Abstract
Transcription initiation is a major control point for the precise regulation of gene expression. Our knowledge of this process has been mainly derived from protein-centric studies wherein cis-regulatory DNA sequences play a passive role, mainly in arranging the protein machinery to coalesce at the transcription start sites of genes in a spatial and temporal-specific manner. However, this is a highly dynamic process in which molecular motors such as RNA polymerase II (RNAPII), helicases, and other transcription factors, alter the level of mechanical force in DNA, rather than simply a set of static DNA–protein interactions. The double helix is a fiber that responds to flexural and torsional stress, which if accumulated, can affect promoter output as well as change DNA and chromatin structure. The relationship between DNA mechanics and the control of early transcription initiation events has been under-investigated. Genomic techniques to display topological stress and conformational variation in DNA across the mammalian genome provide an exciting new insight on the role of DNA mechanics in the early stages of the transcription cycle. Without understanding how torsional and flexural stresses are generated, transmitted, and dissipated, no model of transcription will be complete and accurate.
Keywords: Transcription initiation, DNA mechanics, Chromatin, Topoisomerase, Molecular motors, DNA topology
The basics—the DNA
The topological and conformational problems intrinsic to a B-DNA double helix were immediately apparent to Watson and Crick (Watson and Crick 1953a, b). Because the chemical functional groups that instruct genetic processes by base pairing are protected by the double helix, unwinding is required for polymerization of nascent nucleic acid or for recognition by an RNA effector (gRNA, micro RNA, etc.). In a segment of DNA with fixed ends, with each 10.4-bp right-handed helical turn, the anti-parallel strands become progressively intertwined. In such a topological domain the strands are physically linked and are inseparable without breaking the phosphodiester backbone of at least one of the strands (Vinograd and Lebowitz 1966). The number of helical turns in such a topologically closed segment of DNA is referred to as twist (Tw). Untwisting or overwinding DNA, which alters the number of base pairs per helical turn within such domains, mathematically and physically must be compensated for by deformations of the central axis of the double helix, known as ‘supercoils’. Any three-dimensional (3D) undulation in the central DNA axis not confined to the x–y plane is termed writhe (Wr) (Bates and Maxwell 2005; Cozzarelli and Wang 1990; Sinden 1994). When writhe is sustained over 360° in a circle, a ‘supercoil’ is formed in the DNA. Linking number (Lk) is the number of helical turns in a bound segment of DNA when stressed, and Lk0 is the number of turns when that same segment is unstressed; the difference between Lk and Lk0 is equal to the segment’s supercoils (ΔLk) (Bates and Maxwell 2005; Sinden 1994).
Supercoils may be constrained or unconstrained (Cozzarelli and Wang 1990). Constrained supercoils are fixed upon a surface, leaving the stresses and strains within the fixed DNA unable to equilibrate or communicate with the unbound segments. Supercoils come in two flavors: (1) plectonemic (interwound) supercoils akin to braids, with right-handed plectonemic braids being negative supercoils that impart an unwinding stress on the involved DNA; (2) solenoidal (also called toroidal) coils. Solenoidal coils may be free or may be immobilized by wrapping upon a spool—such as nucleosome (Fig. 1a). Left-handed solenoidal wraps introduce negative supercoils (4 & 7 in Fig. 1b) (Bates and Maxwell 2005; Sinden 1994). A typical nucleosome constrains ∼1.05 negative supercoils that are accounted for by ∼1.7 left-handed solenoidal wraps and ∼0.65 turns of right-handed overwinding. (Luger et al. 1997).
Fig. 1.
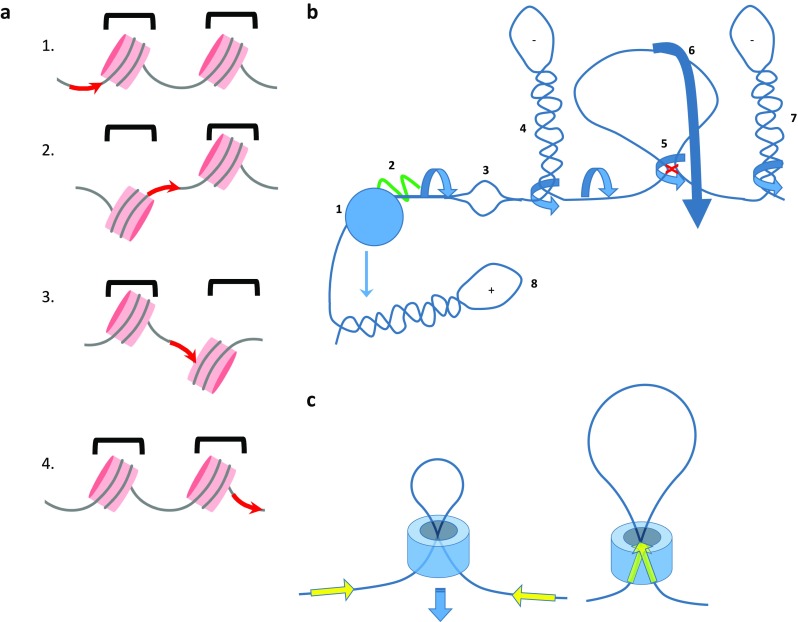
Static, constrained, and dynamic supercoils are context-dependent. a Pink disks indicate nucleosomes. Each nucleosome includes approximately ∼1.7 left-handed (negative) wraps of DNA that due to slight over-twisting (positive) of the helical axis yields a net −1.05 constrained supercoils/nucleosome. 1, 2 A dynamic pulse of torque pumped into the DNA (red arrow) must be accommodated either by propagation through the DNA, disrupting DNA–histone interactions (1), or by an en bloc rotation of the nucleosome that transmits the stress in a salutatory manner to the subsequent linker (2). 3, 4 In a similar manner the dynamic torsional stress is propagated along the chromatin fiber, etc. b The dynamic transmission of torsional stress is likely to be highly context-sensitive. Counter-rotation of RNA polymerase II (RNAPII) (1) and the template pumps torque into the chromatin fiber and—unless the RNAP is immobilized—may entwine the nascent RNA (2, green). Upstream negative supercoils may focally melt DNA (3) that can then adopt other non-B DNA conformations. Torsional stress may extrude plectonemes (4, 7) that compete for formation and growth in a static situation (the dynamics in vivo have not been described). A loop between DNA-bound upstream factors (red x) creates a topological domain insulated from dynamic supercoils (5); in such a situation a rotation of the entire loop about a stem (5) would create a plectoneme from segments embracing the domain, or a translational rotation (6) would transmit the stress as twist to the adjacent segment. If the loop were too encumbered to rotate or translocate, it would comprise a true topological boundary confining the torsional stress. c A chromatin loop can be contained within a collar that creates a topographic, but not true topological domain. For example, sliding of a cohesin (blue) ring along chromatin could help to juxtapose CTCF sites (yellow arrow) at the boundaries of a topologically associated domain. Whether the molecular architecture at these CTCF sites fixes linking number to define a topological domain has not yet been established
These topological considerations dictate that alteration of DNA secondary structure and conformation is inexorably coupled with genetic activity. Yet DNA is a tough, Hookean spring-like molecule that resists bending or twisting away from its most relaxed state. DNA is actually stiffer to twisting than to bending; it takes twice the length to dissipate the same angular displacement of a twist than of a bend (Lavelle et al. 2010). Like a spring, when bent or twisted, potential energy is distributed throughout the molecule. Superhelical density (σ) is defined as ΔLk/Lk0 and is a measure of the over- or under-winding of the double helix. In actively proliferating prokaryotes, σ of ∼ −0.05 are common. At σ = ∼ −0.1, the onset of broadening zones of melting buffers further increase in torsion (Ma et al. 2013; Strick et al. 2000); in the regime of +σ, the Hookean approximation extends well beyond +0.1 and only breaks down at much higher levels than are likely to be achieved in vivo when a major structural phase change occurs.
