Abstract
Background
The burden of subclinical atherosclerosis in asymptomatic individuals is heritable and associated with elevated risk of developing clinical coronary heart disease (CHD). We sought to identify genetic variants in protein-coding regions associated with subclinical atherosclerosis and the risk of subsequent CHD.
Methods and Results
We studied a total of 25,109 European ancestry and African-American participants with coronary artery calcification (CAC) measured by cardiac computed tomography and 52,869 with common carotid intima media thickness (CIMT) measured by ultrasonography within the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium. Participants were genotyped for 247,870 DNA sequence variants (231,539 in exons) across the genome. A meta-analysis of exome-wide association studies was performed across cohorts for CAC and CIMT. APOB p.Arg3527Gln was associated with four-fold excess CAC (P = 3×10-10). The APOE ε2 allele (p.Arg176Cys) was associated with both 22.3% reduced CAC (P = 1×10-12) and 1.4% reduced CIMT (P = 4×10-14) in carriers compared with non-carriers. In secondary analyses conditioning on LDL cholesterol concentration, the ε2 protective association with CAC, although attenuated, remained strongly significant. Additionally, the presence of ε2 was associated with reduced risk for CHD (OR 0.77; P = 1×10-11).
Conclusions
Exome-wide association meta-analysis demonstrates that protein-coding variants in APOB and APOE associate with subclinical atherosclerosis. APOE ε2 represents the first significant association for multiple subclinical atherosclerosis traits across multiple ethnicities as well as clinical CHD.
Keywords: Genome Wide Association Study, exome, coronary artery calcification, carotid intima-media thickness, genomics
Background
Coronary heart disease (CHD) remains the leading cause of death and infirmity in developed countries.1 Atherosclerosis is the underlying pathology of CHD.2 The presence of atherosclerosis in individuals without clinical CHD, termed “subclinical atherosclerosis,” is associated with increased risk of developing clinical CHD independent of traditional risk factors prior to the onset of symptoms.3-6 Subclinical atherosclerosis is a heritable7-9 clinical phenotype that can be ascertained non-invasively as coronary artery calcification (CAC) by cardiac computed tomography (CT) and common carotid intima media thickness (CIMT) by carotid ultrasound.10
Genome-wide association studies (GWAS) within the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium have discovered sites of common non-coding genetic variation associated with both CAC11, 12 and CIMT7, 11 among those of European ancestry. Non-coding single nucleotide polymorphisms (SNPs) at the 9p21 and 6p24 regions, near the CDKN2A and PHACTR1 genes, respectively, are strongly associated with both CAC burden and myocardial infarction (MI).12 The 8q24 (ZHX2), 19q13 (APOC1), and 8q23 (PINX1) loci are strongly associated with CIMT.7 Observed associations for subclinical atherosclerosis among individuals of European ancestry, however, have not been replicated in those of African ancestry.13, 14 Furthermore, since the biologic implications of non-coding variation are not as readily interpreted as with coding variation, the roles of such variants in human atherosclerosis remain unclear.15 Protein-coding variation tends to be infrequently observed and is often inadequately catalogued on earlier GWAS arrays.16 Rare genomic variation is not well-imputed and exome sequencing to detect such uncommon variation across large populations remains a costly endeavor. Here, we leverage the Illumina HumanExome BeadChip array, enriched for protein-coding variation.17 We investigated whether there is evidence for associations of protein-coding variation with two measures of subclinical atherosclerosis across individuals of European and of African ancestry. And we further determine whether such DNA sequence variations may influence CHD risk.
Methods
Study Populations
The Illumina HumanExome Beadchip v1.0 or v1.1 (also known as the “exome chip”) was used to genotype participants across 19 cohorts of the CHARGE Consortium (Supplement).18 Participants with a diagnosis of CHD at the time of CAC phenotyping were excluded from CAC analysis. Participants who underwent carotid endarterectomy prior to CIMT phenotyping were excluded from CIMT analysis. 25,109 participants had CAC measured and 52,869 participants had CIMT measured. Each study received institutional review board approval, participants provided written informed consent, and respective governing ethics committees approved each study.
Measures
CAC Measurement
Cohorts used different CT scanners to ascertain CAC scoring (Table S1). CAC scoring by multidetector CT and by electron beam CT have been previously described to be highly concordant and are both recognized as valid tools to estimate CAC score.19-21 Total CAC score was quantified by the sum of CAC area weighted by density within individual coronary arteries by the Agatston method and the continuous score was used for analysis.22
CIMT Measurement
Common carotid intima media thickness was derived by bilateral longitudinal common carotid artery analysis (imaging and measurement methods are described in the Table S2). The mean of the maximum thickness for each common carotid artery was the analytical variable.
Statistical Analyses
According to prespecified analysis plans, association analyses and meta-analyses were performed using the seqMeta package (http://cran.r-project.org/web/packages/seqMeta/index.html) in the R statistical software as has previously been performed for exome chip-based analyses.23 To reduce skewness, CAC was natural log transformed after adding 1 and CIMT was natural log transformed. Each cohort performed an analysis for each genomic variant with the trait of interest independently and separately for individuals of European and African ancestry to minimize population biases. Covariates in the models included age, sex, and principal components of ancestry derived using EIGENSTRAT.24 For studies with related samples, the pairwise kinship matrix was computed and accounted for in the regression model. Score statistics and genotypic covariance matrices were computed for each cohort and used for additive single variant and gene-based analyses, respectively.
