Abstract
Mutations in RNA splicing factors are the single most common class of genetic alterations in myelodysplastic syndrome (MDS) patients. Although much has been learned about how these mutations affect splicing at a global- and transcript-specific level, critical questions about the role of these mutations in MDS development and maintenance remain. Here we present the questions to be addressed in order to understand the unique enrichment of these mutations in MDS.
Introduction
Myelodysplastic syndromes (MDSs) represent clonal disorders of hematopoietic stem cells (HSCs) whereby hematopoietic cell-intrinsic genetic alterations as well as non–cell autonomous factors contribute to an aberrant HSC that produces morphologically abnormal progeny and has impaired ability to generate mature blood cells. Due to the wide clinical and morphologic heterogeneity of MDS, substantial effort has been placed on characterizing the genetic alterations present in MDS cells in hopes of improving our ability to understand, diagnose, and treat the disease. Extensive targeted mutational analysis of protein coding genes has identified that MDS is characterized by a high frequency of mutations in genes encoding messenger RNA (mRNA) splicing factors as well as epigenetic modifiers.1-3 The discovery of mutations in RNA splicing factors in particular was quite unexpected as these mutations are generally uncommon in cancer yet are present in 50% to 60% of MDS patients. Interestingly, there are clear associations between specific mutated splicing factors and subtypes of MDS, most notably mutations in the RNA splicing factor SF3B1 in >90% of patients with MDS with ring sideroblasts.1,2,4 Moreover, the nature of these mutations is quite conspicuous as they occur as heterozygous point mutations at highly recurrent residues. Finally, within MDS patients, these mutations are almost always mutually exclusive; an MDS patient almost never has >1 RNA splicing factor mutation at the same time1-3 (although rare individuals with mutations in >1 RNA splicing factor have been noted,5 these occur far less frequently than expected by chance in MDS and it is unclear if both mutations in such cases are present within the same cell and/or how the mutations may impact splicing if coexpressed in the same cell).
The discovery of RNA splicing factor mutations has led to intense efforts to understand their biological impact on hematopoiesis,6-9 their genomic and biochemical effects on RNA splicing,10-21 their structural effects,22 and potential means to therapeutically target cells bearing these mutations.8,23-25 Although there have been major advances in understanding the role of RNA splicing factor mutations in MDS pathogenesis and therapy (as reviewed recently26-28), major questions about their biological consequences within cells, requirement in disease initiation vs maintenance, and cell autonomous vs nonautonomous roles remain. In addition, numerous questions about the structural and biochemical effects of the mutations on protein-protein and protein-RNA interactions. Here we present these questions and highlight what is known to date about how and why mutations in RNA splicing factors actually promote MDS development.
Do RNA splicing factor mutations contribute to MDS by changing pre-mRNA splicing and/or by directly affecting other biological processes?
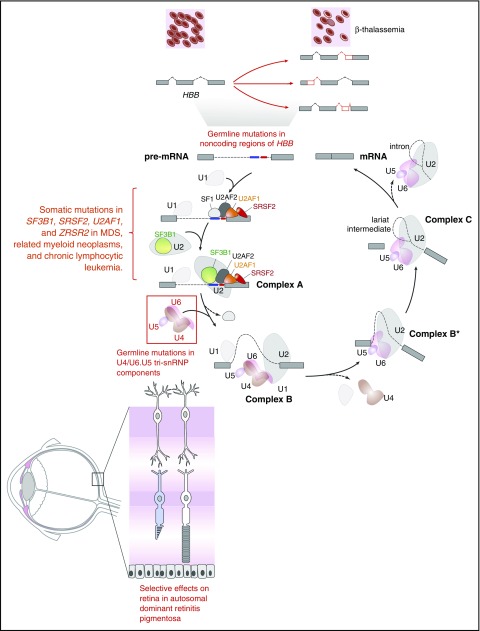
To begin to understand how mutant splicing factors contribute to disease pathogenesis in MDS, it is helpful to consider the role of mutations that affect splicing in other diseases. Splicing represents the key link between transcription and translation, and >90% of human protein-coding genes produce multiple mRNA isoforms.29-31 Mutations that occur in cis in specific genes have been identified in a number of diseases, the oldest example of which are mutations in the β-globin locus that result in β+-thalassemia (reviewed in Ho and Thein32) (Figure 1). Dysregulation of splicing in trans, due to mutations in splicing factors, has been previously discovered in diseases other than MDS, including inherited forms of retinitis pigmentosa (RP). Although most RP-associated mutations affect genes (>60) that are expressed specifically in the retina, RP is also associated with inherited autosomal dominant mutations in genes encoding 6 small nuclear ribonucleoprotein particle (snRNP)–specific proteins (PRPF3, PRPF4, PRPF6, PRPF8, PRPF31, and SNRNP200) and 2 non-snRNP splicing factors (RP9 and DHX38) that are expressed in a wide variety of tissues (reviewed in Růžičková and Staněk33). Despite the global expression of these proteins, effects in non–retinal tissues due to these mutations are limited, and it is postulated that retinal metabolism is uniquely dependent on specific splicing dynamics.
