Abstract
Src homology 3 (SH3) domains are involved in the regulation of important cellular pathways, such as cell proliferation, migration and cytoskeletal modifications. Recognition of polyproline and a number of noncanonical sequences by SH3 domains has been extensively studied by crystallography, nuclear magnetic resonance and other methods. High-affinity peptides that bind SH3 domains are used in drug development as candidates for anticancer treatment. This review summarizes the latest achievements in deciphering structural determinants of SH3 function.
Keywords: Three-dimensional structure, SH3 domain, Cell signaling, Src-family, Myosin, Protein conformation
Introduction
Many proteins, such as protein tyrosine kinases (PTKs) of the Src-family, myosin, cortactin, amphiphysin and spectrin, carry small modules named Src homology 3 (SH3) domains comprising approximately 60 amino acids. SH3 domains are present in all eukaryotes (Gmeiner and Horita 2001; Kaneko et al. 2008) and viruses, and SH3-like domains are found in prokaryotes (Whisstock and Lesk 1999) as, for example, part of Photosystem I (Falzone et al. 1994) or as Thermotoga maritima histidine kinase CheY (Bilwes et al. 1999).
As one of six domains of Src-family PTKs, the SH3 domain carries out significant roles in substrate recognition, membrane localization and regulation of the kinase activity (Gmeiner and Horita 2001; Sriram et al. 2011). SH3 domains bind proline-rich sequences, particularly those carrying the PxxP motif that have the left-handed polyproline 2 (PPII) conformation (Mayer 2001; Musacchio 2002). Two classes of peptide ligands have been identified: class I binds with the consensus sequence RXLPPXP, and class II binds with the consensus sequence XPPLPXR (Feng et al. 1994)—in opposite orientations. In addition, some sequences have been found to deviate from canonical binding. The specificity of the peptide-binding pocket of SH3 in SH3–peptide complexes has been studied by crystallography and nuclear magnetic resonance (NMR) to identify atomic groups critical for interaction. This review summarizes the achievements made in elucidating the structure and function of SH3 domains with an emphasis on the role of the SH3 domain in the deregulation of signaling pathways.
Structure
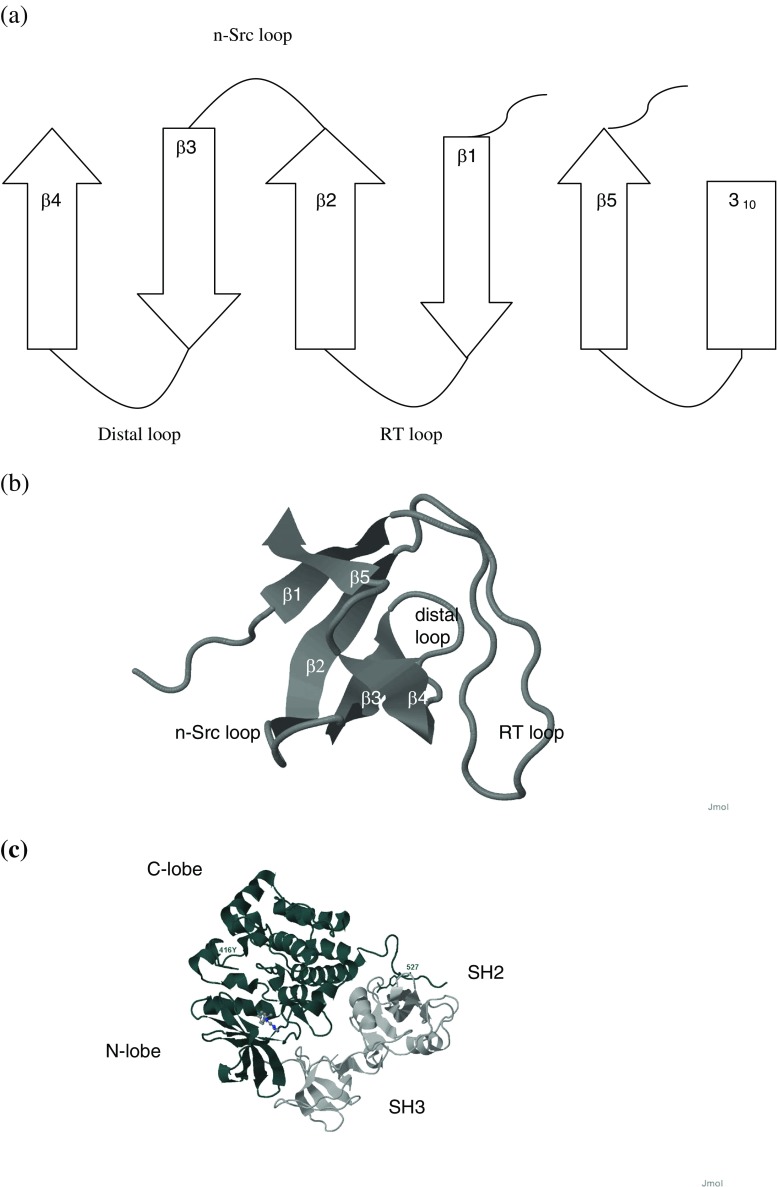
The structure of the SH3 domain in isolated state and surrounded by protein, ligand conformation and specificity of the peptide-binding pocket of SH3 have been extensively studied by crystallography and NMR. The structure of SH3 domain comprises five to eight β-strands arranged into two antiparallel β-sheets or in a β-barrel (for review, see Kaneko et al. 2008). Loops, denoted as RT, Src, and distal loops, respectively, play important roles in the recognition of binding partners (Fig. 1).
Fig. 1.
Structure of the Src homology 3 (SH3) domain. a Topology of secondary structure elements, b tertiary fold, c hematopoetic cell kinase (Hck) (Protein Database code: 1qcf): kinase domain (dark gray) with tandem SH2 and SH3 domains (light gray)
Secondary structure packing subdivides protein β-sheet arrangements into two major categories. The distribution of angles between the strands of the two β-sheets shows peaks around −30 ° or approximately 90 ° (Chothia and Janin 1981). The first group of proteins includes concanavalin A, plastocyanin, γ-crystallin, superoxide dismutase, prealbumin and immunoglobulin, and the second group comprises such proteins as trypsin and staphylococcal nuclease. Two β-sheets of SH3 domains that are almost perpendicular to each other belong to the second group. Although many SH3 domains are soluble, some are difficult to isolate. For example, the Bem1 SH3 domain is insoluble and forms a functional module only together with its C-terminal CI region (Takaku et al. 2010). The amino-terminal β-strand can be aggregation-prone, and the absence of the carboxyl terminus leads to fibril formation (Neudecker et al. 2012).
Ligands bind the SH3 domain in the PPII conformation. Polyproline helix is a major structural motif of collagen that contains three residues per turn (Pauling and Corey 1951; Shoulders and Raines 2009). Its unique feature is the presence of twofold rotational pseudo-symmetry that influences the ability of ligands to bind the SH3 domain in two orientations (Lim et al. 1994; Kaneko et al. 2008). Proline-rich sequences are the most abundant sequences of the proteome (Chandra et al. 2004) and in addition to SH3 are also recognized by the WW, Enabled/vasodilator-stimulated protein homology EVH1, GYF, profilin, ubiquitin E2 variant UEV and CYP domains (Shawn 2005). Some ligands also bind SH3 domains in the 310 helix conformation (Kaneko et al. 2008) or involve both PPII and the 310 helix (Ghose et al. 2001). The packing properties of secondary structure elements have a significant impact on protein assemblies (Kurochkina 2010) and give rise to new questions concerning the involvement of protein modules in the regulation of cellular processes.
