ABSTRACT
For therapeutic monoclonal antibodies (mAbs), detailed analysis of the structural integrity and heterogeneity, which results from multiple types of post-translational modifications (PTMs), is relevant to various processes, including product characterization, storage stability and quality control. Despite the recent rapid development of new bioanalytical techniques, it is still challenging to completely characterize the proteoform profile of a mAb. As a nearly indispensable tool in mAb analysis, mass spectrometry (MS) provides unique structural information at multiple levels. Here, we tested a hybrid strategy for the comprehensive characterization of micro-heterogeneity by integrating 2 state-of-the-art MS-based approaches, high-resolution native MS and targeted glycan profiling, to perform complementary analysis at the intact protein level and released glycan level, respectively. We compared the performance of these methods using samples of engineered half-body IgG4s and a panel of mAbs approved for human use. The glycosylation characterization data derived from these approaches were found to be mutually consistent in composition profiling, and complementary in identification and relative-quantitation of low-abundant uncommon glycoforms. In addition, multiple other sources of micro-heterogeneity, such as glycation, lack of glycosylation, and loss of light chains, could be detected by this approach, and the contribution of multiple types of modifications to the overall micro-heterogeneity could be assessed using our superposition algorithm. Our data demonstrate that the hybrid strategy allows reliable and thorough characterization of mAbs, revealing product characteristics that would easily be missed if only a single approach were used.
KEYWORDS: Bevacizumab, Eculizumab, glycosylation, IgG, Infliximab, mAb structural integrity, native mass spectrometry, Ofatumumab, Panitumumab, Rituximab, Trastuzumab
Introduction
Therapeutic monoclonal antibodies (mAbs) are inherently heterogeneous in structure due to numerous factors, including post-translational modifications (PTMs), incomplete processing, susceptibility to degradation, and disulfide shuffling.1-4 In particular, the glycosylation at Asn297 in the Fc-region contributes greatly to the micro-heterogeneity, affecting the conformation, half-life in serum, efficacy and safety of mAbs.5-7 The glycan chains stabilize the CH2 domain, and removal of certain carbohydrate residues often leads to conformational changes, a decrease in thermal stability, and loss of effector functions.8 For instance, removal of terminal galactose reduces complement-dependent cytotoxicity (CDC),9,10 while a decreased level of core fucosylation enhances antibody-dependent cell-mediated cytotoxicity (ADCC).11-13 Other modifications made to the protein backbone, such as C-terminal lysine clipping,14 deamidation, and oxidation also contribute to the heterogeneity of mAb products.15,16 In some cases, more substantial changes in the protein scaffold can be introduced by either incomplete processing or degradation,17 influencing the antigen binding capability18 or the in vivo clearance rate of mAbs.19 These biologic consequences make comprehensive characterizations of heterogeneity critical for the design, production and clinical use of mAbs.
Currently, mass spectrometry (MS)-based techniques are widely used for the analysis of mAb heterogeneity with special emphasis on glycosylation. It is technically possible to characterize mAb glycosylation at several levels: the intact protein level, the glycopeptide level and the released glycan level.20-24 MS analysis of released glycans is still the method of choice for obtaining structural information on the glycome. Glycan analysis allows for rapid, high-throughput characterization of mAb samples by matching the light chain retention time and accurate mass, providing in-depth structural information on the glycans, including even linkage details.25 Glycopeptide analysis provides simultaneous identification of the glycoproteins and their glycans, and localization, occupancy and micro-heterogeneity can be evaluated by using tandem mass spectrometry (MS/MS) techniques.20,24,26
Recently, site-specific glycosylation analysis of mAbs was shown to benefit from the sensitivity and specify achievable by targeted approaches using multiple reaction monitoring (MRM).27 At the other end of the spectrum, by directly analyzing the intact protein, it is possible to simultaneously and quantitatively profile the distribution of the main glycoproteoforms, which is an important indication for product integrity and consistency.28-30 Although these approaches have proven powerful in providing structural information, no single approach is sufficient for an in-depth characterization of all aspects of heterogeneity. In a recent comprehensive analysis of cetuximab, Ayoub et al. combined multiple schemes (intact analysis, middle-down, middle-up and bottom-up) to reveal distinct glycosylation profiles on the Fab and Fc region, as well as a sequence error in the reported sequence of the light chain.31 This study provided a good example of the benefit of integrating information at multiple levels in dissection of a mAb product.
Here, we combined 2 cutting-edge MS-based approaches, i.e., high-resolution native MS for the global profiling of co-existing proteoforms and targeted glycan analysis of released N-glycans for the structural analysis of individual glycoforms, to characterize 12 therapeutic mAbs, 7 of which are marketed. Combining the data obtained at the intact protein level and released glycan level, we were able to better resolve the overall heterogeneity exhibited by a mAb and quantitatively profile them with satisfactory confidence. In addition, other sources of micro-heterogeneity, such as incomplete lysine clipping, lack of glycosylation, loss of could be inferred simultaneously from the native MS data. The complementary structural information provided by the 2 approaches allow the micro-heterogeneity and structural integrity of mAbs to be characterized in unprecedented detail, useful for pharmaceutical development and mAb quality control, especially when these differentiated proteoforms exhibit difference in potency. In addition, our strategy can be used to detect structural alterations induced by storage-induced degradation of biopharmaceuticals.
Results
Comparative analysis of IgG4-Δhinge half-body N-glycosylation at the intact protein and released glycan levels
To evaluate the performance of native MS and targeted glycan profiling in the analysis of N-glycosylation at the intact protein level and released glycan level, we compared the data provided by these 2 approaches. Three IgG4-Δhinge mutants, i.e., F405T/Y407E, Y407Q and Y407K, were selected as model analytes because their defined half-molecule state harbors only one N-glycosylation site and thus allows unpaired glycan chains to be characterized at the intact protein level. Moreover, as suggested by previous studies,32 each of these 3 mutants exhibits high levels of galactose, sialic acids and branching, providing a wide range of glycoforms valuable for our method validation.