For our primary analyses, we tested the association of each genomic variant with CAC and with CIMT across all samples by meta-analysis that included all cohorts, irrespective of ancestry. We performed single variant analyses on variants that had a minor allele count of at least 20 and gene-based analyses for genes with combined minor allele frequency (MAF) of nonsynonymous variants at least 0.2% to reduce the likelihood of false positive results. We also performed two gene-based tests: 1) T1, where nonsynonymous variants with minor allele frequency (MAF) <1% were collapsed into a gene-based statistic, and 2) sequence kernel association test (SKAT) with MAF <5% for nonsynonymous variants to better account for collapsed variants with bidirectional phenotypic consequences. Regional association plots were generated using LocusZoom.25 For our secondary analyses, we tested the association of each genomic variant with CAC and CIMT by meta-analysis separately among cohorts of European and African ancestry.
Given the 238,065 variants on the array that passed quality control, the Bonferroni-adjusted level of significance for single variant tests was 0.05/238,065 = 2.10×10-7. Given the 17,574 genes with nonsynonymous variants on the array, the Bonferroni-adjusted level of significance for gene-based tests was 0.05/17,574 = 2.85×10-6. For CAC, we had >90% power to detect a variant (MAF <1%) with effect size 0.31 standard deviations, or a gene (combined MAF <1%) with effect size 0.28 standard deviations at a sample size of 25,000. For CIMT, we had >90% power to detect a variant (MAF <1%) with effect size 0.21 standard deviations or a gene (combined MAF <1%) with effect size 0.20 standard deviations with a sample size of 52,000. Power calculations were performed using the Genetic Power Calculator.26
Methods for the secondary analyses are presented in the Supplement.
Results
Study Participants
19 cohorts participated in the meta-analyses of these two subclinical atherosclerotic traits and the clinical characteristics are summarized in Table S1 and Table S2. A total of 25,109 participants were genotyped with the array and had CAC assessed; of these participants, 19,980 were of European ancestry and 5,129 were of African ancestry. 52,869 participants were genotyped and had CIMT assessed; 44,963 were of European ancestry and 7,906 were of African ancestry. 222,701 (93.5%) of the 238,065 variants were polymorphic in the CAC meta-analysis; of polymorphic variants, 193,373 (97.1%) were annotated as nonsynonymous or splice-site variants. Similarly, 227,344 (95.5%) of array variants were polymorphic in the CIMT meta-analysis and, of these, 217,235 (95.6%) were nonsynonymous or splice-site variants.
Coronary Artery Calcification Association
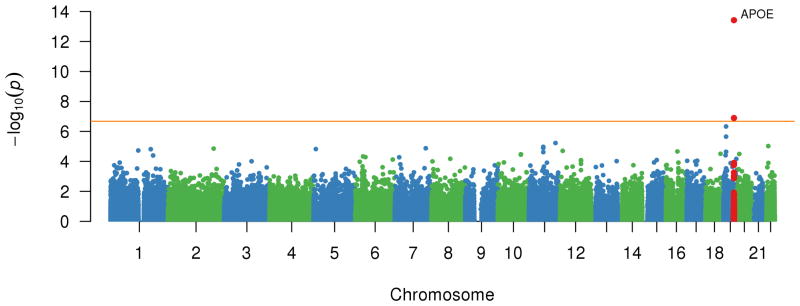
Figure 1 plots the meta-analysis CAC association P-value by genomic locus for each variant. The top loci with lead variants associated with CAC among all participants are listed in Table 1. No systematic association inflation was observed across the set of statistical tests performed (Figure S1).
Figure 1.
Association of each genotyped variant with CAC quantity. Plot of -log10(P) for association of genotyped variants by chromosomal position for all autosomal polymorphisms analyzed in the age-, sex-, and principal components- adjusted model of coronary artery calcification quantity in the meta-analysis. The genes associated with the top associated variants are displayed.
Table 1. Top meta-analysis variant associations for coronary artery calcification quantity.
| All | EA | AA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Variant | Consequence | Nearest Gene* | Chrom:Pos† | Minor Allele | MAF | Beta‡ | SE | P | MAF | P | MAF | P |
| rs10757278 § | intergenic | (CDKN2B) | 9:22124477 | G | 0.43 | 0.21 | 0.020 | 3.14×10-24 | 0.48 | 2.9×10-25 | 0.21 | 0.20 |
| rs9349379‖ | intronic | PHACTR1 | 6:12903957 | G | 0.34 | 0.19 | 0.020 | 4.93×10-20 | 0.39 | 1.28×10-19 | 0.094 | 0.088 |
| rs7412# | missense | APOE | 19:45412079 | T | 0.081 | -0.25 | 0.036 | 1.19×10-12 | 0.074 | 4.43×10-6 | 0.11 | 5.36×10-10 |
| rs1412829§ | intronic | CDKN2B | 9:22043926 | C | 0.34 | -0.14 | 0.021 | 1.56×10-11 | 0.41 | 5.58×10-12 | 0.072 | 0.84 |
| rs5742904** | missense | APOB | 2:21229160 | T | 2.1×10-3 | 1.41 | 0.22 | 2.93×10-10 | 2.7×10-3 | 2.93×10-10 | 0 | NA |
| rs9369640‖ | intronic | PHACTR1 | 6:12901441 | A | 0.43 | -0.11 | 0.019 | 4.91×10-8 | 0.38 | 5.04×10-9 | 0.36 | 0.71 |
| rs769449# | intronic | APOE | 19:45410002 | A | 0.10 | 0.14 | 0.032 | 7.93×10-6 | 0.11 | 1.86×10-6 | 0.024 | 0.19 |
| rs1801696** | missense | APOB | 2:21232044 | T | 4.6×10-3 | 0.63 | 0.14 | 1.44×10-5 | 5.7×10-3 | 9.77×10-6 | 0 | NA |
Genes for SNPs that are outside the transcript boundary of the protein-coding gene are shown in parentheses [eg, (CDKN2B)].
