Abstract
In the body, the brain is the most cholesterol-rich organ. Despite this, remarkably little is known about the mechanisms in the brain that regulate cholesterol homeostasis. Due to the blood-brain barrier, plasma lipoproteins are unable to traverse, and instead cholesterol must be synthesized de novo from within the central nervous system. Thyroid hormone receptors, activated in response to thyroid hormone (T3), are known to modulate the level of serum cholesterol via complex regulatory pathways. By screening for T3-regulated genes we have identified Disp3, a sterol-sensing domain-containing protein that is related to the Dispatched family of proteins. Analysis by RT-PCR and immunohistochemistry demonstrated that DISP3 is predominately expressed in specific cell types of the brain, retina, and testis. Using the model of hyperthyroidism in vivo, we observed the modulation of Disp3 expression in the retina. Furthermore, in vitro analysis of Disp3 expression in cells treated with T3 revealed both positive and negative regulation. DISP3 localizes within the endoplasmic reticulum and was further found to colocalize with cholesterol. Ectopic expression of DISP3 in fibroblasts resulted in elevated cholesterol levels combined with an altered cholesterol distribution. Given that DISP3 is highly expressed in Purkinje cells, hippocampal neurons, and retinal ganglion cells and that its overexpression results in increased cholesterol levels, it is tempting to postulate that DISP3 may contribute to cholesterol homeostasis in neural cell types. Taken together, we propose that DISP3 represents a new molecular link between thyroid hormone and cholesterol metabolism.
Dispatched 3 is a new thyroid hormone receptor-regulated gene that is expressed in neural tissues and modulates both the amount and distribution of cellular cholesterol.
In higher organisms, thyroid hormones (T3 and its precursor T4) are pleiotropic regulators of growth, differentiation, and tissue homeostasis that act by modulating target gene expression (1, 2). Most, if not all, major thyroid hormone functions are mediated by specific high-affinity nuclear receptors that are encoded by two genes, TRα and TRβ (TRs) (2, 3, 4).
Cholesterol is an essential constituent of most biological membranes and is also a precursor of bile acids, steroid hormones, and certain vitamins. Animals rely on two mechanisms to maintain a pool of cholesterol sufficient to meet these requirements; de novo cholesterol synthesis from acetyl coenzyme A and absorption of cholesterol from dietary sources. Thyroid hormone is an important regulator of cholesterol metabolism with many reports demonstrating an inverse correlation between serum cholesterol and thyroid hormone levels (5). Moreover, TRβ-selective agonists have recently been shown to lower serum cholesterol (6). The fundamental organ responsible for controlling cholesterol metabolism is the liver. It maintains cholesterol homeostasis through the coordinate regulation of three pathways, all of which are regulated at the transcriptional level by thyroid hormone (7). The de novo synthesis of cholesterol is regulated by hepatic 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase, whereas the uptake of existing cellular cholesterol is modulated by the low-density lipoprotein receptor (LDLR). Finally, the rate-limiting enzyme 7α-hydroxylase (CYP7A1) functions to eliminate excess cholesterol by regulating the synthesis of bile acids. To monitor the level of membrane sterols, cells employ two sterol-sensing domain (SSD)-containing proteins, sterol regulatory element-binding protein (SREBP), cleavage-activating protein (SCAP) and HMG-CoA reductase that are localized within the endoplasmic reticulum (ER). Under low sterol conditions, SCAP binds to SREBPs to escort them from the ER to the Golgi apparatus where they are processed into functional transcription factors that activate the expression of genes involved in the synthesis of cholesterol (8). When sterols accumulate, the rate-controlling enzyme HMG-CoA reductase, is rapidly degraded, resulting in the termination of sterol synthesis (9).
In addition to regulating processes involving cellular metabolism, studies in both rodents and humans have illustrated how vitally important thyroid hormone is for the growth, development, and function of the central nervous system (CNS). Interestingly, the specific action of T3 differs dramatically across both developmental time and brain region (10, 11). In the brain, cellular uptake of T3 and its precursor T4 occurs with the aid of several transporter proteins via the blood-brain barrier and the choroid plexus-cerebrospinal fluid barrier (12). Compared with other tissues, the brain is highly enriched in cholesterol with approximately 25% of total cholesterol residing in this organ, most of it present in myelin. Due to the restrictive nature of the blood-brain barrier that prevents cholesterol uptake from the peripheral circulation, the majority of cholesterol present in the brain is synthesized de novo from within the CNS (13). Efflux of cholesterol from the brain is controlled by a mechanism that involves the conversion of cholesterol into the oxysterols, 27- and 24S-hydroxycholesterol that readily cross the blood-brain barrier and transit to the liver for excretion in bile (14). Perturbation of pathways modulating cholesterol synthesis and turnover are linked to the development of several neurological disorders that include Alzheimer’s, Huntington’s, and Niemann-Pick type C (NPC) disease (13). NPC disease is a devastating autosomal recessive neurodegenerative disorder that is caused by a mutation in either the npc1 or npc2 genes (15, 16). The NPC1 protein is a large late endosomal/lysosomal protein that contains 13 transmembrane domains, five of which are homologous to the SSDs contained in proteins associated with cholesterol metabolism or cholesterol-linked signaling (17). Pathologically, it is characterized by an accumulation of unesterified cholesterol and other lipids in the CNS that result in a decline in patient neurological function. The exact role of NPC1 and NPC2 in regulating lipid homeostasis and maintaining neurological function is not yet clear, but evidence to date suggests that these proteins are involved in transporting cholesterol out of late endosomes/lysosomes (15). Several other SSD-containing proteins [HMG-CoA reductase, SCAP, NPC1, NPC1L, 7-dehydrocholesterol reductase (7DHCR), Patched 1 and 2, and Dispatched 1 and 2] have also been shown to function in various aspects of cholesterol homeostasis or cholesterol-linked signaling (18). For instance, HMG-CoA reductase is the rate-controlling enzyme in cholesterol synthesis (9), and NPC1L1 is involved in intestinal cholesterol absorption (19), whereas SCAP complexes with SREBPs in response to cholesterol deprivation to enable ER to Golgi transport, subsequently leading to cholesterol synthesis (9). Finally, both Dispatched and Patched, a tumor suppressor, participate in the Hedgehog signaling pathway by either distributing or receiving, respectively, the cholesterol-modified morphogen, Hedgehog (20).
In a screen for T3-regulated genes, we have identified Disp3 as a potential TR target gene. Here we report on the cloning and expression of chicken Disp3. Analysis of the deduced DISP3 protein sequence revealed that it contains a SSD and is closely related to the Dispatched family of proteins. We have validated the regulation of DISP3 by T3 both in vivo and in vitro and demonstrated that its ectopic expression leads to not only an increase in cholesterol but also an altered distribution pattern, suggesting that DISP3 may contribute to regulating cholesterol homeostasis.
