Abstract
Here I review the molecular mechanisms by which water-soluble monomeric amyloid-β (Aβ) peptides are transformed into well-organized supramolecular complexes called amyloid fibrils. The mechanism of amyloid formation is considered theoretically on the basis of experimental results, and the structural and mechanistic similarities of amyloid fibrils to three-dimensional crystals are highlighted. A number of important results from the literature are described. These include the observation that a correct ratio of monomer association and dissociation rate constants is key for formation of well-organized amyloid fibrils. The dynamic nature of the amyloid-β structure is discussed, along with the possibly obligate requirement of the transient formation of a hairpin-like fold prior to its incorporation into amyloid fibrils. Many rounds of monomer association and dissociation events may be present during an apparently silent lag-period. Amongst these association/dissociation events, interaction between the C-terminal regions of the Aβ peptide seems to be more favored. Such association and dissociation events occurring in a “trial-and-error” fashion may be an important requirement for the formation of well-organized amyloid fibrils.
Keywords: Amyloid fibril, Dynamic equilibrium, Nucleation-dependent polymerization model, NMR, Amyloid-β peptide
Introduction
A growing number of proteins have been shown to assemble into unbranched fibrillar structures that are collectively referred to as “amyloid fibrils”. Amyloid fibrils were first found in various serious diseases such as Alzheimer’s disease, spongiform encephalopathies (Creutzfeldt–Jakob disease), and dialysis-related amyloidosis (Sipe 1992; Rochet and Lansbury 2000). Each disease is characterized by the formation and accumulation of amyloid fibrils composed of a particular protein or polypeptide such as amyloid-β peptide, prion protein, and β2-microglobulin. However, many other proteins that are not directly related to diseases have also been shown to form similar fibrillar structures in vitro, suggesting the idea that the formation of these amyloid-like fibrils under certain conditions may be a general characteristic of polypeptide chains (Dobson 2001).
The nature of amyloid fibrils (as non-crystalline, insoluble, and large molecular assemblies) makes it difficult to access them by established structural techniques such as X-ray crystallography or high-resolution solution NMR. Nevertheless, considerable efforts have been made to obtain structural information of the amyloid fibrils using various techniques including X-ray diffraction (Sunde et al. 1997), electron microscopy (Jiménez et al. 1999), solid-state NMR, and others (Williams et al. 2004). From these studies, a “cross-β structure” has been proposed as a common model for the amyloid fibrils, in which the amyloid fibrils are composed predominantly of β-sheet structures, where the β-strands are arranged perpendicular to the fibrillar axis. In addition, short fibril-forming peptide segments of amyloidogenic proteins have been shown to grow into three-dimensional microcrystals that provide the structural details for a cross-β spine, which might also be composed of amyloid fibrils (Nelson et al. 2005; Sawaya et al. 2007; Ivanova et al. 2009).
Whereas details on the molecular structure of amyloid fibrils is gradually accumulating, and many theoretical models of the aggregation process have been so far proposed (Hall & Hirota, 2009; Morris et al. 2009), nevertheless the molecular mechanisms by which a soluble monomer organizes into a large supramolecular complex have not been fully unraveled. In this review, I discuss literature concerned with the molecular mechanism by which amyloid-β (Aβ) peptides organize into well-defined amyloid fibrils with unbranched filamentous morphologies. I argue the case that the organization of water-soluble monomeric peptide into a large supramolecular amyloid complex is dependent upon a subtle balance of the association and dissociation events that occur repeatedly at the tip of the amyloid fibrils. Whereas this review is mostly concerned with the Aβ peptide, a similar discussion could also be applied to the mechanism of amyloid fibril formation by other proteins.
Nucleation-dependent polymerization model
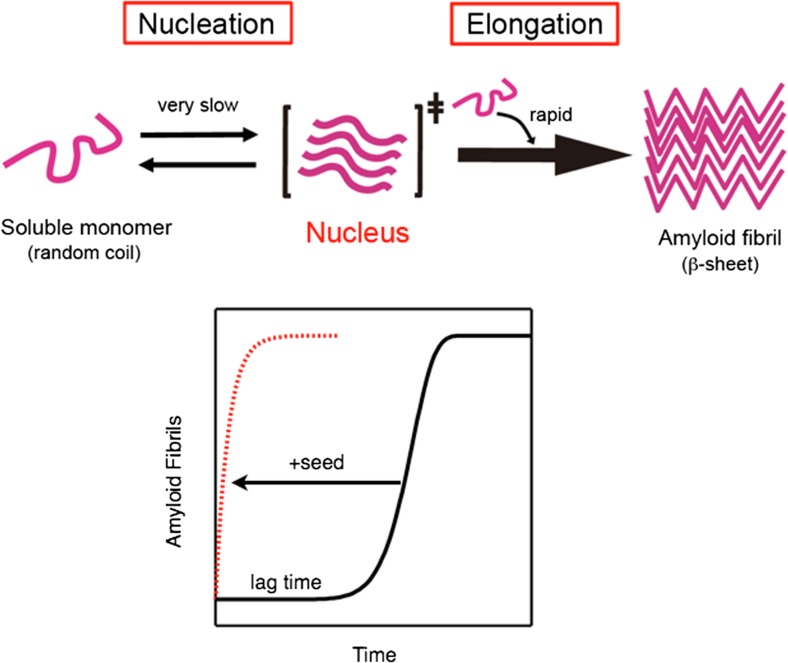
The most convincing model for the formation of amyloid fibrils is the “nucleation-dependent polymerization model”, in which the process of fibrillation is separated into a nucleation phase and an elongation phase (Jarrett and Lansbury 1992, 1993; Naiki and Nakakuki 1996; Naiki and Gejyo 1999). According to this model, the overall fibrillation process is rate-limited by the formation of a “nucleus” of amyloid fibrils that consists of several or more molecules organized into a fibril-compatible structural state. The process of nucleation is thermodynamically unfavorable, and the newly formed nucleus will dissociate to monomers until a certain number of molecules are packed together. On the other hand, once the number of molecules exceeds the threshold value, the equilibrium between the association and dissociation changes dramatically toward the binding reaction (Fig. 1). As a result, the formation of amyloid fibrils is preceded by a long lag period that is followed by a rapid growth of the fibrils. Other models such as the Dock–Lock model (Esler et al. 2000) and secondary nucleation mechanisms (Knowles et al. 2009) have also been proposed, which suggests that nucleation and elongation processes may sometimes be more intricate than the simplest nucleation-dependent polymerization model. Although the detailed mechanisms remain to be elucidated, it is evident that the formation of amyloid fibrils are well described by a characteristic sigmoidal kinetics, indicating the presence of a long lag period that is followed by a rapid extension of fibrils.