Function
SH3 domains carry out a variety of activities. The SH3 domain of Src-family PTKs, which regulate many cellular functions, such as cell proliferation and differentiation, survival, migration and cytoskeletal modifications, is mainly involved in substrate recognition and downregulation of the kinase activity. Phosphorylation of the RT loop of SH3 domain in an adaptor protein can positively regulate kinase activity (Sriram et al. 2011). In myosins, the SH3 domain regulates stability and motility. Interaction of the N-terminal proline-rich extension of the essential light chain with the actin SH3 domain during the ATPase cycle modulates myosin II kinetics (Lowey et al. 2007). The binding of the amphiphysin SH3 domain to the proline-rich region of dynamin regulates the multimerization cycle of dynamin in endocytosis, mainly its ring assembly. This process may be similar to the mediation of actin polymerization by profilin (Owen et al. 1998). The generation of multimeric complexes, such as SLP-76, VAV, and Nck1 association via the C-terminal SH3 domain of Nck1 and the N-terminal SH3 domain of VAV induced by T-cell activation, is critical for actin polymerization (Barda-Saad et al. 2010). Translocation of the regulatory Phox complex to the catalytic core of the NADPH oxidase leads to the assembly and activation of the holoenzyme and occurs through co-operative interactions mediated by the SH3 and PX domains (Shawn 2005). Many adaptor molecules, such as the cas ligand, with multiple SH3 domains (Yao et al. 2007) form a scaffold for the assembly of proteins. SH3 domains of pathogens are possibly associated with invasiveness and survival (Whisstock and Lesk 1999; Lim et al. 2011).
Specificity of binding
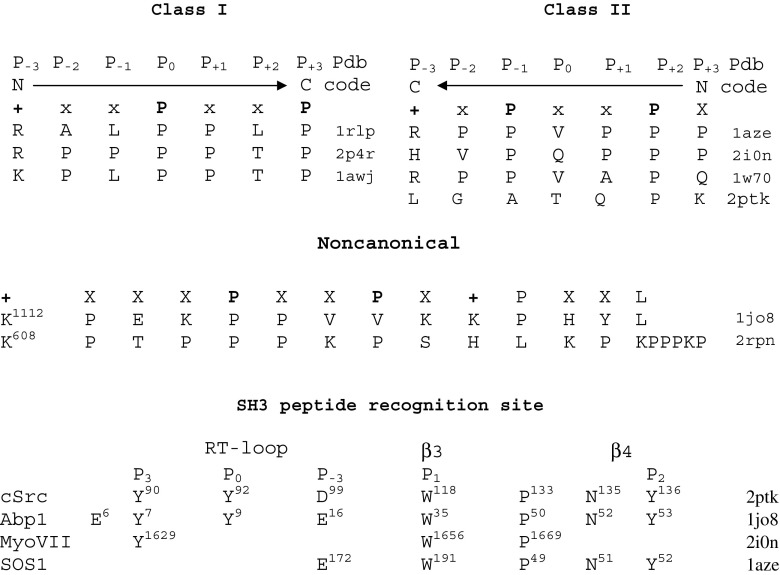
SH3 domains have been identified as protein modules that recognize proline-rich sequences (Mayer et al. 1988; Kaneko et al. 2008). The peptide sequence qPxqP, which binds to the SH3 domain, can be represented as two qP dipeptides linked by any residue x where q is a hydrophobic residue (Kay et al. 2000). Proline-rich sequences binding SH3 carry the PxxP motif which has the left-handed PPII conformation (Musacchio 2002). Flanking residues have been shown to enhance interactions (Feng et al. 1995). Hydrophobic residues flanking PxxP residues contribute to the binding of the Csk SH3 domain and proline-enriched phosphatase (PEP) protein (Gregorieff et al. 1998; Ghose et al. 2001). Two classes of peptide ligands have been identified (Fig. 2): class I peptide ligands bind with the consensus sequence RXLPPXP, and class II ligands bind with the consensus sequence XPPLPXR in the opposite orientation. There is a preference for proline in positions P0 and P3 of class I ligands and in positions P-1 and P2 of class II ligands (Lim et al. 1994; Dalgarno et al. 1998). The canonical binding site of the SH3 domain involves the region between the RT loop and the Src loop. The binding surface can be subdivided into three or four pockets (Lim et al. 1994; Kay et al. 2000; Yao et al. 2007), with each of two pockets recognizing one ɸP (or xP) dipeptide unit, and the third/forth pocket binding positively charged flanking residues (Kay et al. 2000; Shi et al. 2010). Recognition of PxxP mainly occurs with the participation of conserved hydrophobic residues, such as Y1629, W1656 and P1669 of myosin VII (Wang et al. 2007) that correspond to Y90 (P3), W118 (P1), and P133 of Src, respectively. Similarly, a comparison between spectrin and FGR- and yes-related novel kinase (Fyn), members of the Src-family PTKs, shows that they possess a conserved hydrophobic patch composed mainly of aromatic residues, but differ in terms of amino acid sequence and conformation of the two important loops that likely mediate binding specificity of a patch region (Noble et al. 1993). In some proteins, the SH3 site involves the tip of RT loop.
Fig. 2.
Specificity of the SH3 binding pocket. Class I and Class II peptides bind in opposite orientations. Position P-3 involves the positively charged residue (R/K/H) responsible for orientation. Noncanonical binding may involve flanking positive residues from both sides. The recognition site of SH3 carries mainly hydrophobic residues and contains three to four pockets
The mutation of each of the prolines of the PxxP sequence to alanine impairs binding (Kaneko et al. 2008), while the mutation of proline to N-substituted glycine (sarcosine) or N-(S)-phenylethyl preserves or even improves binding (Nguyen et al. 1998; Kaneko et al. 2008). The cyclic side chain that is covalently bound to backbone nitrogen, rigidity of backbone conformation and restrictions on the torsion angles of the preceding residue, preference for several prolines to adopt the characteristic polyproline conformation, which places prolines on the same face of the helix all explain the importance of proline as a key residue in SH3 recognition (Kay et al. 2000).
Noncanonical binding is observed in 53BP2, in which ankyrin repeats and the SH3 domain form a module that binds to p53 (Gorina and Pavletich 1996), immune cell adaptor SKAP55 interacting with the RKxxYxxY motif (Kang et al. 2011), EPS8 recognizing PxxDY (Mongiovi et al. 1999; Kesti et al. 2007), GADS RxxK (Harkiolaki et al. 2003), CIN85/CMS SH3 (Moncalián et al. 2006) and βPIX SH3 (Janz et al. 2007) interacting with PXXXPR. More sequences have been identified, including cortactin SH3 binding RxxPxxxP found in the cytoplasmic region of the calcium activated potassium (BK) channel (Tian et al. 2006) and FUS1 binding Arg–Ser-rich sequences (Tong et al. 2002). Two new sequences have been found for Saccharomyces cerevisiae actin binding protein involved in the organization of actin cytoskeleton Abp1p-SH3, +PX(P/V/L)XXPX + and PXXPXRPXW(L/I). SH3 mediates the Abp1p association with Ser/Thr kinases Prk1p and Ark1p that plays a central role in their localization to actin cortical patches (Fazi et al. 2002).
Additional factors may influence SH3–peptide interactions. All SH3 residues of the Islet brain 1 (IB1) protein involved in dimerization are also those that usually interact with PPII ligands (Kristensen et al. 2006). In contrast, Dictyostelium discoideum myosin VII interacts with its own adjacent proline-rich (PxxP) region at the site distant from the conserved PxxP binding site (Wang et al. 2007).
Recognition of non-PxxP peptide ligands (Kami et al. 2002) occurs in several modes. The binding site of Grb2 SH3 partially overlaps the PxxP site, but not the p67phox and Pex13 sites (Kami et al. 2002). C-terminal Src kinase (Csk) interacts with the proline/glutamic acid/serine/threonine-rich domain (PEST) domain of PEP via its SH3 domain. The PxxP canonical binding site of SH3 domains in the PPII conformation is a part of the PEST domain. In addition, the hydrophobic sequence C-terminal to the Pro-rich site is required, and the region of the PEP peptide bound to the Csk–SH3 in the PPII conformation is followed by a 310 helix that is critical for the association (Ghose et al. 2001).