In the native MS measurements, signals corresponding to various glycoproteoforms could be baseline-resolved based on their unique molecular weight (MW) accurately measured using the Orbitrap EMR mass analyzer (Fig. 1A). Then, the composition of the corresponding species, i.e., the stoichiometry of carbohydrate residues, was calculated using the MW of individual residues. In targeted glycan profiling (Fig. 1B), the glycans released from the glycoproteins were identified by mapping their chromatographic elution profiles to those of the common glycosylations on commercial mAb products covered by a library.25 The glycan compositions were then further verified by the MW measured using the MS coupled with LC. Both approaches yielded results in aspects of accurate MW, glycan composition and relative abundance. To quantitatively compare the 2 data sets, for each mutant we plotted the relative abundances of detected glycans against their MW in a mirror-like fashion (Fig. 1C). Illustratively for IgG4-Δhinge-Y407Q, with 25 and 24 glycoforms mass-assigned by native MS and targeted glycan profiling, respectively, 14 species could be assigned by both approaches, accounting for 92% of the total glycan abundance (Fig. 1C; Table S2). The Pearson correlation coefficient (R) of the full-range mirror-plots is 0.84. In addition to the common glycoforms, the more sophisticated di- and tri-sialylated species such as 6,5,1,2 (annotated as numbers of Hex,HexNAc,Fuc,Sia incorporated, sic passim) and 6,5,1,3 were also detected in similar abundances by both approaches. Similar observations were made in analysis of Y407K and F405T/Y407E (see also Fig. 1). These distribution maps and the R values calculated for the comparisons suggest an overall good agreement between the 2 approaches in terms of detection and identification of the predominant glycoforms, as well as many low abundant ones.
Figure 1.

N-glycosylation on 3 IgG4-Δhinge mutants are quantitatively profiled at the intact protein and the released glycan level. (A) Deconvoluted native mass spectra of the intact IgG4-Δhinge proteins with all glycoproteoforms baseline-resolved, separated by their MW. Asterisks indicate observed glycine truncations in the mAb backbone. (B) Total ion current (TIC) chromatograms of the released glycans that are separated based on their chromatographic elution time. Signal peaks are color-coded using the same scheme as in (A). (C) Direct comparison of the relative abundances of glycans with different compositions determined by the 2 individual approaches, whereby the consistency between the 2 methods was evaluated using Pearson correlation scores. Quantification data of native MS has been adjusted for glycine truncation.
Between the 2 data sets, the discrepancies in abundances of certain glycoforms may partially be attributed to artifacts induced by either approach. Particularly, native MS reported higher abundances of most of the glycoforms containing 5 HexNAc residues compared with glycan profiling (Table S2), suggesting the potential presence of a systematic bias. In native MS, all glycoforms are separated and assigned solely based on MW, and thus the accuracy of quantitation for certain species may be compromised by the occasional overlapping of signals of different glycoforms whose MW difference is smaller than the peak widths, in spite of the instruments' resolving power. Roughly, in our native MS analysis a minimum MW difference of 20 Da is necessary for unambiguous assignments of different glycoforms. For instance, since the MW of glycoform G1 (4,4,0,0) is only 16 Da heavier than that of G0F (3,4,1,0), in the native MS data the signal peak of G1 are merged into that of G0F, resulting in an overestimated abundance of G0F, and false negative detection of G1 (Fig. 1; Table S2). In sharp contrast, targeted profiling provides the released glycans with more efficient separation (based on the chromatographic elution time and MW) and composition verification based on tandem MS patterns when necessary, allowing signals to be assigned with higher confidence. However, biases in the released glycan analysis may exist, for instance due to difference in ionization and detection response that may affect quantitation.36 Moreover, MS analysis of released glycans can result in undesired fragmentation,37 leading to diminished quantitative detection of the species of origin and false positive detection of fragments, especially if these fragment species are not covered by the reference library. As exemplified by the elution profiles of glycans released from F405T/Y407E (Fig. 1B), while 2 of the 5,4,1,2 species were eluted at 25.08 and 25.91 min, respectively, 2 minor species mass-assigned as 5,4,1,1 were observed at the same elution positions, implying that these 5,4,1,1 species were yielded by loss of a sialic acid residue from a subpopulation of 5,4,1,2 at the MS detection stage.
The efficient separation provided by the glycan profiling also allows the exact linkages of carbohydrate residues to be assigned, benefiting from the distinguishable LC-retention and gas-phase fragmentation behaviors exhibited by the structural isomers. As exemplified by the glycan G2FS (5,4,1,1), 3 structural isomers could be identified by matching with the accurate masses and the calibrated chromatographic retention times from the library (Table S3).
Resolving other types of modification enhancing mAb micro-heterogeneity
Next, using a similar workflow, we characterized 7 mAbs approved for human use with high resolution native MS to complement previously performed analyses of their released glycans.25 Use of intact protein samples instead of the released glycans increases not only the total MW of the analytes, but also increases the overall heterogeneity of the sample molecules due to the presence of other modifications, as exemplified in Fig. 2. To best extract the non-N-glycosylation-induced heterogeneity, we additionally measured the N-deglycosylated format of these intact mAbs. As illustrated by 3 examples, mAb backbones can undergo different types of modification that can occur to various extents (Fig. 2). Upon removal of the N-glycan, trastuzumab exhibits a quite uniform MW, suggesting minimal other modifications, and complete C-terminal Lys processing (Fig. 2A). In sharp contrast, both infliximab and bevacizumab exhibit a broad range of intact masses, resulting largely from incomplete C-terminal Lys processing and multiple glycation events, respectively (Fig. 2B, C).
Figure 2.
