Abstract
Androgen signaling plays an important role in many biological processes. Androgen Responsive Gene Database (ARGDB) is devoted to providing integrated knowledge on androgen-controlled genes. Gene records were collected on the basis of PubMed literature collections. More than 6000 abstracts and 950 original publications were manually screened, leading to 1785 human genes, 993 mouse genes, and 583 rat genes finally included in the database. All the collected genes were experimentally proved to be regulated by androgen at the expression level or to contain androgen-responsive regions. For each gene important details of the androgen regulation experiments were collected from references, such as expression change, androgen-responsive sequence, response time, tissue/cell type, experimental method, ligand identity, and androgen amount, which will facilitate further evaluation by researchers. Furthermore, the database was integrated with multiple annotation resources, including National Center for Biotechnology Information, Gene Ontology, and Kyoto Encyclopedia of Genes and Genomes pathway, to reveal the biological characteristics and significance of androgen-regulated genes. The ARGDB web site is mainly composed of the Browse, Search, Element Scan, and Submission modules. It is user friendly and freely accessible at http://argdb.fudan.edu.cn. Preliminary analysis of the collected data was performed. Many disease pathways, such as prostate carcinogenesis, were found to be enriched in androgen-regulated genes. The discovered androgen-response motifs were similar to those in previous reports. The analysis results are displayed in the web site. In conclusion, ARGDB provides a unified gateway to storage, retrieval, and update of information on androgen-regulated genes.
Androgen Responsive Gene Database provides a unified gateway to storage, retrieval and update of information on androgen-regulated genes.
Androgen signaling is critical to the development and maintenance of male sexual characteristics, such as muscle mass, strength, bone mineral density, prostate growth, spermatogenesis, hair pattern, and neuron remodeling (1, 2). It also plays a pivotal role in the female physiology and reproduction (1, 3). Androgen signaling has been linked with many diseases, including prostate cancer, breast cancer, diabetes, metabolic syndrome, and Alzheimer’s disease (1, 4, 5, 6, 7). Thus, fully understanding androgen action is of fundamental significance.
The androgen receptor (AR) is the key mediator of androgen signaling. Upon androgen binding, AR translocates from the cytoplasm to the nucleus. It can directly associate with the androgen-responsive sequences in the transcription-regulatory regions of various target genes and then regulate gene expression (4, 8). The androgen-responsive sequence was thought to be characterized by the six-nucleotide half-site consensus sequence 5′-TGTTCT-3′ spaced by three random nucleotides. However, use of this motif model did not effectively predict AR-binding regions or androgen target genes (9, 10, 11). Liganded AR also interacts with other transcriptional regulators in the nucleus to stimulate or repress transcription, during the process of which AR is indirectly associated with the regulatory sequences (12, 13). In addition, androgen can induce rapid activation of the Src/Raf-1/ERK pathway through AR in the cytoplasm, independently of receptor-DNA interactions (4). Gene expression profiles responding to the androgen stimulation depend on the cell context, including cell type, androgen amount, and exposure time, etc. (14, 15, 16). In conclusion, the androgen-signaling pathway is complex.
PubMed is one of the central literature repositories, covering more than 17 million publications (http://www.ncbi.nlm.nih.gov/sites/entrez?db=PubMed). So far, there are more than 90,000 papers on androgen in PubMed. Facing such a large amount of information, to the best of our knowledge, there is no database focused on the androgen-regulated genes. AndrogenDB, the only database directed toward androgen, focuses on the AR mutations (17), and existing gene regulation databases such as the famous TRANSFAC still lack information on androgen-responsive genes (18).
To complement the existing databases and facilitate androgen research, we constructed the Androgen-Responsive Gene Database (ARGDB). We manually collected the experimentally proved androgen-regulated genes (ARGs) and related evidence from the original research articles. The database was integrated with three well-established annotation systems: National Center for Biotechnology Information (NCBI), Gene Ontology (GO) (19), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (20).