Figure 1.
Diagram of the complexes involved in RNA splicing and how trans-acting mutations in splicing factors as well as mutations in sites required for splicing of genes in cis may result in tissue-specific phenotypic effects. The individual steps and components of each spliceosomal complex have been described recently in other reviews.58-62 Germ line mutations in the genes encoding 6 different members of the U4/U6.U5 tri-small nuclear ribonucleoprotein particle (tri-snRNP; red box) result in the retinal degenerative disorder known as autosomal dominant retinitis pigmentosa. Despite the ubiquitous expression of these proteins and their role in core RNA splicing function required in every cell, overt phenotypic effects of these mutations are only apparent within the retina. In contrast to the U4/U6.U5 tri-snRNP mutations in autosomal dominant retinitis pigmentosa, mutations in the RNA splicing factors SF3B1, U2AF1, SRSF2, and ZRSR2 are enriched in leukemias and subsets of epithelial malignancies. Again here, how mutations in core RNA splicing factors expressed in numerous cell types are enriched in diseases of specific lineages remains to be addressed. In addition to mutations in RNA splicing proteins, mutations in coding or noncoding regions are required for RNA splicing of a gene in cis. The earliest examples of such an alteration are the mutations within HBB that are well known to be associated with β-thalassemia. Despite the fact that such mutations may occur in the germ line, the direct phenotypic effects of these mutations are specific to the hematopoietic system given the importance of hemoglobin β to red blood cell function.
The above data from RP illustrate that different splicing factors exhibit context-dependent functions resulting in cell- and tissue-specific penetrance and expressivity of disease. Why then do 4 splicing factors out of the >300 proteins within the spliceosome account for the majority of splicing factor mutations in MDS? Why is each mutation enriched in specific subtypes of MDS? Why does each mutation occur in a handful of other diseases? Specific mechanisms of how mutations in the key splicing factors SRSF2, SF3B1, U2AF1, and ZRSR2 alter splicing have been identified. Hotspot mutations in SRSF2 alter its RNA binding affinity and specificity.6 SF3B1 mutations may alter its branch point recognition, but distinct mutations within SF3B1 predominate in different diseases (MDS with ring sideroblast, uveal melanoma, chronic lymphocytic leukemia).12,15,34,35 U2AF1 mutations alter exon inclusion/exclusion in a sequence-specific manner,3,10 and last, loss of function mutations in ZRSR2 specifically affects assembly of the minor spliceosome.19 However, many more potential target sequence elements are present in the genome/pre-mRNA than appear to be regulated at any 1 time by these splicing factors, and only a subset of target pre-mRNAs are affected by the mutant proteins. Hence, other factors likely determine when and where these splicing factors are required, thus making their effects highly context dependent. Our understanding of the hematopoietic hierarchy over the past several years has shed light on the fluidity of cell fate decisions and differentiation, and splicing is likely to play a major role in this fluidity.36 Further RNA sequencing of highly purified cell populations across hematopoiesis may shed light on the splice events and cooperating regulators that drive splicing factor–induced disease pathogenesis. Given that splicing factor mutations preferentially occur in those >70 years of age,5 special attention should also be given to aging-related changes in splicing in stem and progenitor cell subsets.37,38