Genomic positions correspond to GRCh37.p13 reference, forward strand.
β-Coefficients are estimated for natural log transformation of total Agatston CAC score+1.
CDKN2B lead variants show modest correlation among EA (r2=0.24) and no correlation among AA.
PHACTR1 lead variants show modest correlation among EA (r2=0.36) and AA (r2=0.05)
APOE lead variants show minimal correlation among EA (r2=0.01) and no correlation among AA.
APOB lead variants are not observed to be correlated.
Abbreviations: AA=African ancestry; AF=minor allele frequency; Chrom:Pos=hg19 build chromosome:position; EA=European ancestry; SE=standard error
We identified previously-described common non-coding variant associations at the 9p21 and 6p24 loci. A 9p21 haplotype marked by lead SNP rs10757278-G (MAF 43%), an intergenic variant, was replicated and associated with increased CAC quantity (23.4%; 95% CI: 18.6, 28.3%; P = 2×10-24). Similarly, rs9349379-G (MAF 34%), an intronic variant within PHACTR1, was associated with increased CAC quantity (20.9%; 95% CI: 16.3, 25.8 %; P = 5×10-20). While these associations were robust for those of European ancestry, there was no apparent evidence for association in those of African ancestry (Figure S2, Figure S3). Both loci display locus heterogeneity, or multiple independent associations, for CAC in those of European ancestry (Table 1). We did not discover non-coding variants at other loci on the exome chip that met our stringent Bonferroni alpha threshold. Previously, rs3809346, an intronic variant of COL4A2, had a suggestive association with CAC,12 but now in our European ancestry sample size that is twice as large, genome-wide significant association was not observed (P = 2×10-3).
Among functional variants, a nonsynonymous APOB (rs5742904-T; MAF 0.2%; NM_000384.2:c.10580G>A; NP_000375.2:p.Arg3527Gln) variant was significantly associated with CAC quantity. Carriers of the rare APOB missense variant had markedly increased CAC (4.1-fold; 95% CI: 2.6-, 6.4-fold; P = 3×10-10). In our meta-analysis, the Old Order Amish cohort primarily accounted for the strong association, and the variant was extremely rarely observed within other cohorts. Furthermore, the variant was not seen among individuals of African ancestry (Figure S4). We also discovered a distinct rare APOB missense variant (rs1801696-T; MAF 0.6% European ancestry; NM_000384.2:c.7696G>A; NP_000375.2:p.Glu2566Lys), detected in individuals of European ancestry in most cohorts, that was moderately associated with increased CAC (1.9-fold; 95% CI: 1.6-, 2.1-fold; P = 9×10-6). This variant was not observed in individuals of African ancestry.
Additionally, a missense 19q13 variant within the APOE gene (rs7412-T; MAF 7.4% European ancestry, 10.8% African ancestry; NM_000041.2:c.526C>T; NP_000032.1:p.Arg176Cys) was associated with diminished CAC quantity (-22.3%; 95% CI, -27.6- -16.7%; P = 1×10-12) (Figure S5). This association was consistent in those of both European ancestry (-17.3%; 95% CI: -23.7, -10.3%; P = 4×10-6) and African ancestry (-35.2%; 95% CI: -43.6, -25.7%; P = 5×10-10) without significant heterogeneity (P = 0.53) (Figure 2). Additionally, an independent variant (rs769449-A; MAF 11% European ancestry, 2.4% African ancestry) within an intron of APOE also had nominal evidence of association with increased CAC quantity only in individuals of European ancestry (+15.0%; 95% CI: 7.9, 22.6%; P = 2×10-5).
Figure 2.
Forest plot of relative CAC quantity for APOE ε2 carriers. CAC quantity for APOE ε2 carriers relative to non-carriers is displayed for all cohorts stratified by European and African ancestries to demonstrate consistency across diverse cohorts and ethnicities.
To improve power of discovery for rare protein-coding variants, we conducted gene-based analyses by aggregating such variants within a gene into a single statistical unit to increase the exposure rate. However, collapsing nonsynonymous variants on the exome chip within a gene did not yield genome-wide significant results (Figure S6).
Carotid Intima Media Thickness Association
There was no systematic inflation of CIMT associations with any variant (Figure S7). The top meta-analysis association findings are listed in Table 2 and a Manhattan plot of all associations is presented in Figure 3.
