Abstract
The ever-increasing capacity of computing resources has extended ribosome calculations from the study of small-scale fluctuations to large-scale barrier-crossing processes. As the field of computational/theoretical biophysics shifts focus to large-scale conformational transitions, there is a growing need for a systematic framework to interpret and analyze ribosome dynamics. To this end, energy landscape principles, largely developed for the study of biomolecular folding, have proven to be invaluable. These tools not only provide a foundation for describing simulations but can be used to reconcile experimental results, as well. In this review, I will discuss recent efforts to employ computational methods to reveal the characteristics of the ribosome’s landscape and how these studies can help guide a new generation of experiments that more closely probe the underlying energetics. As a result of these investigations, general principles about ribosome function are beginning to emerge, including that: (1) small-scale fluctuations are the result of structure, rather than detailed energetics, (2) molecular flexibility leads to entropically favored rearrangements, and (3) tRNA dynamics may be accurately described as diffusive movement across an energy landscape.
Keywords: Simulations, Molecular machine, tRNA, Elongation
Abbreviations
- tRNA
transfer RNA
- aa-tRNA
aminoacyl-transfer RNA
- EF-Tu
Elongation Factor Tu
- EF-G
Elongation Factor G
- SMOG
Structure-based models in Gromacs
- MDFF
Molecular dynamics flexible fitting
- PCA
Principal component analysis
- NMA
Normal mode analysis
Introduction
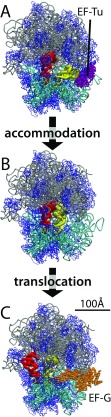
Inspired by decades of investigations into protein dynamics (Frauenfelder et al. 1991; Onuchic et al. 1995; Thirumalai and Hyeon 2005), an increasingly adopted view of biomolecular function is that large-scale conformational rearrangements may be described as movement across an underlying energy landscape (Hyeon and Thirumalai 2011; Whitford et al. 2012; Hills and Brooks 2009; Chan et al. 2011). With exponential growth in the scale of computing (Vendruscolo and Dobson 2011), it is now possible to use simulations to learn about the energy landscapes of complex machines, such as the ribosome. In this review, I will discuss recent advances in molecular biophysics that have provided insights into features of the landscapes that govern ribosome function. The discussion will focus on the merits and limitations of computational techniques that have been applied to study large-scale movement (10–100Å) of transfer RNA (tRNA) inside of the ribosome, specifically tRNA accommodation and translocation (Fig. 1). The aim is that the reader will be left with an understanding of how computational methods provide quantitative frameworks for describing ribosome dynamics.
Fig. 1.
The ribosome and the elongation cycle. The 23S rRNA (grey), 16S rRNA (cyan), ribosomal proteins (blue), EF-Tu (purple), and EFG (orange) are shown in tube representation. The tRNA molecules (yellow, red) are shown in the A) A/T-P/P, B) A/A-P/P, and C) P/P-E/E configurations. During translation of mRNA sequences into protein sequences, the tRNA molecules and ribosome undergo a range of conformational transitions. Two of the largest-scale conformational transitions are aa-tRNA accommodation, where the aa-tRNA enters the ribosome (panel A to B), and tRNA translocation, where the tRNA molecules move between binding sites (panel B to C). PDB IDs: 2WRN, 2WRO(Schmeing et al. 2009), 3I8F, 3I8G (Jenner et al. 2010), 2WRI, 2WRJ (Yong-Gui et al. 2009)
As progress is made to understand the ribosome from kinetic approaches (Johansson et al. 2008; Rodnina and Wintermeyer 2011), structural techniques (Korostelev and Noller 2007; Schmeing and Ramakrishnan 2009; Frank and Spahn 2006), and single-molecule methods (Petrov et al. 2011), the field strives to integrate diverse findings into a cohesive and consistent picture. Similar to experimental approaches, each type of computation is suited to address specific questions, and each possesses limitations. Using the energy landscape description as a common framework for interpreting experiments and simulations, I provide my perspective on how theoretical techniques can provide insights that are integral to establishing a comprehensive understanding of the ribosome.
This review is focused on computational techniques applied to the elongation cycle of the ribosome. Elongation is the process by which tRNA molecules are recruited to translate messenger RNA (mRNA) sequences into protein sequences, and it encompasses numerous large-scale rearrangements. First, aminoacyl-tRNA (aa-tRNA) is delivered by elongation factor Tu (EF-Tu), which is followed by accommodation of the aa-tRNA into the ribosomal A site (≈ 100Å). Next, a peptide bond is formed between the nascent chain and the aa-tRNA. Subsequently, the tRNA molecules move between the ribosomal A, P, and E binding sites, a process known as translocation (≈ 20 − 50Å). During translocation, there are also global rearrangements in the ribosome, which are facilitated by elongation factor G (EF-G). Finally, tRNA is released from the ribosomal E-site.
To illustrate the relative strengths and limitations of computational approaches, the review is organized by types of landscape features, where examples of computational techniques that can characterize each feature are described. Within this context, examples of applications to the ribosome are presented. The discussion will begin with approaches for structural modeling, where individual structures describe minima on the energy landscape. Next, I will discuss the local shape of the landscape about these low-energy (stable) configurations. Finally, I will discuss the dynamics of large-scale rearrangements, where tRNA molecules interconvert between competing energetic minima. The progression from single-basin to barrier-crossing dynamics roughly mirrors the chronology of scientific progress, reflecting the increased challenges when addressing larger-scale features. By describing advances in terms of their contributions to our understanding of energy landscapes, I aim to illustrate the connections between small-scale and large-scale motions, as well as the trajectory of ongoing research.
