Abstract
Background
Mutations in isocitrate dehydrogenase (IDH)1 or -2 are found in ~50% of conventional central chondrosarcomas and in up to 87% of their assumed benign precursors enchondromas. The mutant enzyme acquires the activity to convert α-ketoglutarate into the oncometabolite d-2-hydroxyglutarate (d-2-HG), which competitively inhibits α-ketoglutarate dependent enzymes such as histone- and DNA demethylases.
Methods
We therefore evaluated the effect of IDH1 or -2 mutations on histone modifications (H3K4me3, H3K9me3 and H3K27me3), chromatin remodeler ATRX expression, DNA modifications (5-hmC and 5-mC), and TET1 subcellular localization in a genotyped cohort (IDH, succinate dehydrogenase (SDH) and fumarate hydratase (FH)) of enchondromas and central chondrosarcomas (n = 101) using immunohistochemistry.
Results
IDH1 or -2 mutations were found in 60.8% of the central cartilaginous tumours, while mutations in FH and SDH were absent. The mutation status did not correlate with outcome. Chondrosarcomas are strongly positive for the histone modifications H3K4me3, H3K9me3 and H3K27me3, which was independent of the IDH1 or -2 mutation status. Two out of 36 chondrosarcomas (5.6%) show complete loss of ATRX. Levels of 5-hmC and 5-mC are highly variable in central cartilaginous tumours and are not associated with mutation status. In tumours with loss of 5-hmC, expression of TET1 was more prominent in the cytoplasm than the nucleus (p = 0.0001).
Conclusions
In summary, in central chondrosarcoma IDH1 or -2 mutations do not affect immunohistochemical levels of 5-hmC, 5mC, trimethylation of H3K4, -K9 and K27 and outcome, as compared to wildtype.
Electronic supplementary material
The online version of this article (doi:10.1186/s13569-017-0074-6) contains supplementary material, which is available to authorized users.
Keywords: 5-Hydroxymethylcytosine, 5-Methylcytosine, Histone methylation, Chondrosarcoma, Isocitrate dehydrogenase, Bone tumour, Enchondroma
Background
Mutations in isocitrate dehydrogenase (IDH)-1 or -2 (which we commonly refer to as IDH) are found in gliomas (60–80% [1, 2], acute myeloid leukemia (~20%) [3], cholangiocarcinomas (7–28%) [4–6] and in benign and malignant central cartilaginous tumours [7–9]. Isocitrate dehydrogenase (IDH) is an enzyme involved in the conversion of isocitrate to α-ketoglutarate in the tricarboxylic acid (TCA) cycle (Fig. 1). Mutations are exclusively found on the arginine residues R132 of IDH1 and in R140 and R172 of IDH2.
Fig. 1.

Schematic representation of the effect of the mutations in metabolic enzymes IDH, SDH, and FH on histone modification and DNA methylation. Accumulation of d-2-hydroxyglutarate (d-2-HG), succinate and fumarate competitively inhibit TET2, which converts 5-mC to 5-hmC, which is anticipated to cause loss of 5-hmC and global CpG island hypermethylation. In addition, competitive inhibition of histone demethylases causes altered histone modification
Cartilaginous neoplasias affect the bone and can be roughly divided into benign (enchondromas and osteochondromas), and malignant (chondrosarcomas of different subtypes) [10]. Enchondromas occur in the medulla of bone [10], and occasionally they can present with multiple lesions in non-hereditary Ollier disease or Maffucci syndrome [11]. Conventional central chondrosarcoma is the most common chondrosarcoma subtype, affecting the medulla of bone, which can arise from a pre-existing benign enchondroma. Peripheral conventional chondrosarcoma affects the surface of the bone and arises from a preexisting benign osteochondroma. The best prognostic marker for chondrosarcoma so far is histological grading, since atypical cartilaginous tumour/grade I chondrosarcomas behave locally aggressive and only metastasize in exceptional cases. Grade II and III chondrosarcomas are more cellular and the chance of metastasis is increased (up to 70% of the patients for grade III) [12, 13].
Mutations in IDH are found in ~50% (38–86%) of conventional central chondrosarcomas, in ~54% of dedifferentiated and ~15% of periosteal chondrosarcoma, and in up to 87% of benign enchondromas [7–9], while being absent in peripheral, mesenchymal and clear cell chondrosarcoma. Patients with Ollier disease and Maffucci syndrome carry IDH mutations in a somatic mosaic fashion [8, 9]. While in gliomas the IDH mutations are associated with a more favorable outcome [2, 14, 15], in intrahepatic cholangiocarcinoma, as well as in leukemia, its prognostic value could not unequivocally be shown [16, 17].
The mutant enzyme acquires the ability to convert α-ketoglutarate into the oncometabolite d-2-hydroxyglutarate (d-2-HG), which shows structural similarities with α-ketoglutarate. Indeed, increased levels of d-2-HG have been found in cartilage tumours with an IDH mutation [8]. Elevated levels of d-2-HG were shown to competitively inhibit α-ketoglutarate dependent enzymes such as histone demethylases [18] (Fig. 1), specifically affecting trimethylation of the transcriptionally permissive histone mark H3K4, and the repressive histone marks H3K9 and H3K27 [19–21]. In addition, d-2-HG inhibits the ten-eleven translocation (TET) enzymes [18] (Fig. 1). TET enzymes are involved in DNA demethylation by catalyzing the conversion of 5-methylcytosine (5-mC) into 5-hydroxymethylcytosine (5-hmC) [22, 23]. Indeed, methylation arrays confirmed global hypermethylation in IDH1 mutant enchondromas [9], and decreased levels of 5-hmC were detected in myeloproliferative disorders harboring TET2 mutations [24]. Furthermore, nuclear exclusion of TET1 was shown to be related to loss of 5-hmC in gliomas without IDH mutations [25]. Wiestler et al. described that loss of ATRX (an ATP-dependent helicase that belongs to the SWI/SNF family of chromatin remodeling proteins and facilitates access to nucleosomal DNA) was almost exclusively found in gliomas harboring IDH mutations [26]. Germline mutations in ATRX are associated with X-linked intellectual disability with alpha-thalassemia (ATRX) syndrome, and generally cause loss of protein expression [27].
