SUMMARY
Mammals have extremely limited regenerative capabilities; however, axolotls are profoundly regenerative and can replace entire limbs. The mechanisms underlying limb regeneration remain poorly understood, partly because the enormous and incompletely sequenced genomes of axolotls have hindered the study of genes facilitating regeneration. We assembled and annotated a de novo transcriptome using RNA-sequencing profiles for a broad spectrum of tissues that is estimated to have near-complete sequence information for 88% of axolotl genes. We devised expression analyses that identified the axolotl orthologs of cirbp and kazald1 as highly expressed and enriched in blastemas. Using morpholino anti-sense oligonucleotides, we find evidence that cirbp plays a cytoprotective role during limb regeneration while manipulation of kazald1 expression disrupts regeneration. Our transcriptome and annotation resources greatly complement previous transcriptomic studies and will be a valuable resource for future research in regenerative biology.
INTRODUCTION
The limited capacity of humans to regenerate many tissues, organs, and appendages is a formidable clinical hurdle (Ziegler-Graham et al., 2008). Conversely some animals, including invertebrates such as planaria, and vertebrates such as amphibians, have remarkable regenerative capacity. Among those, many salamanders, including axolotls, can regenerate entire limbs throughout life (reviewed in (Whited and Tabin, 2009). Elucidating the molecular mechanisms that enable such profound regenerative capacity may provide key insights relevant to human regenerative medicine.
The axolotl community has made significant strides in advancing our understanding of limb regeneration, but our knowledge of the molecular mechanisms that underlie axolotl regeneration is still very limited. Unbiased genomics and transcriptomics can often unlock the molecular components of systems that have not been genetically tractable. Unfortunately, the axolotl genome remains mostly unsequenced and poses major challenges at ~32 Gigabases in size (Keinath et al., 2015; Smith et al., 2009; Straus, 1971). RNA-sequencing followed by de-novo transcriptome assembly (Haas et al., 2013; Robertson et al., 2010; Schulz et al., 2012) has offered investigators an alternative for identifying near-full-length transcripts and performing differential gene expression analyses without genome mapping. Recent axolotl transcriptome studies (Knapp et al., 2013; Li et al., 2014; McCusker et al., 2015; Monaghan et al., 2009; Stewart et al., 2013; Voss et al., 2015; Wu et al., 2013) have focused on and significantly advanced our understanding of the changes in transcription over time in the regenerating portion of the limb. However, an important missing component of the existing data sets is deep transcriptional information about each of the presumed parent tissue types within the limb, which contribute progenitors and serve as the template for the future regenerate limb. Thus, examining the transcripts that define them in the differentiated, homeostatic state will be critical for future comparisons with progenitor cells along the temporal path of regeneration.
Here, we combined RNA-Seq of diverse tissues with de novo transcriptome assembly, computational analysis, and experimental validation to develop a systematic map of the axolotl transcriptome. This assembly facilitated identification of specific transcripts and classes of genes whose expression is associated with successful limb regeneration. We experimentally validated our transcriptome’s accuracy by analyzing mRNA expression of identified transcripts using in situ hybridization. Furthermore, we experimentally modified the expression of cirbp and kazald1 (two blastema-enriched transcripts) and uncovered functional roles for these genes in axolotl limb regeneration. The transcriptome and analyses that we provide significantly complement prior research and will be an important resource for future studies of limb regeneration, as well as for inquiries using the axolotl that extend beyond those of limb regeneration.
RESULTS
An RNA-Seq catalog for limb tissues and regeneration
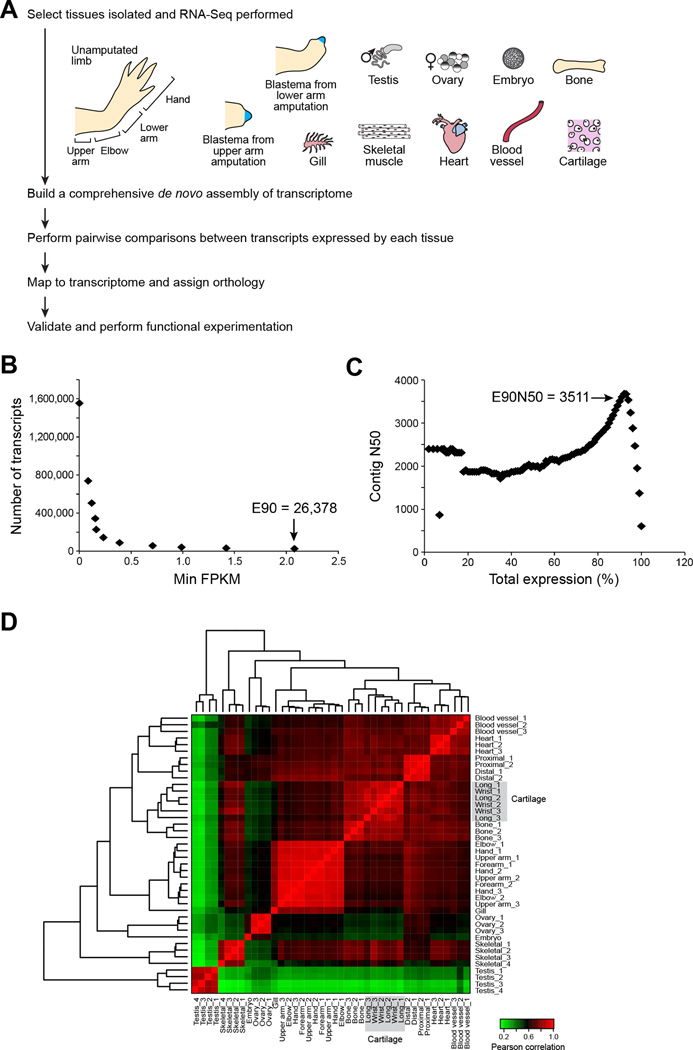
To build a reference map of axolotl limb regeneration, we profiled 42 samples across 16 different tissues (Figure 1A). First, we profiled intact, unamputated limbs to reflect the starting—and end— point for limb regeneration. We sampled four positions along the proximal (shoulder) to distal (fingertip) axis to identify any location-specific transcriptional differences. Second, we sequenced the blastema: a bud-like outgrowth at the tip of the regenerating limb that contains activated progenitor cells that regenerate the internal structures of the limb. To identify transcripts whose regulation distinguishes blastema cells, we removed the regenerate epithelium of blastemas at the medium bud stage before it began to differentiate into the various tissues of the regenerating limb. Third, it is thought that axolotls are able to redeploy transcriptional programs that guide embryogenesis during limb regeneration, and a recent study demonstrated that genes with roles in germline cell renewal are required for axolotl limb regeneration (Zhu et al., 2012). To facilitate the systematic identification of germline and embryonic transcriptional programs that are reactivated during regeneration, we generated transcriptional profiles for testes, ovaries, and embryos (1-cell to pre-hatch stage). Fourth, following amputation, individual tissues within the limb, including skeletal muscle, cartilage, bone, and blood vessels, may contribute activated progenitor cells to the regenerating limb ((Kragl et al., 2009; Muneoka et al., 1986; Sandoval-Guzman et al., 2014); reviewed in (Knapp and Tanaka, 2012)). We therefore sampled and analyzed each of these tissues to define tissue-specific expression and marker transcripts for the differentiated tissue types in limbs. We also included transcripts from heart and gill filaments in our assembly to increase the comprehensiveness of the transcriptome and to provide resources for researchers interested in pursuing questions outside of limb regeneration. Fifth, the blastema is an autonomous unit, programmed from the onset with spatial coordinates that instruct the regrowth of precisely the portion of the limb that has been lost (Crawford and Stocum, 1988a, b; Echeverri and Tanaka, 2005; McCusker and Gardiner, 2013; Mercader et al., 2005; Stocum and Melton, 1977), such that amputation of the hand (a “distal” amputation) results in regeneration of a hand, but amputation of an entire arm (a “proximal” amputation) results in regeneration of an arm. To assess how information is differentially encoded in proximal versus distal blastemas, we sequenced medium bud stage blastemas derived from both proximal and distal amputations.
Figure 1. Pipeline and assessment of read representation for de novo axolotl transcriptome.
(A) Strategy for deriving a tissue-coded de novo transcriptome for axolotl. (B) The count of most highly expressed transcripts is plotted as a function of minimum expression value. 90% of the total expression (E90) is accounted for by the 26,378 most highly expressed transcripts. (C) The contig N50 value is computed for cumulative sets of most highly expressed transcripts. (D) Expression values for all tissue types were compared and Pearson correlation values were computed. Samples were clustered according to Pearson correlation values, indicating high similarity among sample replicates and between similar tissue types. See also Figure S1, Table S1, File S1 and File S2.
A de novo assembled axolotl transcriptome
We assembled 42 RNA-Seq samples totaling ~1.3 billion 100 base paired-end reads, (Table S1) using Trinity (Grabherr et al., 2011; Haas et al., 2013). We first combined reads from all samples and performed in silico normalization, retaining 6.6% of reads for assembly (86 million paired-end reads). We then used Trinity (Table S1) to generate an assembly of 1,554,055 transcript contigs clustered into 1,388,798 “gene” groupings (File S1A, median transcript length: 288 bases, N50 of 606 bases). Although our in silico normalization filtered out the vast majority of the reads prior to assembly, 80% of the original reads mapped back to the assembly, with most mapping as properly paired reads (Figure S1A). The majority of the transcriptome corresponds to lowly expressed transcript contigs (Figure 1B), as 90% of the total transcription is represented by an “E90 transcript set” of 26,378 transcripts (20,506 genes, E90-N50 of 3,511) (Table S1, Figure 1C).
