Abstract
The nuclear receptor-type transcription factor retinoic acid receptor-related orphan receptor α (RORα) is a multifunctional molecule involved in tissue development and cellular function, such as inflammation, metabolism, and differentiation; however, the role of RORα during adipocyte differentiation has not yet been fully understood. Here we show that RORα inhibits the transcriptional activity of CCAAT/enhancer-binding protein β (C/EBPβ) without affecting its expression, thereby blocking the induction of both PPARγ and C/EBPα, resulting in the suppression of C/EBPβ-dependent adipogenesis. RORα interacted with C/EBPβ so as to repress both the C/EBPβ-p300 association and the C/EBPβ-dependent recruitment of p300 to chromatin. In addition to the inhibitory effect on C/EBPβ function, RORα also prevents the expression of the lipid droplet coating protein gene perilipin by peroxisome proliferators-activated receptor γ (PPARγ), acting through the specific mechanism of its promoter. We identified a suppressive ROR-responsive element overlapping the PPAR-responsive element in the perilipin promoter and verified that RORα competitively antagonizes the binding of PPARγ. RORα inhibits PPARγ-dependent adipogenesis along with the repression of perilipin induction. These findings suggest that RORα is a novel negative regulator of adipocyte differentiation that acts through dual mechanisms.
The orphan nuclear receptor RORα restrains adipocyte differentiation through a reduction of C/EBPβ activity and gene expression of lipid droplet-coating protein perilipin.
Adipose tissue serves as an important organ in the control of whole-body energy homeostasis by storing excess energy as triglycerides and releasing fatty acids in response to energy demand (1). An increase in both adipocyte number and size results in obesity, which is a major risk factor for metabolic disorders including diabetes, hypertension, hyperlipidemia, and cardiovascular disease.
Several key transcription factors that regulate adipocyte development have been identified by genetic studies using knockout mice and in vitro analysis using established mouse preadipocyte cell lines, such as 3T3-L1 and 3T3-F442A (2, 3, 4, 5). These central molecules consist of CCAAT/enhancer-binding proteins (C/EBPs) that are a family of leucine zipper transcription factors, and peroxisome proliferators-activated receptor γ (PPARγ), which is a member of the nuclear hormone receptor superfamily and is also known to be a master regulator of adipogenesis. These transcription factors play a central role in the differentiation of preadipocytes into adipocytes that occurs in response to adipogenic stimuli (6, 7).
In cell culture models, preadipocyte differentiation can be triggered by hormonal stimuli including insulin, cAMP, and glucocorticoids (8). The early response to insulin and cAMP is transient expression and activation of C/EBPβ and C/EBPδ, and glucocorticoids potentiate these events (9, 10, 11). The enhanced activity of C/EBPβ and C/EBPδ initiates the expression of the two critical regulators of terminal adipogenesis, C/EBPα and PPARγ. In addition, the expression of both C/EBPα and PPARγ is cooperatively self-regulated, and PPARγ induces the expression of adipocyte-specific genes, such as perilipin, which is an adipocyte-specific lipid droplet coating protein, resulting in the promotion of intracellular fat storage (7, 12, 13, 14, 15). On the other hand, C/EBPα seems to be mainly required for the activation and maintenance of PPARγ expression and the establishment of insulin sensitivity in the mature adipocyte (12, 16). Insulin can also stimulate adipogenesis through another pathway, via induction of sterol regulatory element-binding protein (SREBP)-1c that is a basic helix-loop-helix protein (17). SREBP1c promotes triglyceride synthesis, increases fatty acid synthesis, and facilitates the activity of PPARγ through a production of its endogenous ligands (18, 19).
Overexpression of C/EBPβ can stimulate the forced differentiation of preadipocytes without the addition of hormone inducers by up-regulating the expression of PPARγ and C/EBPα (2, 20). However, the loss of C/EBPβ action results in only partial repression of adipogenesis because of the compensation by C/EBPδ; therefore, the crucial defects in adipogenesis are seen only upon deficits in both C/EBPβ and C/EBPδ (21). Upon the induction of differentiation, C/EBPβ also regulates the cell division of preadipocytes before differentiation, which is termed mitotic clonal expansion (MCE) (22).
In addition to PPARγ, several members of the nuclear receptor family, such as glucocorticoid receptor and estrogen receptor, have been reported to contribute to the regulation of adipocyte differentiation (23, 24). The retinoic acid receptor-related orphan receptor α (RORα) is a nuclear orphan receptor and is involved in various physiological phenomena, including cerebellar development, muscle differentiation, inflammation, and bone metabolism (25, 26). Furthermore, RORα is also implicated in lipid metabolism by inducing the gene expression of apolipoproteins (25), but the role of RORα during adipogenesis is poorly understood. Here we demonstrate that RORα plays a critical role as a novel negative regulator of adipogenesis through dual mechanisms.
Results
RORα expression is induced during adipocyte differentiation
We first analyzed the daily changes of RORα expression during adipocyte differentiation using mouse 3T3-L1 fibroblasts as an in vitro model. Real-time PCR analysis revealed that RORα mRNA level gradually increases from the middle (d 4) to the late (d 10) stage of differentiation (Fig. 1A). We also measured the mRNA levels of the fatty acid binding protein aP2 (known to be an adipocyte differentiation marker) and the transcription factors that stimulate differentiation, C/EBPβ, C/EBPδ, SREBP1, PPARγ, and C/EBPα (supplemental Fig. 1A, which is published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals. org). The mRNA levels of C/EBPβ, C/EBPδ, and SREBP1, the key factors in the early stage of differentiation, reached a peak on d 2 or 4. In contrast, both PPARγ and C/EBPα expression began to increase from d 2 and reached a peak on d 8. Distinct from the other transcription factors, the up-regulation of RORα mRNA was not yet observed on d 2 (Fig. 1A). Next, we investigated the expression pattern of RORα during the first 36 h after the initiation of differentiation (Fig. 1B). Along with a decline in the mRNA level of CHOP, which is a negative regulator of adipocyte differentiation, the RORα mRNA level declined for the first 6 h and then maintained a steady level to 36 h.
Fig. 1.
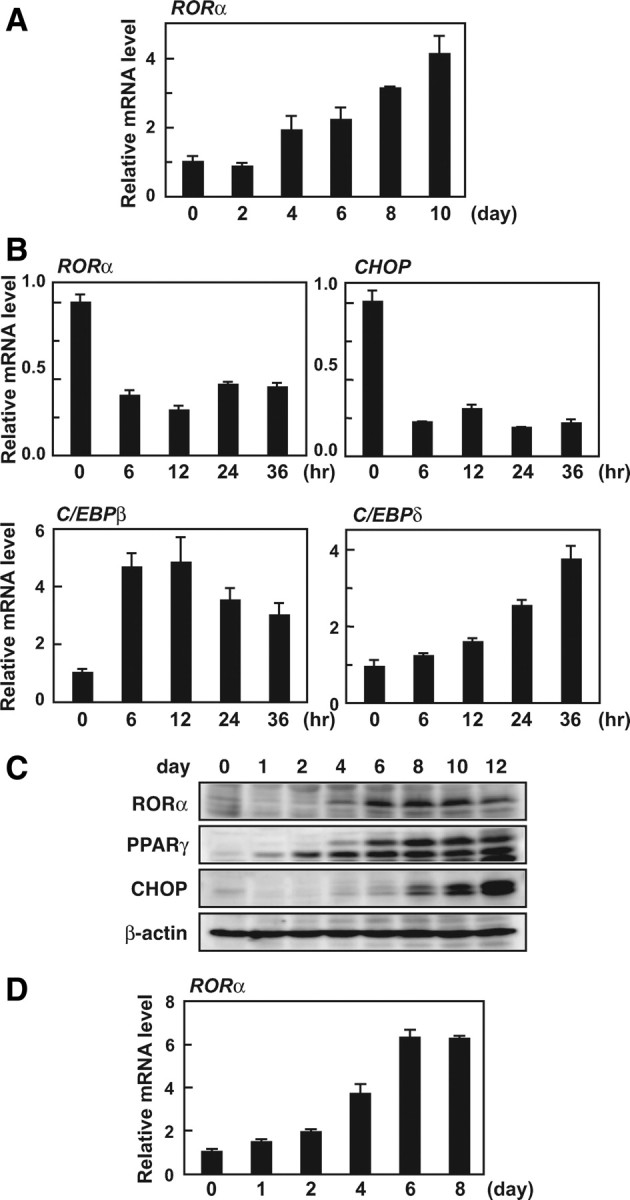
Expression of RORα during adipocyte differentiation. A–C, 3T3-L1 preadipocytes were induced to differentiate as described in Materials and Methods. A and B, Total RNA was isolated from cells on the indicated days (A) or hours (B) after induction. The mRNA levels of RORα, CHOP, C/EBPβ, and C/EBPδ were measured by quantitative RT-PCR and normalized to the amounts of 18S. C, 3T3-L1 cells were harvested on the indicated days after induction, and the cell lysates were analyzed by immunoblotting using anti-RORα, anti-PPARγ, anti-CHOP, and anti-β-actin antibodies. D, The primary preadipocytes were isolated and induced to differentiate as described in Materials and Methods. Total RNA was isolated from cells on the indicated days after induction. The mRNA levels of RORα was measured by quantitative RT-PCR and normalized to the amount of 18S.
To identify which components of the adipogenic cocktail [insulin, 3-isobutyl-1-methylxanthine (MIX), and dexamethasone (DEX)] were responsible for the transient suppression of RORα expression during the initial 36 h, 2 d after confluence, we treated 3T3-L1 preadipocytes with insulin, MIX, or DEX, either separately or in combination, for 24 h (supplemental Fig. 1B). RORα expression was decreased by the treatment with MIX, but not DEX, as was the expression of CHOP (and its target gene, TRB3) (27, 28). As in previous studies (29, 30, 31), insulin treatment induced up-regulation of both CHOP and TRB3 mRNA levels, whereas insulin slightly suppressed RORα expression. These results suggest that RORα expression is suppressed for the initial stage of differentiation by cAMP and insulin signaling, and this regulation is caused through a pathway distinct from regulation of CHOP. We further confirmed the expression pattern of RORα at the protein level by Western blotting (Fig. 1C). RORα expression was initially decreased on d 1 and gradually increased in the late stage of differentiation as well as at the protein level.
To assess the expression of RORα during adipogenesis under more physiological conditions, we isolated primary preadipocytes from the stromal-vascular fraction of mouse sc white adipose tissues and induced differentiation into adipocytes, and the daily changes of RORα, PPARγ and aP2 mRNA levels were analyzed in these cells (Fig. 1D and supplemental Fig. 2A). RORα expression was enhanced during adipogenesis in the primary cells; however, the down-regulation in the early phase was not observed, unlike CHOP expression (supplemental Fig. 2B).