Modeling the structures at energetic minima
Before discussing landscapes, it is important to make a clear distinction between structural modeling and simulations. Here, “modeling” refers to methods that use experimental quantities as input in order to generate a set of atomic coordinates. For example, in X-ray crystallographicy studies, a scattering pattern is collected and then the coordinates of a structural model are refined until there is satisfactory agreement between the “theoretical” and observed patterns. For NMR, resonance peaks provide atomic distance/angle estimates, which are then imposed on a structural model through the use of energetic restraints. For both methods, one seeks to construct a structural model that is consistent with the experimental quantities. Typically, experimental measurements describe low-energy, highly populated configurations, though marginally stable configurations can sometimes be captured (Bouvignies et al. 2011). In the context of energy landscapes, these approaches search for a molecular configuration that is representative of the lowest free-energy ensemble of the system. While these methods are most commonly employed to obtain a single structure from a single experiment, modeling can encompass any number of observables (Alber et al. 2007) and may generate ensembles of structures (Yang et al. 2010). In contrast to structural modeling, the term “simulation” describes methods where the time evolution of the coordinates is obtained for a given energetic model. It should be stressed that the term simulation does not imply a particular level of resolution (Fig. 2), such as all-atom explicit-solvent. Rather, a simulation can be highly detailed (e.g., including quantum mechanical effects), less detailed (e.g., coarse grained), have implicit or explicit solvent, or be composed of any combination thereof.
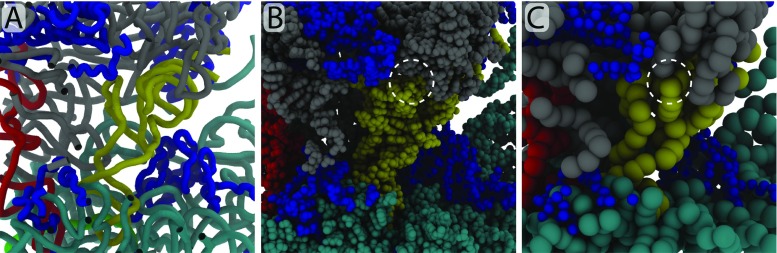
Fig. 2.
Different levels of resolution are suited to address different questions. A pre-accommodation (A/T) configuration of aa-tRNA, shown in A) tube, B) all-atom, and C) coarse-grained representation. The sizes of the spheres in panels B and C reflect the typical effective size used in models of each resolution. When simulating smallscale fluctuations, coarse-grained and higher-resolution models often provide consistent descriptions. For large-scale conformational transitions, such as accommodation, the large number of steric interactions suggests that higher-resolution models are more appropriate. For example, use of a coarse-grained model can easily introduce steric interactions that are not present in the all-atom system (dashed circles). In addition to choosing the most appropriate resolution, different energetic models are available at each resolution. For example,models with all-atom resolution may or may not include explicit-solvent. Additionally, all-atom models may employ semi-empirical or simplified descriptions of the energetics
With recent advances in structural modeling techniques, the distinction between modeling and simulations has become less transparent. A general strategy that has been applied to the ribosome is to build energy functions that encode experimentally obtained information, such as the proximity of specific atoms, interface interactions, or electron densities from cryo-EM (Trabuco et al. 2010; Whitford et al. 2011). These experimentally guided restraints are designed to ensure that the lowest-energy configuration is consistent with the observable. By performing a simulation with this modeling energy function, the atomic coordinates relax from their initial configuration to a lower-energy configuration. When the modeling simulation is completed, each individual restraint should be at its respective minimum. While the relaxation process is technically a simulation, there is no physical justification for why an arbitrary modeling energy function would provide an accurate description of the energy landscape associated with a conformational transition.
Using computation to extract atomic models of the ribosome from cryo-EM data
Simulation-based modeling has been particularly effective at determining structural details from cryoelectron microscopy (cryo-EM) data. As an example, we will consider one specific energy function used for constructing atomic models from cryo-EM densities:
| 1 |
where V theory can be any energy function for the molecule (e.g., all-atom explicit-solvent (Trabuco et al. 2010), or simplified/coarse-grained energetics (Whitford et al. 2011)), and V map is a biasing potential. The biasing potential is designed such that configurations that are more consistent the cryo-EM reconstruction are lower in potential energy. V map can take many forms, where an intuitive definition is the overlap of the simulated and experimental electron densities (Orzechowski and Tama 2008):
| 2 |
W map is the energetic weight of the bias towards the EM density, which ensures the system rapidly adopts a configuration consistent with ρ exp. While the lowest-energy configuration in V model is consistent with the low-energy configuration probed experimentally, introduction of V map can result in high-energy pathways being accessed during a modeling simulation that are functionally irrelevant. Therefore, only the final configuration in a modeling simulation is reflective of the molecule’s energy landscape.
Pioneering efforts in cryo-EM applied to the ribosome (Frank and Agrawal 2000; Valle et al. 2003a; Mikel et al. 2002; Mikel et al. 2003b; Frank and Spahn 2006) have led to an abundance of cryo-EM reconstructions of the complex at various stages of function. Accordingly, simulation-based modeling strategies have been able to help extract atomic-resolution information of stable configurations from these, often lower-resolution, electron densities. To study tRNA accommodation, Schulten and coworkers developed a simulation-based approach that uses a slightly different definition of V map than used by Orzechowski and Tama, which is called molecular dynamics flexible fitting (MDFF). With MDFF the team built models of aa-tRNA in complex with EF-Tu (Villa et al. 2009). The Schulten and Beckmann groups later applied these algorithms to describe trapped configurations during ribosome stalling (Frauenfeld et al. 2011) and membrane translocation of a nascent peptide chain (Becker et al. 2009; Seidelt et al. 2009). By adopting the V map function of Orzechowski and Tama (2008) and an all-atom model with simplified energetics (Whitford et al. 2009), we used simulation-based modeling to generate models of the ribosome during translocation with either one (Ratje et al. 2010), or two (Whitford et al. 2011), tRNA molecules and EF-G. More recently, Bock et al. (2013) used a non-simulation-based iterative-refinement approach to construct atomic models for 13 different cryo-EM reconstructions obtained from a reverse translocation assay. In all of these studies, medium-to-high resolution cryo-EM reconstructions were available. Since each reconstruction is obtained by collecting images for highly populated configurations, each structural model describes a minimum on the free-energy landscape of the ribosome under the given experimental conditions.