Table 2. Top meta-analysis variant associations for carotid intima media thickness.
| All | EA | AA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Variant | Consequence | Nearest Gene* | Chrom:Pos† | Minor Allele | MAF | Beta‡ | SE | P | MAF | P | MAF | P |
| rs7412 | missense | APOE | 19: 45412079 | T | 0.083 | -0.014 | 0.0022 | 3.79×10-14 | 0.079 | 1.97×10-10 | 0.11 | 1.43×10-5 |
| rs11668477 | intergenic | (LDLR) | 19:11195030 | G | 0.27 | -0.0064 | 0.0016 | 4.69×10-7 | 0.20 | 5.26×10-6 | 0.34 | 0.030 |
| rs7188 | 3′UTR | (KANK2) | 19:11275139 | G | 0.29 | 0.0054 | 0.0011 | 2.23×10-6 | 0.33 | 1.36×10-6 | 0.079 | 0.98 |
| rs1712790 | intergenic | (FAM55B) | 11:114621469 | C | 0.47 | -0.0048 | 0.0011 | 5.93×10-6 | 0.48 | 1.89×10-6 | 0.21 | 0.89 |
| rs2298375 | missense | C22orf15 | 22:24106448 | A | 0.086 | 0.0082 | 0.0019 | 9.51×10-6 | 0.085 | 5.64×10-6 | 0.091 | 0.061 |
| rs174547 | intronic | (FADS1) | 11:61570783 | C | 0.30 | -0.0049 | 0.0011 | 1.07×10-5 | 0.34 | 3.84×10-5 | 0.082 | 0.062 |
Genes for SNPs that are outside the transcript boundary of the protein-coding gene are shown in parentheses [eg, (LDLR)].
Genomic positions correspond to GRCh37.p13 reference, forward strand.
β-Coefficients are estimated for natural log transformation of CIMT.
Abbreviations: AA=African ancestry; AF=minor allele frequency; Chrom:Pos=hg19 build chromosome:position; EA=European ancestry; SE=standard error; UTR=untranslated region
Figure 3.
Association of each genotyped variant with CIMT. Plot of -log10(P) for association of genotyped variants by chromosomal position for all autosomal polymorphisms analyzed in the age-, sex-, and principal components- adjusted model of carotid intima media thickness in the meta-analysis. The genes associated with the top associated variants are displayed.
We noted that, in addition to diminished CAC, the rs7412-T APOE ε2 allele was associated with diminished CIMT (-1.4%; 95% CI: -1.8, -1.0%; P = 4×10-14). There was consistency of association across European and African ancestry cohorts (Figure 4 and Figure S8). There was no significant heterogeneity among the cohorts for this association (P heterogeneity = 0.23)
Figure 4.
Forest plot of relative CIMT for APOE ε2 carriers. CIMT for APOE ε2 carriers relative to non-carriers is displayed for all cohorts stratified by European and African ancestries to demonstrate consistency across diverse cohorts and ethnicities.
There were two additional independent suggestive associations at 19q13 at non-coding variants. A variant 5kb upstream of LDLR (rs11668477) was associated with diminished CIMT (P = 5×10-7) primarily among those of European ancestry. This variant has previously been associated with reduced LDL cholesterol.27 The nearby rs7188-G variant (MAF 33% European ancestry, 7.9% African ancestry) within the 3′UTR region of KANK2 was associated with CIMT in those of European ancestry (P = 1×10-6). Additionally, a rare missense variant (rs143873045-A; MAF 0.5% African ancestry; NM_001136191.2:c.1274C>T; NP_001129663.1:p.Ser425Leu) in KANK2 only observed in individuals of African ancestry showed suggestive association with increased CIMT (P = 4×10-4). Lastly, in gene-based analyses, collapsing nonsynonymous variants within a gene did not yield significant associations (Figure S9).
APOE ε2's Effect Conditional on LDL Cholesterol
We sought to determine whether LDL cholesterol concentration accounted for the observed ε2 association with CAC. First, when restricting the original analysis only to participants with LDL cholesterol measurements (n = 20,527), ε2 remained significantly associated with reduced CAC quantity (-22.3%; 95% CI: -25.1, -19.3%; P = 2×10-11) (Table S3). When further adjusting for medication-adjusted LDL cholesterol, the effect estimate was diminished yet the association remained genome-wide significant (-17.0%; 95% CI: -19.7, -14.2%; P = 2×10-8).
APOE ε2's Effect Conditional on ε3 and ε4
Given the absence of ε4 from the array, we sought to determine whether ε2's apparent effect on reduced CAC quantity was due to a referent that includes a previously described risk allele (ε3 + ε4). 5,872 participants had CAC and the major APOE genotypes assessed by PCR. Each APOE genotype's association with CAC (to the ε3/ε3 referent) was performed by cohort and ethnicity and subsequently meta-analyzed with fixed effects. ε2/ε3 was associated with 10.8% reduced CAC (95% CI: -19.6, -0.01%; P = 0.03) and ε2/ε2 with 27.4 % reduced CAC (95% CI: -45.2, -0.04%; P = 0.03) (Figure S10).
Concordance of CHD Variants with Subclinical Atherosclerosis Associations
Of the 57 loci previously associated with CHD mainly in individuals of European or South Asian descent, 40 published variants were on the array and available for analysis. 32 of the 40 variants have the same effect direction for CAC and CHD (P = 1.8×10-4) whereas only 23 variants were concordant for CIMT (P = 0.43) in European ancestry participants (Table S4). When restricting the analysis to variants with at least nominal association (P < 0.05) with CAC, all 17 had concordant effect directions (P = 4.8×10-7). A similar analysis with variants at least nominally associated with CIMT showed that 6 of 11 had concordant effect directions for CHD (P = 0.56).
