Abstract
Despite decades of intense research, the complex biology of glioblastoma (GBM) is not completely understood. Progression-free survival and overall survival have remained unchanged since the implementation of the STUPP regimen in 2005 with concomitant radio-/chemotherapy and adjuvant chemotherapy with temozolomide.
In the context of Hanahan and Weinberg's six hallmarks and two emerging hallmarks of cancer, we discuss up-to-date status and recent research in the biology of GBM. We discuss the clinical impact of the research results with the most promising being in the hallmarks ‘enabling replicative immortality’, ‘inducing angiogenesis’, ‘reprogramming cellular energetics’ and ‘evading immune destruction’.
This includes the importance of molecular diagnostics according to the new WHO classification and how next generation sequencing is being implemented in the clinical daily life. Molecular results linked together with clinical outcome have revealed the importance of the prognostic biomarker isocitratedehydrogenase (IDH), which is now part of the diagnostic criteria in brain tumours. IDH is discussed in the context of the hallmark ‘reprogramming cellular energetics’. O-6-methylguanine-DNA methyltransferase status predicts a more favourable response to treatment and is thus a predictive marker. Based on genomic aberrations, Verhaak et al have suggested a division of GBM into three subgroups, namely, proneural, classical and mesenchymal, which could be meaningful in the clinic and could help guide and differentiate treatment decisions according to the specific subgroup.
The information achieved will develop and improve precision medicine in the future.
Keywords: Glioblastoma WHO classification Hallmarks of cancer Precision medicine Three subgroups, classical, mesenchymal, proneural
Introduction
Glioblastoma (GBM) has a complex biology and despite decades of research, much is still unknown. The incidence is 3.2/100 0001, and GBM is the most malignant brain tumour.
GBM separates from lower grade gliomas by expressing necrosis and/or microvascular proliferation2 and is characterised by rapid, infiltrating growth. GBM can arise either as a primary tumour or as a secondary tumour, the latter as a malignant transformation from a lower grade brain tumour and/or with mutation in the isocitrate dehydrogenase (IDH) gene.
Treatment in patients with a good performance status is multimodal with surgery, radiation and chemotherapy. However, in spite of the intensive treatment, patients have a poor prognosis with a progression-free survival (PFS) of 7–8 months, a median survival of 14–16 months and 5-year overall survival (OS) of 9.8%.3 4
The diagnosis of gliomas has historically been by histopathological examination. Recent advances have indicated the importance of molecular subtyping. As such, a new WHO classification of GBM into GBM, IDH-wildtype, GBM, IDH-mutant and GBM not otherwise specified (NOS) was recently presented.2 This raises the dilemma of contradiction between the histological/phenotypic diagnosis and the molecular/genomic diagnosis. The genomic diagnosis will then overrule and dictate the diagnosis. There has been suggestion of a further subdivision based on the molecular changes by Verhaak et al 5 6 and table 1. However, today, this subdivision has no role in diagnostics and treatment decisions but might help overcome some of the heterogeneity in GBM and improve treatment.
Table 1.
Subclassification in glioblastoma.
| Classical | Mutated in EGFR with high expression. Lacks P53 mutation. CDKN2A deleted which causes inactivation of the RB pathway. Amplification of chromosome 7 and deletion of chromosome 10. Classical O-6-methylguanine-DNA methyltransferase (MGMT)-methylated tumours respond significantly better to aggressive treatment as compared with non-MGMT-methylated classical tumours. Astrocytic-like. |
| Mesenchymal | Mutated in NF which activates the PI3K/Akt pathway. Mutated in PTEN which activates the RAS pathway. Expression of YKL-40 and MET which can cause epithelial-to-mesenchymal transition. Inflammatory and necrotic. MGMT-methylated mesenchymal tumours seem to respond better to aggressive treatment than non-MGMT-methylated tumours, but this is not significant. Astrocytic-like. |
| Proneural | Mutated in PDGFRA which activates the PI3K pathway and the RAS pathway. Mutated in P53, IDH and PDGFRA. If PDGFRA is mutated, then IDH will not be mutated and opposite. No difference in response to aggressive treatment when stratified for MGMT status. Often secondary glioblastoma. Oligodendrocytic-like. |
Modified and simplified from Verhaak et al,5 Cancer Genome Atlas Research Network,9 Murat et al,32
The predictive factor O-6-methylguanine-DNA methyltransferase (MGMT) is now used in treatment decisions due to results with median OS in patients with MGMT-methylated tumour of 22–26 months compared with non-MGMT-methylated tumours of 12–15 months, respectively.7
Hanahan and Weinberg8 have made an impressive work, investigating the similarities in cancer and explaining these from six hallmarks and two emerging hallmarks. In the following below, we discuss these hallmarks in the context of GBM.
Methods
We searched PubMed with no limitation to time. Only articles in English were used.
Sustaining proliferative signalling
Normal cell growth is regulated through a number of growth signals and paracrine signalling that sustains a cell in a healthy, normal homeostasis.
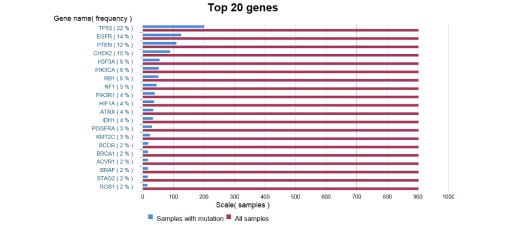
A cancer cell has evolved mechanisms to sustain this proliferative signalling by aberrations in the gene signature. Examples of activating and inactivating mutations can be seen in table 2, and a list of the top 20 mutated genes in GBM can be seen in figure 1.
Table 2.