Results
Chicken DISP3 cDNA and protein sequence
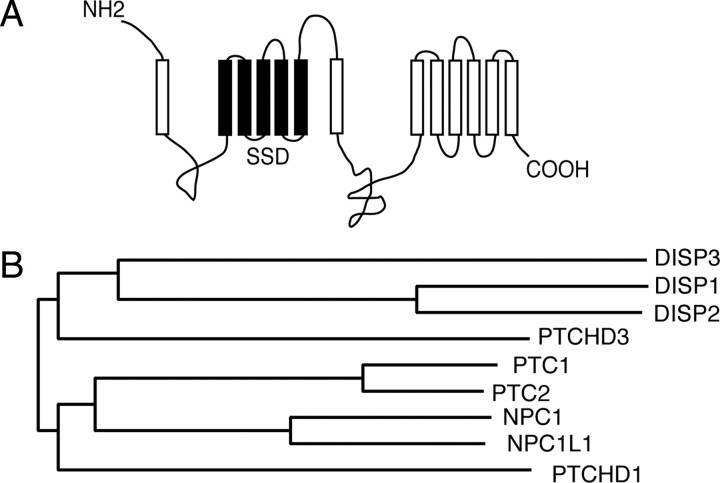
In an attempt to identify novel T3-regulated genes, we performed a differential display screen in avian erythroid progenitor cells ectopically expressing TRα (21). Representational difference analysis of T3-treated cells identified, among others, several gene fragments that were identified within the human genomic sequence KIAA1337 (accession no. AB037758) (Pajer P., M. Zikova, M. Zenke, and P. Bartunek, in preparation). The chicken ortholog of human KIAA1337 was cloned from the brain as described in Materials and Methods and named Trup1 (thyroid hormone receptor up-regulated 1). The complete 4017-bp coding sequence encodes a protein of 1338 amino acids with a calculated molecular mass of about 150 kDa. The consensus sequence of chicken TRUP1/KIAA1337 cDNA has been deposited in GenBank (accession no. EU429800). Different descriptions for KIAA1337 orthologs have been identified in various databases (PTCHD2, DISP3, and RNDEu-2). Recently, Katoh and Katoh (22) identified and bioinformatically characterized in silico the KIAA1337/PTCHD2/RNDEu-2/DISP3 gene by assembling the BU170953 expressed sequence tag and AB037758 sequences. Alignment of the chicken TRUP1/KIAA1337 gene with the mouse and human orthologs revealed 87% homology and 80% identity and 84% homology and 78% identity with the mouse and human variants, respectively (supplemental Fig. 1, published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). Topological predictions revealed that DISP3 is a multispan transmembrane protein. Within the N-terminal section of the protein lies a five-transmembrane region that phylogenetically shares conservation with a domain known as the SSD (18) (Fig. 1A). To further reveal the phylogenetic relationship of TRUP1/KIAA1337/RNDEu-2/PTCHD2/DISP3 to other members of the SSD-containing protein family, amino acid sequences from various family members were aligned and a dendrogram generated. This analysis placed the human TRUP1/KIAA1337 gene in close proximity to the family of Dispatched proteins; thus, in agreement with Katoh and Katoh (22), we have initiated the use of the name DISP3 (Fig. 1B).
Fig. 1.
DISP3 protein and its phylogeny. A, The predicted topology of chicken DISP3 generated by the SOSUI system. The transmembrane and SSD are depicted as open and closed boxes, respectively. B, An evolutionary tree generated from a ClustalW alignment of the DISP, PTC, and NPC1 SSD-containing protein families.
DISP3 mRNA is highly expressed in the brain, retina, and testis
To determine the expression pattern of chicken Disp3, RNA was prepared from a variety of adult chicken tissues and subjected to Northern blot analysis. The approximate 8.4-kb Disp3 transcript was abundantly expressed in the brain with lower expression detected in the heart, testis, and thymus (Fig. 2A). To analyze a wider spectrum of adult chicken and mouse tissues by a more sensitive method, RT-RCR was employed. In agreement with previous data, chicken Disp3 was found highly expressed in the brain, retina, testis, and thymus, with less abundant expression in the spleen and kidney (Fig. 2B). Analysis of mouse Disp3 expression revealed an expression pattern similar to that of the chicken variant, except expression was predominantly restricted to neural tissues such as the brain and retina, with only weak expression detected in the bone marrow and testis (Fig. 2B).
Fig. 2.
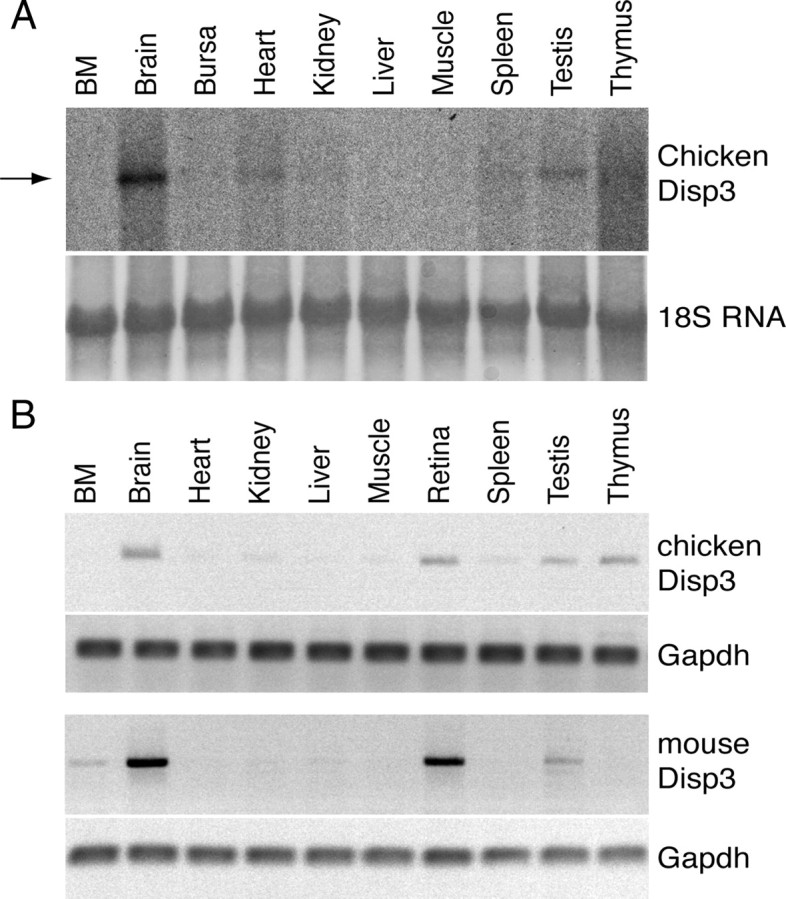
Disp3 mRNA expression in adult chicken and mouse tissues. A, Northern blot analysis of Disp3 expression using RNA extracted from chicken tissues. An arrow indicates the 8.4-kb Disp3 mRNA transcript. 18S rRNA (18S RNA) stained with methylene blue is shown as the loading control. B, RT-PCR analysis of Disp3 expression in selected chicken and mouse tissues. Gapdh served as a control.
DISP3 is expressed in adult neural tissues
To detect DISP3 expression in neural tissues, we have generated a rabbit polyclonal antibody directed against an N-terminal region of chicken DISP3 that is highly conserved from fish to humans (see supplemental Figs. 1 and 2A). To determine antibody specificity, two variants of green fluorescent protein (GFP)-tagged DISP3 (full-length or DISP3-N) were ectopically expressed in chicken embryonic fibroblasts (CEFs) and the resulting protein extracts subjected to Western blotting. In both cases, a specific band corresponding in size to full-length or truncated DISP3 was detected at 180 and 80 kDa, respectively (supplemental Fig. 2B). Additionally, when protein extracts from chicken retina were immunoblotted, a protein of about 165 kDa was visualized that was not detected in liver extracts (supplemental Fig. 2B). Next, the antibody was tested in immunofluorescence experiments. CEF cells transfected with DISP3 or DISP3 tagged with either a GFP or myc epitope along with cultured primary chicken retina cells were examined for DISP3 expression. We observed specific staining in the cytoplasm of cells suggesting that this antibody is not only suitable for immunofluorescence studies but is also sensitive enough to detect endogenous DISP3 (Fig. 3D; supplemental Fig. 2C).
Fig. 3.