Adaptor protein Bem1 provides an example of where the binding site of the proline-rich region extends to the C-terminus and, in addition to the SH3 domain, utilizes the CI region (Takaku et al. 2010). Interestingly, the tyrosine kinase, spectrin and myosin SH3 show similar structures and mode of peptide binding, with the formation of several hydrogen bonds and the interactions exhibiting a mainly hydrophobic character (Fig. 1). Myosin 3 SH3 in complex with Abo-peptide BP1 (type I) and Abl kinase in complex with the same peptide provide examples of just such a similarity (Musi et al. 2006).
The binding of SH3 domains is highly specific. Therefore, proline-rich sequences confer target specificity to regulate protein–protein interactions.
Tyrosine kinase SH3
Src-family PTKs regulate many cellular functions, such as cell proliferation and differentiation, survival, migration and cytoskeletal modifications. They are also highly active in epithelial cancers. Nine members of the Src-family PTKs are Frk, Src, Fyn, Yes (most human tissues), Blk, Fgr, hematopoetic cell kinase (Hck), Lck, and Lyn (hematopoietic cells, Lck and Lyn also in neurons) (Manning et al. 2002). Four domains of Src family PTKs include the N-terminal membrane-targeting SH4, SH3 and SH2 protein–protein interactions modules, the SH1 catalytic domain and the C-terminal regulatory tail with a phosphotyrosine. The important role of the SH3 domain in these proteins has been demonstrated in mutational studies in which mutations in SH3 domains—as well as deletions of the entire SH3 domain of Abl or Src—resulted in activation of the transforming potential of the kinases (Jackson and Baltimore 1989; Kato et al. 1986; Seidel-Dugan et al. 1992; Noble et al. 1993). Several of these SH3 and SH2 interactions were found to be modulated by tyrosine phosphorylation. Phosphorylated C-terminal tyrosine of the kinase binds to its own SH2 domain, and this event results in an inactive state of the kinase. Activation occurs upon dephosphorylation of the tyrosine or conformational change induced by the binding of another protein. Carboxyl-terminal Src kinase, Csk, which contains the kinase, SH2 and SH3 domains, phosphorylates a regulatory tyrosine and serves as a negative regulator of Src-family PTKs. SH2 kinase and SH2–SH3 linkers interact with the N-lobe and stabilize the active conformation of the kinase, whereas in the inactive molecule this interaction is absent due to rotation of the SH2 domain (Ogawa et al. 2002). Phosphorylation of Y251 in the RT loop of the SH3C domain of Abl kinase results in transactivation of the Abl kinase (Sriram et al. 2011). Phosphotyrosine-mediated signaling involves numerous targets that can be identified by such methods as global phosphoproteomic based on quantitative mass spectrometry (Guha et al. 2008). Such unbiased approaches have the potential to identify sites of phosphorylation and quantify the degree of phosphorylation at specific sites of the interaction interface of protein modules, such as the SH3 and SH2 domains.
The three-dimensional (3D) structure of the SH3 domains from tyrosine kinases Lyn (Bauer et al. 2005), Lck (Yamaguchi and Hendrickson 1996; Nasertorabi et al. 2006), Hck (Horita et al. 1998), Src (Xu et al. 1997; Witucki et al. 2002; Cowan-Jacob et al. 2005), Fyn (Lee et al. 1996, Csk (Ghose et al. 2001; Ogawa et al. 2002) and Abl (Nam et al. 1996) have been determined.
Many non-receptor protein tyrosine kinases, such as Src, Janus and ZAP70, use SH2 and/or SH3 modules to regulate their activity via mediation of protein–protein interactions. The activity of Src, PI3 and Itk kinases, for example, is regulated by intramolecular interactions between their SH3 domain and internal proline-rich sequences (Koch et al. 1991; Kapeller et al. 1994; Wang et al. 2007).
Activated CDC42-associated kinase (Ack1) is a non-receptor protein tyrosine kinase activated via tyrosine phosphorylation upon cell stimulation with epidermal growth factor (EGF) or platelet-derived growth factor (PDGF) in response to the activation of integrins by cell adhesion on fibronectin. This kinase contains a membrane-targeting domain and a CDC42 binding (CRIB) domain necessary for its autophosphorylation, a catalytic kinase domain, an autoinhibitory SH3 domain, a Ralt homology region and a proline-rich region. The site of Ack1 tyrosine phosphorylation interacts with the SH2 domain of the adaptor protein Nck. Ack1 binds CDC42 (Galisteo et al. 2006), a member of the Rho family of Ras-related GTP-binding proteins that also include Rho and Rac. These proteins function as molecular switches between GDP-bound (inactive) and GTP-bound (active) states involved in the regulation of actin cytoskeleton (Rudolph et al. 1999). CDC42 induces autophosphorylation of the activation loop and membrane targeting of mixed lineage kinase 3, a kinase that is autoinhibited by its SH3 domain, and a proline-rich region located between its leucine zipper and CRIB domains (Du et al. 2005). RhoU, an atypical member of the CDC42 family, contains proline-rich N-terminal and C-terminal extensions. Co-localization of RhoU with EGFR upon EGF stimulation requires both extensions and GRB2. Mutations within proline-rich sequences impair the association of RhoU with EGFR. Growth factor receptor signaling is regulated by the RhoU interaction with SH3 adaptor proteins (Zhang et al. 2011).
Many scaffold and adaptor proteins also utilize SH3 domains to enhance or inhibit activity of their kinase partners. S. cerevisiae Bem1, a scaffold protein essential for the establishment of cell polarity, binds to CDC42 via a module comprising its second SH3 domain and C-terminal flanking region CI. By binding both the Ste20 proline-rich region (via SH3) and CDC42 (via CI), Bem1 facilitates CDC42 binding to Ste20 CRIB and kinase activation. The Bem1 SH3 domain is insoluble, and the CI region alone does not interact with CDC42; only together they make a functional binding module (Takaku et al. 2010).
Myosin SH3
Many myosins contain SH3 domain and for some function is not known (Foth et al. 2005). The tail regions of myosins I, IV, VII, XII, and XV each contain a putative SH3 domain that may be involved in protein–protein interactions (Wang et al. 2007).
The crystallographic structure of the SH3 domains of several myosins has been determined, including that of myosin 6 (Ménétrey et al. 2008), myosin S1 (Himmel et al. 2002), myosin II (Bauer et al. 2000), muscle myosin (Rayment et al. 1993), smooth muscle myosin (Dominguez et al. 1998; Wendt et al. 2001), myosin S1 (Yang et al. 2007), myosin 3, myosin 5, myosin VII (Wang et al. 2007), myosin 1E (Protein Database code 2xmf), myosin IB and myosin VIIa (Wu et al. 2011).
The motor domain of D. discoideum myosin IE, in contrast to that of smooth muscle myosin and myosin II, lacks the N-terminal SH3-like domain, possibly leading to the increase in the angle of the lever arm by 30 ° and to a larger power stroke. Removal of the SH3 domain affects the interaction with the nucleotide and actin, leading to the reduction of in vitro motility and the loss of actin-activated ATPase activity (Fujita-Becker et al. 2006).
Three domains of myosin VIIa coiled coil, FERM, and SH3 contribute to dimerization of its heavy chain (Kiehart et al. 2004). Some of the mutations that involve the SH3 domain of myosin VII are associated with hearing defects in humans (Wu et al. 2011). Structural studies of S. cerevisiae myosin 3 and homologous myosin 5 SH3 domains have been performed since mutations in human orthologs of these proteins lead to Wiskott–Aldrich syndrome, which is linked to deficiencies in actin polymerization (Musi et al. 2006).
Regulation
The SH2 and SH3 domains are involved in the downregulation or upregulation of the activity of c-Src family PTKs (Gmeiner and Horita 2001; Sriram et al. 2011). The mechanism of regulation of several kinases, including Lck, Hck and c-Src, has been studied by crystallography and NMR (Gaul et al. 2000), and the results suggest that along with phosphorylation of the activation loop, kinases may be regulated by binding or dissociation of their regulatory domains (Ghose et al. 2001; Huse and Kuriyan 2002). The relative movement of the kinase N-lobe and C-lobe results in open (active) and closed (inactive) conformations with the respective changes in the position of two α-helices, namely the C-helix (N-lobe) and G-helix (C-lobe). In active conformation, the N – and C-lobes of the kinase domain allow the γ-phosphate of ATP to be aligned with the substrate acceptor group. In Hck, α-helix C interacts with the divalent metal ions bound to ATP. The active conformation of the kinase is acquired via phosphorylation of Tyr416 (Gmeiner and Horita 2001).
