ABSTRACT
TNF-α (TNF), a pro-inflammatory cytokine is synthesized as a 26 kDa protein, anchors in the plasma membrane as transmembrane TNF (TmTNF), and is subjected to proteolysis by the TNF-α converting enzyme (TACE) to release the 15 kDa form of soluble TNF (sTNF). TmTNF and sTNF interact with 2 distinct receptors, TNF-R1 (p55) and TNF-R2 (p75), to mediate the multiple biologic effects of TNF described to date. Several anti-TNF biologics that bind to both forms of TNF and block their interactions with the TNF receptors are now approved for the treatment of a variety of immune-mediated diseases. Several reports suggest that binding of anti-TNFs to TmTNF delivers an outside-to-inside ‘reverse’ signal that may also contribute to the efficacy of anti-TNFs. Some patients, however, develop anti-TNF drug antibody responses (ADA or immunogenicity). Here, we demonstrate biochemically that TmTNF is transiently expressed on the surface of lipopolysaccharide-stimulated primary human monocytes, macrophages, and monocyte-derived dendritic cells (DCs) and expression of TmTNF on the cell surface is enhanced following treatment of cells with TAPI-2, a TACE inhibitor. Importantly, binding of anti-TNFs to TmTNF on DCs results in rapid internalization of the anti-TNF/TmTNF complex first into early endosomes and then lysosomes. The internalized anti-TNF is processed and anti-TNF peptides can be eluted from the surface of DCs. Finally, tetanus toxin peptides fused to anti-TNFs are presented by DCs to initiate T cell recall proliferation response. Collectively, these observations may provide new insights into understanding the biology of TmTNF, mode of action of anti-TNFs, biology of ADA response to anti-TNFs, and may help with the design of the next generation of anti-TNFs.
KEYWORDS: Anti-drug antibodies (ADA), anti-TNFs, clathrin, endocytosis, membrane TNF, Transmembrane TNF, TmTNF reverse signaling, tetanus toxin
Abbreviations
- TNF
Tumor necrosis factor
- sTNF
Soluble TNF
- mTNF
Membrane bound TNF
- TmTNF
Transmembrane TNF
- TNF-R1
TNF-Receptor 1
- TNF-R2
TNF-Receptor 2
- ADA
Anti-drug antibody
- mAb
Monoclonal antibody
- DCs
Dendritic cells
- TACE
TNF α converting enzyme
- ICD
Intracytoplasmic domain
- IBD
Inflammatory bowel disease
- R-L
Receptor-ligand
- BSA
Bovine serum albumin
- ELISA
Enzyme-linked immunosorbent assay
- SDS-PAGE
Sodium dodecyl sulfate-Polyacrylamide gel electrophoresis
- PMSF
Phenylmethylsulfonyl fluoride
- FACS
Fluorescence-activated cell sorting
- kDa
Kilodalton
- SEC
Size exclusion chromatography
Introduction
Tumor necrosis factor-α (TNF) is a pro-inflammatory cytokine with diverse biologic functions.1,2 Activation of Toll-like receptors in immune cells has been demonstrated to play a role in the synthesis of various pro-inflammatory cytokines, including TNF. TNF belongs to the family of type II cytokines, and is initially expressed as a 233 aa protein consisting of a 35 amino acid (aa) N-terminal intracellular domain (ICD), a conserved 21 aa sequence that spans the plasma membrane, and a large 177 aa C-terminal extracellular domain.3 TNF is synthesized as an ∼25 kDa pro-protein, and, following post-translational modification,4 anchors into the plasma membrane as transmembrane TNF (TmTNF). TmTNF is proteolytically processed by TNF-α converting enzyme, TACE, between residues 76Alanine and 77Valine in the extracellular domain, to release the 157 aa (∼15 kDa) soluble TNF cytokine (sTNF).3 Cleavage of the residual intramembrane amino acid residues by homologues of signal peptide peptidases, SPPL2a and SPPL2b, releases the ICD of TNF, which is suggested to trigger expression of pro-inflammatory cytokine interleukin (IL)-12 in activated dendritic cells (DCs).5,6 The highly conserved ICD of TmTNF is also implicated in a phenomenon termed ‘Reverse’ signaling, whereby the membrane-anchored TNF form transduces cellular signals upon binding to anti-TNF antibodies7 or to its membrane bound or soluble cognate receptors, such as TNF-R1 and TNF-R2.8 TmTNF has been demonstrated to be phosphorylated in the ICD at serine residues in the consensus sequence 2STES5 by casein kinase 1, triggering ‘Reverse’ signaling in mammalian cells.9
TNF is produced by immune cells such as monocytes/macrophages,10 DCs,11 T cells,12 B cells,13 NK cells14 and neutrophils,15 as well as some non-immune cells such as endothelial cells,16 adipocytes,17 fibroblasts18 and osteoclasts.19 The diverse biologic functions of TNF are mediated by binding to, and signaling via, 2 cell surface receptors, TNF-R1 and TNF-R2. TNF-R1 is known to be expressed constitutively on nearly all cell types,20 while TNF-R2 is inducibly expressed by cells of the myeloid lineage, peripheral T cells, alveolar macrophages and lymphocytes.21 While cellular responses to sTNF are dominated by TNF-R1, TmTNF has been reported to strongly stimulate TNF-R2.22 Therefore, the functional outcomes of such interactions may depend upon multiple factors such as the cell types expressing TNF-R1 or TNF-R2, stage of cellular differentiation, rates of pro-TNF synthesis, post-translational modification, and TACE-mediated cleavage of TmTNF to generate sTNF. Thus, the diversity of sTNF/TmTNF effects can be controlled through differential sensitivities of the receptors, resulting in important physiologic outcomes in local or systemic inflammatory responses.
Regardless of the precise understanding of the biologic and pathological roles of the 2 forms of TNF (sTNF and TmTNF), TNF is a clinically validated target. Anti-TNF therapeutics have revolutionized the treatment of many chronic inflammatory diseases, such as rheumatoid arthritis (RA), inflammatory bowel disease (IBD), ankylosing spondylitis, and psoriasis. Anti-TNF biologics are structurally classified into 3 types: (1) bi-valent monoclonal antibodies (mAbs), e.g., infliximab, golimumab; (2) engineered TNF-R2-Fc fusion, etanercept, and (3) mono-valent Fab (lacking the Fc and hinge region) of an anti-TNF mAb covalently linked to polyethylene glycol, certolizumab pegol. While all these anti-TNF antagonists bind to and block interactions of sTNF with the 2 TNF receptors equally well and are known to exert potent clinical effects in RA,23,24 not all show equal efficacy in certain diseases, such as Crohn's. For example, etanercept did not show efficacy in Crohn's disease,25 while infliximab was effective for such granulomatous disease.26 Despite their success in improving patients' quality of life, anti-TNF therapeutics may elicit anti-drug antibodies (ADA) in some patients, and ADA may affect both efficacy and pharmacokinetics. Emerging evidence suggests that the differences in clinical outcome of anti-TNF treatment cannot be explained solely by their capacity to neutralize soluble TNF. Other properties of anti-TNFs, such as interactions with TmTNF and FcγR, may also affect efficacy, potentially ADA responses and overall clinical outcomes. Given that TmTNF is expressed on the surface of many cell types, including antigen-presenting cells, and has the capacity to induce a “reverse signal” in response to binding by an anti-TNF antagonist, we hypothesized that this interaction, in a manner similar to a ligand-receptor interaction, could cause the internalization of the TmTNF-bound anti-TNFs into the cell and subsequent processing for antigen presentation to cognate T cells. And as such, this could be a contributing mechanism leading to the development of an ADA response to the biologic.
TNF protein can be present on cell surfaces either as the 26 kDa TmTNF or as the 15 kDa soluble TNF bound to its receptors. The presence of these forms may depend upon the nature and duration of the stimulus, cell type, environmental factors and the methods used to detect surface TNF. Over the past several years, the biology of TmTNF has been studied using biochemical and functional methods. These methods have included transient and stable overexpression of wild type or TACE-resistant mutant forms of the 26 kDa full length TNF,1 fluorescence-activated cell sorting (FACS), enzyme-linked immunosorbent assay (ELISA), Western blotting, immunoprecipitation, co-culture of stimulated normal cells, cell lines or TNF transfectants and treatment of cells with inhibitors of metalloproteases such as BB1101 and TAPI-2 to block the processing and shedding of TmTNF.27,28
Here, we describe the biologic consequences of anti-TNF-TmTNF interaction. We demonstrate the kinetics of TmTNF expression on freshly isolated human monocytes, monocyte-derived macrophages, and DCs using cell surface biotinylation approach. We further demonstrate that binding of anti-TNF to TmTNF on DCs results in rapid internalization of the anti-TNF-TmTNF complex, first entering the endosomes and then the lysosomes. The internalized anti-TNF is processed and its peptides displayed on the surface of DCs, which mount a T cell proliferation response. These observations may provide additional insights into understanding the biology of TmTNF, the potential mechanism of action of anti-TNFs, ADA responses, and may help in the design and development of the next generation of anti-TNF therapeutics.
Results
Kinetics of TmTNF protein expression in human monocytes, monocyte derived macrophages and DCs
To study the biology of TmTNF in primary human monocytes, as well as monocyte-derived macrophages and DCs, we first defined biochemically the kinetics of TNF (TmTNF, cytosolic and sTNF) expression profiles following lipopolysaccharide (LPS) stimulation in multiple donors. The kinetics of TNF expression was similar in all donors analyzed; however, the absolute levels of expression varied. The data presented in Figs. 1 and 2 are from monocytes, monocyte-derived macrophages and DCs obtained from a single donor. Monocytes from human peripheral blood mononuclear cells (PBMCs) and monocyte-derived macrophages and DCs were stimulated with LPS in the presence or absence of TAPI-2 (for details, see Materials & Methods). For biochemical analysis of TmTNF, extracellular domains of all cell surface proteins were labeled with a cell-impermeable biotin analog. Biotinylated cell surface proteins were enriched on immobilized streptavidin-agarose beads and subjected to immunoblot analysis using anti-TNF IgG. Among the cell surface proteins, and in comparison to untreated cells, LPS activation of monocytes and monocyte-derived macrophages (Fig. 1A, 1B) and DCs (Fig. 2A, 2B) induced a robust expression of an ∼25 kDa anti-TNF reactive protein. The absence of the cytoplasmic protein GAPDH in the cell surface biotinylated proteins enriched by streptavidin precipitation demonstrates that cytosolic proteins were not biotinylated by the surface biotinylation protocol (Fig. 1A, 1B; Fig. 2A, 2B). The results show that in monocytes, monocyte-derived macrophages, and DCs, TmTNF and cytosolic TNF can be detected by these methods for up to 6 h post LPS stimulation, and during this time soluble TNF continues to accumulate in the cell culture supernatant (Fig. 1C, 1D; Fig. 2C). In addition, treatment of monocytes and monocyte-derived macrophages and DCs with TAPI-2, a TACE inhibitor, enhances cell surface TmTNF detection and reduces sTNF accumulation in the supernatant (Figs. 1 and 2). We have repeatedly observed that, although monocyte-derived macrophages produce copious amounts of sTNF, low levels of TmTNF are detected in these cells in the absence of TAPI-2 treatment. This observation suggests that macrophages may efficiently process TmTNF.
Figure 1.
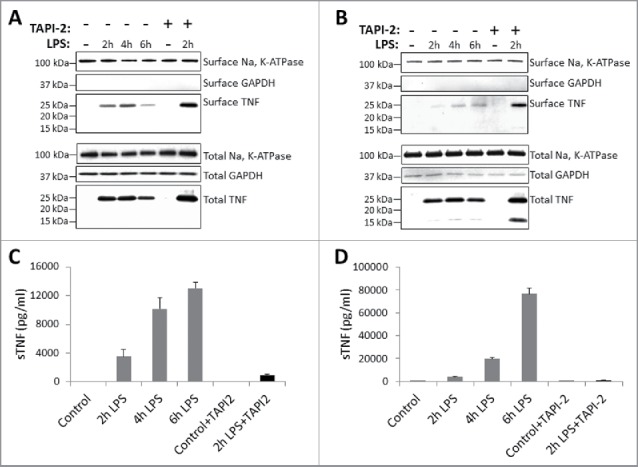
Kinetics of TNF expression in human monocytes and macrophages post LPS treatment. Human CD14+ monocytes (A) or day 7 human CD14+ monocyte-derived macrophages (B) were either untreated or treated with LPS in the presence or absence of TACE inhibitor, TAPI-2, for the indicated periods of time. Cell surface proteins were labeled with cell-impermeable Sulfo-NHS-SS-biotin. Biotinylated surface proteins were precipitated with streptavidin-conjugated agarose beads. Cell surface biotinylated and total proteins were subjected to immunoblotting using anti-TNF IgG. Na, K-ATPase and GAPDH protein expressions were used as total protein loading and cell surface biotinylation controls. Cell-free culture supernatants from monocytes (C) and macrophages (D) as treated in (A) and (B), respectively, were assayed for the presence of soluble TNF (sTNF) by ELISA.
