Abstract
A deranged differentiation is often a landmark of transformed cells. We used a thyroid cell line expressing an inducible Ras oncoprotein in order to study the hierarchy of molecular events leading to suppression of thyroid-specific gene expression. We find that, upon Ras activation, there is an immediate global down-regulation of thyroid differentiation, which is associated with an inhibition of the cAMP signaling pathway. We demonstrate that an unusual negative cross talk between Ras oncogene and the cAMP pathway induces inactivation of the transcription factor Pax8 that we propose as a crucial event in Ras-induced dedifferentiation.
Oncogenic Ras induces dedifferentiation of cultured thyroid cells via an impairment of the cAMP/PKA pathway and a consequent inhibition of the transcription factor Pax8.
Ras proteins play important roles in growth and differentiation of several cell types. However, when deregulated by certain mutations, Ras proteins constitutively activate diverse downstream pathways and can elicit both tumoral transformation and a deranged differentiation phenotype (1, 2). Recent results obtained in animal models indicate cell type-specific mechanisms involved in oncogenic Ras action and suggest that molecular events initiated by Ras activation could either be different or have divergent consequences in diverse cell types (3, 4, 5). Oncogenic Ras proteins have been shown to play important roles in epithelial cell transformation and are indeed associated with 35% of human cancers (1). In particular, mutations on genes encoding Ras proteins have been associated with all types of thyroid malignancies, including anaplastic thyroid cancers, thus suggesting that Ras oncoproteins might have a role in the suppression of thyroid-differentiated phenotype (6). Consistently, experiments performed in immortalized thyroid cell lines showed that transformation by Ras oncogenes inhibits differentiation through still unknown mechanisms (7, 8, 9).
The thyroid-differentiated phenotype is characterized by the expression of a number of proteins that are necessary for biosynthesis of thyroid hormones, such as thyroglobulin (TG), thyroperoxidase (TPO), and the sodium/iodide symporter (NIS). Thyroid-specific gene transcription relies on the coordinated action of a set of transcription factors (Nkx2.1/TITF1, FoxE1/TITF2, Pax8), among which the transcription factor Pax8 appears to play very relevant roles (10, 11). Pax8 transcription factor belongs to the family of paired box (Pax) genes, which consists of tissue-specific transcriptional regulators, essential for normal embryogenesis (12). Loss of function mice models demonstrated that PAX8 is required for the morphogenesis of the thyroid gland (13). Experiments in thyroid cell lines have shown that PAX8 is involved in the maintenance of thyrocyte cell type and is essential for the thyrocyte-specific promoter activation of the FoxE1/TITF2, TPO, TG, and NIS genes (11, 14, 15, 16, 17, 18).
Another well-established regulator of adult thyroid function is the glycoprotein hormone TSH that, through its receptor (TSHr), conveys inside the thyroid cells an elevation of the second messenger cAMP, thus stimulating thyroid growth and function. Gain-of-function mutations of the TSHr gene are associated with congenital hyperthyroidism whereas, on the contrary, TSHr loss-of-function mutations are associated with congenital hypothyroidism (19). Experiments in thyroid cell lines have shown that TSH can regulate, albeit to different degrees, the expression of mRNA of several thyroid-specific genes such as Tg (20, 21, 22), TPO (23, 24), NIS (22, 25), Foxe1/TITF2 (26) and Pax8 (22, 27). Loss-of-function mice models show that TSH/TSHr function is necessary, but not sufficient, for NIS and TPO gene expression (28).
The ability of Ras oncoprotein to negatively regulate differentiation in thyroid cells is of utmost importance in therapy. Cancerous thyroid cells can be killed with radioactive iodide, because they have an exquisite capacity, mediated by the NIS protein, to accumulate this element that is physiologically used for thyroid hormone biosynthesis (29). However, for anaplastic cancer, the most aggressive and inevitably fatal thyroid cancer, the radioactive iodide-based therapy is ineffective because the expression of NIS gene is suppressed in these cells. Investigation of the action exerted by activated Ras on differentiation is of interest to design new, highly required, therapeutic strategies aimed at reexpression of the differentiated phenotype in cancer cells (30, 31).
The availability of cultured, differentiated thyroid cell lines offers an amenable system with which to study the action of activated Ras proteins on the differentiated phenotype of an epithelial cell type. The rat epithelial thyroid cell line FRTL-5 (32), for example, retains in culture the expression of all known thyroid differentiation markers. We have previously shown that FRTL-5-derived stable cell lines expressing H-RasV12 at high levels completely lose their differentiation (33). We also demonstrated that loss of differentiation is an early event induced by Ras oncogene activation, and thus it is not the result of chronic exposure to the activated oncogene (33).
In this study, we used the FRTL-5/ER-RasV12 cell system (33). In these cells, oncogenic Ras can be activated by treatment with tamoxifen (4OHT). We could thus analyze the kinetics of Ras oncogene action on thyroid-specific gene expression to define a molecular hierarchy of events. We demonstrate that Ras oncoprotein inactivates Pax8 transcriptional activity early on in the transformation process and induces inhibition of the TSHr pathway through a double mechanism that involves both down-regulation of TSHr expression itself plus an additional interference with the TSHr signaling, the latter located downstream of cAMP production. We also demonstrate that cAMP pathway inhibition is the cause of Pax8 inactivation and that by restoring a functional cAMP pathway we can restore both Pax8 activity and thyroid-specific gene expression.
Results
Ras activation induces a rapid down-regulation of thyroid-specific gene expression
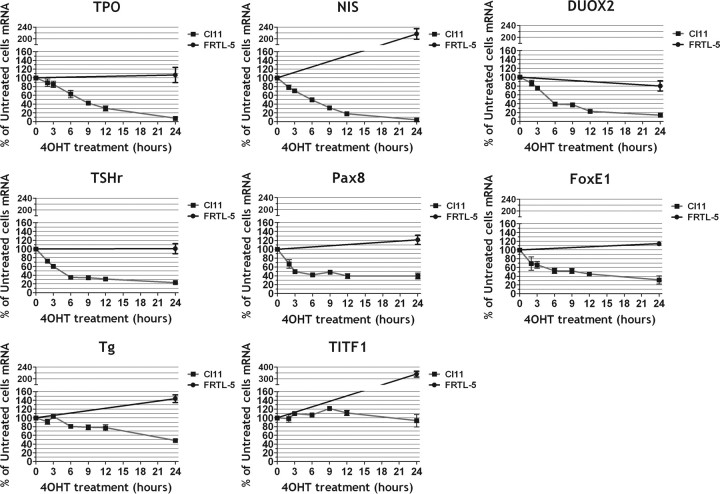
We analyzed, by real-time-PCR, the kinetics of Ras-induced down-regulation of thyroid-specific gene expression in a rat thyroid cell line, called Cl11, expressing an inducible H-RasV12 oncoprotein (ER-RasV12) (33). Shortly after ER-RasV12 activation by 4OHT there is a simultaneous decrease of the mRNAs encoding several thyroid differentiation markers, except for the Tg and Nkx2-1/Titf1 mRNAs that show, at least in the time window used, a small or no decrease, respectively (Fig. 1). The simultaneous down-regulation of several differentiation markers suggests that Ras oncogene might interfere with a regulatory target common to many thyroid-enriched genes. No inhibition of gene expression was detected when wild-type cells were treated with 4OHT, thus demonstrating the role of oncogenic Ras in the observed loss of differentiation.
Fig. 1.
Kinetic of Ras oncogene-induced dedifferentiaton of FRTL-5 cells. Cl11 cells were treated with 4OHT for increasing times as indicated. For each time point mRNA was extracted from triplicate plates of Cl11 or FRTL-5 cell lines and analyzed by real-time RT-PCR for each of the genes indicated.
NIS down-regulation is achieved through a Ras oncogene-mediated impairment of NUE (NIS upstream enhancer) transcriptional activity
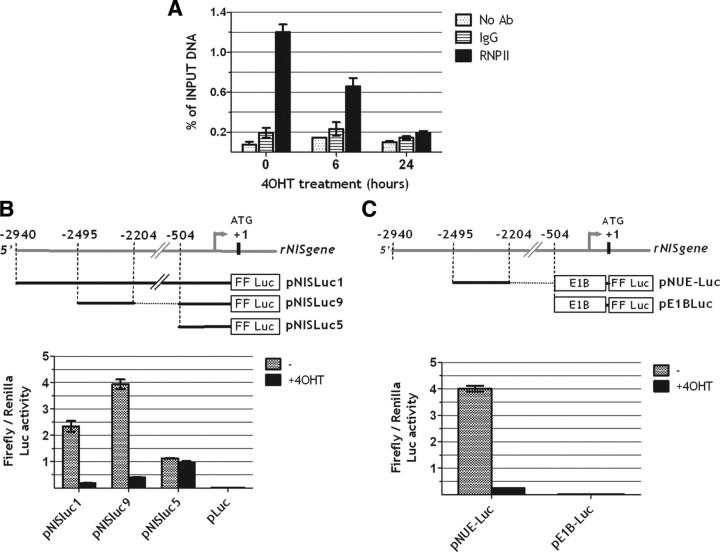
To approach the mechanisms responsible for the oncogenic Ras-induced down-regulation of thyroid-specific gene expression, we focused our attention on the gene encoding the sodium-iodide symporter (NIS), the regulatory elements of which have been extensively studied. First, to test whether the decrease in NIS mRNA levels resulted from decreased transcription, we measured, by chromatin immunoprecipitation (chromatin IP), RNA polymerase II occupancy of the NIS gene coding sequence in Cl11 cells after activation of Ras oncoprotein. This assay shows that the amount of RNA polymerase II bound to the NIS gene is already reduced 6 h after Ras oncogene activation and almost completely abolished at 24 h. These results are consistent with the observed mRNA decrease and indicate that Ras activation rapidly inhibits NIS gene transcription (Fig. 2A).