Results
Database overview
More than 6000 PubMed records were retrieved in total. After manual selection, 950 records were found about androgen regulation of ARG expression or ARG’s androgen-responsive regions. Original publications of those records were investigated, leading to 1785 human genes, 993 mouse genes, and 583 rat genes that were finally included in ARGDB.
ARGDB records are indexed by gene identity (ID). The IDs of 1745 human ARGs, 993 mouse ARGs, and 583 rat ARGs in ARGDB are equal to their Entrez Gene ID. The Gene ID of the remaining ARGs, all of which were noncoding RNA genes, could not be determined. Their IDs in ARGDB are equal to their GenBank accession numbers or their miRBase IDs. Each individual gene record is organized into three parts:
-
1.
Biological annotations, which generally include organism, symbol, name, cytoband, a schematic representation of chromosomal location, summary, GO function, GO process, GO component, and KEGG pathway. Live links to the relevant databases are provided, including NCBI, NURSA, ONCOMINE, BOND, and BioGPS.
-
2.
Experimental details of androgen regulation at the RNA or protein level, which generally include regulation direction, response time, experimental method, tissue/cell type, ligand identity, and androgen amount. The response time is the length of time it takes to detect a significant change in an ARG’s RNA or protein level after androgen stimulation. A colorimetric scale was provided for researchers to quickly see how many publication records report an up-regulation vs. a down-regulation of a given gene.
-
3.
Information of the assayed androgen-responsive sequence, which generally includes original sequence, location in the ARG genomic region, distance from the ARG transcription start site, and an experiment summary. If a mutagenesis assay was performed, the mutated sequence and the result are also displayed.
Parts 2 and 3 both contain experimental evidence to support androgen regulation of ARGs, which was manually extracted from references. Most ARGs only have Part 2 evidence. Parts of ARGs have both parts of evidence. A small number of ARGs only have Part 3 evidence.
The ARGDB web site is mainly composed of four modules: Browse, Search, Element Scan, and Submission (Table 1). The Browse module is the largest and consists of seven submodules: A–Z List, Chromosome Map, GO Function, GO Process, GO Component, KEGG pathway, and Noncoding RNA Gene. Its goal is to display ARGs by classification. Functions of each submodule are summarized in Table 1. As an example, consider the KEGG Pathway submodule (Fig. 1). It incorporates both species-specific and reference KEGG pathways (KEGG link listed in Table 2) (Fig. 1A); the latter pathway is applicable to all organisms. This submodule not only displays ARGs by pathway but also highlights them in the pathway maps by the pathway-coloring tool of KEGG (Fig. 1B). In each individual reference pathway ARGs are displayed by orthology, allowing users to investigate androgen regulation of the ortholog groups (Fig. 1B).
Table 1.
Functions of ARGDB modules
| Module | Function |
|---|---|
| A–Z List | Provide a straightforward list of ARGs, organized alphabetically by their official symbols |
| Chromosome Map | Schematically represent the chromosomal location of ARGs in human, mouse, and rat, respectively |
| GO Function | Provide a graphic tree view of the molecular function ontology and give an overview of the ARG distribution on the tree |
| Provide live links to the GO web site | |
| Display ARGs by molecular function | |
| GO Process | Provide a graphic tree view of the biological process ontology and give an overview of the ARG distribution on the tree |
| Provide live links to the GO web site | |
| Display ARGs by biological process | |
| GO Component | Provide a graphic tree view of the cellular component ontology and give an overview of the ARG distribution on the tree |
| Provide live links to the GO web site | |
| Display ARGs by cellular component | |
| KEGG Pathway | Give an overview of the ARG distribution in the human, mouse, rat, and reference pathways |
| Display ARGs by pathway and highlight them in the pathway maps | |
| List androgen-regulated ortholog groups | |
| Noncoding RNA Gene | List the noncoding RNA genes including miRNA, miscRNA, rRNA, scRNA, snoRNA, snRNA, and tRNA |
| Search | Include the following search fields: gene ID, gene symbol, gene alias, taxonomy, chromosome, GO, KEGG pathway, assayed tissue/cell type, identity of androgen, experiment type (in vivo/in vitro), regulation level (RNA/protein), response time, and evidence for AR target |
| Support a batch search of gene IDs or symbols | |
| Element Scan | Search the genomic regions of the collected ARGs (from 5 kb upstream to 5 kb downstream) for the input sequences |
| Support regular expressions | |
| Submission | Provide an interface for submitting the work on ARGs |
| Generate a temporary text file for a check-up |
miRNA, microRNA; miscRNA, miscellaneous RNA; scRNA, small cytoplasmic RNA; snoRNA, small nucleolar RNA; snRNA, small nuclear RNA.