Other TCA cycle enzymes involved in cancer are succinate dehydrogenase (SDH) in hereditary paragangliomas [28–31] and gastrointestinal stromal tumour (GIST) [32], and fumarate hydratase (FH) in hereditary leiomyomas and renal cell cancer (HLRCC) [33]. Mutations in either SDH or FH lead to loss of function, resulting in accumulation of succinate and fumarate, respectively. Similar to d-2-HG, elevated levels of succinate and fumarate inhibit α-ketoglutarate dependent enzymes [34–36] (Fig. 1). In SDH mutant paragangliomas and GIST, and in FH deficient smooth muscle tumours, 5-hmC was low to absent [37, 38].
In this study, our aim was to evaluate the effect of IDH mutations on outcome, and on histone modifications (H3K4me3, H3K9me3 and H3K27me3), DNA modifications (5-hmC and 5-mC), chromatin remodeling (ATRX), and subcellular localization of TET1 in a cohort of enchondromas and central chondrosarcomas for which we determined mutation status of IDH, SDH and FH.
Methods
Patient series [tissue microarray (TMA)]
The first cohort of cartilaginous tumours used for the tissue microarray (TMA) has been described previously [39] and includes nine enchondromas, 11 osteochondromas, 92 central chondrosarcomas (of which 42 atypical cartilaginous tumours/grade I; 36 grade II and 14 grade III) and 45 peripheral chondrosarcomas (of which 31 atypical cartilaginous tumours/grade I; 11 grade II and 3 grade III). Thus, this TMA contains a cohort of 101 central cartilaginous tumours. Cores from skin, colon, tonsil, prostate, breast carcinoma, spleen and liver were included for control and orientation purposes.
A second cohort of enchondromatosis related tumours was used for which details were also described previously [40]. This second TMA was exclusively used for 5mC and 5hmC immunohistochemistry as a second separate independent series, and was analyzed in the same way as the first cohort. In total, the TMA contains cores from 86 tumours, of which 65 were enchondromatosis related (51 Ollier disease, 13 Maffucci syndrome, 1 polyostotic chondrosarcoma) and 21 solitary enchondromas and chondrosarcomas. Cores from growth plate, articular cartilage, breast carcinoma, prostate, colon, skin and tonsil were included for control and orientation purposes.
Whole sections derived from normal articular cartilage (n = 3) and growth plate (n = 3) were also taken along as controls. All controls were acquired from pathological resections unrelated to cartilaginous tumours. All samples were handled in a coded manner according to the ethical guidelines as described in the Code for Proper Secondary Use of Human Tissue in The Netherlands of the Dutch Federation of Medical Scientific Societies. Follow-up data were available for all subjects (range 7–344 months, mean 150, 9 months).
Analysis of IDH, SDH and FH mutation status
Immunohistochemistry to detect the IDH1 R132H mutation was previously reported for the first cohort and 6 out of 101 central cartilaginous tumours were shown to be positive [9]. For the 95 cases that were negative, we isolated the corresponding DNA. When available, we used frozen tissue, otherwise DNA was isolated from formalin-fixed, paraffin embedded tissue (FFPE). DNA isolation from fresh frozen material was performed using the wizard genomic DNA purification kit (Promega, Madison, WI) according to the manufacturer’s instructions. DNA isolation from paraffin embedded tissue was performed as described [41]. Hydrolysis probes assay for IDH1 R132C and R132H was performed as described previously [9]. Subsequently, PCR amplification of exon 4 of IDH1 was performed for 76 samples that were negative in the hydrolysis probes assay. After each PCR run, melting curves were inspected in order to check the formation of a single product. The PCR products were purified using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) according to the manufacturer’s manual and Sanger sequencing was performed, as described previously [9]. For the remaining 64 samples without an IDH1 mutation, exon 4 of IDH2 was amplified and sequenced for mutations. Oligonucleotide sequences for PCR are shown in Additional file 1: Table S1. Bidirectional Sanger sequencing was performed by Macrogen Europe (Amsterdam, The Netherlands). The sequence results were evaluated using Mutation Surveyor software (Soft-Genetics). To exclude that IDH wildtype cartilage tumours harbor mutations in SDH or FH, TMAs were stained for SDHB (Atlas antibodies, HPA002868) and S-2-succinocysteine (2-SC) [42, 43] as described [37]. Mutations in SDH subunits destabilize the complex, which leads to degradation and loss of staining for SDHB, which is therefore widely used as a marker to screen for mutations in the SDH subunits [44]. Mutations in FH lead to accumulation of fumarate leading to aberrant succination of proteins. Positive staining for (S)-2-succinocysteine (2SC) was shown to be a reliable marker to screen for mutations in FH [42, 43].
Immunohistochemistry
Immunohistochemistry was performed as described previously [37], for histone modifications (H3K4me3; Millipore, 07-473; H3K9me3, Abcam, ab8898; H3K27me3, Millipore, 07-449); a chromatin remodeler (ATRX; Sigma, HPA001906); DNA modifications (5-hmC; Active Motif, 39,769 and 5-mC; Millipore, 33D3) as well as for TET1 (Genetex, N3C1).
Evaluation and scoring of immunohistochemistry
All slides were evaluated by two observers (JVMGB, JS, AHGC, GA in variable combination) independently and discrepancies were discussed to reach consensus. Scoring for both intensity (0 = negative, 1 = weak, 2 = moderate, 3 = strong) and the percentage of positive tumour cells (0 = 0%, 1 = <25%, 2 = 25–50%, 3 = 50–75%, 4 = 75–100%) were added up to a total sum score [45]. The average was taken of the three different cores for further analysis. Since nuclear exclusion of TET1 was associated with loss of 5-hmC in IDH wildtype gliomas, we specifically scored the subcellular localization of TET1 (N = nuclear, C = cytoplasmic, N>C=more nuclear staining than cytoplasmic, C>N=more cytoplasmic staining than nuclear) as described [25].