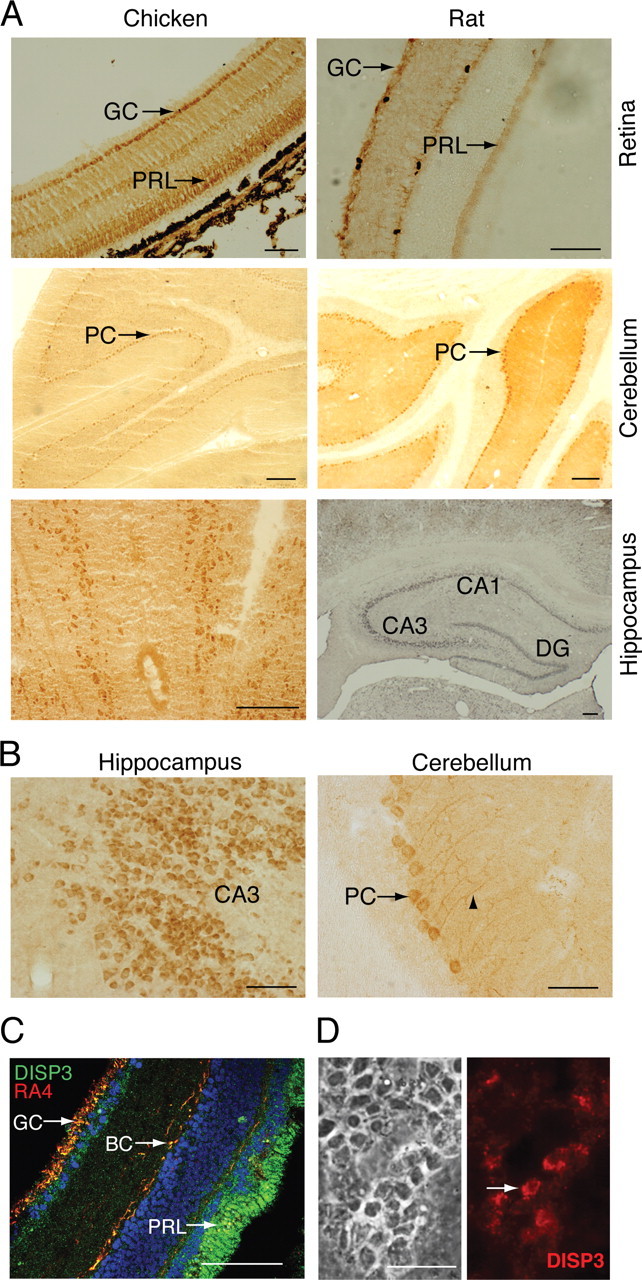
DISP3 is expressed in vertebrate neural cells. A, Representative micrographs of DISP3 immunoreactive cells in the chicken and rat retina, cerebellum, and hippocampus (brown staining). Specific regions of the hippocampus regions CA1, CA3, and dentate gyrus (DG) are shown. GC, Ganglion cells; PRL, photoreceptors; PC, Purkinje cells. B, Magnified images of A, highlighting individual DISP3-positive cells. C, Chicken retina sections double stained with an anti-DISP3 antibody (green) and the RA4 antibody (red) that identifies ganglion cells (GC) and bipolar cells (BC), respectively. 4′,6-Diamidino-2-phenylindole (blue) was used to stain nuclei. Yellow cells highlight areas of colocalization. D, Indirect immunofluorescence performed on primary chicken retina cultures using the purified polyclonal anti-DISP3 antibody. Scale bar, 100 μm for all panels.
Immunohistochemical (IHC) staining of both chicken and rat retinal sections revealed DISP3 is expressed in ganglion and photoreceptor cells of the ganglion and outer nuclear layers, respectively (Fig. 3A). We also observed staining of cells within the inner nuclear layer. To resolve which cells are stained within the inner nuclear layer, we employed the RA4 antibody that identifies ganglion and bipolar cells (23). As clearly shown in Fig. 3C, DISP3 staining colocalized with RA4 staining, implying DISP3 is expressed in the ganglion and bipolar cells of the inner nuclear layer of the retina. Analysis of ex vivo cultured retina cells also displayed distinct cytoplasmic staining of DISP3 (Fig. 3D). Similar analysis of DISP3 expression in the brain revealed prominent staining both in the hippocampus and cerebellum with scattered expression in the cortex (Fig. 3, A and B, and data not shown). In the rat hippocampus, staining appeared in neural cells located within the CA1–CA3 region and dentate gyrus (DG) field (Fig. 3, A and B). Although the anatomical organization of the chicken hippocampus is different from its mammalian counterpart, we were still able to visualize DISP3-positive cells within the hippocampal field of the forebrain. Moreover, in the cerebellum, we were able to distinguish strong DISP3 expression in Purkinje cells, shown previously to express TRα1 and respond to T3 (24) (Fig. 3, A and B).
Thyroid hormone regulates DISP3 expression in vivo
To determine whether T3 regulates DISP3 expression in vivo, we analyzed by IHC 16-d-old developing chicken embryos after hormone administration. Among the tissues screened, the retina exhibited the most considerable difference in DISP3 expression with a significant down-regulation observed in the retinal ganglion cell layer and a mild reduction in the inner and outer nuclear retinal layers (Fig. 4A). To ensure hormone treatment did not perturb retinal ganglion and inner nuclear layer development, we performed RA4 antibody staining. No differences in RA4 staining were detected between control and T3-treated embryos, suggesting that the retinal ganglion cells that normally express DISP3 are still present after T3 treatment. The IHC data was further confirmed by quantitatively measuring the intensity of DISP3 staining in different layers of the retina after hormone treatment. Statistical analysis of these data demonstrated a significant difference (P <0.01) in the retinal ganglion but no other cell layers after hormone treatment (Fig. 4B). Consistent with the aforementioned results, analysis of Disp3 mRNA levels by RT-PCR in total retina also confirmed a decrease in Disp3 after T3 treatment (data not shown). Analysis of other chicken tissues known to express high levels of DISP3, such as the cerebellum and hippocampus, showed no significant differences in expression after T3 treatment at this stage of embryonal development. We also performed a similar study in adult animals but were unable to discern any obvious differences in DISP3 expression after T3 treatment (not shown). Collectively, these results indicate that during embryonal development, DISP3 expression in the chicken retina is down-regulated by T3.
Fig. 4.
Thyroid hormone regulates DISP3 expression. A, Analysis of DISP3 expression by immunohistochemistry in the retina of 16-d-old chicken embryos treated at d 14 either with PBS (as a control) or an initial dose of thyroid hormone (T3, 3 μg) for 24 h before a second dose was administered. Sections were also stained with the control antibody RA4 to monitor the presence of retinal ganglion and bipolar cell layers at this developmental stage. Arrows mark the retinal ganglion cell layer. Scale bar, 100 μm. GCL, Ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; RPE, retinal pigment epithelium. B, Quantification of DISP3 immunostaining shown in Fig. 4A. Intensity of staining was expressed as relative OD. The intensity of background staining was measured at the outer plexiform layer where no specific DISP3 staining was observed. The results shown are from three experimental animals. Triplicate spots from three parallel sections in each cell layer were measured and presented as the mean ± sd. Statistical analysis of the data was carried out using the ANOVA method (F = 27,2; P < 0.01). C, RT-PCR analysis of Disp3 expression in various cell lines. RGC5, Rat retinal ganglion cell line; Y79, human retinoblastoma cell line; HW3.5, mouse hippocampal progenitor cell line; C17.2, mouse cerebral neuronal cell line; TM4, mouse testis cell line. For all cell lines analyzed, 31 cycles of PCR were performed except for the TM4 cell line where 33 cycles were required. Gapdh was used as a control. D, Regulation of Disp3 mRNA by T3 in TM4 and Y79 cells. Cells were grown in standard medium (−) or medium containing the thyroid hormone receptor antagonist KB044146 (Inh) or thyroid hormone (T3) at the final concentration of 5 × 10−7 or 10−7 m, respectively. The qPCR results shown are from three representative experiments assayed in triplicate and are presented as the mean ± sd.