The SH3 and SH2 domains are often present in tandem as, for example, in Fyn (Arold et al. 2001), Abl (Jones et al. 2000), Grb2 (Maignan et al. 1995), Lck (Nasertorabi et al. 2006), p21ras GTPase activating protein (GAP) (Trahey et al. 1988), phospholipase Cγ (Stahl et al. 1988), subunit of phosphatidyl inositol 3′ kinase (Otsu et al. 1991) and Sem5 (Clark et al. 1992). In the absence of the kinase domain the linker region possibly does not adopt a PPII conformation since it does not bind SH3 (Alvarado et al. 2010).
EGF receptor (EGFR) signaling regulates cell motility, differentiation, proliferation and survival. Ligand-induced asymmetric dimerization of EGFR activates the EGFR tyrosine kinase and trans-phosphorylation that turns on specific kinase signaling cascades. An adaptor protein, P130CAS, mediates integrin–EGFR interactions. The SH3 domain of P130CAS interacts with proline-rich regions of dynamin, leading to inhibition of dynamin phosphorylation and EGFR internalization, the major pathway by which EGFR is downregulated (Kang et al. 2011).
SH3 domains are regulated by association with other proteins, phosphorylation, dimerization, and cis–trans isomerization (Sarkar et al. 2011). The link between the structural motif and regulation by phosphorylation highlights four motifs: WW2, SH2, SH3, and PDZ (Akiva et al. 2012). Regulation by the association of SH3 domains also occurs via the formation of homodimers (IB1 protein; Kristensen et al. 2006) or heterodimers (VAV N-terminal and GRB2 C-terminal SH3 domains complex; Nishida et al. 2001). In IB1 protein, the dimer interface and PxxP binding occupy the same site even though the IB1 protein does not contain the PxxP motif; therefore, ligands compete with IB1 protein. All of the SH3 residues of IB1 involved in dimerization are also those that usually interact with PPII ligands (Kristensen et al. 2006).
Vav, a guanine exchange factor for the Rho/Rac family in hematopoietic cells, binds via its SH3 domain to the Grb2 SH3 domain. The RT loop of Vav contains a tetraproline sequence in PPII conformation that can bind to the Vav recognition site for proline-rich peptides and therefore blocks access of ligands (Nishida et al. 2001).
Phosphorylation of the SH3 domain and adjacent regions correlates with the transient activation and hyperphosphorylation of PTKs (Park et al. 1996; Broome and Hunter 1997). Enzymes (c-Src, Btk, Abl) and adaptor proteins (Crk, Nck) carrying SH3/SH2 domains exhibit similarities in their mode of interaction (Broome and Hunter 1997). Overexpression of the CT10 regulator of kinase (CRKII), which has been found in several human cancers, is associated with an aggressive phenotype of oral squamous cell carcinoma. CRKII regulates cell migration, morphogenesis, invasion, phagocytosis and survival (Yamada et al. 2011). Two isoforms, CrkI (SH2–SH3) and CrkII (SH2–SH3–SH3), differ in their transforming activity, which is high for CrkI and low for CrkII. Abl phosphorylates Y221 of CrkII in the linker region between two SH3 domains, thereby creating a binding site for the SH2 domain. Phosphorylated CrkII shows no activity, possibly due to the detachment of CrkII from the SH3- and SH2- binding proteins (Kobashigawa et al. 2007). Conformational change induced by phosphotyrosine recognition promotes binding of the SH3 domain of Abl tyrosine kinase to the DE loop of the SH2 domain (Donaldson et al. 2002). The proposed role of the C-terminal SH3 domain of CrkII is negative regulation of CrkII activity (Muralidharan et al. 2006). The C-terminal SH3 of Crk is phosphorylated in its RT loop at Y251 by Abl kinase; it binds to Abl SH2 and promotes Abl kinase transactivation (Sriram et al. 2011). The conserved sequence P248NAY in the RT loop combined with some mutations might play a role in the activation of Abl kinase (Reichman et al. 2005). Since CrkII also carries the SH2 domain which recruits downstream targets and the Abl family of kinases, a delayed feedback loop is formed (Antoku and Mayer 2009). Abl interactor proteins Abi1 and Abi2 regulate c-Abl kinase activity by interacting with the C-terminal PxxP sequences and c-Abl SH3/SH2 domains. Phosphorylation of the Y213 of Abi1 by c-Abl kinase is followed by the binding of Abi1 to the Abl SH2 domain and subsequent inhibition of c-Abl kinase activity (Xiong et al. 2008). Abi2 binds to c-Abl via dual proline-rich sequence/SH3 domain interaction and contacts between phosphorylated tyrosine of Abi2 and c-Abl SH2 (Dai and Pendergast 1995; Shi et al. 2010). Phosphorylation of the SH3 domain of Abi-1 mediates Abl–Abi1 interaction and contributes to altered cell adhesion (Sato et al. 2011).
To summarize, the importance of SH3 in cell biology is demonstrated by its key role as a mediator of signal regulation. Such vital processes as migration and invasiveness (Yamada et al. 2011), actin reorganization induced by extracellular signals (Antoku and Mayer 2009), shaping spines (Sheng and Kim 2000; Ehlers 2002), among others take a central role when new treatments rely on an understanding of cellular control mechanisms.
Significance and potential for drug design
As one of six domains of the Src-family of PTKs, the SH3 domain plays a role in substrate recognition, membrane localization, and regulation of the kinase activity (Gmeiner and Horita 2001; Sriram et al. 2011). In addition to its role as an adaptor in signal transduction, SH3 is involved in the regulation of enzymatic activity via conformational changes and the recruitment of substrates in cellular compartments, very often acting together with SH2 domain (Vidal et al. 2001).
Malfunction of the SH3 domain has a significant impact on such important processes as p53-mediated apoptosis and DNA repair (Jiang et al. 2001), adhesion-dependent regulation by tyrosine phosphorylation (Moore and Winder 2010), stimulation of mesenchymal stem cell migration, which is important for hypoxic solid tumor development (Proulx-Bonneau et al. 2011), osteoblast maturation and consequently bone formation (Levaot et al. 2011), the assembly of amyloid fibrils and determination of their morphology (Morel et al. 2010), processes associated with a number of neurodegenerative diseases, such as Alzeimer’s (Morel et al 2010), tyrosine phosphatase signaling that affects SH3 binding in patients with rheumatoid arthritis (Bilwes et al. 1999), bacteria adhering to host cells (Queval et al. 2011) and contraction-induced injuries (Banks et al. 2010). Mutations in the SH3 domain of unconventional myosin VIIa have recently been shown to cause deafness in humans, with one mutation, A1628S, located directly in its SH3 domain (Wu et al. 2011). The participation of modular protein domains in the regulation of signaling cascades of G protein-coupled receptors and receptor tyrosine kinases facilitates cross-talk between receptor families (Janz et al. 2007). The acquisition of new SH3 domains leads to a new function (Jefferson et al. 2007).
The concept of exploiting protein–protein interaction for drug design has been contemplated by scientists for decades. SH3 and SH2 domains have been studied as the targets of anti-proliferative agents. As the domains of adaptor proteins and such enzymes as kinases, phosphatases and lipases, they serve as recognition modules that are important for cell signaling. Since these domains can also be found in oncoproteins and proteins overexpressed in deregulated signaling pathways of tumors, they are important candidates for the design of antitumor drugs that potentially could be targeted to disrupt the interaction of SH3 and SH2 modules. Sequences designed for SH2/SH3 recognition may be used for inhibiting proliferative signaling activated by tyrosine kinases (Vidal et al. 2001). Such an approach has been undertaken in the design of anti-proliferative drugs against chronic myelogenous and acute lymphoblastic leukemias (BCR/Abl) and breast and ovarian cancer (Her-2/Neu) (Smithgall 1995). The inhibitors designed for SH2 domains appear to be more specific compared to those of SH3 domains. The attempt to compensate for this shortcoming of low specificity of SH3 domains was made for GRB2 by designing a dimeric peptide recognizing a dimeric SH3 domain. Other candidates for anticancer treatment based on high-affinity peptides that bind the SH3 domain include the Crk adaptor (Feller and Lewitzky 2006) and the Grb2 and Crk SH2/SH3 (Kardinal et al. 1999; Kadaveru et al. 2009). Further elucidation of the structure and function of SH3 domains interacting with their partners and the development of new peptide-based drug design approaches will facilitate the exploration of these important protein modular interaction domains as drug targets.