Statistical analysis
Statistical significance between groups was determined using ANOVA with Tukey as posthoc analysis and Chi square test as appropriate. Kaplan–Meier curves were plotted for the different stainings to determine the effect on overall survival and metastasis free survival, statistical significance was determined using Log Rank test. Data was analyzed using the IBM SPSS Statistics 23 software.
Results
IDH mutations in 60.8% of central cartilaginous tumours
In total, the IDH mutation status could be determined for 74 out of 101 central cartilaginous tumours of the first cohort. Thirty-seven tumours contained a mutation in IDH1 at the R132 position, for which the mutational spectrum is shown in Fig. 2a. In addition, eight samples harbored an IDH2 R172S mutation. Furthermore, 27 tumours were confirmed to lack IDH hotspot mutations, two of which contained an IDH1 G105G polymorphism. These two cases were excluded from analysis since this polymorphism was recently suggested to be a possible prognostic marker in leukemia [46]. Mutation analysis failed for 27 samples due to poor quality of the DNA derived from decalcified FFPE tissue. The clinicopathological data of the genetically confirmed IDH mutant versus wildtype patients are shown in Table 1.
Fig. 2.
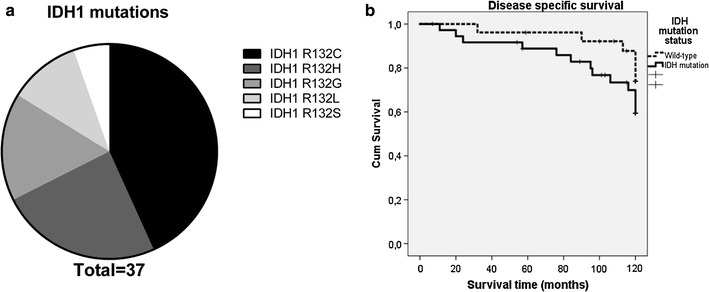
Genotyping of central cartilaginous tumours. a The mutational spectrum for IDH1 in the central chondrosarcomas from the TMA. In total 37 samples harbored an IDH1 mutation, of which the most prevalent mutation is the R132C. Eight samples harbored an IDH2 R172S mutation. b Disease specific survival of patients with IDH mutated (R132C, R132H, R132G, R132l, R132S, R172S) chondrosarcomas compared to patients with wild-type IDH chondrosarcomas revealed no statistical significant difference (p = 0.183)
Table 1.
Clinicopathological data of the IDH mutant versus wild-type group
| IDH wild-type (n = 29) | IDH mutation (n = 45) | |
|---|---|---|
| Male | 9 (31%) | 19 (42%) |
| Median age at diagnosis | 51 (21–79) | 51 (22–85) |
| Histology | ||
| Enchondroma | 1 (15%) | 6 (85%) |
| ACT/grade I | 14 (50%) | 14 (50%) |
| Grade II | 9 (34%) | 18 (66%) |
| Grade III | 5 (42%) | 7 (58%) |
| Metastasis | 4 (45%) | 5 (55%) |
| Median follow-up (months) | 137 (7–278) | 121 (11–312) |
ACT atypical cartilaginous tumour
No association between IDH mutation and outcome
There was no significant difference in disease specific survival between IDH mutant and IDH wildtype chondrosarcomas in the first cohort (p = 0.183, Fig. 2b), independent from grade (data not shown). Also, when the different mutations were analyzed separately (R132C, R132H, R132G, R132l, R132S, R172S and wild-type) or in combination (the most common R132C and R132G versus the others) no difference in outcome was found (Additional file 2: Figure S1, p = 0.726), although numbers are small. Likewise, metastasis free survival was not associated with IDH mutation status (p = 0.96, data not shown).
SDH and FH mutations were absent in chondrosarcoma
We excluded mutations in other metabolic enzymes in the 27 cartilaginous tumours that were wildtype for IDH, since all tumours were positive for SDHB immunohistochemical staining, suggesting an intact SDH complex and thereby excluding mutations in the different SDH subunits [44] (Additional file 3: Figure S2A; Additional file 4: Table S2). Furthermore, all primary tumours were negative for the presence of 2-SC (Additional file 3: Figure S2B; Additional file 4: Table S2), which was shown to be a robust biomarker for mutations in FH [42, 43].
Trimethylation of H3K4, H3K9 and H3K27 was highly abundant in chondrosarcoma
The histone modification marks H3K4me3, H3K9me3 and H3K27me3 were abundantly expressed in the vast majority of central chondrosarcomas in the first cohort (Fig. 3a, c, e). Therefore, no difference in the levels of trimethylated H3K4, H3K9 or H3K27 could be detected between IDH mutant and wildtype central cartilaginous tumours (Fig. 3b, d, f, p = 0.54, 0.46 and 0.78, respectively), nor between central and peripheral chondrosarcomas (data not shown). Furthermore, studying the low grade and high grade central chondrosarcomas in separate groups revealed no significant differences, and there was no correlation with histological grade (p = 0.24, 0.51 and 0.89, respectively) (data not shown). Also, levels of trimethylation of H3K4, H3K9 and H3K27 were not associated with overall or metastasis free survival (data not shown).
Fig. 3.