Thyroid hormone regulates DISP3 expression in vitro
To gain further insight into the regulation of DISP3 by thyroid hormone, a panel of cell lines was screened for DISP3 expression. Consistent with the in vivo expression pattern of DISP3, prominent expression was observed in the human retinoblastoma (Y79) and mouse hippocampal progenitor (HW3.5) cell lines. DISP3 was also detected in the mouse testis Sertoli-like cell line (TM4) albeit at a much lower level (Fig. 4C). Interestingly, quantitative PCR (qPCR) analysis of cells treated with T3 revealed differential regulation of DISP3 expression depending on the cell line analyzed (Fig. 4D). In TM4 cells, DISP3 expression was down-regulated nearly 3-fold after treatment with T3 compared with cells treated with the TR antagonist (KB044146) alone. In contrast, Y79 cells displayed a 5.6-fold up-regulation after treatment with T3. In both cell lines, the regulation of DISP3 by T3 was found to be concentration dependent at levels ranging from physiological (1–10 nm) to pharmacological (10 μm) (supplemental Fig. 4A). T3-dependent regulation is likely to be specific given that the TR antagonist is capable of reversing the regulatory activity of TR (activated by a pharmacological concentration of T3) in a dose-responsive manner (supplemental Fig. 4B). Taken together, these results confirm Disp3 mRNA is positively or negatively regulated by T3 in vitro depending on the cell type analyzed.
DISP3 protein localizes to the ER, and its overexpression leads to an accumulation of cholesterol
To investigate the subcellular localization of DISP3, we performed indirect immunofluorescence on cells expressing DISP3 and its GFP- or myc-tagged variants (see supplemental Fig. 2C). Similar analysis of COS7 cells transfected with GFP-DISP3 revealed a punctate staining pattern within a perinuclear cytoplasmic structure that most likely represents the ER. In addition, weak expression was also observed in the nuclear membrane. To determine whether DISP3 localizes within the ER, we costained cells with an ER-specific marker and were able to establish extensive colocalization between GFP-DISP3 and the ER tracker (Fig. 5A). Similar results were obtained when myc-tagged or nontagged versions of DISP3 were evaluated (data not shown).
Fig. 5.
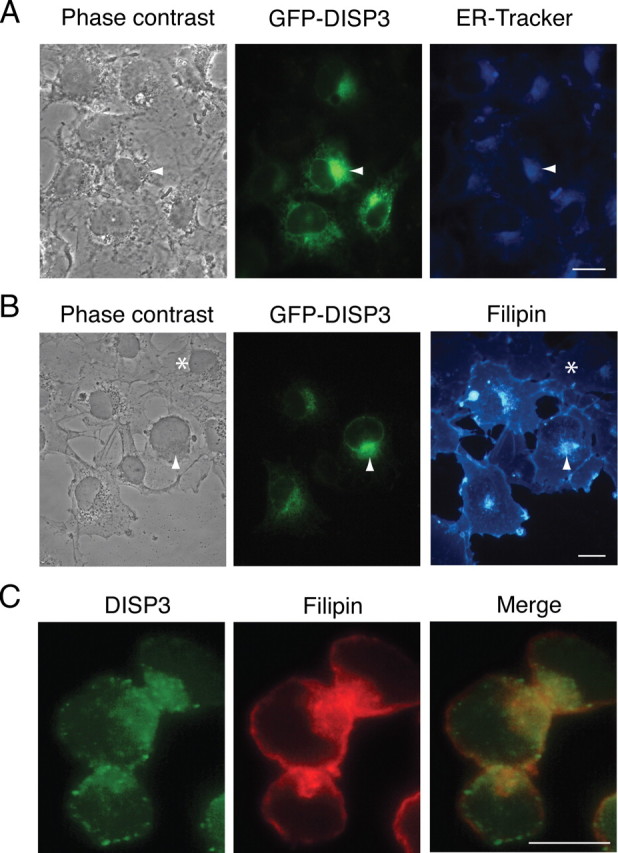
DISP3 localizes to the ER and colocalizes with cholesterol. COS7 cells transiently transfected with GFP-DISP3 were labeled with an ER-Tracker dye to show the sub-cellular localization of DISP3 (A) and by filipin to visualize cellular cholesterol (B). Arrowheads highlight selected DISP3-GFP-transfected cells; asterisks mark nontransfected cells. C, Y79 cells were stained with Disp3 antibody (green) and filipin (red-pseudocolored). Yellow staining represents areas of colocalization. Scale bar, 10 μm.
To establish the influence of DISP3 on intracellular cholesterol distribution, cells transiently expressing GFP-DISP3 were examined by staining with filipin, a fluorescent polyene antibiotic that specifically binds unesterified, free cholesterol. Nontransfected COS7 cells displayed weak filipin staining that was localized to the perinuclear region and plasma membrane (PM). Cells transfected with DISP3 still exhibited weak filipin staining at the PM; however, very strong staining was observed in areas that contain DISP3 (Fig. 5B). Moreover, the intensity of filipin staining in transfected cells highly exceeded that of nontransfected cells, suggesting that the cellular cholesterol content in these cells was increased. To further validate the expression pattern of DISP3 and its potential interaction with cholesterol, we analyzed Y79 retinoblastoma cells, previously shown to express high levels of endogenous Disp3 (Fig. 4C). In agreement with results obtained from cells ectopically expressing DISP3, we observed strong DISP3 staining in ER-like and nuclear membrane structures. Of importance, DISP3 was shown to colocalize with cholesterol in these cells, further substantiating a possible role for DISP3 in cholesterol homeostasis (Fig. 5C).
To study the influence of DISP3 on cholesterol quantity and distribution, we used a retroviral expression system to establish primary CEF cells that stably express DISP3. Consistent with previous filipin staining, control cells displayed only faint filipin staining around the perinuclear region and PM, whereas cells ectopically expressing DISP3 exhibited intense labeling of cytoplasmic punctate vesicular structures, suggesting DISP3 is able to modulate cholesterol levels and localization in the cell (Fig. 6A). To precisely quantify the cellular cholesterol content, we extracted lipids from cells that had been labeled with [3H]cholesterol. Remarkably, compared with control cells, we found a 35% increase in cholesterol uptake and accumulation in cells that ectopically express DISP3 (Fig. 6B). Likewise, total cholesterol measurements obtained using a cholesterol/cholesteryl ester quantification kit showed an average increase in cholesterol of 30% in DISP3-expressing cells (Fig. 6C). Similar results were obtained when cells transiently expressing DISP3 were analyzed (data not shown).
Fig. 6.