RORα expression suppresses the differentiation of 3T3-L1 cells into adipocytes
To examine whether RORα affects adipocyte differentiation, we overexpressed RORα in subconfluent 3T3-L1 preadipocytes with the lentiviral expression vector for 3Flag-RORα. These cells were grown to confluence and then induced to differentiate into adipocytes. At 10 d after initiation of differentiation, control (mock-infected) cells harbored lipid droplets as visualized by Oil red O staining, whereas lipid accumulation was effectively impaired in the cells expressing RORα (Fig. 2A). Lipid droplets were almost equally formed in both the mock-infected and noninfected cells, indicating that lentiviral infection itself does not affect lipid droplet formation (data not shown).
Fig. 2.
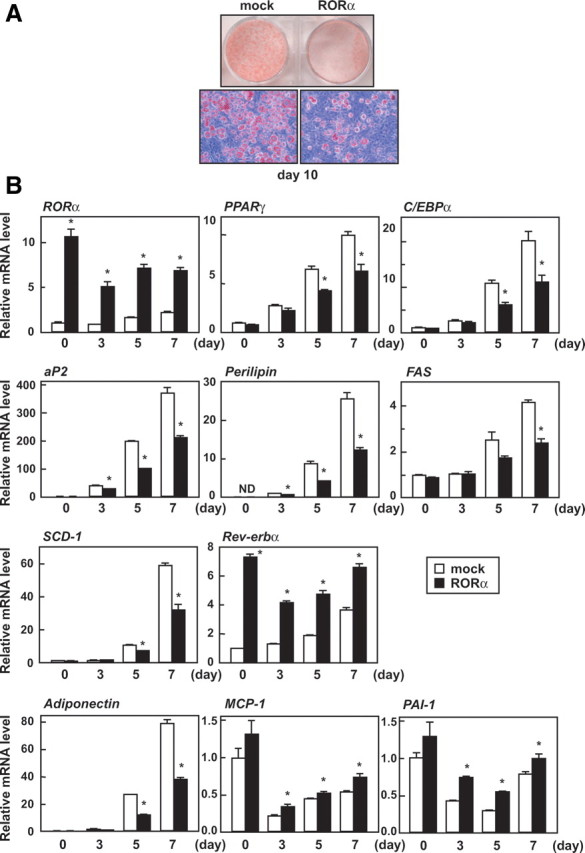
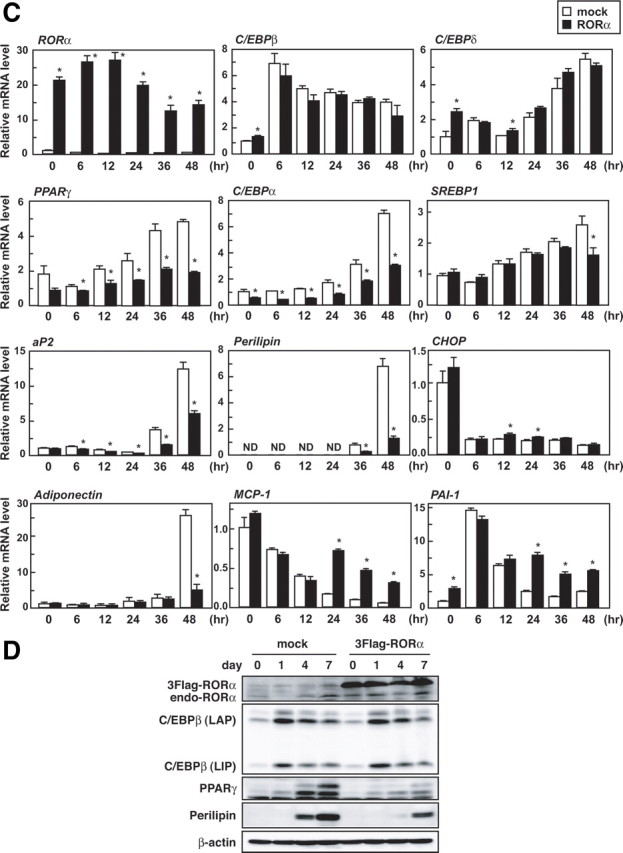
RORα overexpression blocks 3T3-L1 adipogenesis. Subconfluent 3T3-L1 cells were infected with an empty (mock-infected) or RORα-expressing lentiviral vector, and after 3 d, the cells were induced to differentiate. A, Oil red O staining of the cells was performed on d 10 of differentiation. B and C, The cells were harvested on the indicated days (B) or hours (C) after induction, and the relative mRNA levels of the indicated genes were measured by quantitative RT-PCR and normalized to the amount of 18S. All assays were performed in triplicate. The data represent means ± se. *, P < 0.01 compared with mock-infected cells. D, The cells were harvested on the indicated days after induction, and the cell lysates were analyzed by immunoblotting using anti-RORα, anti-C/EBPβ, anti-PPARγ, anti-perilipin, and anti-β-actin antibodies.
Next, we confirmed the inhibitory effect of RORα on the expression of differentiation marker genes by real-time PCR analysis (Fig. 2B). RORα overexpression repressed expression of two principal regulators of terminal adipogenesis, PPARγ and C/EBPα, and their target gene, aP2, and a PPARγ target gene, perilipin. In addition, expression of the SREBP1 target genes, FAS and SCD-1, were also decreased by RORα. Up-regulation of expression of both RORα and its target gene, Rev-erbα, were observed throughout differentiation. These results suggest that RORα inhibits or delays adipocyte differentiation. In addition, during adipocyte differentiation, RORα overexpression down-regulated the expression of adiponectin, which is an antiinflammatory adipocytokine, and up-regulated the expression of macrophage chemoattractant protein-1 (MCP-1) as well as plasminogen activator inhibitor type 1 (PAI-1), which are proinflammatory adipocytokines.
In an attempt to identify potential targets of RORα, we investigated the inhibitory effect of RORα on the gene expression program at the early stage of differentiation (Fig. 2C). Both PPARγ and C/EBPα mRNA expression were suppressed by RORα at the initial stage of differentiation, whereas in the case of the expression of the upstream genes of PPARγ and C/EBPα, C/EBPβ and C/EBPδ, hardly any changed. SREBP1 mRNA level was not affected until 36 h and then declined, suggesting that this reduction results from a delay of differentiation. RORα did not affect the mRNA level of CHOP, which is a dominant-negative regulator of C/EBPs. Taken together, it is likely that RORα prevents adipocyte differentiation by inhibiting the induction of PPARγ and C/EBPα by C/EBPβ and/or C/EBPδ. In addition, RORα suppressed PPARγ and perilipin protein expression without affecting C/EBPβ [liver-enriched transcriptional activating protein (LAP) and liver-enriched transcriptional inhibitory protein (LIP)] protein expression (Fig. 2D).
In the above experiments, 3T3-L1 preadipocytes were infected with a lentiviral expression vector for RORα 3 d before the initiation of differentiation. To lessen the possibility that RORα overexpression directly affects the functions of preadipocytes, we infected 3T3-L1 preadipocytes 2 d after confluence and immediately induced them to start differentiation. As shown in supplemental Fig. 3A, RORα suppressed lipid accumulation under this experimental condition as well. Moreover, RORα expression repressed the protein level of PPARγ and perilipin (supplemental Fig. 3B). These results indicate that RORα functions as a negative regulator independently of the timing of its expression during differentiation. Furthermore, in the differentiation into primary adipocytes, RORα overexpression repressed lipid accumulation as well (supplemental Fig. 4A).
RORα knockdown facilitates the differentiation of 3T3-L1 cells into adipocytes
To clarify the role of endogenous RORα in adipocyte differentiation, we knocked down the expression of RORα in subconfluent 3T3-L1 preadipocytes with a lentiviral vector expressing short hairpin RNA (shRNA). At 3 d after infection, a significant reduction of endogenous RORα expression was confirmed in the cells expressing RORα shRNA as compared with the control cells at both the mRNA (Fig. 3A, left) and protein levels (Fig. 3A, right). Next, to investigate the effect of RORα knockdown in the process of adipocyte differentiation, we infected 3T3-L1 preadipocytes 2 d after confluence and immediately induced them to start differentiation. On d 8, the RORα-knockdown cells accumulated greater amounts of lipids than the control cells, as shown by Oil red O staining (Fig. 3B). Consistent with this observation, the mRNA levels of PPARγ, C/EBPα, aP2, and perilipin were elevated by RORα knockdown on d 4 (Fig. 3C). Moreover, RORα knockdown augmented PPARγ and perilipin protein without altering the C/EBPβ protein level (Fig. 3D). In the differentiation into primary adipocytes, RORα knockdown augmented lipid accumulation and elevated the mRNA levels of C/EBPα as well as perilipin (supplemental Fig. 4, B and C). It is therefore concluded that endogenous RORα acts as a negative regulator of adipogenesis by suppressing both PPARγ and C/EBPα expression.
Fig. 3.