Melting of a DNA segment in response to −σ drains stress and focuses strain into the single-stranded segment; the relaxation of the rest of the region helps to provide the energy for strand separation (Zhabinskaya and Benham 2012, 2013). Unless mitigated, the accruing topological stresses within a topological domain would rapidly rise to levels that impede or arrest genetic processes.
The basics—the enzymes
RNA polymerases (RNAPs) are the principal molecular engines that generate supercoils in metazoans (Kouzine et al. 2013; Naughton et al. 2013; Teves and Henikoff 2014). Transcription requires the screwing of the downstream DNA through the active site of RNAP. Translocation of RNAPs relative to the template occurs via a Brownian-ratchet with hydrolysis of nucleotide triphosphates to nucleotide monophosphates and pyrophosphate (PPi). Subsequent hydrolysis of PPi to monophosphate secures the directionality of elongation (Maoileidigh et al. 2011; Sekine et al. 2015). It has been debated for decades whether the template is threaded through an immobile RNAP tethered to nuclear structures or, alternatively, whether a freely elongating enzyme tracks along and around a DNA fiber. Most likely Newton is right, and they each counter-rotate about their effective center of mass. The distinction here is not just semantic. If RNAPs are immobile, severe topological constraints are imposed on the DNA of an active gene, especially if a transcription unit includes several concurrently elongating transcription complexes. Immobilization of RNAPs might prevent the nascent RNA from becoming entwined with the template, but would augment the accumulation of torsional stress within the DNA (Cook and Gove 1992; Papantonis and Cook 2013). Above a resistance of σ = ∼ ±0.1, torque stalls transcription (Ma et al. 2013; Ma and Wang 2014). Though it is perhaps easier to imagine RNAP stalling when attempting to elongate against a stiffened, over-wound template, the underwound DNA trailing the engaged RNAP also impedes forward extension of the transcription bubble by sucking the polymerase backwards (Ma et al. 2013; Ma and Wang 2014; Strick et al. 2000). So if RNAPs and their templates counter-rotate, transcription of just 10 % of the DNA in a topological domain would arrest transcription unless the torsional stress was relieved. Indeed, in bacteria the phenomenon of transcriptional bursting in which multiple mRNAs are generated in sharp pulses separated by variable periods of inactivity has been attributed to the dyssynchrony of the rapid removal of upstream negative supercoils by topoisomerase 1A (Top1A) versus the slower and sporadic removal of the polymerase-arresting positive supercoils by DNA gyrase (Chong et al. 2014).
In a closed topological domain, strand-breakage is required to change the linking number and relieve torsional stress (Bates and Maxwell 2005; Sinden 1994). This activity is performed by specific enzymes, topoisomerases, that remove supercoils and modulate the topology of DNA in the cell (Baranello et al. 2013). In general, topoisomerases cleave one or both DNA strands, using a tyrosine to attack a phosphodiester bond. The tyrosine–DNA mixed phosphodiester intermediate is charged for attack by a terminal hydroxyl group to reestablish the integrity of the backbone, thus completing the reaction cycle. There are two general categories of topoisomerases, Top1 and Top2 (Champoux 2001). Top1s break one strand and then exploit one of two mechanisms to relieve torsional stress. In one of these mechanisms, Top1a cleaves at single-stranded bubbles and transports the unbroken strand through the enzymatically charged nicked strand, that is subsequently resealed. Because Top1a operates at a single-stranded bubble, its activity is restricted to negatively supercoiled DNAs that unwind easily; overwinding hinders the formation of such bubbles and so positively supercoiled DNA is not effectively relaxed. Using the second mechanism, Top1b cleaves one strand and so enables one or more cycles of right- or left-handed spinning around the unbroken strand, according to prevailing torsion, followed by closure of the enzyme-charged nick. Top1s act directly to change twist—any influence on the plectonemic or solenoidal trajectory of the helical axis requires a repartitioning between twist and writhe. Both Top1a and Top1b release potential energy stored in a torsionally stressed segment of DNA to reduce ∣ΔLk∣. Top2s are ATP-dependent enzymes that act directly on writhe by first locally forming a loop and then at a crossing point, transporting one double helical segment through a transient enzyme-charged double-stranded break in the other, followedby resealing the break, thus reversing the handedness of the loop, and thereby changing the linking number in steps of two. Mechanistically operating on writhe, Top2 requires repartitioning of strain along the helical axis to relax twist. Though relaxation is an energetically favored event, the extra energy from ATP-hydrolysis sharpens the distribution of relaxed products more than can be explained by a thermal Boltzmann partition function (Bates and Maxwell 2005; Podtelezhnikov et al. 1999; Rybenkov et al. 1997; Vologodskii 2009; Yan et al. 1999). There are two major Top2s, Top2A and Top2B (Nitiss 2009). The differing kinetics of Top1s and Top2s may underlie particular patterns of gene expression. In bacteria, the phenomenon of transcriptional bursting, in which multiple mRNAs are generated in sharp pulses separated by variable periods of inactivity, has been attributed to the dyssynchrony of the rapid removal of upstream negative supercoils by Top1A versus the slower and sporadic removal of the polymerase-arresting positive supercoils by DNA gyrase (Chong et al. 2014). In eukaryotes, topoisomerases are relatively abundant enzymes, and when freely diffusing, soluble Top1s, and Top2s would be anticipated to remove rapidly unrestrained supercoils, leaving constrained supercoils on nucleosomes as the primary source of torsional stress, although any capacity to use this stress for work would be obligatorily coupled with chromatin remodeling. Similarly, enzymes that unwind (helicases) or track around (translocases) the DNA helix can generate equal positive and negative torsional stress in closed topological domains; coupled with topoisomerases, these enzymes can be used to introduce positive or negative supercoiling.