Fig. 1.
Amyloid fibril formation predicted by the nucleation dependent polymerization model. A large energy barrier between water-soluble monomers and insoluble amyloid fibrils is present. As a result, the spontaneous formation of amyloid fibrils from highly purified monomers is usually preceded by a long lag-period. The addition of small amount of preformed fibrils lowers the energy barrier, by acting as a template (also called as nucleus or seed) for amyloid formation, resulting in a rapid elongation of fibrils without lag-period
Association–dissociation equilibrium during the lag period
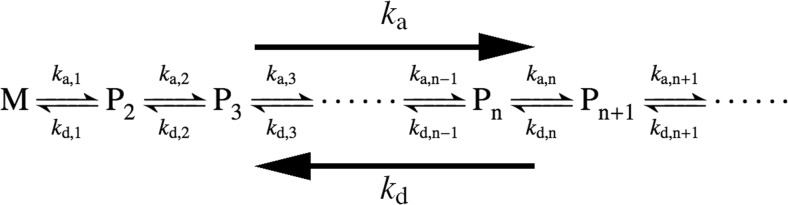
The overall fibrillation process can be represented by a series of microscopic association and dissociation rate constants (Fig. 2). During the time course of amyloid fibril formation, it is evident from experimental observation that the direction of the reaction changes from dissociation-dominant to association-dominant phases. While the equilibrium is dominated by the dissociation reaction during the nucleation phase, the association reaction becomes more preferred in the growth phase. Two opposite mechanisms are possible to explain this macroscopically observed change in the direction of the reaction. One possible mechanism involves a general “increase” in the microscopic association rate constant of the two molecules with increasing aggregate size. The other possible mechanism involves a “decrease” in the microscopic dissociation rate constant, with increasing aggregate size. Here I will argue that the second mechanism, the “decrease” in the microscopic dissociation rate constant seems to be more reliable to explain the macroscopically observed change in the direction of the reaction.
Fig. 2.
Kinetic model of amyloid fibril formation. Macroscopic association and dissociation rate constants (k a and k d, respectively) are represented by a series of microscopic association and dissociation rate constants (k a,n and k d,n, where the subscript n represents the polymerizatin number). M and P n denote monomer and polymer respectively. Incoming and outgoing monomers were omitted for clarity
As per Fig. 2, the microscopic association rate constant, k a,n, is considered to be determined by the microscopic affinity and the probability of collision between two molecules. Therefore, the association rate constant depends on the pH, temperature, protein concentration, and shearing forces (agitation) in the solution. However, if these initial conditions are constant during the time course of amyloid fibril formation, all of the microscopic association rate constants will be the same, regardless of the polymerization number. That is,
| 1 |
On the other hand, the value of the microscopic dissociation rate constant between two transiently associating molecules is determined by the microscopic stability of the complex. The microscopic stability of the complex is, in turn, depended on the number and the rigidity of hydrogen bonds, electrostatic attraction, and hydrophobic interaction between two molecules. These interactions are considered to become stronger if the polymerization number increases because the degree of packing between the molecules and the rigidity of these interactions are also increased. Therefore,
| 2 |
Equations (1) and (2) provide us with two predictions capable of being tested at the macroscopic level of observation. One of the predictions is as follows. The microscopic association rate constant k a may be much smaller than the dissociation constant k d while the polymer size is small. However, the dissociation constant k d will continuously decrease as the polymerization number increases. Therefore, the association constant k a eventually becomes larger than the dissociation constant k d when the aggregate reaches a certain size. At this point, where the size of the aggregates exceeds the “critical aggregation number”, the direction of the overall reaction will change from a dissociation-dominant to an association-dominant situation. It should be noted that, in this case, the reversing of the direction of apparent equilibrium is caused not by the increase of the association, but rather by the decrease of the dissociation rate constant.
The second prediction is as follows. From experiment, it is evident that a rapid growth of amyloid fibrils can only be observed during the elongation phase. However, the equalities in Eq. (1) tell us that a considerable number of the association events are always occurring during the course of the formation of amyloid fibrils. Therefore, the same frequency of association events is also occurring during the apparently silent, long, lag-phase of amyloid fibril formation. The reason why we don’t see any elongation of amyloid fibrils during the nucleation phase is simply that the microscopic dissociation constant is much larger than the association rate constant when the polymerization number is smaller than the threshold value (critical aggregation number). Nevertheless, a considerable number of association and dissociation events must be occurring during the apparently silent nucleation phase in the course of amyloid fibril formation. Indeed, repeated dissociation and re-association of molecules is evidenced by the H/D exchange combined with NMR and mass-spectroscopy in the amyloid fibril formed by SH3 domain (Carulla et al. 2005). Such a “dynamic equilibrium” is considered to be important for the formation of well-organized assembly of molecules, as discussed below.
