Abstract
Bladder cancer is the fifth most common cancer with significant morbidity and mortality. Recently, numerous studies demonstrated that microRNAs are emerging as diagnostic biomarkers for bladder cancer. Specific miRNA profiles have been identified for several samples from patients with bladder cancer. MicroRNAs are noncoding RNA molecules of approximately 23 nucleotides that play important roles in multiple steps during the progression of bladder cancer. Here, we review the expression profiles of miRNAs and their biological functions, regulation, and clinical implications in bladder cancer. Either downregulation or upregulation of miRNAs occurs in bladder cancer through epigenetic changes or defects of the biogenesis apparatus. Deregulation of miRNAs is involved in cell cycle arrest, apoptosis, proliferation, metastasis, drug resistance, and other functions in bladder cancer. A number of miRNAs, have been associated with tumor type, stage, or patient survival, and miRNAs might be developed as diagnostic or prognostic markers. A better understanding of the roles of miRNAs in bladder cancer will shed light on the molecular mechanisms of bladder cancer.
Keywords: Bladder cancer, Biomarkers, MicroRNA, Diagnostic
Introduction
Bladder cancer (BC) most commonly refers to carcinoma of the epithelial lining of the urinary bladder, the urothelium. Its symptoms are nonspecific and include hematuria, discomfort during urination, and higher frequency and urgency of urination (1). Urothelial bladder cancer constitutes two distinct clinical phenotypes. The common tumors are low grade and non-invasive which may relapse locally but development infrequently; other tumors which are muscle invasive often develop rapidly and have a poor prognosis (2,3). In the United States, an estimated 54,610 new cases of bladder cancer were expected to occur in men in 2013, and 17,960 women presumably acquired this malignancy. The probability of developing this disease seems to increase with age, growing from a 0.02% chance of having bladder cancer by the age of 39 years to a 3.69% (1 in 27) chance of having this malignancy over the age of 70 years (1,4). In the most recent national cancer report in Iran, the age-specific incidence rates for bladder cancer were 13.03 in males and 3.32 in females per 100,000 population (Report of National Cancer Registration Iran, 2008) (5). Early diagnosis is crucial and patients with bladder cancer must be followed every 3 to 6 month. The current modalities for diagnosis and follow-up of the bladder cancer are cystoscopy and urine cytology; one an invasive high-cost method and the other a low-sensitive and operator-dependent method (6,7). In our recent project, we are determining the common mutations in FGFR3 and HRAS genes in Iranian patients with bladder cancer. These genes mutations associated with bladder cancer and have been extensively studied. In recent years, there has been a tremendous and growing interest among researchers to investigate the role of miRNAs in bladder cancer.
Diagnosis, grading, and staging
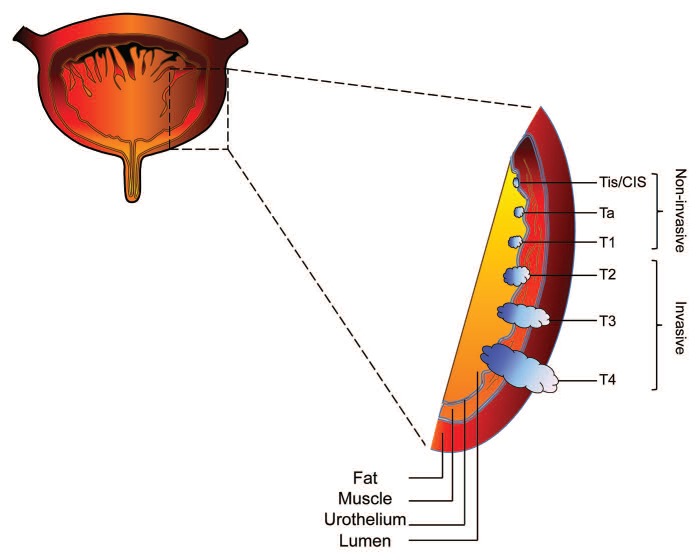
Initial diagnostic workup usually involves cystoscopy and urine cytology. Cystoscopy is the gold standard for the initial diagnosis and staging of bladder cancer. Tumors are classified as low-grade or high-grade (8). Grading refers to the degree of aggressiveness of the cancer cells, with low-grade cancer being the less aggressive cells, which grow slower, look quite normal, and act similar to healthy cells, and with high-grade cancer being characterized by fast-growing cells, which look and act in a disorganized manner and are more likely to progress into the muscle layer of the bladder (1). The most widely used classification for grading of nonmuscle invasive urothelial neoplasms has been the 1973 World Health Organization (WHO) classification. This system has designations for papilloma and Grades 1, 2, and 3 carcinomas. In 2004, members of the WHO and International Society of Urologic Pathologists published and recommended a revised consensus classification for papillary neoplasms (9,10). T-stages 1 through 4 represent the degree to which the cancer has grown in the adjacent layers of tissue, from the connective tissue just beneath the urothelium (T1) to tissue structures located outside the bladder (T4) (Fig. 1) (1). The vast majority (90%) of bladder cancer cases are transitional cell carcinomas (TCC), of which 75–85% present as non-muscle invasive tumors at the time of first diagnosis (Tis/CIS, Ta, and T1) (11-13).
