Abstract
Many genetic mutations in sarcomeric proteins, including the cardiac myosin regulatory light chain (RLC) encoded by the MYL2 gene, have been implicated in familial cardiomyopathies. Yet, the molecular mechanisms by which these mutant proteins regulate cardiac muscle mechanics in health and disease remain poorly understood. Evidence has been accumulating that RLC phosphorylation has an influential role in striated muscle contraction and, in addition to the conventional modulation via Ca2+ binding to troponin C, it can regulate cardiac muscle function. In this review, we focus on RLC mutations that have been reported to cause cardiomyopathy phenotypes via compromised RLC phosphorylation and elaborate on pseudo-phosphorylation rescue mechanisms. This new methodology has been discussed as an emerging exploratory tool to understand the role of phosphorylation as well as a genetic modality to prevent/rescue cardiomyopathy phenotypes. Finally, we summarize structural effects post-phosphorylation, a phenomenon that leads to an ordered shift in the myosin S1 and RLC conformational equilibrium between two distinct states.
Keywords: Cardiomyopathy, Hypertrophy, Myosin light chains, Phosphorylation, Pseudo-phosphorylation
Introduction
Diseases of the heart and circulatory system are leading causes of death worldwide, with heart failure (HF) being highly prevalent in the most affluent parts of the world (Kannel 2000). Familial hypertrophic cardiomyopathy (HCM), which often leads to HF, is an autosomal dominant disorder manifested by ventricular and septal hypertrophy, myofibrillar disarray, abnormal ECG findings and sudden cardiac death (SCD). HCM is the most common cause of SCD among young individuals, particularly competitive athletes (Maron 2002). The disease originates from mutations in genes encoding major contractile proteins of the heart, such as β-myosin heavy chain (β-MHC) (44%), myosin binding protein C (MyBP-C) (35%), regulatory light chain (RLC) (2%), essential light chain (ELC) (1.6%), α-tropomyosin (α-Tm) (2.5%), troponin T (TnT) (7%), troponin I (TnI) (5%), troponin C (TnC) (∼1%), α-actin (1%), and titin (<1%) (Alcalai et al. 2008). The characteristic pathologic features of HCM include cardiac hypertrophy, extensive disorganization of the myocyte structure (myocyte disarray) and myocardial fibrosis (Binder et al. 2005; Ho et al. 2010). Unfortunately, there is currently no cure for HCM, and it is now becoming evident that any effective therapy must target the underlying mechanisms involved in the pathogenesis of the disease (Granzier and de Tombe 2015; Moore et al. 2012; Tardiff et al. 2015). Current approaches solely rely on the symptomatic treatments of diastolic dysfunction, a hallmark of HCM, and include the use of β-blockers and Ca2+ channel blockers to slow heart rate and increase the diastolic filling time (Ho et al. 2002; Olivotto et al. 2009). Clinicians and researchers all agree that target-specific therapies hold the most future promise for treating HCM (Bers and Harris 2011; Olivotto et al. 2009). The recent discovery of omecamtiv mecarbil and MYK-461, small molecules specifically targeting the function of myosin, the major force-generating protein of the heart, highlights the importance of such approaches (Green et al. 2016; Malik et al. 2011).
Cardiac muscle contraction is driven by the ATP-dependent cyclic attachments and detachments of the myosin cross-bridges to actin-containing thin filaments (Geeves and Holmes 2005). RLC is a major regulatory subunit of myosin and a modulator of the troponin/tropomyosin and Ca2+-controlled regulation of cardiac muscle contraction (Szczesna 2003). It is attached to MHC at the head–rod junction and, together with ELC, stabilizes the α-helical neck region of the myosin head, also called the lever arm (Burghardt and Sikkink 2013; Geeves 2002; Rayment et al. 1993). The N-terminal domain of the RLC contains a divalent cation binding site and a myosin light chain kinase (MLCK)-specific phosphorylation site at Ser-15 (Fig. 1). In the beating heart under basal physiological conditions, RLC was seen to be phosphorylated at about 0.4 mol of phosphate (Pi)/mol of RLC in a variety of animal species, including humans (Huang et al. 2008; Kamm and Stull 2011). More recently, rodent hearts were shown to be basally phosphorylated at less than 0.05 mol Pi/mol RLC in vivo (Kampourakis and Irving 2015), creating an absence of a clear consensus. In murine striated cardiac muscle, RLC may exist in an unphosphorylated, singly phosphorylated or doubly phosphorylated state (Ser 14/15). However, human ventricular RLC may only be singly phosphorylated (Ser-15) (Fig. 1) or singly deamidated (Asn-14 to Asp-14) (Scruggs et al. 2010; Scruggs and Solaro 2011). These sites are mostly phosphorylated by a cardiac-specific MLCK (cMLCK), encoded by MYLK3, which, in addition to its role in Ca2+ sensitization of myofilaments (Sweeney and Stull 1986), may serve as an adaptive mechanism to rapidly facilitate sarcomere organization in cardiac myocytes in response to hypertrophic stimuli associated with increased intracellular Ca2+ (Aoki et al. 2000). A significant role for cMLCK has been shown in cardiogenesis (Seguchi et al. 2007) and proper myofibrillogenesis (Terry et al. 2006). Upregulation of cMLCK was considered as a mechanism to promote sarcomere reassembly and enhanced contractility of the failing heart (Seguchi et al. 2007).
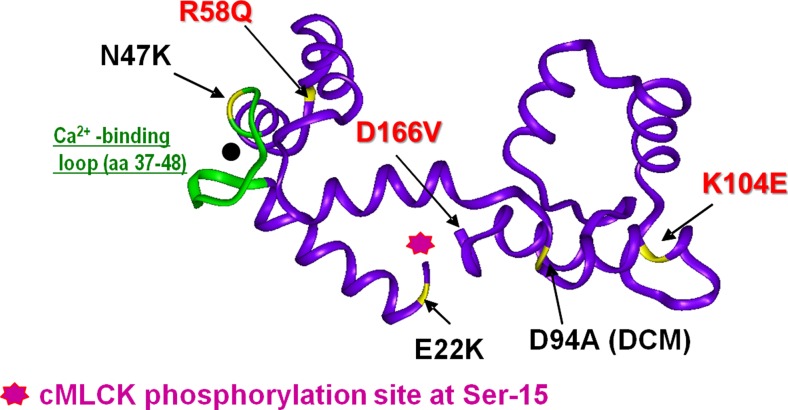
Fig. 1.