Fig. 2.
Oncogenic Ras down-regulates NIS transcription by inactivating NUE (NIS upstream enhancer). A, Cl11 cells were treated with 4OHT for the indicated times (0, 6 h, 24 h). For each time point NIS transcription was measured by performing a chromatin-IP with RNA polymerase II Antibody (RNPII) as described in Materials and Methods. For each time point a chromatin-IP was performed also, as negative controls, with nonspecific rabbit IgG (IgG) or in the absence of antibody (no Ab). Immunoprecipitated DNA was measured by real-time PCR and is reported as percentage of input DNA for each time point. B and C, Transient transfection assays of firefly luciferase reporter (FF luc) constructs containing NIS-regulatory regions in Cl11 cells treated or not with 4OHT. pLuc in panel B is the empty vector.
The NIS gene-regulatory elements affected by Ras oncoprotein were investigated using previously described chimeric constructs (15) in which either the entire upstream region of the rNIS gene or deletion derivatives are fused to the LUC reporter gene (Fig. 2B). Each construct was transiently transfected into Cl11 either in the presence or absence of 4OHT. The 2.9-kb DNA fragment from the rNIS regulatory region (pNIS-Luc1) is significantly down-regulated by Ras activation. The same extent of inhibition is also seen with the pNIS-Luc9 construct in which the sequence located between positions −2495 and −2264, corresponding to the NUE (15) is fused to the proximal NIS promoter (between positions −564 and +1). In contrast, no Ras-induced inhibition is observed on the activity of the proximal NIS promoter (pNIS-Luc5). We conclude that Ras oncoprotein reduces NIS expression mainly through inhibition of NUE activity.
To further assess the ability of oncogenic Ras to inhibit NUE activity independently from other surrounding NIS gene sequences, we cloned the NUE sequence upstream of the E1B TATA box in pGL3-basic and tested its transcriptional activity in Cl11 either in the presence or absence of 4OHT (Fig. 2C). Again, a severe impairment of transcriptional activity is observed after Ras activation.
Taken together, these data demonstrate that Ras oncogenic activity inhibits NIS gene expression at the transcriptional level and that such inhibition is mediated, at least in part, by interference with the stimulatory activity of the NUE-regulatory element.
Activation of Ras reduces both the amounts of Pax8 protein and its transcriptional activity
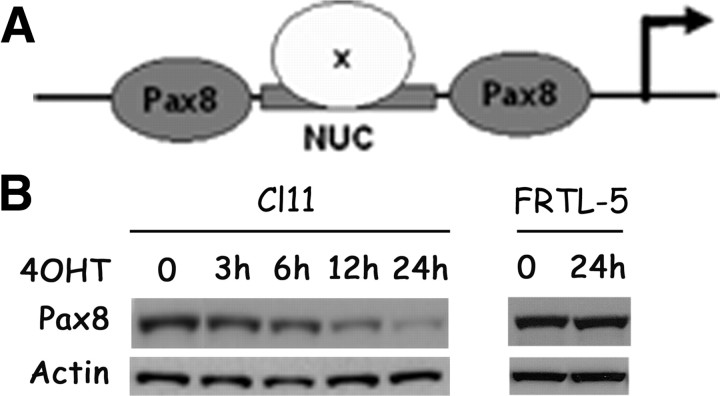
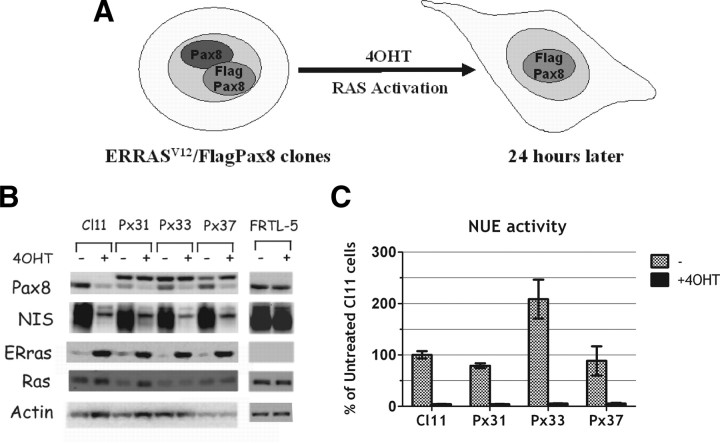
The NIS enhancer NUE contains at least three functionally relevant protein-binding sites: a central cAMP-response element-like element called NUC surrounded by two Pax8 binding sites (Ref. 15 and Fig. 3A). The two Pax8-binding sites have been shown to be necessary for NUE activity (34). In Cl11 cells, Ras activation induces an early and significant down-regulation of Pax8 protein levels (Fig. 3B). To analyze the role of Pax8 down-regulation in Ras oncoprotein-induced dedifferentiation, we stably transfected into Cl11 cells a Pax8 expression vector that encodes a flagged Pax-8 protein (Flag-Pax8) under the control of a Ras-independent promoter. We selected three stable clones (Px31, Px33, and Px37 in Fig. 4) in which the Pax8 protein levels are unaffected by Ras activation, because the transfected, flagged protein maintains its expression whereas the endogenous protein is down-regulated (Fig. 4, A and B). However, irrespective of Pax8 protein levels, expression of the endogenous NIS gene was still down-regulated by Ras activation as in the parental cell line (Fig. 4B). Consistently, also the activity of a transfected NUE enhancer was down-regulated by oncogenic Ras, both in the presence and in the absence of the Pax8 protein (compare results in Cl.11 with those obtained in cell lines Px31, Px33, and Px37 in Fig. 4C). These results suggest that the Ras oncoprotein interferes with transcriptional activity of Pax8, in addition to the observed negative effect on Pax8 protein levels. To further test this hypothesis, we used a reporter construct driving the expression of Luciferase under the control of an artificial promoter made by a pentamer of Pax8-binding sites [Cp5 (33)] located upstream of the E1B TATA box (Fig. 5A). Cp5 is stimulated by both wild-type and flagged-Pax8 in nonthyroid cells, demonstrating that the addition of the flag does not have any adverse effect on Pax8 transcriptional activity (Fig. 5B). However, data in Fig. 5C show that Cp5 activity is equally down-regulated by the activated Ras both in the absence (Cl11) and in the presence of Pax8 (Px31, Px33, Px37). Furthermore, given that there is no interference by the activation of Ras on the DNA-binding activity of Pax8 (Fig. 5D), we conclude that the inhibition of NIS expression by oncogenic Ras is mediated by an interference with Pax8 transcriptional activity. Such an inhibition presumably precedes the observed decrease in Pax8 protein levels. No decrease of either Pax8 levels or transcriptional activity were ever observed in wild-type FRTL-5 cells treated with 4OHT, demonstrating that both are elicited by oncogenic Ras (data not shown).
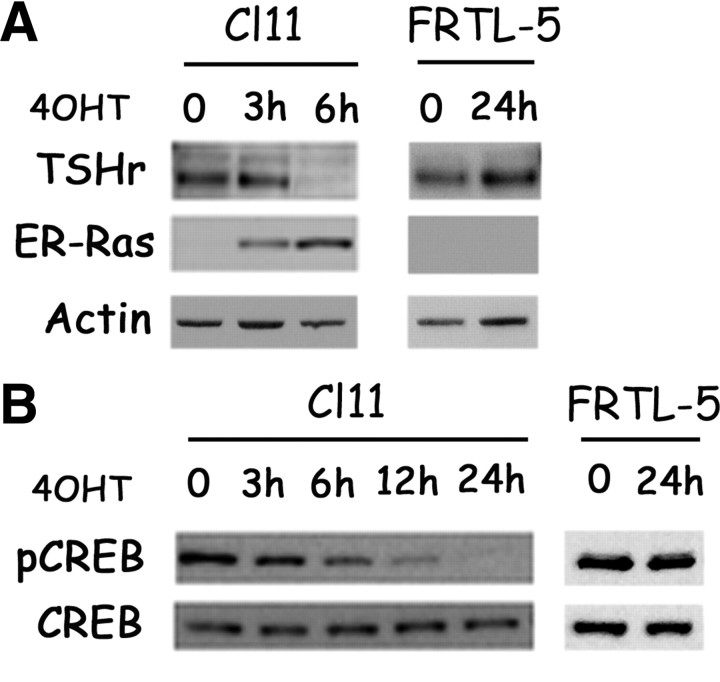
Fig. 3.
Kinetic of Ras oncogene-induced Pax8 protein down-regulation. A, Schematic representation of NIS enhancer NUE. B, Western blot analysis of Pax8 protein levels in Cl11 and FRTL-5 cells treated with 4OHT for the reported increasing time. Levels of actin are used to normalize for protein loading.
