Abstract
Resistance to xenobiotic nucleosides used to treat acute myeloid leukemia (AML) and other cancers remains a major obstacle to clinical management. One process suggested to participate in resistance is reduced uptake into tumor cells via nucleoside transporters, although precise mechanisms are not understood. Through transcriptomic profiling, we determined that low expression of the ergothioneine transporter OCTN1 (SLC22A4; ETT) strongly predicts poor event-free survival and overall survival in multiple cohorts of AML patients receiving treatment with the cytidine nucleoside analog cytarabine. Cell biological studies confirmed OCTN1-mediated transport of cytarabine and various structurally-related cytidine analogs, such as 2′deoxycytidine and gemcitabine, occurs through a saturable process that is highly sensitive to inhibition by the classic nucleoside transporter inhibitors dipyridamole and nitrobenzylmercaptopurine ribonucleoside (NBMPR). Our findings have immediate clinical implications given the potential of the identified transport system to help refine strategies that could improve patient survival across multiple cancer types where nucleoside analogs are used in cancer treatment.
Keywords: Nucleosides, Transporter, Leukemia, Cytarabine, OCTN1
Introduction
All endogenous and xenobiotic nucleosides are polar hydrophilic compounds that are poorly membrane permeable and require functional transporters to enter cells. The two major classes of mammalian nucleoside transporters consist of equilibrative nucleoside transporters (ENTs) and concentrative nucleoside transporters (CNTs). Two proteins of the former class, ENT1 (SLC29A1) and ENT2 (SLC29A2), mediate the transport of purine and pyrimidine nucleosides down their concentration gradients. These transporters exhibit broad permeant selectivity and are subdivided based on their sensitivity (ENT1) or resistance (ENT2) to inhibition by nanomolar amounts of nitrobenzylmercaptopurine ribonucleoside (NBMPR) (1).
One of the first pyrimidine nucleosides to be developed as an anticancer drug was cytarabine (cytosine arabinoside; Ara-C), a derivative of 2′-deoxycytidine (2). Intracellular accumulation of cytarabine depends on plasma concentrations of the drug and the compound enters cells exclusively by transporter-mediated processes (3). Once cytarabine has entered cells, it is phosphorylated by deoxycytidine kinase to the mono-phosphorylated, 5′-derivative Ara-CMP and subsequently by nucleotide kinases to the pharmacologically-active, tri-phosphorylated metabolite Ara-CTP (4). Cytarabine is used clinically in the treatment of hematological malignancies, in particular in acute myeloid leukemia (AML), although intrinsic insensitivity and/or the development of resistance remains a major obstacle to successful long-term treatment outcome (5). Reduced uptake into leukemic cells has been proposed as a process underlying most instances of clinical resistance to cytarabine, although the responsible mechanism remains poorly understood (6).
Previous investigations demonstrated that low cytarabine uptake in AML cells predicts poor response to therapy (7), that cytarabine uptake is required for its conversion to Ara-CTP by leukemic blast cells, and that myeloblasts form more Ara-CTP than lymphoblasts because of higher nucleoside transport (8). Interestingly, cytarabine uptake in AML cells is completely inhibited by nanomolar concentrations of NBMPR and dipyridamole (9). Because the uptake of cytarabine in leukemic blasts is highly sensitive to NBMPR, it has been assumed and asserted for several decades that cytarabine enters cells through an ENT1-dependent mechanism (1). This conclusion is supported by some (10, 11) but not all statistical association studies (12–14), indicating that increased ENT1 mRNA abundance is correlated with increased cellular sensitivity or clinical responsiveness to cytarabine. More importantly, however, published studies to date employing heterologous expression systems have shown that cytarabine is either very weakly transported by ENT1 (<2-fold vs control) (15, 16) or is not transported by ENT1 at all (17). Against this background, the elucidation of novel, ENT1-independent transport mechanisms leading to anti-leukemic effects following cytarabine treatment is considered of high clinical relevance. In the present study, we report that the ability of cytarabine and several structurally-related nucleosides to enter cells is facilitated by the ergothioneine uptake transporter OCTN1 (SLC22A4; ETT), and that low expression of OCTN1 in leukemic cells is a strong predictor of poor survival in multiple cohorts of patients with AML treated with cytarabine-based regimens.
Materials and Methods
Additional details of materials and methods are reported in the Supplementary Methods.
Clinical association studies
Gene expression data were collected from 168 pediatric patients with de novo AML enrolled on the AML02 trial (18, 19), of whom 134 were included in the analysis [2 were not randomized; 14 were excluded due to lack of induction I minimal residual disease (MRD) assessment; 1 was excluded due to lack of FLT3-ITD status; and 17 were excluded due to lack of French-American-British (FAB) classification status]. Total RNA was isolated from primary blast samples, as previously described (18, 19). Data on all 392 SLC (uptake) transporter probe sets from the Affymetrix U133Av2 expression array were extracted, log2 transformed, normalized by Z-score, visualized using R software package (R Studio v0.98.1091), and tested for statistical association with overall survival (OS) and event-free survival (EFS). Of the transporter probe sets, 22 were associated with both OS and EFS (P < 0.05), among which 14 were associated with a hazard ratio of less than 1 and improved survival. After exclusion of genes encoding transporter proteins not localized to the outer membrane, SLC22A4 (OCTN1) was identified as the top-ranking gene for further consideration (Supplementary Table S1). The association between SLC22A4 expression and OS and EFS was tested by 3 different Cox regression models. The first model was only stratified by treatment arms, the second was adjusted by risk and stratified by treatment arms, and the third was adjusted by age, MRD, FLT3-ITD, core binding factor (CBF), FAB category AML-M7 without t(1;22) and other 11q23, and stratified by treatment arm. To illustrate the association between gene expression and survival outcome in Kaplan-Meier plots, SLC22A4 expression values were categorized into 3 groups based on quantile ranking. Other cut-off points for grouping were considered, for example on the basis of possible gaps within the dataset after plotting expression data within their empirical distribution function. However, no gaps were observed, suggesting that quantile raking is an appropriate strategy to cluster expression values. Complete remission (CR) status was designated as less than 5% of blasts in the bone marrow, and induction failure was defined as the absence of CR within 3 months of the start of treatment.
External validation was performed on published data from 54 pediatric patients with AML receiving cytarabine-based chemotherapy on whom gene expression (Affymetrix U95Av2 microarray) and clinical data (EFS and CR) were available [see Table S2 in (14)]. Gene expression values provided in the published report were transformed to percentiles such that the hazard ratio can be explained as a change of event rate per one percentile change of expression from the lowest expression value observed. Additional external validation studies on OS and EFS involved gene expression analyses on the AML gene expression dataset from The Cancer Genome Atlas (TCGA), consisting of RNAseq data from Illumina Hi-seq analyses on 172 samples from adult patients with de novo AML (20). The information of interest was extracted using the cBioPortal for Cancer Genomics (21, 22).