Examples of oncogenes and tumour suppressor genes
| Activating (oncogenes) | Inactivating (tumour suppressor genes) |
| B-RAF | Tumour protein (TP53) |
| Phosphoinositide 3-kinase (PI3K) isoforms | Retinoblastoma associated (RB) |
| Rat sarcoma (RAS) | Mammalian target of rapamycin (mTOR) |
| Epidermal growth factor receptor (EGFR) | Phosphatase and tensin homolog (PTEN) |
| Platelet derived growth factor receptor A (PDGFRA)
|
Breast cancer1 (BRCA1) |
Figure 1.
Top 20 mutated genes in glioblastoma based on 712 samples of astrocytoma grade IV. 16
The Cancer Genome Atlas (TCGA) group first investigated the genomic characterisation in GBM in 2008.9 Two hundred and six specimens of GBM tissue were analysed, and significant findings were done in the following three core pathways: receptor tyrosine kinase (RTK)/rat sarcoma (RAS)/PI3K, p53 and RB with alterations in 88%, 78% and 87%, respectively.
The most significant alteration in the RTK/RAS/PI3K pathway was in epidermal growth factor receptor (EGFR) in 45%. EGFR can be altered in a number of ways10, and mutation in the EGFR gene results in overexpression of EGFR in GBM as seen in the classical subtype. In total, activating alterations were found in 70% in the RTKs.
The most significant inactivating alteration was found in the PTEN gene in 36%, thereby losing the negative feedback to PI3K causing proliferation and decreased apoptosis. RAS was only mutated in 2% of the specimens, but this is of importance due to the role as a key activator and the influence on several proteins downstream.
In the p53 pathway, the most significant findings were in 49% of CDKN2A and 35% in TP53.
In the RB pathway, CDKN2A and CDKN2B were inactivated in 52% and 47%, respectively and the gene RB was homozygote deleted in 11%.
In 2013, the same specimens were analysed with next-generation sequencing (NGS) and another 337 specimens were added, ending up with 543 tumours.11 Seventy-one significantly mutated genes (SMGs) were found, which many of them corresponded with previous findings. In long-term survivors, aberrations in EGFR, CDK4 and CDKN2A were less frequent. Focus should therefore be on the three pathways described.
Evading growth suppressors
Loss of function in tumour suppressor genes such as NF2, LKB1, RB or TP53 is essential. The latter two genes play a crucial role in the G1 phase in the cell cycle, having the ability to delay entrance into the S-phase to repair the damage detected or ultimately cause apoptosis of the cell.
NF2 codes for Merlin which cause binding of cell adhesion molecules such as E-cadherin on the transmembrane RTK in the cytoplasm, making the cell adhesion stronger and more dense thereby limiting the ability to bind growth factors.12 13
LKB1 is a key activator of mTOR and acts by altering and stabilising the epithelial architecture.14
Mutation in the RB gene is not as common as mutations in the protein (p)RB. RB is mutated in most other cancers but only 6%–11% in GBM.15 16 PRB acts on extracellular signals, and inactivation of pRB can happen in a number of ways in the malignant cell; CDK 4 and 6 can phosphorylate pRB, making it inactive and thus allowing the cell to enter the G1 phase in the cell cycle or the gene can be deleted by mutation. The proteins of the gene CDKN2A work by inhibiting CDK4 and 6. When CDKN2A function is lost by mutation, this indirectly inhibits the function of pRB. It is therefore important to know whether RB insufficiency is due to RB mutation or due to aberrations in the pathway. The latter makes the cancer cell responsive to anti-pathway treatment, whereas the first makes it resistant to the same drugs. Studies in breast17 and bladder18 cancers have shown that loss of pRB makes the cancer more susceptible to radiotherapy and chemotherapy, suggesting pRB loss as a predictive marker of response to such.
The p53 pathway acts on intracellular signals. TP53 is mutated in 27%–33.8% of GBM16 19 and is more correlated to astrocytomas than to oligodendrogliomas. A total of 10 isoforms of TP53 have been identified resulting in different expression of p5320–22, and it seems that mutations in TP53 are not tumour-type specific but are shared across tumour types.23 It also seems that mutations in TP53 do not change with chemotherapy.24 Other isoforms have been identified in patients with breast and ovary cancers and acute myeloid leukaemia.20 The different isoforms showed different response to treatment and PFS.
This suggests that TP53 may be used as a prognostic biomarker in some cancers but not in GBM. In the COSMIC-database, the prognostic value has been investigated. In brain cancer, three studies found a positive predictive value, whereas two studies did not and four studies were not related to outcome. Only studies with more than 50 patients were included.16
The above shows that knowledge is being obtained concerning tumour suppressor genes, but it is yet too scarce to target these in clinical protocols.
Activating invasion and metastasis
The ability of communication between cancer cells and the periphery, the neoplastic stroma, is proving more important in terms of invasive growth and metastases.
Since extracranial metastases are extremely rare in GBM,25 invasion and migration are the main features of GBM spreading.
Three major ways of invasion, migration and metastases have been identified, which will be discussed next; the collective invasiveness, where the cancer cell invades to nearby tissue through existing interstices in the extracellular matrix, thereby expanding from the primary tumour but without directly detachment. Connexin 43 (cnx43) plays a role in the tight junctions between cells. GBM cells can downregulate cnx43, thereby causing lesser adherence and communication between the cells and making possible invasion to nearby tissue.26
Another way is invasion by inflammatory cells where protumoural immune cells produce extracellular matrix degrading enzymes in the periphery, making way for the cancer cell and creating an imbalance between tissue inhibitor of metalloproteinases and metalloproteinases. Hypoxia also causes an increase in proinflammatory proteins and cancer stem cells.27 The proinflammatory proteins allow for cancer stem cells to differentiate, causing gliomagenesis.