Describing structural fluctuations about a minimum
After one identifies a configuration that is representative of an energetic minimum, the next step is to determine the shape of the landscape about that point. As discussed in the previous section, modeling techniques use experimental measurements to determine a structure. The subsequent discussion focuses on calculations that use a structural model as input. These experimentally obtained configurations are frequently assumed to represent energetic minima, about which the local landscape is extrapolated. While these approximations have limits, it has become clear that the structure is a major determinant of the landscape about a minimum.
Coarse-grained models predict correlated rearrangements of ribosomal stalks and subunits
To describe fluctuations about a minimum, it is convenient to consider the Taylor expansion of the energy about the point {x i0}:
| 3 |
With regards to the shape of the landscape, one may disregard the constant V(x 01, .. x 0n). Further, at a minimum, the first derivatives are equal to zero. Finally, when considering small displacements, terms of O(3) may be neglected, leaving only:
| 4 |
Accordingly, for small fluctuations about a minimum, the landscape may be approximated as a sum of second-order terms. Assuming that the scale of intra-molecular interactions is correlated with the spatial proximity of each atom pair, then a reasonable approximation is that for atom pairs beyond a distance threshold. To obtain a more intuitive description of the landscape, one may convert to normal coordinates Q i, allowing the potential energy to be expressed as:
| 5 |
where ω i is the frequency associated with mode i. The landscape is then described in terms of displacements along orthogonal directions in which the potential energy is quadratic.
Normal mode analysis (NMA) was first applied to a coarse-grained elastic-network model of the ribosome by Tama et al. (2003), where they found that the lowest-frequency modes (i.e., smallest curvature, or largest-scale fluctuations) correspond to rotations of the subunits and rearrangements of the stalks. In a subsequent application of NMA using a coarse-grained elastic-network model, Kurkcuoglu et al. (2008) found that the presence/absence of specific ribosomal proteins perturbs the modes, implicating changes in the local landscape. While NMA is a powerful tool for deducing the local curvature of the landscape, and thereby inferring the directions of small-scale fluctuations, a limitation is that it is unknown how far the approximation is valid.
An alternative to NMA is principal component analysis (PCA). PCA is a general method by which a simulation is performed and the covariance matrix for atomic displacements is calculated and diagonalized. This yields a set of orthogonal eigenvectors, where each describes specific correlated motions within the system. Similar to NMA, PCA can be applied to any energetic model, regardless of the resolution and energetic detail. In contrast to NMA, PCA is not based on a second-order expansion of the energy, allowing it to be used to identify correlated motions that may involve barrier-crossing events. For the ribosome, Trylska et al. (2005) performed PCA analysis for a coarse-grained model and found anti-correlated movement of the stalk regions. By using a coarse-grained model, they were able to obtain a sufficiently long trajectory, allowing the covariance matrix to converge. It is now also possible to calculate PCs for an all-atom model with simplified energetics, which also implicates subunit rotation (Fig. 3). For explicit-solvent simulations of the ribosome, interpreting PCs is often less straightforward. We recently attempted to calculate the PCs from a 1.4-microsecond all-atom explicit-solvent simulation (Whitford et al. 2013), and found that the covariance matrix did not converge, due to a conformational rearrangement after ≈500 ns. In a separate study, Bock et al. (2013) calculated PCs from all-atom explicit-solvent simulations that were 200–300 ns in duration and they found reasonable convergence. Since shorter simulations only sample phase space in the immediate vicinity of each minimum, the local fluctuations may converge, whereas longer simulations often interconvert between minima a small number of times. Similar to NMA, PCA implicates the direction and scale of significant fluctuations by considering the covariance of coordinates. Thus, while PCA does not rely on a harmonic approximation to the landscape, higher-order correlations may not be detected.
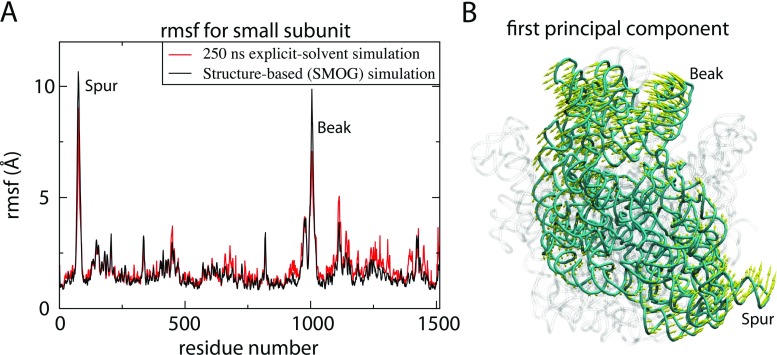
Fig. 3.