Fig. 1.
Diagram of progressive stages of transitional cell carcinoma of the bladder. Non-muscle invasive transitional cell carcinoma is staged at Tis/CIS, Ta, and T1, whereas muscle invasive transitional cell carcinoma is staged at T2, T3 and T4. Tis/CIS, carcinoma in situ.
The standard for the initial diagnosis and prognostic assessment of bladder cancer is cystoscopy and histopathological analysis of biopsy specimens. However, current prognosticators such as tumor grade, size, and multifocality do not accurately reflect clinical outcome and have limited usefulness for a reliable risk-adjusted therapy decision (14,15). Therefore, the identification of new biomarkers to improve the diagnosis and prognosis of different bladder cancer entities is currently a challenge of special interest (6,12,13). Accumulating evidence suggests aberrant miRNA expression patterns in most human malignancies, and some highly expressed miRNAs might function as oncogenes by repressing tumor suppressors; conversely, miRNAs expressed at low levels might function as tumor suppressors by negatively regulating oncogenes (12,13,15). Based on reports describing miRNA signatures, several down-regulated and up-regulated miRNAs have been discovered in bladder cancer (12,13,15,16). Some of those miRNAs are thought to be potential biomarkers for bladder cancer in diagnosis and prognosis prediction, as well as a treatment target (13,15). Epigenetics has been shown to play a much greater role than previously thought in the initiation and propagation of tumors. The use of epigenetics for the diagnosis and treatment of bladder cancer appears to be a promising field. Drugs using epigenetic targets are undergoing a test for their toxicity, safety, and potential synergistic effects with other drugs. Epigenetic profiling also has the potential to identify patients at high risk of bladder cancer through non-invasive urine tests. Such testing could help identify patients at an earlier stage of the disease. The possibility of epigenetic changes being “ reversed” makes such epigenetic modification therapy a powerful potential option for cancer treatment in the future (17).
miRNA biogenesis and function
MicroRNAs (miRNAs) are small, non-coding RNA molecules of 21-25 nucleotides in length that regulate the gene expression by base–pairing with the transcripts of their targets i.e. protein-coding genes, leading to down-regulation or repression of the target Genes (18,19). miRNAs are transcribed from the long primary transcript (pri-miRNAs) by RNA polymerase II, in the nucleus and processed into characteristic stem-loop precursor miRNAs (pre-miRNAs) by the enzyme Drosha. Then pre-miRNAs are transported into the cytoplasm, where they are transformed into small, single-stranded miRNAs with the help of Dicer (13,16). Once processed into their mature form, miRNAs enter the miRNA silencing complex (miRISC) to degrade or silence mRNAs. miRNAs regulate gene expression by imperfect or near-perfect base pairing to mostly the 3´ untranslated region (UTR) of target mRNAs, thereby inhibiting protein synthesis or causing mRNA degradation(16,20). The 5´ end sequence of miRNA is called ‘‘seed’’ and has a length of 6-8 nucleotides which is energetically favorable for the miRNA target interaction (Fig. 2) (13,20).
Fig. 2.