Acknowledgments
Conflict of interest
None.
Abbreviations
- Ack-1
Activated CDC42-associated kinase
- Csk
Carboxyl-terminal Src kinase
- Fyn
FGR and yes-related novel kinase
- Hck
Hematopoetic cell kinase
- IB1
Islet brain 1
- PEP
Proline-enriched phosphatase
- PEST
Proline/glutamic acid/serine/threonine-rich domain
- PTK
Protein tyrosine kinase
- SH3
Src homology 3
References
- Akiva E, Friedlander G, Itzhaki Z, Margalit H. A dynamic view of domain-motif interactions. PloS Comp Biol. 2012;8:e1002341. doi: 10.1371/journal.pcbi.1002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado JJ, Betts L, Moroco JA, Smithgall TE, Yeh JI. Crystal structure of the Src family Kinase Hck SH3-SH2 linker regulatory region supports an SH3-dominant activation mechanism. J Biol Chem. 2010;285:35455–35461. doi: 10.1074/jbc.M110.145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoku S, Mayer BJ. Distinct roles for Crk adaptor isoforms in actin reorganization induced by extracellular signals. J Cell Sci. 2009;122:4228–4238. doi: 10.1242/jcs.054627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arold ST, Ulmer TS, Mulherni TD, Werner JM, Ladbury JE, Campbell ID, Noble MEM. The role of the Src homology 3-Src homology 2 interface in the regulation of Src kinases. J Biol Chem. 2001;276:17199–17205. doi: 10.1074/jbc.M011185200. [DOI] [PubMed] [Google Scholar]
- Banks P, Franks NP, Dickinson R. Competitive inhibition at the glycine site of the N-methyl-D-aspartate receptor mediates xenon neuroprotection against hypoxia-ischemia. Anesthesiology. 2010;112:614–622. doi: 10.1097/ALN.0b013e3181cea398. [DOI] [PubMed] [Google Scholar]
- Barda-Saad M, Shirasu N, Pauker MH, Hassan N, Perl O, Balbo A, Yamaguchi H, Houtman JCD, Appella E, Schuck P, Samelson LE. Cooperative interactions at the SLP-76 complex are critical for actin polymerization. EMBO J. 2010;29:2315–2328. doi: 10.1038/emboj.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer CB, Holden HM, Thoden JB, Smith R, Rayment I. X-ray structures of the apo and MgATP-bound states of Dictyostelium discoideum myosin motor domain. J Biol Chem. 2000;275:38494–38499. doi: 10.1074/jbc.M005585200. [DOI] [PubMed] [Google Scholar]
- Bauer F, Schweimer K, Meiselbach H, Hoffmann S, Rösch P, Sticht H. Structural characterization of Lyn-SH3 domain in complex with a herpes viral protein reveals an extended recognition motif that enhances binding affinity. Prot Sci. 2005;14:2487–2498. doi: 10.1110/ps.051563605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilwes AM, Alex LA, Crane BR, Simon MI. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/S0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- Broome MA, Hunter T. The PDGF receptor phosphorylates Tyr 138 in the c-Src SH3 domain in vivo reducing peptide ligand binding. Oncogene. 1997;14:17–34. doi: 10.1038/sj.onc.1200798. [DOI] [PubMed] [Google Scholar]
- Chandra BR, Gowthaman R, Akhouri RR, Gupta D, Sharma A. Distribution of proline-rich (PxxP) motifs in distinct proteomes: functional and therapeutic implications for malaria and tuberculosis. Protein Eng Des Sel. 2004;17:175–182. doi: 10.1093/protein/gzh024. [DOI] [PubMed] [Google Scholar]
- Chothia C, Janin J. Relative orientation of close-packed,8-pleated sheets in proteins. Proc Natl Acad Sci USA. 1981;78:4146–4150. doi: 10.1073/pnas.78.7.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SG, Stern MJ, Horvitz HR. C. Elegans cell-signalling gene sem-5 encodes a protein with SH2 and SH3 domains. Nature. 1992;356:340–344. doi: 10.1038/356340a0. [DOI] [PubMed] [Google Scholar]
- Cowan-Jacob SW, Fendrich G, Manley PW, Jahnke W, Fabbro D, Liebetanz J, Meyer T. The crystal structure of a C-Src complex in an active conformation suggests possible steps in C-Src activation. Structure. 2005;13:861–871. doi: 10.1016/j.str.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Dai Z, Pendergast AM. Abi2, a novel SH3-containing protein interacts with the c-Abl tyrosine kinase and modulates c-Abl transforming activity. Genes Dev. 1995;9:2569–2582. doi: 10.1101/gad.9.21.2569. [DOI] [PubMed] [Google Scholar]
- Dalgarno DC, Botfield MC, Rickles RJ. SH3 domains and drug design: ligands, structure, and biological function. New York: John Wiley & Sons; 1998. [DOI] [PubMed] [Google Scholar]
- Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell. 1998;94:559–571. doi: 10.1016/S0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- Donaldson LW, Gish G, Pawson T, Kay LE, Forman-Kay JD. Structure of a regulatory complex involving the Abl SH3 domain, the Crk SH2 domain, and aCrk-derived phosphopeptide. Proc Natl Acad Sci USA. 2002;99:14053–14058. doi: 10.1073/pnas.212518799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, BC Bock, Schachter KA, Chao M, Gallo KA. CDC42 Induces activation loop phosphorylation and membrane targeting of mixed lineage kinase 3. J Biol Chem. 2005;280:42984–42993. doi: 10.1074/jbc.M502671200. [DOI] [PubMed] [Google Scholar]
- Ehlers MD. Molecular morphogens for dendritic spines. Trends Neurosci. 2002;25:64–67. doi: 10.1016/S0166-2236(02)02061-1. [DOI] [PubMed] [Google Scholar]
- Falzone CJ, Kao Y-H, Zhao J, Bryant DA, Lecomte JTJ. Three-dimensional solution structure of PsaE from the Cyanobacterium synechococcus sp. strain PCC 7002, a photosystem I protein that shows structural homology with SH3 domains. Biochemistry. 1994;33:6052–6062. doi: 10.1021/bi00186a004. [DOI] [PubMed] [Google Scholar]
- Fazi B, Jamie M, Cope TV, Douangamath A, Ferracuti S, Schirwitz K, Zucconi A, Drubin DG, Wilmanns M, Cesareni G, Castagnoli L. Unusual binding properties of the SH3 domain of the yeast actin-binding protein Abp1. J Biol Chem. 2002;277:5290–5298. doi: 10.1074/jbc.M109848200. [DOI] [PubMed] [Google Scholar]
- Feller SM, Lewitzky M. Potential disease targets for drugs that disrupt protein-protein interactions of Grb2 and Crk family adaptors. Curr Pharmacol Des. 2006;12:529–548. doi: 10.2174/138161206775474369. [DOI] [PubMed] [Google Scholar]
- Feng S, Chen JK, Yu H, Simon JA, Schreiber SL. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- Feng S, Kasahara C, Rickles RJ, Schreiber SL. Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands. Proc Natl Acad Sci USA. 1995;92:12408–12415. doi: 10.1073/pnas.92.26.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proc Natl Acad Sci USA. 2005;103:3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita-Becker S, Tsiavaliaris G, Ohkura R, Shimada T, Manstein DJ, Sutoh K. Functional characterization of the N-terminal region of myosin-2. J Biol Chem. 2006;281:36102–36109. doi: 10.1074/jbc.M605171200. [DOI] [PubMed] [Google Scholar]
- Galisteo ML, Yang Y, Ureña J, Schlessinger J. Activation of the nonreceptor protein tyrosine kinase Ack by multiple extracellular stimuli. Proc Natl Acad Sci USA. 2006;103:9796–9801. doi: 10.1073/pnas.0603714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaul BS, Harrison ML, Geahlen RL, Burton RA, Post CB. Substrate recognition by the Lyn protein-tyrosine kinase. J Biol Chem. 2000;275:16174–16182. doi: 10.1074/jbc.M909044199. [DOI] [PubMed] [Google Scholar]