Fig. 4.
Ras oncogene down-regulates NIS expression and NUE activity even if Pax8 protein is expressed to wild-type levels. A, Schematic representation of Flag-Pax8 stable clones (Px clones), which keep the expression of the ectopic Flag-Pax8 even after ER-RasV12 activation. B, Cl11, three representative Flag-Pax8 clones (Px31, Px33, and Px37) and FRTL-5 were treated 24 h with or without 4OHT and analyzed by Western blot for several genes as indicated. In Px clones the ectopic Flag-Pax8 is identified by the band migrating above the endogenous Pax8 protein visible in Cl11. C, Transient transfection assay of NUE reporter activity in Cl11 cells and in Px clones (Px31, 33, and 37) with or without 4OHT. NUE activity (normalized for transfection efficiency) is expressed as percent of activity obtained in untreated Cl11 cells.
Fig. 5.
Oncogenic Ras impairs Pax8 transactivation activity. A, Schematic representation of Cp5 artificial promoter. B, Transient transfection assay of Cp5 reporter activity in Hela cells cotransfected, respectively, with an empty vector (Cp5), with 100 ng of Pax8 (+Pax8) or with 100 ng of the flagged Pax8 (+FlagPax8). C, Transient transfection assay of Cp5 reporter activity in Cl11- and Flag Pax8-expressing clones (Px31, Px33, and Px37) performed in the absence or presence of 4OHT. Cp5 reporter activity (normalized for transfection efficiency) is expressed as percentage of activity obtained in untreated Cl11 cells. D, Parallel bandshift analysis of nuclear Pax8 DNA-binding activity in Cl11 and Flag Pax8-expressing clones (Px31, Px33, and Px37). Lower panel, Reported binding of nuclear extract to the unrelated SP1 binding sequence.
Role of the TSH-receptor and of cAMP levels in oncogenic Ras-induced dedifferentiation
Pax8 transcriptional activity has been suggested to be dependent upon TSH stimulation of the cAMP pathway (22, 35). TSHr mRNA is rapidly reduced after Ras activation (Fig. 1). Such a decrease is immediately reflected in a reduction in TSHr protein level (Fig. 6A) and in a functional impairment of the cAMP pathway, as demonstrated by a significant decrease in the levels of phosphorylated cAMP response element-binding protein (CREB) (Fig. 6B). Again, none of these effects could be observed in FRTL-5 cells treated with 4OHT, thus demonstrating that they are truly a consequence of Ras activation (Fig. 6, A and B; FRTL-5). In addition, TSH has been previously established to be necessary for maintaining thyroid differentiation (36, 37) and, in particular, for NIS expression (28, 38). We thus hypothesized that inhibition of the TSHr/cAMP pathway could be a crucial event in Ras oncoprotein-induced NIS down-regulation.
Fig. 6.
Oncogenic Ras early inhibits TSHr pathway. A and B, Cl11 and FRTL-5 cells were treated with 4OHT for the reported time and analyzed by Western blot. A, TSHr expression analysis; ERRAS shows accumulation of the inducible oncoprotein. Actin is used to normalize for protein loading. B, Total CREB and phospho-CREB Ser133 (pCREB) analysis.
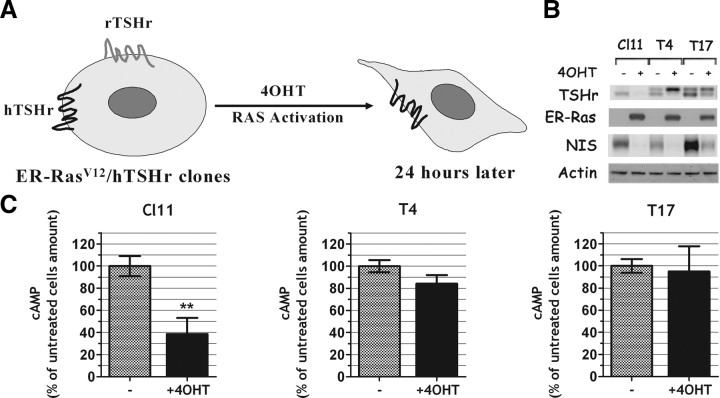
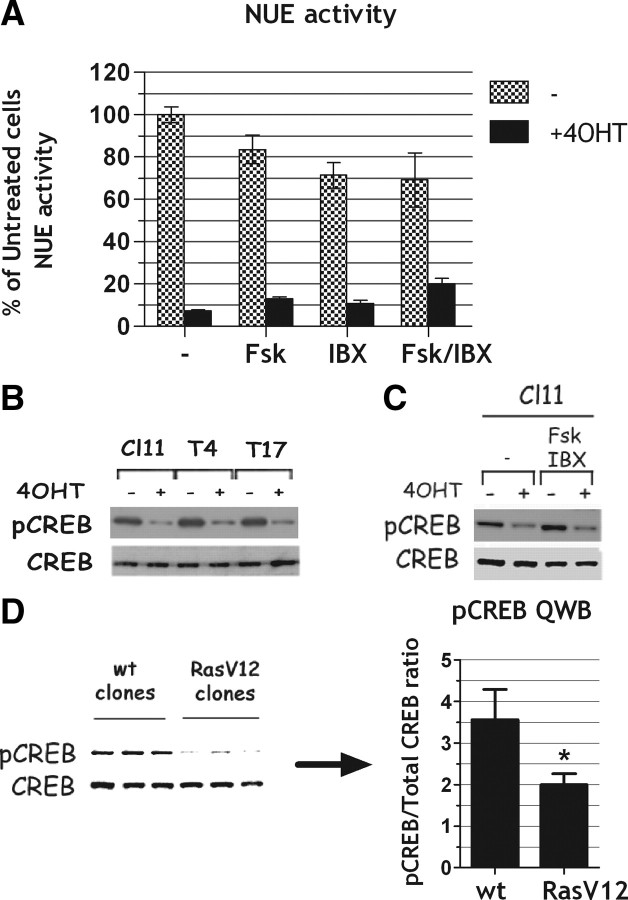
To test whether oncogenic Ras was down-regulating NIS through inhibition of TSHr expression, we used an approach similar to the one illustrated above for Pax8. Thus, we stably transfected into Cl11 an expression vector encoding the human TSHr (39) under the control of a Ras-independent promoter (Fig. 7A). In the stable hTSHr-expressing clones (T4 and T17 in Fig. 7B), expression of the ectopic TSHr is maintained after Ras activation. However, regardless of TSHr protein levels, the expression of the endogenous NIS gene was still down-regulated by Ras activation as in the parental cell line (Fig. 7B). We conclude that TSHr expression down-regulation is not the crucial event in oncogenic Ras-induced dedifferentiation of FRTL-5 cells. Measurements of intracellular cAMP demonstrate that whereas in the parental cell line Ras activation decreases cAMP levels, in clones T4 and T17 such a decrease is not observed. This evidence is in support of the persistent expression of a functional TSHr in clones T4 and T17 (Fig. 7C). In keeping with these observations, reagents capable of increasing cAMP levels, such as forskolin (FSK) and the phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine (IBX), were ineffective in contrasting the inhibitory effect of activated Ras on the endogenous NIS expression (data not shown) or on NUE-transcriptional activity (Fig. 8A). We conclude that Ras oncoprotein-induced NIS down-regulation is not due either to the decreased levels of TSH receptor or to a reduced production of cAMP, because restoring either of them does not prevent the negative effect exerted by activated Ras. Furthermore, also CREB phosphorylation is impaired by Ras activation in T4 and T17 clones, as in the parental cell line Cl11 (Fig. 8B) even though TSHr expression is maintained in T4 and T17 and greatly reduced in Cl11 (Fig. 7B). This observation suggests that oncogenic Ras acts downstream of TSHr. Given that FSK and IBX could not rescue CREB phosphorylation in the presence of activated Ras (Fig. 8C), we conclude that Ras oncogene inactivates the cAMP pathway through a mechanism downstream of those regulating intracellular cAMP levels. Interestingly, using cells stably transfected with a constitutively active Ras oncoprotein, we demonstrate that the observed down-regulation of phosphorylated CREB (pCREB) is a persistent effect of oncogenic Ras (Fig. 8D).
Fig. 7.
TSHr down-regulation is not the crucial event in Ras oncogene-induced dedifferentiation. A, Schematic representation of SPRT-TSHr-expressing clones, which keep the expression of the ectopic TSHr (hTSHr) even after ER-RasV12 activation. B and C, Cl11- and SPRT-TSHr-expressing clones were treated 24 h with or without 4OHT. B, Total proteins were analyzed by Western blot for the reported genes. In T4 and T17 clones the ectopic hTSHr is identified by the band migrating above the endogenous rTSHr visible in the untreated Cl11. C, cAMP was quantitatively measured in each clone before and after the 4OHT treatment. **, P < 0.01.
Fig. 8.