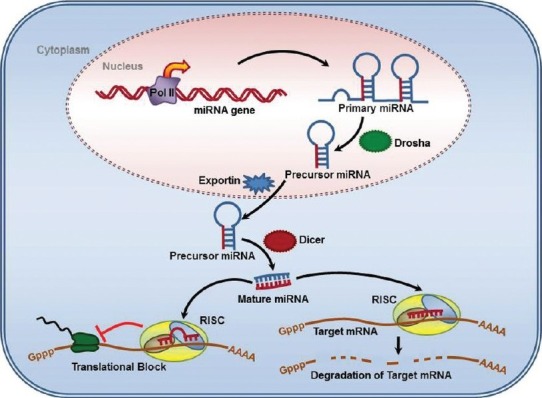
MicroRna processing pathway. miRs are processed through multiple steps, starting in the nucleus and continuing in the cytoplasm
Bladder cancer associated miRNAs
Many diagnostic and prognostic biomarkers of BC have been identified over the past decade. Most of these are the biomarkers for gene transcription, DNA methylation, or protein expression (21). Each miRNA has the potential to target a large number of genes (on average about 500 for each miRNA family) (22-24). Alteration in miRNA expression may occur earlier in BC and affect carcinogenesis and tumor behavior (20,25,26). Gottardo et al. firstly reported 10 up-regulated miRNAs in BC (25,27). Altered expression of many miRNAs has been identified in Dyrskjot et al. study (25,28). Some miRNAs are related to epithelial mesenchymal transition (25,26,29). Therefore miRNAs expression has a noticeable role in bladder tumor genesis (25). More than 50% of miRNA genes are located in cancer-associated genomic regions or in fragile sites of the genome. Many types of cancer are associated with aberrantly expressed miRNAs. Both losses and gains of miRNA function contribute to cancer development and continued tumor growth (30,31). miRNAs may act as both oncogenes or tumor suppressors (30,32). Furthermore, different cancer types, stages, and differentiation grades may have unique miRNA expression profiles, which make miRNAs potent biomarkers for cancer diagnosis (30,33,34). Among the studies examined in a review, the most commonly up-regulated miRNA in BC was miR- 129 (30,35). In contrast, miR-145 and miR-133a were reported to be down-regulated in cancer tissues, and these two markers allowed the authors to distinguish cancer cells from non-cancer cells with a sensitivity >70% and specificity >75% (30,36). The authors found that the levels of miR-200 family members – miR-141, miR-155, and miR-429 – were lower in urine sediment in BC patients. Levels of miR-200a, miR-200b, miR-200c, miR-141, miR-429, miR-205, miR-192, and miR-146a increased significantly after surgery. These findings suggest that miR-200-family miRNAs (miRs-141, -429, -192, -146a, -141, and others) are promising as non-invasive, diagnostic, and prognostic markers. In another study (28,37), miRNA levels were measured in the plasma of patients with or without BC. The authors identified a total of 79 differentially expressed plasma miRNAs. Some diagnostically relevant miRNAs, such as miR-200b, were up-regulated in the BC patients, whereas others, such as miR-92 and miR-33, were inversely correlated with the clinical stage of cancer. These findings support the notion that cell-free circulating miRNAs in the blood can be released in the bladder, as well as in many other tissues (30,38). By comparing the miRNA panel in nonmalignant and malignant bladder tissue, the researchers identified seven significantly up-regulated and eight down-regulated miRNAs. miR-100, miR-125b,miR-130a,miR-139-5p,miR-145*,miR-199a-3p,miR-214 and miR-222 were found to be down-regulated in invasive bladder tumors compared to superficial tumors, as well as in other tumor entities. The expressions of miR-20a, miR-106b, miR-141, miR-130b, miR-200a, miR-200a* and miR-205 were found to be up-regulated in bladder cancer (14). miR-10b, 19a, 126, 145, 221, 296-5p and 378 were significantly down-regulated in bladder cancer compared to adjacent normal urothelium. miR-145 was the most down-regulated microRNA of this group. miR-19b, 21, 205 and 210 showed no significant difference between the 2 tissue types. High miR-21 expression correlated with worse overall patient survival (p =0.0099). Multivariate analysis revealed that miR-21, 210 and 378 may serve as independent prognostic factors for overall patient survival (p= 0.005, 0.033 and 0.012, respectively). miR-21 and 378 may serve as independent prognostic factors for recurrence (p=0.030 and 0.031, respectively) (Fig. 3) (Table 1) (39). To date, the majority of miR studies in BC use profiling to describe dynamic changes in miR expression across stage and grade. Generalized down-regulation of miRs, including those that target the fibroblast growth factor 3 (FGFR3) pathway such as miR-145, miR-101, miR-100 and miR-99a, has been observed in low-grade, non-muscle invasive bladder cancer (NMIBC). In contrast, generalized increased expression of miRs is observed in high grade, muscle invasive bladder cancer (MIBC) compared with adjacent normal bladder urothelium, including miRs predicted to target p53, such as miR-21 and miR-373. Furthermore, p53 suppresses transcriptional factors which promote mesenchymal differentiation, ZEB-1 and ZEB-2, through regulation of the miR200 family (40).