- Ghose R, Shekhtman A, Goger MJ, Ji H, Cowburn D. A novel, specific interaction involving the Csk SH3 domain and its natural ligand. Nat Struct Biol. 2001;8:997–1004. doi: 10.1038/nsb1101-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gmeiner WH, Horita DA. Implications of SH3 domain structure and dynamics for protein regulation and drug design. Cell Biochem Biophys. 2001;35:127–140. doi: 10.1385/CBB:35:2:127. [DOI] [PubMed] [Google Scholar]
- Gorina S, Pavletich NP. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- Gregorieff A, Cloutier JF, Veillette A. Sequence requirements for association of protein-tyrosine phosphatase PEP with the Src homology 3 domain of inhibitory tyrosine protein kinase p50(csk) J Biol Chem. 1998;273:13217–13222. doi: 10.1074/jbc.273.21.13217. [DOI] [PubMed] [Google Scholar]
- Guha U, Chaerkady R, Marimuthu A, Patterson AS, Kashyap MK, Harsha HC, Sato M, Bader JS, Lash AE, Minna JD, Pandey A, Varmus HE. Comparisons of tyrosine phosphorylated proteins in cells expressing lung cancer-specific alleles of EGFR and KRAS. Proc Natl Acad Sci USA. 2008;105:14112–14117. doi: 10.1073/pnas.0806158105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkiolaki M, Lewitzky M, Gilbert RJC, Jones EY, Bourette RP, Mouchiroud G, Sondermann H, Moare I, Feller SM. Structural basis for SH3 domain-mediated high affinity binding between Mona/Gads and SLP-76. EMBO J. 2003;22:2571–2582. doi: 10.1093/emboj/cdg258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmel DM, Gourinath S, Reshetnikova L, Shen Y, Szent-Gyorgyi A-G, Cohen C. Crystallographic findings on the internally uncoupled and near-rigor states of myosin: further insights into the mechanics of the motor. Proc Natl Acad Sci USA. 2002;99:12645–12650. doi: 10.1073/pnas.202476799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horita DA, Baldisseri DM, Zhang W, Altieri AS, Smithgall TE, Gmeiner WH, Byrd RA. Solution structure of the human Hck SH3 domain and identification of its ligand binding site. J Mol Biol. 1998;278:253–265. doi: 10.1006/jmbi.1998.1690. [DOI] [PubMed] [Google Scholar]
- Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/S0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- Jackson P, Baltimore D. N-terminal mutations activate the leukemogenic potential of the myristoylated form of c-Abl. EMBO J. 1989;8:449–456. doi: 10.1002/j.1460-2075.1989.tb03397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janz JM, Sakmar TP, Min KC. A novel interaction between atrophin-interacting protein 4 and p21-activated kinase-interactive exchange factor is mediated by an SH3 domain. J Biol Chem. 2007;28:28893–28903. doi: 10.1074/jbc.M702678200. [DOI] [PubMed] [Google Scholar]
- Jefferson JJ, Ciatto C, Shapiro L, Liem RKH. Structural analysis of the plakin domain of bullous pemphigoid Antigen1 (BPAG1) suggests that plakins are members of the spectrin superfamily. J Mol Biol. 2007;366:244–257. doi: 10.1016/j.jmb.2006.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Axe T, Holgate R, Rubbi CP, Okorokov AL, Mee T, Milner J. P53 binds the nuclear matrix in normal cells: binding involves the proline-rich domain of p53 and increases following genotoxic stress. Oncogene. 2001;20:5449–5458. doi: 10.1038/sj.onc.1204705. [DOI] [PubMed] [Google Scholar]
- Jones RJ, Brunton VG, Frame MC. Adhesion-linked kinases in cancer; emphasis on Src, focal adhesion kinase and PI 3-kinase. Eur J Cancer. 2000;36:1595–1606. doi: 10.1016/S0959-8049(00)00153-2. [DOI] [PubMed] [Google Scholar]
- Kadaveru K, Vyas J, Schiller MR. Viral infection and human disease—insights from minimotifs. Front Biosci. 2009;13:6455–6471. doi: 10.2741/3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami K, Takeya R, Sumimoto H, Kohda D. Diverse recognition of non-PxxP peptide ligands by the SH3 domains from p67phox, Grb2 and Pex13P. EMBO J. 2002;21:4268–4276. doi: 10.1093/emboj/cdf428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Shawn LL, Li SC. The SH3 domain—a family of versatile peptide- and protein-recognition module. Front Biosci. 2008;13:4938–4952. doi: 10.2741/3053. [DOI] [PubMed] [Google Scholar]
- Kang YS, Kim W, Huh YH, Bae J, Kim JS, Song WK. P130Cas attenuates epidermal growth factor (EGF) receptor internalization by modulating EGF-triggered dynamin phosphorylation. PloSOne. 2011;6:e20125. doi: 10.1371/journal.pone.0020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapeller R, Prasad KVS, Janssen O, Hou W, Schaffhausen BS, Rudd CE, Cantley LC. Identification of Two SH3-binding motifs in the regulatory subunit of phosphatidylinositol 3-kinase. J Biol Chem. 1994;269:1927–1933. [PubMed] [Google Scholar]
- Kardinal C, Posern G, Zheng J, Knudsen BS, Moarefi I, Feller SM. Rational development of cell penetrating high affinity SH3 domain binding peptides that selectively disrupt the signal transduction of Crk family adapters. Ann N Y Acad Sci USA. 1999;886:289–292. doi: 10.1111/j.1749-6632.1999.tb09439.x. [DOI] [PubMed] [Google Scholar]
- Kato J, TakejaT GC, Iba H, Levy JB, Hanafusa H. Amino acid substitutions sufficient to convert the nontransforming p60csrc protein to a transforming protein. Mol Cell Biol. 1986;6:4155–4160. doi: 10.1128/mcb.6.12.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- Kesti T, Ruppelt A, Wang JH, Liss M, Wagner R, Tasken K, Saksela K. Reciprocal regulation of SH3 and SH2 domain binding via tyrosine phosphorylation of a common site in CD3epsilon. J Immunol. 2007;179:878–885. doi: 10.4049/jimmunol.179.2.878. [DOI] [PubMed] [Google Scholar]
- Kiehart DP, Franke JD, Chee MK, Montague RA, T-l C, Roote J, Ashburner M. Drosophila crinkled, mutations of which disrupt morphogenesis and cause lethality, encodes Fly myosin VIIA. Genetics. 2004;168:1337–1352. doi: 10.1534/genetics.104.026369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobashigawa Y, Sakai M, Naito M, Yokochi M, Kumeta H, Makino Y, Ogura K, Tanaka S, Inagaki F. Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK. Oncogene. 2007;14:503–510. doi: 10.1038/nsmb1241. [DOI] [PubMed] [Google Scholar]
- Koch CA, Anderson D, Moran MF, Ellis C, Pawson T. SH2 and SH3 domains: elements that control interactions of cytoplasmic signaling proteins. Science. 1991;252:668–674. doi: 10.1126/science.1708916. [DOI] [PubMed] [Google Scholar]
- Kristensen O, Guenat S, Dar I, Allaman-Pillet N, Abderrahmani A, Ferdaoussi M, Roduit R, Maurer F, Beckmann JS, Kastrup JS, Gajhede M, Bonny C. A unique set of SH3–SH3 interactions controls IB1 homodimerization. EMBO J. 2006;25:785–797. doi: 10.1038/sj.emboj.7600982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurochkina N. Helix-helix interactions and their impact on protein motifs and assemblies. J Theor Biol. 2010;264:585–592. doi: 10.1016/j.jtbi.2010.02.026. [DOI] [PubMed] [Google Scholar]