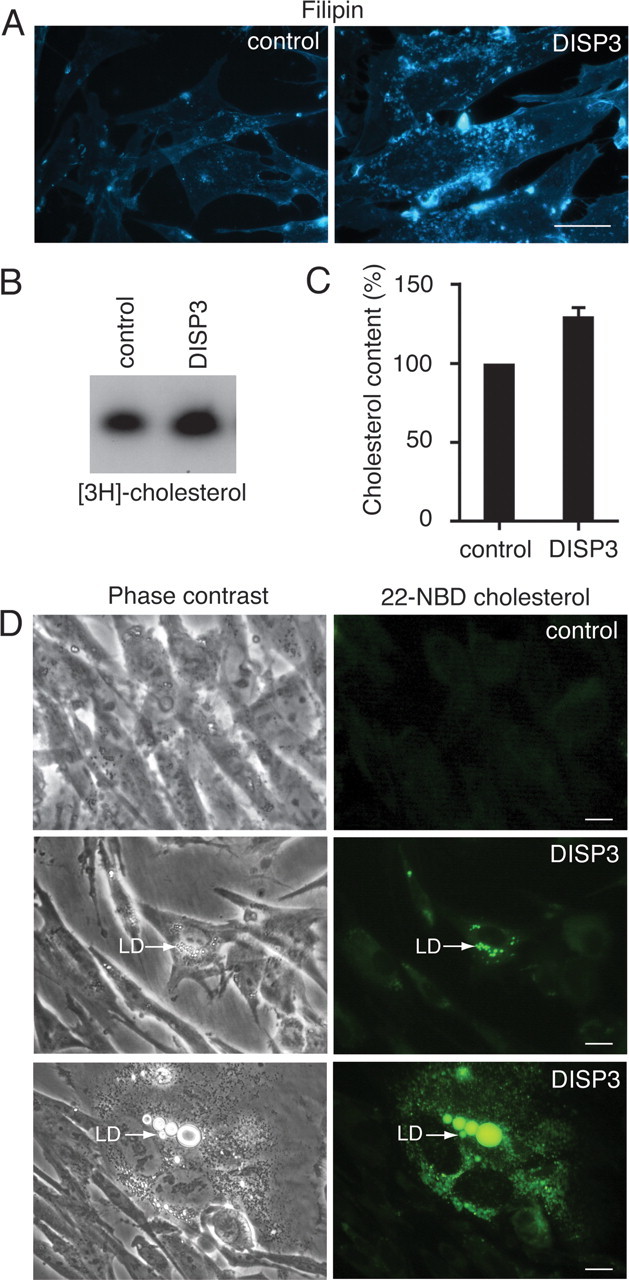
Ectopic expression of DISP3 leads to an accumulation of cholesterol. CEFs stably transfected with a vector encoding DISP3 or an empty vector control were analyzed by filipin staining (A), thin-layer-chromatography (B), and cholesterol/cholesteryl ester quantification kit (C). The results shown in C are presented as the mean ± sd. D, Accumulation of exogenous NBD-cholesterol in lipid droplets (LD) as indicated by the arrows. Scale bar, 10 μm.
To further demonstrate a link between DISP3 and cholesterol, we created a mutant version that lacks the SSD (ΔSSD). In comparison with cells expressing an empty vector control, cells expressing ΔSSD-DISP3 displayed only a slight increase in filipin staining around the nucleus and only an approximate 10% increase in total cholesterol content (supplemental Fig. 5). Together, these results indicate that expression of DISP3 leads to an increase in cellular cholesterol and that the SSD is important for this function.
Interestingly, from our ectopic expression experiments with wild-type DISP3, we also noted a marked increase in the number of cells that contained intracellular lipid droplets. In eukaryotic cells, lipid droplets form the main store of lipids and primarily contain triacylglycerol, diacylglycerol, and cholesteryl esters (25). To confirm the accumulation of cholesterol in these droplets, we analyzed the uptake of fluorescent 22-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-23,24-bisnor-5-cholen-3β-ol (NBD)-cholesterol into cells expressing DISP3. After incubation with NBD-cholesterol, we were able to visualize highly fluorescent areas of the cell whose pattern overlapped perfectly with the lipid droplets visible by phase-contrast microscopy (Fig. 6D; supplemental Fig. 6A). In an attempt to quantify the increase in lipid droplet size and number in DISP3-expressing cells, we stained cells with Nile Red, a dye that specifically associates with the neutral lipids contained within lipid droplets (supplemental Fig. 6B), and measured the intensity of staining by flow cytometry. From this analysis, we observed an increase in fluorescence intensity in DISP3-expressing cells relative to control cells (supplemental Fig. 6C). Therefore, these results suggest that ectopic expression of DISP3 not only increases total cholesterol levels in cells but also induces lipid droplet formation.
Discussion
In this study, we have identified, cloned, and characterized chicken Disp3. Analysis of the full-length cDNA sequence revealed that it encodes a 13-transmembrane domain-containing protein that is related to the Dispatched family of proteins. Sequence alignments with structurally related proteins also demonstrated that DISP3 contains a putative SSD. Previous analysis of other SSD-containing proteins has shown that the presence of an SSD confers an ability to interact with cholesterol that in turn allows these proteins to participate in cholesterol homeostasis or cholesterol-linked signaling. For instance, the SSD in SCAP and NPC1 have been shown to bind cholesterol and/or oxysterols with high affinity in vitro (26, 27). Consistent with the above observations, our experiments also demonstrate that DISP3 when ectopically expressed is able to affect cholesterol homeostasis, resulting in an increase in total cholesterol that relocates and accumulates within the ER and other punctate structures within the cytoplasm. Furthermore, deletion of the SSD results in a diminished ability of DISP3 to modulate cholesterol. Interestingly, deletion of this domain did not completely abolish cholesterol accumulation and redistribution, suggesting that the mechanisms controlling this phenotype cannot be explained solely as a function of the SSD.
In addition, we have observed in fibroblasts that stably express DISP3 an increase in the number of cytosolic lipid droplets that was confirmed by immunofluorescence and flow cytometry. Lipid droplets are thought to arise when cells accumulate toxic levels of free cholesterol. To overcome the toxicity associated with high cholesterol, cells convert cholesterol to cholesteryl esters that are subsequently stored in lipid droplets (28). The higher level of free cholesterol present in cells provide a possible explanation for the increased number of lipid droplets, further implicating a function for DISP3 that involves modulating cholesterol homeostasis.
We have screened a panel of mouse and chicken tissues for DISP3 expression and found high expression in neural tissues. Our DISP3 expression data in the brain was recently confirmed in the mouse by a genome-wide in situ hybridization study whose results are available from the Allen Institute for Brain Science web site (shown also in supplemental Fig. 3) (29). In addition to confirming high Disp3 expression in the hippocampus and cerebellum and diffuse expression in the cortex, the study also revealed strong Disp3 expression in the mitral cell layer of the olfactory bulb (29). Interestingly, the tissues identified as expressing Disp3 are also known to be affected in a number of neurodegenerative disorders. Studies have reported that the level of cholesterol in certain areas of the hippocampus are significantly altered in patients suffering from Alzheimer’s disease and that cholesterol metabolism in the brain is central to Alzheimer’s disease pathophysiology (30). Because DISP3 is expressed in neural tissues that are also affected by neurodegenerative disorders and has a role in modulating cholesterol levels, it is tempting to speculate that DISP3 may play a role in the development or progression of these diseases. Although a definite function for DISP3 has not been elucidated, we can speculate about its many possible roles in the cell. For instance, steroid hormones act on brain tissues to regulate several neuronal functions. Not only is the brain a target for peripheral steroid hormone action but it is also a site for de novo steroid synthesis. These neurosteroids derived from cholesterol are synthesized in Purkinje cells of the cerebellum as well as pyramidal neurons of the hippocampus (31). In ganglion cells of the rodent retina, cholesterol is known to enhance many aspects of synapse development (32). Because DISP3 is highly expressed in the cell types mentioned, it is possible that by regulating cholesterol homeostasis in these tissues, DISP3 may support either neurosteroid synthesis or synapse development and function.