Three-dimensional representation of human ventricular RLC (Swiss-Prot: P10916) and cardiomyopathy-linked mutations (pdb: 1WDC). In red, HCM phenotypes of decreased RLC phosphorylation in transgenic mouse myocardium. Adapted from Szczesna et al. (2001)
The Kasahara group previously demonstrated that cMLCK plays a functional role in cardiac adaptations as well as mediating the transition from compensated to decompensated hypertrophy (Warren et al. 2012). The study reported that heart failure under pressure overload induced by transaortic constriction (TAC) in wild-type (WT) mice reduces cMLCK and consequently decreases the phosphorylated cardiac RLC. This adverse effect on heart function was attenuated by cMLCK overexpression in the absence of increased loading. Similarly, Massengill et al. (2016) reported that an acute, inducible reduction of cMLCK leads to sarcomeric disorganization, fibrosis, reduced contractility and cell death, and causes severe systolic and diastolic dysfunction and rapid progression to heart failure with reduction of fractional shortening. In addition, Mylk3-KO mouse hearts, lacking expression of cMLCK, displayed a significant reduction in Ca2+ amplitude and Ca2+ decay rates as well as SERCA2a mRNA, implying impaired calcium handling (Massengill et al. 2016).
Genetic mutations in myosin regulatory light chain lead to cardiomyopathy
Recent genetic studies have revealed that mutations in MYL2, encoding the human ventricular RLC, are more common than previously reported (for review, see Muthu et al. 2012a; Szczesna 2003) and in just the last few years, new RLC mutations have been identified (Caleshu et al. 2011; Santos et al. 2012), with some detected multiple times and in different ethnic populations (Andersen et al. 2009; Garcia-Pavia et al. 2011). Multiple reports of MYL2 genetic variations have been published to date. They are A13T (Andersen et al. 2001; Hougs et al. 2005; Poetter et al. 1996), F18L (Flavigny et al. 1998; Richard et al. 2003), M20L (Olivotto et al. 2008), E22K (Garcia-Pavia et al. 2011; Kabaeva et al. 2002; Poetter et al. 1996); I44M (Santos et al. 2012), N47K (Andersen et al. 2001, 2009), G57E (Caleshu et al. 2011), R58Q (Flavigny et al. 1998; Kabaeva et al. 2002; Morner et al. 2003; Olivotto et al. 2008; Richard et al. 2003), P95A (Poetter et al. 1996), K104E (Andersen et al. 2001), E134A (Di Donna et al. 2010; Olivotto et al. 2008); D166V, IVS5-2 (an A > G transversion in intron 5 that leads to a premature termination codon) (Richard et al. 2003), and IVS6-1 (a G > C transversion in the acceptor splice site of intron 6) (Andersen et al. 2001). Recently, a novel sarcomeric protein mutation in the MYL2 gene was identified by exome sequencing in a pedigree with familial dilated cardiomyopathy (DCM) (Huang et al. 2015). The missense variant occurred at Asp-94, mutating it to Ala (D94A), and was not observed in 5400 Exome Variant Server control DNAs (Huang et al. 2015) (Fig. 1).
Myosin regulatory light chain phosphorylation in health and disease
In the early 1970s, S.V. Perry suggested that myosin light chains could exist as charged variants (Perrie et al. 1972, 1973), shortly after which phosphorylation was established as a significant mechanism in cardiac and skeletal muscle (Frearson and Perry 1975). Myosin RLC phosphorylation-induced enhancement of isometric force and increased rates of muscle contraction have been reported in a wealth of studies from different laboratories (Colson et al. 2010; Davis et al. 2001; Greenberg et al. 2009; Scruggs and Solaro 2011; Sheikh et al. 2012; Sweeney et al. 1993; Warren et al. 2012). Specifically, RLC phosphorylation was shown to accelerate the rate of cross-bridge entry into the force generating state by regulating the proximity as well as the interaction of myosin with actin, thereby increasing Ca2+ sensitivity and amplitude of force at all activation levels (Colson et al. 2010; Szczesna et al. 2002).
In the human heart, Davis and Epstein proposed that myosin RLC phosphorylation is high in the epicardium and gradually decreases from the apex to the base, contributing to the twisting motion (torsion) of the ventricles during systole, and thus regulating diastolic and systolic function of the heart (Davis et al. 2001). This claim of a spatial gradient of RLC phosphorylation, however, has been challenged by the Stull group and quantitative measurements have revealed no evidence for its existence in the hearts of WT and transgenic mice overexpressing cMLCK (Huang et al. 2008; Kamm and Stull 2011). Significantly depressed Ser-15 RLC phosphorylation was reported in HF patients (van der Velden et al. 2003a, b) and was also observed in experimental animal models of cardiac disease (Scruggs et al. 2009; Sheikh et al. 2012; Yuan et al. 2015). Studies in mice showed that reduced RLC phosphorylation resulted in abnormal heart performance, presumably through morphological and/or myofibrillar functional alterations (e.g., change in force, myofilament calcium sensitivity, ATPase activity, cross-bridge kinetics) (Huang et al. 2008; Kerrick et al. 2009b; Warren et al. 2012). Likewise, attenuation of RLC phosphorylation in cMLCK knock-out mice was demonstrated to cause ventricular hypertrophy, fibrosis and dilated cardiomyopathy (Ding et al. 2010). Scruggs et al. (2009) determined the effects of RLC phosphorylation on the in situ cardiac systolic mechanics and in vitro myofibrillar mechanics using a transgenic mouse model expressing a cardiac specific non-phosphorylatable RLC [Tg-RLC(P-)]. The group showed a decrease in base-line load-independent measures of contractility and power as well as an increase in ejection duration together with a depression in phosphorylation levels of MyBP-C and TnI. Interestingly, following β1-adrenergic stimulation, non-transgenic controls were significantly phosphorylated at RLC Ser-15, which was abrogated in Tg-RLC(P-). The study showed the importance of RLC phosphorylation in maintenance of normal ejection fraction and β1-adrenergic responsiveness (Scruggs et al. 2009). These reports indicate that maintaining the physiological levels of RLC phosphorylation is critical for the normal function of the heart and suggest that the myocardium containing dephosphorylated myosin, the molecular motor of muscle contraction, has a reduced ability to produce force and maintain cardiac function at physiological levels.