Local fluctuations are consistent between highly detailed and simplified models. A) Spatial RMSF of the 16S rRNA calculated from a 250-ns explicit-solvent simulation (> 3 × 106 atoms) and an all-atom structure-based SMOG model (≈150,000 atoms) (Whitford et al. 2010a). While the SMOG model uses a simplified description of the energetics, the local fluctuations are consistent in scale and distribution. B) The first principal component (PC) calculated from a simulation with an all-atom SMOG model (simulation described in Noel et al. (2014) is shown, where the arrows indicate the direction and scale of movement of each P atom in the small subunit (cyan). The first PC captures rotation of the small subunit, consistent with NMA and PCA results with coarse-grained models (Trylska et al. 2005; Tama et al. 2003), demonstrating that local fluctuations are not strongly influenced by energetic details. For reference, the 23S rRNA is shown in transparent representation in the background
Detailed simulations predict the scale of local fluctuations
Coarse-grained approaches are often effective at identifying large-scale correlated movements, though most methods require the scale of the fluctuations to be fit to an independent metric. That is, the energy and temperature in coarse-grained models have effective values (i.e., averaged over the excluded degrees of freedom). Thus, it is necessary to tune the effective temperature or energy to yield structural fluctuations that are of the same scale as in solution. One way to address this is to calculate the fluctuations about a minimum using all-atom explicit-solvent simulations and then reparameterize the coarse-grained (or simplified) simulations until there is a satisfactory level of agreement. In contrast to coarse-grained models, or models that employ simplified energetics, explicit-solvent models assign non-specific van der Waals and electrostatic parameters to each atom, and the only experimental information explicitly included in the simulation is the initial configuration. Since these models have been parameterized so that absolute temperature scales are used, when sufficient computing resources are available, they can help verify the distribution of small-scale fluctuations observed with simplified models.
As all-atom explicit-solvent simulations have been extended to hundreds of nanoseconds (Whitford et al. 2010b; Bock et al. 2013), and even a microsecond (Whitford et al. 2013), it has recently become possible to use these simulations as reliable benchmarks for parameterizing coarse-grained/simplified models. To construct coarse-grained models for which the localized fluctuations are consistently described, Voth and coworkers have developed general dynamics-based coarse-graining approaches (Saunders and Voth 2013). With these methods, they used a 100-ns simulation of a ribosome in explicit solvent as a benchmark for constructing a minimal model composed of 480 beads that interact through harmonic potentials (Zhang et al. 2011). The strength of each bead–bead interaction was adjusted such that the fluctuations in the coarse-grained system were consistent with the fluctuations in the explicit-solvent simulation. In principle, this model could be extended to study conformational rearrangements associated with elongation, though it is unclear what effect coarse-graining will have on large-scale movements of tRNA (Fig. 2). In a separate study, we performed multiple 200–300-ns explicit-solvent simulations of a ribosome and calculated the RMSF of each atom (Whitford et al. 2010a). We then compared these values to those obtained using an all-atom (all non-hydrogen atom) structure-based SMOG model. The SMOG model defines the crystallographic (or cryo-EM) structure as the lowest-energy configuration (Whitford et al. 2009; Noel et al. 2010). However, in contrast to the harmonic approximation made for NMA calculations, non-bonded interactions in the SMOG model can be broken at high temperatures. While the SMOG model lacks non-specific stabilizing interactions, when all native interactions were given equal energetic weight, the scale and distribution of the RMSF were consistent between both simulation approaches (Fig. 3). The values from both simulations were also consistent with estimates obtained from anisotropic B-factors (Korostelev et al. 2008), converted to RMSF values according to the methods of García et al. (1997). Agreement between models of multiple resolutions and energetic representations, as well as with experiments, indicates that the direction and scale of local fluctuations are dominated by the shape of the molecule, and they are not strongly dependent on the energetic details.
Traversing the landscape: conformational rearrangements between minima
With an understanding of the fluctuations about stable configurations, we turn our attention towards describing large-scale motions, where the tRNA and ribosome interconvert between energetic minima. Sanbonmatsu et al. (2005) were the first to attempt to address this via computation by performing a 2-ns explicit-solvent simulation that employed targeting protocols to induce tRNA movement. While kinetic and thermodynamic data could not be obtained at that time, those calculations did suggest the presence of a sterically accessible pathway along which aa-tRNA may enter the ribosome. With increases in computing capacity, this simulation can now be performed on a desktop computer, and large supercomputers (1024–16384 cores) can produce 15–170 ns per day (Whitford et al. 2013; Bock et al. 2013; Kutzner et al. 2014). In addition to increased computing power, we made a significant shift in the simulation of ribosomes by adopting all-atom models with simplified energetic schemes (Whitford et al. 2010a; Whitford and Sanbonmatsu KY 2013; Noel et al. 2014). Motivated by the study of protein folding, where analysis of energy landscapes is common practice (Frauenfelder et al. 1991; Onuchic et al. 1995; Thirumalai and Hyeon 2005; Hyeon and Thirumalai 2011; Whitford et al. 2012), these simpler models allowed us to explore different realizations of the energy landscapes that govern large-scale conformational transitions. A fortuitous property of these models is that they are computationally less demanding than explicit-solvent simulations, which enables stochastic barrier-crossing processes to be simulated. Together, these advances are allowing simulations to begin to provide descriptions of how tRNA molecules diffuse across the landscape. Drawing significantly from our own work, I will highlight the most recent computational/theoretical studies that have employed the energy landscape description to quantitatively analyze large-scale transitions of the ribosome.