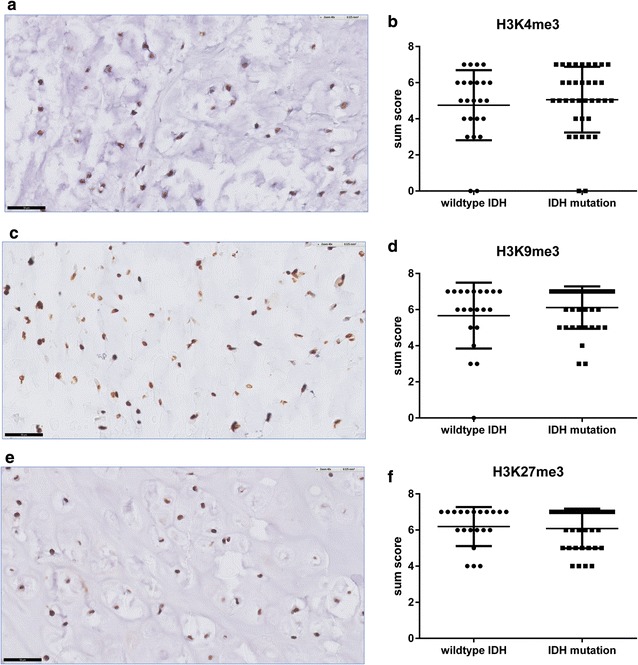
No difference in trimethylation of histone marks H3K4, H3K9 and H3K27 in central chondrosarcomas. Strong nuclear positivity for H3K4me3 (a), H3K9me3 (c) and H3K27me3 (e) in IDH wildtype central chondrosarcomas as well as central chondrosarcomas harboring an IDH mutation. Staining was scored as the sum of the intensity and the percentage of positive tumour cells (b, d, f). (Scores were rounded to zero decimal places and black bar indicates 50 µm)
Loss of ATRX in 5.6% of conventional central chondrosarcomas
Since in gliomas loss of the chromatin remodeler ATRX can be found in IDH mutant tumours [26], we evaluated a possible co-occurrence in chondrosarcoma. Two out of 36 (5.6%) conventional chondrosarcomas in the first cohort for which staining was evaluable show complete loss of ATRX (data not shown). One of these two tumours harbored an IDH1 R132 mutation, whereas the other tumour was confirmed to be wildtype for IDH.
Variable levels of 5-hydroxymethylcytosine (5-hmC) and 5-methylcytosine (5-mC) in chondrosarcomas
Since d-2-HG competitively inhibits the TET enzymes [18], which catalyze the conversion of 5-mC into 5-hmC, we evaluated the distribution of 5-mC as well as 5-hmC. Levels of 5-hmC (Fig. 4a) and 5-mC (Fig. 4c) were highly variable in chondrosarcoma. In most tumours only a fraction of the tumour cells were positive. However, no significant difference could be observed in the levels of 5-hmC (Fig. 4b) or 5-mC (Fig. 4d) between IDH wildtype and IDH mutant chondrosarcomas, considering sum score, or considering intensity and percentage of positive tumour cells separately. Interestingly, in high grade chondrosarcomas the percentage of 5-mC positive tumour cells was significantly higher as compared to low grade chondrosarcomas, which was independent of the IDH mutation status (p = 0.013) (Fig. 4e). This was however not reflected by a detectable decrease in 5hMC (data not shown). To increase the number of enchondromas and to compare enchondromatosis and solitary tumours, we additionally evaluated the second cohort of enchondromatosis related cartilaginous tumours for 5mC and 5hmC levels. Levels of 5mC were significantly higher in solitary tumours compared to tumours occurring in the context of enchondromatosis (p = 0.012) (Fig. 4f), which was independent of histological grade. No difference was detectable for 5hmC (data not shown).
Fig. 4.
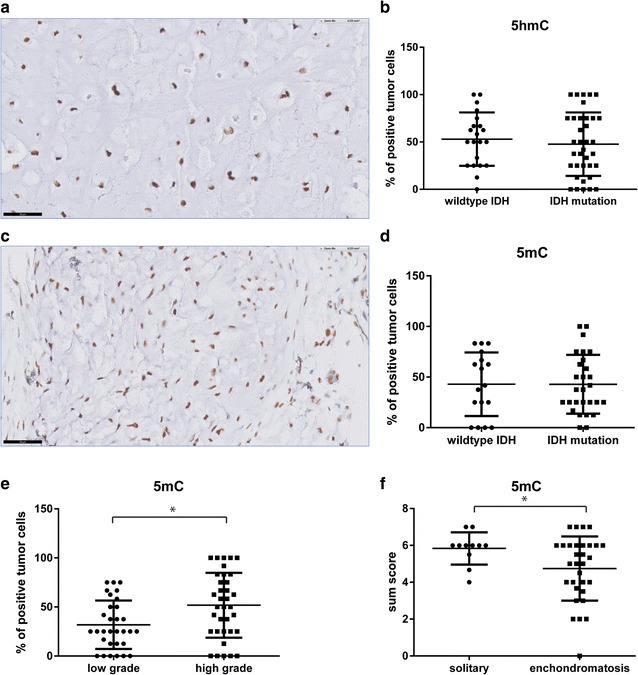
Variable levels of 5-hydroxymethylcytosine (5-hmC) and 5-methylcytosine (5-mC) in chondrosarcomas. Percentage of tumour cells positive for 5hmC (a, b) and 5mC (c, d) in IDH mutant versus IDH wildtype tumours (black bar indicates 50 µm). The percentage of 5-mC positive tumour cells was significantly higher in high grade chondrosarcomas as compared to low grade, which was independent of the IDH mutation status (e). Solitary chondrosarcomas showed significantly higher levels of 5mC compared to enchondromatosis related chondrosarcomas, independent of the histological grade (f)
Nuclear exclusion of TET1 is associated with loss of 5hmC in chondrosarcoma
Since we detected loss of 5-hmC in a subset of chondrosarcomas, which was not correlated with IDH mutation status, we evaluated whether, similar to gliomas [26], loss of 5-hmC was associated with nuclear exclusion of TET1. The majority of chondrosarcomas showed predominantly nuclear staining (N>C). No complete exclusion from the nucleus was seen in any of the chondrosarcomas (Fig. 5a). Chondrosarcomas with predominantly cytoplasmic expression of TET1 (C>N) showed significantly more often loss of 5hmC (p = 0.0001)(Fig. 5b).