A number of mutations known to cause HCM reside in the region of the Ser-15 phosphorylation site. Szczesna et al. (2001) reported that A13T (in close proximity to Ser-15) led to decreased phosphorylation levels, impaired secondary structure and reduced calcium binding properties of RLC, which were normalized following phosphorylation. DCM-linked D94A-RLC mutation was shown to significantly alter the N-terminal α-helical region of the RLC, and trigger intramolecular rearrangements in the RLC molecule that ultimately resulted in alterations of RLC function (Huang et al. 2015). Other studies from Szczesna et al.’s laboratory have also revealed compromised RLC phosphorylation in several different HCM models (Tg-R58Q and Tg-D166V), occurring concurrently with diminished maximal tension and altered Ca2+ sensitivity of contraction (Abraham et al. 2009; Kerrick et al. 2009a; Muthu et al. 2012b). Studies with HCM causing mutations in the myosin RLC suggest a correlation between the severity of cardiomyopathy phenotype and the level of RLC phosphorylation in vivo (Huang et al. 2014; Kerrick et al. 2009b; Muthu et al. 2012b). The compromised phosphorylation of RLC mutants compared to WT suggests that diminished RLC phosphorylation observed in HCM hearts may have an important physiological role and is central to the understanding of the mutation-elicited detrimental cardiomyopathy phenotype. It is likely that a mutation-induced conformational change in the α-helical MHC lever arm domain contributes to diminished RLC phosphorylation. The mutation may also affect the incorporation of the mutant RLC into the MHC and impose changes on the interaction of RLC with myosin heavy chain and possibly with the ELC modulating myosin unitary step-size (Wang et al. 2013).
Therapeutic potential of myosin RLC pseudo-phosphorylation
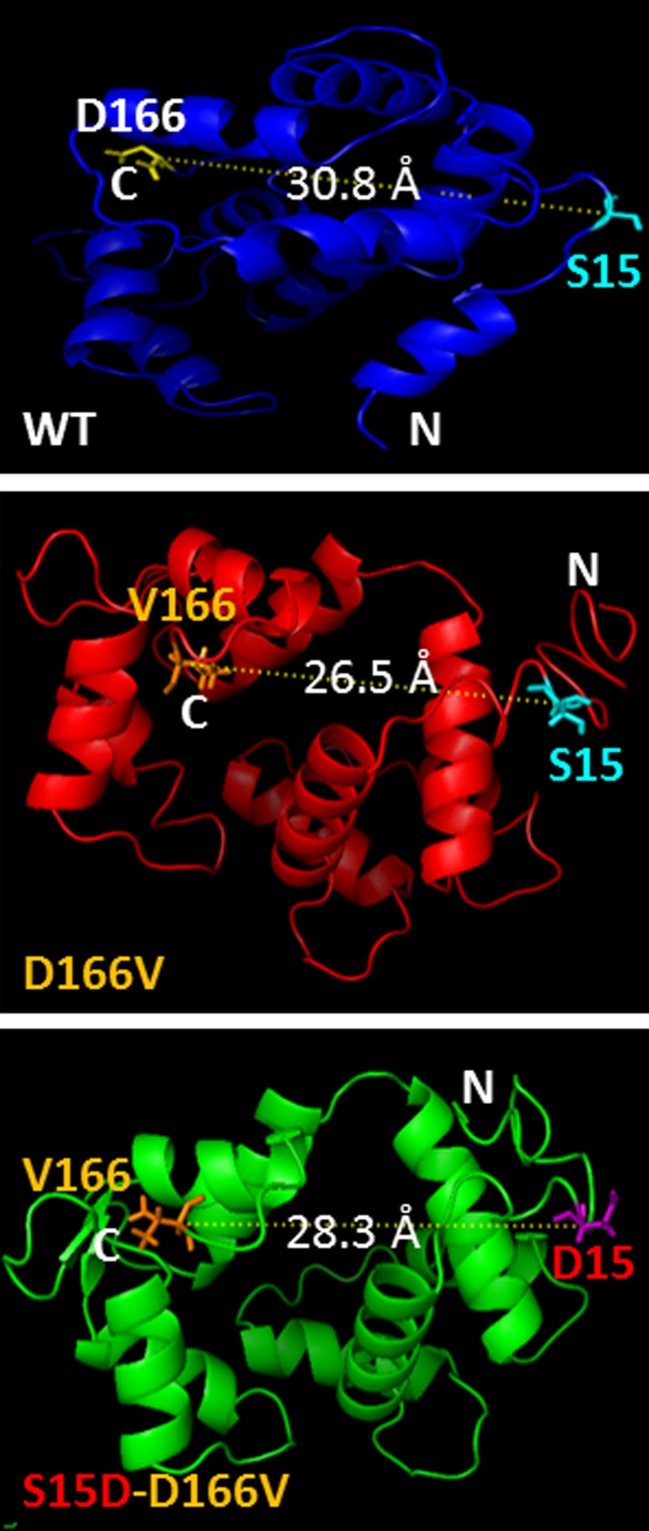
Pseudo-phosphorylation of myosin RLC has been used both in vitro and in vivo as a genetic approach to normalize RLC phosphorylation (Yu et al. 2016). Yuan et al. (2015) recently showed that hypertrophic cardiomyopathy caused by the D166V RLC mutation could be prevented/rescued by cardiac-specific expression of the pseudo-phosphorylated variant (S15D) of the human ventricular D166V-RLC. Using transgenic S15D-D166V murine models, the group reported significant improvements of intact heart function in S15D-D166V mice compared with D166V alone with the systolic and diastolic indices reaching those monitored in WT mice. A largely reduced maximal tension and abnormally high myofilament Ca2+ sensitivity observed in D166V-mutated hearts were reversed in S15D-D166V mice (Yuan et al. 2015). Previously, Muthu et al. (2014) examined in vitro the effectiveness of pseudo-phosphorylation of cardiac myosin RLC, using recombinant phosphomimetic RLC mutants reconstituted in porcine cardiac muscle preparations. They showed an S15D-induced rescue of both the enzymatic and binding properties of D166V-myosin to actin as well as an increase in the force production capacity in the in vitro motility assays (Muthu et al. 2014). These results demonstrated for the first time that RLC pseudo-phosphorylation can be used as a novel target to reverse the majority of the mutation-induced phenotypes, the idea that was ultimately tested in transgenic mice (Yuan et al. 2015). Mechanistically, they proposed that S15D-mediated structural changes in the RLC molecule may augment acto–myosin interactions and the generation of force, thus improving cardiac contractility and averting detrimental heart remodeling (Fig. 2). The study suggested that a serine-15 to aspartic acid substitution may be capable of correcting the mutation-induced inhibitory conformation of the RLC that is rendered unphosphorylatable in the D166V-HCM hearts (Fig. 2) (Yuan et al. 2015).
Fig. 2.