Fig. 3.
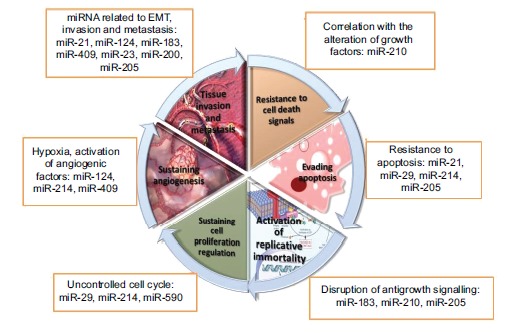
Hallmarks of bladder cancer beyond miRNAs perspective
Table 1. miRNAs associated with Bladder cancer .
| MicroRNA | Chromosome location | Level | Sample | Target gene | Biological role | Clinical significance |
| Mir-21 | Chr 17:57918627-57918698 | Increase | Fresh tissue and bladder cell lines | P53, AKT, PTEN | Apoptosis, mesenchymal transition | Diagnostic, prognostic |
| Mir-18396-182 cluster | Chr 7: 129414745-129414854 Chr 7: 129414532-129414609 Chr 7: 129410223-129410332 | Increase | Fresh tissue ,urine and bladder cell lines | PI3K/AKT/mTOR pathway | Cell growth, migration, apoptosis | Diagnostic, prognostic |
| Mir-29c | Chr 1: 207975197-207975284 | Decrease | Fresh tissue and FFPE | methyltransferase | Maintain normal methylation profile | Stratificationfor high and low risk groups,goodprognosis |
| Mir-210 | Chr 11: 568089-568198 | Increase | Tissue and bladder cell lines | E2F3, FGFRL1, HOXA1 | Cell growth, migration, apoptosis | Diagnostic, prognostic |
| Mir-124 | Chr 20: 61809852-61809938 | Decrease | Tissue and bladder cell lines | ROCK1 | Migration and invasion | Biomarker for Diagnostics or prediction the response to therapy |
| Mir-409 | Chr 14: 101531637-101531715 | Decrease | Tissue and bladder cell lines | c-Met and c-Fos | Migration and invasion | prognostic |
| Mirs-23bfamily | chr 9: 9784749097847586 | Decrease | Fresh tissue and FFPE | ZEB1 | Epithelial-mesenchymal transition | Therapeutic target |
| Mirs200famil y | Chr 12: 7072862-7072929 | Decrease | Tissue,serum and urine | ZEB1, ZEB2 and ERFF-1 | Epithelial-mesenchymal transition | Diagnostic, prognostic |
| Mir-205 | Chr 1: 209605478-209605587 | Increase | Tissue and urine | PTEN, AKT, VEGF | Apoptosis cell cycle invasion, Epithelial-mesenchymal transition | Prognostic, therapeutic target |
| Mir-214 | Chr 1: 172107938-172108047 | Decrease | Tissue,serum and urine | B-catenin | Inhibition of cell proliferation, migration and invasion | Diagnostic, prognostic |
| Mir-5903p | Chr 7: 73605528-73605624 | Decrease | Fresh tissue | TFAM, (PI3K), AKT, MMP2 and MMP9 | Tumorogenesis | Therapeutic target |
Abbreviation: Chr: Chromosome. FFPE: Formalin-fixed paraffin embedded
Database of miRNAs related to BC progression
miRNAs can potentially be used as biomarkers for BC diagnosis, prognosis, and to inform treatment strategies. For this purpose, we performed a search in the MedLine database for papers using the search terms microRNA, miRNA, bladder, urothelial, and cancer (30). miRNA-target interactions were extracted from TarBase 5.0 (41,42) and miRTarBase (41,43). Different databases use varying symbols to represent miRNAs and genes. miRNAs and their host genes were extracted from miRBase (41,44) and NCBI. miRBase provides a collection of confirmed human miRNAs. Official symbols and IDs from NCBI were used to verify host genes and their miRNAs. Differentially expressed genes of BC were extracted from the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway database (45) and the Cancer Genetics Web which can be accessed online at http://www.cancerindex. org (see some miRNAs related databases in Tables 2 &3).