Source of cell lines
HEK293 (Invitrogen), HeLa and CHO (ATCC) were obtained from commercial sources and used without further authentication by the authors. The PK15 and PK15NTD cell lines were provided by Chung-Ming Tse (Johns Hopkins University, Baltimore, MD) for providing The AML cell line CHRF288-11 cells were obtained from Tanja Gruber (St. Jude Children’s Research Hospital, Memphis, TN); CMS from Yubin Ge (Karmanos Cancer Institute, Detroit, MI); M07e, MOLM-13, ML-2, and NB4 from DSMZ; MV4-11, HL-60, KG-1, THP-1, and U937 from ATCC; and OCI-AML3 from Brian Sorrentino (St. Jude Children’s Research Hospital, Memphis, TN). All cell lines are tested regularly for mycoplasma contamination using a commercially available kit (Lonza). Passages are kept to a minimum and cells are not used beyond passage 30.
Transfection of HEK293 cells
The human embryonic kidney cell line HEK293 (Invitrogen) was used for the generation of OCTN1- and ENT1-overexpressing models, because these cells had the lowest native uptake of cytarabine compared with other cells commonly used for transfection, such as the cervical cancer cell line HeLa or Chinese hamster ovary (CHO) cells (0.94 vs 5.4 vs 4.6 pmol/mg/min, respectively). Interestingly, OCTN1 has been immune-detected in HeLa cells (23), and the relatively high uptake of cytarabine in CHO cells was previously connected with the number of NBMPR-binding sites (24). Reconstructed OCTN1 and ENT1 cDNAs (Origene) with a Flag-tag were subcloned into the pMIG II vector, engineered from a MSCV-IRES-GFP vector. Empty vector (VC) or the vector encoding OCTN1-Flag or ENT1-Flag were co-transfected with pEQ-Pam3(-E) and pVSVg into 293T cells using Fugene transfection reagent (Roche). Virus-containing medium was collected at 72 hours and used to infect HEK293 cells in the presence of hexadimethrine bromide (polybrene; 6 μg/ml). At 7–10 days after infection, GFP-positive cells were sorted by FACS to ~95% purity. Cells were maintained in DMEM (Life Technologies) supplemented with 10% fetal bovine serum (Hyclone) at 37°C with 5% CO2.
The expression levels of OCTN1 in the transfected HEK293 cells were confirmed by RT-PCR and western blot (Supplementary Fig. S1A), and plasma membrane localization of OCTN1 was demonstrated by confocal microscopy (Supplementary Fig. S1B). Additional characterization of the model system with gene expression arrays revealed detectable transcripts in HEK293 cells of other putative cytarabine transporter genes, including those encoding CNT1, CNT2, CNT3, ENT1, ENT2, ENT3, and ENT4 (Supplementary Fig. S1C), as well as OCTN1-related genes, including those encoding OAT1, OAT2, OAT3, OCTN2, OCT1, OCT2, and OCT3 (Supplementary Fig. S1D). Quantitative RT-PCR with Transporter RT2 Profiles PCR arrays (Supplementary Fig. S1E), western blot (Supplementary Fig. S1F), and functional analysis (Supplementary Fig. S1G), using uridine as a substrate for ENT1 and carnitine as a substrate for OCTN2, confirmed that these transporters were not differentially expressed in cells transfected with empty vector as compared with cells overexpressing OCTN1. Functionally, the OCTN1-overexpressed HEK293 cells were further characterized by their ability to efficiently accumulate the known OCTN1 substrates ergothioneine, a naturally-occurring thiourea derivative of histidine (25), as well as carbon 14-labeled tetraethylammonium (TEA), a small molecule organic cation (26). The observed transport efficiencies for ergothioneine (62 ± 8 μl/mg/min) and TEA (1.1 ± 0.12 μl/mg/min) were similar to those reported previously (25). Phenotypic characterization of the heterologous expression model of ENT1 indicated efficient accumulation of the known substrates uridine (3.3-fold vs control cells) and gemcitabine (1.5-fold). These observations suggest that the overexpressed HEK293 cells represent a bona fide model to evaluate a possible contribution of OCTN1 and ENT1 to the transport of AML-directed chemotherapeutics, such as cytarabine.
Real-time PCR
RNA was reverse transcribed using SuperScript III first strand synthesis supermix for real-time RT-PCR (Invitrogen), according to manufacturer’s recommendations. Gene transcripts were quantified using SYBR Green PCR mastermix (Qiagen) and primers obtained from Applied Biosystems that were specific to OCTN1, ENT1 and other transporters of interest. Reactions were carried out in triplicate, with transcripts normalized to GAPDH.
Gene-chip arrays
RNA was extracted from HEK293 cells using the RNEasy mini kit (Qiagen), samples were amplified, and then analyzed using the Affymetrix GeneChip arrays. Data on 293 solute carrier genes were extracted, normalized by the RMA algorithm, and analyzed using the Partek Genomics Suite 6.4 software.
Immunoblot analysis
For protein expression of select transporters, cells were lysed, and crude membrane fractions prepared using the ProteoExtract Native Membrane Protein Extraction Kit (Calbiochem), according to the manufacturer’s protocol. Protein concentration of the membrane preps were determined using the Pierce BCA Protein Assay Kit (ThermoScientific) in microplates. Membrane samples (20 μg) were separated by SDS-PAGE and transferred to PVDF. Western blot analysis was performed using α-OCTN1 (1:500; Sigma-Aldrich), α-ENT1 (1:1,000, Invitrogen), α-OCTN2 (1:1,000, Invitrogen), or α-transferrin receptor (TfR1; 1:1,000; Invitrogen). Secondary α-rabbit or α-mouse antibodies (1:2,000) conjugated to peroxidase (Jackson Immuno Research) were used and proteins were visualized by chemiluminescence using the SignalFire ECL Reagent (Cell Signaling) on an Odyssey Fc Imaging System (Li-Cor). Immunoblots were performed a minimum of two times on samples collected from different experiments.
Site-directed mutagenesis
The QuikChange II XL Site-Directed Mutagenesis Kit (Agilent) was utilized to generate the OCTN1 variant rs1050152 (c.1507C>T; L503F), according to suggested methods. The mutagenesis primers were designed using the QuikChange Primer Design program and synthesized by Integrated DNA Technologies. The plasmid was sequenced to confirm successful mutagenesis and then used to generate stably transfected HEK293 cells.
Cellular accumulation studies
Cells were seeded at 5×105 cells/well in 6-well plates. At 70% confluence, cells were washed with phosphate buffered saline (PBS) and replaced with fresh, serum-free medium containing varying concentrations of radiolabeled or unlabeled compound. At the end of the incubation period, the medium was removed, and cells were washed twice with ice-old PBS. A 25-μl aliquot of the lysate was used to estimate protein concentration using a BCA protein assay kit. Radioactivity was measured on a Perkin Elmer Topcount or a Beckman LS 6500 liquid scintillation counter, after mixing the sample with Scintisafe 30% scintillation fluid (PerkinElmer). Cytarabine and phosphorylated metabolites were measured in cell extracts by liquid chromatography-tandem mass spectrometry, as previously described (27). Cellular accumulation was expressed as pmol per mg of protein (pmol/mg), as a percentage of control cells (%VC) with vector set to 100%, or as a concentration-normalized transport efficiency (μl/mg/min). At least two independent experiments were performed using multiple replicates.