Modeled structure of the human ventricular RLC-WT, D166V and S15D-D166V mutant proteins. Note the mutation-rendered changes in the C-α distance between the site of HCM mutation and the myosin RLC phosphorylation site at Serine15 (S15). The predicted structures of RLCs were based on pdb structures 3jvtB, 1prwA, 4ik1A, 2mysA, 4i2yA and 2w4aB. The predicted structures were then modeled using the PyMOL (www.pymol.org) molecular visualization system to allow determination of the C-α distances (Å) between neighboring amino acid residues. From Yuan et al. (2015)
Pseudo-phosphorylation has also been explored to investigate regulatory roles of phosphorylation in other sarcomeric proteins. Pseudo-phosphorylated Tm was shown to increase maximal myosin ATPase activity without altering the steady-state Ca2+ sensitivity of the myofilament, thereby regulating muscle relaxation dynamics (Nixon et al. 2013). Similarly, viral-based gene transfer studies show that replacing myofilament cardiac TnI with phosphomimetic cTnI S43/S45D may serve as modulatory brakes by slowing/reducing contraction as well as leading to adaptive changes to modulate cross-bridges cycling and fine-tune contractile performance (Lang et al. 2015). Finally, phosphorylation in myosin essential light chain has also been studied. Scheid et al. (2016) investigated the role of the highly conserved Ser-195 phosphorylation site of the ELC using heterozygous adult zebrafish lazy susan (laz m647) in regulating contractile function in normal physiology and disease. After physical stress, heart function of laz heterozygous zebrafish severely deteriorated, causing heart failure and sudden death. They showed that, upon physical stress, ELCs become phosphorylated and the lack of Ser-195 dominant-negatively impairs ELC phosphorylation, resulting in altered acto-myosin sliding velocities and myosin binding cooperativity, and causing reduced force generation and organ dysfunction. Interestingly, the group previously showed that phosphomimetic S195D-ELC could rescue the phenotype of homozygous laz m647 (Meder et al. 2009). Similarly, S195D pseudo-phosphorylation of ELC mutants linked to HCM was recently shown to rescue the abnormally high Ca2+-sensitivity of force and compromised ATPase activity in ELC-exchanged porcine cardiac muscle preparations (Huang and Szczesna-Cordary 2015; Szczesna-Cordary and de Tombe 2016).
Structural effects associated with myosin RLC pseudo/phosphorylation
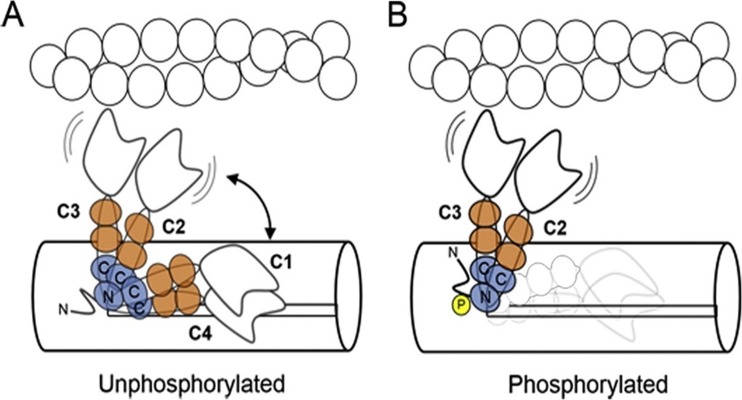
Structural studies on muscle filaments implicated the negative charge of myosin RLC phosphorylation in repelling the myosin heads away from the thick filament toward actin as a mechanism with an increase in the likelihood of cross-bridge binding and force production (Colson et al. 2010; Levine et al. 1995, 1998; Metzger et al. 1989). Electron microscopy studies of isolated myosins and thick filaments suggested a conserved molecular mechanism in which RLC phosphorylation activates or potentiates contractility by disrupting a compact ‘OFF’ conformation of myosin in which the myosin heads are folded back on the myosin tail (Jung et al. 2008; Wendt et al. 2001; Woodhead et al. 2005). Using bifunctional rhodamine probes on the cardiac RLC, Kampourakis et al. have reported that phosphorylation enhances active force and its Ca2+-sensitivity and alters thick filament structure, with the myosin head domains becoming more perpendicular to the filament axis (Fig. 3) (Kampourakis and Irving 2015; Kampourakis et al. 2016). RLC phosphorylation is thus believed to alter the conformational equilibrium towards states in which the heads can more readily interact with the thin filament. In the absence of phosphorylation, myosin S1 and RLC regions are in conformational equilibrium between binding to the thick filament backbone (OFF state) and moving away from the backbone (ON state), while RLC phosphorylation shifts this equilibrium towards the ON state (Kampourakis and Irving 2015) (Fig. 3). Interestingly, the authors reported that cardiac RLC phosphorylation does not lead to a disordered structure and an ordered RLC–thick filament backbone interaction is maintained post-phosphorylation even in the calcium-activated systolic state (Kampourakis and Irving 2015). This evidence challenges the previous studies that (1) implicated “weakening/breaking” mechanism by which RLC phosphorylation controls myosin head conformation by disrupting a compact OFF conformation of myosin (Jung et al. 2008) or (2) maintained that RLC phosphorylation produces an order-to-disorder transition in skeletal muscle thick filaments (Levine et al. 1995; Stewart et al. 2010). The study also suggested a rather global change in RLC environment associated with changes in tertiary structure/intermolecular interactions, which argues against a previously reported unchanged conformation post-phosphorylation in scallop catch muscle myosin (Kampourakis and Irving 2015; Kumar et al. 2011).
Fig. 3.
Schematic model for the effect of cRLC phosphorylation on myosin head orientation. a Unphosphorylated myosin heads are in a conformational equilibrium between more parallel OFF-states (peaks C1 and C4,) and more perpendicular ON-states (peaks C2 and C3), controlled by the interaction of the cRLC with the thick filament surface. The cRLC and ELC are shown in blue and orange, respectively. The N- and C-terminal lobes of the cRLC are labeled accordingly. b Phosphorylation of cRLC destabilizes the parallel OFF-conformations (C1 and C4) promoting attachment of the catalytic domain of myosin to actin. From Kampourakis and Irving (2015)
Using a Q-dot assay and single molecule approach, the Burghardt group showed that RLC phosphorylation is a significant modulator of myosin activation mechanism facilitating the movement of actin filaments with the largest 8-nm step-size compared to a non-phosphorylated 5-nm step-size translation (Wang et al. 2014). The authors argued that the longer working stroke upon myosin RLC phosphorylation is due to facilitation of the direct N-terminus ELC–actin interaction, another interesting mechanism for RLC pseudo/phosphorylation-mediated enhancement of cardiac function (Wang et al. 2014). Using molecular dynamics simulations, the Thomas group studied the effect of pseudo-phosphorylation mutants of smooth muscle RLC on the structural dynamics and showed that a simple charge replacement at position Ser-19 does not mimic the activating effect of myosin phosphorylation (Espinoza-Fonseca et al. 2014). They showed that phosphorylation produces a unique conformational balance in the RLC, which most likely plays a key role in smooth muscle activation (Espinoza-Fonseca et al. 2014).