Fig. 1.
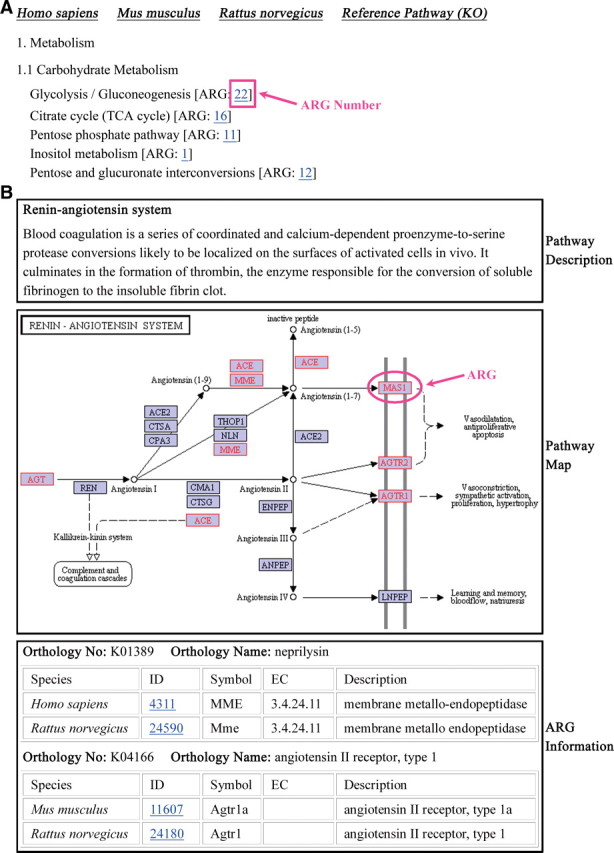
Overview of the KEGG pathway submodule. A, It incorporates the human, mouse, rat, and reference KEGG pathways (KEGG link listed in Table 2). For each pathway we count the ARG number, which is clickable and will take users to the corresponding pathway page. B, The following information is displayed on a pathway page: pathway description, pathway map, and ARGs involved in the selected pathway. ARGs are marked with red box and text in the pathway map by using the KEGG pathway-coloring service. Additionally, ARGs are displayed by orthology on a reference pathway page (panel B).
Table 2.
Reference databases by ARGDB
| Name | Link |
|---|---|
| PubMed | http://www.ncbi.nlm.nih.gov/sites/entrez?db=PubMed |
| Entrez Gene | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene |
| Genome | http://www.ncbi.nlm.nih.gov/sites/entrez?db=genome |
| Nucleotide | http://www.ncbi.nlm.nih.gov/sites/entrez?db=nuccore |
| BLAST | http://blast.ncbi.nlm.nih.gov/Blast.cgi |
| GO | http://www.geneontology.org/ |
| KEGG Pathway | http://www.genome.ad.jp/kegg/pathway.html |
| NURSA | http://www.nursa.org/ |
| ONCOMINE | http://www.oncomine.org |
| BOND | http://bond.unleashedinformatics.com |
| BioGPS | http://biogps.gnf.org |
| miRBase | http://microrna.sanger.ac.uk/ |
BLAST, Basic local alignment search tool.