Cytotoxicity assays
To evaluate cytotoxicity in response to cytarabine and related nucleoside analogs, HEK293 cells were seeded at 2,000 cells per well in 96-well filtered plates and cultured for 20 hours. Cells were then treated with nucleosides of interest at increasing concentrations (0.313–100 μM). Cell viability was determined using a CellTiter-Glo luminescent cell viability assay at 72 hours (Promega). The concentration of drug that inhibited cell proliferation by 50% (IC50) was determined using the software program GraphPad Prism version 5.0 (GraphPad Software).
Silencing of OCTN1 expression
The on-target plus SMART pool human SLC22A4 siRNA for OCTN1 silencing (Dharmacon) and the Mission siRNA Negative Non-targeting Control (NT) #1 siRNA (Sigma) were used in all experiments. OCI-AML3 cells were transfected with OCTN1 siRNA or negative control SiRNA using Nucleofector II and nucleofector Kit T (Amaxa), according to the manufacturer’s protocol. Briefly, OCI-AML3 cells (3 × 106) were treated with 3 μg of siRNA for each sample, and OCTN1 suppression was evaluated using crude membrane fractions extracted 48 hours later using ProteoExtract Native Membrane Protein Extraction Kit (Calbiochem). Next, immunoblots were performed with 10 μg membrane samples using SDS-PAGE and then transferred to PVDF membranes. The membrane was probed using SLC22A4 (1:500; ThermoFisher Scientific) or α-transferrin receptor (1:1,000; Invitrogen), and a secondary α-rabbit or α-mouse IgG conjugated to peroxidase (Jackson ImmunoResearch) was used at 1:10,000 or 1:1,000. Proteins were visualized by chemiluminescence with the Signal Fire ECL Reagent (Cell Signaling), and quantitated by Imagine J software. Immunoblots were performed a minimum of two times on samples collected from different experiments.
Cellular uptake assay in OCI-AML3 cells was also determined after 48 hours transfection with OCTN1 siRNA or control. Cells were pre-incubated with DMSO or NBMPR (10 μM) for 15 min, followed by addition of [3H]cytarabine (1 μM), and uptake assays were performed with 5- min incubation times. After incubation, cells were washed three times with ice-cold PBS, lysed with 1 N NaOH, and agitated for 1 hour. Lysate aliquots of 25 μL were used to estimate protein concentration using a BCA protein assay kit (Thermo-Scientific). Radioactivity was measured on a liquid scintillation analyzer Tri-Carb 4810 TR (Perkin Elmer) after mixing the sample with 400 μL of Emulsifier-safe scintillation fluid (Perkin Elmer). Cytarabine uptake levels were normalized to total protein levels in each group, and expressed relative to the uptake observed in cells transfected with the NT siRNA.
Statistics
Data are presented as mean values and SEM, unless stated otherwise. An unpaired two sided Student’s t-test or one-way analysis of variance with a Tukey post-hoc test were used to evaluate statistical significance, using P < 0.05 as a cut-off.
Results and Discussion
Identification of OCTN1 as a predictor of survival in AML
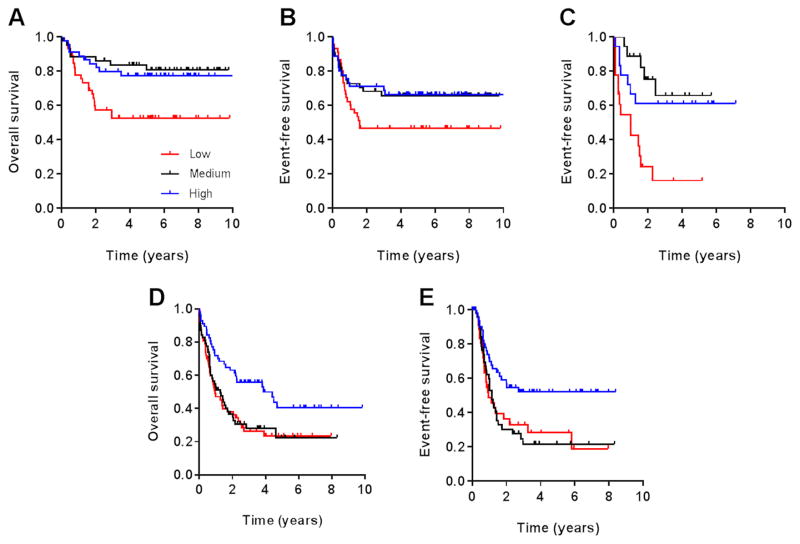
Since the commonly-held contention of ENT1 playing an important or even exclusive role in cytarabine transport into AML cells is poorly supported by experimental evidence, we postulated the existence on myeloblasts of a presently unknown, highly-efficient cytarabine carrier. To identify such a new putative cytarabine uptake transporter, we performed comprehensive transcriptomic profiling of blast samples from patients with AML receiving cytarabine-based chemotherapy. This analysis was based on the expectation that such an approach would identify genes that, when overexpressed on AML blasts, increase mediated intracellular accumulation of cytarabine, and consequently associate with improved responsiveness. Using the Affymetrix U133Av2 platform, we interrogated 392 transporter probe sets captured on the microarray in 134 pediatric patients with de novo AML treated on the AML02 multicenter trial (18, 19). After adjusting for known prognostic factors, including age, risk group, and treatment arm, the gene most significantly associated with both overall survival (P = 0.0074; hazard ratio, 0.56) and event-free survival (P = 0.024; hazard ratio, 0.67) was SLC22A4, a gene encoding the ergothioneine uptake transporter OCTN1 (ETT) (Fig. 1A–B and Table 1). In particular, we observed that increased expression of SLC22A4 (probe set ID, 205896_at) was associated with a reduction in the rate of relapse, resulting in an improved survival rate. This finding was independently replicated in a second cohort of 54 pediatric patients with AML for whom event-free survival data were available (P = 0.0074; hazard ratio, 0.50), and who received a similar cytarabine-based treatment regimen (Fig. 1C) (14). In addition, increased expression of SLC22A4 was associated with improved overall survival (P = 0.0048) and event-free survival (P = 0.0091) in 172 clinically-annotated adult cases of de novo AML (Fig. 1D–E). Recent genetic association studies have revealed the molecular genetic heterogeneity of AML and have shown that mechanisms of resistance are highly heterogeneous (28). In this connection, it is of interest to points out that the expression of SLC22A4 was significantly higher in patients entering complete remission as compared with those experiencing induction failure or relapse within the first year of complete remission in both the AML02 trial (P=0.029) and the pediatric replication cohort (P=0.0014). This finding suggests that the observed association of OCTN1 with survival is unrelated to heterogeneity of the disease but rather with it being a predictor of treatment response.
Fig. 1. Association of OCTN1 gene expression with survival in AML.
Kaplan-Meier plots show rates of overall survival and event-free survival according to leukemic blast expression of the OCTN1 transporter gene SLC22A4 in a cohort of 134 pediatric patients with AML (A, B), an independent cohort of 54 pediatric patients with AML (C), and a cohort of 172 adult patients with AML (D, E), all receiving cytarabine-based chemotherapy. Gene expression ranks represent low expressors (bottom 1/3; shown in RED), intermediate expressors (middle 1/3; shown in BLACK) and top expressors (top 1/3; shown in BLUE).
Table 1.