Intranuclear topology, topography, and cartography
A large number of studies have begun to characterize chromatin domains, sometimes referred to as TADs (topologically associating domains). TADs partition chromosomes into blocks of DNA at scales of hundreds of kilobases to megabases of DNA and are frequently bounded at inverted CTCF-binding sites, suggesting that CTCF-mediated loops comprise a major organizing principle for the genome (Cook and Gove 1992; Dekker and Heard 2015; Dekker and Misteli 2015; Dixon et al. 2012; Guo et al. 2015; Ji et al. 2016; Pope et al. 2014; Smith et al. 2016; Tang et al. 2015; Valton and Dekker 2016). CTCF and cohesin are enriched at TAD boundaries (Guo et al. 2015; Ji et al. 2016; Katainen et al. 2015; Rao et al. 2015; Sofueva et al. 2013; Tang et al. 2015; Xiao et al. 2011; Zuin et al. 2014). The requirement for inverted CTCF sites is perplexing (Guo et al. 2015; Rao et al. 2014; Rao et al. 2015); loops occurring on this scale would be expected to include many statistical segments and therefore to be insensitive to CTCF-site orientation. The orientation-dependence likely is a clue to an organizing architectural or mechanistic (tracking?) principle of chromosome structure (Rao et al. 2015). Formally, the boundaries of TADs have not been shown to comprise true topological boundaries that fix the linking number of the included DNA. If the protein–protein and protein–DNA interactions involved in the juxtaposition of two remotely situated, CTCF–cohesin–DNA complexes were to flicker on and off while maintaining a loose-fitting cohesin collar, no true topological domain would persist (Fig. 1c). Because cohesin forms a ring large enough to include two double helices (Gligoris and Lowe 2016), torsional stress could flow between TADs even while maintaining the topography inferred from looping assays.
The general view of the arrangement of topological domains along a chromosome fiber is of a chain of serially looped/linked domains separated by topological boundaries (Lieberman-Aiden et al. 2009; Rao et al. 2015). A far more complicated situation may be imagined; protein–protein, protein–DNA, and nucleic acid–nucleic acid (DNA-RNA, RNA-RNA, and DNA-DNA) linkages all have the potential to create a complex meshwork of interlocking loops, creating domains within domains ranging in size from dozens of bases to megabase scales. An example of a micro domain would be the 147 bp of DNA fixed upon the surface of a nucleosome that constrains −1.05 supercoils (Luger et al. 1997). Stresses imparted along a segment of chromatin are likely to be preferentially partitioned into the linker regions separating the nucleosomes; the stereochemistry of the nucleosome determines the trajectory of the encompassing DNA and is likely to shield that DNA (at least transiently) from excessive strain. En bloc rotation of a nucleosome would transmit torsional strain in a saltatory manner from linker to linker skipping over the nucleosome (Fig. 1a). The crossing or safety-pin-like opening of the DNA stems entering and exiting the nucleosome would have a major impact on ΔWr of a chromatin segment (Bancaud et al. 2006). In this context, it is easy to appreciate how modifications of histone tails that alter linker-DNA length and/or the strength of the engagement of the linker-DNA at nucleosome entry and exit sites change the elastic moduli of linker regions and modify the distribution and transmission of torsional and flexural stresses. Linker histones (H1) and non-histone chromosomal proteins (HMGs) would also be expected to modify how chromatin accommodates and transmits torsional stress (Ivanchenko et al. 1996; Sheflin and Spaulding 1989). Stable protein–protein loops connecting DNA-bound factors would segregate torsional stresses within and outside of the loop, forcing external stresses either to accumulate proximal to the loop boundary or to bypass the loop via a large-scale translational rotation of the loop transmitting stress to the next linked segment of the DNA fiber. The relaxation of torsional stress within a meshwork of interwoven and interlocking loops may prove to be a complicated matter. Topoisomerases must find and act within each domain or microdomain to minimize the overall torsional stress of the genome. Whether there is sufficient topoisomerase activity in a cell to relax the genome is dependent on the frequency and time-scale of force generation versus torque removal, as well as upon the architecture, stability and distribution of topological boundaries. Molecular mechanisms may be envisioned that enhance or restrict topoisomerase access to torsionally strained regions of the genome.
Dynamic supercoils—in vitro
The topological state of the DNA can be deconstructed into two types of supercoils, stable and dynamic (Droge and Nordheim 1991; Kouzine et al. 2004; Liu and Wang 1987; Wu et al. 1988). Stable supercoils can be exemplified by the topological state of a plasmid with a fixed, non-zero ΔLk. In heterochromatin where gene activity is infrequent and molecular interactions are relatively unchanging, it is reasonable to expect that the flexural and torsional stresses are balanced and have reached steady state, if not equilibrium. In this situation unrestrained supercoils likely will have been minimized in those domains that are accessible to Top1 and Top2. Dynamic supercoils reflect strain responding to the sustained application of torque by active cranking of a DNA or chromatin fiber, even under topologically open conditions. In such a non-equilibrium system, the linking number is not defined, and there is no fixed relationship of linking number with twist or with writhe. In much the same way that a drive shaft is strained only while being torqued by an engine with its strain relaxing almost instantaneously when disengaged from the engine, so dynamic supercoils demand ongoing genetic activity, especially transcription, to sustain torsional stresses and strains. Because of the transient and perhaps evanescent nature of dynamic supercoils, their significance and even existence in vivo and in vitro have proven difficult to measure and assess.