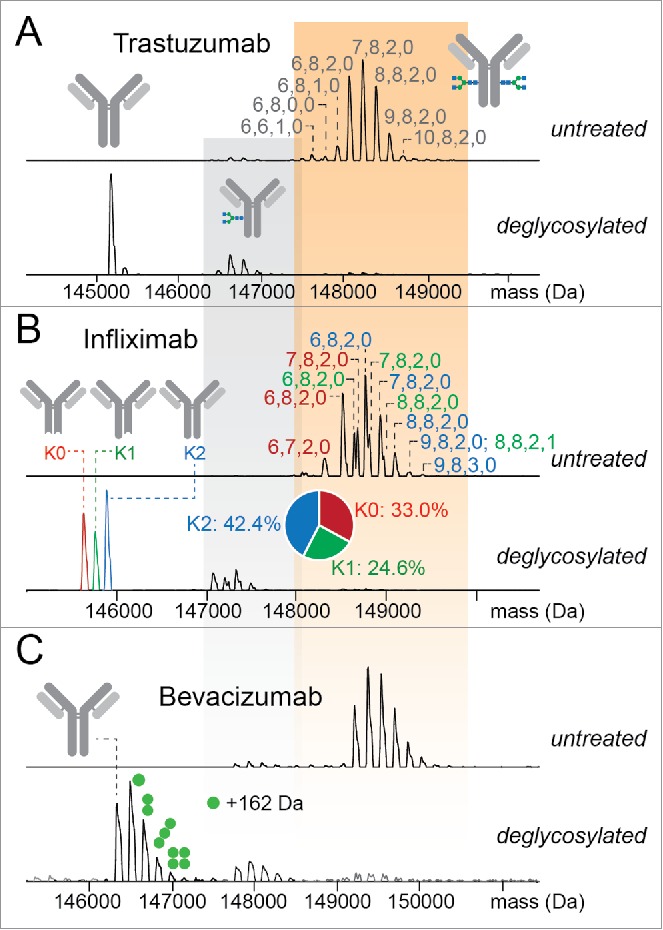
Resolving N-glycosylation and other types of modification increasing mAb micro-heterogeneity on therapeutic mAbs. (A) Deconvoluted native MS spectra of N-glycosylated and deglycosylated trastuzumab, revealing only minor modifications besides N-glycosylation. (B) Deconvoluted native MS spectra of infliximab revealing incomplete C-terminal lysine processing of this mAb. The annotations of the glycoforms are color-coded according to the number of clipped Lysine residues. (C) Deconvoluted native MS spectra of bevacizumab suggesting 1–4 hexose residues are still attached after N-deglycosylation revealing the occurrence of extensive glycation (annotated with the green dots).
Taking advantage of its relatively monodisperse protein backbone, the glycosylation profiling for trastuzumab appears to be nearly as straightforward as for the IgG4-Δhinge mutants. However, as the carbohydrate content of the intact IgG1 molecule originates from 2 glycan chains, the analysis at the glycan level is needed to determine the detailed glycan composition. Based on the MW and relative abundances of individual N-glycan chains determined at the released glycan level, we simulated the distribution of MW of the intact mAb assuming random pairing of the 2 glycan chains, and compared it with the experimentally measured native MS spectrum. In this spectrum 14 unique MWs corresponding to different glycan compositions could be assigned (Fig. 2A, Fig. S1), including both abundant glycans, such as 6,8,2,0 (G0F/G0F), 7,8,2,0 (G0F/G1F), 8,8,2,0 (G0F/G2F or G1F/G1F), and lower abundant glycans, such as 6,6,0,0 to 10,8,2,2. As illustrated in Fig. 3A, a high correlation coefficient (R = 0.92) was obtained between the 2 sets of data, demonstrating the validity of direct profiling of the paired glycan composition on IgGs by native MS. However, since in the native MS data a single composition may originate from multiple possible combinations of glycan pairs, analysis at the intact protein level alone is insufficient to determine the contribution of each pair. Also, it is noteworthy that the assumption of random glycan pairing we made for the simulation may not represent the actual situation,38 which could contribute to the remaining differences in abundance between the measured and simulated data sets.
Figure 3.

Comparison of experimental and simulated spectra. For trastuzumab (A) and bevacizumab (B), the experimental deconvoluted intact protein mass spectra (blue) are compared with the simulated spectra, based on the detected quantitative profile of N-glycosylation and/or other backbone modifications (red). The correlation between data sets is evaluated by means of Pearson Correlation and is 0.92 for trastuzumab and 0.95 for bevacizumab. For bevacizumab, simulation excluding the contribution of glycation (white) results in a significantly lower correlation coefficient (R = 0.69).
Clipping of the C-terminal Lys occurs to a variable extent in most mAb products. Although manufacturers usually consider these variants to have no considerable effect on the potency or safety profile, a recent study demonstrated that C-terminal Lys may interfere with the ordered oligomerization of IgG at the cell surface, leading to suboptimal C1q binding and CDC of opsonized cells.14 Providing an alternative to charge-based separation techniques such as isoelectric focusing (IEF) electrophoresis (either capillary- or slab-based) and ion exchange chromatography,39,40 MS analysis of deglycosylated mAb allows simple evaluation of the amount of C-terminal Lys retained (0, 1 or 2, annotated as K0, K1 and K2, respectively). Unlike trastuzumab whose C-terminal Lysine residues were completely processed (i.e., only K0), infliximab displayed K0, K1 and K2 variants in comparable abundances (Fig. 2B, lower panel), resulting in a relatively complicated mass spectrum even in its deglycosylated form (Fig. 2B, upper panel). Based on the MW and distribution of the K0, K1 and K2 backbones, we were able to determine the compositions and abundances of the N-glycan chain pairs attached to infliximab, by subtracting the contributions of K0, K1 and K2 backbones according to their ratios (Fig. 2B, upper panel). The extracted quantitative profile of infliximab is consistent with a previous study using capillary electrophoresis (CE),41 demonstrating the comparable effectiveness of native MS in separating charge variants of mAbs.
Glycation caused by exposure of mAb to reducing sugars is another common type of modification that occurs either under physiologic conditions or during cell culture.42,43 In many cases, glycation alters the charge profile of pharmaceutical products and may thus affect its quality.44,45 Because the net contribution of a monosaccharide to the total MW of protein is equivalent regardless of its site of attachment through a glycan chain or to the backbone, in the intact-MW analysis the removal of N-glycosylation is required to distinguish the glycation from the extended N-glycosylation. Indeed, although the glycosylated format of bevacizumab showed a profile similar in heterogeneity to trastuzumab, removal of the N-glycans demonstrated a significantly higher extent of glycation for bevacizumab (∼1.3 copies of hexose residues on average) (Fig. 2C, lower panel).