Correlation of SLC22A4 expression with survival in AML.
| Model* | P value | HR | lower 95% CI | upper 95% CI |
|---|---|---|---|---|
| Overall survival | ||||
| 1 | 0.0077 | 0.5939 | 0.4048 | 0.8712 |
| 2 | 0.0033 | 0.5540 | 0.3738 | 0.8212 |
| 3 | 0.0074 | 0.5611 | 0.3677 | 0.8562 |
| Event-free survival | ||||
| 1 | 0.0184 | 0.6645 | 0.4731 | 0.9333 |
| 2 | <0.0001 | 0.2089 | 0.1000 | 0.4368 |
| 3 | 0.0240 | 0.6694 | 0.4724 | 0.9484 |
Model 1 is only stratified by treatment arms; Model 2 is adjusted by risk and stratified by treatment arms; Model 3 is adjusted by age, MRD, FLT3-ITD, CBF, M7 without t(1;22) and other 11q23, which are the factors that were found to be statistically significant in the original association analyses (18, 19), and stratified by treatment arm. Note that independent of the model, SLC22A4 expression is significantly associated with both overall survival and event free survival outcomes, with higher expression levels being associated with better survival outcomes (ie, hazard ratios significantly less than 1).
Abbreviation: HR, hazard ratio; CI, confidence interval.
Identification of OCTN1 as a high-affinity cytarabine transporter
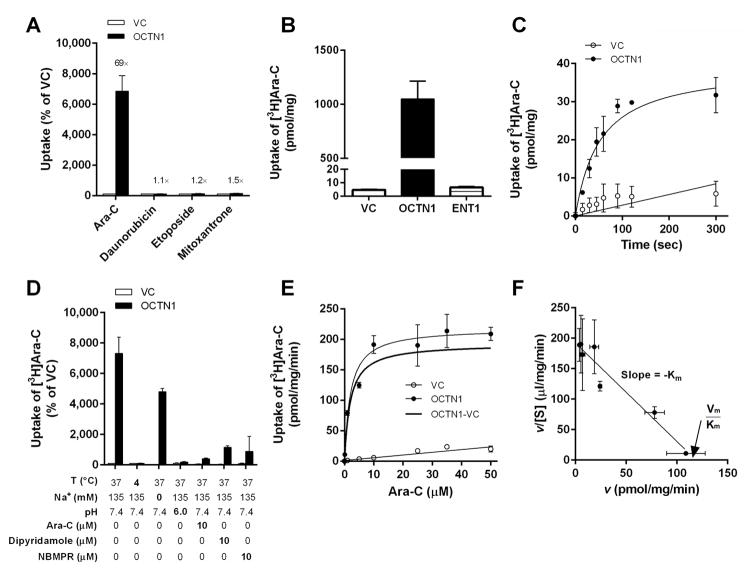
In the gene-expression association study that led to the identification of SLC22A4 as a predictor of survival in AML, patients received combination chemotherapy involving cytarabine, daunorubicin, etoposide, and mitoxantrone (19). An initial examination of these 4 compounds in our heterologous expression model revealed that cytarabine uptake was increased by 60 to 70 fold in HEK293 cells engineered to overexpress OCTN1 compared with control cells, whereas the increases ranged only 1.1 to 1.5 fold for the 3 other drugs (Fig. 2A). The weak interaction observed here between OCTN1 and etoposide or mitoxantrone is in line with previously reported uptake data (29, 30), and with the notion that etoposide can competitively inhibit the specific binding of NBMPR to an unidentified cytarabine carrier on AML blasts (31). Importantly, in a comparative analysis, we found that the efficiency of cytarabine transport by OCTN1 was greater than that observed for ENT1 (Fig. 2B) in the same HEK293 cell-based model system. Subsequent studies indicated that OCTN1-mediated cytarabine transport was time-dependent (Fig. 2C), sensitive to temperature, pH-dependent as observed for TEA transport by OCTN1 (32), saturable (Fig. 2D), and was associated with a Michaelis-Menten constant (Km) of 1.95 ± 0.37 μM and a maximum velocity (Vmax) of 193 pmol/mg/min (Fig. 2E). In addition, an Eadie-Hofstee analysis revealed that the transporter interaction with cytarabine involved a single, saturable binding-site on the OCTN1 protein (Fig. 2F). Interestingly, whereas the OCTN1-mediated transport of ergothioneine is dependent on sodium (33), no pH dependence was found for the transport of cytarabine by OCTN1, although the absolute intracellular drug uptake was decreased in the absence of sodium (Fig. 2C). However, a proportionally similar decrease in cytarabine uptake was observed in vector control cells in the absence of sodium, indicating that the net OCTN1-mediated uptake is independent of sodium. This unusual phenomenon has been reported previously for pyrilamine, verapamil, and etoposide in cells transfected with OCTN2 (29), although the mechanism underlying this phenomenon remains unclear.
Fig. 2. In vitro transport of cytarabine by OCTN1.
Characterization of the transport of various AML-directed therapeutics (concentration, 1 μM; 5 min uptake) was performed in HEK293 cells transfected with an empty vector (VC) or OCTN1 (A). A comparison of OCTN1- and ENT1-mediated transport of cytarabine was done in HEK293 cells (B). Time-dependence of cytarabine (Ara-C) transport by OCTN1 at early time points (range, 10–300 s) (C). Sensitivity of OCTN1-mediated cytarabine transport to temperature, sodium, pH, and inhibitors (D). Concentration-dependent transport of cytarabine (1–50 μM; 5 min uptake) by OCTN1 (E), and these data shown as an Eadie-Hofstee transformation (F). Data are shown as mean values (symbols) and SEM (error bars), using 9–60 observations per group. Solid lines represent a fit of the experimental data to a non-linear maximum-effect model or linear regression. Abbreviations: v, transport velocity; [S], substrate concentration; Km, Michaelis-Menten constant; Vmax, maximal velocity.