In the Search module, researchers can search ARGDB for target genes by gene identifiers, biological information, or experimental details (Table 1).
The Element Scan module is designed to search the genomic regions of the collected ARGs for the androgen-response elements (Table 1). Users can get the following information on the input sequence after scanning: location in the genomic structure of the longest transcriptional variants of the nearby ARGs (upstream, exon, intron, or downstream), strands relative to the ARGs, and distance from the ARG transcription start site.
The Submission module offers a user-friendly interface for researchers to report their findings on ARGs (Table 1). After careful examination, the submitted data will be put into the database.
GO function, GO process, and KEGG pathway enrichment in ARGs
In all, 79 GO functions, 519 GO processes, and 28 KEGG pathways were enriched in human, mouse, and rat (listed on the Statistics webpage). Many of them were overrepresented in all three species, such as the prostate cancer pathway. It is notable that many disease pathways were enriched in ARGs, such as prostate cancer, lung cancer, melanoma, bladder cancer, glioma, myeloid leukemia, colorectal cancer, and thyroid cancer.
Motifs of androgen-responsive sequences
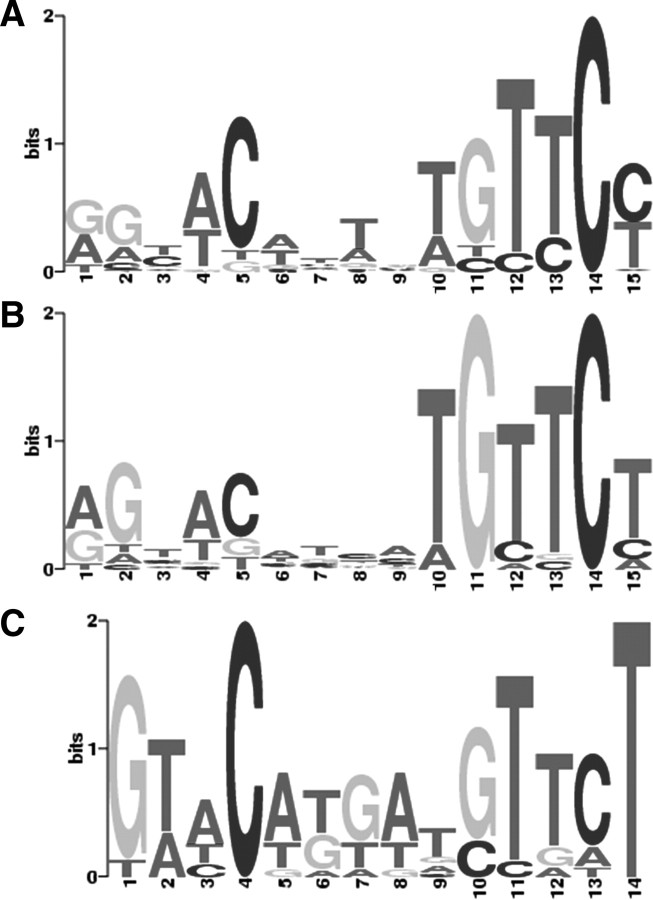
Both human and mouse motifs are 15 bp and are characterized by a three-nucleotide spaced partial palindromic or direct repeat of 5′-TGTTCT-3′ (Fig. 2, A and B). The rat motif is similar, except for the lack of the leftmost nucleotide (Fig. 2C). Across three species, the right six nucleotides, 5′-TGTTCT-3′, are more conserved than the left.
Fig. 2.
Motifs of androgen-responsive sequences identified by MEME. Motifs identified in human, mouse, and rat were represented by MEME LOGO (http://meme.sdsc.edu/meme4/cgi-bin/meme.cgi). E-value of motif A (human), B (mouse), and C (rat) is 2.1E-20, 1.4E-15, and 1.6E-4, respectively.