Table 2. MicroRNA target prediction tools .
| Algorithm | Web link |
| TargetScan | http://genes.mit.edu/targetscan |
| miRanda | http://www.microrna.org |
| PicTar | http://pictar.bio.nyu.edu |
| RNAhybrid | http://bibiserve.techfak.uni-bielefeld.de/rnahybrid |
| Diana-microT | http://www.diana.pcbi.upenn.edu/cgi-bin/micro_t.cgi |
| Target Boost | http://demo1.interagon.com/demo |
| Rna 22 | http://cbcsrv.watson.ibm.com/rna22_target.html |
| MicroTar | http://tiger.dbs.nus.edu.sg/microtar/ |
| NBmiRTar | http://wotan.wistar.upenn.edu/NBmiRTar |
| miRecords | http://mirecords.umn.edu/miRecords/ |
Table 3. Databases for microRNA and targets .
| Database | Web link |
| MiRBase | http://microrna.sanger.ac.uk/ |
| TarBase | http://diana.cslab.ece.ntua.gr/tarbase/ |
| Argonaute | http://www.ma.uni-heidelberg.de/apps/zmf/argonaute/ |
| miRecords | http://mirecords.umn.edu/miRecords/ |
Conclusion
The field of miR research has advanced significantly within the last decade, and more recent investigations are focusing on the role of miRs in BC. Aberrations in miR expression identified in NMIBC and MIBC support and provide insight into molecular alterations known to distinguish the two parallel pathways of bladder carcinogenesis. miRs that target the FGFR3 pathway, specifically miR-99a, miR-100, miR-101, and miR-145, are among the most altered in non-muscle invasive disease with reduced expression. In contrast, miRs predicted to target p53, including miR-21 and miR-373, are the most up-regulated in MIBC, and p53 suppresses transcriptional factors which promote mesenchymal differentiation, ZEB-1, and ZEB-2, through regulation of the miR-200 family. The discovery of additional changes in miR expression in BC may ultimately expand what is known about molecular drivers of BC pathogenesis outside of the FGFR3 and p53 pathways. A number of questions remain regarding factors that regulate miR expression, including changes in miR processing machinery and potentially dynamic alterations in response to stressors such as chemotherapy. Thus far, a complex network of interactions is emerging that implicates novel mechanisms of gene modulation in BC. It is hoped that better understanding of these mechanisms will lead to unique opportunities for the development of diagnostic biomarkers and targeted therapy across the spectrum of BC. The heterogeneity of tumor specimens and research methods limits the reproducibility of changes in miRNAs expression profiles between studies and underscores the importance of in vivo validation in a field that utilizes in silico miR target prediction models. In this review we summarized the current knowledge and concept concerning the involvement of miRNAs in bladder cancer.
Cite this article as: Homami A, Ghazi F. MicroRNAs as biomarkers associated with bladder cancer. Med J Islam Repub Iran 2016 (29 December). Vol. 30:475.
References
- 1.Braicu C, Cojocneanu-Petric R, Chira S, Truta A, Floares A, Petrut B. et al. Clinical and pathological implications of miRNA in bladder cancer. International journal of nanomedicine. 2015;10:791. doi: 10.2147/IJN.S72904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mashhadi R, Pourmand G, Mehrsai A, Saeed P, Dialameh H, Ahmadi A. et al. Effect of PTEN Gene Mutations and Environmental Risk Factors on the Progression and Prognosis of Bladder Cancer. Iranian Journal of Public Health. 2014;43(1):56–61. [PMC free article] [PubMed] [Google Scholar]
- 3.Cairns P, Evron E, Okami K, Halachmi N, Esteller M, Herman JG. et al. Point mutation and homozygous deletion of PTEN/MMAC1 in primary bladder cancers. Oncogene. 1998;16(24):3215–8. doi: 10.1038/sj.onc.1201855. [DOI] [PubMed] [Google Scholar]
- 4. Browse the SEER Cancer Statistics Review 1975-2012.