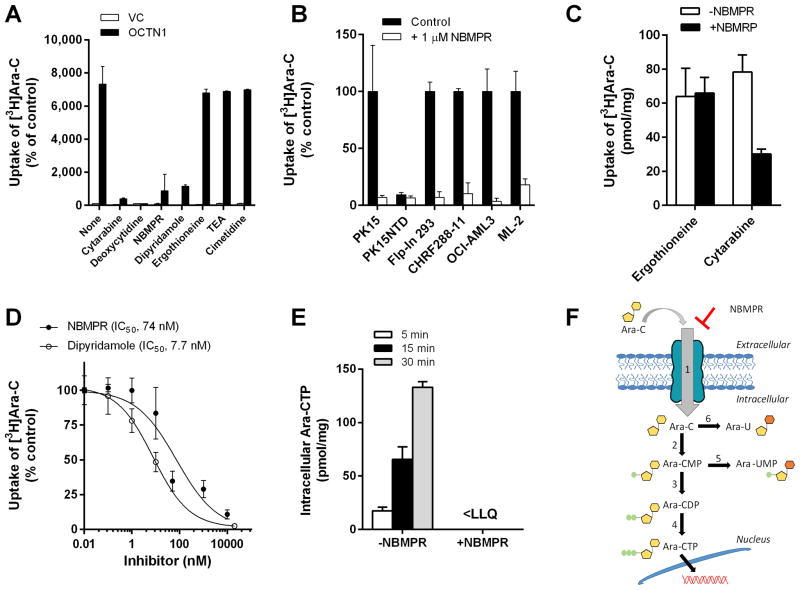
Next, we found that OCTN1-mediated transport of cytarabine was highly sensitive to a 10 to 50 fold molar excess of cytarabine, its structurally-related endogenous nucleoside 2′-deoxycytidine, or the classic nucleoside transporter inhibitors NBMPR and dipyridamole (Fig. 3A). The transport of cytarabine could also be inhibited by NBMPR in the Sus scrofa epithelial kidney cell line PK15 (34), Flp-In 293 cells engineered to overexpress OCTN1, and several AML cell lines (Fig. 3B). However, cytarabine transport was insensitive to the presence of the known OCTN1 substrates ergothioneine and TEA, or to the known OCTN1 inhibitor cimetidine (Fig. 3A) (35). The notion that compounds such as ergothioneine and TEA do not competitively inhibit cytarabine transport suggests the possible existence of a substrate recognition site on OCTN1 for cytarabine that is distinct from that used by ergothioneine and other low-molecular weight cations. This is consistent with the observation that even high micromolar concentrations of NBMPR did not inhibit OCTN1-mediated transport of ergothioneine in HEK293 cells or in human primary epidermal keratinocytes (Fig. 3C), cells that are known to take up nucleosides such as cytarabine and clofarabine (36), and that functionally express OCTN1 (37). Further examination in HEK293 cells revealed that the NBMPR- and dipyridamole-mediated inhibition of cytarabine transport by OCTN1 was associated with low nanomolar IC50 values (Fig. 3D), which are in the same range as levels required to inhibit ENT1 function or cytarabine uptake in human myeloblasts (38).
Fig. 3. Inhibition of OCTN1-mediated cytarabine transport.
Characterization of the transport of cytarabine (Ara-C; concentration, 1 μM; 5-min uptake) in the absence or presence of putative OCTN1 inhibitors (concentration, 10–50 μM) was performed in HEK293 cells transfected with an empty vector (VC) or OCTN1 (A). Influence of NBMPR on the transport of cytarabine in various cell lines (B) and primary human epidermal keratinocytes (C). Concentration-dependence of the inhibitory properties of NBMPR and dipyridamole on OCTN1-mediated transport of cytarabine was assessed over a 0.01–10 μM concentration range (5-min uptake) (D). The time-dependent influence of NBMPR (10 μM) on the formation of the pharmacologically-active metabolite cytarabine triphosphate (Ara-CTP) in cells exposed to cytarabine (10 μM; 5–30 min uptake) was determined by liquid chromatography-tandem mass spectrometry (E). Data are shown as mean values (bars or symbols) and SEM (error bars), using 6–15 observations per group. Solid lines represent a fit of the experimental data to an inverse non-linear maximum-effect model (panel D). Abbreviations: NBMPR, nitrobenzylmercaptopurine ribonucleoside; IC50, concentration required to inhibit OCTN1-mediated cytarabine transport by 50%; <LLQ, lower than the lower limit of quantitation of the analytical assay. A schematic of OCTN1-mediated transport, metabolism, and (in)activation of cytarabine in cells is shown in (F). 1, NBMPR-sensitive transport by OCTN1; 2, deoxycytidine kinase; 3, deoxycytidine monophosphate kinase; 4, nucleoside diphosphate kinase; 5, cytoplasmic 5′-nucleotidase; 6, cytidine deaminase.
The formation of Ara-CTP in HEK293 cells overexpressing OCTN1 increased in a time-dependent manner, and could be completely prevented by NBMPR (Fig. 3E), suggesting that its mechanism of interaction involves a restriction of cytarabine access to the intracellular compartment (Fig. 3F). In contrast, the presence of MK571, an inhibitor of ATP-binding cassette transporters involved in the cellular efflux of cytarabine and its mono-phosphorylated metabolite Ara-CMP (39), did not substantially influence the extent of radioactivity accumulating following a 5-min exposure of OCTN1-expressing HEK293 cells to cytarabine (Supplementary Fig. S2A). The lack of substantial efflux occurring during the experimental conditions applied was confirmed by measuring the time-course of extracellular cytarabine-derived radioactivity after loading of cells with cytarabine (Supplementary Fig. S2B).
OCTN1 expression in myeloid cells affects cytarabine uptake
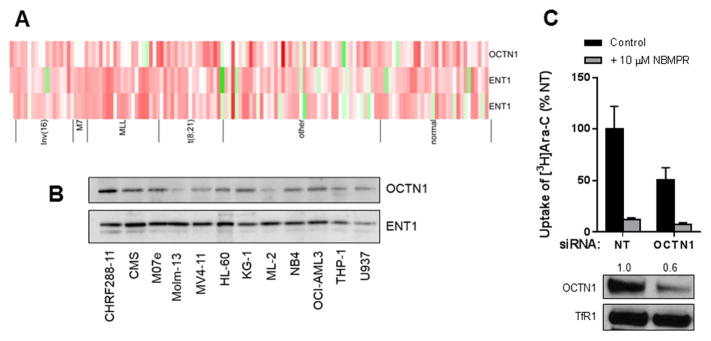
Before evaluating the contribution of OCTN1 to cytarabine transport in AML cells, we confirmed that the OCTN1 gene was present on AML blasts, and found that it was highly variable between patients and among different AML subtypes (Fig. 4A). Moreover, the OCTN1 protein was expressed in a majority of commonly used AML cell lines (Fig. 4B). We next compared the uptake of cytarabine in one of these cells lines with reduced OCTN1 expression after siRNA. Compared to cells transfected with a non-targeting control siRNA, partial silencing of OCTN1 resulted in significantly decreased uptake of cytarabine (Fig. 4C). These results are consistent with the involvement of OCTN1 as an uptake transporter of cytarabine that can confer drug sensitivity in AML cells.
Fig. 4. Influence of OCTN1 on cytarabine uptake in AML cells.
Gene expression of OCTN1 and ENT1 (two probe sets) in primary AML blast samples from pediatric patients used in the survival analysis shown in Fig. 1A–B, as determined by microarray analysis (A). Each column represents an individual primary sample, and columns are categorized by cytogenetic AML subtypes of prognostic relevance. (B) Protein expression of OCTN1 in a panel of 12 AML cell lines. (C) Cellular uptake of cytarabine (Ara-C; 1 μM; 5 min uptake) with or without NBMPR pre-incubation was measured in OCI-AML3 cells 48 hours after transfection with a non-targeting (NT) control siRNA or a siRNA targeting OCTN1. Results are shown as cytarabine uptake as compared to cells transfected with NT siRNA. Data are representative of two independent experiments done in triplicate. OCTN1 protein expression was determined by western blot from membrane extraction of OCI-AML3 cells 48 hours post-transfection, and the transferrin receptor served as loading control. The relative expression difference of OCTN1 after RNAi is indicated by the numbers above the lanes.
