesins: localization-dependent functions and regulation during cell division. Biochem Soc Trans. 2011;39:1154–1160. doi: 10.1042/BST0391154. [DOI] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Varga V, Leduc C, Bormuth V, Diez S, Howard J. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell. 2009;138:1174–1183. doi: 10.1016/j.cell.2009.07.032. [DOI] [PubMed] [Google Scholar]
- Vinogradova MV, Malanina GG, Reddy AS, Fletterick RJ. Structure of the complex of a mitotic kinesin with its calcium binding regulator. Proc Natl Acad Sci USA. 2009;106:8175–8179. doi: 10.1073/pnas.0811131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova MV, Malanina GG, Reddy VS, Reddy AS, Fletterick RJ. Structural dynamics of the microtubule binding and regulatory elements in the kinesin-like calmodulin binding protein. J Struct Biol. 2008;163:76–83. doi: 10.1016/j.jsb.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Vinogradova MV, Malanina GG, Waitzman JS, Rice SE, Fletterick RJ. Plant kinesin-like calmodulin binding protein employs its regulatory domain for dimerization. PLoS One. 2013;8:e66669. doi: 10.1371/journal.pone.0066669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova MV, Reddy VS, Reddy AS, Sablin EP, Fletterick RJ. Crystal structure of kinesin regulated by Ca(2+)-calmodulin. J Biol Chem. 2004;279:23504–23509. doi: 10.1074/jbc.M400741200. [DOI] [PubMed] [Google Scholar]
- Visscher K, Schnitzer MJ, Block SM. Single kinesin molecules studied with a molecular force clamp. Nature. 1999;400:184–189. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- Wade RH, Kozielski F. Structural links to kinesin directionality and movement. Nat Struct Biol. 2000;7:456–460. doi: 10.1038/75850. [DOI] [PubMed] [Google Scholar]
- Wagenbach M, Domnitz S, Wordeman L, Cooper J. A kinesin-13 mutant catalytically depolymerizes microtubules in ADP. J Cell Biol. 2008;183:617–623. doi: 10.1083/jcb.200805145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waitzman JS, Larson AG, Cochran JC, et al (2011) The loop 5 element structurally and kinetically coordinates dimers of the human kinesin-5, Eg5. Biophys J 101:2760–2769. doi:10.1016/j.bpj.2011.10.032 [DOI] [PMC free article] [PubMed]
- Walker RA, Salmon ED, Endow SA. The Drosophila claret segregation protein is a minus-end directed motor molecule. Nature. 1990;347:780–782. doi: 10.1038/347780a0. [DOI] [PubMed] [Google Scholar]
- Weaver LN, Ems-McClung SC, Stout JR, LeBlanc C, Shaw SL, Gardner MK, Walczak CE. Kif18A uses a microtubule binding site in the tail for plus-end localization and spindle length regulation. Curr Biol. 2011;21:1500–1506. doi: 10.1016/j.cub.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White HD, Belknap B, Webb MR. Kinetics of nucleoside triphosphate cleavage and phosphate release steps by associated rabbit skeletal actomyosin, measured using a novel fluorescent probe for phosphate. Biochemistry. 1997;36:11828–11836. doi: 10.1021/bi970540h. [DOI] [PubMed] [Google Scholar]
- Wickstead B, Gull K, Richards TA. Patterns of kinesin evolution reveal a complex ancestral eukaryote with a multifunctional cytoskeleton. BMC Evol Biol. 2010;10:110. doi: 10.1186/1471-2148-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehlke G. A look into kinesin's powerhouse. FEBS Lett. 2001;508:291–294. doi: 10.1016/s0014-5793(01)03064-2. [DOI] [PubMed] [Google Scholar]
- Woehlke G, Ruby AK, Hart CL, Ly B, Hom-Booher N, Vale RD. Microtubule interaction site of the kinesin motor. Cell. 1997;90:207–216. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
- Wood KW, Lad L, Luo L, et al (2010) Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proc Natl Acad Sci USA 107:5839–5844. doi:10.1073/pnas.0915068107 [DOI] [PMC free article] [PubMed]
- Xiol J, Spinelli P, Laussmann MA, et al (2014) RNA clamping by vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 157:1698–1711. doi:10.1016/j.cell.2014.05.018 [DOI] [PubMed]
- Yajima J, Mizutani K, Nishizaka T. A torque component present in mitotic kinesin Eg5 revealed by three-dimensional tracking. Nat Struct Mol Biol. 2008;15:1119–112. doi: 10.1038/nsmb.1491. [DOI] [PubMed] [Google Scholar]
- Yamada K, Kunishima N, Mayanagi K, et al (2001) Crystal structure of the Holliday junction migration motor protein RuvB from Thermus thermophilus HB8. Proc Natl Acad Sci USA 98:1442–1447. doi:10.1073/pnas.031470598 [DOI] [PMC free article] [PubMed]
- Yan Y, Sardana V, Xu B, et al (2004) Inhibition of a mitotic motor protein: where, how, and conformational consequences. J Mol Biol 335:547–554 [DOI] [PubMed]
- Yardimci H, van Duffelen M, Mao Y, Rosenfeld SS, Selvin PR. The mitotic kinesin CENP-E is a processive transport motor. Proc Natl Acad Sci USA. 2008;105:6016–6021. doi: 10.1073/pnas.0711314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Gennerich A, Vale RD. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- Yun M, Bronner CE, Park CG, Cha SS, Park HW, Endow SA. Rotation of the stalk/neck and one head in a new crystal structure of the kinesin motor protein, Ncd. EMBO J. 2003;22:5382–5389. doi: 10.1093/emboj/cdg531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M, Zhang X, Park CG, Park HW, Endow SA. A structural pathway for activation of the kinesin motor ATPase. EMBO J. 2001;20:2611–2618. doi: 10.1093/emboj/20.11.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YC, Kull FJ, Cochran JC. Modulation of the kinesin ATPase cycle by neck linker docking and microtubule binding. J Biol Chem. 2010;285:25213–25220. doi: 10.1074/jbc.M110.123067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuravleva A, Clerico EM, Gierasch LM. An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell. 2012;151:1296–1307. doi: 10.1016/j.cell.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuravleva A, Gierasch LM. Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones. Proc Natl Acad Sci USA. 2011;108:6987–6992. doi: 10.1073/pnas.1014448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerle CT, Frieden C. Analysis of progress curves by simulations generated by numerical integration. Biochem J. 1989;258:381–387. doi: 10.1042/bj2580381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 127 kb)