- Lee CH, Saksela K, Mirza UA, Chait BT, Kuriyan J. Crystal structure of the conserved core of HIV-1 NEF complexed with a SRC family SH3 domain. Cell. 1996;85:931–942. doi: 10.1016/S0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- Levaot N, Simoncic PD, Dimitriou JD, Scotter A, La Rose J, Ng AHM, Willett TL, Wang CJ, Janmohamed S, Grynpas M, Reichenberger E, Rottapel R. 3BP2-Deficient mice are osteoporotic with impaired osteoblast and osteoclast functions. J Clin Invest. 2011;121:3244–3257. doi: 10.1172/JCI45843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WA, Richards FM, Fox RO. Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature. 1994;372:375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- Lim DC, Cooke BM, Doerig C, Saeij JPJ. Toxoplasma and plasmodium protein kinases: roles in invasion and host cell remodeling. Int J Parasitol. 2011;42:21–32. doi: 10.1016/j.ijpara.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowey S, Saraswat LD, Liu H, Volkmann N, Hanein D. Evidence for an interaction between the SH3 domain and the nterminal extension of the essential light chain in class II myosins. J Mol Biol. 2007;37:902–913. doi: 10.1016/j.jmb.2007.05.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maignan S, Guilloteau JP, Fromage N, Arnoux B, Becquart J, Ducruix A. Crystal structure of the mammalian Grb2 adaptor. Science. 1995;268:291–293. doi: 10.1126/science.7716522. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Mayer BJ. SH3 domains: complexity in moderation. J Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- Mayer BJ, Hamaguchi M, Hanafusa H. A novel viral oncogene with structural similarity to phospholipase C. Nature. 1988;332:272–275. doi: 10.1038/332272a0. [DOI] [PubMed] [Google Scholar]
- Ménétrey J, Llinas P, Cicolari J, Squires G, Liu X, Li A, Sweeney HL, Houdusse A. The post-rigor structure of myosin VI and implications for the recovery stroke. EMBO J. 2008;27:244–252. doi: 10.1038/sj.emboj.7601937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncalián G, Cárdenes N, Deribe YL, Spínola-Amilibia M, Dikic I, Bravo J. Atypical polyproline recognition by the CMS N-terminal Src homology 3 domain. J Biol Chem. 2006;281:38845–38853. doi: 10.1074/jbc.M606411200. [DOI] [PubMed] [Google Scholar]
- Mongiovi AM, Romano PR, Panni S, Mendoza M, Wong WT, Musacchio A, Cesareni G, Di Fiore PP. A novel peptide-SH3 interaction. EMBO J. 1999;18:5300–5309. doi: 10.1093/emboj/18.19.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CJ, Winder SJ. Dystroglycan versatility in cell adhesion: a tale of multiple motifs. Cell communication and signaling. 2010;8:3–15. doi: 10.1186/1478-811X-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel B, Varela L, Azuaga AI, Conejero-Lara F. Environmental conditions affect the kinetics of nucleation of amyloid fibrils and determine their morphology. Biophys J. 2010;99:3801–3810. doi: 10.1016/j.bpj.2010.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan V, Dutta K, Cho J, Vila-Perello M, Raleigh DP, Cowburn D, Muir TW. Solution structure and folding characteristics of the C- terminal SH3 domain of c-Crk-II. Biochemistry. 2006;45:8874–8884. doi: 10.1021/bi060590z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A. How SH3 domains recognize proline. Adv Protein Chem. 2002;61:211–268. doi: 10.1016/S0065-3233(02)61006-X. [DOI] [PubMed] [Google Scholar]
- Musi V, Birdsall B, Fernandez-Ballester G, Guerrini R, Salvatori S, Serrano L, Pastore A. New approaches to high-throughput structure characterization of SH3 complexes: the example of myosin-3 and myosin-5 SH3 domains from S. cerevisiae. Protein Sci. 2006;4:795–807. doi: 10.1110/ps.051785506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam HJ, Haser WG, Roberts TM, Frederick CA. Intramolecular interactions of the regulatory domains of the Bcr-Abl kinase reveal a novel control mechanism. Structure. 1996;4:1105–1114. doi: 10.1016/S0969-2126(96)00116-5. [DOI] [PubMed] [Google Scholar]
- Nasertorabi F, Tars K, Becherer K, Kodandapani R, Liljas L, Vuori K, Ely KR. Molecular basis for regulation of Src by the docking protein p130Cas. J Mol Recognit. 2006;19:30–38. doi: 10.1002/jmr.755. [DOI] [PubMed] [Google Scholar]
- Neudecker P, Robustelli P, Cavalli A, Walsh P, Lundström P, Zarrine-Afsar A, Sharpe S, Vendruscolo M, Kay LE. Structure of an intermediate state in protein folding and aggregation. Science. 2012;336:362–366. doi: 10.1126/science.1214203. [DOI] [PubMed] [Google Scholar]
- Nguyen JT, Turck CW, Cohen FE, Zuckermann RN, Lim WA. Exploiting the basis of proline recognition by SH3 and WW domains: design of N-substituted inhibitors. Science. 1998;282:2088–2092. doi: 10.1126/science.282.5396.2088. [DOI] [PubMed] [Google Scholar]
- Nishida M, Nagata K, Hachimory Y, Horiuchi M, Ogura K, Mandiyan V, Schlessinger J, Inagaki F. Novel recognition mode between VAV and GRB2 SH3 domains. EMBO J. 2001;20:2995–3007. doi: 10.1093/emboj/20.12.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble MEM, Musacchio A, Saraste M, Courtneidge SA, Wierenga RK. Crystal structure of the SH3 domain in human Fyn; comparison of the three-dimensional structures of SH3 domains in tyrosine kinases and spectrin. EMBO J. 1993;12:2617–2624. doi: 10.2210/pdb1shf/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa A, Takayama Y, Sakai H, Chong KT, Takeuchi S, Nakagawa A, Nada S, Okada M, Tsukihara T. Structure of the carboxylterminal Src kinase, Csk. J Biol Chem. 2002;277:14351–14354. doi: 10.1074/jbc.C200086200. [DOI] [PubMed] [Google Scholar]
- Otsu M, Hiles I, Gout I, Fry MJ, Ruiz-Larrea F, Panayotou G, Thompson A, Dhand R, Hsuan J, Totty N, et al. Characterization of two 85 kd proteins that associate with receptor tyrosine kinases, middle-T/pp 60c-src complexes, and PI3-kinase. Cell. 1991;65:91–104. doi: 10.1016/0092-8674(91)90411-Q. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Wigge P, Vallis Y, Moore JDA, Evans PR, McMahon HT. Crystal structure of the amphiphysin-2 SH3 domain and its role in the prevention of dynamin ring formation. EMBO J. 1998;17:5273–5285. doi: 10.1093/emboj/17.18.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Wahl MI, Afar DEH, Turck CW, Rawlings DJ, Tam C, Scharenberg AM, Kinet J-P, Witte ON. Regulation of Btk function by a major autophosphorylation site within the SH3 domain. Immunity. 1996;4:515–525. doi: 10.1016/S1074-7613(00)80417-3. [DOI] [PubMed] [Google Scholar]
- Pauling L, Corey RB. The structure of fibrous proteins of the collagen-gelatin group. Proc Natl Acad Sci USA. 1951;37:272–281. doi: 10.1073/pnas.37.5.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proulx-Bonneau S, Guezguez A, Annabi B. A concerted HIF-1a/MT1-MMP signalling axis regulates the expression of the 3BP2 adaptor protein in hypoxic mesenchymal stromal cells. PloSOne. 2011;6:e21511–e21520. doi: 10.1371/journal.pone.0021511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval CJ, Nicolas V, Beau I. Role of Src kinases in mobilization of glycosylphosphatidylinositol-anchored decay-accelerating factor by Dr fimbria-positive adhering bacteria. Infect Immun. 2011;79:2519–2534. doi: 10.1128/IAI.01052-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Reichman C, Singh K, Liu Y, Singh S, Li H, Fajardo JF, Fiser A, Birge RB. Transactivation of Abl by the Crk II adapter protein requires a PNAY sequence in the Crk C-terminal SH3domain. Oncogene. 2005;24:8187–8189. doi: 10.1038/sj.onc.1208988. [DOI] [PubMed] [Google Scholar]