Disp3 was originally identified in a screen for T3-regulated genes. Interestingly, the specific cell types identified as expressing DISP3 within the retina and brain were shown to be perturbed in TRα or TRβ knockout mice, further supporting the regulation of Disp3 by TR (1, 4, 5). Whether DISP3 expression is directly regulated by T3 in vivo remains unclear, although in vitro validation experiments using erythroid progenitor cells suggest a direct mechanism (data not shown). To date, we have described only the in vivo down-regulation of DISP3 in response to T3 in the embryonal (d 16) chicken retina. We cannot exclude that T3 treatment may affect DISP3-expressing cells at different stages of embryonic and/or postnatal development. So far, only a limited number of T3-regulated genes in the brain have been described (33, 34, 35, 36, 37), which is in contrast to the number of T3-regulated genes identified in the liver. One possible explanation is that some target genes may be activated in a region- and stage-specific manner, making their identification difficult.
Similarly, we observed in vitro that DISP3 can be either positively or negatively regulated in response to T3 treatment, depending on the cellular context. Such regulation of DISP3 may be due to a number of factors that include the spatiotemporal pattern of expression of the different thyroid receptors (38), the interplay with region-specific coactivators or corepressors, and/or the local availability of T3 that is regulated by specific deiodinases (39). Taken together, both the in vivo and in vitro results suggest that the regulation of DISP3 is complex and that further studies are required to confirm the regulation of DISP3 by T3 in the brain.
In the current study, we have identified the novel T3-regulated gene Disp3 that is predominately expressed in neural tissues. Given DISP3 contains an SSD and ectopic expression induces an accumulation of cholesterol and lipid droplet formation, we believe that DISP3 is able to modulate cholesterol homeostasis. In the liver, it is established that T3 is a regulator of serum cholesterol levels; however, until now, there has been no evidence that there is a connection between cholesterol levels and T3 in the brain. Therefore, based on the tissue expression of DISP3 in the brain and its ability to modulate cholesterol at a cellular level, we propose that this protein might represent a new molecular link between thyroid hormone action and cholesterol metabolism in neural tissues.
Materials and Methods
Animals
To induce hyperthyroidism, chicken embryos were treated with T3 (Sigma, St. Louis, MO) at d 14 of development (stage Hamburger-Hamilton 40). Eggs were injected twice at 24-h intervals with 100 μl of a T3 solution (3 μg T3 in PBS). At d 16 of development, embryos (stage Hamburger-Hamilton 42) were euthanized for tissue collection. All experiments conducted on Wistar rats and Brown Leghorn chickens (IMG, Prague, Czech Republic) were performed in accordance with the Animal Protection Law of the Czech Republic (Section 11 No. 207/2004 Coll.) and approved by the Institute’s bioethical committee (Project No. 112/2006).
Cell culture
The human retinoblastoma Y79, mouse testis TM4, rat retinal ganglion RGC5, quail fibroblast QT6, and monkey fibroblast COS7 cell lines were cultured under standard conditions. Transient transfections were performed using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) as described by the manufacturer. Stable transfectants were obtained as described previously (40) by transfecting CEFs with the retroviral vector SFCV-hygro encoding full-length Disp3 together with the helper virus RCAN.
Primers
The following primers were used: K1337N, 5′-GCGCTGGACATGGACACAGAGGATG-3′; K1337C, 5′-TCGTGTGGCTCTTGGTGTAGGGTC-3′; K1337/1, 5′-GAGTTCTGCTGGAAGCCCCATGAGG-3′; K1337/2, 5′-CCAAAGAATGACAGGAACACTGAG-3′; K1337/4, 5′-CTCATCTACATACCAGTAGAACTCG-3′; K1337/5, 5′-TCACCACCGTGATTGCCACCGTC-3′; K1337/6, 5′-TCCCAGCCAGACCAGTACATGATG-3′; K1337/7, 5′-ACAGCTGCACCTCAACGCTTCC-3′; K1377/8, 5′-ATGTTGCTGGCCTTCATCAGCAG-3′; K1337/9, 5′-GTGCACCAAGAAGAGCACTTTGCAC-3′; K1337/10, 5′-CTGCCCATGTCTCCATGACGTG-3′ K1337/11, 5′-CGGGATCCGCCAACACGCAGACACACGCAC-3′; K1337/12, 5′-CGGGATCCTCATCTCACTTCATAGTCAAACA-3′; K1337/13, 5′-CCCAAGCTTACCATGGACACAGAGGATGACCC-3′; K1337/22 (qPCR), 5′-CAGCAGCTTTGACCTCTTCA-3′; K1337/23 (qPCR), 5′-GCAACATCTGCAGGAAGGA-3′; GAPDH (sense), 5′-CCATGACAACTTTGGCATTG-3′; and GAPDH (antisense), 5′-TCCCCACAGCCTTAGCAG-3′.
Phylogenetic analysis
Multiple sequence alignments were performed with the ClustalW algorithm using the MacVector software package (Accelrys, Cambridge, UK). Topology prediction was generated by the SOSUI system (41).
Cloning, 5′- and 3′-rapid amplification of cDNA ends (RACE), sequencing, and DNA constructs
The primary chicken Disp3 cDNA sequence was determined by using a combination of RT-PCR and 5′- and 3′-RACE reactions. The sequence data have been submitted to GenBank under accession no. EU429800.
The proposed coding sequence of the gene was assembled/predicted based on the published chicken genomic sequence, chicken expressed sequence tag data and homology comparisons with the human and mouse gene orthologs. The RT-PCR primers K1337N and K1337C were designed to amplify the full-length coding sequence of chicken Disp3. The AccuTaq polymerase mix (Sigma) and the following cycling conditions were used: 35 cycles of 94 C for 5 sec, 62 C for 30 sec, and 68 C for 5 min. The nucleotide sequence of Disp3 was determined by direct sequencing of RT-PCR products generated using the primers K1337N and K1337C from different tissues (adult and embryonic brain). The following set of sequencing primers was then used: K1337/7, K1337/1, K1377/8, K1337/9, K1337/10, and K1337/6. The furthest 5′- and 3′-untranslated region sequences were obtained by 5′- and 3′-SMART RACE procedures using the primers K1337/5, K1337/2, and K1337/4 as described by the manufacturer (Clontech, Mountain View, CA). Sequencing reactions were performed using the BigDye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) as directed by the manufacturer.
To create the Disp3-GFP fusion construct, full-length Disp3 was cloned into the pEGFP-C1 vector (Clontech). The ΔSSD-DISP3 construct was generated by removing the region comprising amino acids 307–587.
Northern blot analysis, RT-PCR, and real-time qPCR
Total RNA was prepared using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. For Northern blotting, 10 μg total RNA was electrophoresed and Disp3 detected using a 32P-labeled probe (622–1200 bp from EU429800). For RT-PCR, 2 μg total RNA was reverse transcribed using random hexamer primers (Invitrogen) and Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI). All qPCR were performed in triplicate using the SYBR Green I Master mix and the LightCycler 480 system (Roche Diagnostics, Mannheim, Germany).
Expression of recombinant proteins in Escherichia coli
The His-DISP3 expression vector encoding amino acids 208–400 (amplified by primers K1337/11 and K1337/12) of chicken DISP3 was transformed into BL21(DE3) E. coli cells. Bacteria were induced to express His-DISP3 using isopropyl β-d-1 thiogalactopyranoside and the resulting recombinant His-tagged proteins were purified using a Ni2+-nitriloacetic acid agarose column as recommended by the manufacturer (QIAGEN, Hilden, Germany) and described previously (42).
Production of polyclonal antibodies
New Zealand rabbits were immunized intradermally three times at 6-wk intervals with affinity-purified, recombinant His-DISP3 antigen. Each immunization contained 150 μg antigen in complete Freund’s adjuvant (Sigma, St. Louis, MO). Bordetella pertussis agglutinogen was also injected im with each immunization. Immune sera were collected 3 wk after the final immunization.