Replication of Convergent Subclinical Atherosclerosis Finding with CHD
21,182 individuals of European ancestry, independent of the sample for subclinical atherosclerosis investigations, were genotyped by the Illumina HumanExome BeadChip array, of whom 9,472 had CHD.28 In cross-sectional analyses, meta-analysis of rs7412-T confirmed a significantly lower odds of CHD (odds ratio 0.77; 95% CI: 0.71, 0.84; P = 1.47×10-10).
Discussion
In our exome-wide association analysis for subclinical atherosclerosis in two distinct ethnicities, we find that protein-coding mutations in APOB and APOE are associated with subclinical atherosclerosis. While the association for APOB was driven by a founder mutation in the Amish, a missense mutation in APOE (ε2) was associated with both reduced CAC and CIMT in individuals of European ancestry and African ancestry, even when adjusting for LDL cholesterol concentration. Furthermore, carriers of the ε2 allele had a reduced risk of coronary heart disease. Here, we provide evidence for the first exome-wide association across multiple subclinical atherosclerosis traits and multiple ethnicities for APOE ε2.
Both CAC and CIMT have been proposed as proximal clinical phenotypes of atherosclerosis that may identify individuals at high risk for developing clinical CHD. However, we see that alleles that associate with increased CHD risk also appear to largely result in increased CAC, which is less consistently observed with CIMT. This is concordant with the prior observation that CAC outperforms CIMT in predicting cardiovascular events.5, 29 Recently, post hoc analyses in statin trials to prevent cardiovascular disease observed that those with a higher burden of CHD-predisposing alleles are more likely to derive clinical benefit from preventive statin therapy.30
The APOB p.Arg3527Gln (also known as p.Arg3500Gln) has been previously been shown to lead to increased concentrations of LDL cholesterol and premature CHD.31 Our association signal for this variant was nearly exclusively driven by the Old Order Amish, where it is known to be a founder mutation (MAF 12%) predisposing to increased LDL cholesterol concentrations and CAC quantity through disruption of the LDL receptor binding domain.32 We also observed a distinct APOB missense mutation, p.Glu2566Lys, with borderline association with increased CAC quantity. Unlike p.Arg3527Gln, p.Glu2566Lys does not occur within the LDL receptor binding domain but occurs within a conserved amphipathic motif of the β2 domain predicted to influence the conversion of VLDL to LDL.33
Furthermore, we demonstrated that APOE p.Arg176Cys (ε2 allele) was associated with reduced CAC and reduced CIMT in both individuals of European and African ancestry. APOE is an essential mediator of the catabolism and clearance of triglyceride-rich and cholesterol-rich lipoproteins. The major alleles, ε2, ε3, and ε4, have been previously linked to cardiovascular disease, from the candidate gene era, and ε2 is the least common allele.34, 35 Previously, CHD risk predisposition from ε4 was primarily thought to be mediated by LDL cholesterol raising effects but observations with ε2 have been mixed.35 Similarly, ε4, unlike ε2, has been generally linked to ischemic stroke risk.36 Major reasons for the lack of association of the major APOE alleles with cardiovascular traits in prior genome-wide association studies include the notable absence of rs7412 and rs429358 on population-based genotyping arrays as well as poor imputation of these variants. Similarly, rs429358 is not included on the array used for this study.
ApoE is a major ligand of LDL receptor and a key mediator of remnant lipoprotein particle clearance.37, 38 The ε2 allele is believed to result in less efficient LDL receptor binding by altering the positive potential.39 Using publicly available data, ε2 does not impact expression of nearby genes in GTEx nor does it demonstrate enhancer or promoter chromatin marks in ENCODE HepG2 liver cells supporting ε2's direct impact on ApoE itself. ApoE ε2 can alternatively clear lipoproteins via cell-surface heparan sulfate proteoglycan and LDL receptor-related protein.40-42 ApoE ε2 transgenic mice crossbred with ApoB transgenic mice have lower LDL cholesterol.42 Furthermore, ApoE ε2 transgenic mice lacking LDL receptor still had lower LDL cholesterol suggesting that hypocholesterolemia appears independent of ε2's effects on LDL receptor.35, 43 ApoE ε2 impairs lipoprotein lipase-mediated metabolism of VLDL to LDL potentially through the displacement of ApoCII, an activator of lipoprotein lipase.43 The consequent diminished hepatic cholesterol may subsequently increase LDL receptors for ApoB-containing lipoproteins like LDL.
Interestingly, despite accounting for LDL cholesterol or serum triglycerides, we observe that ε2 still is highly associated with reduced CAC quantity. It is likely that single cross-sectional measure of lipoproteins, while correlates with, does not fully account for lifelong lipoprotein exposures. ApoE ε2 homozygotes who develop type III hyperlipoproteinemia have a marked increase in remnant lipoprotein particles unlike heterozygotes. Analogously, ApoE ε2-overexpressing mice have increased hepatic VLDL production.42 Thus, while ApoE ε2 heterozygotes may have an increase in VLDL production and decreased triglyceride catabolism via lipoprotein lipase, the observation of similar triglyceride levels compared to non-carriers suggests preservation of, or enhanced, clearance of remnant lipoprotein particles. We hypothesize that ApoE ε2's association with reduced subclinical atherosclerosis may be due to increased clearance of both atherogenic LDL and remnant lipoprotein particles through LDL receptor-dependent and -independent pathways. Further work is needed to test this hypothesis.