- Rudolph MG, Wittinghofer A, Vetter IR. Nucleotide binding to the G12V-mutant of CDC42 investigated by X-ray diffraction and fluorescence spectroscopy: Two different nucleotide states in one crystal. Protein Sci. 1999;8:778–787. doi: 10.1110/ps.8.4.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar P, Saleh T, Tzeng S-R, Birge RB, Kalodimos CG. Structural basis for regulation of the Crk signaling protein by a proline switch. Nature Chem Biol. 2011;7:51–57. doi: 10.1038/nchembio.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Maruoka M, Yokota N, Kuwano M, Matsui A, Inada M, Ogawa T, Ishida-Kitagawa N, Takeya T. Identification and functional analysis of a new phosphorylation site (Y398) in the SH3 domain of Abi-1. FEBS Lett. 2011;585:834–840. doi: 10.1016/j.febslet.2011.02.012. [DOI] [PubMed] [Google Scholar]
- Seidel-Dugan C, Meyer BE, Thomas SM, Brugge JS. Effects of SH2 and SH3 deletions on the functional activities of wild-type and transforming variants of c-Src. Mol Cell Biol. 1992;12:1835–1845. doi: 10.1128/mcb.12.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawn SCL. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 2005;390:641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Kim F. The shank family of scaffold proteins. J Cell Sci. 2000;113:1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- Shi X, Opi S, Lugar A, Restouin A, Coursindel T, Parrot I, Perez J, Madore E, Zimmermann P, Corbeil J, Huang M, Arold ST, Collette Y, Morelli X. Identification and biophysical assessment of the molecular recognition mechanisms between the human haemopoietic cell kinase Src homology domain 3 and ALG-2-interacting protein X. Biochem J. 2010;431:93–102. doi: 10.1042/BJ20100314. [DOI] [PubMed] [Google Scholar]
- Shoulders MD, Raines RT. Collagen structure and stability. Annu Rev Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithgall TE. SH2 and SH3 domains: potential targets for anti-cancer drug design. J Pharmacol Toxicol Methods. 1995;34:125–132. doi: 10.1016/1056-8719(95)00082-7. [DOI] [PubMed] [Google Scholar]
- Sriram G, Reichman C, Tunceroglu A, Kausha N, Saleh T, Machida K, Mayer B, Ge Q, Li J, Hornbeck P, Kalodimos CG, Birge RB. Phosphorylation of Crk on tyrosine 251 in the RT loop of the SH3C domain promotes Abl kinase transactivation. Oncogene. 2011;30:4645–4655. doi: 10.1038/onc.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl ML, Ferenz CR, Kelleher KL, Kriz RW, Knopf JL. Sequence similarity of phospholipase C with the non-catalytic region of Src. Nature. 1988;332:269–272. doi: 10.1038/332269a0. [DOI] [PubMed] [Google Scholar]
- Takaku T, Ogura K, Kumeta H, Yoshida N, Inagaki F. Solution structure of a novel CDC42 binding module of Bem1 and its interaction with Ste20 and CDC42. J Biol Chem. 2010;285:19346–19353. doi: 10.1074/jbc.M110.116749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Chen L, McClafferty H, Sailer CA, Ruth P, Knaus HG, Shipston MJ. A noncanonical SH3 domain binding motif links BK channels to the actin cytoskeleton via the SH3 adapter cortactin. FASEB J. 2006;20:2588–2590. doi: 10.1096/fj.06-6152fje. [DOI] [PubMed] [Google Scholar]
- Tong AH, Drees B, Nardelli G, Bader GD, Brannetti B, Castagnoli L, Evangelista M, Ferracuti S, Nelson B, Paoluzi S, Quondam M, Zucconi A, Hogue CW, Fields S, Boone C, Cesareni G. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science. 2002;295:321–324. doi: 10.1126/science.1064987. [DOI] [PubMed] [Google Scholar]
- Trahey M, Wong G, Halenbeck R, Rubinfeld B, Martin GA, Ladner M, Long CM, Crosier WJ, Watt K, Koths K, et al. Molecular cloning of two types of GAP complementary DNA from human placenta. Science. 1988;242:1697–1700. doi: 10.1126/science.3201259. [DOI] [PubMed] [Google Scholar]
- Vidal M, Gigoux V, Garbay C. SH2 and SH3 domains as targets for anti-proliferative agents. Crit Rev Oncol Hematol. 2001;40:175–186. doi: 10.1016/S1040-8428(01)00142-1. [DOI] [PubMed] [Google Scholar]
- Wang Q, Deloia MA, Kang Y, Litchke C, Zhang N, Titus MA, Walters KJ. The SH3 domain of a M7 interacts with its C-terminal proline-rich region. Protein Sci. 2007;16:189–196. doi: 10.1110/ps.062496807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci USA. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisstock JC, Lesk AM. SH3domains in prokaryotes. Trends Biochem Sci. 1999;24:32–33. doi: 10.1016/S0968-0004(99)01366-3. [DOI] [PubMed] [Google Scholar]
- Witucki LA, Huang X, Shah K, Liu Y, Kyin S, Eck MJ, Shokat KM. Mutant tyrosine kinases with unnatural nucleotide specificity retain the structure and phospho-acceptor specificity of the wild-type enzyme. Chem Biol. 2002;9:25–33. doi: 10.1016/S1074-5521(02)00091-1. [DOI] [PubMed] [Google Scholar]
- Wu L, Pan L, Wei Z, Zhang M. Structure of MyTH-FERM domains in myosin VIIa tail bound to cargo. Science. 2011;331:757–760. doi: 10.1126/science.1198848. [DOI] [PubMed] [Google Scholar]
- Xiong X, Cui P, Hossain S, Xu R, Warner B, Guo X, An X, Debnath AK, Cowburn D, Kotula L. Allosteric inhibition of the nonMyristoylated c-Abl tyrosine kinase by phosphopeptides derived from Abi1/Hssh3bp1. Biochim Biophys Acta. 2008;1783:737–747. doi: 10.1016/j.bbamcr.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase C-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- Yamada S, Yanamoto S, Kawasaki G, Rokutanda S, Yonezawa H, Kawakita A, Nemoto TK. Overexpression of CRKII increases migration and invasive potential in oral squamous cell carcinoma. Cancer Lett. 2011;303:84–91. doi: 10.1016/j.canlet.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Hendrickson WA. Structural basis for activation of human lymphocyte kinase Lck upon tyrosine phosphorylation. Nature. 1996;384:484–489. doi: 10.1038/384484a0. [DOI] [PubMed] [Google Scholar]
- Yang Y, Gourinath S, Kovacs M, Nyitray L, Reutzel R, Himmel DM, O'Neall-Hennessey E, Reshetnikova L, Szent-Gyorgyi A-G, Brown JH, Cohen C. Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor. Structure. 2007;15:553–564. doi: 10.1016/j.str.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Yao B, Zhang J, Dai H, Sun J, Jiao Y, Tang Y, Wu J, Shi Y. Solution structure of the second SH3 domain of human CMS and a newly identified binding site at the C-terminus of c-Cbl. Biochim Biophys Acta. 2007;177:35–43. doi: 10.1016/j.bbapap.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Zhang JS, Koenig A, Young C, Billadeau DD. GRB2 couples RhoU to epidermal growth factor receptor signaling and cell migration. Mol Biol Cell. 2011;22:2119–2130. doi: 10.1091/mbc.E10-12-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]