Fig. 5.

Lower 5hmC levels in tumours with decreased nuclear staining for TET1. a There is no significant difference in subcellular localization of TET1 in IDH mutant versus IDH wildtype chondrosarcomas, and complete nuclear exclusion of TET1 was absent. b In tumours in which the staining was predominantly cytoplasmic, 5-hmC levels were significantly lower as compared to tumours in which staining was predominantly nuclear (scores were rounded to zero decimal places)
Discussion
We here report the analysis of mutations in genes encoding the metabolic enzymes IDH, SDH and FH in a cohort of ~100 central cartilaginous tumours, and demonstrate that the prevalence of mutations in IDH1 or -2 is ~60%, which is comparable to previously published data [8, 9]. As approximately 40% of the central chondrosarcomas lack detectable mutations in IDH, we investigated the possible involvement of two additional TCA cycle enzymes that are mutated in tumours, SDH and FH. Mutations in components of the SDH-complex and FH cause upregulation of succinate and fumarate, respectively, which, similar to d-2-HG, inhibit the TET enzymes [34–36] (Fig. 1). Using immunohistochemistry as a surrogate for mutation analysis, we show that SDH and FH mutations are not involved in the subset of chondrosarcomas that are wildtype for IDH. Moreover, it is unlikely that mutations in TET2 are playing a prominent role in chondrosarcoma, since Tarpey et al. demonstrated TET2 mutations in only one out of 49 (2%) chondrosarcomas subjected to whole-exome sequencing [47].
In contrast to gliomas, we here show that in chondrosarcoma, mutations in IDH are not significantly correlated with outcome. Disease specific survival and metastasis free survival did not differ between wild type and IDH mutant tumours of 63 patients. In a previous, separate series, we also found no prognostic value of these mutations [9]. Interestingly, patients with gliomas harboring IDH mutations have a more favorable outcome, independent of grade, as compared to gliomas that are wildtype for IDH [2, 14, 15]. In intrahepatic cholangiocarcinoma, as well as in leukemia, the prognostic significance of IDH mutations has remained controversial. The most recent studies however fail to demonstrate prognostic significance of the IDH mutation in these two tumour types [16, 17], which is comparable to chondrosarcoma.
Our aim was to evaluate the effect of IDH mutations on histone modifications, DNA modifications, chromatin remodeling, and subcellular localization of TET1 in a cohort of central cartilaginous tumours with known IDH mutation status. Immunoreactivity for the histone modification marks H3K4me3, H3K9me3 and H3K27me3 was observed in the majority of chondrosarcomas, irrespective of mutation status or histological grade. In contrast, we previously showed increased H3K9me3 in SDH mutant paragangliomas and FH mutant smooth muscle tumours using the same immunohistochemical methods. This was supported using SDH knockdown in cell lines, which demonstrates that our approach can detect differences in the methylation of these lysine residues [37]. Lu et al. showed increased H3K9me3 levels in IDH1 mutant gliomas as compared to the IDH wildtype counterparts, which were almost negative for H3K9me3 [48]. Rohle et al. reported removal of the repressive H3K9me3 and H3K27me3 marks after inhibition of mutant IDH1 using AGI-5198 in IDH1 mutant glioma cells [49]. In contrast to these findings in other tumour types, we showed previously that inhibition of mutant IDH1 in chondrosarcoma cell lines did not alter trimethylation of H3K4, H3K9 and H3K27 [50], which is in concordance with the present immunohistochemical results.
In addition to these covalent histone modifications, ATP-dependent chromatin remodeling complexes facilitate access of nucleosomal DNA. ATRX is an example of such a chromatin remodeler. We investigated expression of ATRX, since in gliomas loss of ATRX can be found in IDH mutant tumours [26]. Again, results are different from glioma as we found a low prevalence of loss of ATRX (5.6%), without any correlation to IDH mutation status in chondrosarcoma. The observed differences between IDH mutated chondrosarcomas versus IDH mutated gliomas on DNA and histone modifications, is likely attributable to additional genetic alterations that cooperate with mutant IDH to initiate cancer, e.g. ATRX and TP53 mutations in IDH mutant gliomas and COL2A1, YEATS2, NRAS, TP53, Rb- and Hh- signaling mutations in chondrosarcomas [47, 51–53]. d-2-HG competitively inhibits the TET enzymes [18] which is expected to result in inhibition of the conversion of 5-mC to 5-hmC [54]. Thus, loss of 5hmC expression is expected in tumours harbouring mutations in IDH, FH, SDH, similar to leukemias with mutations in TET2 [54]. Indeed, we previously showed loss of 5-hmC in SDH mutant paragangliomas and FH mutant smooth muscle tumours, again verifying our methodology [37]. Also in SDH deficient GIST, 5-hmC was low to absent [38]. For IDH mutant gliomas, however, reports have been conflicting [55, 56] and we now show that also in chondrosarcomas a correlation between mutations in IDH and loss of 5-hmC is absent. Despite this, we confirmed a correlation between loss of 5-hmC and diminished nuclear staining for TET1, which was also found in IDH wildtype gliomas [25] and SDH deficient paragangliomas [37].
Thus, overall we did not detect any differences in trimethylation of histone marks H3K4, -K9 and K27, and in 5-mC and 5-hmC levels between IDH mutant and IDH wildtype chondrosarcomas. Recently, Thienpont and colleagues reported that tumour hypoxia causes DNA hypermethylation and loss of 5hmC by reducing TET activity [57]. Interestingly, cartilage tissue and chondrosarcomas are known to have a hypoxic microenvironment [58, 59]; [60]; [61]. They also demonstrated a large overlap between genes hypermethylated in hypoxic versus IDH1 mutant glioblastomas [57]. Thus, as tumour hypoxia may have the same effect as an IDH mutation in chondrosarcoma, this may explain why we did not detect any differences using immunohistochemistry.