In summary, RLC phosphorylation has been shown to be an important modulator of muscle cell dynamics and function, and that targeting this post-translational modification may be of great importance for clinical medicine aimed at effective and target-specific treatments for HCM/HF patients. In cardiac hypertrophy, the heart responds to a multitude of intrinsic and extrinsic stimuli that increase biomechanical stress (Frey and Olson 2003). RLC phosphorylation may play a role in an adaptive cardiac response by facilitating the acto-myosin interaction and thereby enhancing contractility. This involves a regulated conformational shift and possibly a facilitated longer working stroke. As a result, regulating RLC phosphorylation levels is a major therapeutic target in the field of cardiomyopathy and HF subjects. We also report on studies that have successfully examined the efficacy of pseudo-phosphorylation as a potential rescue mechanism, both in vitro and in vivo, and therefore may form the basis of a working proof-of-principle concept for future therapeutic interventions.
Compliance with ethical standards
Funding
This work was supported by NIH-HL123255 (DSC).
Conflict of interest
Sunil Yadav declares that he has no conflict of interest. Danuta Szczesna-Cordary declares that she has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Abraham TP, Jones M, Kazmierczak K, Liang HY, Pinheiro AC, Wagg CS, Lopaschuk GD, Szczesna-Cordary D. Diastolic dysfunction in familial hypertrophic cardiomyopathy transgenic model mice. Cardiovasc Res. 2009;82:84–92. doi: 10.1093/cvr/cvp016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcalai R, Seidman JG, Seidman CE. Genetic basis of hypertrophic cardiomyopathy: from bench to the clinics. J Cardiovasc Electrophysiol. 2008;19:104–110. doi: 10.1111/j.1540-8167.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- Andersen PS, Havndrup O, Bundgaard H, Moolman-Smook JC, Larsen LA, Mogensen J, Brink PA, Børglum AD, Corfield VA, Kjeldsen K, et al. Myosin light chain mutations in familial hypertrophic cardiomyopathy: phenotypic presentation and frequency in Danish and South African populations. J Med Genet. 2001;38 doi: 10.1136/jmg.38.12.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen PS, Havndrup O, Hougs L, Sorensen KM, Jensen M, Larsen LA, Hedley P, Thomsen AR, Moolman-Smook J, Christiansen M, et al. Diagnostic yield, interpretation, and clinical utility of mutation screening of sarcomere encoding genes in Danish hypertrophic cardiomyopathy patients and relatives. Hum Mutat. 2009;30:363–370. doi: 10.1002/humu.20862. [DOI] [PubMed] [Google Scholar]
- Aoki H, Sadoshima J, Izumo S. Myosin light chain kinase mediates sarcomere organization during cardiac hypertrophy in vitro. Nat Med. 2000;6:183–188. doi: 10.1038/72287. [DOI] [PubMed] [Google Scholar]
- Bers DM, Harris SP. Translational medicine: to the rescue of the failing heart. Nature. 2011;473:36–39. doi: 10.1038/473036a. [DOI] [PubMed] [Google Scholar]
- Binder WD, Fifer MA, King ME, Stone JR. Case records of the Massachusetts General Hospital. Case 26-2005. A 48-year-old man with sudden loss of consciousness while jogging. N Engl J Med. 2005;353:824–832. doi: 10.1056/NEJMcpc059021. [DOI] [PubMed] [Google Scholar]
- Burghardt TP, Sikkink LA. Regulatory light chain mutants linked to heart disease modify the cardiac myosin lever arm. Biochemistry. 2013;52:1249–1259. doi: 10.1021/bi301500d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caleshu C, Sakhuja R, Nussbaum RL, Schiller NB, Ursell PC, Eng C, De Marco T, McGlothlin D, Burchard EG, Rame JE. Furthering the link between the sarcomere and primary cardiomyopathies: restrictive cardiomyopathy associated with multiple mutations in genes previously associated with hypertrophic or dilated cardiomyopathy. Am J Med Genet A. 2011;155:2229–2235. doi: 10.1002/ajmg.a.34097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson BA, Locher MR, Bekyarova T, Patel JR, Fitzsimons DP, Irving TC, Moss RL. Differential roles of regulatory light chain and myosin binding protein-C phosphorylations in the modulation of cardiac force development. J Physiol. 2010;588:981–993. doi: 10.1113/jphysiol.2009.183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JS, Hassanzadeh S, Winitsky S, Lin H, Satorius C, Vemuri R, Aletras AH, Wen H, Epstein ND. The overall pattern of cardiac contraction depends on a spatial gradient of myosin regulatory light chain phosphorylation. Cell. 2001;107:631–641. doi: 10.1016/S0092-8674(01)00586-4. [DOI] [PubMed] [Google Scholar]
- Di Donna P, Olivotto I, Delcre SD, Caponi D, Scaglione M, Nault I, Montefusco A, Girolami F, Cecchi F, Haissaguerre M, et al. Efficacy of catheter ablation for atrial fibrillation in hypertrophic cardiomyopathy: impact of age, atrial remodelling, and disease progression. Europace. 2010;12:347–355. doi: 10.1093/europace/euq013. [DOI] [PubMed] [Google Scholar]
- Ding P, Huang J, Battiprolu PK, Hill JA, Kamm KE, Stull JT. Cardiac myosin light chain kinase is necessary for myosin regulatory light chain phosphorylation and cardiac performance in vivo. J Biol Chem. 2010;285:40819–40829. doi: 10.1074/jbc.M110.160499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Fonseca LM, Colson BA, Thomas DD. Effects of pseudophosphorylation mutants on the structural dynamics of smooth muscle myosin regulatory light chain. Mol BioSyst. 2014;10:2693–2698. doi: 10.1039/C4MB00364K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavigny J, Richard P, Isnard R, Carrier L, Charron P, Bonne G, Forissier JF, Desnos M, Dubourg O, Komajda M, et al. Identification of two novel mutations in the ventricular regulatory myosin light chain gene (MYL2) associated with familial and classical forms of hypertrophic cardiomyopathy. J Mol Med (Berl) 1998;76:208–214. doi: 10.1007/s001090050210. [DOI] [PubMed] [Google Scholar]