Functional annotation with Trinotate and assessment of transcriptome completeness
Annotating de novo assembled transcripts in the absence of an assembled genome, as in the case of the axolotl, remains a bioinformatics challenge with few available tools. We therefore developed Trinotate (http://trinotate.github.io), an annotation protocol and toolkit for de novo assembled transcriptomes. Trinotate extracts predicted coding regions using TransDecoder (http://transdecoder.github.io), searches both the entire transcripts and the predicted coding regions separately for protein homology by BLAST search against SwissProt, and annotates the coding regions for domain content, signal peptides, and transmembrane domains. Trinotate reports its annotations in a convenient tab-delimited file (File S1B).
We applied Trinotate to the 1.6 million transcripts in our assembly, finding a large number of likely full-length transcripts enriched in the E90 set. Trinotate reported 109,180 transcripts matching 29,529 unique SwissProt proteins, with 27,056 Trinity transcripts matching 13,501 proteins across at least 80% of the matching protein’s length. These transcripts likely represent fully or near fully reconstructed transcripts with detectable homologs in other species, of which over a third (9,950) are significantly enriched (22-fold, P-value<2.2×10−16, Fisher’s exact test) among the E90 transcript set.
By subjecting our transcriptome to BUSCO analysis (Simao et al., 2015), we identified by this measure that our transcriptome contains near-complete gene sequence information for 88% of the genes in the axolotl genome (Table 1). We finally applied the Core Eukaryotic Gene Mapping Approach (CEGMA) (Parra et al., 2007), which further indicated that our transcriptome is more than 98% complete (Table S1).
Table 1.
BUSCO analysis of transcriptome completeness.
| Reference | Species | Summary in BUSCO annotation |
|---|---|---|
| Described in this manuscript | A. mexicanum | C:88%[D:53%],F:4.5%,M:7.3%,n:3023 |
| (Jiang et al., 2016) | A. mexicanum | C:89%[D:42%],F:2.7%,M:7.8%,n:3023 |
| www.ambystoma.org/genome-resources/5-gene-and-est-database | A. mexicanum | C:42%[D:0.9%],F:17%,M:40%,n:3023 |
| (Li et al., 2014) | A. mexicanum | C:48%[D:21%],F:10%,M:41%,n:3023 |
| (Stewart et al., 2013) | A. mexicanum | C:17%[D:1.6%],F:15%,M:67%,n:3023 |
| (Knapp et al., 2013) | A. mexicanum | C:9.7%[D:0.7%],F:8.5%,M:81%,n:3023 |
| (Abdullayev et al., 2013) | N. viridescens | C:84%[D:41%],F:4.6%,M:11%,n:3023 |
| http://sandberg.cmb.ki.se/redspottednewt/ | N. viridescens | C:87%[D:56%],F:4.1%,M:8.3%,n:3023 |
| (Looso et al., 2013) | N. viridescens | C:30%[D:7.0%],F:10%,M:58%,n:3023 |
| (Nakamura et al., 2014) | C. pyrrhogaster | C:82%[D:34%],F:4.6%,M:13%,n:3023 |
Differential expression analysis identified tissue-specific gene expression and splicing
We estimated the expression profiles for all transcripts across all samples, and found excellent correlation between biological replicates (average Pearson correlation between replicates: 0.90; Figure 1D). As expected, samples derived from similar tissues (e.g., sections of the arm segments; bone and cartilage; proximal and distal blastemas) had more highly correlated expression than more distinct tissues such as the testes, which exhibited the most divergent transcriptional profile.
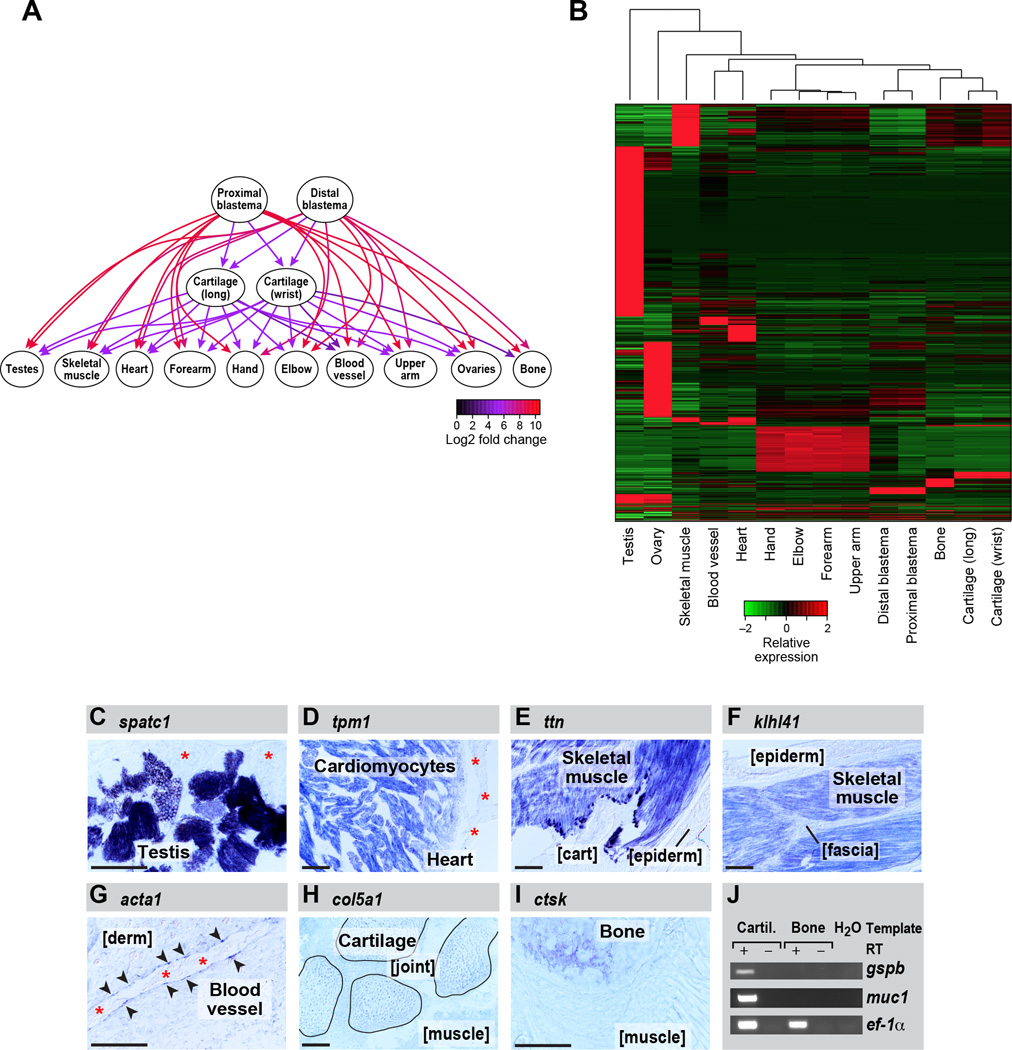
We next identified differentially expressed transcripts, conservatively focusing on transcripts identified as differentially expressed by multiple methods (Figure S1B) and restricted to samples with biological replicates (this excludes gill filament and embryos which were used only for assembly). We identified 60,355 transcripts corresponding to 41,697 genes as at least 2-fold significantly differentially expressed (FDR <= 0.05) between at least two tissue type comparisons (File S2). Finally, we used a graph-based analysis to identify genes that are differentially expressed across each set of tissues (Figure 2A). In this approach, a graph is constructed for each differentially expressed transcript such that each tissue type is represented by a node, and an edge is drawn from a significantly up-regulated tissue node to the down-regulated tissue node. The graph is then partitioned to identify the maximal set of upregulated tissues for that transcript. Visualization of the graphical output for a given transcript intuitively presents the tissue(s) with the highest expression level for the transcript of interest (Figure 2A).
Figure 2. Differential gene expression analysis across each set of tissues identifies transcripts most enriched in specific tissue types.
(A) Graph illustrating the methodology for the identification of genes that are tissue-enriched in the context of all tissue pairwise comparisons using kazal-type serine peptidase inhibitor domain 1 (kazald1) as an example. Directed edges are drawn from upregulated to downregulated tissues, and fold changes in expression are indicated by the edge colors. (B) Heatmap showing all transcripts that are enriched in specific tissue types. (C–J) RNA in situ hybridization performed on tissue sections. (C) speriolin (speri) is enriched in the germ cells in testis but not detectable in adjacent support cells (asterisks). (D) tropomyosin 1 (tpm1) is enriched in cardiomyocytes within the heart and is not detectably expressed by other heart cell types such as epicardium (asterisks). (E) titin is enriched in limb skeletal muscle but is not detectable in adjacent cartilage (cart) and epidermis (epiderm). (F) kelch repeat and BTB domain-containing protein 10 (kbtba) is highly enriched in skeletal muscle and not detectable in adjacent tissues such as epidermis (epiderm) and fascia. (G) actin, alpha1, skeletal muscle (acts) mRNA is enriched in the very thin layer of vascular endothelial cells lining the blood vessels (arrowheads) and absent from adjacent dermis (derm). Asterisks mark red blood cell clumps in the vessel lumen. (H) collagen type V alpha 1 (co5a1) expression is highly enriched in cartilage; shown are four carpals (outlined) within the wrist. Expression in joint (between carpals) and in adjacent muscle is diminished. (I) A bone-enriched marker, cathepsin k (catk), is highly expressed in ossified portions of the humerus, and low in adjacent muscle. (J) platelet binding protein GspB (gspb) and mucin 1 (muc1) are detected in cartilage but not bone by RT-PCR. eukaryotic translation elongation factor 1 alpha 1 (ef-1a) serves as the loading control. Scale bars (bottom left of each panel) are 100µm. See also Figure S7, Table S2, Table S3, File S1 and File S2.
