To the Editor
At present, no clinical tests are widely used to assess mutational burden in cutaneous T cell lymphoma (CTCL) with leukemic (L-CTCL) involvement, such as in Sézary syndrome (SS), and routine blood analysis offers limited guidance for assessing prognosis and optimizing treatment within the highly variable population of affected patients. For now, L-CTCL contrasts sharply with more common and better-characterized cancers, such as chronic lymphocytic leukemia and ductal carcinoma of the breast, for which mutation analysis stratifies prognostic subgroups and can influence disease management (Nuciforo et al., 2016; Van Dyke et al., 2016). We have developed and validated a panel of 11 fluorescence in situ hybridization (FISH) probes designed to capture gene copy number alterations (GCNAs) present in 97.5% of patients with L-CTCL as elucidated in a recent exome study (Choi et al., 2015). The panel includes 5 probes previously developed for TP53, MYC, RB1, CDKN2A, and ATM, as well as 6 newly designed probes for STAT3/5B, ARID1A, ZEB1, FAS, CARD11, and DNMT3A (Figure 1a). FISH technology particularly suits CTCL, a cancer whose mutational landscape is characterized by a preponderance of gene copy number amplifications and deletions compared to single nucleotide variants relatively overrepresented in many other forms of cancer. In this study, we used the 11-probe panel to assess for genetic abnormalities in sorted or unsorted peripheral blood from 24 patients (patient characteristics in Supplementary Materials) with a range of disease presentations including SS, patch/plaque mycosis fungoides (MF), follicular mycosis fungoides (F-MF), tumor-stage mycosis fungoides, and follicular mucinosis –with evidence of blood involvement (Gibson et al., 2016).
Figure 1. L-CTCL FISH panel composition and probe microscopy.
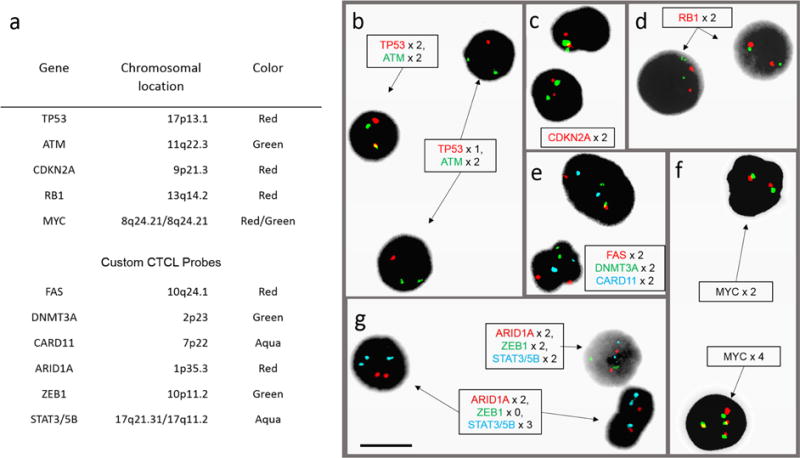
(a) Table of genes, chromosomal locations, and fluorescent colors for 11 clinically validated probes used in the CTCL FISH panel. (b–g) Post-hybridization microscopy images using 11-probe FISH panel on peripheral blood samples from patients with L-CTCL and abnormal FISH results. (b) Field with cell showing normal copy number of two TP53 (red) signals and two ATM (green) signals, and two cells with deletion of one copy of TP53. (c) Cells with two copies of CDKN2A (red) and 9q12 control probe (green). (d) Two cells with two copies of RB1 (red) and 13q control (green). (e) Signals for DMNT3A (green), CARD11 (aqua), and FAS (red), both cells showing two copies of each. (f) Cell on top with two copies of MYC stained with the red and green probes (both for MYC); cell below showing four copies of MYC. (g) Signals for ARID1A (red), ZEB1 (green), and STAT3/5B (aqua); one cell with normal copy numbers and two cells with homozygous deletion of ZEB1 and amplification of STAT3/5B. Black bar = 10 μm.