As a non-enzymatic process, glycation introduces a reducing sugar at a primary amine on the protein, either the lysine (Lys) side-chain epsilon amino group or the α-amino N-terminus.46,47 While the precise localization of glycation sites requires the analysis at the peptide level using a bottom-up approach,48-51 the distribution of glycated mAbs and the average glycation level are unique information provided by analysis at the intact protein level. Based on the distribution of glycan chains profiled at the glycan level, the MW of protein backbone and the glycation profile measured at the intact protein level, we simulated a profile reflecting the overall heterogeneity including contributions from both glycosylation and glycation, and compared the simulated profile with the experimental profile (Fig. 3B). The consistent patterns of the profile envelopes (R = 0.95) demonstrates the accuracy of profiling results of individual types of modifications, and the validity of reconstitution of overall heterogeneity using these data. We made another simulation, now excluding glycation, as shown in Fig. 3B. The resulting simulated spectrum exhibited a distinct peak distribution compared with the experimental MS data and the correlation decreased from 0.95 to 0.69, demonstrating the effect of glycation on the overall heterogeneity of this mAb molecule.
Monitoring the global structural integrity of mAbs
Because some of the studied mAbs had been stored for extended periods and their expiration date had passed, we examined the global stability and structural integrity of the samples, using native MS as primary read-out. Next to the full mAb structures (denoted L(2)H(2)g(2) where L, H and g denote the light, heavy, and glycan chains, respectively) we also detected mAb-like products lacking the light chains or one or 2 glycan chains. Even for the structurally most stable trastuzumab, which exhibited the least structural heterogeneity of the mAbs studied here, we detected a low abundant species lacking one full N-glycan chain (L(2)H(2)g(1); Fig. 4A). The profile of the retained N-glycan chain resembles the distribution of the ensemble of glycan chains profiled for the overall population of trastuzumab (Fig. S2). Such single site-glycosylation was detected for all investigated mAbs, with the relative abundance ranging from 0.8% to 2.8% (Table 1). Singly glycosylated IgGs have been shown to display a decrease in thermal stability, C1q binding affinity, Fc gamma receptors (FcγRs) binding affinity and ADCC.52
Figure 4.
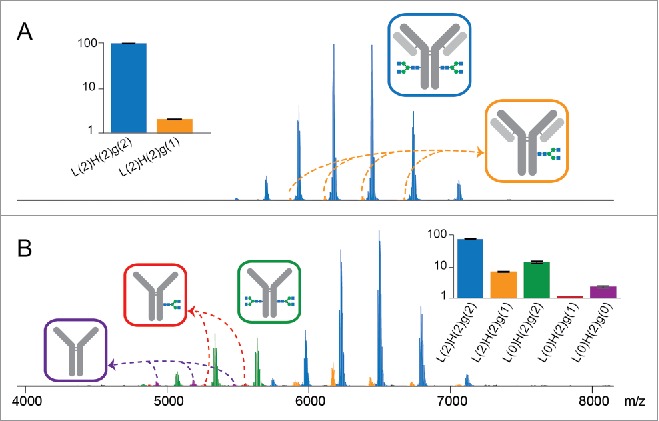
mAb structural integrity probed by native MS at the intact protein level. (A) Native mass spectrum of trastuzumab, which populates predominantly the intact format L(2)H(2)g(2) (blue; L, H and g denote light chain, heavy chain and N-glycan chain, respectively, with numbers indicated in the parentheses) and marginally the single site-glycosylated format L(2)H(2)g(1) (orange). (B) For bevacizumab, in addition to the 2 formats detected in (A), the light-chain-free species with 2, 1 and 0 N-glycan chains attached (green, red and purple, respectively) are detected. Relative abundances of each species are estimated using summed ion intensities corresponding to all detected charge states, with data normalized to the ensemble of species and plotted on a logarithmic scale in the insets.
Table 1.
Key characteristics of the therapeutic mAbs investigated.
| ID | Number of glycan chains | Number of glycoforms assigneda | Single site-glycosylation (%) | Light chain loss | C-terminal K retainedb (%) | Glycation levelc |
|---|---|---|---|---|---|---|
| Panitumumab | 2 | 12 | 2.8 | ND | ND | < 0.05 |
| Rituximab | 2 | 19 | 1.6 | ND | ND | < 0.05 |
| Bevacizumab | 2 | — | 1.6 | 17.3 | ND | 1.3 |
| Trastuzumab | 2 | 14 | 2.0 | ND | ND | < 0.05 |
| Infliximab | 2 | — | 0.8 | ND | 54.7 | < 0.05 |
| Ofatumumab | 2 | 12 | 1.9 | 17.7 | ND | < 0.05 |
| Eculizumab | 2 | 11 | 1.6 | ND | ND | < 0.05 |
Number of glycoforms assigned by the complementary approaches. Confident assignments of Bevacizumab and Infliximab via measurements at the intact level are hampered by the co-occurence and interference with other backbone modifications.
Calculated as , where is the abundance of species.
Reported as the average copies of monosaccharide residues attached to the mAb. A minimum abundance of 5% of the singly glycated species (relative to the non-glycated species) was set as the threshold for calculation.
Among the mAb samples stored for long periods, bevacizumab and ofatumumab exhibited additional partial loss of their light chains. For bevacizumab, we detected species missing both light chains, with 2, 1 or 0 N-glycan chains attached. Notably, in both cases, no mAb molecules lacking only one light chain were observed, suggesting the removal of the light chains is not a random or sequential process (Fig. 4B). Species of low abundance lacking both light chains were also detected in ofatumumab, yet no further glycan loss was observed (Fig. S3). In healthy mammals, failure of light chain association usually results in rapid intracellular degradation of the heavy chains due to the inefficient transport from ER to Golgi,53,54 preventing the expression of heavy chain antibodies (L(0)H(2)). The production of natural L(0)H(2) with VH/CH1 domains partially removed was found in humans with H chain disease.55 All the L(0)H(2) species observed in this work retained heavy chain integrity, implying that the absence of light chains is likely the result of degradation. The exact nature of the reactions responsible for such release of the light chain remains to be investigated. We rule out in-source dissociation of the products during the MS measurement, since no free light chain ions were detected under the gentle ionization conditions of data acquisition. In addition, the charge density carried by the L(0)H(2) species is in line with the empirical expectation,56 suggesting these species are not the products of gas phase fragmentation.57 Native MS provides a convenient means to characterize the heterogeneity-inducing degradation products of proteins in solution without any front-end separation.