Despite intense studies and advances in understanding the effects of splicing factor mutations on splicing, the question still remains unanswered of how these specific splicing factor mutations cause MDS/acute myeloid leukemia. If it is “simply” alteration of splicing, would one not expect mutations in many splicing factors to occur in MDS? It is possible that functions other than splicing contribute to the pathogenic effects of these mutant proteins. Given the frequency of mutations in epigenetic regulators in MDS, it is an intriguing possibility that additional non–splicing roles for mutated splicing factors in transcriptional regulation play a role in disease pathogenesis. Single reports have described a role for SRSF2 in transcriptional elongation,39 whereas SF3B1 has been reported to interact with chromatin via binding to nucleosomes located at exonic positions.40 Dissection of the role of these non–splicing mechanisms may shed further light on the role of these splicing factor mutations in MDS. It is also important to note that mutations in individual splicing factors appear to coexist with distinct mutations in epigenetic modifiers (reviewed recently27). For example, mutations in U2AF1 tend to significantly coexist with those in ASXL1, whereas mutations in SF3B1 frequently coexist with those in DNMT3A.41-43 Further efforts to understand the mechanistic and biological contributions of these coexisting mutations on splicing and hematopoiesis may greatly facilitate our understanding of how mutations in RNA splicing promote MDS development.
What is the role of RNA splicing factors in MDS development vs maintenance?
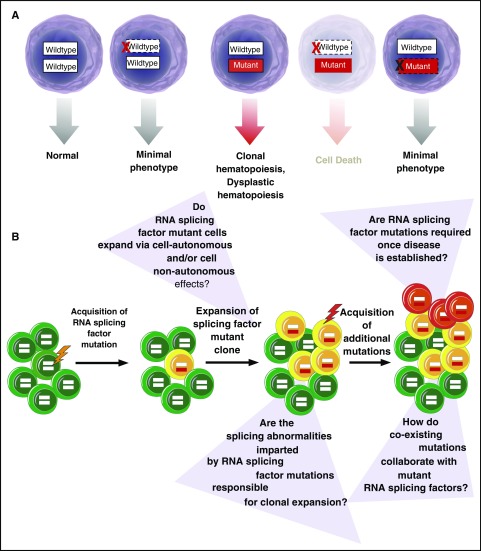
Mutations in SF3B1, SRSF2, and U2AF1 are consistently expressed in the heterozygous state concomitant with expression of the wild-type allele.1-3 Although the nature of these alterations as heterozygous missense mutations was initially assumed to suggest that they confer a gain of function, it is also now clear that the wild-type allele is absolutely required for cell survival in the setting of expression of the mutant allele11,23,44 (Figure 2). In contrast, several recent studies have identified that ablation of the mutant RNA splicing factor allele appears to have no effect on cancer maintenance.11,25,44 This was demonstrated in 2 studies where deletion of the mutant U2AF1S34F or SF3B1R625G allele in the context of human lung epithelial11 and uveal melanoma44 cells, respectively, had no effect on cell proliferation or survival in vitro. In contrast, deletion of the wild-type U2AF1 or SF3B1 allele resulted in death of cells expressing only the mutant allele.11,44 Similarly, work from our group demonstrated that expression of the hemizygous SRSF2P95H/− is incompatible with hematopoietic cell survival, a finding distinct from that seen with simply heterozygous deletion of wild-type SRSF2.23
Figure 2.