After design and production of FISH probes for STAT3/5B, ARID1A, ZEB1, FAS, and CARD11, probe hybridization sites were validated on metaphase chromosome spreads. Written informed consent of 24 patients who had a confirmed or suspected diagnosis of CTCL was obtained in accordance with protocols approved by the Institutional Review Board of Yale School of Medicine. Populations enriched for abnormal CD3+CD4+ lymphocytes, most frequently also CD26− and/or CD7−, were purified from Ficoll-isolated PBMC by either flow cytometric sorting or by magnetic bead isolation. Samples underwent fixation and overnight FISH hybridization with the panel of 11 probes, and probe signals were quantified by fluorescence light microscopy at 100 or 200 nuclei per probe (Figure 1b). CD8+ T cells collected as control populations were used to generate cutoffs (Supplementary Materials) for positive FISH abnormalities at the 99th percentile of binomial distribution of proportions calculated for each probe (Wiktor et al., 2006).
GCNAs were detected by FISH (Figure 2a) in 10 of 10 patients meeting International Society of Cutaneous Lymphoma (ISCL) criteria for SS and B2 stage blood involvement, 1 patient with F-MF not meeting B2 criteria, 1 patient with tumor-stage MF meeting B2 criteria, and 2 patients with MF one of whom met B2 criteria. No abnormalities were detected by FISH in the other 10 patients, of whom only 1 patient (with MF) met B2 involvement by one criterion (41.5% CD7− of CD4+ T cells; sorted CD7− cells). Among the 9 remaining patients with no detected GCNAs, 6 had a diagnosis of MF; 1 had a diagnosis of F-MF; 1 had a diagnosis of follicular mucinosis in association with Crohn’s disease (elevated CD4/8 ratio of 5.7, negative T cell receptor (TCR) PCR analysis for clonality); and 1 had atypical angioedematous plaques with initially positive (but subsequently negative) TCR clonality by PCR. Overall, of patients meeting revised 2007 ISCL criteria for B2 blood involvement, 12 of 13 patients (92%) had GCNAs detected by FISH.
Figure 2. Distribution of deletions and amplifications detected by FISH.
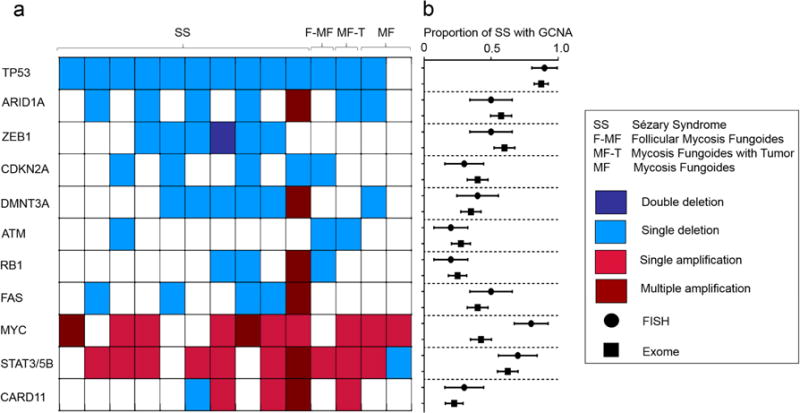
(a) Heatmap of deletions and amplification in CTCL driver genes detected by the 11-probe CTCL FISH panel among 10 patients with SS, 1 patient with F-MF, 1 patient with MF-T, and 2 patients with MF. Each column shows FISH status for one patient. (b) Comparison of proportion (+/− standard error) of SS patients with abnormal copy numbers in each gene by FISH (n=10) versus CTCL exome sequencing (n=40) on non-overlapping patient cohorts. No significant differences were found for any gene (Fisher’s exact test, p > 0.1). *For STAT3/5B FISH probe, comparator gene from exome data is STAT5B.