Dimeric Aβ (1–40) failed to form typical amyloid fibrils
Extensive efforts to elucidate the molecular organization in the amyloid fibrils have been performed, and a variety of models have been proposed (Nelson and Eisenberg 2006; Perutz et al. 2002). Among them, the “cross-β” structure seems to be one of the most convincing models, where each molecule assumes a predominantly β-sheet structure that runs perpendicular to the fibril axis (Dobson 1999; Inouye et al. 1993; Sunde et al. 1997). In addition, a detailed analysis by solid-state NMR spectroscopy has suggested that the amyloid fibrils formed by partial or full-length Aβ-(1–40) and Aβ-(1–42) are predominantly composed of in-register, parallel β-sheets, although several variations in molecular organizations have been identified (Antzutkin et al. 2000; Balbach et al. 2002; Benzinger et al. 1998; Lührs et al. 2005; Tycko et al. 2009). These studies suggest that whereas the C-terminal part of the molecule assumes a well-organized structure that is stabilized by multiple hydrogen bonds, the N-terminal regions seems to be relatively flexible. For example, Petkova et al. (2002, 2006) showed that the C-terminal part of Aβ-(1–40) adopts a strand–turn–strand conformation, in which two β-strands composed of V12–V24 and A30–V40 were connected by a turn at G25–G29. On the other hand, the signals from the N-terminal 10 residues were significantly weak, suggesting that these regions are structurally disordered in the fibrils. This is also supported by analyses of H/D exchange detected with solution NMR spectroscopy (Olofsson et al. 2006; Whittemore et al. 2005).
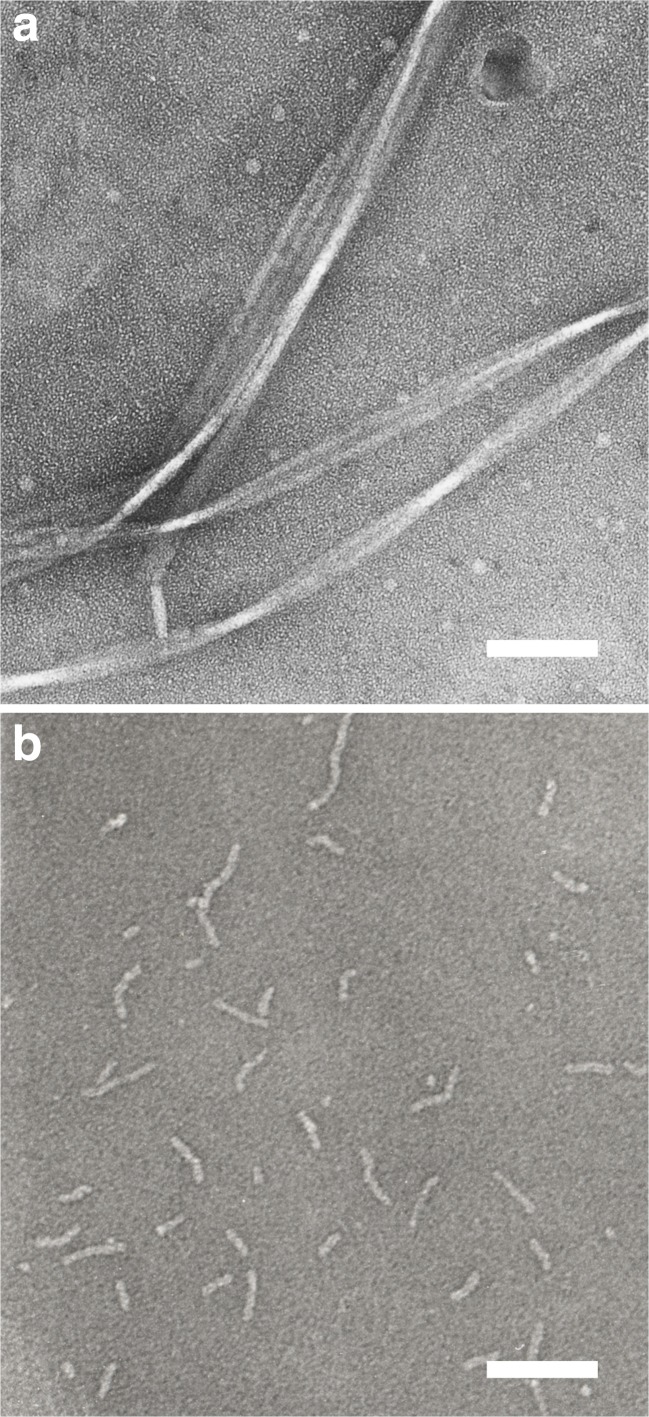
Based on the structural models for the amyloid fibrils formed by amyloid-β peptides, it has been hypothesized that amyloid fibril formation would be facilitated by covalently linking two Aβ molecules in a parallel orientation. We prepared cystein-substituted mutants of Aβ-(1–40) peptides and homodimer of them, in which two Aβ mutants were covalently linked through disulfide bonds at the N- or C-termini (Yamaguchi et al. 2010). We analyzed kinetics of the amyloid fibril formation of monomeric and covalently-linked dimeric form of cystein mutant of Aβ-(1–40), and compared with those of wild-type Aβ peptide. We found that the covalent-linking of two peptides greatly facilitated the fibril formation by these Aβ mutants, suggesting that typical amyloid fibrils were stabilized by a stack of parallel in-register β-strands. However, careful observation by transmission electron microscope revealed that the morphology of aggregates formed by disulfide-linked Aβ dimer was completely different from that formed by wild-type Aβ molecules (Fig. 3). Wild-type Aβ-(1–40) formed a long, unbranched, and straight fiber that resembles a typical amyloid fibril found in the literature. The disulfide-linked dimeric mutant was found to form fibrillar aggregates but with completely different morphology of a much shorter and curvy shape.