Discussion
Gene records in ARGDB were all manually selected, evaluated, and completed, although there are many automatic text-mining tools available (21). Overdependence on the automatic methods can cause loss and misrepresentation of information, because a significant amount of information in the biomedical literature was encoded in the form of natural language that is not unified or structured. For example, representations of androgen-responsive sequences are generally spread over an article, thus causing difficulty when one attempts to extract useful information from the article automatically.
ARGDB attempts to set appropriate standards for ARG selection. For example, genes only affected by castration or antiandrogen were excluded. Castration affects not only androgen but also other testicular factors, and the function of antiandrogen is not limited to androgen antagonist (22). Genes with protein expression regulated by androgen were collected in the database because protein abundance correlates with the transcriptional level to some extent. However, it is clear that fluctuation in the level of a protein response to androgen only provides indirect evidence for androgen regulation of the encoding gene, because protein abundance is also affected by many other factors, such as the rate of translation.
A few genes included in the database were only proved to contain the androgen-responsive regions; researchers should treat these genes cautiously for several reasons. First, an in vitro assay cannot represent what actually occurs in vivo. Additionally, growth factors or other cellular signaling pathways can cause ligand-independent activation of AR (23, 24); thus a gene containing AR-binding sites does not necessarily require androgen for its regulation. The Search Module can help researchers exclude those genes if desired.
ARGDB provides comprehensive information on ARGs. It collected important details of the androgen regulation experiments that will facilitate further evaluation and selection by researchers according to their own standards. ARGDB was integrated with multiple annotation resources including NCBI, GO, and KEGG pathway, to reveal the biological characteristics and roles of ARGs. Furthermore, a part of noncoding RNA genes was manually curated. Much of the information in ARGDB can be represented graphically, such as ARG distribution on a chromosome or in a KEGG pathway.
ARGDB can be used as a research aid. By using its data, we analyzed the GO and KEGG pathway enrichment in ARGs. Many disease pathways were enriched, such as prostate cancer, which was overrepresented in all concerned species. These facts indicate the important role of androgen signaling.
Because the previous TGTTCT motif model of the androgen-responsive sequences cannot effectively predict AR-binding regions or androgen target genes (9, 10, 11), androgen-responsive sequences assayed in experiments were collected in the database for motif discovery. However, motifs refound in human, mouse, and rat by us were essentially consistent with the previous model (8), except that across three species the right six nucleotides, 5′-TGTTCT-3′, were more conserved than the left.
In conclusion, ARGDB provides reliable and integrated knowledge on androgen signaling. As we know, cross talk between different pathways plays an important role in carcinogenesis (25). ARGDB will facilitate not only androgen studies but also pathway-cross talk studies when combined with other databases such as ERGDB (http://datam.i2r.a-star.edu.sg/ergdbV2/) (26).
Materials and Methods
Information retrieval
To efficiently obtain as complete information as possible, three independent PubMed (Table 2) query sets were performed as follows:
-
1.
1) This set of queries consisted of androgen names (androgen, testosterone, R1881, or dihydrotestosterone), regulation and information on a single gene. The gene information, including symbols, aliases, and names, was extracted from NCBI deposits (Table 2) and then processed by a Perl program that removed terms shorter than two characters. Because of the large number of genes in human, mouse, and rat, the gene-by-gene queries were conducted by MySQL and a self-developed Perl program, which made use of the NCBI eUtils service to access the PubMed database.
-
2.
2) This set consisted of androgen-response element.
-
3.
3) Queries in this set consisted of high-throughput methods (array or Serial Analysis of Gene Expression) and androgen.
All queries searched publications dating from January 1987 to May 2008. Reviews and other nonoriginal research publications were excluded.