Finally, epithelial-to-mesenchymal transition (EMT)28 can occur. The heterogenous GBM cell can display epithelial features, and EMT has been observed in GBM.29 For EMT to happen, E-cadherin is often lacking.30 E-cadherin normally forms junctions between adjacent epithelial cells, thereby assisting senescence and diminishing the ability to grow and invade. When impaired, this causes disruption of the normal cell–cell contact and cell polarity, enabling cell motility. The cell can then undergo epigenetic changes, resulting in dedifferentiation and acquisition of stem cell features.
Hypoxia can also recruit myeloid cells. They cause upregulation of transforming growth factor (TGF)-β, epithelial growth factor, platelet-derived growth factor (PDGFRA) and TWIST, which secretes transcription factors like N-cadherin that is necessary for EMT. Expression of TGF-β and TWIST is higher in necrotic areas and so is the expression of the stem cell marker CD133. Expression of TGF-β in necrotic areas and expression of CD133 are correlated to poorer survival.31 32
Enabling replicative immortality
The gene telomerase reverse transcriptase (TERT) causes expression of telomerase that can add lengths to the telomeres. Telomerase is almost absent in normal cells but is abundantly represented in cancer cells, and the gene has been found mutated in 51% of GBM.16 This enables the cell to avoid telomere shortening and causing an otherwise doomed cell to reverse into immortality.
In the 2013 TCGA study,11 expression of TERT was found in 21/25 cases accessible for investigation. In the remaining four samples, mutations were found in the transcriptional regulator gene alpha thalassaemia mental retardation (ATRX). These alterations were not expressed concurrently. They implied that either TERT or ATRX is responsible for the telomere lengthening. ATRX was expressed concurrently with mutations in TP53 and IDH1, representing secondary GBM.
Ceccarelli et al found that ATRX mutation was associated with lengthening of telomeres while, on the other hand, TERT mutated tumours did not have a difference in telomere length compared with normal tissue controls.33
To support the latter, the authors of a recently published abstract presented at American Society of Oncology (ASCO) 2016 investigated 303 patients and found human TERT (hTERT) mutation in 75% of the patients. In substratification based on hTERT status and MGMT methylation, patients with hTERT mutation lived significantly longer with median OS of 28.3 months in methylated tumours and 15.9 months in non-MGMT-methylated tumours. No difference was observed with hTERT non-mutation regardless of MGMT methylation. This was validated in a TCGA cohort.34 Whether this means that TERT plays a role in the better prognosis in MGMT-methylated tumours, is yet to be fully discovered.
Inducing angiogenesis
Vascular endothelial growth factor (VEGF) A gene stimulates angiogenesis and is rather constant. Angiogenesis can be stimulated in a variety of ways, for example, by oncogenes such as RAS and MYC or by inflammatory reactions. Bone marrow–derived cells (BMDCs) such as macrophages, neutrophils, mast cells and myeloid progenitors are recruited due to the peritumoural oedema. Some of the recruited vascular progenitor cells can transform into pericytes or endothelial cells, protecting and stabilising the newly formed vessel.35 Bevacizumab (BEV) is a humanised monoclonal antibody that targets the vascular endothelial growth factor receptor.36 It can only cross the blood–brain barrier (BBB) where this is destroyed, as seen in GBM. BEV was approved in 2009 by the Food and Drug Administration (FDA) for treatment of recurrent GBM.
A meta-analysis,37 including two large phase III studies, the AvaGlio38 and the RTOG085,39 have demonstrated an increase in PFS but no influence on OS in newly diagnosed GBM patients treated with BEV.
Mechanisms of resistance to BEV are partly due to immunogenic disturbances. The BEV-induced hypoxia recruits proangiogenic BMDCs, mainly tumour-associated macrophages,40 thereby ignoring the effect of BEV.
Hypoxia and BMDCs can enable EMT, as mentioned above, causing a transformation to the more infiltratory mesenchymal subtype.41 Urup et al investigated whether proneural and mesenchymal subtype showed predictiveness towards response to BEV and found that this was not the case. They found that low gene expression of angiotensinogen and high expression of human leucocyte antigen (HLA) class II were predictive markers of response to BEV,42 but this needs to be validated.
Resisting cell death
There are three mechanisms of cell death: apoptosis, autophagy and death by necrosis. The mechanisms are listed hierarchically.
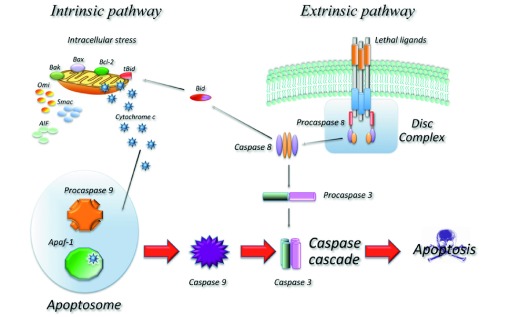
In a normal cell, apoptosis can be divided into an extrinsic part (death receptor mediated) and an intrinsic part (mitochondria-mediated) (figure 2). Aberrations in these subtle mechanisms can lead to avoidance of apoptosis, which can also be achieved by loss of TP53 and RB. PI3K, AKT and mTOR can block apoptosis and autophagy when survival signals are abundant.
Figure 2.
The intrinsic apoptotic pathway is a balance between proapoptotic proteins, for example, Bax and Bak and antiapoptotic proteins. The latter works by inhibiting Bax and Bak. When the inhibition stops, Bax and Bak change the mitochondrial outer membrane, causing release of cytochrome C, which triggers the Caspases and causes apoptosis.87
Another aspect is autophagy, which happens with metabolic stress, nutrient limitation or dysfunction of organelles, thereby decreasing the activity of the cell. The metabolites produced can be used as energy, also in a cancer cell where energy is sparse. This represents a dilemma as autophagy in the early stage cancer can be tumour degrading and in late-stage cancer, can be tumour enhancing.