Proportions of FISH-identified GCNAs present per gene in patients with Sézary syndrome did not significantly differ from those observed in a recent large-scale exome study on a separate patient cohort (p-values 0.27–1.00, Fisher’s exact test, Figure 2b). One patient had an unanticipated single deletion of CARD11 in 92% of cells scored. CARD11 promotes the T cell receptor-mediated activation cascade and has been found amplified in CTCL exome data (Choi et al., 2015). However separate comparative genomic hybridization (CGH) data have shown a more balanced frequency of amplification and deletion in CARD11 (Kiel et al., 2015), and its functional significance in CTCL cells remains incompletely explored. Multiple atypical amplifications, including 4x amplifications in ARID1A, DNMT3A, RB1, and FAS, were seen in a separate patient with longstanding disease.
Much research remains to be completed relating patient outcomes and treatment susceptibilities to the status of driver genes in CTCL. Within our cohort of patients, specific GCNAs of note include 2 of 10 SS patients with double amplifications of the well-characterized oncogene MYC (Dang, 2012) found within broad amplifications on chromosome 8q in CTCL; joint amplification of several genes has complicated the interpretation of the significance of individual components in this region (Choi et al., 2015). Recent progress on investigational anti-Myc therapeutics (Stellas et al., 2014) may offer a targeted approach to assess the role of this oncogene. Also notable was 1 SS patient with homozygous deletion of ZEB1 and peripheral blood T cell count above 20,000/μL by 8 months after initial clinical presentation with erythroderma. ZEB1, a transcriptional repressor of IL-2 (Wang et al., 2009) and contributor to TGF-β1 mediated growth inhibition in adult T cell leukemia/lymphoma (Nakahata et al., 2010), has been found homozygously deleted in 10% of exome-sequenced L-CTCL patient samples; homozygous deletion in a mouse model has given rise to fatal T cell lymphomas in 84% of affected animals (Hidaka et al., 2008). Yet the relative response of CTCL harboring these and other mutations to immune-modulating therapies, biologic entities, or traditional chemotherapy is currently unknown.
We believe that the presented 11-probe FISH panel may offer the capacity to facilitate the diagnosis of L-CTCL, while also providing genetic status based on many of the most commonly represented GCNAs reported in CTCL. Since many of the genes represented have only recently been published as oncologic drivers of CTCL, we are currently unable to correlate these GCNAs with clinical outcomes; however, as outcomes data from genetic studies accumulate in the near future, this panel may also provide a helpful tool for prognosis and treatment stratification with advantages including rapid turnaround and ease of clinical implementation in hospitals performing FISH studies. We suggest the use of this 11-probe FISH panel to enhance the efficient testing of patients with L-CTCL to the standards of many other cancers of the blood, and suggest the potential utilization of FISH in personalized medicine for CTCL patients. Applying the panel to an expanded patient cohort will be necessary before the full utility, sensitivity, and specificity of FISH analysis in CTCL diagnostics can be conclusively determined. For investigative purposes, correlating GCNAs with expression levels and other biomarkers may reveal critical pathways underpinning disease behavior and new targets for therapeutic intervention. For clinical use, a practical and cost-effective strategy to consider would be to utilize a four probe subset consisting of the two most informative probes assessing deletion (TP53, ARID1A) and amplification (MYC, STAT3/5B), consistent with a previously published study (Vermeer et al., 2008). Combining exome data (Choi et al., 2015) with our patient cohort, GCNAs have been found present in this subset of genes in 96.2% of CTCL patients with stage B2 blood involvement (two-sided 95% Clopper Pearson confidence interval 87.0%–98.8%).
Supplementary Material
Acknowledgments
We thank Lesley Devine (Yale Cancer Center) for flow cytometric sorting, and Jennifer Boyle (Yale Genetics) for microscopy images. The study was supported by the Yale SPORE in Skin Cancer and NIH/NCI P50 CA121974 and the Drs Martin & Dorothy Spatz Charitable Foundation.