Discussion
Here, we demonstrated that glycoprotein analysis at the intact protein and released glycan level using high-resolution native MS and targeted glycan profiling, respectively, can provide mutually consistent information on the composition and abundance of glycoprotein proteoforms. However, careful optimization and proper data processing are needed to minimize the artifacts induced in either approach. Both methods provide highly complementary data, but also have their own niche. The multiple-dimensional separation of glycoforms and the tandem MS data gathered by targeted profiling of released glycans enables the differentiation of structural isomers, as well as elucidation of the linkages within carbohydrate residues. Native MS analysis at the intact protein level provides a global snapshot of the glycoform distribution with high confidence, and can concomitantly reveal co-occurring modification on the protein, such as truncations and glycations.
In the combined analysis of a sample panel of model proteins that included aged therapeutic mAbs, we demonstrated the power of using these hybrid MS-based approaches concomitantly. The combined analysis provided detailed structural information, both qualitative and quantitative, and better resolved the overall heterogeneity caused not only by N-glycosylation, but also by C-terminal Lys processing, glycation, single site-glycosylation, and loss of specific polypeptide chains. A comparison of simulated and experimental data, using dedicated mathematical algorithms, allowed the heterogeneity to be further assessed and cross-validated. Both approaches provide reasonable high-throughput because they are compatible with automated sampling, and neither of them requires any time-consuming stages. Therefore, we conclude that the combination of these 2 workflows will provide an ideal platform to measure micro-heterogeneity, not only in IgGs, but also in all other kind of industrial and plasma glycoproteins.
Materials & methods
Materials
The monoclonal hinge-deleted IgG4 antibody (IgG4-Δhinge-WT) and its mutants (Y407A, Y407E, Y407Q and Y407K) used in this study were gifts from Genmab (Utrecht, The Netherlands). These proteins were expressed in HEK-293F cells and purified as described previously.32 The 7 therapeutic mAbs used in this work (with specifications are listed in Table S1) were kind gifts from the UC Davis Medical Center, all representing expired batches (lot number unknown). PNGase F was obtained from Roche (Indianapolis, USA). Dithiothreitol (DTT) was purchased from Sigma-Aldrich (Steinheim, Germany).
High-resolution native MS analysis using an Orbitrap EMR
Prior to native MS measurement, the aqueous environment of all mAb stock solutions (IgG4-Δhinge antibodies were prepared in PBS; the powder of therapeutic mAbs was reconstituted in Milli-Q water) were substituted with 150 mM aqueous ammonium acetate (pH adjusted to 7.5 using ammonium hydroxide) by ultrafiltration using a 10 kDa cut-off filter (Merck Millipore, Germany). Protein concentrations were measured by UV absorbance at 280 nm and adjusted to 2–3 µM for native MS injection. PNGase F was used to remove the N-glycans following the procedure supplied by the manufacturer.
1–3 uL of samples loaded in home-made gold-coated borosilicate capillaries were directly infused into an Exactive Plus Orbitrap instrument with extended mass range (EMR) (Thermo Fisher Scientific, Germany) using an m/z range of 500–10,000 Th. The voltage offsets on transport multi-poles and ion lenses were manually tuned to achieve optimal transmission of protein ions at elevated m/z. Nitrogen was infused in the higher-energy collisional dissociation (HCD) cell at a gas pressure of 6–8 × 10−10 bar. MS parameters were adjusted as follows: spray voltage 1.2–1.3 V; source fragmentation 30 V; source temperature 250°C; collision energy 30 V; resolution (at m/z 200) 17,500. The instrument was mass-calibrated using CsI clusters, as described previously.33
The raw mass spectra were deconvoluted using Protein Deconvolution v2.0 (Thermo Fisher Scientific, Germany) and BioAnalyst v1.1 (AB/Sciex, Canada). The deconvoluted data were further processed as described previously to solve the composition of glycosylation and other possible PTMs.29 Average MW values of individual carbohydrate residues used for MW calculation include: hexose/mannose/galactose (Hex/Man/Gal) – 162.1424 Da; N-acetylhexosamine/N-acetylglucosamine (HexNAc) – 203.1950 Da; deoxyhexose (Fuc) – 146.1430 Da; N-acetylneuraminic acid (Neu5Ac, Sia) – 291.2579 Da; N-glycolylneuraminic acid (Neu5Gc) – 307.2573 Da.
Targeted N-glycan profiling using nanoLC-Chip-Q-TOF MS
N-glycans were released from the proteins before the analysis using a nanoLC-chip-Q-TOF MS system. The release, purification, injection, data acquisition and analysis of N-glycan were performed following the described previously protocols.25 In brief, N-glycans were released from reduced mAb by PNGase F digestion, followed by solid phase extraction (SPE) purification, chemical reduction by NaBH4, and SPE again for the removal of reductant. Purified N-glycans were then loaded to Agilent nanoLC-chip-Q-TOF analyzed with the exact same condition of a previously reported human serum N-glycan structural library. A mixture of serum N-glycans standard was prepared and analyzed together with mAb N-glycans to calibrate chromatographic retention times in the serum N-glycan library as described previously.34 Structures of N-glycan were identified by matching with accurate masses and calibrated chromatographic retention times from the library.
Intergrative data construction of pseudo native MS spectrum
We perform in silico data construction to simulate a zero-charge, native MS spectrum based on the masses and relative abundances of glycans identified using targeted glycan profiling, the basic principle of which has been described previously.35 The data construction is achieved based on the following 3 elements: 1) the mass of the mAb backbone; 2) the masses and relative abundance of the glycans identified by targeted glycan profiling; and 3) extra modifications on mAb backbone.
To compare the simulated result with the native MS spectra, the native MS spectra (acquired in profile mode) are pre-processed by convoluting the ESI spectrum to a zero-charge state spectrum using the Protein Deconvolution package (Thermo Fisher Scientific). Subsequently, the standard Pearson correlation is performed to evaluate the data generated from the 2 platforms.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Y.Y. and A.J.R.H. are supported by the EU funded ITN project ManiFold, grant 317371. A.J.R.H. is further supported by the European Union Horizon 2020 program FET-OPEN project MSmed, grant 686547, and the research and innovation program RELENT, grant 668036. This work was further supported by The Netherlands Organization for Scientific Research (NWO) via the Roadmap Initiative Proteins@Work (project number 184.032.201), and the TOP-Punt Grant 718.015.003 for A.J.R.H.