Fig. 3.
Transmission electron micrograph of “typical” amyloid fibrils formed by wild-type Aβ-(1–40) peptide (a) and short, thin, curvy aggregates formed by a disulfide-linked dimer of A2C-Aβ-(1–40) (b). Scale bar: 100 nm. The figure was redrawn from Yamaguchi et al. (2010) with permission
We also analyzed the interaction between these “curvy aggregates” and wild-type Aβ-(1–40) molecules, and found that the addition of the preformed curvy aggregate did not accelerate the amyloid fibril formation by wild-type Aβ-(1–40) at all. This suggests that the curvy aggregates do not have an ability to act as the template for the amyloid fibril formation, one of the most remarkable criteria for the amyloid fibrils. Therefore, although a parallel dimer formation by a covalent disulfide-linkage facilitated the aggregation process of Aβ molecules, the resultant curvy aggregates formed by these mutant Aβ-(1–40) dimer were different from typical amyloid fibrils. Furthermore, the preformed curvy aggregates formed by the disulfide-linked dimer of Aβ (1–40) mutants were found to reversibly dissociate into the soluble monomers with random coil conformations upon treatment with the reducing reagent, DTT. This indicates that the curvy aggregate is not the thermodynamically most stable state, but rather the kinetically trapped metastable state. We considered the formation of such a metastable state was the result of the loss of appropriate balance between the association and dissociation interactions.
Fibrillation versus crystallization
The nucleation-dependent polymerization model of amyloid fibril formation displays multiple similarities to crystallization. First, both processes require a trace amount of the “nucleus” on which the rest of the molecules stack to form a well-organized rigid structure. In the absence of a preformed nucleus, formation of amyloid fibrils is preceded by a long lag-phase. Crystallization of proteins also takes a long incubation period, typically from days to weeks, even in the presence of precipitating reagents, indicating that the initial “nucleation” is defined by a very large energy barrier. Secondly, the formation of long, straight, and regularly stacked amyloid fibrils and the growth of large three-dimensionally organized crystals both seem to be highly dependent on a subtle balance of the association and dissociation of the protein molecules. This is exemplified by the observation that amorphous aggregates are rapidly formed if the interactions between protein molecules are too strong. These situations are caused by the increase of hydrophobic interaction by the covalent bonding of two molecules, shielding of the electrostatic repulsion by high concentration of salt, and by the addition of inappropriate concentration range of precipitating reagents (Hong et al. 2002; Luft and DeTitta 1997; Yoshimura et al. 2012).
In the above observation, we found that the amyloid-β peptides failed to form “typical” amyloid fibrils, but formed thin, short, and curvy aggregates when two molecules were tied to each other by a disulfide covalent link. This may result from too great an attractive interaction between the molecules because of the increased hydrophobic interactions. We therefore consider that the appropriate balance between association and dissociation rate constants is necessary for the formation of a well-organized regularly stacked “cross-β” structure of amyloid fibrils. It is also important to note that, as discussed above, even the most simple “nucleation-dependent growth model” can predict the presence of repeated association and dissociation events between preformed amyloid fibrils and free monomers in the solution with a considerable value of k d during an apparently “silent” long lag phase before the burst of the growth of amyloid fibrils. If so, then this raises the question, is it possible to experimentally demonstrate the presence of the repeated association and dissociation events during the formation of amyloid fibrils? In relation to this question, I now discuss results from our recent studies which revealed the presence of a “dynamic equilibrium” between several conformations in the monomeric water-soluble molecules, as well as the dynamic association and dissociation events that are occurring repeatedly during the apparently “silent” lag phase.
Transient formation of intermediate conformational states of amyloid-β monomers during the lag-phase
The linewidth of solution NMR signals depends on the overall rotational correlation time of the protein molecule, τ c, as well as the local mobility of the nuclear spins of interest. Both dynamic motions become larger at higher temperature, and therefore signal intensity is also expected to increase with temperature. Contrary to this general idea, we found that the peak intensity of Aβ-(1–40) peptide in both the 1H-15N and 1H-13C HSQC spectra was dramatically decreased upon increasing the temperature. The solution conditions for the NMR measurement (50 μM peptide, 100 mM NaCl, 50 mM Na-phosphate (pH 6.5)) were almost the same as that used for the spontaneous amyloid fibril formation. Therefore, these NMR measurements were performed during the lag-phase which usually precedes the spontaneous amyloid fibril formation. We confirmed that the disappearance of NMR signal upon increasing temperature does not result from the formation of amyloid/aggregate at higher temperature, because the intensity of NMR peaks was recovered by lowering the temperature. Instead, the decrease in signal intensity at higher temperature was caused by chemical exchange between several different magnetic environments experienced by each nuclear spin.
The chemical exchange in NMR measurements can be classified as “slow”, “intermediate” and “fast”, depending on the relative amplitude of the exchange rate constant, k ex = k A + k B, and chemical shift difference between the two states, ΔΩ = |ΩA – ΩB| (Scheme 1).
| Scheme 1 |
If the exchange rate is slow (k ex < < ΔΩ), two distinct signals are observed at resonance frequencies ΩA and ΩB. On the other hand, when the exchange rate is fast (k ex > > ΔΩ), a single resonance is observed at the population-weighted average chemical shift of the nucleus in the two sites. If the exchange rate is in the order of the chemical shift difference between the two sites, k ex ∼ ΔΩ, both resonance lines will apparently “disappear” because of severe line broadening. This is known as chemical exchange line broadening (Cavanagh et al. 2007; Palmer et al. 2001).