Interestingly, we recently reported that the presence of a functional polymorphic germline variant in the OCTN1 gene (rs1050152; c.1507C>T; L503F) (40) is significantly correlated with febrile neutropenia, the major dose-limiting toxicity of cytarabine, in a cohort of 164 pediatric patients with AML receiving cytarabine-based therapy (41). Expression of this mutant OCTN1 in HEK293 cells (Supplementary Fig. S3A–B) revealed that, compared with the reference OCTN1, this variant is associated with increased transport of TEA and increased formation of Ara-CTP after exposure to cytarabine (Supplementary Fig. S3C–D). This finding is consistent with the contention that OCTN1 transports cytarabine into normal bone marrow and blood cells where it is phosphorylated, that these phosphorylated metabolites accumulate in these cells, and produce dose-limiting hematological toxicities.
Role of OCTN1 in cellular sensitivity to nucleoside analogs
Previous investigations demonstrated that certain antiviral nucleosides, including azidothymidine, ganciclovir, and acyclovir, as well as the anticancer nucleosides 2-chloro-2′-deoxyadenosine (cladribine) and cytarabine itself, can be transported to a moderate extent (1.5 to 4-fold vs control cells) by other SLC22A family members that show similarity to OCTN1, such as rat OCT1 (SLC22A1) (42, 43). In heterologous expression models analogous to the one developed for OCTN1, we found, however, that cytarabine was not a transported substrate of the human organic cation transporters OCT1, OCT2 (SLC22A2), and OCT3 (SLC22A3) (Supplementary Fig. S4A), or the carnitine transporter OCTN2 (SLC22A5) (Supplementary Fig. S4B). This suggests that OCTN1 may be unique among human SLC22A family of transporters in its ability to interact with cytarabine.
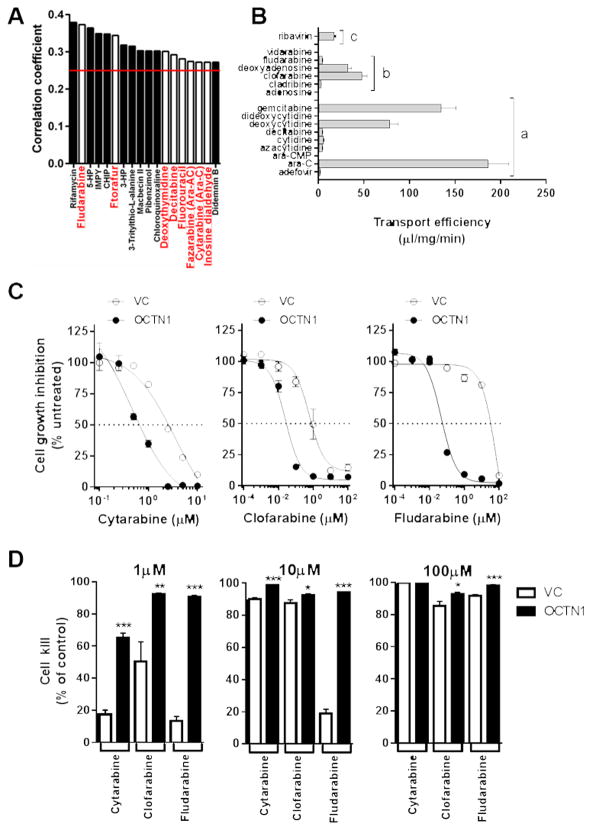
In order to evaluate the selectivity of OCTN1 for the transport of nucleoside analogs, we initially performed pharmacologic profiling of the NCI60 cell panel, which comprises 60 different human tumor cell lines, representing leukemia, melanoma and cancers of the lung, colon, brain, ovary, breast, prostate, and kidney. The 60 cell line dose response produced by a given compound results in a biological response pattern that can be utilized in a recognition algorithm known as COMPARE. Applying this algorithm on 118 well-characterized anticancer drugs, Pearson correlation coefficients (R) were calculated for assessment of OCTN1-drug relationships. Using a cut-off R > 0.25, corresponding to P < 0.03, we found that the cytotoxicity of 18 anticancer drugs of different classes was statistically significantly associated with OCTN1 expression (Fig. 5A). Among these were 8 antimetabolites, including FDA-approved nucleoside analogs (fludarabine, decitabine, and cytarabine), nucleoside dialdehyde derivative (inosine dialdehyde), and nucleobase analogs (ftorafur and fluorouracil). Confirmatory validation studies performed in our transfected HEK293 cells revealed that OCTN1 can transport a remarkably broad range of nucleosides with variable efficiency, primarily depending on the 2′ and 5′ sugar positions, including all tested antimetabolites identified as hits in our COMPARE analysis (Fig. 5B).
Fig. 5. Transport of nucleosides by OCTN1.
(A) Expression of OCTN1 was evaluated in the NCI60 cancer cell line panel, and rank-ordered by observed correlation coefficients (R) between gene expression and cytotoxic potencies of 118 drugs (antimetabolites are in red bold and underlined). Compounds (N = 18) with positive values of R > 0.25 (indicated by red bar) and corresponding to P < 0.03 are shown. The transport of nucleoside analogs (concentration, 1 μM; 5-min uptake; 9–30 observations per group) by OCTN1 was further evaluated in transfected HEK293 cells (B). Bars represent normalized transport divided by substrate concentration. a, cytidine analogs; b, adenosine analogs; c, guanosine analog. The contribution of OCTN1 to nucleoside-induced cytotoxicity was evaluated in HEK293 cells transfected with OCTN1 or empty vector (VC) after continuous drug exposure followed by CellTiter-Glo analysis at 72 hours (12 observations per concentration, per group) with increasing concentrations (C) and at low (1 μM), intermediate (10 μM), and high-doses (100 μM) (D). Data are shown as mean values (bars or symbols) and SEM (error bars). Solid lines represent a fit of the experimental data to an inverse non-linear maximum-effect model. *, p < 0.03; **, p < 0.003; *** p < 0.0001
The newly identified OCTN1 substrates include endogenous nucleosides (eg, 2′-deoxycytidine) and xenobiotic cytidine analogs (eg, gemcitabine), adenosine analogs (eg, clofarabine and fludarabine), and guanosine analogs (eg, ribavirin) (Supplementary Fig. S5). Among the tested non-nucleoside antimetabolites, we found that compared with xenobiotic nucleosides, OCTN1 demonstrated modest transport of fluorouracil (6.5-fold vs control) but this was not observed not for hydroxyurea (1.1×), mercaptopurine (1.3×), methotrexate (1.1×), or a host of anti-cancer drugs from other therapeutic classes (Supplementary Fig. S6). It should be pointed out that a previous investigation that correlated the expression patterns of SLC transporters (including SLC22A4) with growth inhibition data for 1,429 compounds in the NCI60 cell line panel failed to identify cytarabine as a hit (30). The reasons underlying these discrepant findings are not entirely clear, but they may relate to differences in the experimental models and specific conditions applied to the COMPARE analysis.
In addition to increasing accumulation of multiple xenobiotic nucleosides, overexpression of OCTN1 in HEK293 cells was associated with increased sensitivity to cytarabine and other, related nucleoside analogs used in the treatment of AML (Fig. 5C), including clofarabine (44) and fludarabine (45). In particular, we observed that the IC50 values decreased from 2.57 to 0.66 μM (3.9-fold increased sensitivity) for cytarabine, from 0.82 to 0.028 μM (29-fold) for clofarabine, and from 34 to 0.056 μM (608-fold) for fludarabine, respectively. These findings are consistent with the ability of HEK293 cells to activate nucleosides through phosphorylation (Fig. 3E), and confirm that OCTN1 can sensitize cells to xenobiotic nucleoside analogs. Likewise, we observed that deficiency in OCTN1 expression could be overcome at intermediate (10 μM) and high (100 μM) doses of cytarabine and related nucleoside analogs (Fig. 5D). While the mechanism(s) of drug uptake under these conditions require further investigation; these findings are consistent with the observed Km value of 1.95 μM, where at higher concentrations OCTN1 is saturated and not contributing to mechanisms of uptake.
