Abstract
In this issue of Blood, Shi et al1 demonstrate the importance of Ssb1 and Ssb2, 2 highly conserved single-stranded DNA (ssDNA)–binding proteins, in maintaining hematopoietic stem cell (HSC) genome integrity and blood system function. Strikingly, the authors demonstrate that compound loss of Ssb1 and Ssb2 leads to rapid induction of interferons (IFNs) and HSC depletion, underwritten by severe replication stress and genomic instability in HSCs. As many bone marrow failure (BMF) syndromes arise from defects in DNA repair,2 these findings provide new insights into the events and signals that initiate HSC depletion in this context.
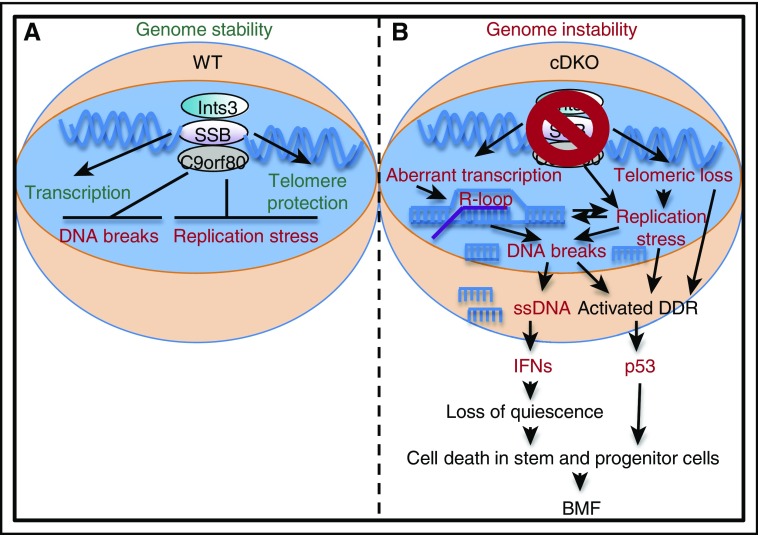
Ssb1 and Ssb2 protect genome integrity, via promoting resolution of R-loops during transcription, and facilitate telomere maintenance (A). Shi et al demonstrate that compound loss of Ssb1 and Ssb2 in HSCs leads to replication stress and DNA breaks at actively transcribed sites prone to R-loop formation. This leads to accumulation of intracellular ssDNA, production of IFNs, and loss of quiescence, which drives HSC depletion and BMF at least in part via a p53-dependent mechanism (B). Collectively, these findings provide insight into the mechanisms by which faulty DNA repair can induce BMF. See the complete Figure 7 in the article by Shi et al that begins on page 2479. DDR, DNA damage response; WT, wild-type.
BMF diseases, characterized by aplasia in one or more blood lineages, are often linked to defects in genome maintenance. Indeed, the inherited disorders Fanconi anemia (FA) and dyskeratosis congenita (DKC) are directly traced to mutations in key DNA repair and telomere maintenance genes, respectively.2 In order to faithfully copy the genome, DNA replication machinery must resolve numerous obstacles, including conflicts with transcriptional machinery that can lead to stalled replication forks and accumulation of ssDNA:RNA hybrid “R-loops” that can lead to DNA double-strand breaks (DSBs).3 Ssb1 and Ssb2 interact with the Integrator complex, a multiprotein structure that binds to R-loop–prone DNA regions and facilitates their resolution.4 Strikingly, the authors find that accumulation of R-loops and DSBs is significantly increased in Ssb1/Ssb2 conditional double knockout (cDKO) HSCs, suggesting that R-loop accumulation may play a role in BMF pathology (see figure). Breaks labeling, enrichment on streptavidin, and next-generation sequencing (BLESS) analyses of bone marrow cells conducted by the investigators showed that DNA breaks occurred in actively transcribed regions prone to R-loop formation, particularly at transfer RNA genes and in CpG islands. Interestingly, R-loop accumulation has also been observed in an FA model,5 suggesting that dysfunctional coordination of replication and transcription, and hence R-loop formation, may be a key initiating event for BMF. Replication stress phenotypes that impair HSC function have also been observed at other genomic sites, such as ribosomal RNA genes (rDNA) in aged HSCs.6 While breaks at rDNA sites were not observed in the BLESS analysis conducted here, unresolved R-loops and/or breaks in such locations could also be potential drivers of BMF, and further studies will be needed to directly demonstrate this relationship.
Notably, Shi et al observe induction of an IFN gene signature, as well as loss of quiescence and induction of p53-dependent apoptosis, in Ssb1/Ssb2 cDKO HSCs (see figure). IFNs are cytokines produced in response to infection by intracellular pathogens, including viruses and bacteria.7 However, IFNs also suppress bone marrow function and have been in widespread clinical use for the treatment of myeloproliferative neoplasms, including chronic myelogenous leukemia.7 IFNs are now viewed as key regulators of HSC function and are capable of forcing quiescent HSCs into the cell cycle, where they can undergo apoptosis in a p53-dependent fashion.8 Recent work has highlighted the pathogenic effects of IFN signaling induced by treatment of Fanca−/− mice with the double-stranded RNA analog poly(I:C). These experiments demonstrated that IFNs can promote BMF by forcing HSCs into the cell cycle, thereby facilitating accumulation of unrepaired DNA damage that culminates in depletion.9 Although IFN-responsive genes are activated in Ssb1/Ssb2 cDKO HSCs, blockade experiments will be needed to definitively establish their role as a causal factor in the BMF phenotype. Nonetheless, these data shine a spotlight on IFNs as pathogenic factors that can force repair-deficient HSCs into cell cycle checkpoints that trigger their depletion or senescence.