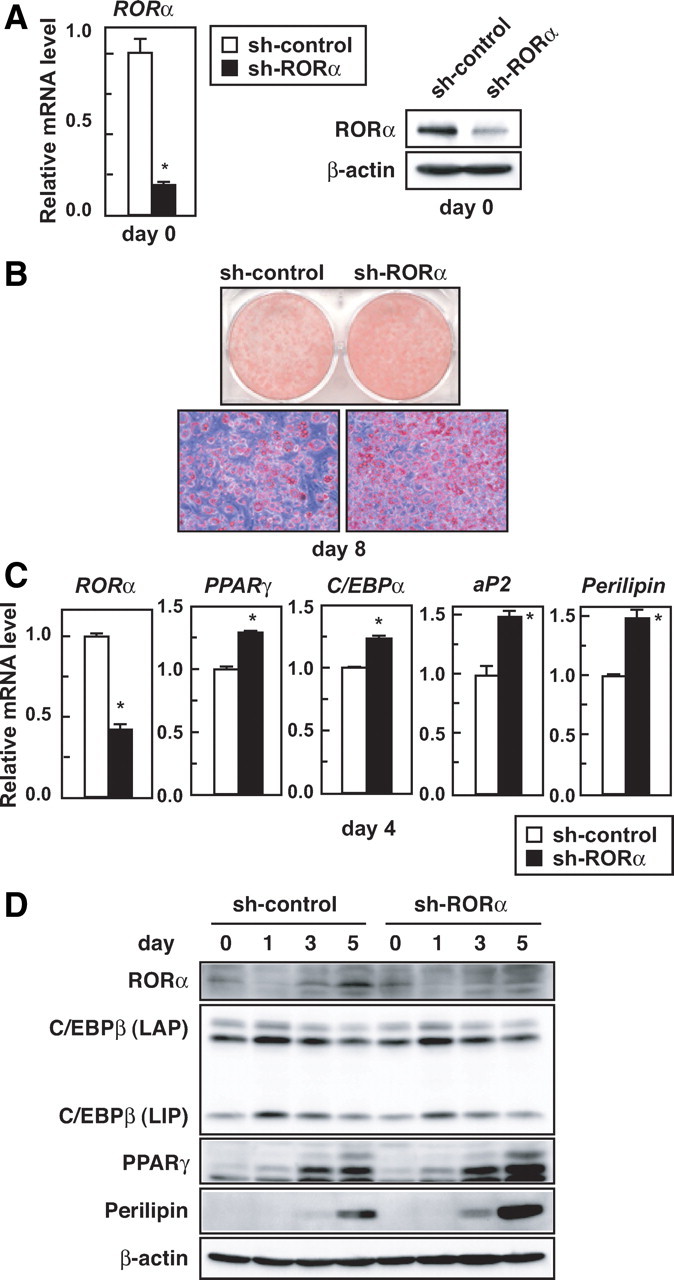
RORα knockdown promotes 3T3-L1 adipogenesis. A, Subconfluent 3T3-L1 cells were infected with a lentiviral vector expressing control shRNA or shRNA for RORα. After 3 d, the knockdown efficiency of RORα expression levels in 3T3-L1 cells on d 0 was measured by quantitative RT-PCR (left) or immunoblotting using anti-RORα and anti-β-actin antibodies (right). The data represent means ± sd. *, P < 0.01 (n = 3) compared with sh-control cells (left). B–D, 3T3-L1 cells 2 d after confluence were infected with a lentiviral vector expressing control shRNA or shRNA for RORα, and the cells were immediately induced to differentiate. B, Oil red O staining of the cells was performed on d 8. C, The cells were harvested on d 4, and the relative mRNA levels of the indicated genes were measured by quantitative RT-PCR and normalized to the amount of 18S. The data represent means ± sd. *, P < 0.01 (n = 3) compared with sh-control cells. D, The cells were harvested on the indicated days after induction, and the cell lysates were analyzed by immunoblotting using anti-RORα, anti-C/EBPβ, anti-PPARγ, anti-perilipin, and anti-β-actin antibodies.
RORα inhibits the transcriptional activity of C/EBPβ
The expression of both PPARγ and C/EBPα are induced by C/EBPβ and C/EBPδ at the early stages of adipogenesis (2, 3). Because RORα repressed the expression of PPARγ and C/EBPα without changing expression of C/EBPβ and C/EBPδ (Fig. 2C), we examined whether RORα directly affects the transcriptional activities of C/EBPβ and C/EBPδ. The effect of RORα on an artificial reporter gene that included four repeats of the C/EBP regulatory element of the C/EBPα promoter was first examined. As shown in Fig. 4A, RORα inhibited the activation of this promoter by C/EBPβ, but not C/EBPδ (Fig. 4A). RORα also repressed the activation of the aP2 promoter reporter gene by C/EBPβ (Fig. 4B). On the other hand, RORα did not affect the transcriptional activity of C/EBPα (data not shown). These results suggest that RORα exclusively suppresses the transcriptional activity of C/EBPβ among these C/EBP family members.
Fig. 4.
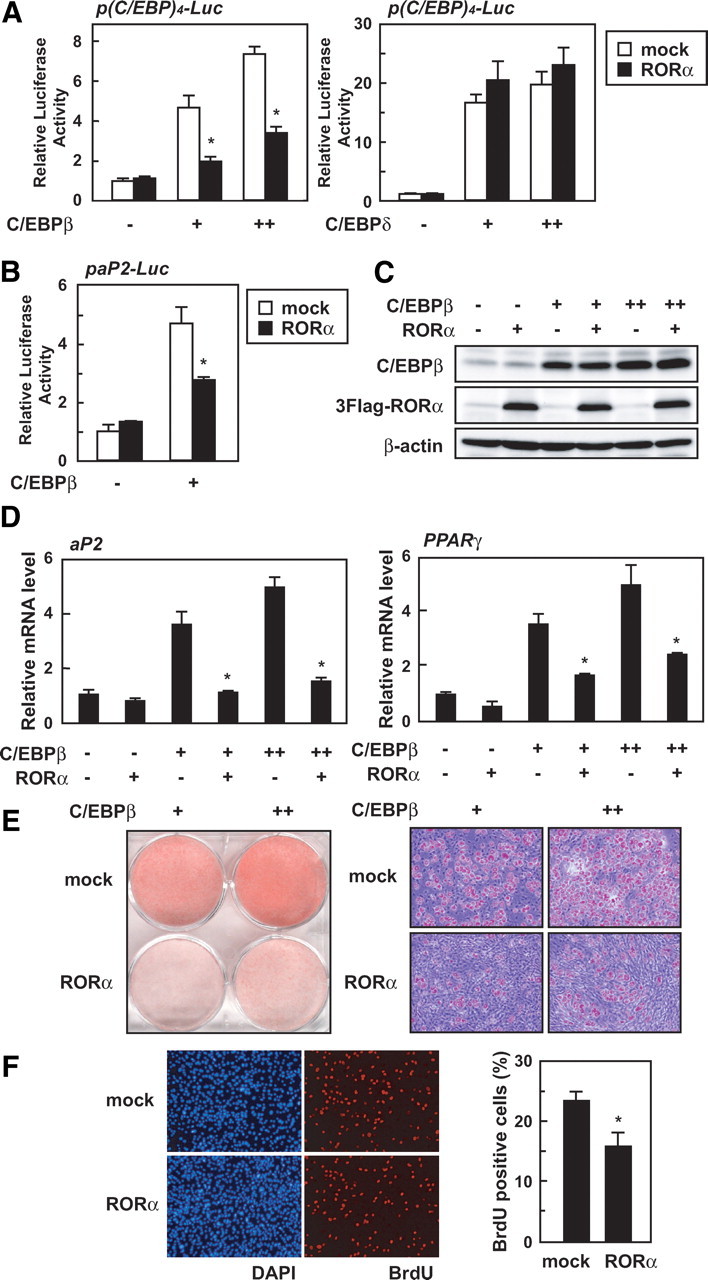
RORα prevents the functional activity of C/EBPβ. A and B, 3T3-L1 cells were transiently transfected with p(C/EBP)4-Luc, paP2-Luc, and/or pCMV-β-gal in the absence or presence of the expression vectors for C/EBPβ, C/EBPδ, and/or RORα. After 48 h, the luciferase activity in cell lysates was measured and was normalized with β-galactosidase activity. The data represent means ± sd. *, P < 0.01 (n = 3) compared with no RORα-transfected cells. C–E, Subconfluent 3T3-L1 cells were infected with an empty (mock-infected), C/EBPβ-, and/or RORα-expressing lentiviral vector. C, After 24 h infection, the cells were harvested and the cell lysates were analyzed by immunoblotting using anti-C/EBPβ, anti-Flag, and anti-β-actin antibodies. D, After 24 h infection, the cells were harvested, and relative mRNA levels of indicated genes were measured by quantitative RT-PCR and normalized to the amounts of 18S. The data represent means ± sd. *, P < 0.01 (n = 3) compared with no RORα-infected cells. E, After 12 d infection, the cells were harvested and Oil red O staining was performed. F, Subconfluent 3T3-L1 cells were infected with an empty (mock-infected) or RORα-expressing lentiviral vector, and after 3 d, the cells were induced to differentiate. After 24 h induction, the cells were labeled with BrdU for 3 h and then fixed as described in Materials and Methods. Percentages of BrdU-positive cells are represented (right). The data represent means ± sd. *, P < 0.01 (n = 6; the counted fields) compared with mock cells.
To further investigate whether RORα inhibits the function of C/EBPβ, we constructed a lentiviral expression vector for C/EBPβ and generated C/EBPβ- and/or RORα-overexpressing 3T3-L1 cells to examine whether RORα would prevent C/EBPβ-dependent adipocyte differentiation. At 24 h after infection, expression of these two proteins was analyzed by Western blotting (Fig. 4C). The mRNA levels of aP2 and PPARγ were increased when only C/EBPβ was expressed, and RORα abolished the increase without affecting C/EBPβ expression (Fig. 4, C and 4D). At 24 h after infection, the C/EBPα mRNA level was not yet elevated by C/EBPβ (data not shown), whereas at 4 d after infection, the C/EBPβ-dependent increase in C/EBPα mRNA was observed and inhibited by RORα (supplemental Fig. 5). Consistent with a previous finding (20), C/EBPβ-expressing 3T3-L1 cells differentiated into adipocytes and accumulated triglyceride without hormonal stimulation, whereas triglyceride accumulation was repressed by RORα expression (Fig. 4E).
C/EBPβ plays an essential role in the MCE at the early stages of adipogenesis (22). To test whether RORα affects MCE, which is a C/EBPβ-dependent process, we induced control and RORα-expressing 3T3-L1 cells to differentiate into adipocytes and monitored cell-cycle progression by determining 5-bromo-2′-deoxyuridine (BrdU) incorporation (Fig. 4F). As shown in Fig. 4F, a reduction in the number of BrdU-positive cells was observed in RORα-expressing cells compared with control cells. These results clearly demonstrate that RORα inhibits the function of C/EBPβ during adipocyte differentiation.