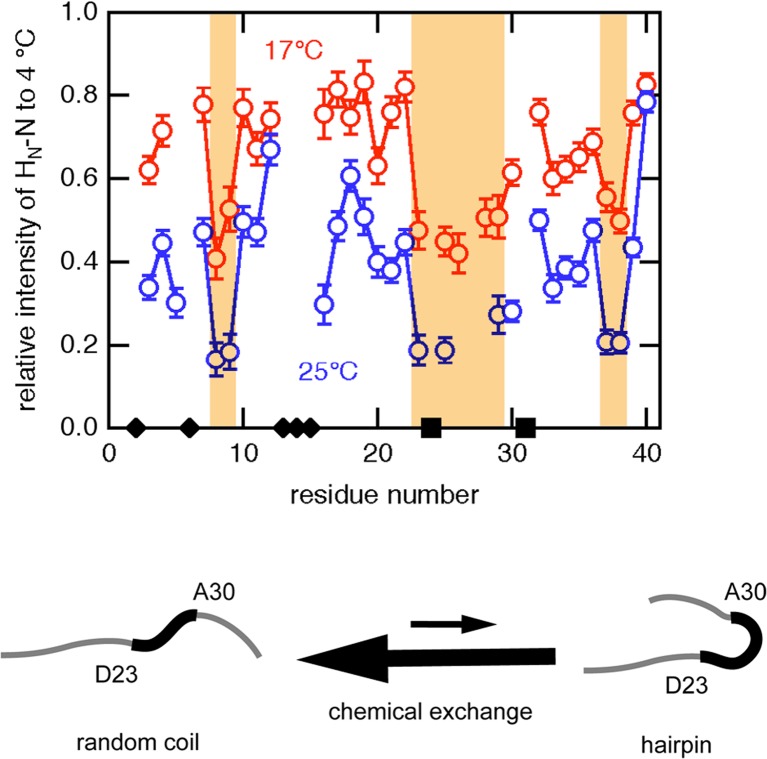
We analyzed the temperature dependency of each resonance peaks in the 1H-15N and 1H-13C HSQC spectra, and found that a remarkable decrease in the peak intensity was observed in the consecutive region around D23–A30 as well as several additional residues such as S8, G9, G37, and G38 (Yamaguchi et al. 2011). A detailed analysis suggested the degree of chemical exchange line broadening depends significantly on the amplitude of chemical shift difference between two states (ΔΩ = |ΩA – ΩB|). Although we do not know the exact value of ΔΩ for each residue because the population of the “minor” conformation is under the detection limit, the residues in the region of D23–A30 may have larger ΔΩ values than other residues. This means that a nuclear spin in this region experiences a drastic change in the chemical environment when the molecule adopts the “minor” conformation.
We also examined the effect of the total Aβ-(1–40) peptide concentration on the peak intensity in the 1H-15N HSQC spectrum. In contrast to the effect of an increase in temperature, the signal intensity was not affected by the total protein concentration, indicating that the chemical exchange was caused by the intramolecular interaction. We therefore considered that an Aβ-(1–40) molecule in solution is in dynamic equilibrium between a random coil conformation and a folded structure, with a turn at around D23–A30 (Fig. 4). This partially folded structure is probably stabilized by the transient formation of intramolecular hydrogen bonds between backbone atoms. We also consider that the conformation of Aβ-(1–40) molecules in the solution is not unique and uniform at all, but is repeatedly changing to such “minor” conformational state(s) with a hairpin-like structure during a long, apparently silent, lag-phase before the formation of amyloid fibrils. The presence of such dynamic equilibrium between several minor conformations during the lag-phase may be important for the formation of well-organized amyloid fibrils which resemble the three-dimensional crystals.
Fig. 4.
A significant decrease in the NMR signal intensity upon increasing temperature in the consecutive region from D23 to A30. Transient formation of a hairpin-like conformation was suggested as a sufficient explanation. The figure was redrawn from Yamaguchi et al. (2011) with permission
Interaction between Aβ molecules demonstrated by the paramagnetic relaxation enhancement of NMR signals
The temperature dependency of 1H-15N and 1H-13C HSQC spectra of Aβ-(1–40) molecules suggested the presence of significant intramolecular dynamic motion during the long lag-period. However, we did not obtain any evidence for the presence of intermolecular interaction during the lag-phase before the amyloid fibril formation, although this would be expected based on the theoretical consideration as mentioned above. Next, we examined the heteromolecular interaction between Aβ-(1–40) and Aβ-(1–42) by using a paramagnetic relaxation enhancement (PRE) of NMR signals (Yamaguchi et al. 2013).
To examine the possible and repeatedly occurring intermolecular association/dissociation events during a long-lag period, we focused on the following experimental observation. A careful kinetic analysis has revealed the difference between homogeneous and heterogeneous fibril formation by two major variants of amyloid-β peptides, Aβ-(1–40) and Aβ-(1–42) (Hasegawa et al. 1999). As shown in Fig. 1, the spontaneous amyloid fibril formation by both Aβ-(1–40) and Aβ-(1–42) exhibits a very long lag-period if the molecules are highly purified. On the other hand, the addition of a small amount of preformed amyloid fibrils decreases the energy barrier between monomers and amyloid fibrils, resulting in the disappearance of a lag-phase. Although the detailed mechanism of how the “seed” fibrils decrease a large energy barrier is unknown, the “seeding” effect of the preformed amyloid fibrils seems to reflect the homology between two molecules. If the added “seed” fibrils were composed of the same molecules as those dissolved in the solution, the effects were maximum and no lag-phase would be exhibited. On the other hand, if the added “seed” were composed of a different kind of protein, e.g., the addition of β2-microglobulin amyloids into soluble amyloid-β peptides, no interaction would occur between them. Such observations make sense by considering the resemblance of amyloid fibrils to the three-dimensional crystals.