Immunoblotting, immunofluorescence, and immunohistochemistry
Protein samples were separated by SDS-PAGE and transferred electrophoretically onto nitrocellulose membranes that were subsequently blocked in a solution of 5% nonfat milk mixed in Tris-buffered saline containing 0.05% Tween 20. Filters were incubated in primary antibody diluted in 1% milk/Tris-buffered saline containing 0.05% Tween 20 overnight before being washed and incubated with secondary antibody (ECL kit; Amersham, Little Chalfont, UK). For detection of DISP3 by immunofluorescence, cells were grown on glass coverslips, fixed in 3% paraformaldehyde, and permeabilized with 0.2% Triton X-100 before being incubated with the polyclonal anti-DISP3 antibody. Cells were then washed in PBS and incubated with the fluorescently labeled goat antirabbit Alexa Fluor 568 secondary antibody (Invitrogen) before being mounted in Mowiol 4-88 (Calbiochem, San Diego, CA). For immunofluorescence of cryosections, tissues were incubated with RA4 and DISP3 antibodies followed by goat antimouse tetramethylrhodamine isothiocyanate and goat antirabbit Alexa Fluor 488 secondary antibodies (Invitrogen). Sections were then mounted in Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA). For detection of unesterified cholesterol, cells were fixed in 3% paraformaldehyde, stained with filipin (50 μg/ml; Sigma), and viewed using a UV filter set 340–380/40 nm. To fluorescently label intracellular lipid droplets, cells were cultivated overnight in medium supplemented with NBD-cholesterol (5 μg/ml; Sigma, St. Louis, MO). For ER labeling experiments, the ER-Tracker Dye (Invitrogen) was used at a concentration of 300 nm. Nile Red staining was carried out either on fixed (for fluorescence microscopy) or unfixed (for flow cytometry) cells. Cells were stained in a solution of Nile Red (final concentration 1 μg/ml) and analyzed by fluorescence microscopy or flow cytometry (583/26 nm) for the quantification of lipid droplets (43, 44). Immunohistochemistry was carried out on 12-μm-thick cryocut sections of tissue that were transferred to SuperFrost Plus slides and stored at −80 C before use. Staining was visualized as described previously (45) by the avidin-biotin method using diaminobenzidine as the chromogen. Slides were developed in diaminobenzidine for exactly the same time to achieve the same intensity of background staining. Photographs were taken with the Leica DMIRB microscope (Leica, Wetzlar, Germany) and processed with the Leica IM500 and Adobe Photoshop software. Statistical analysis of densitometry data was performed using the one-way ANOVA statistical method.
Determination of cellular cholesterol content
Lipids were extracted from cells with hexane-isopropanol (3:2). The total cholesterol content was determined using a cholesterol/cholesteryl ester quantification kit (Calbiochem). Cholesterol uptake was measured by incubating primary fibroblasts in medium containing [3H]cholesterol (1 μCi/μl; Moravek Biochemicals, Brea, CA). Lipids were extracted as described above, and [3H]cholesterol was separated using thin-layer chromatography. Total protein concentrations were measured using a bicinchoninic acid kit (Pierce, Rockford, IL).
Acknowledgments
We thank N. Agarwal for the RGC5 cell line, C. Cepko for the RA4 monoclonal antibody, D. Spengler for RNA from the HW3.5 and C17.2 cell lines, and M.-R. Witt for the KB044146 antagonist. We also thank C. Thiele for help with cholesterol quantification, D. Sedlak for help with flow cytometry, L. Pichlikova for help with antibody production, and M. Dvorak for critical reading of the manuscript.
NURSA Molecule Pages:
Ligands: Thyroid hormone.
Footnotes
This work was supported by Project No. IAA500520705 from the Grant Agency of the Academy of Sciences of the Czech Republic to M.Z. and in part by Projects AV0Z50520514, LSHM-CT-2005-018652 (FP7, CRESCENDO), LC06077, and LC06061 of the Ministry of Education, Youth, and Sports of the Czech Republic and by the EMBO/HHMI Scientist Award to P.B.
Disclosure Summary: The authors have nothing to disclose.
First Published Online January 29, 2009
Abbreviations: CEF, Chicken embryonic fibroblast; CNS, central nervous system; ER, endoplasmic reticulum; GFP, green fluorescent protein; HMG CoA, 3-hydroxy-3-methylglutaryl coenzyme A; IHC, immunohistochemistry; NDP, 22-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-23,24-bisnor-5-cholen-3β-ol); NPC, Niemann-Pick type C; PM, plasma membrane; qPCR, quantitative PCR; RACE, rapid amplification of cDNA ends; SCAP, SREBP cleavage-activating protein; SREBP, sterol regulatory element-binding protein; SSD, sterol-sensing domain.
References
- 1.Forrest D, Vennstrom B2000. Functions of thyroid hormone receptors in mice. Thyroid 10:41–52 [DOI] [PubMed] [Google Scholar]
- 2.Yen PM2001. Physiological and molecular basis of thyroid hormone action. Physiol Rev 81:1097–1142 [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Lazar MA2000. The mechanism of action of thyroid hormones. Annu Rev Physiol 62:439–466 [DOI] [PubMed] [Google Scholar]
- 4.Flamant F, Samarut J2003. Thyroid hormone receptors: lessons from knockout and knock-in mutant mice. Trends Endocrinol Metab 14:85–90 [DOI] [PubMed] [Google Scholar]
- 5.O'Shea PJ, Williams GR2002. Insight into the physiological actions of thyroid hormone receptors from genetically modified mice. J Endocrinol 175:553–570 [DOI] [PubMed] [Google Scholar]
- 6.Baxter JD, Webb P, Grover G, Scanlan TS2004. Selective activation of thyroid hormone signaling pathways by GC-1: a new approach to controlling cholesterol and body weight. Trends Endocrinol Metab 15:154–157 [DOI] [PubMed] [Google Scholar]
- 7.Ness GC, Pendleton LC, Li YC, Chiang JY1990. Effect of thyroid hormone on hepatic cholesterol 7α hydroxylase, LDL receptor, HMG-CoA reductase, farnesyl pyrophosphate synthetase and apolipoprotein A-I mRNA levels in hypophysectomized rats. Biochem Biophys Res Commun 172:1150–1156 [DOI] [PubMed] [Google Scholar]
- 8.Brown MS, Goldstein JL1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331–340 [DOI] [PubMed] [Google Scholar]
- 9.Goldstein JL, DeBose-Boyd RA, Brown MS2006. Protein sensors for membrane sterols. Cell 124:35–46 [DOI] [PubMed] [Google Scholar]
- 10.Chan S, Kilby MD2000. Thyroid hormone and central nervous system development. J Endocrinol 165:1–8 [DOI] [PubMed] [Google Scholar]
- 11.Bernal J2007. Thyroid hormone receptors in brain development and function. Nat Clin Pract Endocrinol Metab 3:249–259 [DOI] [PubMed] [Google Scholar]
- 12.Abe T, Suzuki T, Unno M, Tokui T, Ito S2002. Thyroid hormone transporters: recent advances. Trends Endocrinol Metab 13:215–220 [DOI] [PubMed] [Google Scholar]