Figure 2.
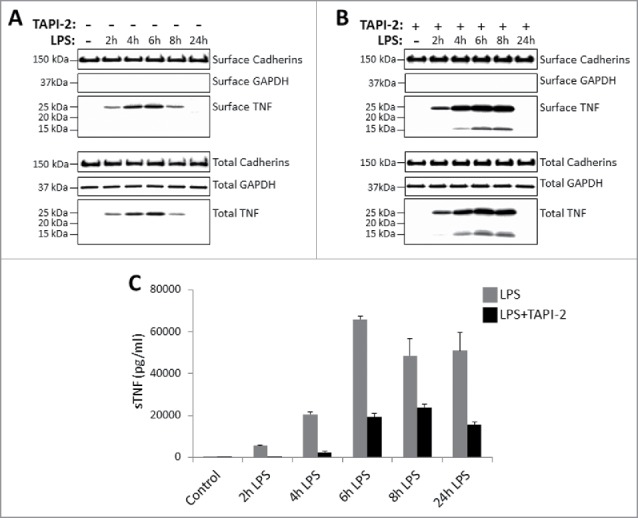
Kinetics of TNF expression in monocyte-derived dendritic cells post LPS treatment. Day 5 human CD14+ monocyte-derived DCs were either untreated or treated with LPS in the absence (A) or presence (B) of 20 μM TACE inhibitor, TAPI-2, for the indicated periods of time. Cell surface proteins were labeled with cell-impermeable Sulfo-NHS-SS-biotin. Biotinylated surface proteins were precipitated with streptavidin-conjugated agarose beads. Cell surface biotinylated and total proteins were subjected to immunoblotting using anti-TNF IgG. Cadherins and GAPDH protein expressions were used as total protein and cell surface biotinylation loading controls. Cell-free culture supernatants of DCs as treated in (A) and (B) above were assayed for the presence of soluble TNF (sTNF) (C) by ELISA.
In addition, and as expected, the data in Fig. 2B clearly demonstrates that TAPI-2 treatment of DCs enhanced the expression of ∼25 TmTNF protein on the cell surface and simultaneously reduced production, and hence, accumulation of sTNF in the supernatant (Fig. 2C). Interestingly, in the presence of LPS and TAPI-2 we also detected a 15 kDa ‘cleaved’ TNF being biotinylated by the cell impermeable biotin analog (Fig. 2B). This 15 kDa ‘cleaved’ form of TNF was not observed in surface biotinylated proteins in the absence of TAPI-2 (Fig. 1A, 1B; Fig. 2A). This observation suggests that perhaps in the presence of TAPI-2, 1 or 2 TNF monomers of the membrane anchored TNF trimer may be cleaved by TACE but remain attached to the membrane anchored un-cleaved component of the TNF trimer, and therefore may not be released into the supernatant. Collectively, data in Figs. 1 and 2 confirm that in monocytes, monocyte-derived macrophages, and DCs, TmTNF transiently anchors in the membrane and is cleaved by TACE to release sTNF in the culture supernatant.
Binding of anti-TNFs to TmTNF on DCs results in internalization of anti-TNF/TmTNF complex
Endocytosis involves the internalization of transmembrane protein receptors and their extracellular ligands into cytoplasmic vesicles that are pinched off from the plasma membrane. The receptor-ligand (R-L) complexes are internalized by multiple mechanisms.29 Internalization and intracellular trafficking are complex processes that require a well-orchestrated array of signaling cascades and cytoskeletal re-organization.30 Internalized proteins can be either re-cycled to the surface or delivered to intracellular vesicles such as endosomes and lysosomes, where in the presence of hydrolytic enzymes or low pH, proteins are degraded and their peptides displayed on the surfaces of cells.31 As TmTNF may serve as a receptor,32 and as DCs are efficient antigen-presenting cells,31 we hypothesized that binding of anti-TNF to TmTNF could result in internalization and delivery of anti-TNF to various antigen processing compartment(s) of DCs.
To assess cellular uptake of antibodies, we used pHrodo dye-conjugated anti-TNF mAbs, which do not fluoresce at neutral pH (outside the cell), but their fluorescence dramatically increases as pH decreases from neutral to acidic in intracellular vesicles like endosomes and lysosomes. Also, the fluorescence emitted by the pHrodo red dye is directly proportional to the drop in pH.33 To determine whether anti-TNF/TmTNF complexes are internalized, we treated DCs with LPS for 2 h to induce TmTNF, in the absence of TAPI-2, followed by incubating the cells with pH-sensitive pHrodo red dye-conjugated anti-TNF mAb. Internalization of the conjugated anti-TNF was monitored using fluorescence microscopy. The pHrodo-conjugated anti-TNF (stained red) could be seen in vesicle-like structures inside the cell, surrounded by the plasma membrane (stained green using anti-HLA class I IgG) in LPS-treated DCs (Fig. 3A). We were unable to detect such fluorescence in untreated, control DCs (data not shown). Furthermore, in contrast to LPS-treated DCs incubated with pHrodo-conjugated isotype control human IgG1, greater than 90% of LPS-treated DCs incubated with pHrodo-conjugated anti-TNF were positive for pHrodo fluorescence emission as assessed by FACS (Fig. 3B). The observed intensity of wavelength emission suggested that the conjugated anti-TNF must be localized in low pH (acidic) compartments. To rule out any nonspecific uptake of anti-TNF, we assessed by FACS the uptake of pHrodo-conjugated isotype control human IgG1 or pHrodo-conjugated anti-TNF in either untreated control DCs or DCs treated with LPS for 2 h to induce TmTNF expression. We observed significantly higher uptake of pHrodo-conjugated anti-TNF mAb in DCs treated with LPS for 2 h compared with the uptake of pHrodo-conjugated isotype control human IgG1 or anti-TNF mAb in control, untreated DCs (Fig. 3C). These data demonstrate TmTNF-mediated internalization of anti-TNF mAb in DCs.
Figure 3.
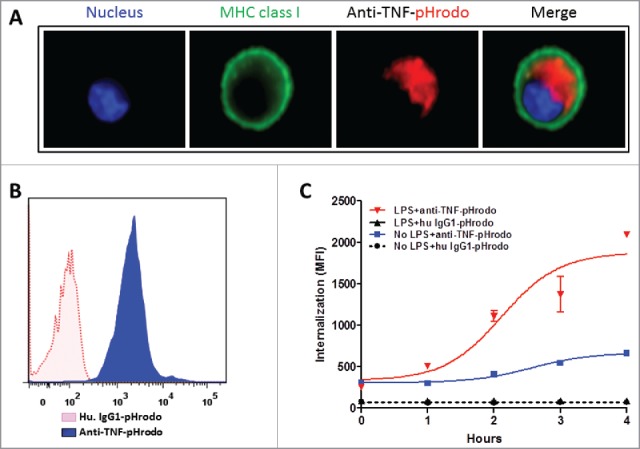
TmTNF-dependent endocytosis of anti-TNFs in dendritic cells. (A) Day 5 human CD14+ monocyte-derived DCs were treated with LPS for 2 h, incubated with pHrodo-conjugated anti-TNF and cellular uptake of labeled anti-TNF was monitored. Plasma membrane was stained using anti-MHC class I IgG (green), the nucleus (blue), and the internalized pHrodo-anti-TNF (red) were visualized using Amnis imaging flow cytometry. (B) Day 5 human DCs were treated with LPS for 2 h and cells were incubated either in the presence of pHrodo-conjugated matched isotype control IgG (red dotted histogram) or pHrodo-conjugated anti-TNF (blue filled histogram). Cellular uptake of the labeled antibodies was monitored for up to 4 h using flow cytometry. (C) To measure the kinetics of endocytosis of anti-TNF into acidic compartments in DCs, cells were either untreated (No LPS) or treated with LPS for 2 h. Cells were incubated with either human IgG1-pHrodo or anti-TNF-pHrodo antibodies. Endocytosis of the internalized anti-TNF was measured as an increase in fluorescence intensity by flow cytometry for up to 4 h.
Next, we determined if TmTNF-mediated internalization (endocytosis) of anti-TNF is associated with recruitment of any adaptor proteins on the ICD of TmTNF. To this end, TmTNF expression was induced in 2 h LPS-treated DCs and cells treated either with human IgG1 isotype control or with an anti-TNF antibodies for 5 min at 37°C. TmTNF/anti-TNF complexes were enriched on Protein A-agarose affinity column and subjected to peptide analysis using liquid chromatography-mass spectrometry (LC-MS)/MS. We were unable to detect any TNF peptides in TmTNF-expressing DCs incubated with isotype control human IgG1. In contrast, anti-TNF enriched immune-complexes revealed the presence of peptide sequences corresponding to TNF, as well as various proteins known to be involved in receptor-mediated endocytosis of ligands (myosin 1G, β tubulin, myosin 1D, IQGAP1; data not shown), along with peptides from clathrin heavy chain (Fig. 4A). To verify a direct association of clathrin heavy chain with TmTNF, we subjected the immune complexes from the above experiment to immunoblotting using either anti-TNF or anti-clathrin heavy chain antibodies. We observed TmTNF-associated clathrin heavy chain from immune-complexes purified from DCs treated with LPS and anti-TNF, and were unable to detect either TmTNF or clathrin heavy chain in LPS-treated DCs incubated with human IgG1 control isotype antibody (Fig. 4B). Furthermore, to determine if endocytosis of TmTNF/anti-TNF was dependent on its association with clathrin, we used Dyngo 4a, a dynamin inhibitor known to inhibit clathrin-mediated endocytosis of various receptor-ligand complexes.34,35 LPS-treated, TmTNF-expressing DCs were either untreated or treated with 5 μM Dyngo 4a and endocytosis of pHrodo-conjugated anti-TNF mAb was monitored using FACS. We observed significantly higher uptake of anti-TNF in DCs in the absence of Dyngo 4a, whereas Dyngo 4a was sufficient to abolish TmTNF/anti-TNF internalization (Fig. 4C). These data clearly demonstrate that in DCs expressing TmTNF, endocytosis of TmTNF/anti-TNF complex was dependent on clathrin.
Figure 4.
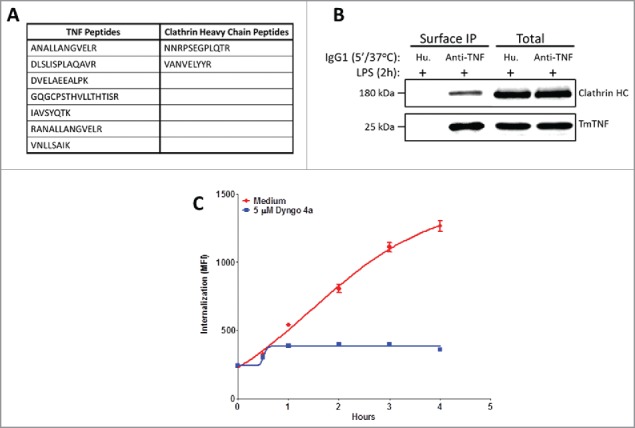
Clathrin-dependent endocytosis of anti-TNFs in dendritic cells. (A) Identification of anti-TNF/TmTNF-associated proteins: Day 5 human CD14+ monocyte-derived DCs were treated with LPS for 2 h and incubated either with isotype matched human IgG1 or with anti-TNF for 5 min at 37°C. Peptide sequences corresponding to TNF or clathrin heavy chain from anti-TNF immunoprecipitated proteins were identified by LC-MS/MS. (B) Identification of anti-TNF/TmTNF-associated clathrin heavy chain: Immunoprecipitated proteins and total cell extracts from DCs as treated in (A) above were subjected to immunoblotting using anti-clathrin heavy chain or anti-TNF antibodies. IP: Immunoprecipitation. (C) Effect of clathrin inhibitor on endocytosis of anti-TNF: Day 5 human CD14+ monocyte-derived DCs were treated with LPS for 2 h either in the absence (Medium) or presence of a 5 μM clathrin inhibitor, Dyngo 4a. Endocytosis of the internalized anti-TNF was measured as an increase in fluorescence intensity by flow cytometry for up to 4 h.