Of the differentially expressed transcripts, 14,594 transcripts (11,523 genes) were most tissue-enriched given all pairwise tissue comparisons (Figure 2B, Table S2). Testes and ovaries had the most tissue-enriched transcripts (7,795 and 2,247 respectively, together 69%), followed by skeletal muscle tissue (961 transcripts), and the combination of all arm-segmented tissues (elbow, forearm, upperarm, and hand, 892) compared to each of the remaining tissues, including those tissues that comprise the arm (e.g., bone). The tissue-specific transcripts were enriched for physiologically relevant functional categories based on enrichment analysis of Gene Ontology (GO) terms assigned from Swissprot annotations of homologous proteins using Trinotate ((e.g. spermatogenesis was enriched in testes specific genes (FDR<5.5e−44), muscle contraction for skeletal muscle (FDR<5.1e−20), etc., Table S2)).
We experimentally validated by in situ hybridization and RT-PCR some of the tissue-enriched expression predictions to demonstrate our ability to correctly predict diverse tissue-specific gene expression patterns (Figure 2C–J). For example, as predicted by our analysis, speriolin (spermatogenesis and centriole associated 1, spatc1) is enriched in germ cells within the testes (Figure 2C). This is consistent with limited studies on Speriolin that demonstrate this protein associates with centrosomes in spermatocytes and exhibits testis-specific expression in mouse (Goto and Eddy, 2004; Goto et al., 2010).
Finally, we identified differentially expressed transcripts that are also associated with differential isoform usage across tissues. Of the 60,355 differentially expressed transcripts (41,697 genes), there are 29,750 transcripts (11,092 genes) with putative alternatively spliced isoforms. For 7,300 genes, the alternative isoforms have different patterns of tissue enrichment, suggestive of alternate functional roles (Table S3). Most of these genes (4,097 of 7,300) have at least one isoform significantly enriched in either testes or ovaries, with far fewer genes with evidence for tissue-specific isoforms in other tissues (e.g., 96 genes involving alternate isoform enrichment between heart and skeletal muscle). Notably, only 346 genes of the 11,523 genes with strong tissue specific expression (Figure 2B) show evidence for tissue specific isoform usage, the vast majority (315 genes) in either testes or ovaries. This data suggests axolotls may be less reliant upon alternative splicing in directing tissue-specific gene expression than has been documented in mammals (for example, (Wang et al., 2008)).
A transcriptional program for blastemas implicates the RNA life cycle in limb regeneration
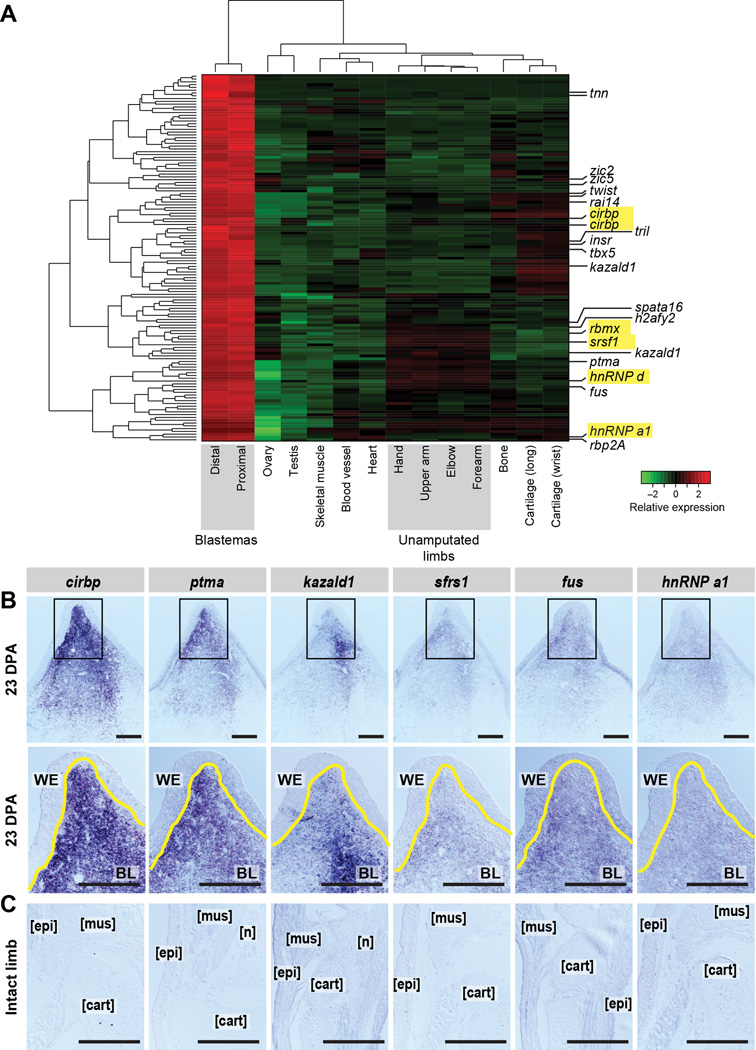
We next analyzed the 159 transcripts (151 genes) differentially induced in all blastema samples (proximal and distal) as compared to all other sampled tissues (Figure 3A, Table S4). Notably, many of the blastema-specific genes (64/151) do not have a similar sequence identified in another organism in the Uniref90 database (E<=10−3, BLASTX). Nonetheless, several blastema-specific genes have known blastema relevance (e.g. twist family bhlh transcription factor 1 (twist1) (Satoh et al., 2008)) or are predicted to encode proteins in pathways implicated in salamander limb regeneration (retinol binding protein 2a (rbp2a) (Maden, 1983), matrix metallopeptidase 11 (mmp11) (Yang et al., 1999)). In particular, we identified the T-box transcription factor tbx5, previously reported as enriched in forelimb blastemas (Khan et al., 2002), discriminating its tissue-specific expression among 13 total tbx genes detected in this axolotl transcriptome. Furthermore, when analyzing forelimb blastemas, our differential expression analysis did not erroneously uncover tbx4, which has been documented to only be upregulated in hindlimb blastemas (Khan et al., 2002).
Figure 3. Identification and validation of blastema-enriched transcripts.
(A) We identified 159 transcripts (151 genes) enriched in blastema (the combination of proximal and distal blastema tissue) as compared to all other tissues. Those predicted to encode proteins with RNA-binding/regulation properties are highlighted yellow. (B–C) In situ hybridization for six highly blastema-enriched transcripts at 23 DPA (top two panels) and on intact limbs (lower panels). Second line of panels in (B) are higher magnifications of the boxed areas in above images. Yellow line marks WE/BL boundary. WE=wound epidermis, BL=blastema, mus= muscle, epi= epidermis, cart=cartilage, n= nerve. Scale bar is 500µm. See also Figure S3, Table S4, File S1 and File S2.
Several blastema-specific genes are predicted to encode RNA-binding proteins, highlighting a potential role for the regulation of the RNA life cycle in regeneration (Figure 3A, highlighted). Among the blastema-induced transcripts are three predicted heterogenous nuclear ribonucleoproteins (hnrnp) transcripts, an RNA-binding motif (rbm3) ortholog, a serine/arginine-rich RNA splicing factor (sfrs1) ortholog, and the axolotl ortholog of cold-inducible RNA-binding protein (cirbp). Interestingly, a newt ortholog of cirbp was recently identified in a subtractive hybridization screen as upregulated in limb blastemas compared to non-amputated limbs (Jiang D, 2014), raising the possibility that cirbp and its homologs may be important regulators in regeneration across species. We also validated several of the blastema-enriched genes by in situ hybridization (Figure 3B), and confirmed they are not appreciably expressed in unamputated limbs (Figure 3C).