Our study has several strengths. First, we perform a genetic association meta-analysis across the largest set of individuals to-date for subclinical atherosclerosis in two distinct ancestries. Second, we characterize the association of protein-coding genomic variation, which has not been well studied at the population level, with subclinical atherosclerosis. Third, we explore mechanisms of association through lipoprotein-mediation analyses. Fourth, we provide novel insights with both cross-ethnicity and cross-atherosclerosis trait observations. Fifth, we relate the associations of these subclinical atherosclerosis genetic variants on risk for CHD.
While our study has several strengths, we note some key limitations. First, not all protein-coding variation is catalogued on the exome chip. Due to purifying selection, disruptive protein-coding variation is rare.44 By potentially not accounting for the totality of disruptive variation not on the array, variance is increased and power is not optimized for gene-based analyses. Whole exome sequencing can better address this limitation as such technologies continue to become more cost-effective for large-scale experiments. Second, our analyses of prior associations at non-coding sites are restricted to sites on the exome chip. We were able to robustly replicate prior non-coding association analyses for CAC at 9p21 and 6p24.12 A prior meta-analysis for CIMT genome-wide association discovered one genome-wide association, an intergenic common variant (rs11781551-A) 385kb from ZHX2 at 8q24.7 No variant with modest linkage disequilibrium with this variant was present on the exome chip thereby limiting ability for replication. An intronic variant in PINX1 at 8q23 and intergenic variant 2.3kb from APOC1 at 19q13 previously had suggestive association but no suitable proxies to replicate association were available on the exome chip. Third, our analysis still demonstrates a paucity of genome-wide associations for these quantitative atherosclerotic traits and highlights an important challenge to ongoing CAC association analyses.
Genetic determinants of CHD have been characterized among individuals of European ancestry but the strongest association signals have not replicated in those of African ancestry which may be due to smaller sample sizes hindering statistical power or different key genetic drivers. But now we demonstrate a cardioprotective genetic mechanism in those of European ancestry and African ancestry through the reduction of subclinical atherosclerosis. We propose potential mechanisms and call for renewed attention to APOE ε2 in the genesis of atherosclerosis underlying clinical cardiovascular disease. Lastly, given the strong concordance of subclinical atherosclerosis measures and clinical CHD, our findings support a future study of genotypes, subclinical atherosclerosis, and incident CHD.
Supplementary Material
Clinical Perspective.
Atherosclerosis is the underlying pathologic substrate for coronary heart disease. The presence of atherosclerosis, termed “subclinical atherosclerosis”, is associated with an increased risk of developing clinical coronary heart disease. Genetic factors influence the development of subclinical atherosclerosis. Prior analyses of single nucleotide variants (SNVs) across the genome, have identified SNVs associated with both coronary artery calcification (CAC) and carotid intima media thickness (CIMT). However, such variants reside within non-coding portions of the genome limiting the interpretation of the biological role of these SNVs. Therefore, we employed a novel genotyping platform focused on densely cataloguing protein-coding SNVs across the genome. We associated these protein-coding SNVs with CAC and CIMT in 25,109 and 52,869 individuals, respectively, of European or African ancestry. We discovered that APOE p.Arg176Cys (APOE ε2 allele) is associated with reduced CAC and CIMT in carriers compared to non-carriers. For the first time, we observed these associations in both individuals of European ancestry and of African ancestry. The association of the variant with CAC is preserved even after accounting for its effects on LDL cholesterol. Finally, carriers are at reduced risk for developing clinical coronary heart disease. These observations represent novel insights about the genetic determinants of atherosclerosis in a multiethnic sample.
Acknowledgments
Sources of Funding: P.N. is supported by the John S. LaDue Memorial Fellowship in Cardiology, Harvard Medical School. M.K. is supported by the NWO VENI grant (VENI, 91616079). Infrastructure for the CHARGE Consortium is supported in part by the National Heart, Lung, and Blood Institute grant R01HL105756. Funding support for the CHARGE Consortium Exome Chip analyses is provided in part by the National Heart, Lung, and Blood Institute grant R01HL120393. Please refer to the Supplementary Materials regarding additional sources of funding.
Disclosures: P.N. reports grant support from Amarin Corporation. I.B.B. is the Executive Director of Analytical Genetics at Regeneron Pharmaceuticals Inc. Ivana Isgum is reports research grants from Pie Medical Imaging BV and the Netherlands Organisation for Health Research and Development (ZonMw). Oscar H. Franco O.H. is employed by ErasmusAGE, a center for aging research across the life course funded by Nestlé Nutrition (Nestec Ltd.), Metagenics Inc., and AXA. Ingrid B. Borecki is employed by Regeneron Pharmaceuticals Inc. Dermot F. Reilly is employed by Merck. Khanh-dung Nguyen is employed by Biogen.
Footnotes
All other authors did not report any relevant disclosures.