Internalized TmTNF-anti-TNF complex undergoes vesicular trafficking, processing, and anti-TNF peptides can be eluted from the cell surface of DCs
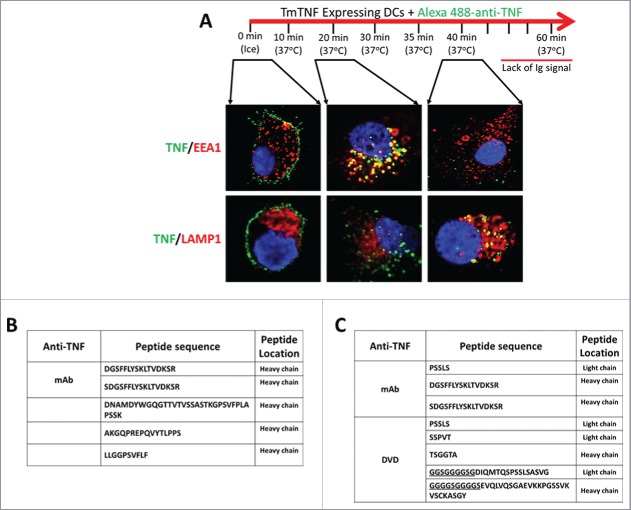
To further understand intracellular compartmentalization/trafficking of the TmTNF/anti-TNF complexes, we performed a pulse-chase (time course) confocal microscopy study using Alexa 488-conjugated humanized anti-TNF mAb. Specific intracellular vesicles were identified by staining cells either with anti-EEA1 (early endosome marker) or anti-LAMP1 (lysosomal marker). These studies (Fig. 5A) demonstrated that early after addition (up to 25 minutes) of anti-TNF mAb to 2 h LPS-treated, TmTNF-expressing DCs, the anti-TNF (green fluorescence) migrates from the cell surface into vesicles that stained with anti-EEA1 (early endosomes; red fluorescence), but not with anti-LAMP1 stained compartments (lysosomes; red fluorescence). However, with time (35–45 minutes), the anti-TNF co-localizes significantly with anti-LAMP1 stained compartment, but not with anti-EEA1 stained vesicles (Fig. 5A). Following about 1 h, the anti-TNF signal could not be detected either on the cell surface or inside the cells. Such observations suggested to us that TmTNF-mediated internalized anti-TNF may not re-cycle back to the cell surface, and at least traffics through early endosomes to lysosomes, where perhaps it is degraded and therefore cannot be detected.
Figure 5.
TmTNF-dependent vesicular trafficking of humanized anti-TNFs and detection of cell surface anti-TNF peptides in dendritic cells. (A) Compartmentalization of anti-TNF: Day 5 human CD14+ monocyte-derived DCs were treated with LPS for 2 h and the cells ‘pulsed’ on ice using Alexa 488-conjugated anti-TNF (0 min, ice). The fluorescent label on the anti-TNF was ‘chased’ for up to 1 h at 37°C. Endosomes and lysosomes were stained with Alexa 647-conjuated anti-EEA1 and anti-LAMP1, respectively. Nucleus was stained with DAPI. Images were acquired using confocal microscopy. (B) Identification of cell surface-associated anti-TNF peptides: Day 5 human CD14+ monocyte-derived DCs were treated with LPS for 2 h and incubated with either an anti-TNF mAb or a DVD-Ig containing one anti-TNF domain for 6 h. Cell surface displayed peptides were eluted under mild acidic conditions and the identity of anti-TNF peptides was obtained using LC-MS/MS. (C) Identification of HLA-DR-associated anti-TNF peptides: Day 5 human CD14+ monocyte-derived DCs were treated with LPS for 2 h and incubated with anti-TNF for up to 8 h. HLA-DR was immunoprecipitated and the identity of HLA-DR-associated anti-TNF peptides was obtained using LC-MS/MS (underlined: linker peptide sequence of anti-TNF DVD-Ig).
To determine the fate of internalized anti-TNFs, we stimulated DCs for 2 h with LPS to induce the expression of TmTNF, followed by incubation with either a humanized anti-TNF mAb or a humanized dual-variable-domain (DVD)-Ig molecule containing anti-TNF domains for 6 h. The DVD-Ig molecule, a tetravalent dual specific antibody format, contains 2 variable domains (VDs) on each Fab linked in tandem by a linker36 and is bivalent for each specified target. The DVD-Ig used in this study contained one set of anti-TNF binding domain and a second set of VDs that recognized a second target, anti-IL-17. The 2 VDs (anti-TNF and anti-17 on each Fab) of this DVD-Ig molecule were linked together by a 14 aa GS linker. Both, the humanized anti-TNF mAb and the humanized anti-TNF DVD-Ig had similar potency for TNF (Table 1). DCs treated with anti-TNFs were washed with mild acid to elute the surface displayed peptides and subjected to LC-MS/MS for peptide identification. LC-MS/MS analysis identified peptides of humanized anti-TNF molecules (Fig. 5B). These data supports the notion that internalized anti-TNF biologics are degraded and processed perhaps in the lysosomes and their peptides can be eluted from the surface of DCs.
Table 1.
Use of various anti-TNF antibodies in the current study.
| Anti-TNF | SEC Profile (% Monomer) | FACS Binding | TNF Neutralization (L929 assay) | Cellular Uptake |
|---|---|---|---|---|
| Anti-TNF-1* | 100 | + | 0.06 nM | + |
| Anti-TNF-2** | 94 | + | 0.05 nM | + |
| Anti-TNF-3** | 95 | + | 0.01 nM | + |
| Anti-TNF-4** | 96 | + | 0.015 nM | + |
| DVD-Ig** | 98 | + | 0.03 nM | + |
: Human IgG1;
: Humanized IgG1
To further establish that the peptides eluted from the surface of humanized anti-TNF mAb-treated DCs were associated with HLA-DR molecules, we treated LPS-stimulated DCs expressing TmTNF with humanized anti-TNF mAb as above and HLA-DR protein was immunoprecipitated as described in Materials & Methods. HLA-DR bound peptides were subjected to LC-MS/MS analysis. The analysis identified the same peptide that we had identified in our surface elution analysis, along with some additional peptides (Fig. 5C). Interestingly, some peptides from the cell surface of DVD-Ig-treated DCs were from the linker region with/without flanking sequences that matched the amino acid sequence of the VDs in the DVDIg molecule (Fig. 5C, underlined peptide sequences). Taken together our data demonstrates that the humanized anti-TNF internalized by TmTNF-expressing DCs are processed and loaded on to HLA-DR for antigen presentation. However, it should be noted that not all peptides presented within the context of HLA-DR are immunogenic, as some mAb-derived peptides may in fact be tolerogenic.37
TmTNF-dependent uptake of anti-TNF conjugated tetanus toxin peptides are processed and presented to T cells by DCs
To determine if anti-TNF-fused peptides can be presented on the surface of DCs and can initiate an immune response, we fused 2 tetanus toxin (TT) peptides38 at the C-terminal heavy chains of a humanized anti-TNF (anti-TNF-TT fusion IgG; Fig. 6A). Addition of TT peptides to the humanized anti-TNF mAb did not interfere with their ability to potently neutralize sTNF in the L929 apoptotic assay (Fig. 6B). Two h LPS-treated, TmTNF-expressing DCs were either left untreated or incubated in the presence of recombinant TT or anti-TNF-TT fusion proteins (anti-TNF-S1 or anti-TNF-S2). T cell proliferation assay was performed by incubating anti-TNF-TT treated cells with autologous T cells. As expected, compared with untreated DCs, the proliferation of T cells significantly increased when they were co-cultured with recombinant TT-pulsed DCs (Fig. 6C). Furthermore, TmTNF-expressing DCs that had been treated with anti-TNF-S1 or anti-TNF-S2 antibodies showed further increase in proliferated T cells compared with untreated or TT-treated DCs (Fig. 6C). Proliferation of both CD4+ and CD8+ T cells was observed along with IL-2/interferon γ production (data not shown). These results demonstrate TmTNF-dependent cellular trafficking of anti-TNF to proteolytic compartments (i.e., lysosomes), their efficient proteolysis, and sufficient display of peptides on the surface of DCs to initiate a T cell recall immune response by PBMCs of individuals who had been previously exposed to TT, and hence possessed TT-specific memory T cells.
Figure 6.
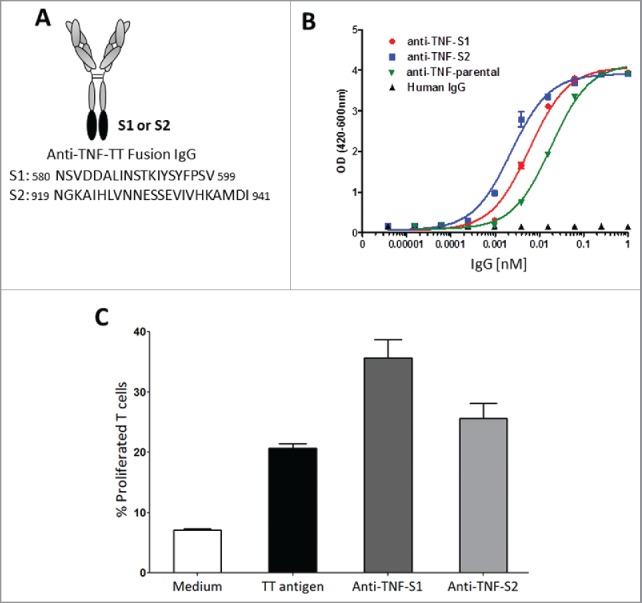
TmTNF-dependent uptake of humanized anti-TNF mAb and generation of a memory T cell recall response by dendritic cells. (A) Scheme of generating anti-TNF-TT fusion IgGs: Sequence of tetanus toxin (TT) peptides (S1 or S2) were fused to the C-terminus of the heavy chains of an anti-TNF to obtain anti-TNF-S1 or anti-TNF-S2 antibodies. (B) Testing the potency of anti-TNF-TT fusion IgGs: The potency of the parental anti-TNF and anti-TT-fusion IgGs (anti-TNF-S1 and anti-TNF-S2) to neutralize soluble TNF were tested using the L929 assay. (C) Measuring T cell recall response in DCs: Day 5 human CD14+ monocyte-derived DCs were treated with LPS for 2 h and incubated either with tetanus toxin (TT antigen), anti-TNF-S1 or anti-TNF-S2. DCs were co-cultured with CFSE-labeled autologous T cells, and T cell proliferation assessed using flow cytometry.
Discussion
Here, we describe some biologic consequences of anti-TNF mAb/TmTNF interaction. We show for the first time that interaction of anti-TNF mAbs with TmTNF on DCs results in rapid endocytosis of the anti-TNF/TmTNF complex, resulting in lysosomal delivery and degradation of anti-TNF. Peptides from anti-TNF molecules can be eluted from the surface of DCs, and may be processed and presented by DCs to initiate a T cell immune (recall) response. These observations may provide additional insights into the biology of TmTNF, the mechanism of action of anti-TNFs, the biology of antibodies to anti-TNF antagonists (immunogenicity of anti-TNF antagonists) observed in some patients, and may help in the design of a next generation of anti-TNFs with greater efficacy and reduced (or no) immunogenicity.