Moreover, we previously demonstrated that mutant IDH1 is not essential for chondrosarcoma cell proliferation and survival, as its inhibition using AGI-5198 decreased levels of d-2-HG without affecting tumourigenic properties of chondrosarcoma cell lines [50]. We also showed that trimethylation of H3K4, H3K9 and H3K27 did not change using AGI-5198 [50]. On the other hand, we and others have shown that mutant IDH1 plays a crucial role in the early development of benign enchondromas, as osteoblast differentiation was inhibited while promoting chondrogenic differentiation of mesenchymal stem cells [62, 63]. Moreover, Jin et al. have shown that during this process the repressive mark H3K9me3 and the active mark H3K4me3 were increased [63]. Thus, taken together, these data suggest that IDH mutations, and the resulting epigenetic changes, are only important for the initiation of enchondroma. However, once these enchondromas have matured, and after progression to chondrosarcoma, other processes are likely involved, since detectable changes in histone marks or 5-hmC are lacking and there is no correlation between IDH mutation and prognosis in central cartilaginous tumours.
Additional files
Additional file 1: Table S1. IDH primer sequences.
Additional file 2: Figure S1. No statistical significant difference was observed in disease specific survival between different IDH mutations (R132C, R132H, R132G, R132 l, R132S, R172S) compared to IDH wild-type chondrosarcomas (n = 63, p = 0.726).
Additional file 3: Figure S2. (A) All cartilage tumours on TMA were positive for SDHB, indicating absence of SDH mutations. (B) All cartilage tumours on TMA lacked detection of succinated protein using 2-SC staining, indicating absence of FH mutations, inset shows positive staining for 2-SC in a leiomyoma derived from a patient with a germline FH mutation as positive control (Scores were rounded to zero decimal places).
Additional file 4: Table S2. Mutation status for SDH (SDHB) and FH (2-SC) defined by immunohistochemistry on the TMA.
Authors’ contributions
AHGC wrote the manuscript, collecting immunohistochemistry data, data analysis. JS collecting immunohistochemistry data, data analysis, reviewed the manuscript. GA collecting immunohistochemistry data, reviewed the manuscript. IHBB performed the experiments, reviewed the manuscript. NF generated and provided the 2SC antibody, reviewed the manuscript. ASH collecting immunohistochemistry data, reviewed the manuscript. PMW-K performed the experiments, reviewed the manuscript. A-MC-J supervision experiments, reviewed the manuscript. JVMGB design of the study, scoring immunohistochemistry, primary supervisor of the project and writer. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thank Jolieke van Oosterwijk and Twinkal Pansuriya for construction of tissue microarrays, Yvonne de Jong for data analysis, Laura Paardekooper for technical assistance during the IDH mutation analysis, and Silvère M. van der Maarel for fruitful discussions.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The manuscript and the additional files contain all data.
Consent for publication
All authors agreed with current manuscript.
Ethics approval and consent to participate
All samples were handled according to the Dutch code of proper secondary use of human material as accorded by the Dutch society of pathology (Federa). The samples were handled in a coded manner. All study methods were approved by the LUMC ethical board (B17.002).
Funding
The study was financially supported by the Dutch Cancer Society (KWF Grant Number UL 2013-6103).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Arjen H. G. Cleven, Email: A.H.G.Cleven@lumc.nl
Johnny Suijker, Email: J.Suijker@lumc.nl.
Georgios Agrogiannis, Email: agrojohn@med.uoa.gr.
Inge H. Briaire-de Bruijn, Email: I.H.Briaire-de_Bruijn@lumc.nl
Norma Frizzell, Email: norma.frizzell@uscmed.sc.edu.
Attje S. Hoekstra, Email: A.S.Hoekstra@lumc.nl
Pauline M. Wijers-Koster, Email: P.M.Wijers-Koster@lumc.nl
Anne-Marie Cleton-Jansen, Email: A.M.Cleton-Jansen@lumc.nl.
Judith V. M. G. Bovée, Phone: ++31 71 5266617, Email: J.V.M.G.Bovee@lumc.nl
References
- 1.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 3.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kipp BR, Voss JS, Kerr SE, Barr Fritcher EG, Graham RP, Zhang L, et al. Isocitrate dehydrogenase 1 and 2 mutations in cholangiocarcinoma. Hum Pathol. 2012;43:1552–1558. doi: 10.1016/j.humpath.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang P, Dong Q, Zhang C, Kuan PF, Liu Y, Jeck WR, et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. 2013;32:3091–3100. doi: 10.1038/onc.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 8.Amary MF, Damato S, Halai D, Eskandarpour M, Berisha F, Bonar F, et al. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat Genet. 2011;43:1262–1265. doi: 10.1038/ng.994. [DOI] [PubMed] [Google Scholar]
- 9.Pansuriya TC, van Eijk R, d’Adamo P, van Ruler MA, Kuijjer ML, Oosting J, et al. Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome. Nat Genet. 2011;43:1256–1261. doi: 10.1038/ng.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucas DR, Bridge JA. Chondromas: enchondroma, periosteal chondroma. In: Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F, editors. WHO Classification of Tumours of Soft Tissue and Bone. Lyon: International Agency for Research on Cancer (IARC); 2013. p. 252–4.