- 5.Salehi A, Khezri AA, Malekmakan L, Aminsharifi A. Epidemiologic status of bladder cancer in Shiraz, southern Iran. Asian Pac J Cancer Prev. 2011;12(5):1323–7. [PubMed] [Google Scholar]
- 6.Ziaee SA, Moula SJ, Hosseini Moghaddam SM, Eskandar-Shiri D. Diagnosis of bladder cancer by urine survivin, an inhibitor of apoptosis: a preliminary report. Urology journal. 2009;3(3):150–3. [PubMed] [Google Scholar]
- 7.Sharp JD, Hausladen DA, Maher MG, Wheeler MA, Altieri DC, Weiss RM. Bladder cancer detection with urinary survivin, an inhibitor of apoptosis. Front Biosci. 2002;7(1):36–41. doi: 10.2741/sharp. [DOI] [PubMed] [Google Scholar]
- 8.Park JC, Citrin DE, Agarwal PK, Apolo AB. Multimodal management of muscle-invasive bladder cancer. Current problems in cancer. 2014;8(3):80–108. doi: 10.1016/j.currproblcancer.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall MC, Chang SS, Dalbagni G, Pruthi RS, Seigne JD, Skinner EC. et al. Guideline for the management of nonmuscle invasive bladder cancer (stages Ta, T1, and Tis): 2007 update. The Journal of urology. 2007;178(6):2314–30. doi: 10.1016/j.juro.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 10. Eble JN, Sauter G, Epstein JI, Sesterhenn IA. Pathology and genetics of tumours of the urinary system and male genital organs. Lyon: IARC 2004:90-109.
- 11.Besaratinia A, Cockburn M, Tommasi S. Alterations of DNA methylome in human bladder cancer. Epigenetics. 2013;8(10):1013–22. doi: 10.4161/epi.25927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Babjuk M, Oosterlinck W, Sylvester R, Kaasinen E, Böhle A, Palou-Redorta J. et al. EAU guidelines on non–muscle-invasive urothelial carcinoma of the bladder, the 2011 update. European urology. 2011;59(6):997–1008. doi: 10.1016/j.eururo.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 13.Prasad SM, DeCastro GJ, Steinberg GD. Urothelial carcinoma of the bladder: definition, treatment and future efforts. Nature Reviews Urology. 2011;8(11):631–42. doi: 10.1038/nrurol.2011.144. [DOI] [PubMed] [Google Scholar]
- 14.Ratert N, Meyer HA, Jung M, Lioudmer P, Mollenkopf HJ, Wagner I. et al. miRNA profiling identifies candidate mirnas for bladder cancer diagnosis and clinical outcome. The Journal of Molecular Diagnostics. 2013;15(5):695–705. doi: 10.1016/j.jmoldx.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Sexton WJ, Wiegand LR, Correa JJ, Politis C, Iravani Dickinson S, Kang LC. Bladder cancer: a review of non-muscle invasive disease. Cancer control. 2010;17(4):256. doi: 10.1177/107327481001700406. [DOI] [PubMed] [Google Scholar]
- 16.Dip N, Reis ST, Timoszczuk LS, Viana NI, Piantino CB, Morais DR. et al. Stage, grade and behavior of bladder urothelial carcinoma defined by the microRNA expression profile. The Journal of urology. 2012;188(5):1951–6. doi: 10.1016/j.juro.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Harb-De la Rosa A, Acker M, Kumar RA, Manoharan M. Epigenetics application in the diagnosis and treatment of bladder cancer. The Canadian journal of urology. 2015;22(5):7947–51. [PubMed] [Google Scholar]
- 18.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. Journal of biomedical informatics. 2011;44(5):839–47. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 20.Puerta-Gil P, García-Baquero R, Jia AY, Ocaña S, Alvarez-Múgica M, Alvarez-Ossorio JL. et al. miR-143, miR-222, and miR-452 are useful as tumor stratification and noninvasive diagnostic biomarkers for bladder cancer. The American journal of pathology. 2012;180(5):1808–15. doi: 10.1016/j.ajpath.2012.01.034. [DOI] [PubMed] [Google Scholar]
- 21.Van Tilborg AA, Bangma CH, Zwarthoff EC. Bladder cancer biomarkers and their role in surveillance and screening. International Journal of Urology. 2009;16(1):23–30. doi: 10.1111/j.1442-2042.2008.02174.x. [DOI] [PubMed] [Google Scholar]
- 22.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 23.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nature reviews Drug discovery. 2010;9(10):775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahdavinezhad A, Mousavi-Bahar SH, Poorolajal J, Yadegarazari R, Jafari M, Shabab N. et al. Evaluation of miR-141, miR-200c, miR-30b Expression and Clinicopathological Features of Bladder Cancer. International journal of molecular and cellular medicine. 2015;4(1):32. [PMC free article] [PubMed] [Google Scholar]