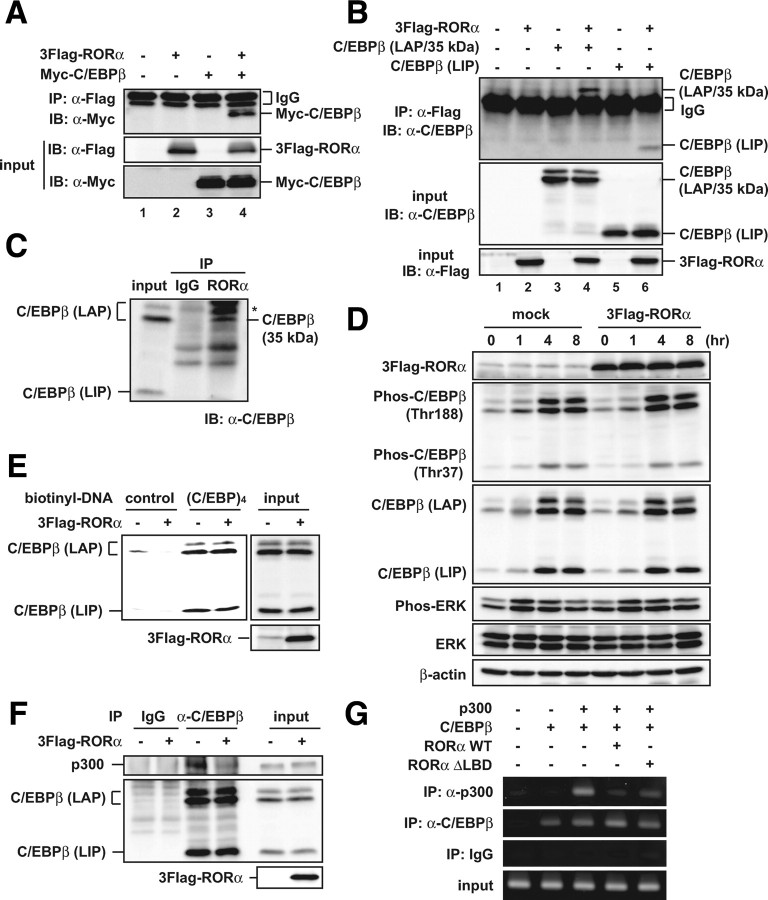
RORα physically interacts with C/EBPβ
Because RORα repressed the transcriptional activity of C/EBPβ without any changes in its expression, we assessed whether the inhibition of C/EBPβ activity by RORα is attributable to a physical interaction between these proteins. As shown in Fig. 5A, Myc-C/EBPβ was co-immunoprecipitated with 3Flag-RORα when both were expressed in 293T cells (Fig. 5A, lane 4). C/EBPβ mRNA generates at least three proteins, a minor isoform of LAP (38 kDa), a major isoform of LAP (35 kDa), and LIP (20 kDa) through alternative translation or proteolytic cleavage, and LIP acts as a dominant-negative regulator of the isoforms of LAPs that act as a transcriptional activator (30, 32, 33). In transfected 293T cells, LIP (20 kDa), unlike LAP (35 kDa), was only slightly coimmunoprecipitated with 3Flag-RORα (Fig. 5B). Similarly, the association between endogenous RORα and a major isoform of LAP (35 kDa) was observed in 3T3-L1 preadipocytes (Fig. 5C). The same results were also obtained in 3T3-L1 cells on d 1 and 7 after the induction of differentiation (data not shown). These results indicate that RORα physically interacts with a major isoform of LAP during adipogenesis.
Fig. 5.
RORα physically interacts with C/EBPβ and inhibits the C/EBPβ-dependent recruitment of p300. A and B, 293T cells were transiently transfected with the indicated constructs. After 48 h, the cell lysates were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-Myc antibody (A) or anti-C/EBPβ antibody (B). The expression level of each protein was assessed by immunoblotting of the cell lysates (input) with the indicated antibodies. C, 3T3-L1 cells were grown to confluence and cultured for another 2 d. The cell lysates were immunoprecipitated with anti-RORα or control IgG and immunoblotted with anti-C/EBPβ antibody. The asterisk in the panel indicates nonspecific bands. D–F, Subconfluent 3T3-L1 cells were infected with an empty (mock-infected) or RORα-expressing lentiviral vector, and after 3 d, the cells were induced to differentiate. D, The cells were harvested at the indicated hours after induction, and the cell lysates were analyzed by immunoblotting using anti-Flag, anti-phospho-C/EBPβ, anti-C/EBPβ, anti-phospho-ERK, anti-ERK, and anti-β-actin antibodies. E, After 24 h induction, the cell lysates were subjected to promoter pull-down assays using the biotin-conjugated nucleotide containing the four repeats of the C/EBP regulatory element of the C/EBPα promoter [(C/EBP)4] or a control sequence from the pGL3-basic vector and immunoblotted with anti-C/EBPβ antibody. The expression level of each protein was assessed by immunoblotting of the cell lysates (input) with the indicated antibodies. F, After 24 h induction, the cell lysates were immunoprecipitated with anti-C/EBPβ antibody or control IgG and immunoblotted with anti-p300 antibody. The expression level of each protein was assessed by immunoblotting of the cell lysates (input) with anti-p300, anti-C/EBPβ, and anti-Flag antibodies. G, 293T cells were transiently transfected with the indicated constructs. After 48 h, the cells were subjected to ChIP assay using the indicated antibodies or control IgG. The immunoprecipitated region containing C/EBP elements within the PPARγ2 promoter was amplified with PCR using specific primers.
RORα does not inhibit the DNA-binding activity and phosphorylation of C/EBPβ at the early stage of adipogenesis
To investigate how RORα inhibits the activity of C/EBPβ via the interaction, we first examined the effect of RORα on the phosphorylation of C/EBPβ. C/EBPβ is sequentially phosphorylated during differentiation of 3T3-L1 adipocytes, and this phosphorylation is critical for the DNA-binding activity of C/EBPβ (34). 3T3-L1 preadipocytes overexpressing RORα were induced to differentiate into adipocytes, and phosphorylated LAP/Thr188 and LIP/Thr37 were analyzed by Western blotting using antibodies against them (Fig. 5D). Enforced RORα expression unexpectedly affected the phosphorylation of neither C/EBPβ LAP nor LIP. Additionally, the upstream signaling, phosphorylation of ERK1/2 (Thr202/Tyr204), was not affected by RORα (Fig. 5D).
To determine whether RORα affects the DNA-binding activity of C/EBPβ, we performed a promoter pull-down assay using a biotin-conjugated probe containing four repeats of the C/EBP regulatory element of the C/EBPα promoter in control and RORα-expressing 3T3-L1 cells. As shown in Fig. 5E, RORα overexpression left unchanged the volume of endogenous C/EBPβ protein bound to C/EBP binding sites at 24 h after induction of differentiation (Fig. 5E). These results suggest that RORα antagonism of the transcriptional activity of C/EBPβ is not by way of a prevention of its phosphorylation and DNA binding.
RORα inhibits the C/EBPβ-dependent recruitment of p300 on the chromatin
Because RORα more tightly interacted with the C/EBPβ LAP isoform than the LIP isoform lacking the transcriptional activation domain, the effect of RORα expression on the association between C/EBPβ and p300, the latter of which is one of the critical factors in the transcriptional coactivator complex for C/EBPβ, was examined. Lentivirus-mediated RORα overexpression prevented the interaction between endogenous C/EBPβ and p300 at 24 h after the induction of differentiation in 3T3-L1 cells (Fig. 5F). In addition, RORα interfered with the recruitment of p300 by C/EBPβ on the endogenous PPARγ promoter (Fig. 5G, lanes 3 and 4). These results indicate that RORα inhibits the C/EBPβ-dependent recruitment of p300 on chromatin, thereby repressing the transcriptional activity of C/EBPβ.
To assess the importance of the ligand-binding domain (LBD) of RORα in terms of its function during adipogenesis, we examined the effect of RORαΔLBD [amino acids (aa) 1–235] lacking the LBD on the C/EBPβ transcriptional activity and C/EBPβ-dependent adipogenesis and found that RORαΔLBD functioned similarly to wild-type RORα, without any induction of RORα target genes, Rev-erbα and Bmal1 (supplemental Fig. 6. A and B). Furthermore, RORαΔLBD inhibited the C/EBPβ-dependent p300 recruitment on the PPARγ promoter as well (Fig. 5G, lanes 3 and 5). These results suggest that both the LBD and ligand-mediated modulation of RORα activity are dispensable for the inhibitory effect of RORα on adipogenesis.
RORα inhibits activation of the perilipin promoter by PPARγ
PPARγ induces the expression of C/EBPα during adipocyte differentiation, thereby dramatically stimulating the induction of adipocyte genes (12, 35). In an attempt to assess whether RORα affects the transcriptional activity of PPARγ as well as C/EBPβ, we found that RORα represses the activation of the perilipin promoter by PPARγ (supplemental Fig. 7A). In contrast, RORα did not affect the promoter activities of two other genes, aP2 and LPL, by PPARγ (supplemental Fig. 7, B and C), suggesting that RORα inhibits the function of PPARγ through a specific mechanism of the perilipin promoter.
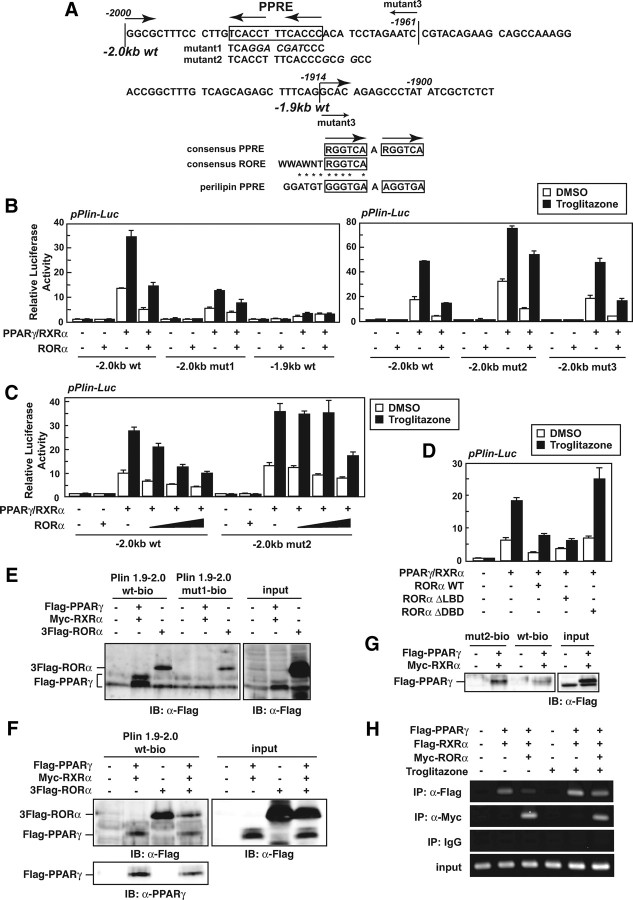
The distal perilipin promoter contains a functional PPARγ-responsive element (PPRE) located in a region of approximately −2.0 kb (region −1986 to −1974) (Fig. 6A, top) (15). Mutation in the PPRE (mutant 1) decreased the PPARγ-dependent activation of the promoter and almost abolished the repressive effect of RORα as well (pPlin-Luc −2.0 kb mutant 1; Fig. 6, A, top, and B, left). In addition, when a shorter promoter gene lacking the PPRE (pPlin-Luc −1.9 kb; −1914 to +50) was used, the promoter did not respond to either PPARγ or RORα (Fig. 6, A, top, and B, left). These results indicate that RORα suppresses the activity of perilipin promoter enhanced by PPARγ via the PPRE.