References
- 1.Walsh G, Jefferis R. Post-translational modifications in the context of therapeutic proteins. Nat Biotechnol 2006; 24:1241-52; PMID:17033665; http://dx.doi.org/ 10.1038/nbt1252 [DOI] [PubMed] [Google Scholar]
- 2.Wang W, Singh S, Zeng DL, King K, Nema S. Antibody Structure, Instability, and Formulation. J Pharm Sci 2007; 96:1-26; PMID:16998873; http://dx.doi.org/ 10.1002/jps.20727 [DOI] [PubMed] [Google Scholar]
- 3.Beck A, Wurch T, Bailly C, Corvaia N. Strategies and challenges for the next generation of therapeutic antibodies. Nat Rev Immunol 2010; 10:345-52; PMID:20414207; http://dx.doi.org/ 10.1038/nri2747 [DOI] [PubMed] [Google Scholar]
- 4.Beck A, Wagner-Rousset E, Ayoub D, Van Dorsselaer A, Sanglier-Cianférani S. Characterization of therapeutic antibodies and related products. Anal Chem 2013; 85:715-36; PMID:23134362; http://dx.doi.org/ 10.1021/ac3032355 [DOI] [PubMed] [Google Scholar]
- 5.Jefferis R. Glycosylation of recombinant antibody therapeutics. Biotechnol Prog 2005; 21:11-6; PMID:15903235; http://dx.doi.org/ 10.1021/bp040016j [DOI] [PubMed] [Google Scholar]
- 6.Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The Impact of glycosylation on the biological function and structure of human Immunoglobulins. Annu Rev Immunol 2007; 25:21-50; PMID:17029568; http://dx.doi.org/ 10.1146/annurev.immunol.25.022106.141702 [DOI] [PubMed] [Google Scholar]
- 7.Sha S, Agarabi C, Brorson K, Lee D-Y, Yoon S. N-Glycosylation design and control of therapeutic monoclonal antibodies. Trends Biotechnol 2016; Oct;34(10):835-46; PMID:27016033; http://dx.doi.org/22123061 10.1016/j.tibtech.2016.02.013 [DOI] [PubMed] [Google Scholar]
- 8.Zheng K, Bantog C, Bayer R. The impact of glycosylation on monoclonal antibody conformation and stability. mAbs 2011; 3:568-76; PMID:22123061; http://dx.doi.org/ 10.4161/mabs.3.6.17922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyd PN, Lines AC, Patel AK. The effect of the removal of sialic acid, galactose and total carbohydrate on the functional activity of Campath-1H. Mol Immunol 1995; 32:1311-8; PMID:8643100; http://dx.doi.org/ 10.1016/0161-5890(95)00118-2 [DOI] [PubMed] [Google Scholar]
- 10.Raju TS. Terminal sugars of Fc glycans influence antibody effector functions of IgGs. Curr Opin Immunol 2008; 20:471-8; PMID:18606225; http://dx.doi.org/ 10.1016/j.coi.2008.06.007 [DOI] [PubMed] [Google Scholar]
- 11.Shields RL, Lai J, Keck R, O'Connell LY, Hong K, Meng YG, Weikert SHA, Presta LG. Lack of fucose on human IgG1 N-Linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. J Biol Chem 2002; 277:26733-40; PMID:11986321; http://dx.doi.org/ 10.1074/jbc.M202069200 [DOI] [PubMed] [Google Scholar]
- 12.Shinkawa T, Nakamura K, Yamane N, Shoji-Hosaka E, Kanda Y, Sakurada M, Uchida K, Anazawa H, Satoh M, Yamasaki M, et al.. The absence of fucose but not the presence of Galactose or Bisecting N-Acetylglucosamine of Human IgG1 Complex-type Oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J Biol Chem 2003; 278:3466-73; PMID:12427744; http://dx.doi.org/ 10.1074/jbc.M210665200 [DOI] [PubMed] [Google Scholar]
- 13.Ferrara C, Grau S, Jäger C, Sondermann P, Brünker P, Waldhauer I, Hennig M, Ruf A, Rufer AC, Stihle M, et al.. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcγRIII and antibodies lacking core fucose. Proc Natl Acad Sci 2011; 108:12669-74; PMID:21768335; http://dx.doi.org/ 10.1073/pnas.1108455108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van den Bremer ET, Beurskens FJ, Voorhorst M, Engelberts PJ, de Jong RN, van der Boom BG, Cook EM, Lindorfer MA, Taylor RP, van Berkel PH, et al.. Human IgG is produced in a pro-form that requires clipping of C-terminal lysines for maximal complement activation. mAbs 2015; 7:672-80; PMID:26037225; http://dx.doi.org/ 10.1080/19420862.2015.1046665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houde D, Peng Y, Berkowitz SA, Engen JR. Post-translational modifications differentially affect IgG1 conformation and receptor binding. Mol Cell Proteomics 2010; 9:1716-28; PMID:20103567; http://dx.doi.org/ 10.1074/mcp.M900540-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brorson K, Jia AY. Therapeutic monoclonal antibodies and consistent ends: terminal heterogeneity, detection, and impact on quality. Curr Opin Biotechnol 2014; 30:140-6; PMID:25022603; http://dx.doi.org/ 10.1016/j.copbio.2014.06.012 [DOI] [PubMed] [Google Scholar]
- 17.Ackermann M, Jäger V, Marx U. Influence of cell- and media-derived factors on the integrity of a human monoclonal antibody after secretion into serum-free cell culture supernatants. Biotechnol Bioeng 1995; 45:97-106; PMID:18623090; http://dx.doi.org/ 10.1002/bit.260450202 [DOI] [PubMed] [Google Scholar]
- 18.Li F, Vijayasankaran N, Shen A, Kiss R, Amanullah A. Cell culture processes for monoclonal antibody production. mAbs 2010; 2:466-77; PMID:20622510; http://dx.doi.org/ 10.4161/mabs.2.5.12720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiorella BL, Winkelhake J, Young J, Moyer B, Bauer R, Hora M, Andya J, Thomson J, Patel T, Parekh R. Effect of culture conditions on IgM antibody structure, pharmacokinetics and activity. Biotechnol Nat Publ Co 1993; Mar;11(3):387-92; PMID:7763441; http://dx.doi.org/19212958 10.1038/nbt0393-387 [DOI] [PubMed] [Google Scholar]
- 20.Huhn C, Selman MHJ, Ruhaak LR, Deelder AM, Wuhrer M. IgG glycosylation analysis. Proteomics 2009; 9:882-913; PMID:19212958; http://dx.doi.org/ 10.1002/pmic.200800715 [DOI] [PubMed] [Google Scholar]