Describing diffusive barrier-crossing events in the ribosome
While the energy landscape perspective provides an elegant conceptual framework for thinking about dynamics, it is also a robust quantitative framework for precisely describing how molecular energetics facilitate large-scale, often collective, dynamics. For example, in the analysis of chemical reactions, interconversion between the reactant and product states is described as being the result of molecular vibrations towards a transition state, which is defined as a saddle point on the energy landscape. Molecular vibrations give rise to characteristic frequencies with which attempts are made to cross the barrier (roughly p s −1), and the probability of successfully crossing is given by exp(−ΔG TSE/k B T), where ΔG TSE is the free-energy barrier height. In the context of biomolecular folding, movement between the folded and unfolded ensembles is not governed by molecular vibrations. Rather, these collective processes are diffusive (Fig. 4A), and attempts to cross a barrier occur with frequencies of ≈ 1μ s −1(Kubelka et al. 2004). The stark difference between the fundamental character of these processes makes it clear that in order systematically study the ribosome, which encompasses chemical reactions and order–disorder transitions, it is essential that the appropriate landscape description be applied to each step. Two aspects of this objective are, (1) What coordinates most accurately describe the rate-limiting barriers? and (2) What are the values of the diffusion coefficients along functionally relevant coordinates? Each of these questions is discussed below.
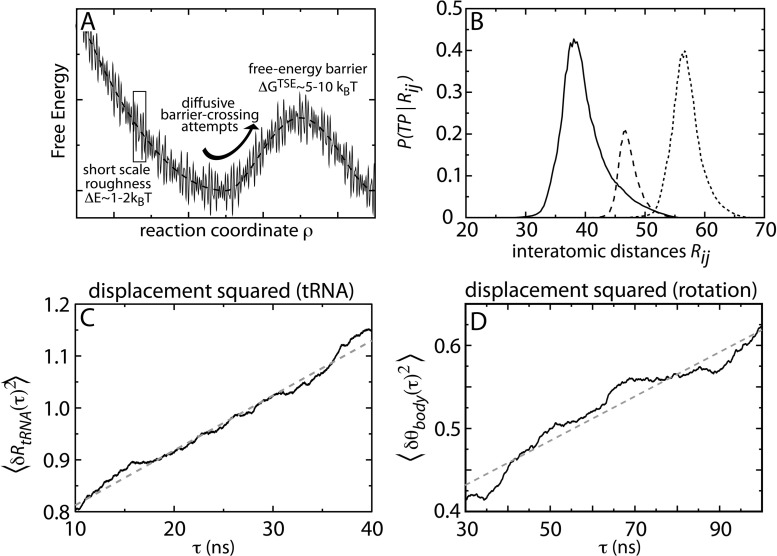
Fig. 4.
Relationship between landscapes, diffusion, and transition paths. A) Schematic illustrating an energy landscape where there is short-scale roughness and a large-length scale barrier. The shortscale roughness determines the effective diffusion coefficient along the coordinate. Diffusive movement leads to barrier crossing attempts, where the probability of successfully crossing is determined by the barrier height ΔG TSE. B) Probability of being on a transition path during accommodation, for multiple coordinates, calculated from a simplified all-atom structure-based SMOG model (Noel et al. 2014). A common coordinate in experiments (large dashed line) is a poor indicator of when the system is crossing the barrier. Two alternate coordinates (solid line, fine dashed line) perform significantly better, nearly reaching the diffusive limit of 0.5. To complement the analysis from simplified models, effective diffusion coefficients can be obtained from explicit-solvent simulations by calculating the slope of the displacement squared as a function of lag time for C) tRNA movements and D) subunit rotations (Whitford et al. 2010b; Whitford et al. 2013)With values for the effective diffusion coefficient, one may interconvert between barrier heights and rates.
Identifying coordinates along which ribosome motion is diffusive
To quantify the energy landscape of a process, one should determine which reaction coordinates ρ (not to be confused with the electron density in earlier discussions) accurately capture the transition state ensembles (TSEs), and whether the motion is diffusive. To address these points, we performed long simulations of aa-tRNA accommodation using an all-atom model with simplified energetics, where many (≈ 100) barrier-crossing events were observed in an individual trajectory. From a phenomenological standpoint, the overall dynamics exhibited by this model were consistent with a wide range of experiments, suggesting that it provides a satisfactory description of the structural properties associated with aa-tRNA accommodation. Further, use of this simplified model was particularly appropriate in this instance, since the barrier we were probing is the result of steric effects, specifically Helix 89 (Whitford et al. 2010a). By properly accounting for the sterics through the explicit representation of all non-hydrogen atoms, the simulated movement across this barrier is expected to mimic tRNA movement in solution. From this trajectory, we projected the dynamics along more than 150 reaction coordinates. We quantified the dynamics along each by calculating the flux between the endpoints, the probability of being on a transition path P(TP|ρ), and by fitting the displacement squared 〈[ρ(t + τ) − ρ(t)]2〉 to . When the underlying dynamics is diffusive, and an appropriate coordinate is measured, P(TP|ρ) will reach a value of 0.5 at the TSE, the flux will be minimized (i.e., no false positives) and α=1. Based on these metrics, our analysis demonstrated that the kinetics of aa-tRNA during accommodation may be accurately described as effective diffusion along a one-dimensional coordinate based on atomic distances between tRNA molecules (Noel et al. 2014). In the same study, we also found that a currently used coordinate in smFRET experiments was a relatively poor indicator of when the tRNA is at the TSE, and numerous higher-performing coordinates were implicated (Fig. 4B). Since all of the coordinates were based on atom-atom pairs, these results suggest that attaching FRET dyes to different residues on the tRNA molecules can lead to improved signals in single-molecule experiments. Specifically, the signals arising from these new labeling sites should be more strongly correlated with accommodation transition events, which may reduce the number of observed false positives and provide more precise information about the location of the transition state.