The biology of TNF is complex. TNF exists as soluble and membrane forms and both forms are known to interact with 2 receptors, TNF-R1 and TNF-R2, each with distinct signaling pathways and biologic outcome.39 Although the 2 forms of TNF can interact with both receptors, it is believed that TNFR1 and TNFR2 may preferentially interact with soluble TNF and membrane TNF, respectively.22 TNF biology, and in particular anti-TNF/membrane TNF interactions, is additionally complicated by the fact that the membrane form of TNF can also exist in 2 forms, either as membrane-anchored transmembrane TNF (TmTNF; ∼25 kDa) or as TACE-cleaved soluble TNF bound to its cognate cell surface receptors40 (mTNF; 15 kDa). MAbs and etanercept (Enbrel®) can bind TmTNF,41 but some mAbs and etanercept may not be able to bind the mTNF. In addition, the biologic consequences of an anti-TNF/TmTNF and anti-TNF/mTNF interaction may be quite distinct. In the former case, the biologic consequences are a result of signaling through TmTNF (“reverse” signaling) and in the latter case the consequences may be a result of signaling through the TNF receptors (R1 or R2) to which the mTNF is bound. The fate of the anti-TNF bound to either TmTNF or mTNF would then be determined by if and how anti-TNF/TmTNF and anti-TNF/mTNF/TNFR1 or TNFR2 complexes are taken up by the cells (endocytosis) and their subsequent intracellular trafficking pathways of re-cycling or degradation (discussed below).
To study the consequences of anti-TNF interactions with TmTNF, we took a systematic approach to first define the kinetics of TmTNF expression in freshly isolated human monocytes, monocyte-derived macrophages, and DCs. Using the surface biotinylation method, only TmTNF was detected early after LPS stimulation (2–6 h post stimulation) (Figs. 1, 2, in the absence of TAPI-2), but mTNF could not be detected. However, using a surface biotinylation approach we detected mTNF on monocytes treated with LPS for 24 h (data not shown). The presence of 15 kDa, TACE-cleaved form of mTNF was detected on the surface of DCs, but only in the presence of TAPI 2, and the potential reasons for this are discussed above (Results section). Therefore, to study the consequences of anti-TNF mAb interaction with TmTNF, we performed studies, as reported here, using 2–4 h LPS-treated DCs in the absence of TAPI-2, a time point when only TmTNF was expressed on the surface of DCs.
To communicate with the external environment and maintain cellular and tissue homeostasis, cells utilize multiple endocytosis pathways to bring external signals inside the cell through receptor-ligand (R-L) interactions. Such R-L interactions induce receptor clustering on the cell membrane, which may depend upon receptor density, ligand concentration, ligand binding properties, and may require a certain amount of time on the surface for internalization (endocytosis) machinery to assemble, e.g., clathrin-coated pit/vesicle formation, cytoskeletal reorganization and recruitment of adaptor proteins.30 These initial events in the R-L interactions determine the fate of the R-L complexes inside the cells, i.e., whether the internalized ligand will be re-cycled back to the surface or be degraded in the lysosomes.30 The data presented in Figs. 3 and 5 clearly demonstrates that the anti-TNF/TmTNF complex is rapidly endocytosed. The anti-TNF internalization is highly dependent upon the expression of TmTNF, as no non-specific uptake of either isotype control human IgG1 by LPS stimulated or untreated control DCs was observed (Fig. 3C). Only minimal uptake of anti-TNF was observed by control, untreated DCs. At this time we do not know if these non-LPS-stimulated DCs that had been cultured for 5 d in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL-4 expressed minute levels of TmTNF protein that could not be detected by the cell surface biotinylation method we used. However, no TACE-cleaved TNF could be detected in the culture supernatants of such cells by ELISA.
As endocytosis is associated with cytoskeletal changes and recruitment of cytoskeletal/adaptor proteins, we attempted to determine if we could identify, with the anti-TNF mAb, any TmTNF-associated proteins at a given time point. As expected, we identified not only several TNF peptides, but also peptides representing clathrin heavy chain and several other structural/cytoskeletal proteins that may be associated with the anti-TNF/TmTNF endocytosis machinery.29 Because we detected clathrin heavy chain peptides in the anti-TNF precipitation of TmTNF-expressing DCs, we wanted to determine if Dyngo 4a, a compound reported to inhibit clathrin-mediated endocytosis, would block TmTNF-mediated internalization of anti-TNF mAb. Indeed, Dyngo 4a treatment of LPS stimulated, TmTNF-expressing DCs almost completely inhibited TmTNF-mediated endocytosis of anti-TNF mAbs. Collectively, our results in Fig. 4 demonstrate that anti-TNF internalization is an active, TmTNF-dependent process.
Internalized R-L complexes, after trafficking through various intracellular compartments can either be re-cycled back to the surface or undergo degradation, in particular in the lysosomes.42 The fate of the internalized complexes, in particular the antibody/receptor complexes, is determined by multiple factors, including structural features of the antibody, the biology of the target receptor and the nature of the antibody/receptor interaction. It is well known that a large proportion of circulating antibodies are taken up by cells non-specifically, and, following interaction with the neonatal Fc receptor (FcRn) at low pH in the endosomes, are re-cycled back into circulation. However, if the antibody molecule fails to interact with FcRn in a productive manner, the antibody molecule follows the degradation pathway in the lysosomes.43 In the case of antibodies that are internalized upon binding to their cell surface targets, the fate of the antibody molecule is determined by the biology of the target receptor and the nature of antibody/target interaction.44 For example, certain receptors re-cycle efficiently and, if the antibody remains bound to the target at low pH in the endosomes, it may re-cycle back to the surface with the receptor.45 In other instances, certain receptors upon internalization deliver their cargo (including an antibody) directly to the lysosomes for degradation.31 To understand if TmTNF-mediated internalized anti-TNF mAb follows the re-cycling or the degradation pathway, we first studied the kinetics of intracellular trafficking of Alexa-488-conjugated humanized anti-TNF mAb using confocal microscopy. Under these experimental conditions (described in Material & Methods), we observed that following the initial clustering of the humanized anti-TNF mAb on the cell surface, it is internalized into early endosomes (as assessed by EEA 1 staining), rapidly transits to lysosomes (as assessed by LAMP 1 staining) and then cannot be detected. These observations suggest that TmTNF-mediated internalized anti-TNF mAbs may not re-cycle, but rather are delivered rapidly to the lysosomes, where they are degraded. These properties of the antibody-target interactions (i.e., internalization, rapid transit to and degradation of the antibody in lysosomes) are desired features for antibody-drug conjugate (ADC) therapeutics, and as such our data suggests that an anti-TNF ADC could form the basis for the next generation of anti-TNF therapeutics with enhanced efficacy and reduced immunogenicity.
It is well known that peptides from proteins degraded in various intracellular compartments can be presented on the cell surface within the context of antigen-presenting molecules (i.e., HLA in humans and MHC in mice). Therefore, we used the method of cell surface peptide elution under mild acidic conditions40 to determine if we can detect humanized anti-TNF derived peptides from the surface of DCs. As described above (Material & Method and Results sections) we were able to identify peptides from mild acid eluates of the DC surface that matched the sequences of our humanized anti-TNF molecule, in particular specific peptides from the humanized DVD-Ig G4S linker region. As the G4S linker is believed to be an “inert” non-immunogenic sequence and is commonly used as a linker peptide in making bi- and multi- specific biologics and other fusion proteins, further studies are required to determine if the high frequency of processed G4S-derived peptides detected in our experiments reflects a property of this sequence, the context within which this linker is placed, or some specific property of the TmTNF intracellular trafficking. At this time we do not know if these G4S-sequence containing peptides or any other anti-TNF-derived peptides are immunogenic.
DCs are professional antigen processing and presenting cells. It is well established that targeting specific receptors on DCs with an antibody to which specific antigenic peptides are fused can initiate specific immune responses to the fused peptides.31,46 To determine if the TmTNF-mediated internalization and proteolytic processing of anti-TNF results in presentation of fused peptides by DCs to initiate a specific T cell (recall) response, we fused known immunogenic TT-derived peptides to a humanized anti-TNF molecule. The TT peptide fused humanized anti-TNF mAbs maintained their functional potency. When co-cultured with DCs treated with TT peptide fused humanized anti-TNF, autologous T cells (from pre-screened donors) that respond to TT peptides induced proliferation of T-cells (a recall response). These data demonstrate that TmTNF-mediated internalized anti-TNF mAb is proteolytically processed and the fused TT peptides can be productively presented by DCs to T cells. Our data may have implications in understanding why some patients treated with anti-TNF antagonists develop an anti-drug antibody (ADA) response. Multiple factors determine an ADA response.47 Although antigen processing and presentation is a critical requirement to initiate an immune response, not all peptides presented by antigen-presenting cells are immunogenic or can initiate an immune response. In fact, some mAb-derived peptides may be tolerogenic.37 The data presented in this study showing that anti-TNF/TmTNF complexes are rapidly internalized and that the endocytosed anti-TNF is rapidly delivered to the lysosomes, processed and anti-TNF peptides presented by DCs suggest that the fate of biologic therapeutics may be determined by the target biology, in this case TmTNF. Our data suggest TmTNF may possess intrinsic features that increase the chances of ADA development. Therefore, the rate of TNF synthesis, the levels of TmTNF expression and the cell types making TmTNF may all be critical factors determining the level and intensity of ADA development under inflammatory conditions in which co-stimulatory signals are available.
The data presented herein regarding TmTNF-dependent internalization, intracellular trafficking and eventually degradation of anti-TNF mAbs may have implications in further understanding the mechanism of action of anti-TNF therapeutic agents. Several potential mechanisms have been proposed to account for the remarkable efficacy and safety profiles of anti-TNFs. These mechanisms include: 1) blocking interactions of both sTNF and TmTNF with their receptors, R1 and R2;48 2) elimination of membrane TNF (both sTNF bound to its receptors and membrane anchored TmTNF) -expressing T cells by antibody-dependent cell-mediated cytotoxicity or complement-dependent cytotoxicity via Fc-effector functions;49 3) generation of regulatory macrophages and T regulatory (Treg) cells upon interaction with mTNF.50,51 In addition to these mechanisms, our data suggest an additional mechanism that may also play a role in determining overall efficacy of anti-TNF molecules, i.e., rapid internalization and degradation of TmTNF. Internalization and degradation of this complex may have several implications. First, reduced TmTNF availability for cleavage by TACE, thereby reducing sTNF release. Second, lack of TACE cleavage due to TmTNF internalization may prevent further cleavage of transmembrane domain of TmTNF by SPPL2a/SPPL2b, and thereby generation of the ICD that has been suggested to induce IL-12 production.5 Third, reduced availability of TmTNF may reduce potential pro-inflammatory signals through R2 or R1. Fourth, receptor internalization (endocytosis) and intracellular signaling may be linked events.30 In this regard, we have observed that interaction of anti-TNF mAb with TmTNF on DCs generates a “reverse” signal that may modulate production of additional pro-inflammatory mediators, including TNF production, suggesting a negative feedback loop (data not shown). Studies are in progress to further understand the interaction/relationship between anti-TNF/TmTNF complex internalization and “reverse” signaling events and if the interaction of TmTNF with cell surface TNF R1/R2 also results in TmTNF internalization and “reverse” signal. Answers to these questions will enhance our understanding of TmTNF biology and experiments are underway to address these questions.
As summarized in Fig. 7, in this study we demonstrated that anti-TNF molecules are actively internalized upon binding to TmTNF on monocyte-derived DCs and rapidly enter the lysosomes where they are degraded. Anti-TNF peptides can be eluted from the surface of DCs and are presented to T cells to initiate a recall response. Although not all displayed peptides are immunogenic, the observations presented here may provide additional insights into the biology of TmTNF, the mechanism of action of anti-TNFs, the biology of antibodies to TNF antagonists (immunogenicity of TNF antagonists) observed in some patients, and may help in the design of next generation of anti-TNFs with greater efficacy and reduced (or no) immunogenicity, such as anti-TNF drug conjugates.
Figure 7.