Fig. 6.
RORα inhibits the activation of the perilipin promoter by PPARγ. A, The mouse perilipin promoter sequence. The transcription start site is +1. The PPRE is boxed. The mutant sequence is shown in italic letters under the individual original sequence (top). Alignment of the PPRE in the perilipin promoter with the consensus PPRE and RORE is shown (bottom). B–D, 3T3-L1 cells were transiently transfected with pPerilipin-Luc (pPlin-Luc) and pCMV-β-gal in the absence or presence of expression vectors for PPARγ, RXRα, RORα WT, RORα ΔLBD, and/or RORα ΔDBD. After 24 h, the cells were treated with 10 μm troglitazone for 16 h. The luciferase activity in cell lysates was measured and was normalized with β-galactosidase activity. E–G, 293T (E and F) or 3T3-L1 (G) cells were transiently transfected with the indicated constructs. After 48 h, the cell lysates were subjected to promoter pull-down assays using the biotin-conjugated nucleotide of the perilipin promoter (−2000 to −1921) and immunoblotted with anti-Flag or PPARγ antibody. The expression level of each protein was assessed by immunoblotting of the cell lysates (input) with the indicated antibodies. H, 293T cells were transiently transfected with the indicated constructs. After 24 h, the cells were treated with 10 μm troglitazone for 16 h. The cells were subjected to ChIP assay using the indicated antibodies or control IgG. The immunoprecipitated region containing the PPRE within the perilipin promoter was amplified with PCR using specific primers.
The consensus PPRE consists of a direct repeat (DR) sequence of (A/G)GGTCA that is separated by one nucleotide (termed DR-1), and RORα binds as a monomer to a ROR-responsive element (RORE) composed of a 5′ 6-bp A/T-rich sequence that precedes a 3′ (A/G)GGTCA core half-site motif (WWAWNT-RGGTCA, where W or R represents A/T or A/G, respectively) (Fig. 6A, bottom) (36, 37). The PPRE in the distal region of the perilipin promoter overlaps a sequence (GGATGT-GGGTGA) similar to the consensus RORE (WWAWNT- RGGTCA) (Fig. 6A, top); therefore, we speculated that RORα binds to the PPRE of the perilipin promoter, thereby interfering with the binding of PPARγ to this element. To test this hypothesis, we first constructed a mutant reporter gene of the perilipin promoter with a substitution of G for A/T in the 5′ flanking region of the PPRE (pPlin-Luc −2.0 kb mutant 2; Fig. 6A). The activity of this promoter was more strongly enhanced by PPARγ than the wild-type promoter, and the repressive effect of RORα was reduced especially in the presence of troglitazone, a potent agonist of PPARγ (Fig. 6B, right). Moreover, RORα dose-dependently repressed the activation of the wild-type perilipin promoter by PPARγ but failed in case of the mutant promoter (mutant 2) at the lower expression levels (Fig. 6C), suggesting that the 5′ flanking A/T of PPRE is critical for the repressive effect of RORα. On the other hand, deletion of the region from −1960 to −1915 did not affect the responses to PPARγ and RORα at all (pPlin-Luc −2.0 kb mutant 3; Fig. 6A, top, and B, right), indicating that the upstream region (−2000 to −1961) containing the PPRE is indispensable for the RORα action. Next, the functional domain of RORα necessary for its repressive effect on the PPARγ-dependent activation of the perilipin promoter was investigated. Although the deletion of LBD (RORαΔLBD) did not affect the RORα repressive function, the deletion of DNA-binding domain [RORαΔDBD (aa 1–72, 140–523)] completely abolished it (Fig. 6D), indicating that the DBD of RORα, unlike the LBD, is indispensable for the repressive effect.
To further investigate the possibility of competition between PPARγ and RORα for the binding to PPRE in the perilipin promoter, we performed a promoter pull-down assay using a biotinyl-oligonucleotide corresponding to the PPRE region (−2000 to −1921). As shown in Fig. 6E, RORα tightly bound to the wild-type oligonucleotide as well as PPARγ; however, the binding of RORα to the PPRE-mutated oligonucleotide (mut1-bio) was significantly decreased (Fig. 6E, lanes 3 and 6), indicating that RORα can bind to the PPRE. We therefore examined the repressive effect of RORα on the binding of PPARγ to the PPRE and found that the amount of PPARγ protein bound to the PPRE was effectively decreased by RORα expression (Fig. 6F, lanes 2 and 4). Moreover, mutation in RORE (mut2-bio) increased the binding of PPARγ to the perilipin promoter oligonucleotide in 3T3-L1 cells (Fig. 6G). The inhibitory effect of RORα on the binding of PPARγ to the perilipin promoter was investigated by chromatin immunoprecipitation (ChIP) assay as well. The treatment with troglitazone increased the binding of PPARγ/RXRα heterodimer to the endogenous perilipin promoter (Fig. 6F, lanes 2 and 5), and RORα expression repressed the binding both in the absence and presence of troglitazone (Fig. 6F, lanes 3 and 6). These data demonstrate that RORα competitively inhibits the DNA binding of PPARγ to PPRE in the perilipin promoter.
RORα inhibits PPARγ-dependent expression of perilipin and adipogenesis
Overexpressed PPARγ can lead fibroblasts into forced differentiation into adipocytes by directly inducing the expression of adipogenic genes, including perilipin (7). Perilipin is a protein that coats lipid droplets of adipocytes and is thought to have a critical role in increasing triacylglycerol storage by blocking hormone-sensitive lipase-mediated lipolysis (38, 39, 40). Because RORα down-regulated the perilipin promoter activity enhanced by PPARγ, we investigated whether RORα would repress the PPARγ-dependent expression of perilipin and triglyceride accumulation in PPARγ-expressing 3T3-L1 cells that were generated by infection with an expression lentiviral vector for PPARγ. Both in the presence and absence of troglitazone, ectopic expression of PPARγ caused up-regulation of perilipin expression at both the mRNA and protein levels, and RORα reduced these up-regulated levels without altering PPARγ expression itself (Fig. 7, A and B). Moreover, RORα interfered with the PPARγ-induced differentiation into adipocytes that occurred without hormonal stimulation (Fig. 7C). These results clearly indicate that RORα inhibits adipogenesis driven by PPARγ, partly due to decreased perilipin gene expression.
Fig. 7.
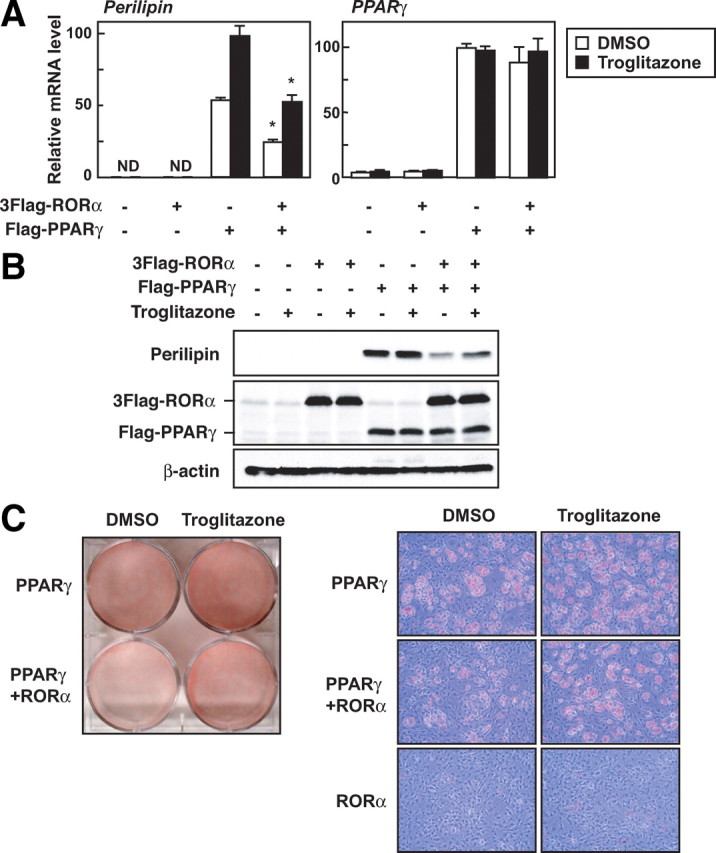
RORα inhibits PPARγ-dependent expression of perilipin and adipogenesis. A–C, Subconfluent 3T3-L1 cells were infected with an empty (mock-infected), PPARγ-, and/or RORα-expressing lentiviral vector. After 3 d infection, the cells were treated with or without 10 μm troglitazone. After 16 h, the mRNA levels of perilipin and PPARγ were measured by quantitative RT-PCR and normalized to the amount of 18S (A). The data represent means ± sd. *, P < 0.01 (n = 3) compared with no RORα-infected cells. After 16 h, the expression level of each protein was assessed by immunoblotting using anti-perilipin, anti-Flag, and anti-β-actin antibodies (B). After 9 d, the cells were harvested and Oil red O staining was performed (C).
Discussion
RORα is a nuclear receptor-type transcription factor that contributes to the regulation of various physiological phenomena, including skeletal muscle and smooth muscle cell differentiation (25), but the precise functions of RORα during adipocyte differentiation remain unknown. In this study, we have revealed that RORα functions as a novel negative regulator of differentiation of preadipocytes into adipocytes through a suppression of the C/EBPβ transcriptional activity and perilipin gene expression.
RORα repressed the expression of the C/EBPβ and C/EBPδ target genes, aP2, PPARγ, and C/EBPα, without affecting the expression of C/EBPβ and C/EBPδ (Fig. 2). In addition, RORα inhibited the transcriptional activation and adipogenesis driven by ectopically expressed C/EBPβ (Fig. 4). These results clearly demonstrate that RORα is a regulatory molecule that blocks adipogenesis by turning down the functions of C/EBPβ. RORα did not affect the expression of positive and negative heterodimerization partners of C/EBPβ such as C/EBPδ or CHOP (Fig. 2) (6). Although the activity of C/EBPβ is also controlled by protein degradation via a proteasome-dependent pathway during adipocyte differentiation (41), RORα left unchanged the expression level of the C/EBPβ protein (Fig. 2). Moreover, RORα did not affect phosphorylation or the DNA-binding activity of C/EBPβ (Fig. 5). During adipogenesis, C/EBPβ activity is regulated by the transcriptional coactivator complex containing p300, GCN5, and PCAF or the transcriptional corepressor complex containing HDAC and mSin3A (11, 42, 43). In this study, RORα prevented the C/EBPβ-dependent recruitment of p300 to chromatin (Fig. 5). It seems likely that RORα disturbs the C/EBPβ activity by affecting the recruitment of p300 and possibly other cofactors as well.