ARG collection
Abstracts of the retrieved PubMed records were manually screened. Records that described androgen regulation of ARG expression or ARG-regulatory sequences or described AR binding to ARG-regulatory sequences were selected. The original publications associated with these records were then further perused for ARG collection. An ARG included in the database satisfies both of the following conditions:
-
1.
1) It belongs to human, mouse or rat.
-
2.
2) It was experimentally proved to be regulated by androgen at the RNA or protein level or to contain one or more transcription regulatory regions that could be bound by AR or mediate the response of reporter genes to androgen. The gene change was caused by androgen alone and was statistically significant. If no statistical test was conducted, the change should be greater than or equal to 1.5-fold. Genes with expressions or regulatory regions only affected by castration, antiandrogen, or a combination of androgen and other agents are not included.
Because protein abundance correlates with the transcriptional level to some extent, genes with protein expressions affected by androgen were included in the database. Changes in protein secretion, modification, or activity were not taken for the evidence of androgen regulation. Genes with transcription-regulatory regions affected by the interaction between AR and other transcriptional regulators, rather than direct AR binding, were also included in the database.
To determine the exact identity of ARGs, the Basic Local Alignment Search Tool (BLAST, Table 2) was applied. If no corresponding sequences were supplied in the references, ARGDB collected extensive information to confirm ARG’s identity, such as gene symbols, gene names, chromosomal location, and so on.
The following experimental information was manually abstracted from original publications:
-
1.
1) Answers to the following questions regarding androgen regulation of the ARG’s RNA or protein level were recorded in the database.
Was ARG up- or down-regulated by androgen?
How long after androgen supplementation was ARG significantly changed?
In what context was ARG changed, including type of cells or tissues, kind of androgen, and amount of androgen?
What methods were employed to prove the androgen regulation?
-
2.
2) Transcription regulatory sequences, which were proved to be directly bound by AR or to mediate the response of reporter genes to androgen, were collected. The corresponding experimental conditions and results were summarized. Location of those sequences in the genomic regions of the relevant ARGs and distance from the ARG transcription start sites were determined by the Element Scan module in the ARGDB web site. If a mutagenesis assay was conducted, the mutated sequence and the result were also collected. For regulatory regions that were indirectly bound by AR, their sequences were not recorded but the evidence in support of androgen influence was summed up. For convenience, the above two types of regulatory sequences were both termed “androgen-responsive sequences.”
Data annotation and data mining
The Entrez Gene ID for the majority of ARGs was determined. Each gene was annotated with information from NCBI, GO, and KEGG (Table 2). Taxon, symbol, name, cytoband, exact genomic position, genomic structure, genomic sequence, and summary were extracted from NCBI deposits. The GO system describes molecular functions, biological processes, and subcellular locations. KEGG provides the pathway information. Live links to the relevant databases were also made. This includes links to NCBI, NURSA (27), ONCOMINE (28), BOND, and BioGPS (Table 2). The Entrez Gene ID could not be determined for some noncoding RNA genes. These ARGs were manually annotated according to references and miRBase (29) (Table 2).
Hypergeometric tests were performed to analyze the GO category or KEGG pathway enrichment in ARGs (30). The Bonferroni method was used for multiple testing corrections. A GO category or a KEGG pathway with adjusted P value ≤ 0.05 was regarded to be overrepresented. Enrichment analysis was performed in human, mouse, and rat separately.
For motif discovery, 51 human, 28 mouse, and 14 rat androgen-responsive sequences collected in the database were analyzed by Multiple EM for Motif Elicitation (MEME) (31, 32). All the above sequences had been validated to be bound by AR in EMSA or DNA footprinting.
Acknowledgments
We thank Yun Gao and Danchen Bao (School of Life Sciences, Fudan University) for their assistance. We also thank Matthew Gaines (University of Oklahoma) for editing the manuscript.
NURSA Molecule Pages:
Ligands: Dihydrotestosterone;
Nuclear Receptors: AR.