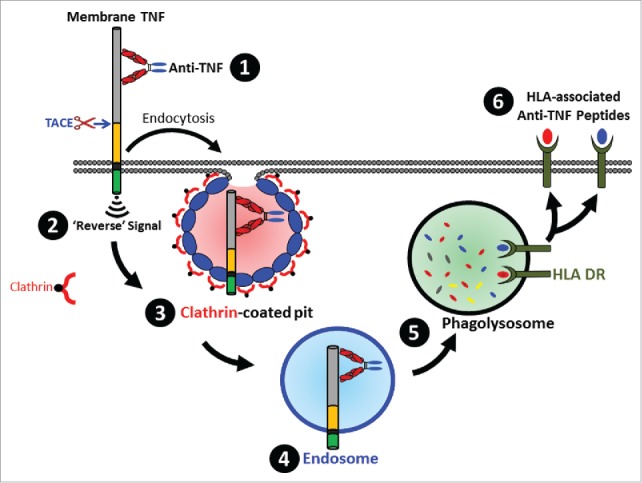
Schematic overview of the fate of TmTNF-bound anti-TNF. Binding of anti-TNF to cell surface expressed TmTNF (1) results in transducing intracellular ‘Reverse’ signal (2) and formation of a clathrin-coated pit (3) resulting in endocytosis of the complex into the endosomes (4) followed by delivery of the complex to the phagolysosome compartment (5) where proteolytically processed anti-TNF peptides are loaded onto HLA-DR and subsequently displayed as HLA-DR-associated peptides on the cell surface (6).
Materials & methods
Anti-TNFs tested
Table 1 lists the anti-TNF antibodies tested in the current study. These antibodies (either fully human or humanized IgG1 isotype) were expressed in human embryonic kidney (HEK)293 cells by transient transfections and purified using Protein-A affinity chromatography at AbbVie. Biophysical characteristics of the purified antibodies were analyzed using size exclusion chromatography (SEC) and all the listed antibodies were found to be greater than 94% monomer. The antibodies were found to have almost similar, sub-nM affinities, in their capacity to neutralize soluble TNF as assessed using the L929 assay52 and cellular uptake of pHRodo-conjugated mAbs as assessed by FACS. A dual variable domain Ig (DVD-Ig) molecule containing 2 TNF binding variable domains and 2 IL17 binding variable domains, and hence bivalent for TNF, with similar potency to the above mentioned anti-TNF mAbs, was also tested. The description of the DVD-Ig format is described in detail elsewhere.36
Isolation of monocytes and their differentiation to macrophages and dendritic cells
PBMCs were isolated from healthy donors by density gradient centrifugation over Ficoll-Paque53 (GE Healthcare). Monocytes were isolated from PBMCs by positive selection using CD14 microbeads and a magnetic cell separator as per the manufacturer's instructions (MACS, Miltenyi Biotec). The purity of the isolated monocytes was greater than 98% as determined by flow cytometry. Monocytes were cultured in cell growth medium (RPMI1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM L-glutamine, 100 μg/ml penicillin and streptomycin) at a density of 1 × 106 cells/ml at 37°C in humidified incubator with 5% CO2. Macrophages were generated by culturing 1 × 106/ml CD14+ monocytes in cell growth medium containing 100 ng/ml of recombinant human GM-CSF (AbbVie) and 2% heat-inactivated human AB serum (Invitrogen) for 7 days54,55 and were found to be larger in size compared with monocytes and were firmly attached to the tissue culture vessel. DCs were generated by culturing 1 × 106 cells/ml CD14+ monocytes in cell growth medium containing 100 ng/ml of recombinant human GM-CSF (AbbVie) and 5 ng/ml of human IL-4 (R&D Systems) for 5 d. To induce TNF, monocytes, macrophages and DCs were stimulated with the indicated concentrations of LPS (E. coli and S. typhimurium, Sigma) for the indicated periods of time, either in the presence or absence of 20 μM TAPI-2 (Sigma), a potent inhibitor of matrix metalloproteinases, including TACE.27
Cell surface biotinylation
Cell surface biotinylation of proteins was performed as described.56 Briefly, after the indicated treatments, cells (3 × 106) were washed twice with ice-cold PBS-CM (phosphate-buffered saline (PBS) containing 1 mM CaCl2 and 1 mM MgCl2). Cell surface proteins were derivatized twice using 1 mg/ml cell-impermeable EZ-Link-Sulfo-NHS-SS-Biotin (Thermo Fisher Scientific) in PBS-CM on ice for 30 min protected from light with gentle agitation. Excess biotin was quenched by incubating the cells for 10 min on ice in 50 mM NH4Cl in PBS-CM. Cells were washed twice with PBS-CM and total proteins extracted in 150 μl lysis buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% n-octyl-β-D-glucoside, 1% Triton X-100, protease inhibitor cocktail and 1 mM phenylmethylsulfonyl fluoride (PMSF)) on ice for 45 min and centrifuged at 14,000 x g for 10 min at 4°C. Clarified supernatant was transferred to a fresh micro-centrifuge tube on ice and total proteins estimated by BCA protein assay reagent (Thermo Fisher Scientific). To enrich cell surface biotinylated proteins, 25–75 μg total proteins in 500 μl lysis buffer were mixed constantly at 4°C overnight with 50 μl streptavidin-conjugated agarose beads (Thermo Fisher Scientific). Agarose beads were collected by centrifugation at 2,500 x g and sequentially washed twice (15 min between each wash) by suspending in 1 ml ice-cold lysis buffer, twice (15 min between each wash) in 1 ml ice-cold 500 mM NaCl, and once in 1 ml 50 mM Tris-HCl, pH 8. Streptavidin-agarose bound, cell surface biotinylated proteins, along with 5–15 μg total proteins in a separate tube, were suspended in SDS-PAGE sample buffer containing 62.5 mM Tris-HCl pH 6.8, 2% SDS, 4 M urea, 5% β-mercaptoethanol containing 10% glycerol. Proteins were separated on 4–20% Novex Tris-Glycine SDS-PAGE (Invitrogen) using 1X gel running buffer (Invitrogen), and transferred onto a 0.2 μm nitrocellulose membrane (Bio-Rad) for 1 h using 1X gel transfer buffer (Invitrogen). The nitrocellulose membrane was incubated in 5% non-fat dry milk in TBS-T (25 mM Tris-HCl, 150 mM NaCl, pH 7.5, containing 0.2% Tween-20) for 30 min at room temperature with gentle agitation, washed once in TBS-T for 5 min at room temperature and incubated overnight with gentle agitation at 4°C with the indicated primary antibodies (anti-TNF (BAF210, Lot ST2813011)) from R&D Systems; anti-GAPDH (2118, Lot 8), anti-Pan-cadherin (4073, Lot 1) and anti-Na, K-ATPase (3010, Lot 4) from Cell Signaling Technology). The membrane was washed twice for 15 min each with TBS-T with vigorous agitation at room temperature and incubated with appropriate horseradish peroxidase (HRP)-conjugated secondary IgG (NIF824, Lot 9564802 and NIF825, Lot 6000560 from GE Healthcare) in 5% non-fat dry milk in TBS-T for 45 min at room temperature with gentle agitation. The membrane was washed as above, and subjected to ECL or ECL Prime Western blotting analysis systems (GE Healthcare).
Antibody internalization assays
To determine if anti-TNF molecules bound to TmTNF are internalized into specific intracellular compartments, we conducted a series of experiments using either pHrodo- or Alexa 488-conjugated anti-TNF antibodies. pHrodo dye-conjugated antibodies are non-fluorescent outside the cell, but their fluorescence dramatically increases as pH decreases from neutral to acidic, as in intracellular vesicles like endosomes and lysosomes, making such dyes ideal tools to study endocytosis of pHrodo-conjugated antibodies.57 Since pHrodo-conjugated probes show pH-dependent fluorescence activation,33 conjugation of anti-TNF antibodies using pHrodo red dye or Alexa 488 was performed as per the manufacturer's protocol (Invitrogen). Cell surface expression of TmTNF was induced by treating DCs for 2 h with LPS at 37° C and the cells extensively washed using ice-cold growth medium to remove soluble TNF generated in the supernatants. Cells were incubated on ice in ice-cold medium supplemented with 2% human AB serum for 15 min to prevent processing of TmTNF to soluble TNF and to block cell surface Fc receptors. Cells were centrifuged under cold conditions and were further incubated on ice for 1 h either with ice-cold pHrodo-conjugated isotype matched control antibody or with pHrodo-conjugated anti-TNF in 2% human AB serum to specifically label cell surface TmTNF and prevent its cellular uptake. Cells were washed twice with ice-cold cell growth medium to remove unbound antibody and transferred to 37°C in growth medium containing 2% human AB serum for up to 4 h to determine kinetics of pHrodo accumulation in the intracellular acidic compartments. To observe distinct intracellular vesicles from the plasma membrane, cells were stained with Alexa 488-conjugated anti-MHC class I antibody (Biolegend). Accumulated fluorescence was recorded either by flow cytometry (BD Fortessa) or fluorescence microscopy (Amnis Imaging Flow Cytometers, EMD Millipore).
To precisely determine intracellular trafficking of TmTNF-bound anti-TNF antibody to either early endosome or lysosome, we performed immunocytochemistry by incubating DCs treated with 1 μg/ml LPS for 2 h to induce TmTNF expression. Cells were washed and incubated on ice for 1 h with Alexa 488-conjugated anti-TNF IgG in ice-cold cell growth medium supplemented with 2% human AB serum to block cell surface Fc receptors and to specifically label cell surface TmTNF and restrict its cellular uptake. After washing cells to remove excess antibody, cells were transferred to 37°C for up to 1 h to directly visualize the trafficking and cellular distribution of the labeled anti-TNF at various time points. Cells were fixed at room temperature using 4% para-formaldehyde for 15 min and permeabilized (PBS containing 0.3% Triton X-100 and 10% donkey serum) for 30 min at room temperature. Early endosomes or lysosomes were stained using rabbit anti-EEA1 (3288, Lot 7 from Cell Signaling Technology; 1:200) or rabbit anti-LAMP1 (9091, Lot 4 from Cell Signaling Technology; 1:200) antibodies, respectively, in antibody dilution buffer (PBS containing 0.1% Triton X-100, 1% BSA and 5% donkey serum). Cells were further stained for 45 min at room temperature using Alexa 647-conjugated anti-rabbit IgG (711–605–152, Lot 110488 from Jackson ImmunoResearch; 1:600 in antibody dilution buffer). Nucleus was stained with Nuce blue or DAPI (Invitrogen) and images acquired by confocal microscopy (Laica TCS SP5).
Identification of TmTNF-associated proteins
To identify TmTNF-associated proteins, DCs (10 × 106) were treated with LPS for 2 h at 37°C to induce cell surface expression of TmTNF. Cells were washed and incubated for 10 min in growth medium containing 5% heat-inactivated human AB serum followed by addition of either human IgG1 (15 μg/ml) or anti-TNF (15 μg/ml) antibodies at 37°C for the indicated period of time. Cells were washed twice using ice-cold PBS and total proteins extracted using cell lysis buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% n-octyl-β-D-glucoside, 1% Triton X-100, protease inhibitor cocktail and 1 mM PMSF) on ice for 45 min. Samples were centrifuged at 14,000 x g for 10 min at 4°C and clarified supernatant transferred to a fresh micro-centrifuge tube on ice. To enrich immune complexes, total proteins in 0.5 ml lysis buffer were mixed end-over-end for 1 h at 4°C by adding 50 μl Protein A-conjugated agarose beads (Cell Signaling Technologies). Agarose beads were collected by centrifugation at 2,500 x g and sequentially washed twice (15 min each) by suspending in 1 ml ice-cold lysis buffer and twice (15 min each) in 1 ml ice-cold PBS. Proteins were eluted from the beads using 100 mM glycine-HCl, pH 2.8.