Effective diffusion coefficients bridge free-energy barriers and ribosome kinetics
One may use explicit-solvent simulations to estimate diffusion-limited attempt frequencies associated with barrier crossing. To this end, we have performed explicit-solvent simulations of the ribosome (200 ns (Whitford et al. 2010b) to 1.4 microseconds (Whitford et al. 2013)), from which we have calculated estimates of the effective diffusion coefficients D eff along putative coordinates (Fig. 4). With knowledge of the length scale of the rearrangement and D eff, one may infer the likely time for barrier-crossing attempts. We found that for tRNA rearrangements the attempt frequencies are ≈ 1μ s −1, while for subunit rotations the frequencies range from 0.02 to 0.5 μ s −1. Comparing D eff with diffusion coefficients in solution (D free) can be used to identify the scale of the short-scale roughness ΔE that is introduced by ribosomal interactions, via the relation: (Bryngelson and Wolynes 1989). We found that ΔE ≈ 1 − 2k B T for both tRNA movements and subunit rotations. This is perhaps a surprisingly small value, considering the large number of interface interactions in the ribosome. Once these quantities are probed experimentally (Hyeon and Thirumalai 2003), it will be interesting to see how similar the roughness is in all-atom explicit-solvent models and the ribosome in solution.
Entropic changes in tRNA can guide elongation dynamics
When quantifying energetics, it is often necessary to consider entropic changes. By utilizing simplified models, spontaneous large-scale conformational transitions are computationally accessible. These types of simulations have made it clear that aa-tRNA molecules possess a high degree of flexibility in the 3’-CCA tail (Whitford et al. 2010a). The flexibility of the 3’-CCA tail is not surprising, as it is the only single-stranded portion of the molecule. In addition to significant flexibility, the range of available configurations of the tail decreases as the aa-tRNA molecule enters the ribosome. This transition to an ordered ensemble indicates that configurational entropy of the aa-tRNA molecule decreases during accommodation. Analogous to folding, where the unfolded ensemble is stabilized by entropy and the folded configuration is stabilized by enthalpy, the A/T (pre-accommodation) ensemble is stabilized by configurational entropy and energetics drive movement to the A/A (post-accommodation) configuration. This interpretation is supported by the fact that as the simulated temperature is increased, the A/T ensemble becomes more thermodynamically favorable (Noel et al. 2014). This physical-chemical property carries significant biological implications. That is, the accommodation step is associated with fidelity of aa-tRNA molecules. If an aa-tRNA is not cognate (i.e., not correct), it can be rejected before fully accommodating. With regards to flexibility, since the change in entropy is centered about the tail, accommodation of the tail is the final substep. By delaying the entry of the 3’-CCA tail, which carries the covalently linked amino acid, proofreading steps may be employed before incorporating a potential mistake into the growing protein chain. In contrast, simulations of hybrid-state formation (Whitford and Sanbonmatsu 2013), a step not implicated in fidelity, suggest at that point there are less significant changes in configurational entropy. While these results provide a clear connection between physical chemistry and biology, care should be taken to not over-interpret the simulated dynamics. That is, the presence of a change in configurational entropy is supported by the fact that the models’ description of molecular flexibility is consistent with experimental observations and theoretical considerations. However, we would not claim an exact value of the change in entropy from these simulations. That is, these calculations clearly indicate that there are significant changes in configurational entropy, though further analysis and measurements are required to determine the precise values.
Piecing together a free-energy profile for tRNA reverse translocation
In a recent study, Bock et al. (2013) used 13 relatively long (≈ 200–300 ns) explicit-solvent simulations of the ribosome at intermediate stages of reverse translocation. Each simulation was initiated at a configuration identified through cryo-EM measurements. Consistent with earlier discussion, these snapshots represent local free-energy minima, under the specific experimental conditions. They next calculated the principal components about each minimum and then used the PCs to extrapolate likely rates of interconversion between minima. With the predicted barriers, they confirmed that the sequence of reconstructions represents an energetically viable pathway during reverse translocation. With regards to the full landscape associated with reverse-translocation (and possibly forward translocation), even this massive amount of sampling is insufficient to rule out the presence of alternate low-energy pathways. In future studies, it will be interesting to see the extent to which the results depend on the specific details of the explicit-solvent model and simulation parameters, and how well the landscape associated with forward translocation is described by the landscape of the reverse process.
Future challenges
The energy landscape approach to studying dynamics of the ribosome continues to be developed and refined. Here, I have focused on a series of theoretical advances that have been directed towards studying the properties of the energy landscapes associated with large-scale conformational rearrangements during elongation. To complement the study of large-scale motions, there has also been a broad range of simulations focused on smaller-scale transitions in the ribosome, chemical reactions, and transitions not directly involved with tRNA movement (Vicatos et al. 2013; Sanbonmatsu 2012). An outstanding challenge is to directly correlate the properties of the landscapes associated with chemical reactions and large-scale tRNA/ribosome rearrangements. An additional challenge facing the computational and experimental communities is that it is largely unknown how the landscape responds to changes in conditions, such as solvent composition, the presence of co-factors and modifications, or fluctuations in temperature. Through the systematic use of an energy landscape perspective, each of these factors may be rationalized, which will then provide a cohesive and comprehensive description of ribosome dynamics in the cell.
Compliance with Ethical Standards
Funding
This work was supported by an NSF CAREER Award (Grant MCB-1350312). This work was supported in part by the National Science Foundation through XSEDE resources provided by TACC under Grant No. TG-MCB110021. We also acknowledge generous support provided by the Northeastern University Discovery Cluster.
Conflict of interest
Paul Whitford declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human or animal subjects performed by the author.