Ras oncogene impairs cAMP signaling by acting downstream of cAMP production. A, Transient transfection assay of NUE reporter activity in Cl11 cells treated or not with 4OHT either in the absence or in the presence of FSK and IBX or a combination of them for 48 h as reported in the figure. NUE activity is expressed as percentage of the activity obtained in untreated cells. B–D, Western blot analysis of CREB phosphorylation at Ser133 (pCREB). Total CREB (CREB) was used to normalize. B, Cl11 and SPRT-TSHr-expressing clones (T4 and T17) treated 24 h with or without 4OHT. C, Cl11 cells treated 24 h with or without 4OHT either in the presence or absence of FSK/IBX. D, QWB analysis of three wild-type FRTL-5 stable clones expressing only the resistance gene (wt clones) and three FRTL-5 stable clones expressing RasV12 (RasV12 clones). pCREB signal quantitation for three independent experiments is reported on the right side. *, P ≅ 0.03. wt, Wild type.
Oncogenic Ras interferes with protein kinase A (PKA) activity
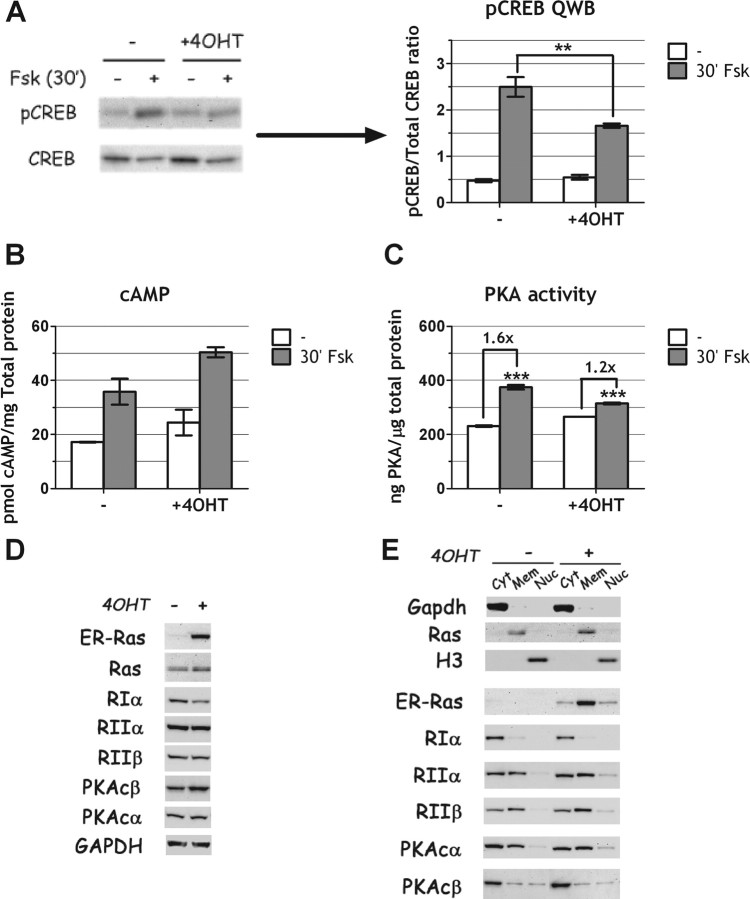
Ras oncoprotein in our cells inhibits CREB phosphorylation at Ser133. However, many signaling pathways and several different kinases converge on CREB Ser133 (40). Given the specific stimulation of PKA, via cAMP, by FSK, we tested whether oncogenic Ras interferes with FSK-induced CREB phosphorylation. To this end, we acutely stimulated Cl11 cells with FSK, in the absence or in the presence of oncogenic Ras and measured CREB phosphorylation by quantitative Western blot (QWB). The results of such an experiment show that Ras significantly reduces FSK-induced CREB phosphorylation (Fig. 9A) even though cAMP levels are equally stimulated both in the presence and in the absence of the oncoprotein (Fig. 9B), strongly suggesting that Ras interferes with PKA activity. In support of this conclusion, we show a reduced PKA activation by FSK in the presence of oncogenic Ras. Such a reduction is not due to alterations in levels or subcellular localization of PKA-regulatory and/or catalytic subunits (Fig. 9, D and E). It should be noted that in the experiments shown in Fig. 9, cells have been starved for TSH, to monitor their response to FSK. For this reason, the effects of oncogenic ras, in the absence of FSK, on CREB phosphorylation (panel A), cAMP (panel B), and PKA (panel C) cannot be observed.
Fig. 9.
Ras oncogene impairs PKA activity but not its expression or localization. A–C, Cl11 cells were starved 48 h in 4H medium (0.2% serum, no TSH, no insulin), cultured an additional 24 h in 4H medium either with or without 4OHT and then stimulated with FSK (10 μm) for 30 min. A, QWB analysis of CREB phosphorylation at Ser133 shows an impairment of CREB phosphorylation in the presence of ras oncogene. B, cAMP measurement shows equal production of cAMP in both conditions. C, PKA activity assay shows reduced PKA activity in Ras oncogene-activated cells. D and E, Cl11 cells were treated 24 h with or without 4OHT in 6H medium. Catalytic (PKAcα, PKAcβ) and regulatory (RIα, RIIα, RIIβ) PKA subunit expression was analyzed by Western blot in total (D) and fractionated (E) cellular extracts. ERRas was also measured to verify Ras oncogene activation by 4OHT. D, Unfractionated extracts. E, Fractionated extracts (Cyt, Cytosolic protein fraction; Mem, membrane/organelle protein fraction; Nuc, nuclear protein fraction). GAPDH (cytosolic), endogenous Ras (membranes), and histone H3 (nuclear) were used to monitor efficacy of the fractionating procedure. **, P < 0.01; ***, P ≅ 0.002.
We conclude that oncogenic Ras in thyroid cells interferes with PKA activity. The mechanism of such interference remains to be elucidated.
The reduction of Pax8 activity by Ras oncogene occurs through inhibition of the cAMP pathway
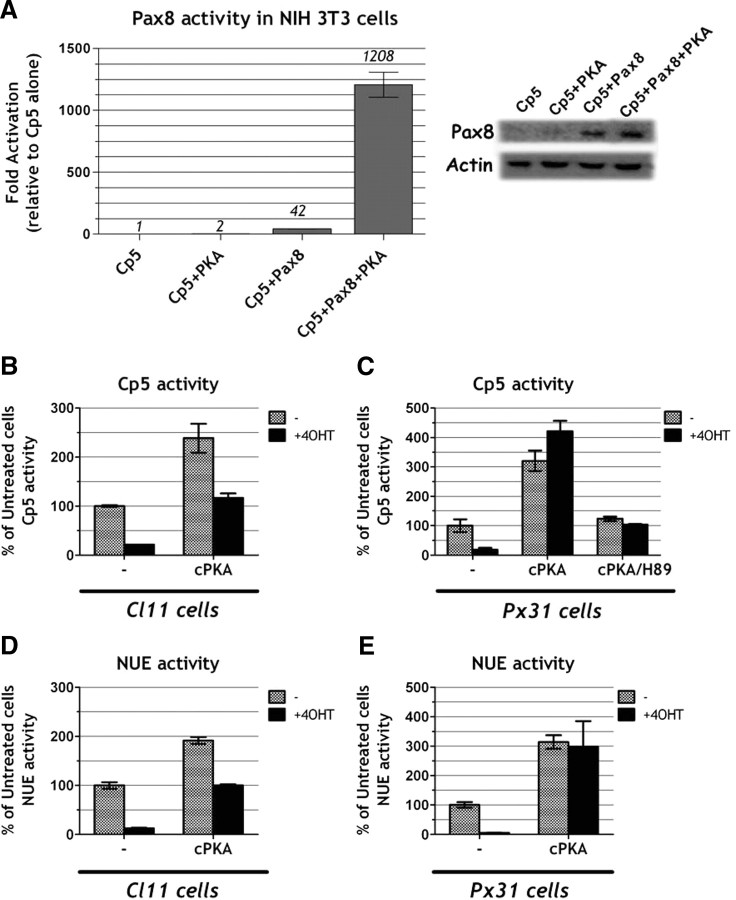
Given that oncogenic Ras negatively interferes with PKA, we wondered whether such interference could explain the observed reduction in Pax8-dependent transcription. First, to test if PKA regulates Pax8 activity, we measured the stimulation of the Cp5 reporter by Pax8 in the NIH 3T3 cell line either in the presence or absence of a PKA catalytic subunit α (cPKA) expression vector. We found, indeed, that Pax8 transcriptional activity is strongly enhanced (∼30-fold) by PKA overexpression, without a significant effect on the levels of Pax8 protein (Fig. 10A). Next, we tested whether PKA could rescue the inhibition exerted by oncogenic Ras on Pax8-dependent NIS transcription. Even though we have shown that in our cells Ras does not impact on the amount of intracellular PKA (Fig. 9D), we reasoned that PKA overexpression might counteract Ras inhibition, whatever the nature of the inhibitory mechanism is. We therefore measured the effect of cPKA overexpression on Ras oncogene-induced inhibition of Cp5 transcription. These experiments were carried out in both Cl11 and Px31 cells because these two cell lines express, after Ras activation, very little (Cl11) or normal levels (Px31) of the Pax8 protein (Fig. 4B). We observed that cPKA greatly stimulates Cp5 activity in the presence of activated Ras, and such an effect is even more pronounced in Px31 cells, in which the levels of Pax8 are close to normal (Fig. 10, B and C). As predicted, the PKA-specific inhibitor H89 completely blocks the observed stimulation of Cp5 transcription, thus demonstrating that the observed effect is PKA dependent (Fig. 10C). Finally, we could demonstrate that also the activity of NUE is rescued by transfection of PKA, again with a better efficiency in Px31 (Fig. 10E) than in Cl11 (Fig. 10D) cells.