The success of affinity purified TmTNF was verified by Western immunoblotting using anti-TNF antibody (R&D systems; BAF210). Remaining samples were subjected to high resolution sequencing mass spectrometry (LC-MS/MS) for identification of TmTNF-associated proteins. 30 μL of the immunoprecipitation eluates were combined with 10 μL of 4x non-reducing LDS-PAGE sample buffer and heated at 80°C for 10 min before loading onto 1.5 mm Bis-Tris 4–12% gradient gels (Invitrogen). Gel electrophoresis was conducted using non-reducing MES running buffer for one hour, followed by staining in Sypro Ruby. Using a Safe Imager™ 2.0 transilluminator (Invitrogen, Carlsbad, CA) to visualize the protein staining, each sample lane was cut into 48 equal pieces using a clean scalpel and the segments were added column-wise to a 96-well conical-bottom polypropylene plate. The plates were placed on a holder held at 50°C throughout the in-gel proteolysis process. The gel pieces were first washed and de-stained by adding 50 μL of 100 mM ammonium bicarbonate and incubating for 10 min before removing and discarding the wash. This washing process was repeated an additional 2 times, before washing once in 50 μL of acetonitrile for 5 min. The gel pieces were allowed to dry for 10 min after the removal of acetonitrile. Reduction and alkylation of the proteins was accomplished through addition of 50 μL of freshly prepared 10 mM dithiothreitol in 100 mM ammonium bicarbonate for 30 min followed by the addition of 50 μL of freshly prepared 55 mM iodoacetamide in 100 mM ammonium bicarbonate for 20 min. An additional 100 μL of acetonitrile was added for 5 min after reduction and alkylation, before removing and discarding the liquid. The samples were further washed in 50 μL of 100 mM ammonium bicarbonate for 10 min, followed by 3 sequential washes in 50 μL of acetonitrile, each lasting 5 min. The gel pieces were allowed to dehydrate for 20 min on the hot plate. Once dried, 25 μL of 6 ng/μL sequencing grade modified trypsin reconstituted in 50 mM ammonium bicarbonate was added to each sample. The plates were covered and allowed to incubate for 3 h at 50°C. After proteolysis was completed, 30 μL of extraction buffer was added (50:50 formic acid:acetonitrile), the plates re-covered and further incubated for 30 min at 50°C. The solution was collected and transferred to a hard shell 96-well thin wall PCR receiving plate and stored at 4°C. A second extraction was conducted using 15 μL of extraction buffer supplemented with an additional 10 μL of acetonitrile. After 30 min incubation at 50°C, the liquid was combined with the first extraction in the receiving plate. The sample plates were lyophilized and the samples reconstituted in 10 μL of 5% acetonitrile and 0.1% formic acid in nanopure water. The plates were briefly centrifuged before injection onto the mass spectrometer.
For LC-MS/MS analysis, samples were injected onto a 15 cm capillary column (I.D. 75 μm, O.D. 360 μm, tip size 15 μm) packed with Halo-Hilic resin (2.7 μm particle size, 90 Å pore size). The peptides were eluted using a Waters NanoAcquity in a 35 min linear gradient (15 to 35% acetonitrile in 0.1% formic acid) at 0.25 μL/min. The eluent was directed into a LTQ-Velos Orbitrap Pro mass spectrometer. Data-dependent scans were collected in the Orbitrap with tandem mass spectra collected in the ion trap. LC-MS/MS data files were searched using Mascot Daemon v.2.2 (Matrix Science, Boston, MA) against the NCBI nr mammalian database (version update on August 7th, 2012). Search parameters included 2+ to 3+ charge states, 2 missed cleavages, oxidized methionine variable modification and mass errors of + 20 ppm for intact spectra and + 0.8 Da for tandem mass spectra. Search results were compiled in Scaffold v.2.0 (Proteome Software, Portland, OR) and exported to Microsoft Excel. Gene identifications were assigned using in-house software with ambiguous protein entries subjected to BLAST searching against the human SWISS-PROT database. All keratin, trypsin and immunoglobulin proteins were eliminated from the data sets as exogenously introduced contaminants.58
Identification of TmTNF-associated clathrin heavy chain
To identify TmTNF-associated heavy chain of clathrin, DCs (3 × 106) were treated with LPS for 2 h at 37°C to induce cell surface expression of TmTNF. Cells were washed and incubated for 10 min in growth medium containing 5% heat-inactivated human AB serum followed by addition of either human IgG1 (15 μg/ml) or anti-TNF (15 μg/ml) antibodies at 37°C for 5 min. Cells were washed twice using ice-cold PBS and total proteins extracted using cell lysis buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% n-octyl-β-D-glucoside, 1% Triton X-100, protease inhibitor cocktail and 1 mM PMSF) on ice for 45 min. Samples were centrifuged at 14,000 x g for 10 min at 4°C and clarified supernatant transferred to a fresh micro-centrifuge tube on ice. To enrich immune complexes, total proteins in 0.5 ml lysis buffer were mixed end-over-end for 1 h at 4°C by adding 50 μl Protein A-conjugated agarose beads (Cell Signaling Technologies). Agarose beads were collected by centrifugation at 2,500 x g and sequentially washed twice (15 min each) by suspending in 1 ml ice-cold lysis buffer and twice (15 min each) in 1 ml ice-cold PBS. Proteins were subjected to SDS-PAGE chromatography followed by Western immunoblotting using either anti-TNF (R&D Systems; BAF210) or anti-clathrin heavy chain antibody (ab21679, Lot GR264025–1 from Abcam).
Cell Surface-associated anti-TNF Peptide Isolation
Mild acid elution
Cell surface-associated peptides were isolated using mild acidic conditions as described with some modifications.40 Briefly, DCs (∼6–8 × 106) were treated with LPS for 2 h at 37°C to induce cell surface expression of TmTNF. Cells were washed and incubated with anti-TNFs in cell growth medium at 37°C for 6–8 h. Cells were washed twice using ice-cold PBS. Cell surface associated peptides were eluted by incubating cells in ice-cold acid elution buffer (100 mM glycine, pH 2.8 containing 150 mM NaCl) on ice for 5 min, and the step repeated once. Pooled cell surface eluates were neutralized using 1/10 (v/v) 1 M Tris-HCl, pH 8 and subjected to LC-MS/MS for anti-TNF peptide identification using high resolution sequencing mass spectrometry.
Immunoprecipitation of HLA-DR
HLA class II (HLA-DR)-associated peptides were isolated as described previously with some modifications.59 Briefly, DCs (∼8–10 × 106) were treated with LPS for 2 h at 37°C to induce cell surface expression of TmTNF. Cells were washed and incubated with anti-TNFs in cell growth medium at 37°C for up to 8 h. Cells were washed twice using ice-cold PBS and total proteins extracted using cell lysis buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% n-Octyl-β-D-glucoside, 1% Triton X-100, protease inhibitor cocktail and 1 mM PMSF) on ice for 45 min. Samples were centrifuged at 14,000 x g for 10 min at 4°C. Clarified supernatant was transferred to a fresh micro-centrifuge tube on ice and total proteins estimated using the BCA protein assay reagent (Thermo Fisher Scientific). To enrich HLA class II protein, 500–1000 μg total proteins in 1 ml lysis buffer were mixed end-over-end overnight at 4°C with 20 μg mouse anti-human HLA-DR IgG (Biolegend). The next day, immune complexes were enriched by adding 50 μl Protein G-conjugated agarose beads (Cell Signaling Technologies) and the sample mixed with constant shaking for 2 h at 4°C. Agarose beads were collected by centrifugation at 2,500 x g and sequentially washed twice (15 min each) by suspending in 1 ml ice-cold lysis buffer and twice (15 min each) in 1 ml ice-cold PBS. Proteins were eluted from the beads using 0.2% trifluoroacetic acid. The success of affinity purified HLA-DR was verified by Western immunoblotting using rabbit anti-human HLA-DR antibody (ab92511, Lot GR2651–7 from Abcam). Remaining samples were subjected to centrifugation using 10 kDa cut-off spin columns (EMD Millipore) and the flow-through subjected to high resolution sequencing mass spectrometry (LC-MS/MS) for identification of anti-TNF peptides.
Anti-TNF peptide detection by LC-MS/MS
Cell surface peptides obtained from DCs, as mentioned above, were subjected to peptide identification using capillary LC-MS system, which consists of an UPLC (Acquity UPLC system, Waters) coupled to LTQ-Orbitrap velos mass spectrometer (Thermo Fisher, Rockford, IL). The column used was C18 reversed-phase column (Waters, BEH, 1 × 50 mm i.d., 1.7 µm particle size). Forty µL of sample was loaded at 98% mobile phase A (0.1% FA in Milli-Q water) and 2% mobile phase B (0.1% FA in acetonitrile), after 5 min, peptides were eluted using gradient as follows: 2% mobile phase B to 20% mobile phase B in 60 min, 20% mobile phase B to 35% mobile phase B in 20 min, 98% mobile phase B for 5 min, 2% mobile phase B for 10 min for equilibration. The flow rate was set at 50 µL/min and column oven temperature was set at 60°C. The mass spectrometer was operated in positive ion mode with a scan range from m/z 400 to 2000 with 60,000 resolution at m/z 400. Peptide fragmentation was achieved using collision-induced dissociation (CID) for the top 10 most intense parent ions. Automated gain control (AGC) values for MS and MS/MS were 1E6 and 5E4, respectively. Ion spray voltage was set at 4500 V and the source temperature was set at ambient.
To identify the peptides from anti-TNF antibody or DVD molecule, the resulting mass spectrometry raw data was searched against antibody sequence using proteome discoverer with the enzyme specificity set as “no enzyme." The mass tolerance for MS was 10 ppm, and MS/MS was 0.2 Da. The positive hits from database search were manually validated. Alternatively, the raw data was searched against human protein peptide sequence database using Mascot™ (Matrix science) or PEAKS™ (Bioinformatics Solutions).
Generation of anti-TNF-TT fusion peptide antibodies
We chose the following 2 peptide sequences from different regions of the tetanus toxin protein60 to be fused to the C-terminus of the heavy chain of an anti-TNF antibody:
S1 Peptide Sequence: 580-NSVDDALINSTKIYSYFPSV-599
S2 Peptide Sequence: 919-NGKAIHLVNNESSEVIVHKAMDI-941
gBlock gene fragments encoding the above peptide sequences of the tetanus toxin were synthesized (IDT) containing the 5′ and 3′-overlapping sequences of the parental plasmid, AB436VH (pHybE-hIgG1,z,non-a, Abbvie). The nucleotide sequences encoding the respective S1 and S2 peptides are underlined below:
S1 Peptide Encoding Nucleotide Sequence:
(caaccactacacgcagaagagcctctccctgtctccgggtaaaaacagcgtggacgacgccctgatcaacagcaccaagatctacagctacttccccagcgtgtgagcggccgctcgaggccggcaaggccggatcccccgacctcgacct)
S2 Peptide Encoding Nucleotide Sequence (ccactacacgcagaagagcctctccctgtctccgggtaaaaacggcaaggccatccacctggtgaacaacgagagcagcgaggtgatcgtgcacaaggccatggacatctgagcggccgctcgaggccggcaaggccggatcccccgacctcga)
AB436VH containing plasmid DNA (pHybE-hIgG1, z, non-a) was digested using NotI (NEB) and purified by Qiaquick PCR purification kit (Qiagen). gBlock fragments were inserted into the linear plasmid by homologous recombination in competent DH5α E. coli (Invitrogen). Proteins were expressed by transfecting the respective plasmids encoding the heavy chain (AB436VH-S1, -S2, encoding nucleotide sequences) and light chain (AB436VL) in HEK293–6E cells (ATCC) using polyethyleneimine (PEI).61 Seven days later, TT-fusion mAbs were purified using Protein A chromatography (GE Healthcare) and dialyzed against PBS. All antibodies were confirmed to be less than 10% aggregates by SEC (AbbVie).
L929 cytotoxic assay
To ascertain the neutralizing efficacy of anti-TNF-TT fusion antibodies compared with the parental anti-TNF, we performed L929 assay as described previously.52 Briefly, murine L929 fibrosarcoma cells in log phase were harvested by trypsinization, washed and suspended in cell growth medium (RPMI containing 10% FBS, 2 mM L-glutamine, 1% Na-pyruvate, 1% non-essential amino acids, 0.1% β-mercaptoethanol and 1% Pen/Strep). Cells were counted and plated in triplicate in 50 μl cell growth medium supplemented with 2 μg/ml actinomycin D (Sigma) in 96-well tissue culture plates at cell density of 1 × 106 cells/ml at 37°C in humidified 5% CO2 incubator. Beginning at 10 nM, 1:3 serial dilutions of anti-TNF antibodies to be tested, and 100 pg/ml recombinant human TNF were performed separately in cell growth medium, mixed and incubated at room temperature for 1 h. 50 μl of the above dilutions of anti-TNF:TNF were added to the wells containing L929 cells to yield a final concentration of 1 μg/ml actinomycin D per well, along with appropriate positive (containing TNF alone) and negative (without TNF) controls. Cells were incubated for ∼18 h at 37°C in humidified 5% CO2 atmosphere. Freshly prepared cell proliferation detection reagent, WST-1 (Roche), was added to each well to evaluate cell viability and the cells further incubated at 37°C in humidified 5% CO2 atmosphere for 4 h. Absorbance was recorded using Spectramax plate reader (Molecular Devices) at 420–600 nm for spectrophotometric quantification of cell viability. A nonlinear regression curve was generated by plotting the antibody concentrations on a logarithmic scale on the x-axis and the OD on the y-axis.