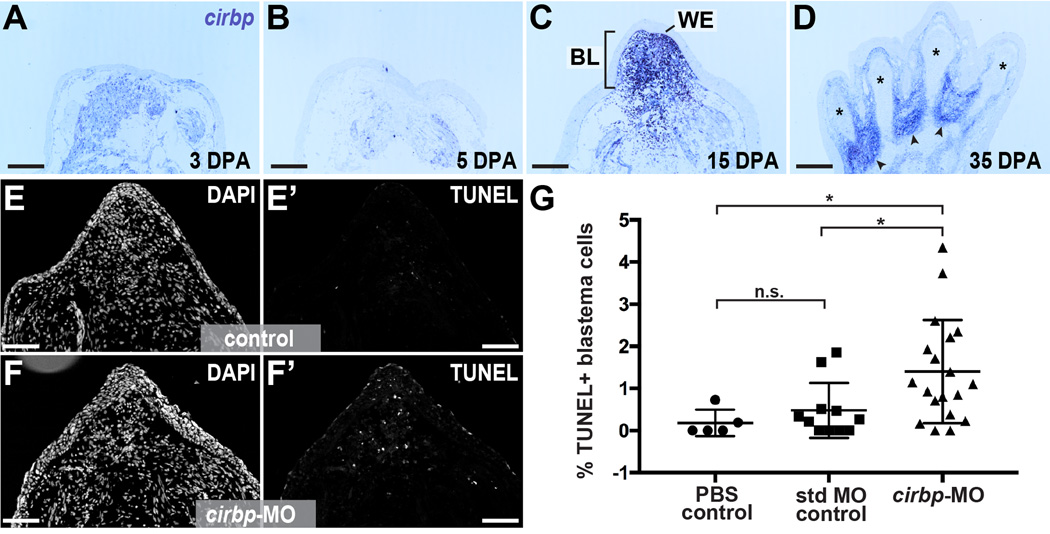
We further explored the expression of cirbp during limb regeneration and found that cirbp expression was apparent as early as 3 days post-amputation (Figure 4A), persisted as the blastema consolidates, and grows through palette formation (Figure 4B–D). Past studies have demonstrated that CIRBP proteins regulate cell fate by inhibiting pro-apoptotic pathways during newt spermatogenesis (Eto et al., 2009) and in the mammalian nervous system (Zhang et al., 2015). To further explore an anti-apoptotic role for axolotl cirbp in limb regeneration, we delivered a translation-blocking morpholino to early blastemas (Figure S2), and assayed cell death using TUNEL staining 4 days later. Administration of cirbp-targeting morpholino caused a significant increase in the percentage of TUNEL-positive blastema cells relative to PBS and standard morpholino controls (cirbp-targeting morpholino: N=19 limbs; standard control morpholino: N=11 limbs; PBS control: N=5 limbs) (Figure 4E–G). These results suggest a conserved role for Cirbp-related proteins in regulating cell death across vertebrates and highlight a cytoprotective role for RNA-binding proteins within blastema cells.
Figure 4. Axolotl Cold-Inducible RNA-binding Protein is a cytoprotective factor for blastema cells.
(A–D) In situ hybridization for cirbp over course of regeneration. * denotes newly-differentiated cartilage in digits, arrowhead denotes interdigital regions. WE=wound epidermis, BL=blastema, mus= muscle, epi= epidermis, cart=cartilage, n= nerve. Scale bar is 500µm. (E-F’) Tissue sections from regenerating limbs treated with standard MO control (E-E’) and cirbp-targeting morpholino (F-F’) stained for nuclei (DAPI, E and F) and TUNEL (E’ and F’). Scale bar is 500µm. (G) Quantification of percentage of TUNEL (+) blastema cell nuclei in control and cirbp-MO-treated limbs. * indicates p < 0.05, n.s. indicates not significant. See also Figure S2 and File S1.
We also examined the 60 transcripts specifically downregulated in blastemas (Figure S3). These included transcripts that encode proteins that likely regulate fatty acid metabolism (prkab2, cyp2a13, and faah), glucose metabolism (pgm1, gp1), cell cycle (ccng1), and mitochondrial respiration (cox7a2 and cox7a2l). The expression of most of these blastema-repressed transcripts is higher in skeletal muscle than in bone and cartilage (e.g. casq1), suggesting that blastema cells may be more actively repressing a muscle program than a skeletal program.
Functional studies indicate a critical role for kazald1 in limb regeneration
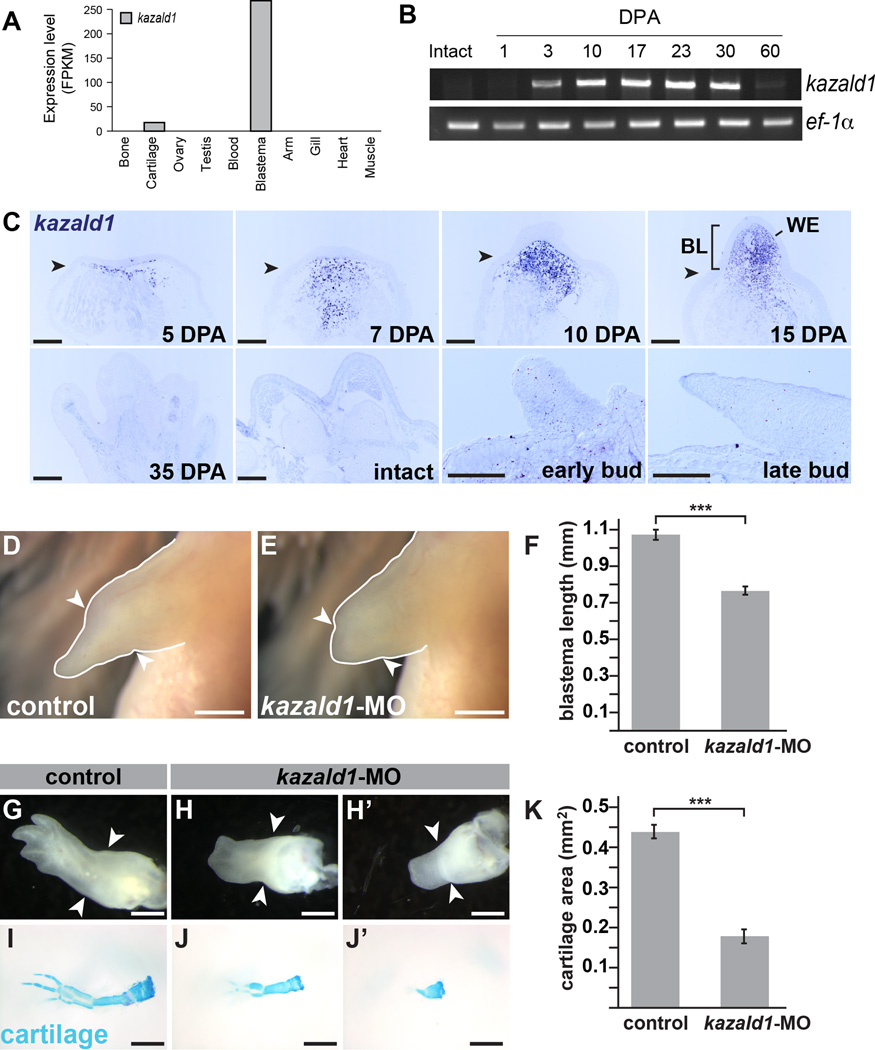
The most blastema-enriched transcript identified by our analyses was kazald1, whose transcription was >10-fold upregulated in blastemas versus all other tissues examined (Figure 5A). Kazd1 is predicted to have a Kazal-type serine protease inhibitor domain, a follistatin-like domain, and an immunoglobulin domain. We find that kazald1 expression is not detectable in the intact limb pre-amputation, increases shortly after amputation, is maintained during the blastemal stages of regeneration, and is dramatically downregulated near the end of regeneration (Figure 5B, C). Notably, kazald1 is not detectable in the developing limb bud within limb bud progenitor cells (Figure 5C). Therefore, its function in blastemas may be regeneration-specific, in contrast to other genes whose expression is similar between limb buds and blastemas (for example, sonic hedgehog (Torok et al., 1999); hoxd-8, -10, and -11 (Torok et al., 1998)).
Figure 5. Kazald1 the most robust blastema marker, is required for limb regeneration.
(A) Differential tissue expression analysis identifies kazald1 as the most blastema-enriched transcript compared to all other tissues sequenced. (B) RT-PCR performed on blastema cDNA samples throughout the course of regeneration for kazald1 expression. Kazald1 was not detected in intact limbs and at 1 DPA, and has dramatically diminished by 60 DPA. (C) In situ hybridization for kazald1 in the blastema over course of regeneration (top panels). Kazald1 is not detectable in regenerated limbs (35 DPA), intact limbs or in developing limb buds (lower panels). (D–E) Regenerating limbs at 19 DPA treated with control (D) or kazald1-targeting morpholino (E); quantified in (F). (G–J’) Regenerating limbs at 28 DPA treated with control (G) or kazald1-targeting morpholino (H-H’). (I–J’) Same specimens stained with Alcian blue to visualize cartilage; quantified in (K). Scale bar for all panels in C is 500µm; scale bar for D-E and H-J’ is 1mm. *** indicates p<0.001 and error bars are SEM. Arrowheads mark amputation plane in each image. BL=blastema, WE=wound epidermis. See also Figure S4, Figure S5, Figure S6, File S1 and File S2.
We hypothesized that diminishing kazald1 expression during regeneration would have deleterious consequences and found that kazald1-targeting morpholinos delayed limb regeneration (Figure 5D–F; Figure S4) and resulted in significantly smaller blastemas as compared to controls (Figure 5F: N=48 limbs per condition; Figure S4B–D: N=48 control MO2-treated limbs, N=44 kazald1-MO2-treated limbs). Further, the kazald1-morpholino-treated animals had delayed chondrification and differentiation of the digits at the palette to early digits stage, with less than 50% total Alcian-blue-staining area relative to controls (Figure 5G–K: N=46 control MO1-treated limbs, N=45 kazald1-MO1-treated limbs; Figure S4E–I: N=48 control MO2-treated limbs, N=44 kazald1-MO2-treated limbs). This data strongly suggests that kazald1 is critical for the proper progression of limb regeneration.
Motivated by the finding that kazald1 is essential for normal regeneration, we leveraged our transcriptome to create kazald1-targeted deletions in both embryos and in unamputated limbs via electroporation (Supplemental Information and Figure S5) to further complement our studies with morpholino knockdown. Although we were able to successfully edit the kazald1 gene, we did not achieve a sufficient level of editing efficiency to study the effect of genetic loss-of-function of kazald1 on limb regeneration with a high degree of confidence (17 – 46% efficiency; Figure S5); we did not observe regenerative defects in mosaic animals or electroporated limbs most likely due to low editing efficiency (data not shown). Nevertheless, future studies with complete genetic loss-of-function of kazald1 are warranted and will prove highly valuable to further examining this gene’s mechanistic role in axolotl limb regeneration.