Several approaches have been used to ask: what does DNA look like during transcription in vitro and in vivo? In vitro this question has been addressed using both biochemical and single-molecule experiments (Kouzine et al. 2004; Ma et al. 2013). The first experiments used transcribing bacteriophage T3 and/or T7 RNAPs as engines to torque DNA templates. The T3 and T7 promoters were divergently oriented and 1.3 kb apart. Between the two promoters, two loxP sites were separated by 1.0 kb. Upon the addition of Cre-recombinase to transcription reactions, recombination rapidly excised a circle trapping any torsional strain passaging through the segment at the moment of ring closure. Amazingly, even in linearized templates, high levels of torsional stress were trapped. Up to 14 supercoils were visualized within this 1-kb segment, a σ of −0.14. The intensity of dynamic supercoiling was sensitive to the transcription rate (controlled in vitro by limiting nucleotide triphosphates), the drag of the nascent RNAs (controlled by transcription unit length and/or the addition of RNase), and the viscosity of the medium. Thus, during transcription, the naked DNA template, even in a topologically open system, behaves as if it were supercoiled. Notably, immediately upon cessation of transcription, the naked DNA template in this system relaxed fully. Transcription-dependent melting of the far upstream element (FUSE) of the human MYC promoter was seen when this element was included in the excised segment, just as previously hypothesized (He et al. 2000). Such transcription-dependent melting licensed the binding to FUSE of FUSE binding protein (FBP) and FBP interacting repressor (FIR) in vitro (Liu et al. 2006). FUSE melting is a premier example of the concordance between the prediction of DNA melting using the thermodynamic-based calculations of the supercoil induced duplex destabilization (SIDDs) algorithm and experiment (Benham 1993; Bi and Benham 2004).
The elastic response to the dynamically applied torque is subject to a host of poorly characterized variables. First, the primary sequence of the DNA may include sequences (such as FUSE) that ‘buckle’ in response to applied torques and adopt other non-B DNA structures (Nelson 1999), such as melted bubbles, Z-DNA, quadruplex, H-DNA (Fig. 1b 3), etc. These structures in turn will have elastic moduli distinct from those of the Watson–Crick double helix and so may alter the transmission of stress to more remote sites (Chou et al. 2014; Kahn et al. 1994; Lu et al. 1992; Thomas and Bloomfield 1983). Second, the trajectory of the helical axis, the configuration of chromatin, the size and shape of the elongating transcription, replication and chromatin complexes, as well as the arrangement, length, and conformations of the nascent polynucleotides sprouting from the transcription machinery will alter both the generation and propagation of mechanical forces (Fig. 1b 4–7). Sequence-dependent bends in DNA generate fixed undulations of the helical axis that resist inversion and so convert the transmission of twist into translational rotation (Nelson 1999). Dynamic supercoils may also drive the local extrusion of a plectonemic braid (Fig. 1b 4, 7, 8). This accommodation entirely by writhe may be favored in the absence of an extending tension (Medalion and Rabin 2016; Wada and Netz 2009).
The generation and propagation of torsional stress during transcription have also been studied in single-molecule studies. Wang and colleagues found that while Escherichia coli RNAP is stalled by the accumulation of downstream, positive supercoils, it is a strong enough engine to plow on despite the accrual of upstream, negative supercoils (Ma et al. 2013). This asymmetry is explained by a melted zone behind the transcribing RNA polymerase that grows due to supercoil-induced duplex destabilization. In contrast, any such downstream conformational accommodation requires torques far exceeding the stall point of RNAPII. It should be noted that almost all single-molecule studies apply extensile force to visualize torsion-dependent changes in DNA length; while low extending force may not alter the physical characteristics of the double helix, they will certainly alter the probability of communication through looping.
Dynamic supercoiling in vivo
The proof of dynamic supercoiling and careful characterization of the mechanics of transcriptionally generated torsion in vitro suggested the potential relevance of dynamic supercoiling in vivo. However issues related to the rate of transcription, the arrangement of chromatin, nuclear organization, among others left the true biological relevance, and indeed the existence of dynamic supercoiling in living cells, unresolved if not in doubt. In a typical diploid cell there are about ∼100,000 mRNAs/cell (http://bionumbers.hms.harvard.edu/) and ∼10,000 expressed genes (so ∼10 mRNAs/gene on average). The half-life of a typical mRNA is ∼7 h, so remembering that cells are diploid, ∼2.5 productive transcription events per promoter are required every 7 h. Therefore, the promoter of a typical gene fires productively every 2–3 h. This rate is likely slow enough to allow passive accommodation of the mechanical stress through polymer folding and baseline topoisomerase activity. In contrast, the most highly expressed genes may demand firing rates several hundred-fold higher. Thus, dynamic supercoiling, if occurring in vivo, is likely to be associated primarily with the most highly transcribed genes. Two approaches have been used to establish the existence of dynamic supercoiling in vivo. First, paralleling their 2004 in vitro study, Kouzine et al. (2008) inserted a loxP-flanked test segment between two metal-inducible metallothionein promoters in an episome in Burkitt lymphoma cells. These cells also expressed a Cre-estrogen receptor fusion protein. Upon the addition of zinc, reporter expression was dramatically increased; then following a short pulse of tamoxifen to activate the recombinase, DNA was recovered, and the topological state of the excised inter-promoter circle was directly assessed by electrophoresis with Southern blotting. Remarkably, after accounting for the restrained supercoils on the five included nucleosomes, the in vivo situation paralleled the in vitro results. If the test segment included the FUSE, in vivo as in vitro, active transcription melted the element and sponsored transcription amplification (Benjamin et al. 2008). Once excised and isolated from further torque generation, torsional stress was detected for more than 30 minutes, and with topoisomerase inhibition the rate of relaxation was dramatically slowed. Thus, the trafficking to and the action of topoisomerases on stressed DNA are neither instantaneous nor especially rapid.
A second approach to assess the level of supercoiling in vivo is the intercalation of psoralen into a test segment of DNA. This photo-reactive tricyclic aromatic compound intercalates poorly between the tightly stacked bases of positively supercoiled DNA, but inserts better and better as the double helix becomes progressively more underwound (Kramer et al. 1999; Sinden 1994; Sinden et al. 1980). Upon UV-irradiation, psoralen reacts with both strands to create an interstrand cross-link, with the density of cross-linking reflecting the degree of negative supercoiling. After purification and fragmentation of the genome, cross-linked and uncross-linked fragments are easily separated and hybridized to microarrays. Separation of cross-linked and uncross-linked DNA fragments can also be achieved due to the anomalous migration of the former. Kouzine et al. (2008) reported that Southern blots demonstrated that transcription enhanced the psoralen-dependent cross-linking of upstream DNA.