Cell death by necrosis mediates a proinflammatory response in the microenvironment. This activates the adaptive immune response with recruitment of BMDCs. Therefore, cell death by necrosis is considered as collateral damage for a tumour cell. Hence, if autophagy is impaired together with a defective apoptosis, a cell can become tumourigenic or die by necrosis and inflammation, causing even more space for tumourigenesis and a poor prognosis.43
In GBM, it seems that the cells are more prone to death by necrosis or autophagy as they are in a large extent resistant to death by apoptosis due to impairment of TP53 or RB.44 45 As an example, temozolomide induces death by autophagy.46 Selective autophagy may be a potential target, and the importance of autophagy was highlighted by the recognition of 2016 Nobel Prize winner in medicine, cell biologist Yoshinori Ohsumi.47
In a study of 350 specimens of astrocytomas WHO grades I–IV, it was found that autophagy is enhanced in astrocytomas regardless of WHO grade and prognosis, casting light once again on the microenvironment.48
Evading immune destruction
The immune system is constantly surveilling the homeostasis and an immune competent person is able to eradicate many cancers in the making.49 Experiments in immune incompetent mice where cytotoxic T-lymphocytes (CTLs) and natural killer cells were depleted showed an increasing tendency towards developing cancer.50
GBM cells are able to avoid an immune response due to a limited number of antigens and an ability to recruit myeloid-derived suppressor cells. However, GBM causes leaks in the astrocytic end feet that are part of the BBB.51
Presently, the main focus is on peptide and dendritic cell vaccines and checkpoint inhibitors.
Rindopepimut is an EGFRvIII peptide vaccine conjugated to an immunogenic carrier protein and admixed with the adjuvant granulocyte macrophage colony-stimulating factor.52 Rindopepimut was investigated in the phase II trial ReACT for patients with relapse of GBM.53 They were randomised to BEV plus Rindopepimut or control. Preliminary results presented at ASCO 2015 showed a significant OS of 11.3 months in the Rindopepimut group compared with 9.3 in the control group and an objective response rate of 23%–30% vs 9%–18%, respectively. In the single arm phase II trial, ACT III, for patients with newly diagnosed GBM, a median PFS and OS of 9.2 and 21.8 months were found, respectively. The results were better for patients with MGMT methylation.54 A double-blind phase III trial, the ACT IV was then initiated. However, the protocol has been stopped, since it was found that the study would not meet its primary OS endpoint. Noteworthy was that the control group lived longer than predicted based on historical-matched control groups and thus might have masked the effect of Rindopepimut. Patients treated with the drug prior to termination of the protocol are still offered treatment in compassionate use programmes.55
Another study investigating actively personalised peptide vaccination (GAPVAC) is being performed in newly diagnosed patients and is now closed for inclusion.56
A dendritic cell vaccine works by acting as an antigen-presenting cell. It is possible either to extract autologous antigen-specific T cells, expand them ex vivo and re-infuse into patients or by vaccination with an antigen together with an adjuvant.57
A phase I/II trial including 22 patients with grade II–IV gliomas, showed a positive immunological response in 13 patients.58 The procedure, though, is time consuming and it seems that only 4% of the injected vaccine arrive at the draining lymph node.59 Other studies have shown increased effect by prestimulation of the injection site.60
CTL-4 and programmed death 1 (PD1) are receptors on the T-cell causing apoptosis of the T-cell when abundant and inappropriate, thereby preventing development of autoimmune diseases. Cancer cells can bind to these receptors, causing apoptosis of the T-cell. A CTL-4 inhibitor revolutionised the treatment of unresectable malignant melanoma when Ipilimumab was approved by FDA in 2011.61
A PD1 inhibitor is a monoclonal antibody that binds to and occupies the PD1 receptor. PD1 inhibitors are now being investigated in first-line settings of GBM.62 63 A combination with CTL-4 and PD1 inhibitors are also being performed and immune checkpoint inhibitors could potentially be more effective with prepriming of the immune system with a dendritic cell vaccine or a peptide vaccine.64
Another promising field of research is in the oncolytic viro therapy where poliovirus is genetically engineered with rhinovirus (PVSRIPO). PVSRIPO binds to the Ig superfamily adhesion molecule CD155 or Necl5, which GBM cells express.65 The effect is local and cytotoxic.66 A trial with 22 GBM patients with relapse is being performed at Preston Robert Tisch Brain Tumor Center at Duke University, USA. The treatment is promising and phase II/III trials are being planned.
Autologous lymphoid effector cells specific against tumour cells (ALECSAT) in recurrent GBM have also been tested. No increase in PFS or OS was found, and the study was stopped prematurely.67 It has been suggested that the negative results could be explained due to the fact that patients in the ALECSAT group started treatment 28 days after standard treatment with BEV and Irinotecan in the control group68 and different set-up may be investigated.
Reprogramming cellular energetics
Cancer cells can reprogram their metabolism into favouring anaerob glycolysis followed by lactate acid fermentation in the cytosol,69 thereby producing only two-three molecules of ATP per molecule glucose instead of the 38 accomplished through mitochondrial oxidative phosphorylation. This is overcome by upregulating the number of glucose uptake receptors, namely GLUT1, a trade achieved by RAS, MYC and TP53. The anaerobic glycolysis produces intermediates used to facilitate other biosynthetic pathways.
Five metabolic IDH genes have been defined, coding for three IDH enzymes. The enzymes are responsible for the oxidative carboxylation of isocitrate to α-ketoglutarate producing nicotinamide adenine dinucleotide phosphate (NADPH).