- 11.Pansuriya TC, Kroon HM, Bovee JVMG. Enchondromatosis: insights on the different subtypes. Int J Clin Exp Pathol. 2010;3:557–569. [PMC free article] [PubMed] [Google Scholar]
- 12.Hogendoorn PCW, Bovée JVMG, Nielsen GP. Chondrosarcoma (grades I-III), including primary and secondary variants and periosteal chondrosarcoma. In: Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F, editors. WHO Classification of Tumours of Soft Tissue and Bone. Lyon: IARC; 2013. pp. 264–268. [Google Scholar]
- 13.Evans HL, Ayala AG, Romsdahl MM. Prognostic factors in chondrosarcoma of bone. A clinicopathologic analysis with emphasis on histologic grading. Cancer. 1977;40:818–831. doi: 10.1002/1097-0142(197708)40:2<818::AID-CNCR2820400234>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 14.Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27:4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 15.Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15:6002–6007. doi: 10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 16.Goyal L, Govindan A, Sheth RA, Nardi V, Blaszkowsky LS, Faris JE, et al. Prognosis and clinicopathologic features of patients with advanced stage isocitrate dehydrogenase (IDH) mutant and IDH wild-type intrahepatic cholangiocarcinoma. Oncologist. 2015;20:1019–1027. doi: 10.1634/theoncologist.2015-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DiNardo CD, Ravandi F, Agresta S, Konopleva M, Takahashi K, Kadia T, et al. Characteristics, clinical outcome, and prognostic significance of IDH mutations in AML. Am J Hematol. 2015;90:732–736. doi: 10.1002/ajh.24072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pronier E, Almire C, Mokrani H, Vasanthakumar A, Simon A, da Mor CRM, et al. Inhibition of TET2-mediated conversion of 5-methylcytosine to 5-hydroxymethylcytosine disturbs erythroid and granulomonocytic differentiation of human hematopoietic progenitors. Blood. 2011;118:2551–2555. doi: 10.1182/blood-2010-12-324707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller T, Gessi M, Waha A, Isselstein LJ, Luxen D, Freihoff D, et al. Nuclear exclusion of TET1 is associated with loss of 5-hydroxymethylcytosine in IDH1 wild-type gliomas. Am J Pathol. 2012;181:675–683. doi: 10.1016/j.ajpath.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 26.Wiestler B, Capper D, Holland-Letz T, Korshunov A, von Deimling A, Pfister SM, et al. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013;126:443–451. doi: 10.1007/s00401-013-1156-z. [DOI] [PubMed] [Google Scholar]
- 27.Argentaro A, Yang JC, Chapman L, Kowalczyk MS, Gibbons RJ, Higgs DR, et al. Structural consequences of disease-causing mutations in the ATRX-DNMT3-DNMT3L (ADD) domain of the chromatin-associated protein ATRX. Proc Natl Acad Sci USA. 2007;104:11939–11944. doi: 10.1073/pnas.0704057104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 29.Niemann S, Muller U. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nat Genet. 2000;26:268–270. doi: 10.1038/81551. [DOI] [PubMed] [Google Scholar]
- 30.Astuti D, Latif F, Dallol A, Dahia PL, Douglas F, George E, et al. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet. 2001;69:49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burnichon N, Briere JJ, Libe R, Vescovo L, Riviere J, Tissier F, et al. SDHA is a tumor suppressor gene causing paraganglioma. Hum Mol Genet. 2010;19:3011–3020. doi: 10.1093/hmg/ddq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janeway KA, Kim SY, Lodish M, Nose V, Rustin P, Gaal J, et al. Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. Proc Natl Acad Sci USA. 2011;108:314–318. doi: 10.1073/pnas.1009199108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 34.Smith EH, Janknecht R, Maher LJ., III Succinate inhibition of alpha-ketoglutarate-dependent enzymes in a yeast model of paraganglioma. Hum Mol Genet. 2007;16:3136–3148. doi: 10.1093/hmg/ddm275. [DOI] [PubMed] [Google Scholar]
- 35.Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letouze E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–752. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 37.Hoekstra AS, de Graaff MA, Briaire-de Bruijn IH, Ras C, Seifar RM, van Minderhout I, et al. Inactivation of SDH and FH cause loss of 5hmC and increased H3K9me3 in paraganglioma/pheochromocytoma and smooth muscle tumors. Oncotarget. 2015;6:38777–38788. doi: 10.18632/oncotarget.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mason EF, Hornick JL. Succinate dehydrogenase deficiency is associated with decreased 5-hydroxymethylcytosine production in gastrointestinal stromal tumors: implications for mechanisms of tumorigenesis. Mod Pathol. 2013;26:1492–1497. doi: 10.1038/modpathol.2013.86. [DOI] [PubMed] [Google Scholar]
- 39.Waaijer CJ, de Andrea CE, Hamilton A, van Oosterwijk JG, Stringer SE, Bovee JVMG. Cartilage tumour progression is characterized by an increased expression of heparan sulphate 6O-sulphation-modifying enzymes. Virchows Arch. 2012;461:475–481. doi: 10.1007/s00428-012-1300-5. [DOI] [PubMed] [Google Scholar]
- 40.Pansuriya TC, Oosting J, Krenacs T, Taminiau AH, Verdegaal SH, Sangiorgi L, et al. Genome-wide analysis of Ollier disease: is it all in the genes? Orphanet J Rare Dis. 2011;6:2. doi: 10.1186/1750-1172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gruis NA, Abeln ECA, Bardoel AFJ, Devilee P, Frants RR, Cornelisse CJ. PCR-based microsatellite polymorphisms in the detection of loss of heterozygosity in fresh and archival tumour tissue. Br J Cancer. 1993;68:308–313. doi: 10.1038/bjc.1993.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bardella C, El-Bahrawy M, Frizzell N, Adam J, Ternette N, Hatipoglu E, et al. Aberrant succination of proteins in fumarate hydratase-deficient mice and HLRCC patients is a robust biomarker of mutation status. J Pathol. 2011;225:4–11. doi: 10.1002/path.2932. [DOI] [PubMed] [Google Scholar]
- 43.Ternette N, Yang M, Laroyia M, Kitagawa M, O’Flaherty L, Wolhulter K, et al. Inhibition of mitochondrial aconitase by succination in fumarate hydratase deficiency. Cell Rep. 2013;3:689–700. doi: 10.1016/j.celrep.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirmani S, Young WF. Hereditary Paraganglioma–Pheochromocytoma Syndromes. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJ, et al., editors. Gene reviews. Seattle: University of Washington; 2008.