Psoralen has proven to be a reliable probe of torsional stress, in vivo (Sinden et al. 1980), and the direct assessment of supercoils by electrophoresis and blotting provides a compelling visualization of the topological state of a single segment of test DNA. To expand the scope of this approach and to develop a genome-wide view, cross-linked versus uncross-linked DNAs have been hybridized with microarrays to which DNA oligonucleotides spanning the genome were attached. One method of separation relies upon the use of the inter-strand cross-link as a barrier to digestion by lambda-exonuclease or exonuclease I; this method was used to demonstrate that supercoiling levels decline near the ends of yeast chromosomes, likely due to the diffusion of torsional stress off of a free end (Bermudez et al. 2010; Roca 2011). Electrophoretic separation after cross-linking has also been followed by hybridization with microarrays. The differential hybridization revealed sites of increased intercalation and was attributed to supercoiling in human cells. Negative supercoiling at promoters was found to be a consequence of the level of ongoing transcription (Kouzine et al. 2013).
A variant of this method precisely maps sites of cross-linking; using the interstrand cross-link first as a barrier to protect a DNA strand from exonuclease digestion, and then as a roadblock to DNA polymerase during amplification of the digested DNA by primer extension, the resulting DNA may be sequenced to map reactive psoralens (Teves and Henikoff 2014). Still other variations exploit biotinylated-psoralen and use streptavidin to recover the cross-linked DNA for hybridization with microarrays or binding with a fluorescent streptavidin probe to visualize unconstrained torsional stress in polytene chromosomes (Matsumoto and Hirose 2004; Naughton et al. 2013). Such analyses have demonstrated variation in supercoiling across multiple scales, from overarching chromosomal domains varying in length from hundreds of kilobases to megabases, down to regions of hundreds to a few thousand bases upstream of regions of active genes (microdomains on the order of several dozens of bases would not be resolvable with the current methods). There has been considerable concordance between the studies that altogether support the notion that upstream negative superhelical stress emanates from transcription start sites at active genes, while positive torsional stress accrues in gene bodies. The authors of all studies concluded that transcription is a major generator of supercoiling in vivo. In contrast, the role of replication in generating supercoils in metazoans remains understudied (Yu and Droge 2014).
Coupling and tuning cis-elements with torsional stress
The forces of both static and dynamic supercoils have the capacity to modify DNA and chromatin structure, and therefore to modulate and regulate the binding and organization of transcription and chromatin regulatory factors. To achieve efficient regulation, torque-driven conformational transitions in DNA and RNA must be tuned to the particular topological domain (Zhabinskaya and Benham 2012, 2013). Because all supercoil-driven transitions compete with each other to relieve stress within a static topological domain, the formation of a given structure is dependent on the number and energetics of every B-DNA to non-B DNA transition within a topological domain. This enables distant elements to communicate through the helical fiber without looping. For example, a short very A-T-rich segment may melt first at the onset of supercoiling, only to revert to duplex upon the opening of a longer slightly less A-T-rich segment as supercoiling increases further. Similarly, extruded plectonemes forming on a torsionally stressed fiber communicate over large distances with no direct contact (Fig. 1b). The growth of one plectoneme may absorb stress from the DNA fiber that sustains another, allowing these braided structures to hop throughout a topological domain (van Loenhout et al. 2012); this communication by hopping is extremely rapid, far more rapid than transcription or chromatin remodeling. Whereas the dynamics and kinetics of these competitive structural transitions in general have been partly characterized in vitro, virtually nothing is known of these processes in vivo. Because the elastic moduli of chromatin are different than those of DNA, (chromatin is softer and more pliable material per unit length), the mechanics of communication between remote structures and extruded plectonemes in vitro are almost certainly different from those in vivo (Bancaud et al. 2006; Lavelle et al. 2010).
The competition between separated structures for dynamic supercoils is likely to be more complicated than that for static supercoils within a closed topological domain. First, the magnitude of the torsional stress is not uniform in an open system, but decays roughly linearly from the position of the torque generator to the open boundary of the domain (presumably bypassing interposed domains and microdomains). Second, dynamic supercoils are vectorially propagated, so that a thermodynamically less favored non-B DNA conformation situated closer to the torque-generator may nevertheless outcompete the formation of a more energetically favored structure that is further away—though this situation has not been formally examined. The arrangement and context of torsion-sensitive elements may dictate the regulatory programs that they impart upon nearby genes; for example, deformation of the CT-element just 5′ of the MYC P1 promoter might disturb the transmission of the torsional stress that melts the FUSE 1.4 kb further upstream (Brooks and Hurley 2009) .
Topoisomerases as agents of gene regulation
There is a growing consensus that ongoing transcription injects dynamic supercoiling into chromatin fibers, genome-wide, and so the generation of torsional stress is inexorably linked to transcriptional regulatory processes. However, at steady-state, the level of torsional stress across the genome is a balance between the generation of supercoiling and its removal. Topoisomerases are commonly presumed to be constitutive enzymes that rapidly drain excess twist (Top1) or writhe (Top2) from all the stressed regions of the genome. Very little topoisomerase activity is required at genes which are expressed at low to moderate levels; the intervals between consecutive productive promoter firing events for these genes is likely to be sufficiently long to accommodate dynamic supercoils by diffusion and transmission of torsional stress away from the site of transcription. Adverse topological circumstances that sporadically impede transcription at these low output promoters would likely prove to be inconsequential for their low net expression. Gene expression should become increasingly dependent on topoisomerases as the rate of promoter firing increases the levels of static and dynamic supercoils. Depending on the local geometry and the dynamics of the partitioning of torsional stress between twist and writhe, the relative demand for Top1 versus Top2 is likely to be highly context-sensitive.