IDH1 is localised in the cytosol and peroxisome, delivering energy to production of peroxisomal enzymes thereby affecting many metabolic pathways. IDH2 and IDH3 are localised in the mitochondria, functioning in the tricarboxylic acid cycle, supporting cell growth.70
Only IDH1 and IDH2 are found mutated in GBM; they exert the same mutagenic effect71 and are settled prognostic markers for lower grade gliomas and secondary GBM.72 IDH is found mutated in 70% of lower grade gliomas and secondary GBM and up to 5% in primary GBM.71
IDH mutations decrease the normal IDH activity by approximately 50%, thereby producing less α-ketoglutarate and NADPH and instead produce the onco-metabolite 2-hydroxyglutarate (2-HG) using NADPH, which lowers NADPH further.73 2-HG is an inhibitor of α-ketoglutarate-dependent dioxygenases, which may cause epigenetic changes, including hypermethylation in human gliomas.74 It can also induce an increased removal of an insulator protein, which enables increased contact to PDGFRA, thereby further inducing gliomagenesis.75
With the impaired function of the mitochondria, the production of bioenergy and intermediates is decreased hence the growth of the cancer cell is lowered when compared with IDH-WT gliomas.76 Preliminary data suggest that inhibition of glutaminase which is necessary for production of 2-HG cause slow-down of glioma cell growth77 78, but the data are still immature for therapeutic use.
Studies are emerging though, with the purpose to target IDH mutations. Hence, preliminary data for the AG12079 trial in the glioma expansion cohort with recurrence or progression of GBM showed a response in 2% and stable disease in 83%. (Mellinghoff IK et al Abstract ACTR-46, SNO 2016). A phase I study AG881 with a pan inhibitor of IDH80 is being evaluated, and another phase I study, the NOA16, is investigating treatment in grade III and IV gliomas with an IDH1 peptide vaccine targeting the IDH1R132H.81
In a study of 105 specimens of GBM, 12% had mutations in IDH1 and in these, 83% had mutations in TP53 as opposed to only 27% in the IDH1-WT tumours. None of the IDH1 mutated tumours had mutations in PTEN, RB1, EGFR or NF1 as opposed to 60% in IDH1-WT tumours. The IDH mutated patients had more favourably clinical features regarding median age at diagnosis, frequency of recurrent GBM, secondary GBM and median OS.82
In a meta-analysis including 24 studies with GBM and IDH1 and −2 status, it was found that IDH mutations were prognostic factors for a better OS and PFS. A total of 15 studies included data for OS. The HR was 0.36 (95% CI 0.26 to 0.49, p<0.001) favouring IDH mutations. Out of the 15 studies, eight included data for PFS, and the HR was 0.32 (95% CI 0.24 to 0.46, p<0.001) favouring IDH mutations.83
IDH mutations have been identified in a number of other cancer types with the highest frequency in GBM and melanoma.84–86
Conclusion and perspectives
The most promising next steps are in the hallmarks ‘enabling replicative immortality’, ‘inducing angiogenesis’, ‘reprogramming cellular energetics’ and ‘evading immune destruction’ where the challenge is not to find differences but to find similarities in GBM. The hallmark with the greatest clinical impact is in ‘evading immune destruction’, where immune therapy might actually represent a similarity in cancer treatment. Promising results have been achieved both in first-line and second-line settings, and development of clinical trials with combination therapy with different immunogenic therapies and/or radiotherapy together with predictive markers might improve results even further. Defining and developing prognostic and predictive markers for better patient selection and treatment response is important. Such could be TERT mutation combined with MGMT methylation, which have showed improved OS or high HLA and low angiotensinogen for treatment response with BEV. Development of liquid biopsies for these markers will increase the clinical usability. The metabolism of GBM is another promising field with the role of IDH which represents epigenetic changes and thus a possibility to target the trunk of GBM instead of the branches where the complexity increases.
More individual treatment is warranted. This is becoming even more evident with the new WHO classification. Research in the three subclasses also represents the molecular focus. The SMGs identified in each subclass has significance in each of the six hallmarks and two emerging hallmarks. Different responses to aggressive treatment together with stratification for MGMT status in each subclass have been demonstrated. This indicates that a patient with, for example, subclass proneural, MGMT non-methylated, perhaps should not be offered STUPP regimen, but rather another 1st line treatment and one might hypothesise that a patient with a mesenchymal tumour and hence more inflammation and death by necrosis might respond better to immune therapy. All this needs further validation but is a clinical meaningful way of thinking.
NGS is expanding, and the handling of and interpretation of big data from these analyses should be carefully evaluated and validated. NGS may provide a more detailed information on GBM to help overcome some of the heterogeneity that challenges today's treatment, as today's treatment is not differentiated according to, for example, molecular aberrations except IDH status and MGMT methylation status. This will have increased importance in the future.
With the economical accessibility, more laboratories will be performing NGS with different equipment and experience. Therefore, development of quality assessments and reproducibility is important, and NGS should only be performed in laboratories with the necessary requirements for this. The challenge is well illustrated by tests for MGMT methylation where this can be performed either by immunohistochemistry or PCR. The latter is considered the most reproducible and independent of interobserver variability, and the first is the most accessible for the community but still there is not consensus on this field.
International cooperation with data sharing is necessary in order to enter the era of precision medicine.
Footnotes
Contributors: DSN is the main author and is responsible for the overall content of the review. HSP and UL contributed equally to the supervision process. All three authors have read the manuscript and participated in the editing process and all three authors have approved of the content.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Ostrom QT, Gittleman H, Fulop J, et al. . CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008-2012. Neuro Oncol 2015;17(suppl 4):iv1–iv62. 10.1093/neuonc/nov189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Louis DN, Perry A, Reifenberger G, et al. . The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 2016;131:803–20. 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 3. Stupp R, Hegi ME, Mason WP, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups National Cancer Institute of Canada Clinical Trials Group. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 2009;10:459–66. 10.1016/S1470-2045(09)70025-7 [DOI] [PubMed] [Google Scholar]
- 4. Michaelsen SR, Christensen IJ, Grunnet K, et al. . Clinical variables serve as prognostic factors in a model for survival from glioblastoma multiforme: an observational study of a cohort of consecutive non-selected patients from a single institution. BMC Cancer 2013;13:402. 10.1186/1471-2407-13-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17:98–110. 10.1016/j.ccr.2009.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. BioRxiv the present server for biology. http://biorxiv.org/content/early/2016/08/13/052076 (accessed on 29 Dec 2016).