T cell proliferation assay
DCs were generated from CD14+ monocytes from PBMCs of healthy human donors which had been immunized with tetanus toxoid and were stimulated with 250 ng/ml of Salmonella typhimurium LPS for 2 h at 37°C to induce the expression of TmTNF. DCs were either treated with TT (20 μg/ml) or TT peptides (S1 or S2) conjugated anti-TNF antibodies (anti-TNF-S1 or anti-TNF-S2; 20 μg/ml each) and incubated in cell growth medium at 37°C for an additional 6 h. Autologous T cells were purified from PBMCs of the same donor from which monocytes were obtained to generate DCs using a pan T cell isolation kit (Miltenyi), and labeled with 2.5 μM CFSE (Invitrogen). DCs were washed twice after antibody pulsing and cultured with autologous T cells at a ratio of 1:50 for up to 7 d. T cell proliferation was assessed by using flow cytometry. The percentage of live T cells that had undergone cell division was determined by gating on DAPI-negative CD3+ cells, and assessing the fraction that showed diminished CFSE fluorescence intensity.62
Disclosure of potential conflicts of interest
Arun Deora, Subramanya Hegde, Jacqueline Lee, Chee-Ho Choi, Qing Chang, Cheryl Lee, Lucia Eaton, Hua Tang, Mark Michalak, Medha Tomlinson, Qingfeng Tao, Bohdan Harvey, Shaun McLoughlin, Boris Labkovsky and Tariq Ghayur are employees of AbbVie Inc. and may own AbbVie stocks or stock options. Nidhi Gaur, David Lee and Dongdong Wang were employees of AbbVie at the time of the study. The authors have no other relevant affiliations or financial involvement with any other organization or entity with a financial interest or conflict with the subject matter or materials discussed in the current manuscript. The design, study conduct, and financial support for this research were provided by AbbVie. AbbVie participated in the collection, analysis and interpretation of the data, review, and approval of the publication.
Acknowledgments
The authors gratefully acknowledge and thank the support and feedback provided by Jijie Gu, Robert Stoffel, Jochen Salfeld and Lisa Olson.
References
- 1.Perez C, Albert I, DeFay K, Zachariades N, Gooding L, Kriegler M. A nonsecretable cell surface mutant of tumor necrosis factor (TNF) kills by cell-to-cell contact. Cell 1990; 63:251-8; PMID:2208285; http://dx.doi.org/ 10.1016/0092-8674(90)90158-B [DOI] [PubMed] [Google Scholar]
- 2.Kriegler M, Perez C, DeFay K, Albert I, Lu SD. A novel form of TNF/cachectin is a cell surface cytotoxic transmembrane protein: ramifications for the complex physiology of TNF. Cell 1988; 53:45-53; PMID:3349526; http://dx.doi.org/ 10.1016/0092-8674(88)90486-2 [DOI] [PubMed] [Google Scholar]
- 3.Horiuchi T, Mitoma H, Harashima S, Tsukamoto H, Shimoda T. Transmembrane TNF-alpha: structure, function and interaction with anti-TNF agents. Rheumatology (Oxford) 2010; 49:1215-28; PMID:20194223; http://dx.doi.org/ 10.1093/rheumatology/keq031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, et al.. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature 2013; 496:110-3; PMID:23552949; http://dx.doi.org/ 10.1038/nature12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedmann E, Hauben E, Maylandt K, Schleeger S, Vreugde S, Lichtenthaler SF, Kuhn PH, Stauffer D, Rovelli G, Martoglio B. SPPL2a and SPPL2b promote intramembrane proteolysis of TNFalpha in activated dendritic cells to trigger IL-12 production. Nat Cell Biol 2006; 8:843-8; PMID:16829952; http://dx.doi.org/ 10.1038/ncb1440 [DOI] [PubMed] [Google Scholar]
- 6.Fluhrer R, Grammer G, Israel L, Condron MM, Haffner C, Friedmann E, Böhland C, Imhof A, Martoglio B, Teplow DB, et al.. A gamma-secretase-like intramembrane cleavage of TNFalpha by the GxGD aspartyl protease SPPL2b. Nat Cell Biol 2006; 8:894-6; PMID:16829951; http://dx.doi.org/ 10.1038/ncb1450 [DOI] [PubMed] [Google Scholar]
- 7.Pallai A, Kiss B, Vereb G, Armaka M, Kollias G, Szekanecz Z, Szondy Z. Transmembrane TNF-alpha reverse signaling inhibits lipopolysaccharide-induced proinflammatory cytokine formation in macrophages by inducing TGF-beta: Therapeutic implications. J Immunol 2016; 196:1146-57; PMID:26729808; http://dx.doi.org/ 10.4049/jimmunol.1501573 [DOI] [PubMed] [Google Scholar]
- 8.Smith CA, Farrah T, Goodwin RG. The TNF receptor superfamily of cellular and viral proteins: activation, costimulation, and death. Cell 1994; 76:959-62; PMID:8137429; http://dx.doi.org/ 10.1016/0092-8674(94)90372-7 [DOI] [PubMed] [Google Scholar]
- 9.Watts AD, Onier-Cherix N, Hunt NH, Chaudhri G. Use of fixed cells in cell contact-dependent cytotoxicity assays for TNF: a cautionary report. J Immunol Methods 1999; 225:179-84; PMID:10365794; http://dx.doi.org/ 10.1016/S0022-1759(99)00046-0 [DOI] [PubMed] [Google Scholar]
- 10.de Waal Malefyt R, Abrams J, Bennett B, Figdor CG, de Vries JE. Interleukin 10(IL-10) inhibits cytokine synthesis by human monocytes: an autoregulatory role of IL-10 produced by monocytes. J Exp Med 1991; 174:1209-20; PMID:1940799; http://dx.doi.org/ 10.1084/jem.174.5.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho LJ, Wang JJ, Shaio MF, Kao CL, Chang DM, Han SW, Lai JH. Infection of human dendritic cells by dengue virus causes cell maturation and cytokine production. J Immunol 2001; 166:1499-506;PMID:11160189;http://dx.doi.org/ 10.4049/jimmunol.166.3.1499 [DOI] [PubMed] [Google Scholar]
- 12.Brehm MA, Daniels KA, Welsh RM. Rapid production of TNF-alpha following TCR engagement of naive CD8 T cells. J Immunol 2005; 175:5043-9; PMID:16210607; http://dx.doi.org/ 10.4049/jimmunol.175.8.5043 [DOI] [PubMed] [Google Scholar]
- 13.Williamson BD, Carswell EA, Rubin BY, Prendergast JS, Old LJ. Human tumor necrosis factor produced by human B-cell lines: synergistic cytotoxic interaction with human interferon. Proc Natl Acad Sci U S A 1983; 80:5397-401; PMID:6193516; http://dx.doi.org/ 10.1073/pnas.80.17.5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fauriat C, Long EO, Ljunggren HG, Bryceson YT. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood 2010; 115:2167-76; PMID:19965656; http://dx.doi.org/ 10.1182/blood-2009-08-238469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coulthard LR, Geiler J, Mathews RJ, Church LD, Dickie LJ, Cooper DL, Wong C, Savic S, Bryer D, Buch MH, et al.. Differential effects of infliximab on absolute circulating blood leucocyte counts of innate immune cells in early and late rheumatoid arthritis patients. Clin Exp Immunol 2012; 170:36-46; PMID:22943199; http://dx.doi.org/ 10.1111/j.1365-2249.2012.04626.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imaizumi T, Itaya H, Fujita K, Kudoh D, Kudoh S, Mori K, Fujimoto K, Matsumiya T, Yoshida H, Satoh K. Expression of tumor necrosis factor-alpha in cultured human endothelial cells stimulated with lipopolysaccharide or interleukin-1alpha. Arterioscler Thromb Vasc Biol 2000; 20:410-5; PMID:10669637; http://dx.doi.org/ 10.1161/01.ATV.20.2.410 [DOI] [PubMed] [Google Scholar]
- 17.Cawthorn WP, Sethi JK. TNF-alpha and adipocyte biology. FEBS Lett 2008; 582:117-31; PMID:18037376; http://dx.doi.org/ 10.1016/j.febslet.2007.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Havell EA, Rogerson BJ. Endotoxin-induced tumor necrosis factor alpha synthesis in murine embryo fibroblasts. Infect Immun 1993; 61:1630-5; PMID:8478050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakao A, Fukushima H, Kajiya H, Ozeki S, Okabe K. RANKL-stimulated TNFalpha production in osteoclast precursor cells promotes osteoclastogenesis by modulating RANK signaling pathways. Biochem Biophys Res Commun 2007; 357:945-50; PMID:17467668; http://dx.doi.org/ 10.1016/j.bbrc.2007.04.058 [DOI] [PubMed] [Google Scholar]
- 20.Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol 2003; 3:745-56; PMID:12949498; http://dx.doi.org/ 10.1038/nri1184 [DOI] [PubMed] [Google Scholar]
- 21.Hijdra D, Vorselaars AD, Grutters JC, Claessen AM, Rijkers GT. Differential expression of TNFR1 (CD120a) and TNFR2 (CD120b) on subpopulations of human monocytes. J Inflamm (Lond) 2012; 9:38; PMID:23039818; http://dx.doi.org/ 10.1186/1476-9255-9-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grell M, Douni E, Wajant H, Lohden M, Clauss M, Maxeiner B, Georgopoulos S, Lesslauer W, Kollias G, Pfizenmaier K, et al.. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell 1995; 83:793-802; PMID:8521496; http://dx.doi.org/ 10.1016/0092-8674(95)90192-2 [DOI] [PubMed] [Google Scholar]
- 23.Bathon JM, Martin RW, Fleischmann RM, Tesser JR, Schiff MH, Keystone EC, Genovese MC, Wasko MC, Moreland LW, Weaver AL, et al.. A comparison of etanercept and methotrexate in patients with early rheumatoid arthritis. N Engl J Med 2000; 343:1586-93; PMID:11096165; http://dx.doi.org/ 10.1056/NEJM200011303432201 [DOI] [PubMed] [Google Scholar]
- 24.Lipsky PE, van der Heijde DM, St Clair EW, Furst DE, Breedveld FC, Kalden JR, Smolen JS, Weisman M, Emery P, Feldmann M, et al.. Infliximab and methotrexate in the treatment of rheumatoid arthritis. Anti-Tumor Necrosis Factor Trial in Rheumatoid Arthritis with Concomitant Therapy Study Group. N Engl J Med 2000; 343:1594-602; PMID:11096166; http://dx.doi.org/ 10.1056/NEJM200011303432202 [DOI] [PubMed] [Google Scholar]
- 25.Targan SR, Hanauer SB, van Deventer SJ, Mayer L, Present DH, Braakman T, DeWoody KL, Schaible TF, Rutgeerts PJ. A short-term study of chimeric monoclonal antibody cA2 to tumor necrosis factor alpha for Crohn's disease. Crohn's Disease cA2 Study Group. N Engl J Med 1997; 337:1029-35. [DOI] [PubMed] [Google Scholar]
- 26.Baert FJ, D'Haens GR, Peeters M, Hiele MI, Schaible TF, Shealy D, Geboes K, Rutgeerts PJ. Tumor necrosis factor alpha antibody (infliximab) therapy profoundly down-regulates the inflammation in Crohn's ileocolitis. Gastroenterology 1999; 116:22-8; PMID:9869598; http://dx.doi.org/ 10.1016/S0016-5085(99)70224-6 [DOI] [PubMed] [Google Scholar]
- 27.Mullberg J, Durie FH, Otten-Evans C, Alderson MR, Rose-John S, Cosman D, Black RA, Mohler KM. A metalloprotease inhibitor blocks shedding of the IL-6 receptor and the p60 TNF receptor. J Immunol 1995; 155:5198-205; PMID:7594530 [PubMed] [Google Scholar]
- 28.Gearing AJ, Beckett P, Christodoulou M, Churchill M, Clements J, Davidson AH, Drummond AH, Galloway WA, Gilbert R, Gordon JL, et al.. Processing of tumour necrosis factor-alpha precursor by metalloproteinases. Nature 1994; 370:555-7; PMID:8052310; http://dx.doi.org/ 10.1038/370555a0 [DOI] [PubMed] [Google Scholar]