- Frearson N, Perry SV. Phosphorylation of the light-chain components of myosin from cardiac and red skeletal muscles. Biochem J. 1975;151:99–107. doi: 10.1042/bj1510099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey N, Olson EN. Cardiac hypertrophy: the good, the bad, and the ugly. Annu Rev Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- Garcia-Pavia P, Vazquez ME, Segovia J, Salas C, Avellana P, Gomez-Bueno M, Vilches C, Gallardo ME, Garesse R, Molano J, et al. Genetic basis of end-stage hypertrophic cardiomyopathy. Eur J Heart Fail. 2011;13:1193–1201. doi: 10.1093/eurjhf/hfr110. [DOI] [PubMed] [Google Scholar]
- Geeves MA. Molecular motors: stretching the lever-arm theory. Nature. 2002;415:129–131. doi: 10.1038/415129a. [DOI] [PubMed] [Google Scholar]
- Geeves MA, Holmes KC. The molecular mechanism of muscle contraction. Adv Protein Chem. 2005;71:161–193. doi: 10.1016/S0065-3233(04)71005-0. [DOI] [PubMed] [Google Scholar]
- Granzier HL, de Tombe PP. Myosin light chain phosphorylation to the rescue. Proc Natl Acad Sci U S A. 2015;112:9148–9149. doi: 10.1073/pnas.1511455112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EM, Wakimoto H, Anderson RL, Evanchik MJ, Gorham JM, Harrison BC, Henze M, Kawas R, Oslob JD, Rodriguez HM, et al. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science. 2016;351:617–621. doi: 10.1126/science.aad3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MJ, Mealy TR, Watt JD, Jones M, Szczesna-Cordary D, Moore JR. The molecular effects of skeletal muscle myosin regulatory light chain phosphorylation. Am J Physiol Regul Integr Comp Physiol. 2009;297:R265–R274. doi: 10.1152/ajpregu.00171.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CY, Sweitzer NK, McDonough B, Maron BJ, Casey SA, Seidman JG, Seidman CE, Solomon SD. Assessment of diastolic function with Doppler tissue imaging to predict genotype in preclinical hypertrophic cardiomyopathy. Circulation. 2002;105:2992–2997. doi: 10.1161/01.CIR.0000019070.70491.6D. [DOI] [PubMed] [Google Scholar]
- Ho CY, Lopez B, Coelho-Filho OR, Lakdawala NK, Cirino AL, Jarolim P, Kwong R, Gonzalez A, Colan SD, Seidman JG, et al. Myocardial fibrosis as an early manifestation of hypertrophic cardiomyopathy. N Engl J Med. 2010;363:552–563. doi: 10.1056/NEJMoa1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hougs L, Havndrup O, Bundgaard H, Kober L, Vuust J, Larsen LA, Christiansen M, Andersen PS. One third of Danish hypertrophic cardiomyopathy patients have mutations in MYH7 rod region. Eur J Hum Genet. 2005;13:161–165. doi: 10.1038/sj.ejhg.5201310. [DOI] [PubMed] [Google Scholar]
- Huang W, Szczesna-Cordary D. Molecular mechanisms of cardiomyopathy phenotypes associated with myosin light chain mutations. J Muscle Res Cell Motil. 2015;36:433–445. doi: 10.1007/s10974-015-9423-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Shelton JM, Richardson JA, Kamm KE, Stull JT. Myosin regulatory light chain phosphorylation attenuates cardiac hypertrophy. J Biol Chem. 2008;283:19748–19756. doi: 10.1074/jbc.M802605200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Liang J, Kazmierczak K, Muthu P, Duggal D, Farman GP, Sorensen L, Pozios I, Abraham T, Moore JR, et al. Hypertrophic cardiomyopathy associated Lys104Glu mutation in the myosin regulatory light chain causes diastolic disturbance in mice. J Mol Cell Cardiol. 2014;74:318–329. doi: 10.1016/j.yjmcc.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Liang J, Yuan CC, Kazmierczak K, Zhou Z, Morales A, McBride KL, Fitzgerald-Butt SM, Hershberger RE, Szczesna-Cordary D. Novel familial dilated cardiomyopathy mutation in MYL2 affects the structure and function of myosin regulatory light chain. FEBS J. 2015;282:2379–2393. doi: 10.1111/febs.13286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HS, Komatsu S, Ikebe M, Craig R. Head–head and head–tail interaction: a general mechanism for switching off myosin II activity in cells. Mol Biol Cell. 2008;19:3234–3242. doi: 10.1091/mbc.E08-02-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabaeva ZT, Perrot A, Wolter B, Dietz R, Cardim N, Correia JM, Schulte HD, Aldashev AA, Mirrakhimov MM, Osterziel KJ. Systematic analysis of the regulatory and essential myosin light chain genes: genetic variants and mutations in hypertrophic cardiomyopathy. Eur J Hum Genet. 2002;10:741–748. doi: 10.1038/sj.ejhg.5200872. [DOI] [PubMed] [Google Scholar]
- Kamm KE, Stull JT. Signaling to myosin regulatory light chain in sarcomeres. J Biol Chem. 2011;286:9941–9947. doi: 10.1074/jbc.R110.198697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampourakis T, Irving M. Phosphorylation of myosin regulatory light chain controls myosin head conformation in cardiac muscle. J Mol Cell Cardiol. 2015;85:199–206. doi: 10.1016/j.yjmcc.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampourakis T, Sun YB, Irving M. Myosin light chain phosphorylation enhances contraction of heart muscle via structural changes in both thick and thin filaments. Proc Natl Acad Sci U S A. 2016;113:E3039–E3047. doi: 10.1073/pnas.1602776113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannel WB. Vital epidemiologic clues in heart failure. J Clin Epidemiol. 2000;53:229–235. doi: 10.1016/S0895-4356(99)00135-3. [DOI] [PubMed] [Google Scholar]
- Kerrick WG, Kazmierczak K, Xu Y, Wang Y, Szczesna-Cordary D. Malignant familial hypertrophic cardiomyopathy D166V mutation in the ventricular myosin regulatory light chain causes profound effects in skinned and intact papillary muscle fibers from transgenic mice. FASEB J. 2009;23:855–865. doi: 10.1096/fj.08-118182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrick WGL, Kazmierczak K, Xu Y, Wang Y, Szczesna-Cordary D. Malignant familial hypertrophic cardiomyopathy D166V mutation in the ventricular myosin regulatory light chain causes profound effects in skinned and intact papillary muscle fibers from transgenic mice. FASEB J. 2009;23:855–865. doi: 10.1096/fj.08-118182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar VS, O’Neall-Hennessey E, Reshetnikova L, Brown JH, Robinson H, Szent-Gyorgyi AG, Cohen C. Crystal structure of a phosphorylated light chain domain of scallop smooth-muscle myosin. Biophys J. 2011;101:2185–2189. doi: 10.1016/j.bpj.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang SE, Schwank J, Stevenson TK, Jensen MA, Westfall MV. Independent modulation of contractile performance by cardiac troponin I Ser43 and Ser45 in the dynamic sarcomere. J Mol Cell Cardiol. 2015;79:264–274. doi: 10.1016/j.yjmcc.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RJ, Kensler RW, Yang Z, Sweeney HL. Myosin regulatory light chain phosphorylation and the production of functionally significant changes in myosin head arrangement on striated muscle thick filaments. Biophys J. 1995;68:224S. [PMC free article] [PubMed] [Google Scholar]