- 45.van Oosterwijk JG, Meijer D, van Ruler MA, van den Akker BE, Oosting J, Krenacs T, et al. Screening for potential targets for therapy in mesenchymal, clear cell, and dedifferentiated chondrosarcoma reveals Bcl-2 family members and TGFbeta as potential targets. Am J Pathol. 2013;182:1347–1356. doi: 10.1016/j.ajpath.2012.12.036. [DOI] [PubMed] [Google Scholar]
- 46.Wagner K, Damm F, Gohring G, Gorlich K, Heuser M, Schafer I, et al. Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. J Clin Oncol. 2010;28:2356–2364. doi: 10.1200/JCO.2009.27.6899. [DOI] [PubMed] [Google Scholar]
- 47.Tarpey PS, Behjati S, Cooke SL, Van LP, Wedge DC, Pillay N, et al. Frequent mutation of the major cartilage collagen gene COL2A1 in chondrosarcoma. Nat Genet. 2013;45:923–926. doi: 10.1038/ng.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suijker J, Oosting J, Koornneef A, Struys EA, Salomons GS, Schaap FG, et al. Inhibition of mutant IDH1 decreases d-2-HG levels without affecting tumorigenic properties of chondrosarcoma cell lines. Oncotarget. 2015;6:12505. doi: 10.18632/oncotarget.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clark O, Yen K, Mellinghoff IK. Molecular pathways: isocitrate dehydrogenase mutations in cancer. Clin Cancer Res. 2016;22:1837–1842. doi: 10.1158/1078-0432.CCR-13-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Totoki Y, Yoshida A, Hosoda F, Nakamura H, Hama N, Ogura K, et al. Unique mutation portraits and frequent COL2A1 gene alteration in chondrosarcoma. Genome Res. 2014;24:1411–1420. doi: 10.1101/gr.160598.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang YX, van Oosterwijk JG, Sicinska E, Moss S, Remillard SP, Van Wezel T, et al. Functional profiling of receptor tyrosine kinases and downstream signaling in human chondrosarcomas identifies pathways for rational targeted therapy. Clin Cancer Res. 2013;19:3796–3807. doi: 10.1158/1078-0432.CCR-12-3647. [DOI] [PubMed] [Google Scholar]
- 54.Pronier E, Almire C, Mokrani H, Vasanthakumar A, Simon A, da Mor BD, et al. Inhibition of TET2-mediated conversion of 5-methylcytosine to 5-hydroxymethylcytosine disturbs erythroid and granulomonocytic differentiation of human hematopoietic progenitors. Blood. 2011;118(9):2551–2555. doi: 10.1182/blood-2010-12-324707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu J, Zuo J, Xu Q, Wang X, Wang Z, Zhou D. Isocitrate dehydrogenase mutations may be a protective mechanism in glioma patients. Med Hypotheses. 2011;76:602–603. doi: 10.1016/j.mehy.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 56.Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, et al. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011;71:7360–7365. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thienpont B, Steinbacher J, Zhao H, D’Anna F, Kuchnio A, Ploumakis A, et al. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature. 2016;537:63–68. doi: 10.1038/nature19081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gibson JS, Milner PI, White R, Fairfax TP, Wilkins RJ. Oxygen and reactive oxygen species in articular cartilage: modulators of ionic homeostasis. Pflugers Arch. 2008;455:563–573. doi: 10.1007/s00424-007-0310-7. [DOI] [PubMed] [Google Scholar]
- 59.Kubo T, Sugita T, Shimose S, Matsuo T, Arihiro K, Ochi M. Expression of hypoxia-inducible factor-1alpha and its relationship to tumour angiogenesis and cell proliferation in cartilage tumours. J Bone Joint Surg Br. 2008;90:364–370. doi: 10.1302/0301-620X.90B3.19806. [DOI] [PubMed] [Google Scholar]
- 60.Ayala G, Liu C, Nicosia R, Horowitz S, Lackman R. Microvasculature and VEGF expression in cartilaginous tumors. Hum Pathol. 2000;31:341–346. doi: 10.1016/S0046-8177(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 61.Boeuf S, Bovee JV, Lehner B, Hogendoorn PC, Richter W. Correlation of hypoxic signalling to histological grade and outcome in cartilage tumours. Histopathology. 2010;56:641–651. doi: 10.1111/j.1365-2559.2010.03528.x. [DOI] [PubMed] [Google Scholar]
- 62.Suijker J, Baelde HJ, Roelofs H, Cleton-Jansen AM, Bovee JVMG. The oncometabolite d-2-hydroxyglutarate induced by mutant IDH1 or -2 blocks osteoblast differentiation in vitro and in vivo. Oncotarget. 2015;20:14832–14842. doi: 10.18632/oncotarget.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin Y, Elalaf H, Watanabe M, Tamaki S, Hineno S, Matsunaga K, et al. Mutant IDH1 dysregulates the differentiation of mesenchymal stem cells in association with gene-specific histone modifications to cartilage- and bone-related genes. PLoS ONE. 2015;10:e0131998. doi: 10.1371/journal.pone.0131998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. IDH primer sequences.
Additional file 2: Figure S1. No statistical significant difference was observed in disease specific survival between different IDH mutations (R132C, R132H, R132G, R132 l, R132S, R172S) compared to IDH wild-type chondrosarcomas (n = 63, p = 0.726).
Additional file 3: Figure S2. (A) All cartilage tumours on TMA were positive for SDHB, indicating absence of SDH mutations. (B) All cartilage tumours on TMA lacked detection of succinated protein using 2-SC staining, indicating absence of FH mutations, inset shows positive staining for 2-SC in a leiomyoma derived from a patient with a germline FH mutation as positive control (Scores were rounded to zero decimal places).
Additional file 4: Table S2. Mutation status for SDH (SDHB) and FH (2-SC) defined by immunohistochemistry on the TMA.
Data Availability Statement
The manuscript and the additional files contain all data.