Recent evidence indicates that the topoisomerase activity at a gene does not passively follow dynamic supercoiling; rather the transcription machinery actively manages the activity of Top1 (Baranello et al. 2016). Top1 is catalytically inactive upon joining pre-initiation complexes at transcription start sites (Fig. 2a) but becomes activated subsequent to pause release (Fig. 2d, e); thus negative torsional stress is preserved at start sites to assist strand separation and consequently facilitate DNA bending as required for initiation. The topoisomerase enzymatic activation coincident or subsequent to pause release removes a torsional–mechanical impediment to transcription (Fig. 2e). This activation is mediated by the carboxyl-terminal domain (CTD) of RNAPII, but requires that the CTD is phosphorylated by the kinase activity of the bromodomain chromatin factor BRD4 (Devaiah et al. 2012) (Fig. 2d). Thus, BRD4 couples topoisomerase activity with chromatin and promoter status. Notably, BRD4, which binds with acetylated lysines, is highly enriched at superenhancers, and superenhancer-bound BRD4 loops to interact with the transcription machinery at highly expressed promoters. These results suggest that accumulated torsional stress may supply a force that resists transcriptional elongation and favors pausing. The absence of Top1 activity at start sites may preserve negative supercoiling that (1) assists promoter melting to stabilize open and early transcription complexes and (2) pulls the RNAPII backwards (perhaps promoting backtracking) and assists pausing. The amount of torsion downstream of paused RNAPII is not known as psoralen is a less effective probe for positive than for negative supercoils. The effects on gene expression of Top1 and Top2 are in considerable measure genetically compensatory (though not fully redundant); consequently, it seems likely that Top2 activity might also be stimulated by RNAPII, though this possibility has not yet been assessed. Isolated reports of topoisomerase binding and/or stimulation by sequence-specific transcription factors, including NKX2.3 and TP53, have been reported, though the mechanism and in vivo consequences of this stimulation on transcription or other genetic processes remain unexplored (Bowen et al. 2007; Gobert et al. 1999; Song et al. 2013; Yuwen et al. 1997). Top1 and Top2 have also been localized to enhancers, upstream regulatory regions, and the bodies of inducible genes, though their role in transcription remains to be elucidated (Ju et al. 2006; Puc et al. 2015). Upstream regulatory regions seem to be associated with topoisomerase-associated DNA breaks, but it is not yet clear whether these breaks are snapshots of mid-catalytic cycle, properly functioning topoisomerases, functional meta-stable breaks associated with transcription, or accidents of these normally reliable enzymes (Ju et al. 2006; Madabhushi et al. 2015; Puc et al. 2015).
Fig. 2.
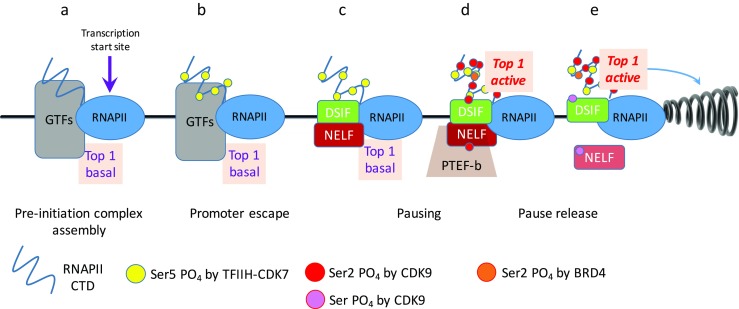
The transcription machinery actively manages topoisomerase activity. General transcription factors (GTFs: TFIIA, B, D, E, F, H) assemble into pre-initiation complexes. Negative elongation factor (NELF) and DRB-sensitivity inducing factor (DSIF) are complexes that are recruited to promoters that cause transcription to pause or to resume elongation, respectively, depending on their phosphorylation status. Positive transcription elongation factor b (PTEB-b) is the CDK9–cyclin T complex that phosphorylates serine 2 (Ser 2) and other targets, including DSIF and NELF. a Pre-initiation complexes assembled at start sites from multiple components include minimally active topoisomerase 1 (Top1); Top1 catalysis is not required for transcription initiation. The carboxyl-terminal domain (CTD) of the RNAPII–CTD complex is at this stage hypophosphorylated. b The kinase activity of TFIIH, CDK7 phosphorylates Ser 5 (yellow circles PO4-Ser 5,) contributing to promoter escape after nucleotide +8. Slow nascent transcription continues until point c, at which the recruitment of DSIF and NELF occurs during pausing. d At this stage Ser 2 is phosphorylated throughout the CTD (red circles PO4-Ser 2), as are DSIF and NELF, by the CDK9 subunit of PTEF-b (pink circles), and selective Ser 2s in the carboxyl terminal half of the CTD are phosphorylated by bromodomain chromatin factor 4 (BRD4; orange circles). e BRD4-phosphorylated CTD activates Top1 catalytic activity, removing torsional stress and allowing elongation to proceed
Any conceptualization of DNA as an instrument for the storage and transmission of genetic information that overlooks the characteristics of the material will prove insufficient to explain and predict the biology of gene expression. Evolution has not just accommodated, but has also exploited the physical and chemical properties of DNA in service of the programs encoded digitally in the DNA sequence. An understanding of the contributions of these ‘analog’ features to genetic transactions will help to explain and—ultimately—control gene activity.
Compliance with ethical standards
Conflict of interest
David Levens declares that he has no conflict of interest.
Laura Baranello declares that she has no conflict of interest.
Fedor Kouzine declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is a contribution to Special Issue “DNA Supercoiling” but has already been published in BREV, September 2016, Volume 8, Issue 3, pp 259–268, DOI 10.1007/s12551-016-0243-5.