Collectively, our findings indicate that OCTN1, a transporter thought to function primarily as a carrier of ergothioneine, can facilitate the intracellular accumulation of cytarabine, a prerequisite to drug-induced anti-leukemic activity. These results identified OCTN1 as an important, previously unrecognized, contributor to the cellular uptake and efficacy of cytarabine, and this process may be relevant for other related nucleoside analogs used in the treatment of AML, such as clofarabine and fludarabine. Our results also suggest a role of OCTN1 in cellular sensitivity to a range of nucleoside analogs with broad implications to additional diseases.
Supplementary Material
Acknowledgments
Financial Support: The project was supported in part by National Institutes of Health grants F32CA180513, P30CA021765, R01CA138744, R01CA151633, and R25CA023944, the American Lebanese Syrian Associated Charities (ALSAC), the Robert-Bosch foundation, and the Interfaculty Center for Pharmacogenomics and Drug Research (ICEPHA). None of the funding bodies had a role in the preparation of the manuscript.
We thank Lara Sucheston-Campbell (The Ohio State University, Columbus, OH) for critical review of the manuscript, Dario Vignali (St. Jude Children’s Research Hospital, Memphis, TN) for providing the MSCV-IRES-GFP vector, and Chung-Ming Tse (Johns Hopkins University, Baltimore, MD) for providing the PK15 and PK15NTD cell lines. The results shown are in part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/).
Footnotes
Conflicts of Interest The authors have no conflicts of interest
References
- 1.Damaraju VL, Damaraju S, Young JD, Baldwin SA, Mackey J, Sawyer MB, et al. Nucleoside anticancer drugs: the role of nucleoside transporters in resistance to cancer chemotherapy. Oncogene. 2003;22:7524–36. doi: 10.1038/sj.onc.1206952. [DOI] [PubMed] [Google Scholar]
- 2.Ellison RR, Carey RW, Holland JF. Continuous infusions of arabinosyl cytosine in patients with neoplastic disease. Clin Pharmacol Ther. 1967;8:800–9. doi: 10.1002/cpt196786800. [DOI] [PubMed] [Google Scholar]
- 3.Wiley JS, Jones SP, Sawyer WH. Cytosine arabinoside transport by human leukaemic cells. Eur J Cancer Clin Oncol. 1983;19:1067–74. doi: 10.1016/0277-5379(83)90029-9. [DOI] [PubMed] [Google Scholar]
- 4.Owens JK, Shewach DS, Ullman B, Mitchell BS. Resistance to 1-beta-D-arabinofuranosylcytosine in human T-lymphoblasts mediated by mutations within the deoxycytidine kinase gene. Cancer Res. 1992;52:2389–93. [PubMed] [Google Scholar]
- 5.Estey EH. Acute myeloid leukemia: 2013 update on risk-stratification and management. Am J Hematol. 2013;88:318–27. doi: 10.1002/ajh.23404. [DOI] [PubMed] [Google Scholar]
- 6.Obata T, Endo Y, Murata D, Sakamoto K, Sasaki T. The molecular targets of antitumor 2′-deoxycytidine analogues. Curr Drug Targets. 2003;4:305–13. doi: 10.2174/1389450033491037. [DOI] [PubMed] [Google Scholar]
- 7.Wiley JS, Jones SP, Sawyer WH, Paterson AR. Cytosine arabinoside influx and nucleoside transport sites in acute leukemia. J Clin Invest. 1982;69:479–89. doi: 10.1172/JCI110472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiley JS, Taupin J, Jamieson GP, Snook M, Sawyer WH, Finch LR. Cytosine arabinoside transport and metabolism in acute leukemias and T cell lymphoblastic lymphoma. J Clin Invest. 1985;75:632–42. doi: 10.1172/JCI111741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White JC, Rathmell JP, Capizzi RL. Membrane transport influences the rate of accumulation of cytosine arabinoside in human leukemia cells. J Clin Invest. 1987;79:380–7. doi: 10.1172/JCI112823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galmarini CM, Thomas X, Calvo F, Rousselot P, Rabilloud M, El Jaffari A, et al. In vivo mechanisms of resistance to cytarabine in acute myeloid leukaemia. Br J Haematol. 2002;117:860–8. doi: 10.1046/j.1365-2141.2002.03538.x. [DOI] [PubMed] [Google Scholar]
- 11.Stam RW, den Boer ML, Meijerink JP, Ebus ME, Peters GJ, Noordhuis P, et al. Differential mRNA expression of Ara-C-metabolizing enzymes explains Ara-C sensitivity in MLL gene-rearranged infant acute lymphoblastic leukemia. Blood. 2003;101:1270–6. doi: 10.1182/blood-2002-05-1600. [DOI] [PubMed] [Google Scholar]
- 12.Abraham A, Varatharajan S, Karathedath S, Philip C, Lakshmi KM, Jayavelu AK, et al. RNA expression of genes involved in cytarabine metabolism and transport predicts cytarabine response in acute myeloid leukemia. Pharmacogenomics. 2015;16:877–90. doi: 10.2217/pgs.15.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu X, Gong S, Monks A, Zaharevitz D, Moscow JA. Correlation of nucleoside and nucleobase transporter gene expression with antimetabolite drug cytotoxicity. J Exp Ther Oncol. 2002;2:200–12. doi: 10.1046/j.1359-4117.2002.01035.x. [DOI] [PubMed] [Google Scholar]
- 14.Yagi T, Morimoto A, Eguchi M, Hibi S, Sako M, Ishii E, et al. Identification of a gene expression signature associated with pediatric AML prognosis. Blood. 2003;102:1849–56. doi: 10.1182/blood-2003-02-0578. [DOI] [PubMed] [Google Scholar]
- 15.Clarke ML, Damaraju VL, Zhang J, Mowles D, Tackaberry T, Lang T, et al. The role of human nucleoside transporters in cellular uptake of 4′-thio-beta-D-arabinofuranosylcytosine and beta-D-arabinosylcytosine. Mol Pharmacol. 2006;70:303–10. doi: 10.1124/mol.105.021543. [DOI] [PubMed] [Google Scholar]
- 16.Zimmerman EI, Huang M, Leisewitz AV, Wang Y, Yang J, Graves LM. Identification of a novel point mutation in ENT1 that confers resistance to Ara-C in human T cell leukemia CCRF-CEM cells. FEBS Lett. 2009;583:425–9. doi: 10.1016/j.febslet.2008.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Endo Y, Obata T, Murata D, Ito M, Sakamoto K, Fukushima M, et al. Cellular localization and functional characterization of the equilibrative nucleoside transporters of antitumor nucleosides. Cancer Sci. 2007;98:1633–7. doi: 10.1111/j.1349-7006.2007.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ross ME, Mahfouz R, Onciu M, Liu HC, Zhou X, Song G, et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104:3679–87. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- 19.Rubnitz JE, Inaba H, Dahl G, Ribeiro RC, Bowman WP, Taub J, et al. Minimal residual disease-directed therapy for childhood acute myeloid leukaemia: results of the AML02 multicentre trial. Lancet Oncol. 2010;11:543–52. doi: 10.1016/S1470-2045(10)70090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research, N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Sign. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pochini L, Scalise M, Indiveri C. Immuno-detection of OCTN1 (SLC22A4) in HeLa cells and characterization of transport function. Int Immunopharmacol. 2015;29:21–6. doi: 10.1016/j.intimp.2015.04.040. [DOI] [PubMed] [Google Scholar]