Past studies using perilipin knockout mice have revealed that perilipin plays a critical role in adipose development in vivo (38, 40). In addition to the inhibitory effect on C/EBPβ functions, RORα further suppresses adipocyte differentiation, particularly the formation of lipid droplets, through a reduction of the perilipin gene expression driven by PPARγ. In the current study, we found that PPRE/DR1 in the distal region of the perilipin promoter functionally overlaps with the RORE. It is likely that RORα represses the perilipin promoter activity by binding to this region competing with PPARγ. The PPRE sequences observed in various PPARγ target genes are listed in supplemental Fig. 8A. The PPREs in the aP2, malic enzyme, LXRα, and adiponectin promoters are highly similar to the consensus RORE; however, RORα barely repressed the PPARγ-dependent activation of the aP2 promoter, which included several PPREs (supplemental Fig. 7B), and did not bind to one of them at all (supplemental Fig. 8B). The PPREs of the aP2 promoter might not possess some as yet unknown key sequence required for the binding of RORα, although they do include a RORE-like sequence. Alternatively, not only the RORE but also the additional sequences in the upstream region of the perilipin promoter (−2000 to −1961) may be necessary for the binding of RORα. It is also possible that RORα might affect adipogenesis by suppressing the promoter activities of malic enzyme, LXRα, adiponectin, and/or other PPARγ target genes through a similar mechanism.
Rev-erbα, which is a nuclear receptor, binds to RORE as a transcriptional repressor, thereby repressing certain transcriptional activation events mediated by RORα (44). Moreover, because Rev-erbα is a RORα target gene, the function of RORα is constitutively regulated by Rev-erbα, forming a negative feedback loop (45, 46). In this study, the effect of Rev-erbα on the C/EBPβ transcriptional activity was examined (supplemental Fig. 9A); however, its suppressive effect was not observed, suggesting that the RORα-dependent repression of the C/EBPβ activity does not result from the transcriptional repressive effect of Rev-erbα. Both Rev-erbα and Bmal1, which is another transcription factor directly induced by RORα, have been reported to promote adipocyte differentiation (47, 48). Therefore, it is possible that the inhibitory effect of RORα on adipogenesis would be regulated in part by the opposite functions of these factors.
A number of transcriptional regulatory factors, such as GATA2/3, KLF2/7, ETO, CHOP, and TRB3, attenuate adipocyte differentiation by acting as anti-adipogenic factors (2, 3, 6, 27, 30). As the expression of these factors diminishes immediately after the initiation of differentiation, it is thought that the reduced gene expression accelerates adipogenesis. Some anti-adipogenic factors, such as GATA2/3, KLF2/7, and ETO, retain a constantly lower expression level throughout adipogenesis. In contrast, the expression of RORα, as well as CHOP and TRB3, was increased as the adipogenesis of both 3T3-L1 cells and primary preadipocytes progressed. These molecules, including RORα, seem likely not only to inhibit the induction of differentiation in preadipocytes, but also to terminate differentiation and control the secretion of adipocytokines in mature adipocytes.
The RORα gene generates at least four isoforms (RORα1–4) through alternative splicing and different promoter use (26, 49). All of these isoforms have been isolated in humans, whereas only RORα1 and RORα4 have been detected in mice. Because these isoforms share common DNA- and ligand-binding domains but differ in their N-terminal sequences, they exert slightly different transcriptional actions. Several tissues and cell types express mainly RORα1 and RORα4, whereas RORα2 and RORα3 are expressed abundantly in the testis. Because RORα1 isoform was used for overexpression of RORα in the current experiments, we further examined the effect of RORα4 on 3T3-L1 adipogenesis, expression of differentiation markers, and the transcriptional activation by C/EBPβ (supplemental Fig. 9, A–C). RORα4 turned out to exert inhibitory effects comparable to RORα1. Taken together with the results of RORα knockdown experiments in Fig. 3, both RORα1 and -α4 evidently restrain adipogenesis.
Staggerer mice contain a spontaneous null mutation consisting of a deletion and frame shift in the region encoding the LBD of RORα, bringing about expression of dysfunctional RORα [RORαsg (aa 1–302)] without its LBD (50, 51). The phenotypes of staggerer mice have been shown to have abnormalities in cerebellar degeneration, immunodeficiencies, muscular atrophy, osteoporosis, and atherosclerosis (52). RORα knockout mice have displayed a similar cerebellar phenotype as the staggerer mice, demonstrating that the LBD of RORα is critical for its function, at least in the cerebellum. In the present study, the importance of the LBD of RORα in terms of its function during adipogenesis was examined (supplemental Fig. 6, Fig. 5G, and Fig. 6D), but the LBD of RORα was found to be dispensable for the inhibitory effect of RORα on adipogenesis. On the other hand, Lau et al. (53) have recently shown that homozygous staggerer mice are characterized by reduced adiposity and are resistant to diet-induced obesity. It was demonstrated that these phenotypes were mainly due to a decrease in plasma cholesterol, triglycerides, and nonesterified fatty acids, resulting from down-regulation of hepatic expression of SREBP1c, and also the reverse cholesterol transporters ABCA1 and ABCG5 (53). Although expression of these genes including SREBP1c in adipose tissues were not shown in that report, we observed only little change in expression of SREBP1c and its target genes (FAS, ACC1, and ACC2) in white adipose tissues among wild-type, homozygous, and heterozygous staggerer mice (supplemental Fig. 10A). Moreover, primary preadipocytes isolated from adipose tissues of staggerer homozygous and heterozygous mice were differentiated to the same degree (supplemental Fig. 10B). These results by our hands suggest that the staggerer dysfunction of RORα (RORαsg) neither reduces adiposity in a direct manner nor affects adipocyte differentiation by itself. To analyze the precise function of RORα in adipose tissues, it would be of great help to develop conditional RORα knockout mice that lack RORα only in adipose tissues.
Recent studies have revealed that obesity induces hypoxia, endoplasmic reticulum stress and an inflammatory reaction, resulting in insulin resistance and dysregulated secretion of adipocytokines (54, 55, 56, 57). Moreover, the expression of RORα has been shown to increase in various cell types in response to hypoxia (49, 58). In the current study, we found that RORα overexpression modulates the expression of antiinflammatory or proinflammatory adipocytokines such as adiponectin, macrophage chemoattractant protein-1, and PAI-1 during adipocyte differentiation (Fig. 2, B and C). Obesity-induced dysregulation of RORα expression may be linked to metabolic diseases and thus have potential as a therapeutic target.
Materials and Methods
Reagents
DMEM, anti-Flag monoclonal antibody (M2), anti-Myc monoclonal antibody (9E10), and anti-β-actin monoclonal antibody (AC-15) were purchased from Sigma Chemical Co. (St. Louis, MO). Anti-BrdU monoclonal antibody (11170376001) was from Roche (Indianapolis, IN). Insulin, MIX, DEX, troglitazone, anti-perilipin polyclonal antibody, and anti-CHOP antiserum were obtained as described previously (27, 59). Anti-RORα polyclonal antibody (H-65), anti-PPARγ polyclonal antibody (H-100), and anti-C/EBPβ (C-19) were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-phospho-C/EBPβ Thr235 (3084), anti-p44/42 ERK polyclonal antibody (9102), and anti-phospho-ERK (Thr202/Tyr204) polyclonal antibody (9101) were from Cell Signaling Technology (Beverly, MA).
Cell culture
Mouse 3T3-L1 preadipocyte cells (obtained from American Type Culture Collection, Manassas, VA) were maintained in DMEM containing 10% calf serum. For the differentiation experiment, the medium was replaced with DMEM containing 10% fetal bovine serum (FBS), 10 μg/ml insulin, 0.5 mm MIX, and 1 μm DEX at 2 d after confluence. After 2 d, the medium was changed to DMEM containing 5 μg/ml insulin and 10% FBS, and then the cells were refed every 2 d. Human 293T embryonic kidney cells were cultured as described previously (27).
Construction of expression plasmids
The lentiviral plasmids pCSII-EF-3Flag-RORα1 and pCSII-EF-3Flag-RORα4 were constructed by inserting a fragment coding for 3Flag-tagged mouse RORα1 and RORα4 into pCSII-EF-MCS-IRES2-Venus (RIKEN, Saitama, Japan), respectively. The lentiviral plasmids for shRNA of mouse RORα or control were constructed by recombining pCS-RfA-EG (RIKEN) with pENTR4-H1 (RIKEN) inserted by oligonucleotide DNA for shRNA expression. The target sequences are as follows: RORα, 5′-GCGCGCTTTGTAGGATTCG-3′ (60), and control (Scramble II Duplex from Dharmacon, Lafayette, CO), 5′-GCGCGCTTTGTAGGATTCG-3′. The plasmids pCMV-LAP and pCMV-LIP were kindly provided by Dr. U. Schibler (Geneva University, Geneva, Switzerland) (64). pCMV-Flag-PPARγ, pCSII-EF-Flag-PPARγ, pCMV-Myc-RXRα, pCMV-Myc-C/EBPβ, pPerilipin-Luc −2.0 kb wt and pPerilipin-Luc −1.9 kb wt were constructed as described previously (15, 27, 62). pCSII-EF-C/EBPβ (LAP) was constructed by ligating rat C/EBPβ cDNA from pCMV-LAP (61). To obtain p(C/EBP)4-Luc, hybridized DNA for four tandem repeats of the C/EBP response element from the mouse C/EBPα promoter were ligated with the pGL3 promoter (Promega, Madison, WI). pCMV-3Flag-RORα1, pCMV-3Flag-RORα1 ΔLBD lacking aa 236–523 (51), pCMV-3Flag-ROR α4, pCMV-3Flag-Rev-erbα, and pPerilipin-Luc −2.0 kb mutants were generated by PCR. All constructs were verified by sequencing.
Lentivirus infection
293T cells were transfected with a lentiviral expression plasmid together with a packaging (pCAG-HIVgp) and a VSV-G- and Rev-expressing plasmid (pCMV-VSV-G-RSV-Rev) by the Chen-Okayama method (28). After 12 h, cells were additionally cultured with a fresh medium containing 10 μm forskolin for 24 h. The medium containing lentiviruses was collected and filtered. 3T3-L1 cells were infected with a medium supplemented with 10 μg/ml polybrene by a centrifugation method (2500 rpm for 90 min). Then the cells were refed with a fresh culture medium (DMEM with 10% calf serum).