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Ws R, Chait BT, Rout MP, Sali A. Determining the architectures of macromolecular assemblies. Nature. 2007;450(7170):683–94. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- Becker T, Bhushan S, Jarasch A, Armache J-P, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, Westhof E, Gilmore R, Mandon EC, Beckmann R. Structure of monomeric yeast and mammalian Sec61 Complexes interacting with the translating ribosome. SCIENCE. 2009;326(5958):1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock LV, Blau C, Schröder GF, Davydov II, Fischer N, Stark H, Rodnina MV, Vaiana AC, Grubmüller H. Energy barriers and driving forces in tRNA translocation through the ribosome. Nat Struct Molecular Bio. 2013;20:1390–1396. doi: 10.1038/nsmb.2690. [DOI] [PubMed] [Google Scholar]
- Bouvignies G, Vallurupalli P, Hansen DF, Correia BE, Lange O, Bah A, Vernon RM, Dahlquist FW, Baker D, Kay LE. Solution structure of a minor and transiently formed state of a T4 lysozyme mutant. Nature. 2011;477(7362):111–114. doi: 10.1038/nature10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryngelson JD,Wolynes PG (1989) Intermediates and barrier crossing in a random energy-model (with applications to protein folding). J Phys Chem-US 93(19):6902–6915
- Chan HS, Zhang Z, Wallin S, Liu Z. Cooperativity, local-nonlocal coupling, and nonnative interactions: principles of protein folding from coarse-grained models. Ann Rev Phys Chem. 2011;62:301–326. doi: 10.1146/annurev-physchem-032210-103405. [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406(6793):318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Frank J, Spahn CMT. The ribosome and the mechanism of protein synthesis. Rep Prog Phys. 2006;69(5):1383–1417. doi: 10.1088/0034-4885/69/5/R03. [DOI] [Google Scholar]
- Frauenfeld J, Gumbart J, O.van der SE, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, Beckmann R. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat Struct Mol Biol. 2011;18(5):614–U127. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254(5038):1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- Gao Y-G, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V (2009) The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326(5953):694–9 [DOI] [PMC free article] [PubMed]
- García AE, Krumhansl JA, Frauenfelder H. Variations on a theme by Debye and Waller: from simple proteins to crystals. Proteins. 1997;29(2):153–60. doi: 10.1002/(SICI)1097-0134(199710)29:2<153::AID-PROT3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Hills RD, Brooks CL. Insights from coarse-grained Gō models for protein folding and dynamics. Int J Mol Sci. 2009;10(3):889–905. doi: 10.3390/ijms10030889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyeon C, Thirumalai D. Can energy landscape roughness of proteins and RNA be measured by using mechanical unfolding experiments. Proc Nat Acad Sci USA. 2003;100(18):10249–53. doi: 10.1073/pnas.1833310100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyeon C, Thirumalai D. Capturing the essence of folding and functions of biomolecules using coarse-grained models. Nat Comms. 2011;2:487. doi: 10.1038/ncomms1481. [DOI] [PubMed] [Google Scholar]
- Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol. 2010;17(5):555–60. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- Johansson M, Lovmar M, Ehrenberg M. Rate and accuracy of bacterial protein synthesis revisited. Curr Op Microbio. 2008;11(2):141–147. doi: 10.1016/j.mib.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Korostelev A, Noller HF. The ribosome in focus: new structures bring new insights. Trends Biochem Sci. 2007;32(9):434–41. doi: 10.1016/j.tibs.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF (2008) Crystal structure of a translation termination complex formed with release factor RF2. Proc Nat Acad Sci USA 105(50):19684–9 [DOI] [PMC free article] [PubMed]
- Kubelka J, Hofrichter J, Eaton WA. The protein folding speed limit. Curr Op Struct Biol. 2004;14(1):76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Kurkcuoglu O, Doruker P, Sen TZ, Kloczkowski A, Jernigan RL. The ribosome structure controls and directs mRNA entry, translocation and exit dynamics. Phys Biol. 2008;5(4):46005. doi: 10.1088/1478-3975/5/4/046005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutzner C, Apostolov R, Hess B, Grubmuller H (2014) Advances in parallel computing volume 25. IOS Press, pp 722–727
- Noel JK, Whitford PC, Sanbonmatsu KY, Onuchic JN (2010) SMOG@ctbp: simplified deployment of structure-based models in GROMACS, pp W657–61 [DOI] [PMC free article] [PubMed]
- Noel JK, Chahine J, Veite VBP, Whitford PC (2014) Capturing transition paths and transition states for conformational rearrangements in the ribosome. Biophys J 107: 2881–2890. doi: 10.1016/j.bpj.2014.10.022. This was the first study to demonstrate that tRNA movement in the ribosome may be accurately described as diffusion along a one-dimensional free-energy profile. These simulations also unambiguously showed that configurational entropy favors the A/T ensemble [DOI] [PMC free article] [PubMed]
- Onuchic JN, Wolynes PG, Luthey-Schulten Z, Socci ND. Toward an outline of the topography of a realistic funnel, protein-folding. Proc Nat Acad Sci USA. 1995;92(8):3626–3630. doi: 10.1073/pnas.92.8.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzechowski M, Tama F (2008) Flexible fitting of high-resolution X-ray structures into cryo electron microscopy maps using biased molecular dynamics simulations. Biophys J 95:5692–5702 [DOI] [PMC free article] [PubMed]