Kazald1’s intense expression within the transient blastema structure suggests that it has a time-delineated function during regeneration. To test this, we used a replication-incompetent retrovirus (Whited et al., 2013) to constitutively overexpress kazald1 in regenerating limbs (Figure S6A). A majority (15/22) of kazald1-infected limbs regenerated abnormally compared to control EGFP infections (0/18) (Figure S6B). The most commonly observed defect following kazald1 overexpression was syndactyly (8/15 abnormal limbs), and other defects included missing digits, truncated digits, severe clinodactyly, and ectopic tissue growth on the palm and digits (Figure S6C).
Our morpholino and retroviral experiments highlight a functional role for kazald1 during limb regeneration, as perturbing its expression during this process adversely affected the progression and outcome of limb regeneration. Another report also discovered enriched expression of kazald1 in a regenerative context in vitro (Athippozhy et al., 2014). However, in this study, the expression of the transcript was not profiled in intact or regenerating limbs, and the bulk of the expression was attributed to nerves. Here, we show that the blastema cells themselves produce large amounts of kazald1 transcript. More studies will be needed to determine if blastema cells are stimulated by the ingrowing regenerating nerve fibers to upregulate this transcript.
Comparison of tissues at different anatomical locations identifies candidate transcripts of positional memory along intact and limb regeneration sites
Axolotl limb tissues maintain a “positional memory” throughout life allowing for the precise regeneration of only lost limb elements following amputation, suggesting that specific genes may be differentially expressed along the length of the limb and control positional memory. To uncover such genes, we assayed samples of similar tissue type but of diverse positional/anatomical context. First, we profiled cartilage from the wrist (carpals) and the ends of long bones (humerus, radius, ulna). Second, we collected samples along the length of the limb, and blastemas arising from either proximal (mid-humerus) or distal (distal radius/ulna) amputations. We then analyzed their expression profiles to identify differentially expressed genes for each context.
Arm segments
We performed two separate analyses to address the transcriptional differences between different locations along the proximal-distal axis of the arm (Figure 6). We first performed pairwise comparisons to identify transcripts significantly differentially expressed between any pair of arm segments. In the second approach, we performed analyses to uncover factors that show graded enrichment across the proximal-distal axis.
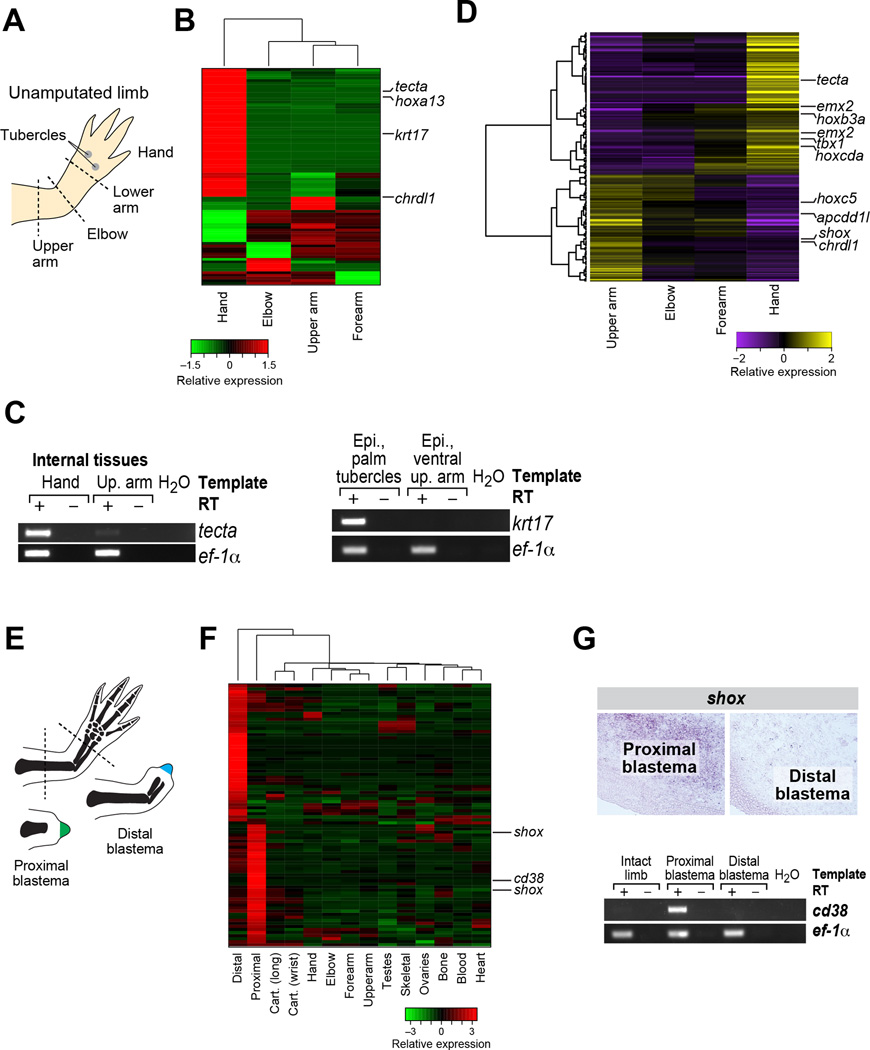
Figure 6. Transcripts differentially expressed in proximal versus distal elements.
(A) Schematic illustrating specific elements of the hand and arm. (B) Differential gene expression analysis identifies transcripts that are enriched in distinct sections of the intact limb. (C) RT-PCR validation of select transcripts identified by differential expression analysis. (D) Gradient gene expression analysis identifies transcripts enriched in a gradient from proximal to distal or distal to proximal. (E) Schematic illustrating amputation planes for sampling of proximal and distal blastemas. (F) Differential expression analysis identifies transcripts that are enriched in proximal versus distal blastemas. (G) In situ and RT-PCR validation of computation predictions of differentially expressed transcripts in proximal and distal blastemas. See also Figure S7, Table S5, File S1 and File S2.
Using the pairwise approach, we uncovered a relatively modest number of transcriptional differences between different segments of the forelimb (“arm”): 636 differentially-expressed transcripts identified between at least one pair of arm segments and 83 transcripts involving all arm segments (Figure 6A,B, Table S5), especially in hand (48/83). Many of these likely reflect differences in the cell type composition of the different arm segments. For example, transcripts enriched in forearm, elbow, and upper arm are also more highly enriched in skeletal muscle and bone tissue; transcripts enriched in upper arm are also enriched in bone and cartilage (Figure S7A). Interestingly, 27 of the 48 transcripts enriched in hand relative to other arm segments are also hand-specific relative to other non-arm tissues. These include keratins and other filamentous proteins (6/27) with known structural roles in skin, consistent with the increased surface area of the hand versus the rest of the arm. Notably, when comparing highly similar tissue types, we found little signal of position-specific expression. In particular, comparing cartilage between wrist and long bone, we found only minor distinctions (Figure S7B), such that the samples were very highly correlated (r=0.98), with only 22 differentially expressed transcripts, mostly in lowly expressed or non-cartilage specific genes.
The arm segment analysis highlighted key transcripts with known function in limb polarity, and candidates with uncharacterized functions in limb polarity (Figure 6B). First, we found hoxa13, which is known to mark distal elements in developing appendages (Haack and Gruss, 1993). Second, we found and validated a specific keratin, krt17, as exclusively expressed in the hand, restricted to the tubercles on the ventral/palmar surface of the hand (Figure 6B,C). Additional hand-enriched transcripts include a homolog to alpha tectorin (tecta), which is required for hearing in mammals (reviewed in (Legan et al., 2014)). Finally, one of the few transcripts that distinguishes upper arm segments from other segments of intact limbs encodes a predicted ortholog of Chordin-like 1 (crdl1) (Figure 6B), a diffusible morphogen (reviewed in (Zakin and De Robertis, 2010)).
We uncovered 875 transcripts demonstrating a significant (FDR <= 0.05) gradient in expression intensity along the proximal-distal axis of the unamputated limb (Figure 6D, Table S5). Among these, we uncovered two transcripts orthologous to emx2 (Empty spiracles homeobox 2) that were most highly expressed in the hand and had diminishing expression toward the proximal sample sites. The newt ortholog of emx2 has previously been shown to exhibit distal-enriched expression in unamputated limbs (Beauchemin et al., 1998). Conversely, we identified apcdd1l (orthologous to Apcdd1-like) as the most significantly graded transcript with high levels in the proximal segments. Apcdd1 is an extracellular membrane-bound signaling protein that can inhibit Wnt signaling (Shimomura et al., 2010).