- 21.Berkowitz SA, Engen JR, Mazzeo JR, Jones GB. Analytical tools for characterizing biopharmaceuticals and the implications for biosimilars. Nat Rev Drug Discov 2012; 11:527-40; PMID:22743980; http://dx.doi.org/ 10.1038/nrd3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beck A, Sanglier-Cianférani S, Van Dorsselaer A. Biosimilar, biobetter, and next generation antibody characterization by mass spectrometry. Anal Chem 2012; 84:4637-46; PMID:22510259; http://dx.doi.org/ 10.1021/ac3002885 [DOI] [PubMed] [Google Scholar]
- 23.Zauner G, Selman MHJ, Bondt A, Rombouts Y, Blank D, Deelder AM, Wuhrer M. Glycoproteomic analysis of antibodies. Mol Cell Proteomics MCP 2013; 12:856-65; PMID:23325769; http://dx.doi.org/ 10.1074/mcp.R112.026005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Cui W, Gross ML. Mass spectrometry for the biophysical characterization of therapeutic monoclonal antibodies. FEBS Lett 2014; 588:308-17; PMID:24291257; http://dx.doi.org/ 10.1016/j.febslet.2013.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song T, Ozcan S, Becker A, Lebrilla CB. In-Depth method for the characterization of glycosylation in manufactured recombinant monoclonal antibody drugs. Anal Chem 2014; 86:5661-6; PMID:24828102; http://dx.doi.org/ 10.1021/ac501102t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Li X, Liu Y-H, Richardson D, Li H, Shameem M, Yang X. Simultaneous monitoring of oxidation, deamidation, isomerization, and glycosylation of monoclonal antibodies by liquid chromatography-mass spectrometry method with ultrafast tryptic digestion. mAbs 2016; 8:1477-86; PMID:27598507; http://dx.doi.org/ 10.1080/19420862.2016.1226715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang N, Goonatilleke E, Park D, Song T, Fan G, Lebrilla CB. Quantitation of site-specific glycosylation in manufactured recombinant monoclonal antibody drugs. Anal Chem 2016; 88:7091-100; PMID:27311011; http://dx.doi.org/ 10.1021/acs.analchem.6b00963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosati S, Yang Y, Barendregt A, Heck AJR. Detailed mass analysis of structural heterogeneity in monoclonal antibodies using native mass spectrometry. Nat Protoc 2014; 9:967-76; PMID:24675736; http://dx.doi.org/ 10.1038/nprot.2014.057 [DOI] [PubMed] [Google Scholar]
- 29.Rosati S, van den Bremer ETJ, Schuurman J, Parren PWHI, Kamerling JP, Heck AJR. In-depth qualitative and quantitative analysis of composite glycosylation profiles and other micro-heterogeneity on intact monoclonal antibodies by high-resolution native mass spectrometry using a modified Orbitrap. mAbs 2013; 5:917-24; PMID:23995615; http://dx.doi.org/ 10.4161/mabs.26282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsons TB, Struwe WB, Gault J, Yamamoto K, Taylor TA, Raj R, Wals K, Mohammed S, Robinson CV, Benesch JLP, et al.. Optimal synthetic glycosylation of a therapeutic antibody. Angew Chem Int Ed Engl 2016; 55:2361-7; PMID:26756880; http://dx.doi.org/ 10.1002/anie.201508723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ayoub D, Jabs W, Resemann A, Evers W, Evans C, Main L, Baessmann C, Wagner-Rousset E, Suckau D, Beck A. Correct primary structure assessment and extensive glyco-profiling of cetuximab by a combination of intact, middle-up, middle-down and bottom-up ESI and MALDI mass spectrometry techniques. mAbs 2013; 5:699-710; PMID:23924801; http://dx.doi.org/ 10.4161/mabs.25423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose RJ, Berkel PHC, van den Bremer ETJ, Labrijn AF, Vink T, Schuurman J, Heck AJR, Parren PWHI. Mutation of Y407 in the CH3 domain dramatically alters glycosylation and structure of human IgG. mAbs 2013; 5:219-28; PMID:23406897; http://dx.doi.org/ 10.4161/mabs.23532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rose RJ, Damoc E, Denisov E, Makarov A, Heck AJR. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat Methods 2012; 9:1084-6; PMID:23064518; http://dx.doi.org/ 10.1038/nmeth.2208 [DOI] [PubMed] [Google Scholar]
- 34.Song T, Aldredge D, Lebrilla CB. A method for in-depth structural annotation of human serum glycans that yields biological variations. Anal Chem 2015; 87:7754-62; PMID:26086522; http://dx.doi.org/ 10.1021/acs.analchem.5b01340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, Liu F, Franc V, Halim LA, Schellekens H, Heck AJR. Hybrid mass spectrometry approaches in glycoprotein analysis and their usage in scoring biosimilarity. Nat Commun 2016; 7:13397; PMID:27824045; http://dx.doi.org/ 10.1038/ncomms13397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cech NB, Enke CG. Practical implications of some recent studies in electrospray ionization fundamentals. Mass Spectrom Rev 2001; 20:362-87; PMID:11997944; http://dx.doi.org/ 10.1002/mas.10008 [DOI] [PubMed] [Google Scholar]
- 37.Bondarenko PV, Second TP, Zabrouskov V, Makarov AA, Zhang Z. Mass measurement and top-down HPLC/MS analysis of intact monoclonal antibodies on a hybrid linear quadrupole ion trap-Orbitrap mass spectrometer. J Am Soc Mass Spectrom 2009; 20:1415-24; PMID:19409810; http://dx.doi.org/ 10.1016/j.jasms.2009.03.020 [DOI] [PubMed] [Google Scholar]
- 38.Masuda K, Yamaguchi Y, Kato K, Takahashi N, Shimada I, Arata Y. Pairing of oligosaccharides in the Fc region of immunoglobulin G. FEBS Lett 2000; 473:349-57; PMID:10818239; http://dx.doi.org/ 10.1016/S0014-5793(00)01557-X [DOI] [PubMed] [Google Scholar]