RNA extraction, RT-PCR, and quantitative real-time RT-PCR
Total cellular RNA was extracted with an RNeasy Mini Kit with an on-column deoxyribonuclease digestion (QIAGEN, Valencia, CA). RNA was reverse transcribed with High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Foster City, CA). Quantitative real-time RT-PCR was performed on an ABI PRISM 7000 sequence detection system using TaqMan universal PCR master mix (Applied Biosystems) or SYBR green PCR master mix (Applied Biosystems) with the primers as follow: Rev-erbα, 5′-CGTTCGCATCAATCGCAACC-3′ (sense) and 5′-GATGTGGAGTAGGTGAGGTC-3′ (antisense), and PAI-1, 5′-AACCCGGCGGCAGATC-3′ (sense) and 5′-CTTGAGATAGGACAGTGCTT-3′ (antisense). Each experiment was performed in triplicate. TaqMan probes/primers were used for mouse RORα, C/EBPα, C/EBPβ, C/EBPδ, C/EBP homologous protein (CHOP), PPARγ, SREBP1, aP2, perilipin, fatty acid synthase (FAS), stearoyl CoA desaturase-1 (SCD-1), acetyl CoA carboxylase1 (ACC1), ACC2, adiponectin, tribbles homolog 3 (TRB3), and 18S (Applied Biosystems).
Immunoprecipitation and Western blot analysis
Cells were transiently transfected and treated as described in the figure legends. The cells were lysed in RIPA buffer [50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 0.1% SDS, 0.5% deoxycholate, and 1% Triton X-100] supplemented with protease inhibitors. The lysates were subjected to immunoprecipitation, and 1–2% of the lysate or co-immunoprecipitate was subjected to SDS-PAGE (8–12.5%), transferred onto polyvinylidene difluoride membrane and probed with antibodies indicated in the figure legends. The immunoreactive proteins were visualized using ECL (GE Healthcare, Piscataway, NJ) or Immobilon (Millipore, Bedford, MA) Western blotting detection reagents, and light emission was quantified with a LAS1000 lumino image analyzer (Fuji, Tokyo, Japan).
Promoter pull-down assay
By using PCR with a biotinylated primer and pGL3-basic, p(C/EBP)4-Luc, pPlin-Luc −2.0 wt, or pPlin-Luc −2.0 kb mutant 1, the promoter was biotinylated at the 5′-end of the lower strand. The cells were lysed in RIPA buffer supplemented with protease inhibitors and 0.2 mg/ml polydeoxyinosinic-deoxycytidylic acid and incubated with a 5 nm biotinylated promoter fragment for 2 h. Fifty milligrams of streptavidin-conjugated magnetic particles (Promega, Madison, WI) were then added and incubated for an additional 1 h. The particles were washed four times, and proteins were eluted with sample buffer and subjected to Western blot analysis.
ChIP assay
ChIP assay was performed using the ChIP assay kit (Upstate Biotechnology Inc., Lake Placid, NY) and the QIAquick PCR purification kit (QIAGEN) according to the manufacturer’s instructions. 293T cells were transfected with expression plasmids by Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA). Soluble chromatin prepared from the cells was immunoprecipitated with the indicated antibodies. PCR was performed with the following primers: perilipin promoter, 5′-CAGGTAGGTACGGAGAAGGAAAGA-3′ (sense) and 5′-AACATAGGGCTTTGTGCCTGAAAG-3′ (antisense), and PPARγ2 promoter; 5′-CCCCTCACAAGACACTGAACATGT-3′ (sense) and 5′-CACCCATGAGTCAAGACATCAGTT-3′ (antisense).
Reporter gene assays
3T3-L1 preadipocytes were transfected with luciferase reporter and expression plasmids by Lipofectamine 2000 reagent (Invitrogen). After 48 h, lysates were prepared and luciferase assays were performed as described previously (27).
Oil red O staining
3T3-L1 adipocytes were fixed with 4% paraformaldehyde in PBS and stained with Oil red O as described previously (27).
BrdU labeling
The 3T3-L1 cells at 24 h after induction of differentiation were labeled for 3 h with 10 μm BrdU. Cells were washed with PBS and fixed in 70% ethanol for 10 min. After denaturation (2 n HCl for 20 min) and neutralization (0.1 m sodium borate, pH 8.5), cells were incubated with anti-BrdU monoclonal antibody and subsequently probed with Cy3-conjugated secondary antibody containing 4′,6-diamidino-2-phenylindole. Two independent experiments were performed. To examine the percentage of BrdU-positive cells, six fields (more than 1000 cells) were counted.
Isolation of primary preadipocytes
Fibroblastic preadipocytes were isolated from stromal-vascular fraction of sc fat pads of 3- to 5-wk-old male mice. The sc fat pads from mice were removed and minced in DMEM containing collagenase (type II; 1 mg/ml) after washing with PBS. After digestion by collagenase (type II; 1 mg/ml) at 37 C for 1 h in a shaking water bath, the digest was filtered through a 100-μm cell strainer. After 10 min centrifugation at 1200 rpm, the mature adipocytes were located on the top white layer and the stromal-vascular fraction containing the preadipocytes was in the pellet. The pellet was resuspended and washed in DMEM with 10% FBS three times. After the pellet was passed through a 25-gauge needle, the cells were plated and cultured for 24 h. The cells were stringently washed with sprinkling medium to remove the cells that did not stick properly and were grown to confluence in DMEM with 10% FBS. The differentiation into adipocytes was stimulated as well as 3T3-L1 cells.
Acknowledgments
We thank Dr. U. Schibler for providing plasmids. We are grateful to Dr. Kevin Boru of Pacific Edit for review of the manuscript.
NURSA Molecule Pages:
Coregulators: p300;
Nuclear Receptors: PPARγ | REV-ERBα | RORα.
Footnotes
This work was supported by research grants from the Ministry of Education, Science, Sports, and Culture of Japan, the Program for the Promotion of Basic Research Activities for Innovative Biosciences, and the Noda Institute for Scientific Research.
Disclosure Summary: The authors have nothing to disclose.
First Published Online March 26, 2009
Abbreviations: aa, Amino acids; BrdU, 5-bromo-2′-deoxyuridine; C/EBP, CCAAT/enhancer-binding protein; ChIP, chromatin immunoprecipitation; DEX, dexamethasone; FBS, fetal bovine serum; LAP, liver-enriched transcriptional activating protein; LBD, ligand-binding domain; LIP, liver-enriched transcriptional inhibitory protein; MCE, mitotic clonal expansion; MIX, 3-isobutyl-1-methylxanthine; PAI-1, plasminogen activator inhibitor type 1; PPARγ, peroxisome proliferators-activated receptor γ; PPRE, PPARγ-responsive element; RORα, retinoic acid receptor-related orphan receptor α; ROR, responsive element; shRNA, short hairpin RNA; SREBP, sterol regulatory element-binding protein.
References
- 1.Spiegelman BM, Flier JS2001. Obesity and the regulation of energy balance. Cell 104:531–543 [DOI] [PubMed] [Google Scholar]
- 2.Farmer SR2006. Transcriptional control of adipocyte formation. Cell Metab 4:263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosen ED, MacDougald OA2006. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 7:885–896 [DOI] [PubMed] [Google Scholar]
- 4.Green H, Kehinde O1979. Formation of normally differentiated subcutaneous fat pads by an established preadipose cell line. J Cell Physiol 101:169–171 [DOI] [PubMed] [Google Scholar]
- 5.Green H, Meuth M1974. An established pre-adipose cell line and its differentiation in culture. Cell 3:127–133 [DOI] [PubMed] [Google Scholar]
- 6.Darlington GJ, Ross SE, MacDougald OA1998. The role of C/EBP genes in adipocyte differentiation. J Biol Chem 273:30057–30060 [DOI] [PubMed] [Google Scholar]
- 7.Tontonoz P, Hu E, Spiegelman BM1994. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell 79:1147–1156 [DOI] [PubMed] [Google Scholar]
- 8.Rubin CS, Hirsch A, Fung C, Rosen OM1978. Development of hormone receptors and hormonal responsiveness in vitro. Insulin receptors and insulin sensitivity in the preadipocyte and adipocyte forms of 3T3-L1 cells. J Biol Chem 253:7570–7578 [PubMed] [Google Scholar]
- 9.Cao Z, Umek RM, McKnight SL1991. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev 5:1538–1552 [DOI] [PubMed] [Google Scholar]
- 10.Green H, Kehinde O1975. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell 5:19–27 [DOI] [PubMed] [Google Scholar]
- 11.Wiper-Bergeron N, Wu D, Pope L, Schild-Poulter C, HacheéRJ2003. Stimulation of preadipocyte differentiation by steroid through targeting of an HDAC1 complex. EMBO J 22:2135–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, McKeon C, Darlington GJ, Spiegelman BM1999. Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell 3:151–158 [DOI] [PubMed] [Google Scholar]
- 13.Freytag SO, Paielli DL, Gilbert JD1994. Ectopic expression of the CCAAT/enhancer-binding protein α promotes the adipogenic program in a variety of mouse fibroblastic cells. Genes Dev 8:1654–1663 [DOI] [PubMed] [Google Scholar]
- 14.Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Spiegelman BM, Mortensen RM1999. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell 4:611–617 [DOI] [PubMed] [Google Scholar]
- 15.Arimura N, Horiba T, Imagawa M, Shimizu M, Sato R2004. The peroxisome proliferator-activated receptor γ regulates expression of the perilipin gene in adipocytes. J Biol Chem 279:10070–10076 [DOI] [PubMed] [Google Scholar]
- 16.Rosen ED, Hsu CH, Wang X, Sakai S, Freeman MW, Gonzalez FJ, Spiegelman BM2002. C/EBPα induces adipogenesis through PPARγ: a unified pathway. Genes Dev 16:22–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JB, Sarraf P, Wright M, Yao KM, Mueller E, Solanes G, Lowell BB, Spiegelman BM1998. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J Clin Invest 101:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fajas L, Schoonjans K, Gelman L, Kim JB, Najib J, Martin G, Fruchart JC, Briggs M, Spiegelman BM, Auwerx J1999. Regulation of peroxisome proliferator-activated receptor γ expression by adipocyte differentiation and determination factor 1/sterol regulatory element binding protein 1: implications for adipocyte differentiation and metabolism. Mol Cell Biol 19:5495–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JB, Wright HM, Wright M, Spiegelman BM1998. ADD1/SREBP1 activates PPARγ through the production of endogenous ligand. Proc Natl Acad Sci USA 95:4333–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeh WC, Cao Z, Classon M, McKnight SL1995. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev 9:168–181 [DOI] [PubMed] [Google Scholar]