Proximal vs. distal blastema
There are 305 transcripts (275 genes) differentially expressed between proximal and distal blastema tissue (File S2), of which 81 transcripts (70 genes, Table S5) are also enriched in blastema tissue relative to other tissues (Figure 6E,F). We validated cd38 and shox, whose expression is significantly enriched in proximal blastemas (Figure 6G). Cd38 is predicted to encode the axolotl ortholog of mammalian CD38, a cell-surface glycoprotein and ADP-ribosyl cyclase (reviewed in (Wei et al., 2014)). CD38 expression at both the mRNA and protein levels can be induced by exposure of cells to retinoic acid (Drach et al., 1993). Since retinoic acid is a proximalizing agent in limb regeneration (Maden, 1983), enrichment of cd38 transcripts in proximal blastemas may reflect activation of retinoic acid signaling. Shox (short-stature homeobox) encodes a predicted homeobox-containing transcription factor. Experimentally removing the most closely-related mouse gene, Shox2, in limb buds results in the near-complete loss of the humerus, a proximal skeletal element, while remaining structures are mostly unaffected (Cobb et al., 2006). Future studies may reveal a functional role for these genes in controlling proximal-distal identity during limb regeneration.
DISCUSSION
Here, we report the assembly and validation of an axolotl transcriptome centered on tissues relevant for limb regeneration that provides near-complete sequence information for exonic regions of ~88% of axolotl genes with a high degree of confidence (Table 1), generating a valuable annotated resource for researchers using axolotl. We used differential expression analysis within the context of a biologically-informed design to identify tissue-specific and anatomically-specific gene expression patterns that we were able to validate experimentally, demonstrating the validity and the breadth of experimental design that can be conducted with our catalog.
Within the blastema, the nexus of limb regeneration, we uncovered an enriched profile of transcripts for members of the heterogenous nuclear ribonucleoprotein complex (hnRNP) such as cirbp, fus, roa1, and hnrnpd. In other biological contexts, RNA binding proteins are capable of modulating large repertoires of mRNA targets, coordinate cellular responses, and maintain cellular phenotypes (Kafasla et al., 2014; Zhang et al., 2013). During limb regeneration, modulation of RNA binding proteins and post-translational regulation of mRNA may constitute major mechanisms for executing the vast cellular changes necessary to regenerate a cmplex structure. Specifically, we highlighted the axolotl ortholog of cirbp as highly blastema-enriched and uncovered a cytoprotective function for this gene during early blastema development. Cirbp encodes a putative RNA-binding protein orthologous to mammalian Cold-Inducible RNA-binding Protein (CIRBP) and highly similar to RBM3 (orthologous axolotl rbmx was also identified as blastema-enriched). Both cirbp and rbm3 have been demonstrated to be transcriptionally upregulated in response to specific cellular stressors including cold shock, hypoxia, and UV irradiation in mammalian cells (Danno et al., 1997; Nishiyama et al., 1997; Wellmann et al., 2004). Our experiments suggest one function of cirbp in regeneration is to protect progenitor cells from cell death, possibly to protect blastema progeniotrs from extensive cellular changes occurring following amputation. We also identified cirbp transcritps at the early to late digit stage (35 dpa) within the interdigital regions, which are presumed to be relatively undifferentiated and will be eliminated as the regenerating digits are sculpted (reviewed in (Montero and Hurle, 2010). Further, fus mRNA has been identified as a major target of mammalian CIRBP (Morf et al., 2012), both of which are significantly enriched in the blastema. Thus, understanding regulatory loops between RNA-binding proteins to modulate cell fate pathways within blastemal cells may be emerging as a theme in limb regeneration, supporting a need for future studies.
We also experimentally tested the consequence of manipulating expression of kazald1, the gene whose transcript best distinguishes blastema cells from all other samples sequenced in our data. We found that reducing expression of kazald1 via morpholinos resulted in an outward lag in the progression of limb regeneration compared to controls while constitutive expression of kazald1 within the blastema resulted in regeneration of deformed limbs, underscoring the importance of temporal and/or spatial regulation of this transcript. Interestingly, a similar gene has been implicated in regeneration in hydra, an invertebrate with extensive regenerative abilities (Chera et al., 2006), raising the possibility that an ancient and conserved role for kazald1 in regeneration exists. Other Kazal-type serine protease inhibitors in mammals serve as biomarkers for various forms of cancer, but their exact functions are only now emerging (reviewed in (Fradet, 2009)). As aspects of limb regeneration share some similarity to cancer (for instance, activation of stem cells and dedifferentiation, cell proliferation, re-activation of developmental pathways, and the wound response) (reviewed in (Pomerantz and Blau, 2013)), links between the two processes warrant further investigation. Future work will be necessary to determine if connections exist between the roles of Kazal-type inhibitors in other organisms and the role of kazald1 in axolotl limb regeneration.
Our assembly and experimentation will serve as a valuable resource for researchers using axolotls in a variety of fields, as well as for those studying regeneration in other models. Our differential gene expression analysis can provide starting points for studying genes that may drive the biological function of particular tissues. It can also be used to identify reliable markers of particular tissue types, and with further experimentation, possibly specific cell types within particular tissues. In many cases, our assembly will provide the full-length coding sequence for future functional studies, including the requisite sequence information to design genome-editing strategies and provide data to minimize potential off-targets. The axolotl transcriptome we report can serve as a basis for mining tissue enriched or repressed transcripts, and will also serve as a powerful reference for more directed future studies. All reconstructed axolotl transcripts, expression data, and feature annotations are available via our web portal at http://portals.broadinstitute.org/axolotlomics.
EXPERIMENTAL PROCEDURES
Trinity de novo RNA-Seq assembly
Trinity version trinityrnaseq_r2013-02-25 was used for de novo transcriptome assembly and analysis as follows. The combined set of 1.3 billion RNA-Seq reads spanning all sampled tissues were combined into a single pair of fastq files, quality trimmed using Trimmomatic (Bolger et al., 2014) (parameters LEADING:5 TRAILING:5 MINLEN:36) and subsequently normalized using the in silico normalization step incorporated into Trinity ($TRINTY_HOME/util/normalize_by_kmer_coverage.pl --seqType fq --JM 100G --left ALL_AXOLOTL_READS.left.fq.P.qtrim.fq –right ALL_AXOLOTL_READS.right.fq.P.qtrim.fq --pairs_together --JELLY_CPU 10 -- PARALLEL_STATS --max_cov 50). The resulting normalized reads were then assembled using Trinity (Trinity.pl --left left.fq --right right.fq --seqType fq --JM 100G --CPU 10).
Transcript annotation using Trinotate
We developed Trinotate as a bioinformatics protocol and software system for annotating putative functional characteristics of transcripts. Similarities to known proteins were detected by a BLASTX search (Camacho et al., 2009) (E <= 1e-5) of a comprehensive protein database formed by merging SwissProt (Boeckmann et al., 2005) with Uniref90 (UniProt, 2015) protein databases downloaded from UniProt (versions available on July 5, 2014). Likely coding regions within transcripts were predicted using TransDecoder (http://transdecoder.github.io), and resulting protein products were searched for sequence similarities against the comprehensive protein database described above (BLASTP, E<=1e-5), and for conserved protein domains using Hmmer (http://hmmer.janelia.org/) and PFam (Finn et al., 2014). Signal peptides were predicted using SignalP (Petersen et al., 2011), and transmembrane region predictions were predicted using TmHMM (Krogh et al., 2001). All results were parsed by Trinotate, stored in a SQLite relational database, and then reported as a tab-delimited transcript annotation summary file (File S1B). Gene Ontology (GO) identifiers were transitively assigned to transcripts based on available GO annotations of best-matching Swissprot entries (File S2).
Transcript abundance estimation and differential expression analysis
RSEM (Li and Dewey, 2011) and mmseq (Turro et al., 2011) software was used to estimate expression values for transcripts, and edgeR (Robinson et al., 2010), EBseq (Leng et al., 2013) and mmdiff (Turro et al., 2014) were used to identify significantly differentially expressed transcripts. Further details can be found in the Supplemental Methods.
Animal experimentation
All animal experiments were performed in accordance with Harvard Medical School’s Institutional Animal Care and Use Committee regulations and in accordance with Animal Experimentation Protocol #04160. For all survival surgeries, animals were anesthetized in 0.1% tricaine and allowed to recover overnight in 0.5% sulfamerazine.
Library preparation and sequencing
Total RNA was purified using either Trizol or Qiagen RNeasy columns. The Illumina TruSeq v2 protocol was used throughout to generate barcoded sequencing libraries. Paired-end, 100-bp sequencing was performed on the Illumina HiSeq 2500 at Harvard Medical School’s Biopolymers facility.
In situ hybridization
Sequences were amplified from cDNA and cloned into pGEM-T-easy vector and sequenced. Depending upon orientation, T7 or Sp6 polymerase was used to transcribe the probe. In situ hybridization was performed as in (Whited et al., 2011).
Statistical analyses
All results from morpholino knockdown and retroviral experiments were quantified by at least one blinded observer. Blastema length and chondrification area were measured using ImageJ. A two-tailed Student’s t-test was used to assess statistical significance. The statistical significance of the phenotypic outcomes from retroviral overexpression of kazald1 was assessed by performing a two-tailed Fisher’s Exact test on a 2×2 contingency table.
Supplementary Material
Acknowledgments
This work was supported by the Harvard Stem Cell Institute (C.J.T.), Brigham & Women’s Hospital (J.L.W.), the Richard and Susan Smith Family Foundation (J.L.W.), NIH/NICHD (1R03HD083434-01 and 1DP2HD087953-01, J.L.W.), NIH/NICHD (R01 HD073104, L.P.); and the Howard Hughes Medical Institute (A.R.). This research was supported by the National Cancer Institute of the National Institutes of Health under Award Number 1U24CA180922-01 (B.J.H. and A.R.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. D.M.B. was supported by an HHMI Gilliam Fellowship. We thank Borja Sese Ballesteros and Yick Fong for their helpful discussions on using CRISPR-Cas9; Jana Hersch for germ cell identification; Josh Gorham and Jim Pancoast for assistance in RNA extraction; Harvard Biopolymers for sequencing; Esther Pearl and Marko Horb for sequencing the embryo samples; Rich Lee for use of shared equipment; Leslie Gaffney for help with figures; and Benjamin Lewis for discussion. A subset of computations in this paper were run on the Orchestra cluster supported by the Research Information Technology Group at Harvard Medical School. We thank the Ambystoma Genetic Stock Center for providing some of the animals (Lexington, KY, NIH grant P40-OD019794).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
For detailed methods, see Supplemental Information.