- 7. Hegi ME, Diserens AC, Gorlia T, et al. . MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005;352:997–1003. 10.1056/NEJMoa043331 [DOI] [PubMed] [Google Scholar]
- 8. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 9. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061–8. 10.1038/nature07385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zandi R, Larsen AB, Andersen P, et al. . Mechanisms for oncogenic activation of the epidermal growth factor receptor. Cell Signal 2007;19:2013–23. 10.1016/j.cellsig.2007.06.023 [DOI] [PubMed] [Google Scholar]
- 11. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network. The somatic genomic landscape of glioblastoma. Cell 2013;155:462–77. 10.1016/j.cell.2013.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guerrero PA, Yin W, Camacho L, et al. . Oncogenic role of merlin/NF2 in glioblastoma. Oncogene 2015;34:2621–30. 10.1038/onc.2014.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Houshmandi SS, Emnett RJ, Giovannini M, et al. . The neurofibromatosis 2 protein, merlin, regulates glial cell growth in an ErbB2- and Src-dependent manner. Mol Cell Biol 2009;29:1472–86. 10.1128/MCB.01392-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer 2009;9:563–75. 10.1038/nrc2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat Rev Cancer 2008;8:714–24. 10.1038/nrc2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bamford S, Dawson E, Forbes S, et al. . The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer 2004;91:355–8. 10.1038/sj.bjc.6601894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Derenzini M, Donati G, Mazzini G, et al. . Loss of retinoblastoma tumor suppressor protein makes human breast cancer cells more sensitive to antimetabolite exposure. Clin Cancer Res 2008;14:2199–209. 10.1158/1078-0432.CCR-07-2065 [DOI] [PubMed] [Google Scholar]
- 18. Agerbaek M, Alsner J, Marcussen N, et al. . Retinoblastoma protein expression is an independent predictor of both radiation response and survival in muscle-invasive bladder cancer. Br J Cancer 2003;89:298–304. 10.1038/sj.bjc.6601063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xiu J, Piccioni D, Juarez T, et al. . Multi-platform molecular profiling of a large cohort of glioblastomas reveals potential therapeutic strategies. Oncotarget 2016;7:21556–69. 10.18632/oncotarget.7722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marcel V, Dichtel-Danjoy ML, Sagne C, et al. . Biological functions of p53 isoforms through evolution: lessons from animal and cellular models. Cell Death Differ 2011;18:1815–24. 10.1038/cdd.2011.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bouaoun L, Sonkin D, Ardin M, et al. . TP53 variations in human cancers: New lessons from the IARC TP53 database and genomics data. Hum Mutat 2016;37:865–76. 10.1002/humu.23035 [DOI] [PubMed] [Google Scholar]
- 22. Marcel V, Palmero EI, Falagan-Lotsch P, et al. . TP53 PIN3 and MDM2 SNP309 polymorphisms as genetic modifiers in the Li-Fraumeni syndrome: impact on age at first diagnosis. J Med Genet 2009;46:766–72. 10.1136/jmg.2009.066704 [DOI] [PubMed] [Google Scholar]
- 23. Petitjean A, Mathe E, Kato S, et al. . Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat 2007;28:622–9. 10.1002/humu.20495 [DOI] [PubMed] [Google Scholar]
- 24. Wong TN, Ramsingh G, Young AL, et al. . Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 2015;518:552–5. 10.1038/nature13968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johansen MD, Rochat P, Law I, et al. . Presentation of two cases with early extracranial metastases from glioblastoma and review of the literature. Case Rep Oncol Med 2016;2016:1–5. 10.1155/2016/8190950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sin WC, Crespin S, Mesnil M. Opposing roles of connexin43 in glioma progression. Biochim Biophys Acta 1818;2012:2058–67 . 10.1016/j.bbamem.2011.10.022 [DOI] [PubMed] [Google Scholar]
- 27. Tafani M, Di Vito M, Frati A, et al. . Pro-inflammatory gene expression in solid glioblastoma microenvironment and in hypoxic stem cells from human glioblastoma. J Neuroinflammation 2011;8:32. 10.1186/1742-2094-8-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hollier BG, Evans K, Mani SA. The epithelial-to-mesenchymal transition and cancer stem cells: a coalition against cancer therapies. J Mammary Gland Biol Neoplasia 2009;14:29–43. 10.1007/s10911-009-9110-3 [DOI] [PubMed] [Google Scholar]
- 29. Iser IC, Pereira MB, Lenz G, et al. . The Epithelial-to-Mesenchymal transition-like process in glioblastoma: an updated systematic review and in silico investigation. Med Res Rev 2016 10.1002/med.21408 [DOI] [PubMed] [Google Scholar]
- 30. Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology 2007;39:305–18. 10.1080/00313020701329914 [DOI] [PubMed] [Google Scholar]
- 31. Iwadate Y, Matsutani T, Hirono S, et al. . Transforming growth factor-β and stem cell markers are highly expressed around necrotic areas in glioblastoma. J Neurooncol 2016;129:101–7. 10.1007/s11060-016-2145-6 [DOI] [PubMed] [Google Scholar]
- 32. Murat A, Migliavacca E, Gorlia T, et al. . Stem cell-related "self-renewal" signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol 2008;26:3015–24. 10.1200/JCO.2007.15.7164 [DOI] [PubMed] [Google Scholar]
- 33. Ceccarelli M, Barthel FP, Malta TM, et al. ; TCGA Research Network. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 2016;164:550–63. 10.1016/j.cell.2015.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nguyen HN, Lie A, Li T, et al. . Human TERT promoter mutation enables survival advantage from MGMT promoter methylation in IDH1 wild-type primary glioblastoma treated by standard chemoradiotherapy. Neuro Oncol 2016. Epub ahead of print: 29 Aug 2016 10.1093/neuonc/now189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Patenaude A, Parker J, Karsan A. Involvement of endothelial progenitor cells in tumor vascularization. Microvasc Res 2010;79:217–23. 10.1016/j.mvr.2010.01.007 [DOI] [PubMed] [Google Scholar]