- 24.Griffith DA, Jarvis SM. Nucleoside and nucleobase transport systems of mammalian cells. Biochim Biophys Acta. 1996;1286:153–81. doi: 10.1016/s0304-4157(96)00008-1. [DOI] [PubMed] [Google Scholar]
- 25.Grundemann D, Harlfinger S, Golz S, Geerts A, Lazar A, Berkels R, et al. Discovery of the ergothioneine transporter. Proc Natl Acad Sci USA. 2005;102:5256–61. doi: 10.1073/pnas.0408624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X, George RL, Huang W, Wang H, Conway SJ, Leibach FH, et al. Structural and functional characteristics and tissue distribution pattern of rat OCTN1, an organic cation transporter, cloned from placenta. Biochim Biophys Acta. 2000;1466:315–27. doi: 10.1016/s0005-2736(00)00189-9. [DOI] [PubMed] [Google Scholar]
- 27.Hu S, Niu H, Inaba H, Orwick S, Rose C, Panetta JC, et al. Activity of the multikinase inhibitor sorafenib in combination with cytarabine in acute myeloid leukemia. J Natl Cancer Inst. 2011;103:893–905. doi: 10.1093/jnci/djr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grimwade D, Ivey A, Huntly BJ. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood. 2016;127:29–41. doi: 10.1182/blood-2015-07-604496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu C, Lancaster CS, Zuo Z, Hu S, Chen Z, Rubnitz JE, et al. Inhibition of OCTN2-mediated transport of carnitine by etoposide. Mol Cancer Ther. 2012;11:921–9. doi: 10.1158/1535-7163.MCT-11-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okabe M, Szakacs G, Reimers MA, Suzuki T, Hall MD, Abe T, et al. Profiling SLCO and SLC22 genes in the NCI-60 cancer cell lines to identify drug uptake transporters. Mol Cancer Ther. 2008;7:3081–91. doi: 10.1158/1535-7163.MCT-08-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White JC, Hines LH, Rathmell JP. Inhibition of 1-beta-D-arabinofuranosylcytosine transport and net accumulation by teniposide and etoposide in Ehrlich ascites cells and human leukemic blasts. Cancer Res. 1985;45:3070–5. [PubMed] [Google Scholar]
- 32.Tamai I, Nakanishi T, Kobayashi D, China K, Kosugi Y, Nezu J, et al. Involvement of OCTN1 (SLC22A4) in pH-dependent transport of organic cations. Mol Pharm. 2004;1:57–66. doi: 10.1021/mp0340082. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura T, Yoshida K, Yabuuchi H, Maeda T, Tamai I. Functional characterization of ergothioneine transport by rat organic cation/carnitine transporter Octn1 (slc22a4) Biol Pharm Bull. 2008;31:1580–4. doi: 10.1248/bpb.31.1580. [DOI] [PubMed] [Google Scholar]
- 34.Ward JL, Sherali A, Mo ZP, Tse CM. Kinetic and pharmacological properties of cloned human equilibrative nucleoside transporters, ENT1 and ENT2, stably expressed in nucleoside transporter-deficient PK15 cells. Ent2 exhibits a low affinity for guanosine and cytidine but a high affinity for inosine. J Biol Chem. 2000;275:8375–81. doi: 10.1074/jbc.275.12.8375. [DOI] [PubMed] [Google Scholar]
- 35.Yabuuchi H, Tamai I, Nezu J, Sakamoto K, Oku A, Shimane M, et al. Novel membrane transporter OCTN1 mediates multispecific, bidirectional, and pH-dependent transport of organic cations. J Pharmacol Exp Ther. 1999;289:768–73. [PubMed] [Google Scholar]
- 36.Lindemalm S, Liliemark J, Larsson BS, Albertioni F. Distribution of 2-chloro-2′-deoxyadenosine, 2-chloro-2′-arabino-fluoro-2′-deoxyadenosine, fludarabine and cytarabine in mice: a whole-body autoradiography study. Med Oncol. 1999;16:239–44. doi: 10.1007/BF02785869. [DOI] [PubMed] [Google Scholar]
- 37.Markova NG, Karaman-Jurukovska N, Dong KK, Damaghi N, Smiles KA, Yarosh DB. Skin cells and tissue are capable of using L-ergothioneine as an integral component of their antioxidant defense system. Free Radic Biol Med. 2009;46:1168–76. doi: 10.1016/j.freeradbiomed.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 38.Wiley JS. Seeking the nucleoside transporter. Nat Med. 1997;3:25–6. doi: 10.1038/nm0197-25. [DOI] [PubMed] [Google Scholar]
- 39.Drenberg CD, Hu S, Li L, Buelow DR, Orwick SJ, Gibson AA, et al. ABCC4 is a determinant of cytarabine-induced cytotoxicity and myelosuppression. Clin Transl Sci. 2016;9:51–9. doi: 10.1111/cts.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urban TJ, Yang C, Lagpacan LL, Brown C, Castro RA, Taylor TR, et al. Functional effects of protein sequence polymorphisms in the organic cation/ergothioneine transporter OCTN1 (SLC22A4) Pharmacogenet Genomics. 2007;17:773–82. doi: 10.1097/FPC.0b013e3281c6d08e.. [DOI] [PubMed] [Google Scholar]
- 41.Drenberg CD, Paugh SW, Pounds SB, Shi L, Orwick SJ, Li L, et al. Inherited variation in OATP1B1 is associated with treatment outcome in acute myeloid leukemia. Clin Pharmacol Ther. 2015;99:651–60. doi: 10.1002/cpt.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen R, Nelson JA. Role of organic cation transporters in the renal secretion of nucleosides. Biochem Pharmacol. 2000;60:215–9. doi: 10.1016/s0006-2952(00)00334-8. [DOI] [PubMed] [Google Scholar]
- 43.Pastor-Anglada M, Molina-Arcas M, Casado FJ, Bellosillo B, Colomer D, Gil J. Nucleoside transporters in chronic lymphocytic leukaemia. Leukemia. 2004;18:385–93. doi: 10.1038/sj.leu.2403271. [DOI] [PubMed] [Google Scholar]
- 44.Ghanem H, Kantarjian H, Ohanian M, Jabbour E. The role of clofarabine in acute myeloid leukemia. Leuk Lymphoma. 2013;54:688–98. doi: 10.3109/10428194.2012.726722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lukenbill J, Kalaycio M. Fludarabine: a review of the clear benefits and potential harms. Leuk Res. 2013;37:986–94. doi: 10.1016/j.leukres.2013.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.