Requirement of RNA splicing factor mutations in disease initiation vs maintenance. (A) RNA splicing factor mutations are consistently expressed in the presence of the wild-type allele.1,3,63 These mutations frequently occur early in the pathogenesis of MDS41-43 and related myeloid neoplasms and are also enriched in clonal hematopoiesis of indeterminate potential,5,45,46,64 suggesting their role in disease initiation. Consistent with a role in disease initiation, expression of these mutations in the heterozygous state in mice in vivo results in an MDS-like phenotype.6-8 Curiously, however, genetic deletion of the mutant allele appears to have no phenotypic effects in multiple cell types in vitro,11,25,44 questioning the role of the mutant allele once a disease is established. In contrast, deletion of the wild-type allele in the setting of expression of the mutant protein is associated with cell lethality.11,23,44 (B) Schematic representation of acquisition of RNA splicing factor mutations early in the pathogenesis of clonal hematopoiesis of indeterminate potential and MDS with questions about the role that these mutations at each stage of disease development.
The concept that mutant RNA splicing factors are not required for disease maintenance is further supported by recent data by Park et al noted above.25 In this study, the authors identified that overexpression of mutant U2AF1S34F is capable of transforming cytokine-dependent Ba/F3 cells. However, once transformed, the cells no longer required the mutant RNA splicing factor for cytokine-independent growth. Overall, these data argue that mutations in RNA splicing factors may not be required once MDS or other forms of cancer are established. The lack of requirement for RNA splicing factor mutations in disease maintenance would suggest that these mutations are critically important in disease initiation, a possibility supported by the fact that mutations in RNA splicing factors appear to be some of the earliest genetic events in MDS development.41-43 However, there are currently no studies evaluating this question in genetically defined cells that are not already in a transformed state. These same mutations are also frequently present in clonal hematopoiesis of indeterminate potential,45,46 a condition highly associated with increased frequency of development of MDS and other myeloid neoplasms.
Overall, more effort is needed to determine the role of RNA splicing factor mutations in disease development vs maintenance. This question is critically important given efforts to therapeutically target cells bearing RNA splicing factor mutations. Data suggesting that the mutant allele is not required for disease maintenance would argue that targeting mutant RNA splicing factor function (as opposed to wild-type splicing) would not yield therapeutic benefit.
Is disruption of RNA splicing a universal disease mechanism in MDS?
The remarkable frequency of RNA splicing factor mutations in MDS begs the question of whether RNA splicing alterations exist across the entire histologic and genetic spectrum of MDS, including those forms of MDS without a known mutated splicing factor. As noted earlier, RNA splicing alterations imparted by mutations in SF3B1,12,15,17,18 SRSF2,6,14 U2AF1,10,11,20 and ZRSR219 are each distinct, and thus, it is not clear what precise splicing changes would be expected in MDS patients lacking these alterations. Recent data suggest that other oncogenes can confer splicing alterations.47,48 Given that splicing occurs cotranscriptionally and given the high frequency of mutations in epigenetic modifiers in MDS patients, disease-associated mutations in epigenetic modifiers may influence splicing of wild-type and/or mutant spliceosomes. Currently, the effects of mutations in any individual epigenetic modifier on splicing or their ability to modulate splicing function of mutant splicing factors in an isogenic context are not defined.
Although there are gene expression microarray data across large numbers of MDS patients,49 the current dearth of comprehensive RNA-seq data evaluating splicing across histologic and genetic subsets of MDS has limited the ability to determine if aberrant splicing exists across subsets of MDS. Given the likelihood of cell-type–specific differences in splice site usage and isoform expression in normal cells,36,50-52 identification of RNA splicing alterations across all MDS is not a trivial undertaking. Results from studies attempting to define alterations in splicing across histologic and genetic subsets of a variety of cancers using publically deposited data have yielded distinct findings likely related to methodological differences.53-56
Conclusion: the importance of rigorously defining aberrant splicing events mediated by RNA splicing factors
Most of the questions posed above about the role of mutated RNA splicing factors in MDS pathogenesis center around the effects of these proteins on RNA splicing at a global or transcript-specific level. It is important to note that currently only a handful of aberrant splicing events have been rigorously molecularly characterized as being aberrantly spliced by mutant RNA splicing factors. Moreover, even fewer of these aberrant splicing events have been studied in detail at a biological level. Rigorous characterization of aberrant splicing events is critically important for reasons that extend beyond identifying these alterations as important in MDS pathogenesis. Identification of bona fide aberrant splicing events mediated by mutant RNA splicing factors facilitate discovery of the precise molecular mechanisms by which mutations cause aberrant splicing events. In addition, knowledge of these events will be essential in generating reagents for use in evaluating drugs, genetic perturbations, and other molecules that might modify the function of mutant splicing factors.