- 36. Poulsen HS, Urup T, Michaelsen SR, et al. . The impact of bevacizumab treatment on survival and quality of life in newly diagnosed glioblastoma patients. Cancer Manag Res 2014;6:373–87. 10.2147/CMAR.S39306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fu P, He YS, Huang Q, et al. . Bevacizumab treatment for newly diagnosed glioblastoma: Systematic review and meta-analysis of clinical trials. Mol Clin Oncol 2016;4:833–8. 10.3892/mco.2016.816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chinot OL, Wick W, Mason W, et al. . Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med 2014;370:709–22. 10.1056/NEJMoa1308345 [DOI] [PubMed] [Google Scholar]
- 39. Gilbert MR, Dignam JJ, Armstrong TS, et al. . A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med 2014;370:699–708. 10.1056/NEJMoa1308573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu-Emerson C, Snuderl M, Kirkpatrick ND, et al. . Increase in tumor-associated macrophages after antiangiogenic therapy is associated with poor survival among patients with recurrent glioblastoma. Neuro Oncol 2013;15:1079–87. 10.1093/neuonc/not082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Iwadate Y. Epithelial-mesenchymal transition in glioblastoma progression. Oncol Lett 2016;11:1615–20. 10.3892/ol.2016.4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Urup T, Michaelsen SR, Olsen LR, et al. . Angiotensinogen and HLA class II predict bevacizumab response in recurrent glioblastoma patients. Mol Oncol 2016;10:1160–8. 10.1016/j.molonc.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Degenhardt K, Mathew R, Beaudoin B, et al. . Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006;10:51–64. 10.1016/j.ccr.2006.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jiang H, White EJ, Conrad C, et al. . Autophagy pathways in glioblastoma. Methods Enzymol 2009;453:273–86. 10.1016/S0076-6879(08)04013-5 [DOI] [PubMed] [Google Scholar]
- 45. Lefranc F, Kiss R. Autophagy, the Trojan horse to combat glioblastomas. Neurosurg Focus 2006;20:E7. 10.3171/foc.2006.20.4.4 [DOI] [PubMed] [Google Scholar]
- 46. Kanzawa T, Germano IM, Komata T, et al. . Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ 2004;11:448–57. 10.1038/sj.cdd.4401359 [DOI] [PubMed] [Google Scholar]
- 47. Ohsumi Y. Historical landmarks of autophagy research. Cell Res 2014;24:9–23. 10.1038/cr.2013.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jennewein L, Ronellenfitsch MW, Antonietti P, et al. . Diagnostic and clinical relevance of the autophago-lysosomal network in human gliomas. Oncotarget 2016;7:20016–32. 10.18632/oncotarget.7910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 2013;14:1014–22. 10.1038/ni.2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fuertes MB, Kacha AK, Kline J, et al. . Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J Exp Med 2011;208:2005–16. 10.1084/jem.20101159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Watkins S, Robel S, Kimbrough IF, et al. . Disruption of astrocyte-vascular coupling and the blood-brain barrier by invading glioma cells. Nat Commun 2014;5:4196. 10.1038/ncomms5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gedeon PC, Choi BD, Sampson JH, et al. . Rindopepimut: anti-EGFRvIII peptide vaccine, oncolytic. Drugs Future 2013;38:147–55. 10.1358/dof.2013.038.03.1933992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. A Study of Rindopepimut/GM-CSF in patients with relapsed EGFRvIII-positive glioblastoma. https://clinicaltrials.gov/NCT01498328 (accessed 29 Dec 2016).
- 54. Schuster J, Lai RK, Recht LD, et al. . A phase II, multicenter trial of rindopepimut (CDX-110) in newly diagnosed glioblastoma: the ACT III study. Neuro Oncol 2015;17:854–61. 10.1093/neuonc/nou348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Phase III Study of Rindopepimut/GM-CSF in patients with newly diagnosed glioblastoma. https://clinicaltrials.gov/NCT01480479 (accessed 29 Dec 2016).
- 56. GAPVAC phase I trial in newly diagnosed glioblastoma patients. https://clinicaltrials.gov/NCT02149225 (accessed 29 Dec 2016).
- 57. Palucka K, Banchereau J. Cancer immunotherapy via dendritic cells. Nat Rev Cancer 2012;12:265–77. 10.1038/nrc3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Okada H, Kalinski P, Ueda R, et al. . Induction of CD8+ T-cell responses against novel glioma-associated antigen peptides and clinical activity by vaccinations with {alpha}-type 1 polarized dendritic cells and polyinosinic-polycytidylic acid stabilized by lysine and carboxymethylcellulose in patients with recurrent malignant glioma. J Clin Oncol 2011;29:330–6. 10.1200/JCO.2010.30.7744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Verdijk P, Aarntzen EH, Lesterhuis WJ, et al. . Limited amounts of dendritic cells migrate into the T-cell area of lymph nodes but have high immune activating potential in melanoma patients. Clin Cancer Res 2009;15:2531–40. 10.1158/1078-0432.CCR-08-2729 [DOI] [PubMed] [Google Scholar]
- 60. MartIn-Fontecha A, Sebastiani S, Höpken UE, et al. . Regulation of dendritic cell migration to the draining lymph node: impact on T lymphocyte traffic and priming. J Exp Med 2003;198:615–21. 10.1084/jem.20030448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hodi FS, O'Day SJ, McDermott DF, et al. . Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711–23. 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Study of nivolumab compared to temozolomide, given with radiation therapy, for newly-diagnosed patients with glioblastoma (GBM, a Malignant Brain Cancer). http://www.clinicaltrials.gov/NCT02617589 (accessed 29 Dec 2016).