- 21.Tanaka T, Yoshida N, Kishimoto T, Akira S1997. Defective adipocyte differentiation in mice lacking the C/EBPβ and/or C/EBPδ gene. EMBO J 16:7432–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang QQ, Otto TC, Lane MD2003. CCAAT/enhancer-binding protein β is required for mitotic clonal expansion during adipogenesis. Proc Natl Acad Sci USA 100:850–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacDougald OA, Mandrup S2002. Adipogenesis: forces that tip the scales. Trends Endocrinol Metab 13:5–11 [DOI] [PubMed] [Google Scholar]
- 24.Pedersen SB, Bruun JM, Hube F, Kristensen K, Hauner H, Richelsen B2001. Demonstration of estrogen receptor subtypes α and β in human adipose tissue: influences of adipose cell differentiation and fat depot localization. Mol Cell Endocrinol 182:27–37 [DOI] [PubMed] [Google Scholar]
- 25.Boukhtouche F, Mariani J, Tedgui A2004. The “CholesteROR” protective pathway in the vascular system. Arterioscler Thromb Vasc Biol 24:637–643 [DOI] [PubMed] [Google Scholar]
- 26.Dzhagalov I, Zhang N, He YW2004. The roles of orphan nuclear receptors in the development and function of the immune system. Cell Mol Immunol 1:401–407 [PubMed] [Google Scholar]
- 27.Takahashi Y, Ohoka N, Hayashi H, Sato R2008. TRB3 suppresses adipocyte differentiation by negatively regulating PPARγ transcriptional activity. J Lipid Res 49:880–892 [DOI] [PubMed] [Google Scholar]
- 28.Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H2005. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J 24:1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyata Y, Fukuhara A, Matsuda M, Komuro R, Shimomura I2008. Insulin induces chaperone and CHOP gene expressions in adipocytes. Biochem Biophys Res Commun 365:826–832 [DOI] [PubMed] [Google Scholar]
- 30.Bezy O, Vernochet C, Gesta S, Farmer SR, Kahn CR2007. TRB3 blocks adipocyte differentiation through the inhibition of C/EBPβ transcriptional activity. Mol Cell Biol 27:6818–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding J, Kato S, Du K2008. PI3K activates negative and positive signals to regulate TRB3 expression in hepatic cells. Exp Cell Res 314:1566–1574 [DOI] [PubMed] [Google Scholar]
- 32.Baer M, Johnson PF2000. Generation of truncated C/EBPβ isoforms by in vitro proteolysis. J Biol Chem 275:26582–26590 [DOI] [PubMed] [Google Scholar]
- 33.Welm AL, Timchenko NA, Darlington GJ1999. C/EBPα regulates generation of C/EBPβ isoforms through activation of specific proteolytic cleavage. Mol Cell Biol 19:1695–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang QQ, Grønborg M, Huang H, Kim JW, Otto TC, Pandey A, Lane MD2005. Sequential phosphorylation of CCAAT enhancer-binding protein β by MAPK and glycogen synthase kinase 3β is required for adipogenesis. Proc Natl Acad Sci USA 102:9766–9771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuo Y, Qiang L, Farmer SR2006. Activation of CCAAT/enhancer-binding protein (C/EBP) α expression by C/EBPβ during adipogenesis requires a peroxisome proliferator-activated receptor-γ-associated repression of HDAC1 at the C/EBPα gene promoter. J Biol Chem 281:7960–7967 [DOI] [PubMed] [Google Scholar]
- 36.Giguère V, Tini M, Flock G, Ong ES, Evans RM, Otulakowski G1994. Isoform-specific amino-terminal domains dictate DNA-binding properties of RORα, a novel family of orphan hormone nuclear receptors. Genes Dev 8:538–553 [DOI] [PubMed] [Google Scholar]
- 37.Giguère V, McBroom LD, Flock G1995. Determinants of target gene specificity for RORα1: monomeric DNA binding by an orphan nuclear receptor. Mol Cell Biol 15:2517–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Botas J, Anderson JB, Tessier D, Lapillonne A, Chang BH, Quast MJ, Gorenstein D, Chen KH, Chan L2000. Absence of perilipin results in leanness and reverses obesity in Lepr(db/db) mice. Nat Genet 26:474–479 [DOI] [PubMed] [Google Scholar]
- 39.Brasaemle DL, Rubin B, Harten IA, Gruia-Gray J, Kimmel AR, Londos C2000. Perilipin A increases triacylglycerol storage by decreasing the rate of triacylglycerol hydrolysis. J Biol Chem 275:38486–38493 [DOI] [PubMed] [Google Scholar]
- 40.Tansey JT, Sztalryd C, Gruia-Gray J, Roush DL, Zee JV, Gavrilova O, Reitman ML, Deng CX, Li C, Kimmel AR, Londos C2001. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc Natl Acad Sci USA 98:6494–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naiki T, Saijou E, Miyaoka Y, Sekine K, Miyajima A2007. TRB2, a mouse Tribbles ortholog, suppresses adipocyte differentiation by inhibiting AKT and C/EBPβ. J Biol Chem 282:24075–24082 [DOI] [PubMed] [Google Scholar]
- 42.Cui TX, Piwien-Pilipuk G, Huo JS, Kaplani J, Kowk R, Schwartz J2005. Endogenous CCAAT/enhancer binding protein β and p300 are both regulated by growth hormone to mediate transcriptional activation. Mol Endocrinol 19:2175–2186 [DOI] [PubMed] [Google Scholar]
- 43.Wiper-Bergeron N, Salem HA, Tomlinson JJ, Wu D, Haché RJ2007. Glucocorticoid-stimulated preadipocyte differentiation is mediated through acetylation of C/EBPβ by GCN5. Proc Natl Acad Sci USA 104:2703–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forman BM, Chen J, Blumberg B, Kliewer SA, Henshaw R, Ong ES, Evans RM1994. Cross-talk among RORα1 and the Rev-erb family of orphan nuclear receptors. Mol Endocrinol 8:1253–1261 [DOI] [PubMed] [Google Scholar]
- 45.Delerive P, Chin WW, Suen CS2002. Identification of Reverbα as a novel RORα target gene. J Biol Chem 277:35013–35018 [DOI] [PubMed] [Google Scholar]
- 46.Raspè E, Mautino G, Duval C, Fontaine C, Duez H, Barbier O, Monte D, Fruchart J, Fruchart J, Staels B2002. Transcriptional regulation of human Rev-erbα gene expression by the orphan nuclear receptor retinoic acid-related orphan receptor α. J Biol Chem 277:49275–49281 [DOI] [PubMed] [Google Scholar]
- 47.Fontaine C, Dubois G, Duguay Y, Helledie T, Vu-Dac N, Gervois P, Soncin F, Mandrup S, Fruchart JC, Fruchar-Najib J, Staels B2003. The orphan nuclear receptor Rev-Erbα is a peroxisome proliferator-activated receptor (PPAR) γ target gene and promotes PPARγ-induced adipocyte differentiation. J Biol Chem 278:37672–37680 [DOI] [PubMed] [Google Scholar]
- 48.Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M, Wada T, Aoyagi T, Tezuka M2005. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci USA 102:12071–12706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miki N, Ikuta M, Matsui T2004. Hypoxia-induced activation of the retinoic acid receptor-related orphan receptor α4 gene by an interaction between hypoxia-inducible factor-1 and Sp1. J Biol Chem 279:15025–15031 [DOI] [PubMed] [Google Scholar]
- 50.Boukhtouche F, Doulazmi M, Frederic F, Dusart I, Brugg B, Mariani J2006. RORα, a pivotal nuclear receptor for Purkinje neuron survival and differentiation: from development to ageing. Cerebellum 5:97–104 [DOI] [PubMed] [Google Scholar]
- 51.Lau P, Bailey P, Dowhan DH, Muscat GE1999. Exogenous expression of a dominant negative RORα1 vector in muscle cells impairs differentiation: RORα1 directly interacts with p300 and myoD. Nucleic Acids Res 27:411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jarvis CI, Staels B, Brugg B, Lemaigre-Dubreuil Y, Tedgui A, Mariani J2002. Age-related phenotypes in the staggerer mouse expand the RORα nuclear receptor’s role beyond the cerebellum. Mol Cell Endocrinol 186:1–5 [DOI] [PubMed] [Google Scholar]
- 53.Lau P, Fitzsimmons RL, Raichur S, Wang SC, Lechtken A, Muscat GE2008. The orphan nuclear receptor, RORα, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J Biol Chem 283:18411–18421 [DOI] [PubMed] [Google Scholar]
- 54.Hosogai N, Fukuhara A, Oshima K, Miyata Y, Tanaka S, Segawa K, Furukawa S, Tochino Y, Komuro R, Matsuda M, Shimomura I2007. Adipose tissue hypoxia in obesity and its impact on adipocytokine dysregulation. Diabetes 56:901–911 [DOI] [PubMed] [Google Scholar]
- 55.Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Görgün C, Glimcher LH, Hotamisligil GS2004. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 306:457–461 [DOI] [PubMed] [Google Scholar]
- 56.Lau DC, Dhillon B, Yan H, Szmitko PE, Verma S2005. Adipokines: molecular links between obesity and atherosclerosis. Am J Physiol Heart Circ Physiol 288:H2031–H2041 [DOI] [PubMed]
- 57.Guzik TJ, Mangalat D, Korbut R2006. Adipocytokines: novel link between inflammation and vascular function? J Physiol Pharmacol 57:505–528 [PubMed] [Google Scholar]
- 58.Chauvet C, Bois-Joyeux B, Berra E, Pouyssegur J, Danan JL2004. The gene encoding human retinoic acid-receptor-related orphan receptor α is a target for hypoxia-inducible factor 1. Biochem J 384:79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohoka N, Hattori T, Kitagawa M, Onozaki K, Hayashi H2007. Critical and functional regulation of CHOP (C/EBP homologous protein) through the N-terminal portion. J Biol Chem 282:35687–35694 [DOI] [PubMed] [Google Scholar]
- 60.Akashi M, Takumi T2005. The orphan nuclear receptor RORα regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol 12:441–448 [DOI] [PubMed] [Google Scholar]
- 61.Descombes P, Schibler U1991. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell 67:569–579 [DOI] [PubMed] [Google Scholar]
- 62.Hattori T, Ohoka N, Inoue Y, Hayashi H, Onozaki K2003. C/EBP family transcription factors are degraded by the proteasome but stabilized by forming dimer. Oncogene 22:1273–1280 [DOI] [PubMed] [Google Scholar]