Further efforts to explore the splicing impact of mutant RNA splicing factors through means other than RNA sequencing may also be incredibly enlightening. For example, evaluation of the effect of these mutations on direct RNA binding targets using techniques such as CLIP-seq (UV cross-linking and RNA immunoprecipitation sequencing) is needed and ongoing.57 In addition, evaluating the transcripts that are actively engaged in translation in isogenic cells wild-type vs mutant for an RNA splicing factor may provide greater resolution on the impact of these mutations on RNA isoforms actually being translated in a cell. Finally, it is important to note that in each of these studies, evaluation of the RNA splicing factor mutations in the correct cellular and genetic context as well as the allelic ratio will be critically important.
Acknowledgments
This work was supported by grants from the Edward P. Evans Foundation (S.H. and O.A.-W.); National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (grant R01 DK102792) and the Frederick A. DeLuca Foundation (S.H.); the Department of Defense Bone Marrow Failure Research Program (BM150092 and W81XWH-12-1-0041), National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (grant R01 HL128239), the Josie Robertson Investigator Program, an award from the Starr Foundation (I8-A8-075), the Leukemia and Lymphoma Society, the Henry & Marilyn Taub Foundation, and the Pershing Square Sohn Cancer Research Alliance (O.A.-W.).
Authorship
Contribution: P.J., S.H., and O.A.-W. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stephanie Halene, 300 George St 786E, New Haven, CT 06511; e-mail: stephanie.halene@yale.edu; and Omar Abdel-Wahab, 1275 York Ave, New York, NY 10065; e-mail: abdelwao@mskcc.org.
References
- 1.Yoshida K, Sanada M, Shiraishi Y, et al. . Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64-69. [DOI] [PubMed] [Google Scholar]
- 2.Papaemmanuil E, Cazzola M, Boultwood J, et al. ; Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365(15):1384-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graubert TA, Shen D, Ding L, et al. . Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2011;44(1):53-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malcovati L, Karimi M, Papaemmanuil E, et al. . SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood. 2015;126(2):233-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKerrell T, Park N, Moreno T, et al. ; Understanding Society Scientific Group. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Reports. 2015;10(8):1239-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim E, Ilagan JO, Liang Y, et al. . SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27(5):617-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shirai CL, Ley JN, White BS, et al. . Mutant U2AF1 expression alters hematopoiesis and pre-mRNA splicing in vivo. Cancer Cell. 2015;27(5):631-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Obeng EA, Chappell RJ, Seiler M, et al. . Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell. 2016;30(3):404-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mupo A, Seiler M, Sathiaseelan V, et al. . Hemopoietic-specific Sf3b1-K700E knock-in mice display the splicing defect seen in human MDS but develop anemia without ring sideroblasts. Leukemia. 2017;31(3):720-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ilagan JO, Ramakrishnan A, Hayes B, et al. . U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 2015;25(1):14-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fei DL, Motowski H, Chatrikhi R, et al. . Wild-type U2AF1 antagonizes the splicing program characteristic of U2AF1-mutant tumors and is required for cell survival. PLoS Genet. 2016;12(10):e1006384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darman RB, Seiler M, Agrawal AA, et al. . Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Reports. 2015;13(5):1033-1045. [DOI] [PubMed] [Google Scholar]