How are IFNs induced in this context? HSCs, like most cells, are replete with intracellular surveillance mechanisms to detect and initiate IFN responses to inappropriately localized or processed nucleic acids that are telltale signs of a virus or other pathogens. Interestingly, intracellular ssDNA resulting from DNA damage in cells of Atm-deficient mice has been shown to trigger cell-autonomous IFN responses via the intracellular DNA sensor stimulator of interferon genes (STING), a transmembrane protein localized in the endoplasmic reticulum that activates TBK1/IRF3 and downstream IFN production.10 Ssb1/Ssb2 cDKO HSCs exhibit increased intracellular ssDNA levels, and in vitro culture experiments demonstrate that activation of IFN-regulated genes is cell autonomous. As unresolved R-loops are potential sources of intracellular ssDNA, they could essentially fuel continued IFN production. Further experiments can establish the necessity of ssDNA and the STING signaling pathway in triggering IFNs in this and other models of BMF. Such findings may cast IFNs in a broader role as enforcers of genome integrity in HSCs and other tissue stem cell populations by forcing the cell cycle entry and checkpoint-mediated elimination of damaged cells.
Collectively, this work provides important insights into mechanisms driving BMF and also identifies Ssb1 and Ssb2 as potential players in BMF syndromes. While it is unlikely that any one human will have compound loss-of-function mutations in SSB1 and SSB2, Ssb1−/− mice are radiosensitive, suggesting that alterations in the function of single SSB genes could play a role in BMF severity or penetrance. Notably, Ssb1/Ssb2 cDKO HSCs also exhibited telomere attrition similar to DKC, suggesting Ssb genes are necessary for numerous genome maintenance functions. This may explain the rapid and penetrant BMF phenotype in Ssb1/Ssb2 cDKO mice relative to other BMF models. Conversely, other tissue dysfunctions, particularly in the gut, of Ssb1/Ssb2 cDKO mice may indirectly contribute to the BMF phenotype by increasing the demand for hematopoietic cells for tissue repair. While a transplantation approach confirmed a cell-autonomous functional defect in Ssb1/Ssb2 cDKO HSCs, further analyses using hematopoietic-specific knockout approaches will provide clearer insight into the roles of Ssb1 and Ssb2 in HSC genome maintenance.
Importantly, these findings synergize with recent work on the role of IFN activation by ssDNA and resultant tissue dysfunction, and this mechanism may also be relevant in aberrant hematopoiesis characteristic of aged HSCs, which are known to exhibit replication stress phenotypes.6 They also point to IFNs as potential targets for blockade therapy to limit HSC depletion. As IFN-blocking drugs are already in clinical use for inflammatory diseases, their efficacy in the context of BMF could be worth detailed exploration. As such, these data highlight the potential role of inflammatory factors in promoting genomic stress phenotypes in HSCs during inflammatory disease and hematological malignancy. Altogether, this work underscores the importance of Ssb1 and Ssb2 in helping HSCs maintain lifelong blood production by tying up their loose DNA ends.
Footnotes
Conflict-of-interest disclosure: The author declares no competing financial interests.
REFERENCES
- 1.Shi W, Vu T, Boucher D, et al. . Ssb1 and Ssb2 cooperate to regulate mouse hematopoietic stem and progenitor cells by resolving replicative stress. Blood. 2017;129(18):2479-2492. [DOI] [PMC free article] [PubMed]
- 2.Wegman-Ostrosky T, Savage SA. The genomics of inherited bone marrow failure: from mechanism to the clinic. Br J Haematol. 2017;44:S431. [DOI] [PubMed] [Google Scholar]
- 3.Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28(13):1384-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richard DJ, Bolderson E, Khanna KK. Multiple human single-stranded DNA binding proteins function in genome maintenance: structural, biochemical and functional analysis. Crit Rev Biochem Mol Biol. 2009;44(2-3):98-116. [DOI] [PubMed] [Google Scholar]
- 5.Schwab RA, Nieminuszczy J, Shah F, et al. . The Fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol Cell. 2015;60(3):351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flach J, Bakker ST, Mohrin M, et al. . Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014;512(7513):198-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trumpp A, Essers M, Wilson A. Awakening dormant haematopoietic stem cells. Nat Rev Immunol. 2010;10(3):201-209. [DOI] [PubMed] [Google Scholar]
- 8.Pietras EM, Lakshminarasimhan R, Techner J-M, et al. . Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J Exp Med. 2014;211(2):245-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walter D, Lier A, Geiselhart A, et al. . Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520(7548):549-552. [DOI] [PubMed] [Google Scholar]
- 10.Härtlova A, Erttmann SF, Raffi FA, et al. . DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 2015;42(2):332-343. [DOI] [PubMed] [Google Scholar]