A more interesting phenomenon is observed if the two molecules are not identical but greatly resemble each other. The chemical structure of Aβ-(1–40) and Aβ-(1–42) is almost the same, but Aβ-(1–42) has two additional residues -Ile-Ala at the C-terminus. This subtle but significant difference between two variants of Aβs resulted in the partial “seeding” effect. When Aβ-(1–40) seed was added to the solution of seed-free Aβ-(1–40), fibril elongation proceeded rapidly without lag time. In contrast, when Aβ-(1–42) seed was added to seed-free Aβ-(1–40), the aggregation profile exhibited a short, but significant lag period (Yamaguchi et al. 2013). This implies that Aβ-(1–40) monomer would first reversibly bind to the end of Aβ-(1–42) seed during lag time, and then become irreversibly associated after conformational change as the Dock–Lock model. To explore the transient and reversible interaction that is possibly occurring during lag time, we performed the paramagnetic relaxation enhancement (PRE)-NMR measurements. The PRE effect is caused by an unpaired electron, which has a high magnetogyric ratio (γe/γH ∼ 600), resulting in a significant decrease in NMR signal intensity of nearby (within ∼ 25 Å) nuclear spins. This method has been successfully used to detect long-range intra- or intermolecular interactions present in low-populated species (Ahn et al. 2006; Bertoncini et al. 2005; Iwahara and Clore 2006; Wu et al. 2008).
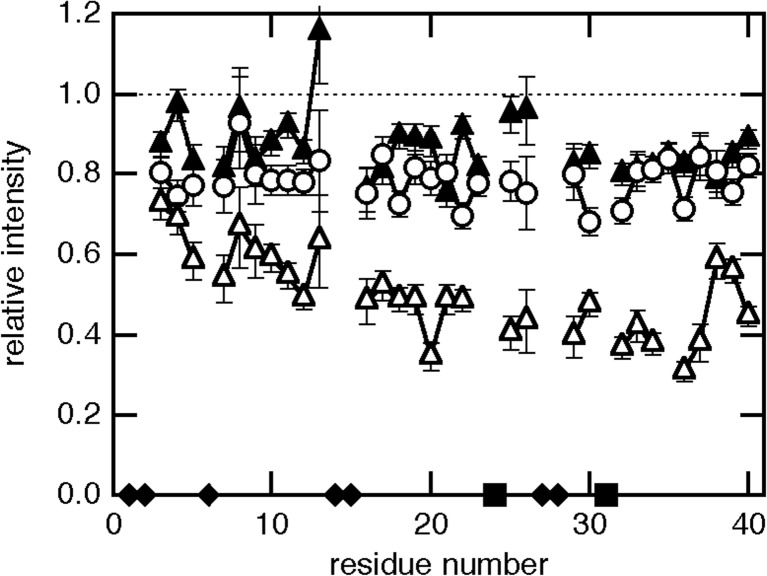
To examine the site-specific interaction between Aβ-(1–42) fibrils and Aβ-(1–40) monomer, two positions, Ala2 and Ala30, were chosen for labeling by PRE reagent. Those Ala residues located at the N-terminal or the center of the molecule were mutated to Cys residues to label by the nitroxiside radical MTSL through the disulfide bond. First, the amyloid seed was prepared by the N-terminally spin-labeled A2C-Aβ-(1–42) and added to 15N-Aβ-(1–40) monomer. In this case, no significant decrease in the intensity of the 1H-15N HSQC spectra was observed, suggesting that the interaction between Aβ-(1–42) seeds and monomeric Aβ-(1–40) was too weak to be detected. In contrast, when the seed prepared by the spin-labeled A30C-Aβ-(1–42) was added to the 15N-Aβ-(1–40) monomer, a remarkable decrease was observed in the peak intensity (Fig. 5). Furthermore, the PRE effect was apparently position-dependent, and the C-terminal region of Aβ-(1–40) exhibited a more remarkable decrease in peak intensity.
Fig. 5.
Paramagnetic relaxation enhancement showing preferable binding through the region from the center to C-terminal of the molecule. Peak intensity of 15N-Aβ-(1–40) in the presence of spin-labeled A2C-Aβ-(1–42) seed (open circle), spin-labeled A30C-Aβ-(1–42) seed (open triangle), or non-labeled A30C-Aβ-(1–42) seed (closed triangle) relative to that in the absence of seeds. The figure was redrawn from Yamaguchi et al. (2013) with permission
The results of PRE experiments emphasize the following aspects: (1) monomeric 15N-Aβ-(1–40) dissolved in solution repeatedly associate and dissociate from the Aβ-(1–42) amyloid seeds during the lag-phase, and (2) the transiently formed “intermediate” seemed to be more stable if Aβ-(1–40) monomer was stacked parallel onto the Aβ-(1–42) seeds. The first point can be reconciled by a simple consideration; the apparent molecular weight of amyloid seeds is too large to be detected by solution NMR. Therefore, the NMR signal would simply disappear if monomeric 15N-Aβ-(1–40) molecule just binds to the amyloid seeds. On the other hand, the intensity of NMR signal would not change at all if there was no interaction between 15N-Aβ-(1–40) and Aβ-(1–42) seeds. The PRE effects exhibited as a decrease in intensity can only be explained through the presence of repeated association and dissociation events during the lag-period. The second point could be deduced by the experimental results that the PRE effects were more significant from the center to the C-terminal of the 15N-Aβ-(1–40) monomer when there was interaction with A30C-Aβ-(1–42) seeds. We consider that there are a great number of association/dissociation events between Aβ-(1–40) monomer and Aβ-(1–42) seeds during the lag-period. The association should be caused by the thermal collision, and many different parts (N-terminal, center and C-terminal) of the Aβ-(1–40) molecule randomly collide to various parts of Aβ-(1–42) molecule on the seeds. The probability of collision between one residue in the Aβ-(1–40) molecule and the PRE-causing residue Cys30 in the Aβ-(1–42) should be the same between residues. Therefore, no positional dependency in the PRE effect should be apparent in the 15N-Aβ-(1–40) molecule if the affinity between the sites is the same as each other. We may therefore conclude that a “parallel” stacking onto the preformed seed is more favorable, and at the same time, many unfavorable association events are also present during the lag-period. Such association and dissociation events in a “trial-and-error” fashion could be important for the formation of well-organized amyloid fibrils in a manner analogous to three-dimensional crystal growth.