Footnotes
This work was supported by National High Technology Research and Development Program of China (863 Program) (Grant 2006AA02Z324), Shanghai Leading Academic Discipline Project (Project No. B111), and National Talent Training Fund in Basic Research of China (Grant J0630643).
Disclosure Summary: The authors have nothing to disclose.
First Published Online September 17, 2009
M.J. and Y.M. are co-first authors. M.J. contributed to the database design and the data collection. Y.M. contributed to the database construction.
Abbreviations: AR, Androgen receptor; ARG, androgen-regulated gene; ARGDB, Androgen-Responsive Gene Database; GO, Gene Ontology; ID, identity; KEGG, Kyoto Encyclopedia of Genes and Genomes; MEME, Multiple EM for Motif Elicitation; NCBI, National Center for Biotechnology Information.
References
- 1.Parker JNPPM2004. Testosterone a medical dictionary, bibliography and annotated research guide to Internet references. San Diego, CA : ICON Health Publications
- 2.Hajszan T, MacLusky NJ, Leranth C2008. Role of androgens and the androgen receptor in remodeling of spine synapses in limbic brain areas. Horm Behav 53:638–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walters KA, Allan CM, Handelsman DJ2008. Androgen actions and the ovary. Biol Reprod 78:380–389 [DOI] [PubMed] [Google Scholar]
- 4.Dehm SM, Tindall DJ2006. Molecular regulation of androgen action in prostate cancer. J Cell Biochem 99:333–344 [DOI] [PubMed] [Google Scholar]
- 5.Nicolás Diaz-Chico B, Germán Rodríguez F, González A, Ramírez R, Bilbao C, Cabrera de León A, Aguirre Jaime A, Chirino R, Navarro D, Díaz-Chico JC2007. Androgens and androgen receptors in breast cancer. J Steroid Biochem Mol Biol 105:1–15 [DOI] [PubMed] [Google Scholar]
- 6.Kalyani RR, Dobs AS2007. Androgen deficiency, diabetes, and the metabolic syndrome in men. Curr Opin Endocrinol Diabetes Obes 14:226–234 [DOI] [PubMed] [Google Scholar]
- 7.Pike CJ, Nguyen TV, Ramsden M, Yao M, Murphy MP, Rosario ER2008. Androgen cell signaling pathways involved in neuroprotective actions. Horm Behav 53:693–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verrijdt G, Haelens A, Claessens F2003. Selective DNA recognition by the androgen receptor as a mechanism for hormone-specific regulation of gene expression. Mol Genet Metab 78:175–185 [DOI] [PubMed] [Google Scholar]
- 9.Masuda K, Werner T, Maheshwari S, Frisch M, Oh S, Petrovics G, May K, Srikantan V, Srivastava S, Dobi A2005. Androgen receptor binding sites identified by a GREF_GATA model. J Mol Biol 353:763–771 [DOI] [PubMed] [Google Scholar]
- 10.Magee JA, Chang LW, Stormo GD, Milbrandt J2006. Direct, androgen receptor-mediated regulation of the FKBP5 gene via a distal enhancer element. Endocrinology 147:590–598 [DOI] [PubMed] [Google Scholar]
- 11.Horie-Inoue K, Takayama K, Bono HU, Ouchi Y, Okazaki Y, Inoue S2006. Identification of novel steroid target genes through the combination of bioinformatics and functional analysis of hormone response elements. Biochem Biophys Res Commun 339:99–106 [DOI] [PubMed] [Google Scholar]
- 12.Lu S, Jenster G, Epner DE2000. Androgen induction of cyclin-dependent kinase inhibitor p21 gene: role of androgen receptor and transcription factor Sp1 complex. Mol Endocrinol 14:753–760 [DOI] [PubMed] [Google Scholar]
- 13.Amir AL, Barua M, McKnight NC, Cheng S, Yuan X, Balk SP2003. A direct β-catenin-independent interaction between androgen receptor and T cell factor 4. J Biol Chem 278:30828–30834 [DOI] [PubMed] [Google Scholar]