Fig. 10.
PKA activity restores oncogenic Ras-mediated impairment of NUE, and Pax8 activity PKA regulates Pax8 activity. A, Transient transfection assay of Cp5 reporter activity in NIH 3T3 cells cotransfected, respectively, with an empty vector (Cp5), with 1 μg of cPKA (Cp5+PKA), with 250 ng of Pax8 (Cp5+Pax8), or with 250 ng of Pax8 and 1 μg of cPKA (Cp5+Pax8+PKA). Western blot analysis of Pax8 expression in the lysates is also shown. B–E, Transient transfection assay of Cp5 (B and C) and NUE (D and E) reporter activity in Cl11 (little Pax8 expression after 4OHT treatment) (B and D) or Px31 cells (Pax8 expressed as wild-type levels after 4OHT treatment) (C and E). Reporter activity was tested with or without 4OHT in the absence of further treatment (−) or with 1 μg of cPKA cotransfecting vector (cPKA) as indicated. The activity of NUE and Cp5 is reported as percentage of activity obtained, respectively, in untreated cells. B, Transient transfection assay of Cp5 reporter activity in Cl11 cells. C, Transient transfection assay of Cp5 reporter activity in Px31 cells. cPKA effects were tested also in the presence (48 h) of PKA inhibitor H-89 12 μm (cPKA/H-89) as indicated. D, Transient transfection assay of NUE reporter activity in Cl11 cells. E, Transient transfection assay of NUE reporter activity in Px31 cells.
Taken together, these data strongly suggest that the negative interference exerted by oncogenic Ras on PKA causes a reduction of Pax8 transcriptional activity that, in turn, provokes a diminished transcription of NIS. It remains to be ascertained what mechanism Ras uses to reduce Pax8 protein levels. Recent data (Di Gennaro, A., M. De Felice, and R. Di Lauro, unpublished results) suggesting that transcription of the Pax8 is autoregulated, might provide an unifying view of these events, and would highlight the block of PKA as the highest in the hierarchy of events leading to Ras-induced dedifferentiation of thyroid cells in culture.
Discussion
In this study, we demonstrate that activation of Ras oncogene in thyroid cells causes loss of activity of the transcription factor Pax8 that can be rescued by overexpression of the PKA catalytic subunit. In keeping with this observation, we show a reduced PKA activity and a significant decrease of phosphorylated CREB in cells expressing oncogenic Ras. These data indicate novel biochemical actions of Ras oncogene that might play important roles in the Ras-induced transformed phenotype.
We have previously demonstrated that such inhibition of differentiation is not an artifact of the chimeric Ras molecule used because stable transfectants expressing oncogenic Ras show a similar phenotype (33). The global down-regulation of thyroid-specific gene expression greatly resembles the effect of TSH starvation on thyroid follicular cells (22, 28, 41). The observations reported here show that among the earliest events after Ras activation are a decrease of TSHr protein and of cAMP signaling, as indicated by a reduced CREB phosphorylation, suggesting that the effects of Ras could be mediated by reduced TSHr signaling.
We tested such a hypothesis by generating a stable cell line that expressed an ectopic TSHr (hTSHr) under the control of a promoter not influenced by Ras activation. We demonstrate that, in these cells, TSHr expression, even though effectively rescued in the presence of activated Ras, is not sufficient to restore a differentiated phenotype. In addition, in hTSHr-expressing cells, oncogenic Ras still impairs CREB phosphorylation, and such an effect cannot be rescued by the cAMP-elevating agent FSK. Furthermore, we demonstrate a clear reduction of PKA activity in cells expressing oncogenic Ras. We conclude that activated Ras inhibits the TSHr pathway at two major levels, one of which is the down-regulation of TSHr expression and a further level that is located downstream of cAMP production and results in reduction of PKA activity, with a consequent block in the transduction of the cAMP signal to the nucleus.
This latter effect by oncogenic Ras indicates the presence of an unusual inhibitory interference exerted by oncogenic Ras on the cAMP-signaling pathway. Although it is well established that cAMP signaling can interfere with Ras action (40, 42), the data presented in this paper represent a strong indication of a block exerted by Ras on cAMP signaling. Our data support the notion that the Ras oncoprotein inactivates the cAMP pathway through a mechanism that is acting downstream of cAMP production and converges on PKA. We propose that in thyroid cells these events lead to the observed global down-regulation of thyroid differentiation.
Previous data on dedifferentiated thyroid-transformed cell lines such as PCpY cells (rat thyroid cell line transformed with polyoma virus middle T antigen) and ARO cells (human anaplastic thyroid carcinoma-derived cell line) have shown that reactivation of Pax8 expression can restore a differentiated thyroid phenotype and, in particular, can up-regulate NIS expression (11, 43).
We thus tested whether Pax8 down-regulation had a key role in oncogenic Ras-induced NIS down-regulation, by generating a stable cell line expressing an ectopic Pax8 under the control of a Ras-independent promoter. However, persistent Pax8 expression does not affect the ability of oncogenic Ras to down-regulate NIS expression. We provide evidence that Ras activation inhibits Pax8 transcriptional activity and that such inhibition is achieved through the inhibition of the TSHr pathway. In fact, we found that Pax8 activity is dependent upon the TSHr/PKA pathway as previously suggested (22, 35) and that PKA overexpression rescues oncogenic Ras-induced Pax8 inhibition. Consistently, we found that PKA overexpression also restores the ability of Pax8 to stimulate NIS transcription mediated by the NUE enhancer.
We thus envision a mechanism in which the initial event after Ras activation is a block of PKA activity. As a consequence, Pax8 activity is impaired, resulting in a decrease of thyroid-specific gene expression. Interestingly, Pax8 transcription itself is under cAMP control (22). Furthermore, preliminary evidence suggests that Pax8 transcription is auto-regulated (Di Gennaro, A., M. De Felice, and R. Di Lauro, unpublished). Thus, the reduced cAMP signaling induced by oncogenic Ras would interfere both with the activity and synthesis of Pax8, resulting in an amplification of the dedifferentiating effect.
Published data support the notion that in thyroid cells oncogenic Ras inhibits cAMP signaling. However, the mechanisms proposed envisage both a delocalization of the PKA catalytic subunit and a down-regulation of the PKA-regulative subunit RIIβ (44, 45, 46, 47). In our system we did not detect either of these effects, because we found no effects of oncogenic Ras on either the intracellular distribution or protein levels for both regulatory and catalytic PKA subunits. However, we demonstrate, for the first time, a clear Ras-induced down-regulation of CREB phosphorylation at Ser133 concomitant with a decreased PKA activity.
Even though CREB Ser133 is the target not only of PKA but also of many other kinases (40, 48), we clearly show that oncogenic Ras blocks FSK-induced CREB phosphorylation in thyroid cells, indicating that PKA is a bona fide target of Ras action. Along the same lines, we show that the stimulation of Pax8 activity by PKA overexpression, in cells expressing oncogenic Ras, is blocked by the PKA-specific inhibitor H89.
We have not identified the mechanism responsible for PKA inhibition by Ras. Because this effect is relieved by PKA overexpression, we hypothesize that we might be titrating out an inhibitory mechanism, the nature of which is at present unknown. Previous data have shown, for example, that up-regulation of PP1 phosphatase activity can down-regulate thyroid differentiation (51). However, further studies are required to define the mechanisms underpinning this Ras oncogene-promoted PKA inhibition.
It should also be noted that, in addition to a potential general inhibition of cAMP pathway induced by Ras oncogene and the consequential effect on Pax8 activity, CREB-compromised function itself could be relevant to thyroid function. It has been reported that CREB activity is required for differentiation of FRTL-5 cells (34, 52) and that mice expressing a dominant-negative CREB transgene show impairment in thyroid differentiation (53).
The understanding of the events causative of loss of thyroid differentiation induced by Ras oncogene in cultured cells might give clues on the genes that could be targeted in human anaplastic thyroid cancers to restore a differentiated phenotype that will allow the ablation of cancerous cells through the well-established therapy protocol based on radioactive iodide.
Materials and Methods
Expression and reporter constructs
The pCEFL-3xFlagPax8 and pCMV-cPKA expression vectors have been previously described (17, 34). The SpRT TSHr coding sequence (Rho-tagged human TSHr) (39) was excised by cleavage with KpnI and XbaI from the original pcDNAIII vector, blunt ended, and cloned into BamH1-blunted sites of pBABEpuro vector. Reporter vectors pNIS-Luc1, NIS-Luc9, and pNIS-Luc5 were previously reported (1). The pE1B-luc construct was obtained by cloning a chemical synthesized oligonucleotide (5′-ctcgagtctagagggtatataatggatcc-3′) containing the E1B TATA box flanked by XhoI site at 5′ and BamH1 site at 3′ into an XhoI/BamH1-cleaved pGL3 basic luciferase reporter vector (Promega Corp., Madison, WI). pNUE-Luc (NUE reporter) construct was obtained by excising with KpnI and XhoI the NUE sequence from the previously reported pNISTKluc3 construct (15) and cloning it into KpnI/XhoI sites of pE1B-luc construct. The pCp5-E1B-Luc (Cp5 reporter) construct was obtained by excising with PvuII and BamHI the Cp5E1b cassette from the previously reported Cp5CAT vector (33) and cloning it into SmaI/BglII-cleaved pGL3 basic luciferase reporter vector (Promega Corp.). The TK-Renilla reporter vector was purchased from Promega Corp. (phRL-TK).