- Levine RJ, Yang Z, Epstein ND, Fananapazir L, Stull JT, Sweeney HL. Structural and functional responses of mammalian thick filaments to alterations in myosin regulatory light chains. J Struct Biol. 1998;122:149–161. doi: 10.1006/jsbi.1998.3980. [DOI] [PubMed] [Google Scholar]
- Malik FI, Hartman JJ, Elias KA, Morgan BP, Rodriguez H, Brejc K, Anderson RL, Sueoka SH, Lee KH, Finer JT, et al. Cardiac myosin activation: a potential therapeutic approach for systolic heart failure. Science. 2011;331:1439–1443. doi: 10.1126/science.1200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron BJ. The young competitive athlete with cardiovascular abnormalities: causes of sudden death, detection by preparticipation screening, and standards for disqualification. Card Electrophysiol Rev. 2002;6:100–103. doi: 10.1023/A:1017903709361. [DOI] [PubMed] [Google Scholar]
- Massengill MT, Ashraf HM, Chowdhury RR, Chrzanowski SM, Kar J, Warren SA, Walter GA, Zeng H, Kang BH, Anderson RH, Moss RL, Kasahara H. Acute heart failure with cardiomyocyte atrophy induced in adult mice by ablation of cardiac myosin light chain kinase. Cardiovasc Res. 2016;111:34–43. doi: 10.1093/cvr/cvw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meder B, Laufer C, Hassel D, Just S, Marquart S, Vogel B, Hess A, Fishman MC, Katus HA, Rottbauer W. A single serine in the carboxyl terminus of cardiac essential myosin light chain-1 controls cardiomyocyte contractility in vivo. Circ Res. 2009;104:650–659. doi: 10.1161/CIRCRESAHA.108.186676. [DOI] [PubMed] [Google Scholar]
- Metzger JM, Greaser ML, Moss RL. Variations in cross-bridge attachment rate and tension with phosphorylation of myosin in mammalian skinned skeletal muscle fibers. Implications for twitch potentiation in intact muscle. J Gen Physiol. 1989;93:855–883. doi: 10.1085/jgp.93.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JR, Leinwand L, Warshaw DM. Understanding cardiomyopathy phenotypes based on the functional impact of mutations in the myosin motor. Circ Res. 2012;111:375–385. doi: 10.1161/CIRCRESAHA.110.223842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morner S, Richard P, Kazzam E, Hellman U, Hainque B, Schwartz K, Waldenstrom A. Identification of the genotypes causing hypertrophic cardiomyopathy in northern Sweden. J Mol Cell Cardiol. 2003;35:841–849. doi: 10.1016/S0022-2828(03)00146-9. [DOI] [PubMed] [Google Scholar]
- Muthu P, Huang W, Kazmierczak K, Szczesna-Cordary D (2012a) Functional consequences of mutations in the myosin regulatory light chain associated with hypertrophic cardiomyopathy. In: Veselka J (ed) Cardiomyopathies – from basic research to clinical management. Ch. 17. InTech, Croatia, pp 383–408
- Muthu P, Kazmierczak K, Jones M, Szczesna-Cordary D. The effect of myosin RLC phosphorylation in normal and cardiomyopathic mouse hearts. J Cell Mol Med. 2012;16:911–919. doi: 10.1111/j.1582-4934.2011.01371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthu P, Liang J, Schmidt W, Moore JR, Szczesna-Cordary D. In vitro rescue study of a malignant familial hypertrophic cardiomyopathy phenotype by pseudo-phosphorylation of myosin regulatory light chain. Arch Biochem Biophys. 2014;552–553:29–39. doi: 10.1016/j.abb.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon BR, Liu B, Scellini B, Tesi C, Piroddi N, Ogut O, Solaro RJ, Ziolo MT, Janssen PM, Davis JP, et al. Tropomyosin Ser-283 pseudo-phosphorylation slows myofibril relaxation. Arch Biochem Biophys. 2013;535:30–38. doi: 10.1016/j.abb.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivotto I, Girolami F, Ackerman MJ, Nistri S, Bos JM, Zachara E, Ommen SR, Theis JL, Vaubel RA, Re F, et al. Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clin Proc. 2008;83:630–638. doi: 10.1016/S0025-6196(11)60890-2. [DOI] [PubMed] [Google Scholar]
- Olivotto I, Girolami F, Nistri S, Rossi A, Rega L, Garbini F, Grifoni C, Cecchi F, Yacoub MH. The many faces of hypertrophic cardiomyopathy: from developmental biology to clinical practice. J Cardiovasc Transl Res. 2009;2:349–367. doi: 10.1007/s12265-009-9137-2. [DOI] [PubMed] [Google Scholar]
- Perrie WT, Smillie LB, Perry SV. A phosphorylated light-chain component of myosin. Biochem J. 1972;128:105P–106P. doi: 10.1042/bj1280105P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrie WT, Smillie LB, Perry SB. A phosphorylated light-chain component of myosin from skeletal muscle. Biochem J. 1973;135:151–164. doi: 10.1042/bj1350151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poetter K, Jiang H, Hassanzadeh S, Master SR, Chang A, Dalakas MC, Rayment I, Sellers JR, Fananapazir L, Epstein ND. Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle. Nat Genet. 1996;13:63–69. doi: 10.1038/ng0596-63. [DOI] [PubMed] [Google Scholar]
- Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, Benaiche A, Isnard R, Dubourg O, Burban M, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- Santos S, Marques V, Pires M, Silveira L, Oliveira H, Lanca V, Brito D, Madeira H, Fonseca E, Freitas A, et al. High resolution melting: improvements in the genetic diagnosis of hypertrophic cardiomyopathy in a Portuguese cohort. BMC Med Gen. 2012;13:17. doi: 10.1186/1471-2350-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid LM, Mosqueira M, Hein S, Kossack M, Juergensen L, Mueller M, Meder B, Fink RH, Katus HA, Hassel D. Essential light chain S195 phosphorylation is required for cardiac adaptation under physical stress. Cardiovasc Res. 2016;111:44–55. doi: 10.1093/cvr/cvw066. [DOI] [PubMed] [Google Scholar]