AUTHOR CONTRIBUTIONS
D.M.B., K.J., T.D., B.J.H., and J.L.W. designed the experiments; D.M.B., K.J., T.D., C.J.T., A.R., B.J.H., and J.L.W. wrote the paper; B.J.H., T.T., and M.B.C. performed the computation and developed related resources; R.M.F. and L.P. performed the embryo sequencing; D.M.B., K.J., T.D., D.P-D., T.J.L., N.D.L., T.K., F.G.D., J.B., S.B., A.R.G., S.L.T., S.C., W.Y., and J.L.W. performed the sample preparations, validations, and functional experimentation. All authors contributed to manuscript editing.
Accession Numbers
Raw sequencing reads were deposited in the NCBI Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA300706) under the accession code PRJNA300706. The transcriptome was deposited in the NCBI Transcriptome Shotgun Assembly Database (TSA) under the accession code GFBM000000000. All data were deposited in the NCBI Gene Expression Omnibus and can be found under accession code GSE92429.
The authors declare no competing interests.
REFERENCES
- Abdullayev I, Kirkham M, Bjorklund AK, Simon A, Sandberg R. A reference transcriptome and inferred proteome for the salamander Notophthalmus viridescens. Exp Cell Res. 2013;319:1187–1197. doi: 10.1016/j.yexcr.2013.02.013. [DOI] [PubMed] [Google Scholar]
- Athippozhy A, Lehrberg J, Monaghan JR, Gardiner DM, Voss SR. Characterization of transcriptional responses of dorsal root ganglia cultured in the presence and absence of blastema cells from regenerating salamander limbs. Regeneration (Oxford) 2014;1:1–10. doi: 10.1002/reg2.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin M, Del Rio-Tsonis K, Tsonis PA, Tremblay M, Savard P. Graded expression of Emx-2 in the adult newt limb and its corresponding regeneration blastema. J Mol Biol. 1998;279:501–511. doi: 10.1006/jmbi.1998.1782. [DOI] [PubMed] [Google Scholar]
- Boeckmann B, Blatter MC, Famiglietti L, Hinz U, Lane L, Roechert B, Bairoch A. Protein variety and functional diversity: Swiss-Prot annotation in its biological context. Comptes rendus biologies. 2005;328:882–899. doi: 10.1016/j.crvi.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chera S, de Rosa R, Miljkovic-Licina M, Dobretz K, Ghila L, Kaloulis K, Galliot B. Silencing of the hydra serine protease inhibitor Kazal1 gene mimics the human SPINK1 pancreatic phenotype. J Cell Sci. 2006;119:846–857. doi: 10.1242/jcs.02807. [DOI] [PubMed] [Google Scholar]
- Cobb J, Dierich A, Huss-Garcia Y, Duboule D. A mouse model for human short-stature syndromes identifies Shox2 as an upstream regulator of Runx2 during long-bone development. Proc Natl Acad Sci U S A. 2006;103:4511–4515. doi: 10.1073/pnas.0510544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K, Stocum DL. Retinoic acid coordinately proximalizes regenerate pattern and blastema differential affinity in axolotl limbs. Development. 1988a;102:687–698. doi: 10.1242/dev.102.4.687. [DOI] [PubMed] [Google Scholar]
- Crawford K, Stocum DL. Retinoic acid proximalizes level-specific properties responsible for intercalary regeneration in axolotl limbs. Development. 1988b;104:703–712. doi: 10.1242/dev.104.4.703. [DOI] [PubMed] [Google Scholar]
- Danno S, Nishiyama H, Higashitsuji H, Yokoi H, Xue JH, Itoh K, Matsuda T, Fujita J. Increased transcript level of RBM3, a member of the glycine-rich RNA-binding protein family, in human cells in response to cold stress. Biochem Biophys Res Commun. 1997;236:804–807. doi: 10.1006/bbrc.1997.7059. [DOI] [PubMed] [Google Scholar]
- Drach J, Zhao S, Malavasi F, Mehta K. Rapid induction of CD38 antigen on myeloid leukemia cells by all trans-retinoic acid. Biochem Biophys Res Commun. 1993;195:545–550. doi: 10.1006/bbrc.1993.2080. [DOI] [PubMed] [Google Scholar]
- Echeverri K, Tanaka EM. Proximodistal patterning during limb regeneration. Dev Biol. 2005;279:391–401. doi: 10.1016/j.ydbio.2004.12.029. [DOI] [PubMed] [Google Scholar]
- Eto K, Eda K, Hayano M, Goto S, Nagao K, Kawasaki T, Kashimura H, Tarui H, Nishimura O, Agata K, et al. Reduced expression of an RNA-binding protein by prolactin leads to translational silencing of programmed cell death protein 4 and apoptosis in newt spermatogonia. J Biol Chem. 2009;284:23260–23271. doi: 10.1074/jbc.M109.018622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic acids research. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradet Y. Biomarkers in prostate cancer diagnosis and prognosis: beyond prostate-specific antigen. Curr Opin Urol. 2009;19:243–246. doi: 10.1097/MOU.0b013e32832a08b5. [DOI] [PubMed] [Google Scholar]
- Goto M, Eddy EM. Speriolin is a novel spermatogenic cell-specific centrosomal protein associated with the seventh WD motif of Cdc20. J Biol Chem. 2004;279:42128–42138. doi: 10.1074/jbc.M403190200. [DOI] [PubMed] [Google Scholar]
- Goto M, O’Brien DA, Eddy EM. Speriolin is a novel human and mouse sperm centrosome protein. Hum Reprod. 2010;25:1884–1894. doi: 10.1093/humrep/deq138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack H, Gruss P. The establishment of murine Hox-1 expression domains during patterning of the limb. Dev Biol. 1993;157:410–422. doi: 10.1006/dbio.1993.1145. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang DZX, Zhao J, Zhou Y, Zhong C, Zhang J, Huang X. Subtractive screen of potential limb regeneration related genes from Pachytriton brevipes. Mol Biol Rep. 2014;41:1015–1026. doi: 10.1007/s11033-013-2946-z. [DOI] [PubMed] [Google Scholar]
- Jiang P, Nelson JD, Leng N, Collins M, Swanson S, Dewey CN, Thomson JA, Stewart R. Analysis of embryonic development in the unsequenced axolotl: Waves of transcriptomic upheaval and stability. Dev Biol. 2016 doi: 10.1016/j.ydbio.2016.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafasla P, Skliris A, Kontoyiannis DL. Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nat Immunol. 2014;15:492–502. doi: 10.1038/ni.2884. [DOI] [PubMed] [Google Scholar]
- Keinath MC, Timoshevskiy VA, Timoshevskaya NY, Tsonis PA, Voss SR, Smith JJ. Initial characterization of the large genome of the salamander Ambystoma mexicanum using shotgun and laser capture chromosome sequencing. Sci Rep. 2015;5:16413. doi: 10.1038/srep16413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan P, Linkhart B, Simon HG. Different regulation of T-box genes Tbx4 and Tbx5 during limb development and limb regeneration. Dev Biol. 2002;250:383–392. [PubMed] [Google Scholar]
- Knapp D, Schulz H, Rascon CA, Volkmer M, Scholz J, Nacu E, Le M, Novozhilov S, Tazaki A, Protze S, et al. Comparative transcriptional profiling of the axolotl limb identifies a tripartite regeneration-specific gene program. PLoS One. 2013;8:e61352. doi: 10.1371/journal.pone.0061352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp D, Tanaka EM. Regeneration and reprogramming. Curr Opin Genet Dev. 2012;22:485–493. doi: 10.1016/j.gde.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of molecular biology. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Legan PK, Goodyear RJ, Morin M, Mencia A, Pollard H, Olavarrieta L, Korchagina J, Modamio-Hoybjor S, Mayo F, Moreno F, et al. Three deaf mice: mouse models for TECTA-based human hereditary deafness reveal domain-specific structural phenotypes in the tectorial membrane. Hum Mol Genet. 2014;23:2551–2568. doi: 10.1093/hmg/ddt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, Haag JD, Gould MN, Stewart RM, Kendziorski C. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Fillmore N, Bai Y, Collins M, Thomson JA, Stewart R, Dewey CN. Evaluation of de novo transcriptome assemblies from RNA-Seq data. Genome Biol. 2014;15:553. doi: 10.1186/s13059-014-0553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhang Z, Xue J, Liu A, Zhang H. Cold-inducible RNA binding protein inhibits H(2)O(2)-induced apoptosis in rat cortical neurons. Brain Res. 2012;1441:47–52. doi: 10.1016/j.brainres.2011.12.053. [DOI] [PubMed] [Google Scholar]
- Looso M, Preussner J, Sousounis K, Bruckskotten M, Michel CS, Lignelli E, Reinhardt R, Hoffner S, Kruger M, Tsonis PA, et al. A de novo assembly of the newt transcriptome combined with proteomic validation identifies new protein families expressed during tissue regeneration. Genome Biol. 2013;14:R16. doi: 10.1186/gb-2013-14-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden M. The effect of vitamin A on the regenerating axolotl limb. J Embryol Exp Morphol. 1983;77:273–295. [PubMed] [Google Scholar]