- 14.DePrimo SE, Diehn M, Nelson JB, Reiter RE, Matese J, Fero M, Tibshirani R, Brown PO, Brooks JD2002. Transcriptional programs activated by exposure of human prostate cancer cells to androgen. Genome Biol 3:RESEARCH0032 [DOI] [PMC free article] [PubMed]
- 15.Shao C, Wang Y, Yue HH, Zhang YT, Shi CH, Liu F, Bao TY, Yang ZY, Yuan JL, Shao GX2007. Biphasic effect of androgens on prostate cancer cells and its correlation with androgen receptor coactivator dopa decarboxylase. J Androl 28:804–812 [DOI] [PubMed] [Google Scholar]
- 16.Yoshioka M, Boivin A, Bolduc C, St-Amand J2007. Gender difference of androgen actions on skeletal muscle transcriptome. J Mol Endocrinol 39:119–133 [DOI] [PubMed] [Google Scholar]
- 17.Gottlieb B, Lehvaslaiho H, Beitel LK, Lumbroso R, Pinsky L, Trifiro M1998. The Androgen Receptor Gene Mutations Database. Nucleic Acids Res 26:234–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E2006. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34:D108–D110 [DOI] [PMC free article] [PubMed]
- 19.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, et al.2004. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 32:D258–D261 [DOI] [PMC free article] [PubMed]
- 20.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484 [DOI] [PMC free article] [PubMed]
- 21.Krallinger M, Valencia A, Hirschman L2008. Linking genes to literature: text mining, information extraction, and retrieval applications for biology. Genome Biol 9(Suppl 2):S8 [DOI] [PMC free article] [PubMed]
- 22.Veldscholte J, Berrevoets CA, Ris-Stalpers C, Kuiper GG, Jenster G, Trapman J, Brinkmann AO, Mulder E1992. The androgen receptor in LNCaP cells contains a mutation in the ligand binding domain which affects steroid binding characteristics and response to antiandrogens. J Steroid Biochem Mol Biol 41:665–669 [DOI] [PubMed] [Google Scholar]
- 23.Sadar MD, Gleave ME2000. Ligand-independent activation of the androgen receptor by the differentiation agent butyrate in human prostate cancer cells. Cancer Res 60:5825–5831 [PubMed] [Google Scholar]
- 24.Culig Z2004. Androgen receptor cross-talk with cell signalling pathways. Growth Factors 22:179–184 [DOI] [PubMed] [Google Scholar]
- 25.Huang H, Tindall DJ2002. The role of the androgen receptor in prostate cancer. Crit Rev Eukaryot Gene Expr 12:193–207 [DOI] [PubMed] [Google Scholar]
- 26.Tang S, Han H, Bajic VB2004. ERGDB: Estrogen Responsive Genes Database. Nucleic Acids Res 32:D533–D536 [DOI] [PMC free article] [PubMed]
- 27.Lanz RB, Jericevic Z, Zuercher WJ, Watkins C, Steffen DL, Margolis R, McKenna NJ2006. Nuclear Receptor Signaling Atlas (www.nursa.org): hyperlinking the nuclear receptor signaling community. Nucleic Acids Res 34:D221–D226 [DOI] [PMC free article] [PubMed]
- 28.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM2004. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34:D140–D144 [DOI] [PMC free article] [PubMed]
- 30.Rivals I, Personnaz L, Taing L, Potier MC2007. Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics 23:401–407 [DOI] [PubMed] [Google Scholar]
- 31.Bailey TL, Elkan C1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36 [PubMed] [Google Scholar]
- 32.Bailey TL, Williams N, Misleh C, Li WW2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369–W373 [DOI] [PMC free article] [PubMed]