Summary
In this review, I have described the underlying mechanisms of amyloid fibril formation of the amyloid-β peptides. I emphasize the similarity between amyloid fibrils and three-dimensional crystals of proteins in several aspects, including a well-ordered molecular organization, the mechanism of formation that depends on a subtle balance between association and dissociation rate constants. I also showed that water-soluble monomers are in dynamic equilibrium between random-coil and transiently formed hairpin-like conformation. Furthermore, it was showed theoretically and experimentally that dynamic equilibrium of association and dissociation events existed between the water-soluble monomers and the seeds of amyloid during an apparently silent long lag-period. Among those randomly occurring association events caused by the thermal collision, the interaction between the C-terminal regions of the Aβ molecules seems to be more favored. Such association and dissociation events occurring in a “trial-and-error” fashion could be important for the formation of well-organized amyloid fibrils.
Compliance with ethical standards
Conflict of interest
Masaru Hoshino declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Ahn HC, Le YT, Nagchowdhuri PS, Derose EF, Putnam-Evans C, London RE, Markley JL, Lim KH. NMR characterizations of an amyloidogenic conformational ensemble of the PI3K SH3 domain. Protein Sci. 2006;15:2552–2557. doi: 10.1110/ps.062154306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antzutkin ON, Balbach JJ, Leapman RD, Rizzo NW, Reed J, Tycko R. Multiple quantum solid-state NMR indicates a parallel, not antiparallel, organization of β-sheets in Alzheimer’s β-amyloid fibrils. Proc Natl Acad Sci U S A. 2000;97:13045–13050. doi: 10.1073/pnas.230315097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbach JJ, Petkova AT, Oyler NA, Antzutkin ON, Gordon DJ, Meredith SC, Tycko R. Supramolecular structure in full-length Alzheimer’s β-amyloid fibrils: evidence for a parallel β-sheet organization from solid-state nuclear magnetic resonance. Biophys J. 2002;83:1205–1216. doi: 10.1016/S0006-3495(02)75244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzinger TL, Gregory DM, Burkoth TS, Miller-Auer H, Lynn DG, Botto RE, Meredith SC. Propagating structure of Alzheimer’s β-amyloid(10–35) is parallel β-sheet with residues in exact register. Proc Natl Acad Sci U S A. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoncini CW, Jung YS, Fernandez CO, Hoyer W, Griesinger C, Jovin TM, Zweckstetter M. Release of long-range tertiary interactions potentiates aggregation of natively unstructured α-synuclein. Proc Natl Acad Sci U S A. 2005;102:1430–1435. doi: 10.1073/pnas.0407146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carulla M, Caddy GL, Hall DR, Zurdo J, Gairí M, Feliz M, Giralt E, Robinson CV, Dobson CM. Molecular recycling within amyloid fibrils. Nature. 2005;436:554–558. doi: 10.1038/nature03986. [DOI] [PubMed] [Google Scholar]
- Cavanagh J, Fairbrother WJ, Palmer AG, III, Rance M, Skelton NJ. Protein NMR spectroscopy. 2. Oxford: Academic; 2007. pp. 333–404. [Google Scholar]
- Dobson CM. Protein misfolding, evolution and disease. Trends Biochem Sci. 1999;24:329–332. doi: 10.1016/S0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- Donon CM. The structural basis of protein folding and its links with human disease. Philos Trans R Soc Lond B. 2001;356(1406):133–145. doi: 10.1098/rstb.2000.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esler WP, Stimson ER, Jennings JM, Vinters HV, Ghilardi JR, Lee JP, Mantyh PW, Maggio JE. Alzheimer’s disease amyloid propagation by a template-dependent dock–lock mechanism. Biochemistry. 2000;39:6288–6295. doi: 10.1021/bi992933h. [DOI] [PubMed] [Google Scholar]
- Hall D, Hirota N. Multi-scale modelling of amyloid formation from unfolded proteins using a set of theory derived rate constants. Biophys Chem. 2009;140:122–128. doi: 10.1016/j.bpc.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Yamaguchi I, Omata S, Gejyo F, Naiki H. Interaction between Aβ(1–42) and Aβ(1–40) in Alzheimer’s β-amyloid fibril formation in vitro. Biochemistry. 1999;38:15514–15521. doi: 10.1021/bi991161m. [DOI] [PubMed] [Google Scholar]
- Hong DP, Gozu M, Hasegawa K, Naiki H, Goto Y. Conformation of β2-microglobulin amyloid fibrils analyzed by reduction of the disulfide bond. J Biol Chem. 2002;277:21554–21560. doi: 10.1074/jbc.M200188200. [DOI] [PubMed] [Google Scholar]
- Inouye H, Fraser PE, Kirschner DA. Structure of β-crystallite assemblies formed by Alzheimer β-amyloid protein analogues: analysis by x-ray diffraction. Biophys J. 1993;64:502–519. doi: 10.1016/S0006-3495(93)81393-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova MI, Sievers SA, Sawaya MR, Wall JS, Eisenberg D. Molecular basis for insulin fibril assembly. Proc Natl Acad Sci U S A. 2009;106:18990–18995. doi: 10.1073/pnas.0910080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- Jarrett JT, Lansbury PT., Jr Amyloid fibril formation requires a chemically discriminating nucleation event: Studies of an amyloidogenic sequence from the bacterial protein OsmB. Biochemistry. 1992;31:12345–12352. doi: 10.1021/bi00164a008. [DOI] [PubMed] [Google Scholar]