Journal Subject Terms: Genetic, Association Studies; Genetics; Computerized Tomography (CT); Primary Prevention; Cardiovascular Disease
References
- 1.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, et al. Heart disease and stroke statistics--2014 update: a report from the american heart association. Circulation. 2014;129:e28–e292. doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–325. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 3.Nambi V, Chambless L, Folsom AR, He M, Hu Y, Mosley T, et al. Carotid intima-media thickness and presence or absence of plaque improves. J Am Coll Cardiol. 2010;55:1600–1607. doi: 10.1016/j.jacc.2009.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polonsky TS, McClelland RL, Jorgensen NW, Bild DE, Burke GL, Guerci AD, et al. Coronary artery calcium score and risk classification for coronary heart disease. JAMA. 2010;303:1610–1616. doi: 10.1001/jama.2010.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeboah J, McClelland RL, Polonsky TS, Burke GL, Sibley CT, O'Leary D, et al. Comparison of novel risk markers for improvement in cardiovascular risk. JAMA. 2012;308:788–795. doi: 10.1001/jama.2012.9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baber U, Mehran R, Sartori S, Schoos MM, Sillesen H, Muntendam P, et al. Prevalence, Impact, and Predictive Value of Detecting Subclinical Coronary and Carotid Atherosclerosis in Asymptomatic Adults: The BioImage Study. J Am Coll Cardiol. 2015;65:1065–1074. doi: 10.1016/j.jacc.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 7.Bis JC, Kavousi M, Franceschini N, Isaacs A, Abecasis GR, Schminke U, et al. Meta-analysis of genome-wide association studies from the CHARGE consortium. Nat Genet. 2011;43:940–947. doi: 10.1038/ng.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peyser PA, Bielak LF, Chu JS, Turner ST, Ellsworth DL, Boerwinkle E, et al. Heritability of coronary artery calcium quantity measured by electron beam computed tomography in asymptomatic adults. Circulation. 2002;106:304–308. doi: 10.1161/01.cir.0000022664.21832.5d. [DOI] [PubMed] [Google Scholar]
- 9.Zhao J, Cheema FA, Bremner JD, Goldberg J, Su S, Snieder H, et al. Heritability of carotid intima-media thickness: a twin study. Atherosclerosis. 2008;197:814–820. doi: 10.1016/j.atherosclerosis.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goff DC, Jr, Lloyd-Jones DM, Bennett G, Coady S, D'Agostino RB, Gibbons R, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation. 2014;129:S49–73. doi: 10.1161/01.cir.0000437741.48606.98. [DOI] [PubMed] [Google Scholar]
- 11.O'Donnell CJ, Cupples LA, D'Agostino RB, Fox CS, Hoffmann U, Hwang SJ, et al. Genome-wide association study for subclinical atherosclerosis in major arterial. BMC Med Genet. 2007;8(Suppl 1):S4. doi: 10.1186/1471-2350-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Donnell CJ, Kavousi M, Smith AV, Kardia SL, Feitosa MF, Hwang SJ, et al. Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation. 2011;124:2855–2864. doi: 10.1161/CIRCULATIONAHA.110.974899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wojczynski MK, Li M, Bielak LF, Kerr KF, Reiner AP, Wong ND, et al. Genetics of coronary artery calcification among African Americans, a meta-analysis. BMC Med Genet. 2013;14:75. doi: 10.1186/1471-2350-14-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Buzkova P, Wassel CL, Roman MJ, North KE, Crawford DC, et al. Lack of associations of ten candidate coronary heart disease risk genetic variants and subclinical atherosclerosis in four US populations: the Population Architecture using Genomics and Epidemiology (PAGE) study. Atherosclerosis. 2013;228:390–399. doi: 10.1016/j.atherosclerosis.2013.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glazier AM, Nadeau JH, Aitman TJ. Finding genes that underlie complex traits. Science. 2002;298:2345–2349. doi: 10.1126/science.1076641. [DOI] [PubMed] [Google Scholar]
- 16.Kiezun A, Garimella K, Do R, Stitziel NO, Neale BM, McLaren PJ, et al. Exome sequencing and the genetic basis of complex traits. Nat Genet. 2012;44:623–630. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grove ML, Yu B, Cochran BJ, Haritunians T, Bis JC, Taylor KD, et al. Best practices and joint calling of the HumanExome BeadChip: the CHARGE Consortium. PLoS One. 2013;8:e68095. doi: 10.1371/journal.pone.0068095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Psaty BM, O'Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daniell AL, Wong ND, Friedman JD, Ben-Yosef N, Miranda-Peats R, Hayes SW, et al. Concordance of coronary artery calcium estimates between MDCT and electron beam tomography. AJR Am J Roentgenol. 2005;185:1542–1545. doi: 10.2214/AJR.04.0333. [DOI] [PubMed] [Google Scholar]
- 20.Mao SS, Pal RS, McKay CR, Gao YG, Gopal A, Ahmadi N, et al. Comparison of coronary artery calcium scores between electron beam computed tomography and 64-multidetector computed tomographic scanner. J Comput Assist Tomogr. 2009;33:175–178. doi: 10.1097/RCT.0b013e31817579ee. [DOI] [PubMed] [Google Scholar]