- 39.Antes B, Amon S, Rizzi A, Wiederkum S, Kainer M, Szolar O, Fido M, Kircheis R, Nechansky A. Analysis of lysine clipping of a humanized Lewis-Y specific IgG antibody and its relation to Fc-mediated effector function. J Chromatogr B 2007 Jun 1; 852(1-2):250-6; PMID:17296336; http://dx.doi.org/18553400 10.1016/j.jchromb.2007.01.024 [DOI] [PubMed] [Google Scholar]
- 40.Dick LW, Qiu D, Mahon D, Adamo M, Cheng K-C. C-terminal lysine variants in fully human monoclonal antibodies: Investigation of test methods and possible causes. Biotechnol Bioeng 2008; 100:1132-43; PMID:18553400; http://dx.doi.org/ 10.1002/bit.21855 [DOI] [PubMed] [Google Scholar]
- 41.Redman EA, Batz NG, Mellors JS, Ramsey JM. Integrated microfluidic capillary electrophoresis-electrospray ionization devices with online MS detection for the separation and characterization of intact monoclonal antibody variants. Anal Chem 2015; 87:2264-72; PMID:25569459; http://dx.doi.org/ 10.1021/ac503964j [DOI] [PubMed] [Google Scholar]
- 42.Lapolla A, Fedele D, Aronica R, Garbeglio M, D'Alpaos M, Seraglia R, Traldi P. The in vivo glyco-oxidation of alpha- and beta-globins investigated by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom RCM 1996; 10:1133-5; PMID:8755240; http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 43.Lapolla A, Tonani R, Fedele D, Garbeglio M, Senesi A, Seraglia R, Favretto D, Traldi P. Non-enzymatic glycation of IgG: an in vivo study. Horm Metab Res Horm Stoffwechselforschung Horm Métabolisme 2002 May; 34(5):260-4; PMID:12063640; http://dx.doi.org/12540626 10.1055/s-2002-32140 [DOI] [PubMed] [Google Scholar]
- 44.Hunter SJ, Boyd AC, O'Harte FPM, McKillop AM, Wiggam MI, Mooney MH, McCluskey JT, Lindsay JR, Ennis CN, Gamble R, et al.. Demonstration of glycated insulin in human diabetic plasma and decreased biological activity assessed by euglycemic-hyperinsulinemic clamp technique in humans. Diabetes 2003; 52:492-8; PMID:12540626; http://dx.doi.org/ 10.2337/diabetes.52.2.492 [DOI] [PubMed] [Google Scholar]
- 45.Banks DD, Hambly DM, Scavezze JL, Siska CC, Stackhouse NL, Gadgil HS. The effect of sucrose hydrolysis on the stability of protein therapeutics during accelerated formulation studies. J Pharm Sci 2009; 98:4501-10; PMID:19388069; http://dx.doi.org/ 10.1002/jps.21749 [DOI] [PubMed] [Google Scholar]
- 46.Garlick RL, Mazer JS. The principal site of nonenzymatic glycosylation of human serum albumin in vivo. J Biol Chem 1983; May 25;258(10):6142-6; PMID:685348012042244 [PubMed] [Google Scholar]
- 47.Shilton BH, Walton DJ. Sites of glycation of human and horse liver alcohol dehydrogenase in vivo. J Biol Chem 1991; Mar 25;266(9):5587-92; PMID:200509912042244 [PubMed] [Google Scholar]
- 48.Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 2002; 12:43R-56R; PMID:12042244; http://dx.doi.org/ 10.1093/glycob/12.4.43R [DOI] [PubMed] [Google Scholar]
- 49.Niwa T. Mass spectrometry for the study of protein glycation in disease. Mass Spectrom Rev 2006; 25:713-23; PMID:16526005; http://dx.doi.org/ 10.1002/mas.20089 [DOI] [PubMed] [Google Scholar]
- 50.Saleem RA, Affholter BR, Deng S, Campbell PC, Matthies K, Eakin CM, Wallace A. A chemical and computational approach to comprehensive glycation characterization on antibodies. mAbs 2015; 7:719-31; PMID:26030340; http://dx.doi.org/ 10.1080/19420862.2015.1046663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Viski K, Gengeliczki Z, Lenkey K, Baranyáné Ganzler K. Parallel development of chromatographic and mass-spectrometric methods for quantitative analysis of glycation on an IgG1 monoclonal antibody. J Chromatogr B Analyt Technol Biomed Life Sci 2016; 1032:198-204; PMID:27179993; http://dx.doi.org/ 10.1016/j.jchromb.2016.04.043 [DOI] [PubMed] [Google Scholar]
- 52.Ha S, Ou Y, Vlasak J, Li Y, Wang S, Vo K, Du Y, Mach A, Fang Y, Zhang N. Isolation and characterization of IgG1 with asymmetrical Fc glycosylation. Glycobiology 2011; 21:1087-96; PMID:21470983; http://dx.doi.org/ 10.1093/glycob/cwr047 [DOI] [PubMed] [Google Scholar]
- 53.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. Naturally occurring antibodies devoid of light chains. Nature 1993; 363:446-8; PMID:8502296; http://dx.doi.org/ 10.1038/363446a0 [DOI] [PubMed] [Google Scholar]
- 54.Bole DG, Hendershot LM, Kearney JF. Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J Cell Biol 1986; 102:1558-66; PMID:3084497; http://dx.doi.org/ 10.1083/jcb.102.5.1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alexander A, Steinmetz M, Barritault D, Frangione B, Franklin EC, Hood L, Buxbaum JN. gamma Heavy chain disease in man: cDNA sequence supports partial gene deletion model. Proc Natl Acad Sci U S A 1982; 79:3260-4; PMID:6808505; http://dx.doi.org/ 10.1073/pnas.79.10.3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fernandez de la Mora J. Electrospray ionization of large multiply charged species proceeds via Dole's charged residue mechanism. Anal Chim Acta 2000; 406:93-104; http://dx.doi.org/ 10.1016/S0003-2670(99)00601-7 [DOI] [Google Scholar]
- 57.Jurchen JC, Williams ER. Origin of asymmetric charge partitioning in the dissociation of gas-phase protein homodimers. J Am Chem Soc 2003 Mar 5; 125(9):2817-26; PMID:12603172; http://dx.doi.org/ 10.1021/ja0211508 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