- 26.Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A. et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clinical Cancer Research. 2009;15(16):5060–72. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, et al., editors. Micro-RNA profiling in kidney and bladder cancers. Urologic Oncology: Seminars and Original Investigations; 2007: Elsevier. [DOI] [PubMed]
- 28. Dyrskjøt L. Classification of bladder cancer by microarray expression profiling: towards a general clinical use of microarrays in cancer diagnostics 2003. [DOI] [PubMed]
- 29. Wszolek MF, Rieger-Christ KM, Kenney PA, Gould JJ, Neto BS, LaVoie AK, et al., editors. A MicroRNA expression profile defining the invasive bladder tumor phenotype. Urologic Oncology: Seminars and Original Investigations; 2011: Elsevier. [DOI] [PubMed]
- 30.Zabolotneva AA, Zhavoronkov A, Garazha AV, Roumiantsev SA, Buzdin AA. Characteristic patterns of microRNA expression in human bladder cancer. Frontiers in genetics. 2012:3. doi: 10.3389/fgene.2012.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S. et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National academy of Sciences of the United States of America. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer and Metastasis Reviews. 2009;28(3-4):369–78. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 33. Cortez MA, Welsh JW, Calin GA. Circulating microRNAs as noninvasive biomarkers in breast cancer. Minimal Residual Disease and Circulating Tumor Cells in Breast Cancer: Springer; 2012:151-61. [DOI] [PMC free article] [PubMed]
- 34.Qi J, Mu D. MicroRNAs and lung cancers: from pathogenesis to clinical implications. Frontiers of medicine. 2012;6(2):134–55. doi: 10.1007/s11684-012-0188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dyrskjøt L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R. et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer research. 2009;69(11):4851–60. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 36.Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K. et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. International Journal of Cance. 2009;125(2):345–52. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 37. Adam L, Wszolek MF, Liu CG, Jing W, Diao L, Zien A, et al., editors. Plasma microRNA profiles for bladder cancer detection. Urologic Oncology: Seminars and Original Investigations; 2013: Elsevier. [DOI] [PMC free article] [PubMed]
- 38.Allegra A, Alonci A, Campo S, Penna G, Petrungaro A, Gerace D. et al. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (review) International journal of oncology. 2012;41(6):1897–912. doi: 10.3892/ijo.2012.1647. [DOI] [PubMed] [Google Scholar]
- 39.Zaravinos A, Radojicic J, Lambrou GI, Volanis D, Delakas D, Stathopoulos EN. et al. Expression of miRNAs involved in angiogenesis, tumor cell proliferation, tumor suppressor inhibition, epithelial-mesenchymal transition and activation of metastasis in bladder cancer. The Journal of urology. 2012;188(2):615–23. doi: 10.1016/j.juro.2012.03.122. [DOI] [PubMed] [Google Scholar]
- 40. Guancial EA, Bellmunt J, Yeh S, Rosenberg JE, Berman DM, editors. The evolving understanding of microRNA in bladder cancer. Urologic Oncology: Seminars and Original Investigations; 2014: Elsevier. [DOI] [PMC free article] [PubMed]
- 41.Li Y, Xu Z, Wang K, Wang N, Zhu M. Network analysis of microRNAs, genes and their regulation in human bladder cancer. Biomedical reports. 2013;1(6):918–24. doi: 10.3892/br.2013.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. Rna. 2006;12(2):192–7. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, et al. miRTarBase: a database curates experimentally validated microRNA–target interactions. Nucleic acids research 2010:gkq1107. [DOI] [PMC free article] [PubMed]
- 44.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic acids research. 2008;36(suppl 1):D154–D8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic acids research. 2010;38(suppl 1):D355–D60. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]