Cell culture
Rat thyroid follicular FRTL-5-derived cell lines were maintained in Coon’s modified F12 medium (EuroClone, Milano, Italy) supplemented with 5% newborn bovine serum (HyClone Laboratories, Logan, UT) and six growth factors (6H), including bovine TSH, 1 mU/ml (Sigma- Aldrich, St. Louis, MO), and insulin, 10 μg/ml (Sigma-Aldrich), as previously described (6H medium) (32). 4OHT treatment, where indicated, was performed by addition of 100 nm 4OHT (Sigma-Aldrich) to the culture medium. Starvation, where reported in the text, was obtained by culturing cells 72 h in 4H medium (0.2% serum, no TSH, no insulin) as previously described (15). Stimulation of the cAMP pathway, where indicated, was performed by adding to the culture medium, respectively, 10 μm FSK (Sigma-Aldrich) to stimulate adenylyl cyclase, and 100 μm IBX (Sigma-Aldrich) to inhibit phosphodiesterases or a combination of them as indicated in the text. Inhibition of PKA activity was obtained by adding to the culture medium 12 μm H-89 (BIOMOL Research Laboratories, Inc., Plymouth Meeting, PA).
Hela cells and NIH 3T3 mouse fibroblasts were grown as previously described (14, 54).
Transfections
All transfections were carried out by the use of FuGene 6 (Roche Molecular Biochemicals, Indianapolis, IN) following the manufacturer’s instructions. For stable transfection experiments, 2 × 106 cells were seeded on 100-mm dishes 24 h before transfection and transfected with 4 μg/dish of the indicated expression vector. Transfected cells were selected 48 h later in the presence of 1 μg/ml of puromycin (Sigma-Aldrich). After 3 wk of continuous selection, single clones were picked, screened for expression of the transgene, and amplified individually.
For transient transfections, 4 × 105 cells were seeded on 60-mm dishes 24 h before transfection. Transfections were performed with 3 μg/dish of total DNA consisting of 1 μg of reporter vector encoding firefly luciferase, 0.5 μg of TK-Renilla vector to follow transfection efficiency, cotransfecting vectors, or empty vector up to 3 μg. Transfection medium was replaced 15 h later with standard culture medium, supplemented or not with additional drugs as indicated in the text, and cells were cultured for additional 48 h.
Cells were lysed in 100 μl/dish PLB buffer 1× (Promega Corp.), and firefly and Renilla luciferase activity were assayed, respectively, on 20 μl of each sample with the Luciferase Assay System (Promega Corp.) and the Renilla Assay System (Promega) following manufacturer’s instructions. Luminescence was measured with LUMAT LB 9507 luminometer (Berthold Technologies, Bad Wildbad, Germany). Firefly luciferase activity was normalized on the activity of TK-Renilla vector to correct each sample for transfection efficiency. Data were obtained from at least two independent experiments with triplicate samples.
Western blotting
Cells (2 ×106) were seeded on 60-mm dishes and cultured 48 h with or without additional drugs for the time indicated in the text. Proteins were extracted in lysis buffer [50 mm Tris (pH 8), 5 m MgCl2, 150 mm NaCl, 1% Triton, 0.1% sodium dodecyl sulfate (SDS), 0.5% deoxycholic acid] supplemented with 10 mm NaF, 1 mm Na3VO4, 1 mm phenylmethylsulfonylfluoride, and protease inhibitor cocktail (Sigma-Aldrich). Protein concentration was measured by the BCA protein assay reagent (Pierce Chemical Co., Rockford, IL), following the manufacturer’s instructions. Fractionated protein extraction was performed using the Proteoextract Subcellulare proteome extraction kit (Calbiochem) and following manufacturer’s instruction. For Western blot analysis 1/20 of each fraction volume was loaded on gel. Western blots were performed as previously described (55). Rabbit polyclonal antibodies against Pax8 and NIS were previously produced in our laboratory and were used at 0.5 μg/ml and 0.2 μg/ml, respectively (7). Mouse monoclonal antibody against Rho-Tag and mouse monoclonal antibody against TSHr (mAb 103) were previously described (39) and were used both at 1:50 diluition. CREB (48H2) rabbit mAb, phospho-CREB (Ser133) mouse mAb, PKA C-α (Cell Signaling Technologies, Beverly, MA), PKAβ cat(C-20) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), PKA RIIβ, PKA RIIα, PKA RIα (BD Transduction Laboratories, Lexington, KY), histone H3 antibody-ChiP grade (ab 1791; Abcam, Inc., Cambridge, MA), GAPDH (clone 6C5) (ImmunoChemical), and RAS (clone Ras10) (Upstate Biotechnology, Lake Placid, NY) antibodies were used as suggested by the manufacturers. Immune complexes were detected by enhanced chemiluminescence as instructed by the manufacturer (Amersham Biosciences, Arlington Heights, IL). Quantitative analysis (QWB), where indicated, was performed on at least three independent experiments by capturing and analyzing chemiluminescence with Chemidoc XRS instrument (Bio-Rad Laboratories, Inc., Hercules, CA) supported by the Quantity One 4.6.5 software (Bio-Rad).
cAMP enzyme immunometric assay
Cells were seeded on 60-mm dishes and cultured either in 4H or 6H medium as indicated in the text for 72 h. After treatments described in the text, cells (∼2 × 106) were lysed in 1 ml of 0.1 m HCl/0.5% Triton. cAMP was quantitatively determined on 100 μl of cell lysate using the cAMP EIA kit (Stressgen Biotechnologies Corp., Victoria, British Columbia, Canada) following manufacturer’s instruction. Briefly, samples were added to wells containing antibodies to cAMP together with a cAMP conjugate to alkaline phosphatase that works as a cAMP competitor in binding the antibody. Alkaline phosphatase activity bound to the antibody is detected and considered inversionally proportional to the amount of cAMP in the sample. A standard curve of cAMP was run in the assay to determine cAMP (pmol/ml) in each sample. These values were then normalized on total protein present in each sample to obtain picomoles of cAMP produced per mg of protein.
PKA kinase activity assay
Cells (5 × 106) were seeded on 100-mm dishes and cultured 72 h in 4H medium. After treatments indicated in the text, cells were lysed in 100 μl of lysis buffer [50 mm Tris, 0.5% Nonidet P-40,150 mm NaCl, 1 mm Na3VO4, 50 mm NaF, 5 mm EGTA, 2 mm EDTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonylfluoride, and protease inhibitor cocktail (Sigma)]. PKA activity was assayed on 0.5 μg of extracted proteins using an ELISA-based commercially available assay kit [PKA kinase activity assay kit (non radioactive) from Stressgen] and following manufacturer’s instructions. Briefly, samples were incubated with ATP, 60 min at 30 C, in the substrate-coated microtiter plate. Phosphorylation of the peptide substrate is then detected through the use of a specific antibody. A standard curve was run in the assay to determine nanograms of active PKA in each sample. These values were then normalized on total protein present in each sample to obtain nanograms of PKA produced per μg of protein.
RNA extraction, and real-time RT-PCR
Cells (1.5 × 106) were seeded on 60-mm dishes and cultured 48 h with or without additional drugs for the time indicated in the text. Total RNA was isolated with RNeasy mini kit (QIAGEN, Chatsworth, CA) following manufacturer’s instructions. Total RNA from each cell line (4 μg) was used as a template for the synthesis of the first-strand cDNA, starting from random hexamers, using the Superscript II Reverse Transcriptase kit (Invitrogen Life Technologies, Carlsbad, CA) according to manufacturer’s instructions. Real-Time RT-PCR was conducted using an ABI Prism 7700 sequence detection system and SYBR Green chemistry (Applied Biosystems, Foster City, CA). Each reaction was carried out in triplicate, on duplicate biological samples, using cDNA obtained from 150 ng of total RNA per reaction as template. Specific primer pairs for each gene analyzed were previously described and used at 130 nm (33). Results were analyzed using α-1 tubulin mRNA (33) as reference gene. Analysis of results was performed following real-time relative-quantitation guidelines through the relative standard curve method as suggested by Applied Biosystems. Briefly, amplification efficiency was calculated from triplicate relative standard curves for each primer pair and then used to convert Ct values obtained from each reaction into relative-expression units (56). Data obtained in this way were then normalized on the relative expression of reference gene. Statistical analysis was performed on normalized relative-expression triplicate values of each experimental point.