- Petrov A, Kornberg G, O’Leary S, Tsai A, Uemura S, Puglisi JD. Dynamics of the translational machinery. Curr Op Struct Biol. 2011;21:137–145. doi: 10.1016/j.sbi.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratje AH, Loerke J, Mikolajka A, Brünner M, Hildebrand PW, Starosta AL, Dönhöfer A, Connell SR, Fucini P, Mielke T, Whitford PC, Onuchic JN, Yu Y, Sanbonmatsu KY, Hartmann RK, Penczek PA, Wilson DN, Spahn CMT. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W. The ribosome as a molecular machine: the mechanism of tRNA-mRNA movement in translocation. Biochem Soc Trans. 2011;39(2):658–62. doi: 10.1042/BST0390658. [DOI] [PubMed] [Google Scholar]
- Sanbonmatsu KY. Computational studies of molecular machines: the ribosome. Curr Op Struct Biol. 2012;22(2):168–174. doi: 10.1016/j.sbi.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanbonmatsu KY, Joseph S, Tung C-S. Simulating movement of tRNA into the ribosome during decoding. Proc Nat Acad Sci USA. 2005;102(44):15854–9. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders MG, Voth GA. Coarse-graining methods for computational biology. Ann Rev Biophys. 2013;42(1):73–93. doi: 10.1146/annurev-biophys-083012-130348. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Ramakrishnan V (2009) What recent ribosome structures have revealed about the mechanism of translation. Nature 461(7268):1234–42 [DOI] [PubMed]
- Schmeing TM, Voorhees RM, Kelley AC, Gao Y-G, Murphy FV, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326(5953):688–94. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache J-P, Villa E, Trabuco LG, Becker T, Mielke T, Schulten K, Steitz TA, Beckmann R. Structural insight into nascent polypeptide chain-mediated stalling translational. Science. 2009;326(5958):1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tama F, Mikel V, Frank J, Brooks CL. Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryo-electron microscopy. Proc Nat Acad Sci USA. 2003;100(16):9319–23. doi: 10.1073/pnas.1632476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumalai D, Hyeon C. RNA and protein folding: common themes and variations. Biochemistry. 2005;44(13):4957–70. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, Schulten K (2008) Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16:673–683 [DOI] [PMC free article] [PubMed]
- Trabuco LG, Schreiner E, Eargle J, Cornish P, Ha T, Luthey-Schulten Z, Schulten K. The role of L1 stalk-tRNA interaction in the ribosome elongation cycle. J Mol Biol. 2010;402(4):741–60. doi: 10.1016/j.jmb.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trylska J, Tozzini V, McCammon JA. Exploring global motions and correlations in the ribosome. Biophys J. 2005;89(3):1455–63. doi: 10.1529/biophysj.104.058495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J (2002) Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. The EMBO J 21(13):3557–3567 [DOI] [PMC free article] [PubMed]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J (2003a) Locking and unlocking of ribosomal motions. Cell 114(1):123–134 [DOI] [PubMed]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J (2003b) Incorporation of aminoacyl-tRNA into the ribosome as seen microscopy by cryo-electron. Nat Struct Mol Biol 10(11):899–906 [DOI] [PubMed]
- Vendruscolo M, Dobson CM. Protein dynamics: Moore’s law in molecular biology. Current Biol. 2011;21(2):R68–R70. doi: 10.1016/j.cub.2010.11.062. [DOI] [PubMed] [Google Scholar]
- Vicatos S, Rychkova A, Mukherjee S, Warshel A. An effective coarse-grained model for biological simulations. Recent refinements and validations. Proteins. 2013;82(7):1168–1185. doi: 10.1002/prot.24482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa E, Sengupta J, Trabuco L, Lebarron J, Baxter W, Shaikh T, Grassucci R, Nissen P, Ehrenberg M, Schulten K, Frank J. Ribosome-induced changes in elongation factor tu conformation control GTP hydrolysis. Proc Nat Acad Sci USA. 2009;106:1063–1069. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford PC, Sanbonmatsu KY. Simulating movement of tRNA through the ribosome during hybrid-state formation. J Chem Phys. 2013;139(12):121919. doi: 10.1063/1.4817212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford PC, Blanchard SC, Cate JHD, Sanbonmatsu KY. Connecting the kinetics and energy landscape of tRNA translocation on the ribosome. PLoS Comp Biol. 2013;9(3):e1003003. doi: 10.1371/journal.pcbi.1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford PC, Noel JK, Gosavi S, Schug A, Sanbonmatsu KY, Onuchic JN (2009) An all-atom structure-based potential for proteins: bridging minimal models with all-atom empirical forcefields. Proteins 75(2):430–441 [DOI] [PMC free article] [PubMed]
- Whitford PC, Geggier P, Altman RB, Blanchard SC, Onuchic JN, Sanbonmatsu KY. Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. RNA. 2010;16:1196–1204. doi: 10.1261/rna.2035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford PC, Onuchic JN, Sanbonmatsu KY. Connecting energy landscapes with experimental rates for aminoacyl-tRNA accommodation in the ribosome. J Am Chem Soc. 2010;132:13170–13171. doi: 10.1021/ja1061399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford PC, Ahmed A, Yu Y, Hennelly SP, Tama F, Spahn CMT, Onuchic JN, Sanbonmatsu KY. Excited states of ribosome translocation revealed through integrative molecular modeling. Proc Nat Acad Sci USA. 2011;108:18943–18948. doi: 10.1073/pnas.1108363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford PC, Sanbonmatsu KY, Onuchic JN. Biomolecular dynamics: order-disorder transitions and energy landscapes. Rep Prog Phys. 2012;75:076601. doi: 10.1088/0034-4885/75/7/076601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Blachowicz L, Makowski L, Roux Benoît. Multidomain assembled states of Hck tyrosine kinase in solution. Proc Nat Acad Sci USA. 2010;107(36):15757–62. doi: 10.1073/pnas.1004569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Sanbonmatsu KY, Voth GA. Key intermolecular interactions in the E. coli 70S ribosome revealed by coarse-grained analysis. J Amer Chem Soc. 2011;133(42):16828–16838. doi: 10.1021/ja2028487. [DOI] [PMC free article] [PubMed] [Google Scholar]