- 29.Mayor S, Pagano RE. Pathways of clathrin-independent endocytosis. Nat Rev Mol Cell Biol 2007; 8:603-12; PMID:17609668; http://dx.doi.org/ 10.1038/nrm2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol 2009; 10:609-22; PMID:19696798; http://dx.doi.org/ 10.1038/nrm2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatterjee B, Smed-Sorensen A, Cohn L, Chalouni C, Vandlen R, Lee BC, Widger J, Keler T, Delamarre L, Mellman I. Internalization and endosomal degradation of receptor-bound antigens regulate the efficiency of cross presentation by human dendritic cells. Blood 2012; 120:2011-20; PMID:22791285; http://dx.doi.org/ 10.1182/blood-2012-01-402370 [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, Yan D, Shi X, Liang H, Pang Y, Qin N, Chen H, Wang J, Yin B, Jiang X, et al.. Transmembrane TNF-alpha mediates "forward" and "reverse" signaling, inducing cell death or survival via the NF-kappaB pathway in Raji Burkitt lymphoma cells. J Leukoc Biol 2008; 84:789-97; PMID:18550789; http://dx.doi.org/ 10.1189/jlb.0208078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miksa M, Komura H, Wu R, Shah KG, Wang P. A novel method to determine the engulfment of apoptotic cells by macrophages using pHrodo succinimidyl ester. J Immunol Methods 2009; 342:71-7; PMID:19135446; http://dx.doi.org/ 10.1016/j.jim.2008.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howes MT, Kirkham M, Riches J, Cortese K, Walser PJ, Simpson F, Hill MM, Jones A, Lundmark R, Lindsay MR, et al.. Clathrin-independent carriers form a high capacity endocytic sorting system at the leading edge of migrating cells. J Cell Biol 2010; 190:675-91; PMID:20713605; http://dx.doi.org/ 10.1083/jcb.201002119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harper CB, Martin S, Nguyen TH, Daniels SJ, Lavidis NA, Popoff MR, Hadzic G, Mariana A, Chau N, McCluskey A, et al.. Dynamin inhibition blocks botulinum neurotoxin type A endocytosis in neurons and delays botulism. J Biol Chem 2011; 286:35966-76; PMID:21832053; http://dx.doi.org/ 10.1074/jbc.M111.283879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu C, Ying H, Grinnell C, Bryant S, Miller R, Clabbers A, Bose S, McCarthy D, Zhu RR, Santora L, et al.. Simultaneous targeting of multiple disease mediators by a dual-variable-domain immunoglobulin. Nat Biotechnol 2007; 25:1290-7; PMID:17934452; http://dx.doi.org/ 10.1038/nbt1345 [DOI] [PubMed] [Google Scholar]
- 37.Cousens LP, Tassone R, Mazer BD, Ramachandiran V, Scott DW, De Groot AS. Tregitope update: mechanism of action parallels IVIg. Autoimmun Rev 2013; 12:436-43; PMID:22944299; http://dx.doi.org/ 10.1016/j.autrev.2012.08.017 [DOI] [PubMed] [Google Scholar]
- 38.Matthews SP, Werber I, Deussing J, Peters C, Reinheckel T, Watts C. Distinct protease requirements for antigen presentation in vitro and in vivo. J Immunol 2010; 184:2423-31; PMID:20164435; http://dx.doi.org/ 10.4049/jimmunol.0901486 [DOI] [PubMed] [Google Scholar]
- 39.Akassoglou K, Douni E, Bauer J, Lassmann H, Kollias G, Probert L. Exclusive tumor necrosis factor (TNF) signaling by the p75TNF receptor triggers inflammatory ischemia in the CNS of transgenic mice. Proc Natl Acad Sci U S A 2003; 100:709-14; PMID:12522266; http://dx.doi.org/ 10.1073/pnas.0236046100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luettig B, Decker T, Lohmann-Matthes ML. Evidence for the existence of two forms of membrane tumor necrosis factor: an integral protein and a molecule attached to its receptor. J Immunol 1989; 143:4034-8; PMID:2556474 [PubMed] [Google Scholar]
- 41.Mitoma H, Horiuchi T, Hatta N, Tsukamoto H, Harashima S, Kikuchi Y, Otsuka J, Okamura S, Fujita S, Harada M. Infliximab induces potent anti-inflammatory responses by outside-to-inside signals through transmembrane TNF-alpha. Gastroenterology 2005; 128:376-92; PMID:15685549; http://dx.doi.org/ 10.1053/j.gastro.2004.11.060 [DOI] [PubMed] [Google Scholar]
- 42.St Pierre CA, Leonard D, Corvera S, Kurt-Jones EA, Finberg RW. Antibodies to cell surface proteins redirect intracellular trafficking pathways. Exp Mol Pathol 2011; 91:723-32; PMID:21819978; http://dx.doi.org/ 10.1016/j.yexmp.2011.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiao SW, Kobayashi K, Johansen FE, Sollid LM, Andersen JT, Milford E, Roopenian DC, Lencer WI, Blumberg RS. Dependence of antibody-mediated presentation of antigen on FcRn. Proc Natl Acad Sci U S A 2008; 105:9337-42; PMID:18599440; http://dx.doi.org/ 10.1073/pnas.0801717105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujimoto K, Ida H, Hirota Y, Ishigai M, Amano J, Tanaka Y. Intracellular dynamics and fate of a humanized anti-interleukin-6 receptor monoclonal antibody, tocilizumab. Mol Pharmacol 2015; 88:660-75; PMID:26180046; http://dx.doi.org/ 10.1124/mol.115.099184 [DOI] [PubMed] [Google Scholar]
- 45.Mellman I, Plutner H, Ukkonen P. Internalization and rapid recycling of macrophage Fc receptors tagged with monovalent antireceptor antibody: possible role of a prelysosomal compartment. J Cell Biol 1984; 98:1163-9; PMID:6715403; http://dx.doi.org/ 10.1083/jcb.98.4.1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonifaz LC, Bonnyay DP, Charalambous A, Darguste DI, Fujii S, Soares H, Brimnes MK, Moltedo B, Moran TM, Steinman RM. In vivo targeting of antigens to maturing dendritic cells via the DEC-205 receptor improves T cell vaccination. J Exp Med 2004; 199:815-24; PMID:15024047; http://dx.doi.org/ 10.1084/jem.20032220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schellekens H. Immunogenicity of therapeutic proteins: clinical implications and future prospects. Clin Ther 2002; 24:1720-40; discussion 19; PMID:12501870; http://dx.doi.org/ 10.1016/S0149-2918(02)80075-3 [DOI] [PubMed] [Google Scholar]
- 48.Rossol M, Meusch U, Pierer M, Kaltenhauser S, Hantzschel H, Hauschildt S, Wagner U. Interaction between transmembrane TNF and TNFR1/2 mediates the activation of monocytes by contact with T cells. J Immunol 2007; 179:4239-48; PMID:17785864; http://dx.doi.org/ 10.4049/jimmunol.179.6.4239 [DOI] [PubMed] [Google Scholar]
- 49.Slevin SM, Egan LJ. New Insights into the mechanisms of action of anti-tumor necrosis factor-alpha monoclonal antibodies in inflammatory bowel disease. Inflamm Bowel Dis 2015; 21:2909-20; PMID:26348448; http://dx.doi.org/ 10.1097/MIB.0000000000000533 [DOI] [PubMed] [Google Scholar]
- 50.Vos AC, Wildenberg ME, Duijvestein M, Verhaar AP, van den Brink GR, Hommes DW. Anti-tumor necrosis factor-alpha antibodies induce regulatory macrophages in an Fc region-dependent manner. Gastroenterology 2011; 140:221-30; PMID:20955706; http://dx.doi.org/ 10.1053/j.gastro.2010.10.008 [DOI] [PubMed] [Google Scholar]
- 51.Nguyen DX, Ehrenstein MR. Anti-TNF drives regulatory T cell expansion by paradoxically promoting membrane TNF-TNF-RII binding in rheumatoid arthritis. J Exp Med 2016; 213:1241-53; PMID:27270893; http://dx.doi.org/ 10.1084/jem.20151255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shiau MY, Chiou HL, Lee YL, Kuo TM, Chang YH. Establishment of a consistent L929 bioassay system for TNF-alpha quantitation to evaluate the effect of lipopolysaccharide, phytomitogens and cytodifferentiation agents on cytotoxicity of TNF-alpha secreted by adherent human mononuclear cells. Mediators Inflamm 2001; 10:199-208; PMID:11577996; http://dx.doi.org/ 10.1080/09629350123139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dispinseri S, Saba E, Vicenzi E, Kootstra NA, Schuitemaker H, Scarlatti G. HIV-1 isolation from infected peripheral blood mononuclear cells. Methods Mol Biol 2014; 1087:187-96; PMID:24158823 [DOI] [PubMed] [Google Scholar]
- 54.Eischen A, Vincent F, Bergerat JP, Louis B, Faradji A, Bohbot A, Oberling F. Long term cultures of human monocytes in vitro. Impact of GM-CSF on survival and differentiation. J Immunol Methods 1991; 143:209-21. [DOI] [PubMed] [Google Scholar]
- 55.Young DA, Lowe LD, Clark SC. Comparison of the effects of IL-3, granulocyte-macrophage colony-stimulating factor, and macrophage colony-stimulating factor in supporting monocyte differentiation in culture. Analysis of macrophage antibody-dependent cellular cytotoxicity. J Immunol 1990; 145:607-15; PMID:2142182 [PubMed] [Google Scholar]
- 56.Deora AB, Kreitzer G, Jacovina AT, Hajjar KA. An annexin 2 phosphorylation switch mediates p11-dependent translocation of annexin 2 to the cell surface. J Biol Chem 2004; 279:43411-8; PMID:15302870; http://dx.doi.org/ 10.1074/jbc.M408078200 [DOI] [PubMed] [Google Scholar]
- 57.Ogawa M, Kosaka N, Regino CA, Mitsunaga M, Choyke PL, Kobayashi H. High sensitivity detection of cancer in vivo using a dual-controlled activation fluorescent imaging probe based on H-dimer formation and pH activation. Mol Biosyst 2010; 6:888-93; PMID:20567775; http://dx.doi.org/ 10.1039/b917876g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 2006; 1:2856-60; PMID:17406544; http://dx.doi.org/ 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- 59.van Haren SD, Herczenik E, ten Brinke A, Mertens K, Voorberg J, Meijer AB. HLA-DR-presented peptide repertoires derived from human monocyte-derived dendritic cells pulsed with blood coagulation factor VIII. Mol Cell Proteomics 2011; 10:M110 002246; PMID:21467215; http://dx.doi.org/ 10.1074/mcp.M110.002246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fairweather NF, Lyness VA. The complete nucleotide sequence of tetanus toxin. Nucleic Acids Res 1986; 14:7809-12; PMID:3774547; http://dx.doi.org/ 10.1093/nar/14.19.7809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gu J, Yang J, Chang Q, Liu Z, Ghayur T, Gu J. Identification of Anti-EGFR and Anti-ErbB3 dual variable domains immunoglobulin (DVD-Ig) proteins with unique activities. PLoS One 2015; 10:e0124135; PMID:25997020; http://dx.doi.org/ 10.1371/journal.pone.0124135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hegde S, Jankowska-Gan E, Roenneburg DA, Torrealba J, Burlingham WJ, Gumperz JE. Human NKT cells promote monocyte differentiation into suppressive myeloid antigen-presenting cells. J Leukoc Biol 2009; 86:757-68; PMID:19465641; http://dx.doi.org/ 10.1189/jlb.0209059 [DOI] [PMC free article] [PubMed] [Google Scholar]