Cross-linked chromatin preparation
Briefly 4 × 106 cells were seeded on 100-mm dishes and cultured 24 h with or without additional drugs for the time indicated in the text. At the end of the treatment, formaldehyde was added to the cells to final 1% for 10 min to cross-link the chromatin and the reaction was stopped by adding glycine to a final concentration of 0.125 m. Cells were washed twice with PBS and collected in cell lysis buffer (85 mm KCl; 0.5% Nonidet P-40; 5 mm HEPES, pH 8.0) (1.5 ml /dish) supplemented with a protease inhibitor cocktail (Sigma), homogenized by Dounce, incubated on ice for 15 min, and centrifuged at 3500 × g for 5 min to pellet the nuclei. The pellet was resuspended in nuclear lysis buffer (10 mm EDTA; 1% SDS; 50 mm Tris-HCl, pH 8.1) in a ratio 400 μl/107cells. Cross-linked chromatin aliquots were either stored at −80 C or processed directly.
Transcription rate measurement
Transcription rate measurement has been performed through an RNA-polymerase II-based chromatin-IP as previously described (57). Cross-linked chromatin (400 μl aliquots) was sonicated on ice with eight pulses of 30% amplitude in a BRANSON 250 sonicator. The average chromatin size of the fragments obtained was about 300 bp. The sonified chromatin was centrifuged at 14,000 × g for 10 min, and the supernatants, containing soluble chromatin fragments, were diluted 10-fold with dilution buffer (165 mm NaCl; 0.01% SDS; 1.1% Triton X-100; 1.2 mm EDTA; 16.7 mm Tris-HCl, pH 8.0) supplemented with protease inhibitor cocktail (Sigma-Aldrich). Diluted samples were precleared with 50 μl/ml of salmon sperm DNA/protein A agarose (Upstate) and left 1 h under rotation at 4 C. After centrifuging 1 min at 3000 × g at 4 C, aliquots from the supernatant of each sample (1 ml each) were incubated with 2.5 μg of RNA polymerase II antibody (Santa Cruz, sc-899), or as negative controls with 2.5 μg of normal rabbit IgG (Upstate) or in the absence of any antibody and allowed to remain overnight at 4 C under rotation. (An aliquot of the supernatant was stored at 4 C to evaluate INPUT DNA for each sample.) The samples were then incubated with 30 μl of salmon sperm DNA/protein A agarose (Upstate) under rotation for an additional period of 1 h. Immunocomplexes were then recovered and washed as suggested by Upstate. Elution was performed with 2 × 100 μl of elution buffer (1% SDS, 100 mm NaHSO3). Both immunoprecipitated chromatin and input chromatin were incubated at 65 C overnight to reverse formaldehyde cross-links. DNA purification was performed as suggested by Upstate and resuspended in 100 μl Tris-EDTA buffer. The entire chromatin-IP procedure has been repeated on independent Tween samples. All obtained DNA samples have been analyzed in triplicate by real-time PCR. Real-time PCR was performed and analyzed as described in the previous paragraph using 5 μl of DNA as template for each reaction. Primer pairs were designed with the Primer express software (Applied Biosystems) to amplify a region of 135 bp in length, corresponding to NIS coding region and located about 2 kb downstream of transcription start site (NisF 5′-cccctcaccctgtctaaccc-3′/NisR 5′-gctgaagagtgaccccagct-3′). For each sample, immunoprecipitated DNA levels have been reported as percent of INPUT DNA, and the average value has been calculated on the two independent chromatin IPs.
Gel mobility shift
Cells (4 × 106) were seeded on 100-mm dishes and cultured 48 h with or without additional drugs for the time indicated in the text. Nuclear extracts were prepared from FRTL-5 cells according to the previously described method (58). The Pax8 probe is the previously reported oligoCπ (5′-TCAGTCACGCGTGACTGGGCAGTG-3′) (18). The Sp1 probe (5′-ATTCGATCGGGGCGGGGCGAG-3′) used to check nuclear extract quality was previously reported (34). The chemically synthesized oligonucleotides were labeled with 32P using polynucleotide kinase and annealed to the antisense complementary sequences. The end-labeled double-strand oligonucleotide probes (80,000 cpm) were mixed with FRTL-5 cell extract (4 μg) in 20 μl of reaction buffer [reaction buffer: 20 mm Tris (pH 7.5), 75 mm KCl, 5 mm MgCl2, 1 mm dithiothreitol, 1 mm EDTA, 3 μg/20 μl poly(deoxyinosine-deoxycytosine), 1 mg/ml BSA, 10% glycerol] and incubated at room temperature for 30 min before loading on the 5% polyacrylamide gel in 0.5× TBE buffer at 200 V. After electrophoresis in a cold room, gels were dried and processed for autoradiography on Kodak (Rochester, NY) OMAX films for 24 h.
Acknowledgments
We thank Gilbert Vassart and Sabine Costagliola for kindly providing us the SpRT-TSHr expression vector and the relative antibodies against TSHr and Rho-Tag.
Footnotes
This work was supported by a grant of Associazione Italiana per la Ricerca sul Cancro (AIRC) to R.D.L. M.G.B. was supported by a fellowship from Federazione Italiana per la Ricerca sul Cancro (FIRC).
Disclosure Summary: The authors have nothing to disclose.
First Published Online March 12, 2009
Abbreviations: Chromatin-IP, Chromatin immunoprecipitation; Cl11, stable cell line derived from FRTL-5, expressing ER-RasV12; Cp5, artificial promoter containing 5 Pax8-binding sites; 8-CPT, cAMP analog 8-(4-chlorophenylthio)adenosine 3′,5′-cyclic monophosphate; CREB, cAMP-response element binding protein; DUOX2, thyroid oxidase; ER-RasV12, oncogenic human h-ras (G12V) fused to the ER ligand-binding domain; FSK, forskolin (stimulator of adenylyl cyclase); hTSHr, Rho-tagged hTSHr; IBX, 3-isobutyl-1-methylxanthine (inhibitor of phosphodiesterases); mAb, monoclonal antibody; NIS, sodium-iodide symporter; NUE, NIS upstream enhancer; 4OHT, tamoxifen; pCREB, phosphorylated CREB; PKA, protein kinase A; Px clones, Cl11-derived stable cell lines expressing Flag-Pax8; QWB, quantitative Western blot; RasV12 clones, stable cell lines derived from FRTL-5, expressing oncogenic human h-ras (G12V); SDS, sodium dodecyl sulfate; T clones, stable cell lines derived from Cl11, expressing hTSHr; TG, thyroglobulin; TPO, thyroperoxidase; TSHr, TSH receptor.
References
- 1.Karnoub AE, Weinberg RA2008. Ras oncogenes: split personalities. Nat Rev 9:517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crespo P, León J2000. Ras proteins in the control of the cell cycle and cell differentiation. Cell Mol Life Sci 57:1613–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guerra C, Mijimolle N, Dhawahir A, Dubus P, Barradas M, Serrano M, Campuzano V, Barbacid M2003. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell 4:111–120 [DOI] [PubMed] [Google Scholar]
- 4.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T2001. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 410:1111–1116 [DOI] [PubMed] [Google Scholar]
- 5.Sansom OJ, Meniel V, Wilkins JA, Cole AM, Oien KA, Marsh V, Jamieson TJ, Guerra C, Ashton GH, Barbacid M, Clarke AR2006. Loss of Apc allows phenotypic manifestation of the transforming properties of an endogenous K-ras oncogene in vivo. Proc Natl Acad Sci USA 103:14122–14127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kondo T, Ezzat S, Asa SL2006. Pathogenetic mechanisms in thyroid follicular-cell neoplasia. Nat Rev 6:292–306 [DOI] [PubMed] [Google Scholar]
- 7.Francis-Lang H, Zannini M, De Felice M, Berlingieri MT, Fusco A, Di Lauro R1992. Multiple mechanisms of interference between transformation and differentiation in thyroid cells. Mol Cell Biol 12:5793–5800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monaco C, Califano D, Chiappetta G, Mineo A, De Franciscis V, Vecchio G, Santelli G1995. Mutated human Kirsten ras, driven by a thyroglobulin promoter, induces a growth advantage and partially dedifferentiates rat thyroid epithelial cells in vitro. Int J Cancer 63:757–760 [DOI] [PubMed] [Google Scholar]
- 9.Kupperman E, Wofford D, Wen W, Meinkoth JL1996. Ras inhibits thyroglobulin expression but not cyclic adenosine monophosphate-mediated signaling in Wistar rat thyrocytes. Endocrinology 137:96–104 [DOI] [PubMed] [Google Scholar]
- 10.Damante G, Tell G, Di Lauro R2001. A unique combination of transcription factors controls differentiation of thyroid cells. Prog Nucleic Acid Res Mol Biol 66:307–356 [DOI] [PubMed] [Google Scholar]