- 63. Study of temozolomide plus radiation therapy with nivolumab or placebo, for newly diagnosed patients with glioblastoma (GBM, a Malignant Brain Cancer). http://www.clinicaltrials.gov/NCT02667587 (accessed 29 Dec 2016).
- 64. Desai R, Suryadevara CM, Batich KA, et al. . Emerging immunotherapies for glioblastoma. Expert Opin Emerg Drugs 2016;21:133–45. 10.1080/14728214.2016.1186643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Merrill MK, Bernhardt G, Sampson JH, et al. . Poliovirus receptor CD155-targeted oncolysis of glioma. Neuro Oncol 2004;6:208–17. 10.1215/S1152851703000577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brown MC, Gromeier M. Cytotoxic and immunogenic mechanisms of recombinant oncolytic poliovirus. Curr Opin Virol 2015;13:81–5. 10.1016/j.coviro.2015.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Randomized phase 2 study to investigate efficacy of ALECSAT in patients with GBM measured compared to avastin/Irinotecan. http://www.clinicaltrials.gov/NCT02060955 (accessed 29 Dec 2016).
- 68. http://cytovac.com/clinical-data/brain-cancer (accessed 29 Dec 2016).
- 69. López-Lázaro M. The warburg effect: why and how do cancer cells activate glycolysis in the presence of oxygen? Anticancer Agents Med Chem 2008;8:305–12. 10.2174/187152008783961932 [DOI] [PubMed] [Google Scholar]
- 70. Xu X, Zhao J, Xu Z, et al. . Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem 2004;279:33946–57. 10.1074/jbc.M404298200 [DOI] [PubMed] [Google Scholar]
- 71. Yan H, Parsons DW, Jin G, et al. . IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360:765–73. 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Brat DJ, Verhaak RG, Aldape KD, et al. ; Cancer Genome Atlas Research Network. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med 2015;372:2481–98. 10.1056/NEJMoa1402121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bleeker FE, Atai NA, Lamba S, et al. . ) mutation is associated with reduced NADP+-dependent IDH activity in glioblastoma. Acta Neuropathol 2010;119:487–94. 10.1007/s00401-010-0645-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Christensen BC, Smith AA, Zheng S, et al. . DNA methylation, isocitrate dehydrogenase mutation, and survival in glioma. J Natl Cancer Inst 2011;103:143–53. 10.1093/jnci/djq497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Flavahan WA, Drier Y, Liau BB, et al. . Insulator dysfunction and Oncogene activation in IDH mutant gliomas. Nature 2016;529:110–4. 10.1038/nature16490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang C, Moore LM, Li X, et al. . IDH1/2 mutations target a key hallmark of cancer by deregulating cellular metabolism in glioma. Neuro Oncol 2013;15:1114–26. 10.1093/neuonc/not087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Guo C, Pirozzi CJ, Lopez GY, et al. . Isocitrate dehydrogenase mutations in gliomas: mechanisms, biomarkers and therapeutic target. Curr Opin Neurol 2011;24:648–52. 10.1097/WCO.0b013e32834cd415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Seltzer MJ, Bennett BD, Joshi AD, et al. . Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res 2010;70:8981–7. 10.1158/0008-5472.CAN-10-1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Study of orally administered AG-120 in subjects with advanced solid tumors, including glioma, With an IDH1 mutation. https://clinicaltrials.gov/NCT02073994 (accessed 29 Dec 2016).
- 80. Study of orally administered AG-881 in patients with advanced solid tumors, including gliomas, with an IDH1 and/or IDH2 mutation. https://clinicaltrials.gov/NCT02481154 (accessed 29 Dec 2016).
- 81. Phase I trial of IDH1 peptide vaccine in IDH1R132H-mutated grade III-IV gliomas. https://clinicaltrials.gov/NCT02454634 (accessed 29 Dec 2016).
- 82. Parsons DW, Jones S, Zhang X, et al. . An integrated genomic analysis of human glioblastoma multiforme. Science 2008;321:1807–12. 10.1126/science.1164382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chen JR, Yao Y, Xu HZ, et al. . Isocitrate dehydrogenase (IDH)1/2 Mutations as Prognostic Markers in Patients With Glioblastomas. Medicine 2016;95:e2583 10.1097/MD.0000000000002583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kang MR, Kim MS, Oh JE, et al. . Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer 2009;125:353–5. 10.1002/ijc.24379 [DOI] [PubMed] [Google Scholar]
- 85. Shen Y, Zhu YM, Fan X, et al. . Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood 2011;118:5593–603. 10.1182/blood-2011-03-343988 [DOI] [PubMed] [Google Scholar]
- 86. Sjöblom T, Jones S, Wood LD, et al. . The consensus coding sequences of human breast and colorectal cancers. Science 2006;314:268–74. 10.1126/science.1133427 [DOI] [PubMed] [Google Scholar]
- 87. Favaloro B, Allocati N, Graziano V, et al. . Role of apoptosis in disease. Aging 2012;4:330–49. 10.18632/aging.100459 [DOI] [PMC free article] [PubMed] [Google Scholar]