Abbreviations in this paper
- CTCL
cutaneous T cell lymphoma
- L-CTCL
leukemic cutaneous T cell lymphoma
- MF
mycosis fungoides
- F-MF
follicular mycosis fungoides
- MF-T
mycosis fungoides with tumor
- SS
Sézary syndrome
- FISH
fluorescence in situ hybridization
- GCNA
gene copy number alteration
- CGH
comparative genomic hybridization
- PBMC
peripheral blood mononuclear cells
- ISCL
International Society of Cutaneous Lymphoma
- IL-2
interleukin-2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Sites of experiments: New Haven, Connecticut, U.S.A.
CONFLICTS OF INTEREST:
JW, JL, and MG are listed as inventors on a provisional patent filed on several probes reported herein.
SUPPLEMENTARY MATERIAL:
Supplementary material is provided in a PDF submitted with this document.
References
- Choi J, Goh G, Walradt T, Hong BS, Bunick CG, Chen K, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet. 2015;47(9):1011–9. doi: 10.1038/ng.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson JF, Huang J, Liu KJ, Carlson KR, Foss F, Choi J, et al. Cutaneous T-cell lymphoma (CTCL): Current practices in blood assessment and the utility of T-cell receptor (TCR)-Vβ chain restriction. Journal of the American Academy of Dermatology. 2016;74(5):870–7. doi: 10.1016/j.jaad.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka T, Nakahata S, Hatakeyama K, Hamasaki M, Yamashita K, Kohno T, et al. Downregulation of TCF8 is involved in the leukemogenesis of adult T-cell leukemia/lymphoma. Blood. 2008;112(2):383–93. doi: 10.1182/blood-2008-01-131185. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Sahasrabuddhe AA, Rolland DC, Velusamy T, Chung F, Schaller M, et al. Genomic analyses reveal recurrent mutations in epigenetic modifiers and the JAK-STAT pathway in Sezary syndrome. Nat Commun. 2015;6:8470. doi: 10.1038/ncomms9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata S, Yamazaki S, Nakauchi H, Morishita K. Downregulation of ZEB1 and overexpression of Smad7 contribute to resistance to TGF-beta1-mediated growth suppression in adult T-cell leukemia/lymphoma. Oncogene. 2010;29(29):4157–69. doi: 10.1038/onc.2010.172. [DOI] [PubMed] [Google Scholar]
- Nuciforo P, Thyparambil S, Aura C, Garrido-Castro A, Vilaro M, Peg V, et al. High HER2 protein levels correlate with increased survival in breast cancer patients treated with anti-HER2 therapy. Mol Oncol. 2016;10(1):138–47. doi: 10.1016/j.molonc.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellas D, Szabolcs M, Koul S, Li Z, Polyzos A, Anagnostopoulos C, et al. Therapeutic effects of an anti-Myc drug on mouse pancreatic cancer. J Natl Cancer Inst. 2014;106(12) doi: 10.1093/jnci/dju320. [DOI] [PubMed] [Google Scholar]
- Van Dyke DL, Werner L, Rassenti LZ, Neuberg D, Ghia E, Heerema NA, et al. The Dohner fluorescence in situ hybridization prognostic classification of chronic lymphocytic leukaemia (CLL): the CLL Research Consortium experience. Br J Haematol. 2016 doi: 10.1111/bjh.13933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer MH, van Doorn R, Dijkman R, Mao X, Whittaker S, van Voorst Vader PC, et al. Novel and highly recurrent chromosomal alterations in Sezary syndrome. Cancer Res. 2008;68(8):2689–98. doi: 10.1158/0008-5472.CAN-07-6398. [DOI] [PubMed] [Google Scholar]
- Wang J, Lee S, Teh CE, Bunting K, Ma L, Shannon MF. The transcription repressor, ZEB1, cooperates with CtBP2 and HDAC1 to suppress IL-2 gene activation in T cells. Int Immunol. 2009;21(3):227–35. doi: 10.1093/intimm/dxn143. [DOI] [PubMed] [Google Scholar]
- Wiktor AE, Van Dyke DL, Stupca PJ, Ketterling RP, Thorland EC, Shearer BM, et al. Preclinical validation of fluorescence in situ hybridization assays for clinical practice. Genet Med. 2006;8(1):16–23. doi: 10.1097/01.gim.0000195645.00446.61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