- 13.DeBoever C, Ghia EM, Shepard PJ, et al. . Transcriptome sequencing reveals potential mechanism of cryptic 3′ splice site selection in SF3B1-mutated cancers. PLOS Comput Biol. 2015;11(3):e1004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Lieu YK, Ali AM, et al. . Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc Natl Acad Sci USA. 2015;112(34):E4726-E4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kesarwani AK, Ramirez O, Gupta AK, et al. . Cancer-associated SF3B1 mutants recognize otherwise inaccessible cryptic 3′ splice sites within RNA secondary structures. Oncogene. 2017;36(8):1123-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alsafadi S, Houy A, Battistella A, et al. . Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat Commun. 2016;7:10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Q, Rodriguez-Santiago S, Wang J, et al. . SF3B1/Hsh155 HEAT motif mutations affect interaction with the spliceosomal ATPase Prp5, resulting in altered branch site selectivity in pre-mRNA splicing. Genes Dev. 2016;30(24):2710-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrocci TJ, Zoerner DM, Paulson JC, Hoskins AA. SF3b1 mutations associated with myelodysplastic syndromes alter the fidelity of branchsite selection in yeast [published online ahead of print 6 January 2017]. Nucleic Acids Res. doi:10.1093/nar/gkw1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madan V, Kanojia D, Li J, et al. . Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat Commun. 2015;6:6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okeyo-Owuor T, White BS, Chatrikhi R, et al. . U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing. Leukemia. 2015;29(4):909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu J, Zhou B, Thol F, et al. . Distinct splicing signatures affect converged pathways in myelodysplastic syndrome patients carrying mutations in different splicing regulators. RNA. 2016;22(10):1535-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cretu C, Schmitzová J, Ponce-Salvatierra A, et al. . Molecular architecture of SF3b and structural consequences of its cancer-related mutations. Mol Cell. 2016;64(2):307-319. [DOI] [PubMed] [Google Scholar]
- 23.Lee SC, Dvinge H, Kim E, et al. . Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat Med. 2016;22(6):672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shirai CL, White BS, Tripathi M, et al. . Mutant U2AF1-expressing cells are sensitive to pharmacological modulation of the spliceosome. Nat Commun. 2017;8:14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park SM, Ou J, Chamberlain L, et al. . U2AF35(S34F) promotes transformation by directing aberrant ATG7 pre-mRNA 3′ end formation. Mol Cell. 2016;62(4):479-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16(7):413-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inoue D, Bradley RK, Abdel-Wahab O. Spliceosomal gene mutations in myelodysplasia: molecular links to clonal abnormalities of hematopoiesis. Genes Dev. 2016;30(9):989-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saez B, Walter MJ, Graubert TA. Splicing factor gene mutations in hematologic malignancies. Blood. 2017;129(10):1260-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang ET, Sandberg R, Luo S, et al. . Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerstein MB, Rozowsky J, Yan KK, et al. . Comparative analysis of the transcriptome across distant species. Nature. 2014;512(7515):445-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413-1415. [DOI] [PubMed] [Google Scholar]
- 32.Ho PJ, Thein SL. Gene regulation and deregulation: a beta globin perspective. Blood Rev. 2000;14(2):78-93. [DOI] [PubMed] [Google Scholar]
- 33.Růžičková Š, Staněk D. Mutations in spliceosomal proteins and retina degeneration [published online ahead of print 14 June 2016]. RNA Biol. doi:10.1080/15476286.2016.1191735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013;45(2):133-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Brooks AN, Fan J, et al. . Transcriptomic characterization of SF3B1 mutation reveals its pleiotropic effects in chronic lymphocytic leukemia. Cancer Cell. 2016;30(5):750-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L, Kostadima M, Martens JH, et al. ; BRIDGE Consortium. Transcriptional diversity during lineage commitment of human blood progenitors. Science. 2014;345(6204):1251033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crews LA, Balaian L, Delos Santos NP, et al. . RNA splicing modulation selectively impairs leukemia stem cell maintenance in secondary human AML. Cell Stem Cell. 2016;19(5):599-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colla S, Ong DS, Ogoti Y, et al. . Telomere dysfunction drives aberrant hematopoietic differentiation and myelodysplastic syndrome. Cancer Cell. 2015;27(5):644-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15(8):819-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kfir N, Lev-Maor G, Glaich O, et al. . SF3B1 association with chromatin determines splicing outcomes. Cell Reports. 2015;11(4):618-629. [DOI] [PubMed] [Google Scholar]