- 13.Vance JE, Hayashi H, Karten B2005. Cholesterol homeostasis in neurons and glial cells. Semin Cell Dev Biol 16:193–212 [DOI] [PubMed] [Google Scholar]
- 14.Bjorkhem I2006. Crossing the barrier: oxysterols as cholesterol transporters and metabolic modulators in the brain. J Intern Med 260:493–508 [DOI] [PubMed] [Google Scholar]
- 15.Ikonen E, Holtta-Vuori M2004. Cellular pathology of Niemann-Pick type C disease. Semin Cell Dev Biol 15:445–454 [DOI] [PubMed] [Google Scholar]
- 16.Carstea ED, Morris JA, Coleman KG, Loftus SK, Zhang D, Cummings C, Gu J, Rosenfeld MA, Pavan WJ, Krizman DB, Nagle J, Polymeropoulos MH, Sturley SL, Ioannou YA, Higgins ME, Comly M, Cooney A, Brown A, Kaneski CR, Blanchette-Mackie EJ, Dwyer NK, Neufeld EB, Chang TY, Liscum L, Strauss 3rd JF, Ohno K, Zeigler M, Carmi R, Sokol J, Markie D, O'Neill RR, van Diggelen OP, Elleder M, Patterson MC, Brady RO, Vanier MT, Pentchev PG, Tagle DA1997. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science 277:228–231 [DOI] [PubMed] [Google Scholar]
- 17.Ioannou YA2000. The structure and function of the Niemann-Pick C1 protein. Mol Genet Metab 71:175–181 [DOI] [PubMed] [Google Scholar]
- 18.Kuwabara PE, Labouesse M2002. The sterol-sensing domain: multiple families, a unique role? Trends Genet 18:193–201 [DOI] [PubMed] [Google Scholar]
- 19.Altmann SW, Davis Jr HR, Zhu LJ, Yao X, Hoos LM, Tetzloff G, Iyer SP, Maguire M, Golovko A, Zeng M, Wang L, Murgolo N, Graziano MP2004. Niemann-Pick C1 like 1 protein is critical for intestinal cholesterol absorption. Science 303:1201–1204 [DOI] [PubMed] [Google Scholar]
- 20.Ingham PW2001. Hedgehog signaling: a tale of two lipids. Science 294:1879–1881 [DOI] [PubMed] [Google Scholar]
- 21.Bartunek P, Zenke M1998. Retinoid X receptor and c-erbA/thyroid hormone receptor regulate erythroid cell growth and differentiation. Mol Endocrinol 12:1269–1279 [DOI] [PubMed] [Google Scholar]
- 22.Katoh Y, Katoh M2005. Identification and characterization of DISP3 gene in silico. Int J Oncol 26:551–556 [PubMed] [Google Scholar]
- 23.McLoon SC, Barnes RB1989. Early differentiation of retinal ganglion cells: an axonal protein expressed by premigratory and migrating retinal ganglion cells. J Neurosci 9:1424–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heuer H, Mason CA2003. Thyroid hormone induces cerebellar Purkinje cell dendritic development via the thyroid hormone receptor α1. J Neurosci 23:10604–10612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy DJ, Vance J1999. Mechanisms of lipid-body formation. Trends Biochem Sci 24:109–115 [DOI] [PubMed] [Google Scholar]
- 26.Radhakrishnan A, Sun LP, Kwon HJ, Brown MS, Goldstein JL2004. Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Mol Cell 15:259–268 [DOI] [PubMed] [Google Scholar]
- 27.Ohgami N, Ko DC, Thomas M, Scott MP, Chang CC, Chang TY2004. Binding between the Niemann-Pick C1 protein and a photoactivatable cholesterol analog requires a functional sterol-sensing domain. Proc Natl Acad Sci USA 101:12473–12478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabas I2002. Consequences of cellular cholesterol accumulation: basic concepts and physiological implications. J Clin Invest 110:905–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176 [DOI] [PubMed] [Google Scholar]
- 30.Poirier J2003. Apolipoprotein E and cholesterol metabolism in the pathogenesis and treatment of Alzheimer’s disease. Trends Mol Med 9:94–101 [DOI] [PubMed] [Google Scholar]
- 31.Tsutsui K, Ukena K, Usui M, Sakamoto H, Takase M2000. Novel brain function: biosynthesis and actions of neurosteroids in neurons. Neurosci Res 36:261–273 [DOI] [PubMed] [Google Scholar]
- 32.Goritz C, Mauch DH, Pfrieger FW2005. Multiple mechanisms mediate cholesterol-induced synaptogenesis in a CNS neuron. Mol Cell Neurosci 29:190–201 [DOI] [PubMed] [Google Scholar]
- 33.Alvarez-Dolado M, Figueroa A, Kozlov S, Sonderegger P, Furley AJ, Munoz A2001. Thyroid hormone regulates TAG-1 expression in the developing rat brain. Eur J Neurosci 14:1209–1218 [DOI] [PubMed] [Google Scholar]
- 34.Alvarez-Dolado M, Ruiz M, Del Rio JA, Alcantara S, Burgaya F, Sheldon M, Nakajima K, Bernal J, Howell BW, Curran T, Soriano E, Munoz A1999. Thyroid hormone regulates reelin and dab1 expression during brain development. J Neurosci 19:6979–6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iglesias T, Caubin J, Stunnenberg HG, Zaballos A, Bernal J, Munoz A1996. Thyroid hormone-dependent transcriptional repression of neural cell adhesion molecule during brain maturation. EMBO J 15:4307–4316 [PMC free article] [PubMed] [Google Scholar]
- 36.Koritschoner NP, Alvarez-Dolado M, Kurz SM, Heikenwalder MF, Hacker C, Vogel F, Munoz A, Zenke M2001. Thyroid hormone regulates the obesity gene tub. EMBO Rep 2:499–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poguet AL, Legrand C, Feng X, Yen PM, Meltzer P, Samarut J, Flamant F2003. Microarray analysis of knockout mice identifies cyclin D2 as a possible mediator for the action of thyroid hormone during the postnatal development of the cerebellum. Dev Biol 254:188–199 [DOI] [PubMed] [Google Scholar]
- 38.Sjoberg M, Vennstrom B, Forrest D1992. Thyroid hormone receptors in chick retinal development: differential expression of mRNAs for α and N-terminal variant β-receptors. Development 114:39–47 [DOI] [PubMed] [Google Scholar]
- 39.Bianco AC, Kim BW2006. Deiodinases: implications of the local control of thyroid hormone action. J Clin Invest 116:2571–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartunek P, Karafiat V, Dvorakova M, Zahorova V, Mandikova S, Zenke M, Dvorak M1997. The Myb leucine zipper is essential for leukemogenicity of the v-Myb protein. Oncogene 15:2939–2949 [DOI] [PubMed] [Google Scholar]
- 41.Hirokawa T, Boon-Chieng S, Mitaku S1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379 [DOI] [PubMed] [Google Scholar]
- 42.Bartunek P, Pichlikova L, Stengl G, Boehmelt G, Martin FH, Beug H, Dvorak M, Zenke M1996. Avian stem cell factor (SCF): production and characterization of the recombinant His-tagged SCF of chicken and its neutralizing antibody. Cytokine 8:14–20 [DOI] [PubMed] [Google Scholar]
- 43.Greenspan P, Fowler SD1985. Spectrofluorometric studies of the lipid probe, nile red. J Lipid Res 26:781–789 [PubMed] [Google Scholar]
- 44.Greenspan P, Mayer EP, Fowler SD1985. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol 100:965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ebling FJP, Maywood ES, Staley K, Humby T, Hancock DC, Waters CM, Evant GI, Hastings MH1991. The role of N-methyl-d-aspartate-type glutamatergic neurotransmission in the photic induction of immediate-early gene expression in the suprachiasmatic nuclei of the Syrian hamster. J Neuroendocrinol 3:641–652 [DOI] [PubMed] [Google Scholar]