- McCusker CD, Athippozhy A, Diaz-Castillo C, Fowlkes C, Gardiner DM, Voss SR. Positional plasticity in regenerating Amybstoma mexicanum limbs is associated with cell proliferation and pathways of cellular differentiation. BMC Dev Biol. 2015;15:45. doi: 10.1186/s12861-015-0095-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker CD, Gardiner DM. Positional information is reprogrammed in blastema cells of the regenerating limb of the axolotl (Ambystoma mexicanum) PLoS One. 2013;8:e77064. doi: 10.1371/journal.pone.0077064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercader N, Tanaka EM, Torres M. Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development. 2005;132:4131–4142. doi: 10.1242/dev.01976. [DOI] [PubMed] [Google Scholar]
- Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95:315–327. doi: 10.1016/j.ygeno.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan JR, Epp LG, Putta S, Page RB, Walker JA, Beachy CK, Zhu W, Pao GM, Verma IM, Hunter T, et al. Microarray and cDNA sequence analysis of transcription during nerve-dependent limb regeneration. BMC Biol. 2009;7:1. doi: 10.1186/1741-7007-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero JA, Hurle JM. Sculpturing digit shape by cell death. Apoptosis. 2010;15:365–375. doi: 10.1007/s10495-009-0444-5. [DOI] [PubMed] [Google Scholar]
- Morf J, Rey G, Schneider K, Stratmann M, Fujita J, Naef F, Schibler U. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science. 2012;338:379–383. doi: 10.1126/science.1217726. [DOI] [PubMed] [Google Scholar]
- Muneoka K, Fox WF, Bryant SV. Cellular contribution from dermis and cartilage to the regenerating limb blastema in axolotls. Dev Biol. 1986;116:256–260. doi: 10.1016/0012-1606(86)90062-x. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Islam MR, Takayanagi M, Yasumuro H, Inami W, Kunahong A, Casco-Robles RM, Toyama F, Chiba C. A transcriptome for the study of early processes of retinal regeneration in the adult newt, Cynops pyrrhogaster. PLoS One. 2014;9:e109831. doi: 10.1371/journal.pone.0109831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama H, Higashitsuji H, Yokoi H, Itoh K, Danno S, Matsuda T, Fujita J. Cloning and characterization of human CIRP (cold-inducible RNA-binding protein) cDNA and chromosomal assignment of the gene. Gene. 1997;204:115–120. doi: 10.1016/s0378-1119(97)00530-1. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Pomerantz JH, Blau HM. Tumor suppressors: enhancers or suppressors of regeneration? Development. 2013;140:2502–2512. doi: 10.1242/dev.084210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, et al. De novo assembly and analysis of RNA-seq data. Nat Methods. 2010;7:909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Guzman T, Wang H, Khattak S, Schuez M, Roensch K, Nacu E, Tazaki A, Joven A, Tanaka EM, Simon A. Fundamental differences in dedifferentiation and stem cell recruitment during skeletal muscle regeneration in two salamander species. Cell Stem Cell. 2014;14:174–187. doi: 10.1016/j.stem.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Satoh A, Bryant SV, Gardiner DM. Regulation of dermal fibroblast dedifferentiation and redifferentiation during wound healing and limb regeneration in the Axolotl. Dev Growth Differ. 2008;50:743–754. doi: 10.1111/j.1440-169X.2008.01072.x. [DOI] [PubMed] [Google Scholar]
- Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura Y, Agalliu D, Vonica A, Luria V, Wajid M, Baumer A, Belli S, Petukhova L, Schinzel A, Brivanlou AH, et al. APCDD1 is a novel Wnt inhibitor mutated in hereditary hypotrichosis simplex. Nature. 2010;464:1043–1047. doi: 10.1038/nature08875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Putta S, Zhu W, Pao GM, Verma IM, Hunter T, Bryant SV, Gardiner DM, Harkins TT, Voss SR. Genic regions of a large salamander genome contain long introns and novel genes. BMC Genomics. 2009;10:19. doi: 10.1186/1471-2164-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart R, Rascon CA, Tian S, Nie J, Barry C, Chu LF, Ardalani H, Wagner RJ, Probasco MD, Bolin JM, et al. Comparative RNA-seq analysis in the unsequenced axolotl: the oncogene burst highlights early gene expression in the blastema. PLoS Comput Biol. 2013;9:e1002936. doi: 10.1371/journal.pcbi.1002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocum DL, Melton DA. Self-organizational capacity of distally transplanted limb regeneration blastemas in larval salamanders. J Exp Zool. 1977;201:451–461. doi: 10.1002/jez.1402010312. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus NA. Comparative DNA renaturation kinetics in amphibians. Proc Natl Acad Sci U S A. 1971;68:799–802. doi: 10.1073/pnas.68.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok MA, Gardiner DM, Izpisua-Belmonte JC, Bryant SV. Sonic hedgehog (shh) expression in developing and regenerating axolotl limbs. J Exp Zool. 1999;284:197–206. [PubMed] [Google Scholar]
- Torok MA, Gardiner DM, Shubin NH, Bryant SV. Expression of HoxD genes in developing and regenerating axolotl limbs. Dev Biol. 1998;200:225–233. doi: 10.1006/dbio.1998.8956. [DOI] [PubMed] [Google Scholar]
- Turro E, Astle WJ, Tavare S. Flexible analysis of RNA-seq data using mixed effects models. Bioinformatics. 2014;30:180–188. doi: 10.1093/bioinformatics/btt624. [DOI] [PubMed] [Google Scholar]
- Turro E, Su SY, Goncalves A, Coin LJ, Richardson S, Lewin A. Haplotype and isoform specific expression estimation using multi-mapping RNA-seq reads. Genome biology. 2011;12:R13. doi: 10.1186/gb-2011-12-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt C. UniProt: a hub for protein information. Nucleic acids research. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss SR, Palumbo A, Nagarajan R, Gardiner DM, Muneoka K, Stromberg AJ, Athippozhy AT. Gene expression during the first 28 days of axolotl limb regeneration I: Experimental design and global analysis of gene expression. Regeneration (Oxf) 2015;2:120–136. doi: 10.1002/reg2.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Graeff R, Yue J. Roles and mechanisms of the CD38/cyclic adenosine diphosphate ribose/Ca(2+) signaling pathway. World J Biol Chem. 2014;5:58–67. doi: 10.4331/wjbc.v5.i1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmann S, Buhrer C, Moderegger E, Zelmer A, Kirschner R, Koehne P, Fujita J, Seeger K. Oxygen-regulated expression of the RNA-binding proteins RBM3 and CIRP by a HIF-1-independent mechanism. J Cell Sci. 2004;117:1785–1794. doi: 10.1242/jcs.01026. [DOI] [PubMed] [Google Scholar]
- Whited JL, Lehoczky JA, Austin CA, Tabin CJ. Dynamic expression of two thrombospondins during axolotl limb regeneration. Dev Dyn. 2011;240:1249–1258. doi: 10.1002/dvdy.22548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whited JL, Tabin CJ. Limb regeneration revisited. J Biol. 2009;8:5. doi: 10.1186/jbiol105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whited JL, Tsai SL, Beier KT, White JN, Piekarski N, Hanken J, Cepko CL, Tabin CJ. Pseudotyped retroviruses for infecting axolotl in vivo and in vitro. Development. 2013;140:1137–1146. doi: 10.1242/dev.087734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Tsai MH, Ho CC, Chen CY, Lee HS. De novo transcriptome sequencing of axolotl blastema for identification of differentially expressed genes during limb regeneration. BMC Genomics. 2013;14:434. doi: 10.1186/1471-2164-14-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang EV, Gardiner DM, Carlson MR, Nugas CA, Bryant SV. Expression of Mmp-9 and related matrix metalloproteinase genes during axolotl limb regeneration. Dev Dyn. 1999;216:2–9. doi: 10.1002/(SICI)1097-0177(199909)216:1<2::AID-DVDY2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome biology. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HT, Xue JH, Zhang ZW, Kong HB, Liu AJ, Li SC, Xu DG. Cold-inducible RNA-binding protein inhibits neuron apoptosis through the suppression of mitochondrial apoptosis. Brain Res. 2015;1622:474–483. doi: 10.1016/j.brainres.2015.07.004. [DOI] [PubMed] [Google Scholar]
- Zhang L, Prak L, Rayon-Estrada V, Thiru P, Flygare J, Lim B, Lodish HF. ZFP36L2 is required for self-renewal of early burst-forming unit erythroid progenitors. Nature. 2013;499:92–96. doi: 10.1038/nature12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Pao GM, Satoh A, Cummings G, Monaghan JR, Harkins TT, Bryant SV, Randal Voss S, Gardiner DM, Hunter T. Activation of germline-specific genes is required for limb regeneration in the Mexican axolotl. Dev Biol. 2012;370:42–51. doi: 10.1016/j.ydbio.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler-Graham K, MacKenzie EJ, Ephraim PL, Travison TG, Brookmeyer R. Estimating the prevalence of limb loss in the United States: 2005 to 2050. Arch Phys Med Rehabil. 2008;89:422–429. doi: 10.1016/j.apmr.2007.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.