- 41.Makishima H, Yoshizato T, Yoshida K, et al. . Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet. 2017;49(2):204-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haferlach T, Nagata Y, Grossmann V, et al. . Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papaemmanuil E, Gerstung M, Malcovati L, et al. ; Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616-3627, quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Q, Derti A, Ruddy D, et al. . A chemical genetics approach for the functional assessment of novel cancer genes. Cancer Res. 2015;75(10):1949-1958. [DOI] [PubMed] [Google Scholar]
- 45.Jaiswal S, Fontanillas P, Flannick J, et al. . Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Genovese G, Jaiswal S, Ebert BL, McCarroll SA. Clonal hematopoiesis and blood-cancer risk. N Engl J Med. 2015;372(11):1071-1072. [DOI] [PubMed] [Google Scholar]
- 47.Hsu TY, Simon LM, Neill NJ, et al. . The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525(7569):384-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koh CM, Bezzi M, Low DH, et al. . MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015;523(7558):96-100. [DOI] [PubMed] [Google Scholar]
- 49.Pellagatti A, Esoof N, Watkins F, et al. . Gene expression profiling in the myelodysplastic syndromes using cDNA microarray technology. Br J Haematol. 2004;125(5):576-583. [DOI] [PubMed] [Google Scholar]
- 50.Pimentel H, Parra M, Gee SL, Mohandas N, Pachter L, Conboy JG. A dynamic intron retention program enriched in RNA processing genes regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 2016;44(2):838-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang G, Huang SC, Wu JY, Benz EJ Jr. An erythroid differentiation-specific splicing switch in protein 4.1R mediated by the interaction of SF2/ASF with an exonic splicing enhancer. Blood. 2005;105(5):2146-2153. [DOI] [PubMed] [Google Scholar]
- 52.Wong JJL, Ritchie W, Ebner OA, et al. . Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154(3):583-595. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, Baran J, Cros A, et al. . International Cancer Genome Consortium Data Portal–a one-stop shop for cancer genomics data. Database (Oxford). 2011;2011:bar026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jung H, Lee D, Lee J, et al. . Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet. 2015;47(11):1242-1248. [DOI] [PubMed] [Google Scholar]
- 55.Sebestyén E, Singh B, Miñana B, et al. . Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res. 2016;26(6):732-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sebestyén E, Zawisza M, Eyras E. Detection of recurrent alternative splicing switches in tumor samples reveals novel signatures of cancer. Nucleic Acids Res. 2015;43(3):1345-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kai Rejeski YL, Tebaldi T, Stefani G, et al. . Integrative genome-wide analysis of RNA binding and splicing reveals complex loss and gain of function alterations by SRSF2 P95 mutations in myelodysplasia [abstract]. Blood. 2015;126(13). Abstract 141. [Google Scholar]
- 58.Daguenet E, Dujardin G, Valcárcel J. The pathogenicity of splicing defects: mechanistic insights into pre-mRNA processing inform novel therapeutic approaches. EMBO Rep. 2015;16(12):1640-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. 2015;84:291-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nguyen TH, Galej WP, Fica SM, Lin PC, Newman AJ, Nagai K. CryoEM structures of two spliceosomal complexes: starter and dessert at the spliceosome feast. Curr Opin Struct Biol. 2016;36:48-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Papasaikas P, Valcárcel J. The spliceosome: the ultimate RNA chaperone and sculptor. Trends Biochem Sci. 2016;41(1):33-45. [DOI] [PubMed] [Google Scholar]
- 62.Sahebi M, Hanafi MM, van Wijnen AJ, et al. . Towards understanding pre-mRNA splicing mechanisms and the role of SR proteins. Gene. 2016;587(2):107-119. [DOI] [PubMed] [Google Scholar]
- 63.Papaemmanuil E, Gerstung M, Bullinger L, et al. . Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xie M, Lu C, Wang J, et al. . Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]