- Scruggs SB, Solaro RJ. The significance of regulatory light chain phosphorylation in cardiac physiology. Arch Biochem Biophys. 2011;510:129–134. doi: 10.1016/j.abb.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scruggs SB, Hinken AC, Thawornkaiwong A, Robbins J, Walker LA, de Tombe PP, Geenen DL, Buttrick PM, Solaro RJ. Ablation of ventricular myosin regulatory light chain phosphorylation in mice causes cardiac dysfunction in situ and affects neighboring myofilament protein phosphorylation. J Biol Chem. 2009;284:5097–5106. doi: 10.1074/jbc.M807414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scruggs SB, Reisdorph R, Armstrong ML, Warren CM, Reisdorph N, Solaro RJ, Buttrick PM. A novel, in-solution separation of endogenous cardiac sarcomeric proteins and identification of distinct charged variants of regulatory light chain. Mol Cell Proteomics. 2010;9:1804–1818. doi: 10.1074/mcp.M110.000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguchi O, Takashima S, Yamazaki S, Asakura M, Asano Y, Shintani Y, Wakeno M, Minamino T, Kondo H, Furukawa H, et al. A cardiac myosin light chain kinase regulates sarcomere assembly in the vertebrate heart. J Clin Invest. 2007;117:2812–2824. doi: 10.1172/JCI30804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh F, Ouyang K, Campbell SG, Lyon RC, Chuang J, Fitzsimons D, Tangney J, Hidalgo CG, Chung CS, Cheng H, et al. Mouse and computational models link Mlc2v dephosphorylation to altered myosin kinetics in early cardiac disease. J Clin Invest. 2012;122:1209–1221. doi: 10.1172/JCI61134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MA, Franks-Skiba K, Chen S, Cooke R. Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers. Proc Natl Acad Sci U S A. 2010;107:430–435. doi: 10.1073/pnas.0909468107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney HL, Stull JT. Phosphorylation of myosin in permeabilized mammalian cardiac and skeletal muscle cells. Am J Phys. 1986;250:C657–C660. doi: 10.1152/ajpcell.1986.250.4.C657. [DOI] [PubMed] [Google Scholar]
- Sweeney HL, Bowman BF, Stull JT. Myosin light chain phosphorylation in vertebrate striated muscle: regulation and function. Am J Phys. 1993;264:C1085–C1095. doi: 10.1152/ajpcell.1993.264.5.C1085. [DOI] [PubMed] [Google Scholar]
- Szczesna D. Regulatory light chains of striated muscle myosin. Structure, function and malfunction. Curr Drug Targets Cardiovasc Haematol Disord. 2003;3:187–197. doi: 10.2174/1568006033481474. [DOI] [PubMed] [Google Scholar]
- Szczesna D, Ghosh D, Li Q, Gomes AV, Guzman G, Arana C, Zhi G, Stull JT, Potter JD. Familial hypertrophic cardiomyopathy mutations in the regulatory light chains of myosin affect their structure, Ca2+ binding, and phosphorylation. J Biol Chem. 2001;276:7086–7092. doi: 10.1074/jbc.M009823200. [DOI] [PubMed] [Google Scholar]
- Szczesna D, Zhao J, Jones M, Zhi G, Stull J, Potter JD. Phosphorylation of the regulatory light chains of myosin affects Ca2+ sensitivity of skeletal muscle contraction. J Appl Physiol. 2002;92:1661–1670. doi: 10.1152/japplphysiol.00858.2001. [DOI] [PubMed] [Google Scholar]
- Szczesna-Cordary D, de Tombe PP. Myosin light chain phosphorylation, novel targets to repair a broken heart? Cardiovasc Res. 2016;111:5–7. doi: 10.1093/cvr/cvw098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff JC, Carrier L, Bers DM, Poggesi C, Ferrantini C, Coppini R, Maier LS, Ashrafian H, Huke S, van der Velden J. Targets for therapy in sarcomeric cardiomyopathies. Cardiovasc Res. 2015;105:457–470. doi: 10.1093/cvr/cvv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry M, Walker DD, Ferrari MB. Protein phosphatase activity is necessary for myofibrillogenesis. Cell Biochem Biophys. 2006;45:265–278. doi: 10.1385/CBB:45:3:265. [DOI] [PubMed] [Google Scholar]
- van der Velden J, Papp Z, Boontje NM, Zaremba R, de Jong JW, Janssen PML, Hasenfuss G, Stienen GJM. The effect of myosin light chain 2 dephosphorylation on Ca2+-sensitivity of force is enhanced in failing human hearts. Cardiovasc Res. 2003;57:505–514. doi: 10.1016/S0008-6363(02)00662-4. [DOI] [PubMed] [Google Scholar]
- van der Velden J, Papp Z, Zaremba R, Boontje NM, de Jong JW, Owen VJ, Burton PBJ, Goldmann P, Jaquet K, Stienen GJM. Increased Ca2+-sensitivity of the contractile apparatus in end-stage human heart failure results from altered phosphorylation of contractile proteins. Cardiovasc Res. 2003;57:37–47. doi: 10.1016/S0008-6363(02)00606-5. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ajtai K, Burghardt TP. The Qdot-labeled actin super-resolution motility assay measures low-duty cycle muscle myosin step size. Biochemistry. 2013;52:1611–1621. doi: 10.1021/bi301702p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ajtai K, Burghardt TP. Ventricular myosin modifies in vitro step-size when phosphorylated. J Mol Cell Cardiol. 2014;72:231–237. doi: 10.1016/j.yjmcc.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren SA, Briggs LE, Zeng H, Chuang J, Chang EI, Terada R, Li M, Swanson MS, Lecker SH, Willis MS, et al. Myosin light chain phosphorylation is critical for adaptation to cardiac stress. Circulation. 2012;126:2575–2588. doi: 10.1161/CIRCULATIONAHA.112.116202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci U S A. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead JL, Zhao F-Q, Craig R, Egelman EH, Alamo L, Padron R. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436:1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- Yu H, Chakravorty S, Song W, Ferenczi MA. Phosphorylation of the regulatory light chain of myosin in striated muscle: methodological perspectives. Eur Biophys J. 2016;45:779–805. doi: 10.1007/s00249-016-1128-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan CC, Muthu P, Kazmierczak K, Liang J, Huang W, Irving TC, Kanashiro-Takeuchi RM, Hare JM, Szczesna-Cordary D. Constitutive phosphorylation of cardiac myosin regulatory light chain prevents development of hypertrophic cardiomyopathy in mice. Proc Natl Acad Sci U S A. 2015;112:E4138–E4146. doi: 10.1073/pnas.1505819112. [DOI] [PMC free article] [PubMed] [Google Scholar]