- Jarrett JT, Lansbury PT., Jr Seeding “one-dimensional crystallization” of amyloid: a pathogenic mechanism in Alzheimer’s disease and scrapie? Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- Jiménez JL, Guijarro JI, Orlova E, Zurdo J, Dobson CM, Sunde M, Saibil HR. Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecuar packing. EMBO J. 1999;18:815–821. doi: 10.1093/emboj/18.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles TP, Waudby CA, Devlin GL, Cohen SI, Aguzzi A, Vendruscolo M, Terentjev EM, Welland ME, Dobson CM. An analytical solution to the kinetics of breakable filament assembly. Science. 2009;326:1533–1537. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- Luft JR, DeTitta GT. Kinetic aspects of macromolecular crystallization. Methods Enzymol. 1997;276:110–131. doi: 10.1016/S0076-6879(97)76053-1. [DOI] [PubMed] [Google Scholar]
- Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Döbeli H, Schubert D, Riek R. 3D structure of Alzheimer’s amyloid-β(1–42) fibrils. Proc Natl Acad Sci U S A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AM, Murielle A, Finke RG. Protein aggregation kinetics, mechanism, and curve-fitting: a review of the literature. Biochim Biophys Acta. 2009;1974:375–397. doi: 10.1016/j.bbapap.2008.10.016. [DOI] [PubMed] [Google Scholar]
- Naiki H, Gejyo F. Kinetic analysis of amyloid fibril formation. Methods Enzymol. 1999;309:305–318. doi: 10.1016/S0076-6879(99)09022-9. [DOI] [PubMed] [Google Scholar]
- Naiki H, Nakakuki K. First-order kinetic model of Alzheimer’s β-amyloid fibril extension in vitro. Lab Investig. 1996;74:374–383. [PubMed] [Google Scholar]
- Nelson R, Eisenberg D. Structural models of amyloid-like fibrils. Adv Protein Chem. 2006;73:235–282. doi: 10.1016/S0065-3233(06)73008-X. [DOI] [PubMed] [Google Scholar]
- Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, Eisenberg D. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson A, Sauer-Eriksson AE, Ohman A. The solvent protection of alzheimer amyloid-β-(1–42) fibrils as determined by solution NMR spectroscopy. J Biol Chem. 2006;281:477–483. doi: 10.1074/jbc.M508962200. [DOI] [PubMed] [Google Scholar]
- Palmer AG, III, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/S0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- Perutz MF, Finch JT, Berriman J, Lesk A. Amyloid fibers are water-filled nanotubes. Proc Natl Acad Sci U S A. 2002;99:5591–5595. doi: 10.1073/pnas.042681399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer’s β-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s β-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochet JC, Lansbury PT., Jr Amyloid fibrillogenesis: themes and variations. Curr Opin Struct Biol. 2000;10:60–68. doi: 10.1016/S0959-440X(99)00049-4. [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, Madsen AØ, Riekel C, Eisenberg D. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- Sipe JD. Amyloidosis. Annu Rev Biochem. 1992;61:947–975. doi: 10.1146/annurev.bi.61.070192.004503. [DOI] [PubMed] [Google Scholar]
- Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, Blake CC. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- Tycko R, Sciarretta KL, Orgel JP, Meredith SC. Evidence for novel β-sheet structures in Iowa mutant β-amyloid fibrils. Biochemistry. 2009;48:6072–6084. doi: 10.1021/bi9002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittemore NA, Mishra R, Kheterpal I, Williams AD, Wetzel R, Serpersu EH. Hydrogen–deuterium (H/D) exchange mapping of Aβ1–40 amyloid fibril secondary structure using nuclear magnetic resonance spectroscopy. Biochemistry. 2005;44:4434–4441. doi: 10.1021/bi048292u. [DOI] [PubMed] [Google Scholar]
- Williams AD, Portelius E, Kheterpal I, Guo JT, Cook KD, Xu Y, Wetzel R. Mapping Ab amyloid fibril secondary structure using scanning proline mutagenesis. J Mol Biol. 2004;335:833–842. doi: 10.1016/j.jmb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Wu KP, Kim S, Fela DA, Baum J. Characterization of conformational and dynamic properties of natively unfolded human and mouse α-synuclein ensembles by NMR: implication for aggregation. J Mol Biol. 2008;378:1104–1115. doi: 10.1016/j.jmb.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Yagi H, Goto Y, Matsuzaki K, Hoshino M. A disulfide-linked amyloid-β peptide dimer forms a protofibril-like oligomer through a distinct pathway from amyloid fibril formation. Biochemistry. 2010;49:7100–7107. doi: 10.1021/bi100583x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Matsuzaki K, Hoshino M. Transient formation of intermediate conformational states of amyloid-β peptide revealed by heteronuclear magnetic resonance spectroscopy. FEBS Lett. 2011;585:1097–1102. doi: 10.1016/j.febslet.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Matsuzaki K, Hoshino M. Interaction between soluble Aβ-(1–40) monomer and Aβ-(1–42) fibrils probed by paramagnetic relaxation enhancement. FEBS Lett. 2013;587:620–624. doi: 10.1016/j.febslet.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Yoshimura Y, Lin Y, Yagi H, Lee YH, Kitayama H, Sakurai K, So M, Ogi H, Naiki H, Goto Y. Distinguishing crystal-like amyloid fibrils and glass-like amorphous aggregates from their kinetics of formation. Proc Natl Acad Sci U S A. 2012;109:14446–14451. doi: 10.1073/pnas.1208228109. [DOI] [PMC free article] [PubMed] [Google Scholar]