- 21.Greenland P, Alpert JS, Beller GA, Benjamin EJ, Budoff MJ, Fayad ZA, et al. 2010 ACCF/AHA guideline for assessment of cardiovascular risk in asymptomatic adults: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2010;56:e50–103. doi: 10.1016/j.jacc.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Agatston AS, Janowitz WR, Hildner FJ, Zusmer NR, Viamonte M, Jr, Detrano R. Quantification of coronary artery calcium using ultrafast computed tomography. J Am Coll Cardiol. 1990;15:827–832. doi: 10.1016/0735-1097(90)90282-t. [DOI] [PubMed] [Google Scholar]
- 23.Peloso GM, Auer PL, Bis JC, Voorman A, Morrison AC, Stitziel NO, et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet. 2014;94:223–232. doi: 10.1016/j.ajhg.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 25.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 27.Sabatti C, Service SK, Hartikainen AL, Pouta A, Ripatti S, Brodsky J, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stitziel NO, Stirrups K, Masca NGD, Erdmann J, Ferrario PG, Konig IR, et al. Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. N Engl J Med. 2016;374:1134–1144. doi: 10.1056/NEJMoa1507652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Folsom AR, Kronmal RA, Detrano RC, O'Leary DH, Bild DE, Bluemke DA, et al. Coronary artery calcification compared with carotid intima-media thickness in the prediction of cardiovascular disease incidence: the Multi-Ethnic Study of Atherosclerosis (MESA) Arch Intern Med. 2008;168:1333–1339. doi: 10.1001/archinte.168.12.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mega JL, Stitziel NO, Smith JG, Chasman DI, Caulfield MJ, Devlin JJ, et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 2015;385:2264–2271. doi: 10.1016/S0140-6736(14)61730-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Innerarity TL, Weisgraber KH, Arnold KS, Mahley RW, Krauss RM, Vega GL, et al. Familial defective apolipoprotein B-100: low density lipoproteins with abnormal receptor binding. Proc Natl Acad Sci U S A. 1987;84:6919–6923. doi: 10.1073/pnas.84.19.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen H, Damcott CM, Rampersaud E, Pollin TI, Horenstein RB, McArdle PF, et al. Familial defective apolipoprotein B-100 and increased low-density lipoprotein cholesterol and coronary artery calcification in the old order amish. Arch Intern Med. 2010;170:1850–1855. doi: 10.1001/archinternmed.2010.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segrest JP, Jones MK, De Loof H, Dashti N. Structure of apolipoprotein B-100 in low density lipoproteins. J Lipid Res. 2001;42:1346–1367. [PubMed] [Google Scholar]
- 34.Hallman DM, Boerwinkle E, Saha N, Sandholzer C, Menzel HJ, Csazar A, et al. The apolipoprotein E polymorphism: a comparison of allele frequencies and effects in nine populations. Am J Hum Genet. 1991;49:338–349. [PMC free article] [PubMed] [Google Scholar]
- 35.Bennet AM, Di Angelantonio E, Ye Z, Wensley F, Dahlin A, Ahlbom A, et al. Association of apolipoprotein E genotypes with lipid levels and coronary risk. JAMA. 2007;298:1300–1311. doi: 10.1001/jama.298.11.1300. [DOI] [PubMed] [Google Scholar]
- 36.Sudlow C, Martinez Gonzalez NA, Kim J, Clark C. Does apolipoprotein E genotype influence the risk of ischemic stroke, intracerebral hemorrhage, or subarachnoid hemorrhage? Systematic review and meta-analyses of 31 studies among 5961 cases and 17,965 controls. Stroke. 2006;37:364–370. doi: 10.1161/01.STR.0000199065.12908.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Innerarity TL, Pitas RE, Mahley RW. Binding of arginine-rich (E) apoprotein after recombination with phospholipid vesicles to the low density lipoprotein receptors of fibroblasts. J Biol Chem. 1979;254:4186–4190. [PubMed] [Google Scholar]
- 38.Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 39.Dong LM, Parkin S, Trakhanov SD, Rupp B, Simmons T, Arnold KS, et al. Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia. Nat Struct Biol. 1996;3:718–722. doi: 10.1038/nsb0896-718. [DOI] [PubMed] [Google Scholar]
- 40.Huang Y, Rall SC, Jr, Mahley RW. Genetic factors precipitating type III hyperlipoproteinemia in hypolipidemic transgenic mice expressing human apolipoprotein E2. Arterioscler Thromb Vasc Biol. 1997;17:2817–2824. doi: 10.1161/01.atv.17.11.2817. [DOI] [PubMed] [Google Scholar]
- 41.de Beer F, Hendriks WL, van Vark LC, Kamerling SW, van Dijk KW, Hofker MH, et al. Binding of beta-VLDL to heparan sulfate proteoglycans requires lipoprotein lipase, whereas ApoE only modulates binding affinity. Arterioscler Thromb Vasc Biol. 1999;19:633–637. doi: 10.1161/01.atv.19.3.633. [DOI] [PubMed] [Google Scholar]
- 42.Mahley RW, Huang Y, Rall SC., Jr Pathogenesis of type III hyperlipoproteinemia (dysbetalipoproteinemia). Questions, quandaries, and paradoxes. J Lipid Res. 1999;40:1933–1949. [PubMed] [Google Scholar]
- 43.Huang Y, Liu XQ, Rall SC, Jr, Mahley RW. Apolipoprotein E2 reduces the low density lipoprotein level in transgenic mice by impairing lipoprotein lipase-mediated lipolysis of triglyceride-rich lipoproteins. J Biol Chem. 1998;273:17483–17490. doi: 10.1074/jbc.273.28.17483. [DOI] [PubMed] [Google Scholar]
- 44.MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.