References
- Bancaud A, Conde e Silva N, Barbi M et al (2006) Structural plasticity of single chromatin fibers revealed by torsional manipulation. Nat Struct Mol Biol 13:444–450. http://www.nature.com/nsmb/journal/v13/n5/suppinfo/nsmb1087_S1.html [DOI] [PubMed]
- Baranello L, Kouzine F, Levens D. DNA topoisomerases beyond the standard role. Transcription. 2013;4:232–237. doi: 10.4161/trns.26598. [DOI] [PubMed] [Google Scholar]
- Baranello L, Wojtowicz D, Cui K, Devaiah B (2016) RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell 165:357–371. doi:10.1016/j.cell.2016.02.036 [DOI] [PMC free article] [PubMed]
- Bates AD, Maxwell A (2005) DNA topology, 2nd edn. Oxford University Press, Oxford
- Benham CJ. Sites of predicted stress-induced DNA duplex destabilization occur preferentially at regulatory loci. Proc Natl Acad Sci USA. 1993;90:2999–3003. doi: 10.1073/pnas.90.7.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin LR, Chung HJ, Sanford S, Kouzine F, Liu J, Levens D. Hierarchical mechanisms build the DNA-binding specificity of FUSE binding protein. Proc Natl Acad Sci USA. 2008;105:18296–18301. doi: 10.1073/pnas.0803279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez I, Garcia-Martinez J, Perez-Ortin JE, Roca J. A method for genome-wide analysis of DNA helical tension by means of psoralen-DNA photobinding. Nucleic Acids Res. 2010;38:e182. doi: 10.1093/nar/gkq687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi C, Benham CJ. WebSIDD: server for predicting stress-induced duplex destabilized (SIDD) sites in superhelical DNA. Bioinformatics (Oxford, England) 2004;20:1477–1479. doi: 10.1093/bioinformatics/bth304. [DOI] [PubMed] [Google Scholar]
- Bowen C, Stuart A, Ju JH et al (2007) NKX3.1 homeodomain protein binds to topoisomerase I and enhances its activity. Cancer Res 67:455–464. doi:10.1158/0008-5472.can-06-1591 [DOI] [PubMed]
- Brooks TA, Hurley LH. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat Rev Cancer. 2009;9:849–861. doi: 10.1038/nrc2733. [DOI] [PubMed] [Google Scholar]
- Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- Chong S, Chen C, Ge H, Xie XS. Mechanism of transcriptional bursting in bacteria. Cell. 2014;158:314–326. doi: 10.1016/j.cell.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou FC, Lipfert J, Das R. Blind predictions of DNA and RNA tweezers experiments with force and torque. PLoS Comput Biol. 2014;10:e1003756. doi: 10.1371/journal.pcbi.1003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PR, Gove F. Transcription by an immobilized RNA polymerase from bacteriophage T7 and the topology of transcription. Nucleic Acids Res. 1992;20:3591–3598. doi: 10.1093/nar/20.14.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzarelli NR, Wang JC (eds) (1990). DNA topology and its biological effects. In: Cold Spring Harbor Monograph Series, vol 20. Cold Spring Harbor Press, Cold Springer Harbor
- Dekker J, Heard E. Structural and functional diversity of topologically associating domains. FEBS Lett. 2015;589:2877–2884. doi: 10.1016/j.febslet.2015.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Misteli T. Long-range chromatin interactions. Cold Spring Harb Perspect Biol. 2015;7:a019356. doi: 10.1101/cshperspect.a019356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Lewis BA, Cherman N et al (2012) BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci USA 109:6927–6932. doi:10.1073/pnas.1120422109 [DOI] [PMC free article] [PubMed]
- Dixon JR, Selvaraj S, Yue F et al (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376–380. doi:10.1038/nature11082 [DOI] [PMC free article] [PubMed]
- Droge P, Nordheim A. Transcription-induced conformational change in a topologically closed DNA domain. Nucleic Acids Res. 1991;19:2941–2946. doi: 10.1093/nar/19.11.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gligoris T, Lowe J. Structural insights into ring formation of cohesin and related Smc complexes. Trends Cell Biol. 2016 doi: 10.1016/j.tcb.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert C, Skladanowski A, Larsen AK. The interaction between p53 and DNA topoisomerase I is regulated differently in cells with wild-type and mutant p53. Proc Natl Acad Sci USA. 1999;96:10355–10360. doi: 10.1073/pnas.96.18.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D et al (2015) CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162:900–910. doi:10.1016/j.cell.2015.07.038 [DOI] [PMC free article] [PubMed]
- He L, Liu J, Collins I, Sanford S, O’Connell B, Benham CJ, Levens D. Loss of FBP function arrests cellular proliferation and extinguishes c-myc expression. EMBO J. 2000;19:1034–1044. doi: 10.1093/emboj/19.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanchenko M, Hassan A, van Holde K, Zlatanova J. H1 binding unwinds DNA. evidence from topological assays. J Biol Chem. 1996;271:32580–32585. doi: 10.1074/jbc.271.51.32580. [DOI] [PubMed] [Google Scholar]
- Ji X, Dadon DB, Powell BE et al (2016) 3D chromosome regulatory landscape of human pluripotent cells. Cell Stem Cell 18:262–275. doi:10.1016/j.stem.2015.11.007 [DOI] [PMC free article] [PubMed]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Kahn JD, Yun E, Crothers DM. Detection of localized DNA flexibility. Nature. 1994;368:163–166. doi: 10.1038/368163a0. [DOI] [PubMed] [Google Scholar]
- Katainen R, Dave K, Pitkänen E et al (2015) CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet 47:818–821. doi:10.1038/ng.3335 [DOI] [PubMed]
- Kouzine F, Liu J, Sanford S, Chung HJ, Levens D. The dynamic response of upstream DNA to transcription-generated torsional stress. Nat Struct Mol Biol. 2004;11:1092–1100. doi: 10.1038/nsmb848. [DOI] [PubMed] [Google Scholar]
- Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- Kouzine F, Gupta A, Baranello L et al (2013) Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol 20:396–403. doi:10.1038/nsmb.2517 [DOI] [PMC free article] [PubMed]
- Kramer PR, Fragoso G, Pennie W, Htun H, Hager GL, Sinden RR. Transcriptional state of the mouse mammary tumor virus promoter can affect topological domain size in vivo. J Biol Chem. 1999;274:28590–28597. doi: 10.1074/jbc.274.40.28590. [DOI] [PubMed] [Google Scholar]
- Lavelle C, Victor JM, Zlatanova J. Chromatin fiber dynamics under tension and torsion. Int J Mol Sci. 2010;11:1557–1579. doi: 10.3390/ijms11041557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L et al (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science (New York, NY) 326:289–293. doi:10.1126/science.1181369 [DOI] [PMC free article] [PubMed]
- Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Kouzine F, Nie Z et al (2006) The FUSE/FBP/FIR/TFIIH system is a molecular machine programming a pulse of c-myc expression. EMBO J 25:2119–2130. doi:10.1038/sj.emboj.7601101 [DOI] [PMC free article] [PubMed]
- Lu M, Guo Q, Kallenbach NR, Sheardy RD. Conformational properties of B-Z junctions in DNA. Biochemistry. 1992;31:4712–4719. doi: 10.1021/bi00134a026. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Ma J, Wang M. Interplay between DNA supercoiling and transcription elongation. Transcription. 2014;5:e28636. doi: 10.4161/trns.28636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bai L, Wang MD. Transcription under torsion. Science. 2013;340:1580–1583. doi: 10.1126/science.1235441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madabhushi R, Gao F, Pfenning AR et al (2015) Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell 161:1592–1605. doi:10.1016/j.cell.2015.05.032 [DOI] [PMC free article] [PubMed]
- Maoileidigh DO, Tadigotla VR, Nudler E, Ruckenstein AE. A unified model of transcription elongation: what have we learned from single-molecule experiments? Biophys J. 2011;100:1157–1166. doi: 10.1016/j.bpj.2010.12.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Hirose S. Visualization of unconstrained negative supercoils of DNA on polytene chromosomes of Drosophila. J Cell Sci. 2004;117:3797–3805. doi: 10.1242/jcs.01225. [DOI] [PubMed] [Google Scholar]
- Medalion S, Rabin Y (2016) Effect of sequence-dependent rigidity on plectoneme localization in dsDNA. J Chem Phys 144(13):135101. doi: 10.1063/1.4945010 [DOI] [PubMed]
- Naughton C, Avlonitis N, Corless S et al (2013) Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol 20:387–395. doi:10.1038/nsmb.2509 [DOI] [PMC free article] [PubMed]
- Nelson P. Transport of torsional stress in DNA. Proc Natl Acad Sci. 1999;96:14342–14347. doi: 10.1073/pnas.96.25.14342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer. 2009;9:327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papantonis A, Cook PR. Transcription factories: genome organization and gene regulation. Chem Rev. 2013;113:8683–8705. doi: 10.1021/cr300513p. [DOI] [PubMed] [Google Scholar]
- Podtelezhnikov AA, Cozzarelli NR, Vologodskii AV. Equilibrium distributions of topological states in circular DNA: interplay of supercoiling and knotting. Proc Natl Acad Sci USA. 1999;96:12974–12979. doi: 10.1073/pnas.96.23.12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V et al (2014) Topologically associating domains are stable units of replication-timing regulation. Nature 515:402–405. doi:10.1038/nature13986 [DOI] [PMC free article] [PubMed]
- Puc J, Kozbial P, Li W et al (2015) Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell 160:367–380. doi:10.1016/j.cell.2014.12.023 [DOI] [PMC free article] [PubMed]
- Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC et al (2015) Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci USA 112:E6456–E6465. doi:10.1073/pnas.1518552112 [DOI] [PMC free article] [PubMed]
- Roca J. The torsional state of DNA within the chromosome. Chromosoma. 2011;120:323–334. doi: 10.1007/s00412-011-0324-y. [DOI] [PubMed] [Google Scholar]
- Rybenkov VV, Ullsperger C, Vologodskii AV, Cozzarelli NR. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science (New York, NY) 1997;277:690–693. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- Sekine S, Murayama Y, Svetlov V, Nudler E, Yokoyama S. The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions. Mol Cell. 2015;57:408–421. doi: 10.1016/j.molcel.2014.12.014. [DOI] [PubMed] [Google Scholar]
- Sheflin LG, Spaulding SW. High mobility group protein 1 preferentially conserves torsion in negatively supercoiled DNA. Biochemistry. 1989;28:5658–5664. doi: 10.1021/bi00439a048. [DOI] [PubMed] [Google Scholar]
- Sinden RR (1994). DNA structure and function. Academic Press, New York
- Sinden RR, Carlson JO, Pettijohn DE. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: analogous measurements in insect and human cells. Cell. 1980;21:773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- Smith EM, Lajoie BR, Jain G, Dekker J. Invariant TAD boundaries constrain cell-type-specific looping interactions between promoters and distal elements around the CFTR locus. Am J Hum Genet. 2016;98:185–201. doi: 10.1016/j.ajhg.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofueva S, Yaffe E, Chan WC et al (2013) Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J 32:3119–3129. doi:10.1038/emboj.2013.237 [DOI] [PMC free article] [PubMed]
- Song LN, Bowen C, Gelmann EP. Structural and functional interactions of the prostate cancer suppressor protein NKX3.1 with topoisomerase I. Biochem J. 2013;453:125–136. doi: 10.1042/BJ20130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick T, Allemand J, Croquette V, Bensimon D. Twisting and stretching single DNA molecules. Prog Biophys Mol Biol. 2000;74:115–140. doi: 10.1016/S0079-6107(00)00018-3. [DOI] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X et al (2015) CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163:1611–1627. doi:10.1016/j.cell.2015.11.024 [DOI] [PMC free article] [PubMed]
- Teves SS, Henikoff S. Transcription-generated torsional stress destabilizes nucleosomes. Nat Struct Mol Biol. 2014;21:88–94. doi: 10.1038/nsmb.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas TJ, Bloomfield VA. Chain flexibility and hydrodynamics of the B and Z forms of poly(dG-dC).poly(dG-dC) Nucleic Acids Res. 1983;11:1919–1930. doi: 10.1093/nar/11.6.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valton AL, Dekker J. TAD disruption as oncogenic driver. Curr Opin Genet Dev. 2016;36:34–40. doi: 10.1016/j.gde.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loenhout MT, de Grunt MV, Dekker C. Dynamics of DNA supercoils. Science. 2012;338:94–97. doi: 10.1126/science.1225810. [DOI] [PubMed] [Google Scholar]
- Vinograd J, Lebowitz J. Physical and topological properties of circular DNA. J Gen Physiol. 1966;49:103–125. doi: 10.1085/jgp.49.6.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vologodskii A. Theoretical models of DNA topology simplification by type IIA DNA topoisomerases. Nucleic Acids Res. 2009;37:3125–3133. doi: 10.1093/nar/gkp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Netz RR (2009) Plectoneme creation reduces the rotational friction of a polymer. Epl 87. doi:10.1209/0295-5075/87/38001
- Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Wu HY, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- Xiao T, Wallace J, Felsenfeld G. Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol Cell Biol. 2011;31:2174–2183. doi: 10.1128/MCB.05093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Magnasco MO, Marko JF. A kinetic proofreading mechanism for disentanglement of DNA by topoisomerases. Nature. 1999;401:932–935. doi: 10.1038/44872. [DOI] [PubMed] [Google Scholar]
- Yu H, Droge P. Replication-induced supercoiling: a neglected DNA transaction regulator? Trends Biochem Sci. 2014;39:219–220. doi: 10.1016/j.tibs.2014.02.009. [DOI] [PubMed] [Google Scholar]
- Yuwen H, Hsia CC, Nakashima Y, Evangelista A, Tabor E. Binding of wild-type p53 by topoisomerase II and overexpression of topoisomerase II in human hepatocellular carcinoma. Biochem Biophys Res Commun. 1997;234:194–197. doi: 10.1006/bbrc.1997.6539. [DOI] [PubMed] [Google Scholar]
- Zhabinskaya D, Benham CJ. Theoretical analysis of competing conformational transitions in superhelical DNA. PLoS Comput Biol. 2012;8:e1002484. doi: 10.1371/journal.pcbi.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhabinskaya D, Benham CJ. Competitive superhelical transitions involving cruciform extrusion. Nucleic Acids Res. 2013;41:9610–9621. doi: 10.1093/nar/gkt733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden M et al (2014) Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci USA 111:996–1001. doi:10.1073/pnas.1317788111 [DOI] [PMC free article] [PubMed]