- 11.Pasca di Magliano M, Di Lauro R, Zannini M2000. Pax8 has a key role in thyroid cell differentiation. Proc Natl Acad Sci USA 97:13144–13149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lang D, Powell SK, Plummer RS, Young KP, Ruggeri BA2007. PAX genes: roles in development, pathophysiology, and cancer. Biochem Pharmacol 73:1–14 [DOI] [PubMed] [Google Scholar]
- 13.Mansouri A1998. The role of Pax3 and Pax7 in development and cancer. Crit Rev Oncog 9:141–149 [DOI] [PubMed] [Google Scholar]
- 14.Zannini M, Francis-Lang H, Plachov D, Di Lauro R1992. Pax-8, a paired domain-containing protein, binds to a sequence overlapping the recognition site of a homeodomain and activates transcription from two thyroid-specific promoters. Mol Cell Biol 12:4230–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohno M, Zannini M, Levy O, Carrasco N, di Lauro R1999. The paired-domain transcription factor Pax8 binds to the upstream enhancer of the rat sodium/iodide symporter gene and participates in both thyroid-specific and cyclic-AMP-dependent transcription. Mol Cell Biol 19:2051–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parlato R, Rosica A, Rodriguez-Mallon A, Affuso A, Postiglione MP, Arra C, Mansouri A, Kimura S, Di Lauro R, De Felice M2004. An integrated regulatory network controlling survival and migration in thyroid organogenesis. Dev Biol 276:464–475 [DOI] [PubMed] [Google Scholar]
- 17.D'Andrea B, Iacone R, Di Palma T, Nitsch R, Baratta MG, Nitsch L, Di Lauro R, Zannini M2006. Functional inactivation of the transcription factor Pax8 through oligomerization chain reaction. Mol Endocrinol 20:1810–1824 [DOI] [PubMed] [Google Scholar]
- 18.Fabbro D, Pellizzari L, Mercuri F, Tell G, Damante G1998. Pax-8 protein levels regulate thyroglobulin gene expression. J Mol Endocrinol 21:347–354 [DOI] [PubMed] [Google Scholar]
- 19.Davies TF, Ando T, Lin RY, Tomer Y, Latif R2005. Thyrotropin receptor-associated diseases: from adenomata to Graves disease. J Clin Invest 115:1972–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avvedimento VE, Tramontano D, Ursini MV, Monticelli A, Di Lauro R1984. The level of thyroglobulin mRNA is regulated by TSH both in vitro and in vivo. Biochem Biophys Res Commun 122:472–477 [DOI] [PubMed] [Google Scholar]
- 21.Santisteban P, Kohn LD, Di Lauro R1987. Thyroglobulin gene expression is regulated by insulin and insulin-like growth factor I, as well as thyrotropin, in FRTL-5 thyroid cells. J Biol Chem 262:4048–4052 [PubMed] [Google Scholar]
- 22.Mascia A, Nitsch L, Di Lauro R, Zannini M2002. Hormonal control of the transcription factor Pax8 and its role in the regulation of thyroglobulin gene expression in thyroid cells. J Endocrinol 172:163–176 [DOI] [PubMed] [Google Scholar]
- 23.Chazenbalk G, Magnusson RP, Rapoport B1987. Thyrotropin stimulation of cultured thyroid cells increases steady state levels of the messenger ribonucleic acid for thyroid peroxidase. Mol Endocrinol 1:913–917 [DOI] [PubMed] [Google Scholar]
- 24.Zarrilli R, Formisano S, Di Jeso B1990. Hormonal regulation of thyroid peroxidase in normal and transformed rat thyroid cells. Mol Endocrinol 4:39–45 [DOI] [PubMed] [Google Scholar]
- 25.Levy O, Dai G, Riedel C, Ginter CS, Paul EM, Lebowitz AN, Carrasco N1997. Characterization of the thyroid Na+/I− symporter with an anti-COOH terminus antibody. Proc Natl Acad Sci USA 94:5568–5573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortiz L, Zannini M, Di Lauro R, Santisteban P1997. Transcriptional control of the forkhead thyroid transcription factor TTF-2 by thyrotropin, insulin, and insulin-like growth factor I. J Biol Chem 272:23334–23339 [DOI] [PubMed] [Google Scholar]
- 27.Van Renterghem P, Vassart G, Christophe D1996. Pax 8 expression in primary cultured dog thyrocyte is increased by cyclic AMP. Biochim Biophys Acta 1307:97–103 [DOI] [PubMed] [Google Scholar]
- 28.Postiglione MP, Parlato R, Rodriguez-Mallon A, Rosica A, Mithbaokar P, Maresca M, Marians RC, Davies TF, Zannini MS, De Felice M, Di Lauro R2002. Role of the thyroid-stimulating hormone receptor signaling in development and differentiation of the thyroid gland. Proc Natl Acad Sci USA 99:15462–15467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robbins RJ, Schlumberger MJ2005. The evolving role of (131)I for the treatment of differentiated thyroid carcinoma. J Nucl Med 46 (Suppl 1):28S–37S [PubMed]
- 30.Cornett WR, Sharma AK, Day TA, Richardson MS, Hoda RS, van Heerden JA, Fernandes JK2007. Anaplastic thyroid carcinoma: an overview. Curr Oncol Rep 9:152–158 [DOI] [PubMed] [Google Scholar]
- 31.Miccoli P, Materazzi G, Antonelli A, Panicucci E, Frustaci G, Berti P2007. New trends in the treatment of undifferentiated carcinomas of the thyroid. Langenbecks Arch Surg 392:397–404 [DOI] [PubMed] [Google Scholar]
- 32.Ambesi-Impiombato FS, Parks LA, Coon HG1980. Culture of hormone-dependent functional epithelial cells from rat thyroids. Proc Natl Acad Sci USA 77:3455–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Vita G, Bauer L, da Costa VM, De Felice M, Baratta MG, De Menna M, Di Lauro R2005. Dose-dependent inhibition of thyroid differentiation by RAS oncogenes. Mol Endocrinol 19:76–89 [DOI] [PubMed] [Google Scholar]
- 34.Chun JT, Di Dato V, D'Andrea B, Zannini M, Di Lauro R2004. The CRE-like element inside the 5′-upstream region of the rat sodium/iodide symporter gene interacts with diverse classes of b-Zip molecules that regulate transcriptional activities through strong synergy with Pax-8. Mol Endocrinol 18:2817–2829 [DOI] [PubMed] [Google Scholar]
- 35.Poleev A, Okladnova O, Musti AM, Schneider S, Royer-Pokora B, Plachov D1997. Determination of functional domains of the human transcription factor PAX8 responsible for its nuclear localization and transactivating potential. Eur J Biochem 247:860–869 [DOI] [PubMed] [Google Scholar]
- 36.De Felice M, Postiglione MP, Di Lauro R2004. Minireview: thyrotropin receptor signaling in development and differentiation of the thyroid gland: insights from mouse models and human diseases. Endocrinology 145:4062–4067 [DOI] [PubMed] [Google Scholar]
- 37.Ledent C, Parmentier M, Maenhaut C, Taton M, Pirson I, Lamy F, Roger P, Dumont JE1991. The TSH cyclic AMP cascade in the control of thyroid cell proliferation: the story of a concept. Thyroidology 3:97–101 [PubMed] [Google Scholar]
- 38.Opitz R, Trubiroha A, Lorenz C, Lutz I, Hartmann S, Blank T, Braunbeck T, Kloas W2006. Expression of sodium-iodide symporter mRNA in the thyroid gland of Xenopus laevis tadpoles: developmental expression, effects of antithyroidal compounds, and regulation by TSH. J Endocrinol 190:157–170 [DOI] [PubMed] [Google Scholar]
- 39.Vlaeminck-Guillem V, Ho SC, Rodien P, Vassart G, Costagliola S2002. Activation of the cAMP pathway by the TSH receptor involves switching of the ectodomain from a tethered inverse agonist to an agonist. Mol Endocrinol 16:736–746 [DOI] [PubMed] [Google Scholar]
- 40.Johannessen M, Delghandi MP, Moens U2004. What turns CREB on? Cell Signal 16:1211–1227 [DOI] [PubMed] [Google Scholar]
- 41.Bruno R, Ferretti E, Tosi E, Arturi F, Giannasio P, Mattei T, Scipioni A, Presta I, Morisi R, Gulino A, Filetti S, Russo D2005. Modulation of thyroid-specific gene expression in normal and nodular human thyroid tissues from adults: an in vivo effect of thyrotropin. J Clin Endocrinol Metab 90:5692–5697 [DOI] [PubMed] [Google Scholar]
- 42.Dumaz N, Marais R 2005 Integrating signals between cAMP and the RAS/RAF/MEK/ERK signalling pathways. Based on the anniversary prize of the Gesellschaft fur Biochemie und Molekularbiologie Lecture delivered on 5 July 2003. at the Special FEBS Meeting in Brussels. FEBS J 272:3491–3504 [DOI] [PubMed] [Google Scholar]
- 43.Presta I, Arturi F, Ferretti E, Mattei T, Scarpelli D, Tosi E, Scipioni A, Celano M, Gulino A, Filetti S, Russo D2005. Recovery of NIS expression in thyroid cancer cells by overexpression of Pax8 gene. BMC Cancer 5:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feliciello A, Giuliano P, Porcellini A, Garbi C, Obici S, Mele E, Angotti E, Grieco D, Amabile G, Cassano S, Li Y, Musti AM, Rubin CS, Gottesman ME, Avvedimento EV1996. The v-Ki-Ras oncogene alters cAMP nuclear signaling by regulating the location and the expression of cAMP-dependent protein kinase IIβ. J Biol Chem 271:25350–25359 [DOI] [PubMed] [Google Scholar]
- 45.Gallo A, Benusiglio E, Bonapace IM, Feliciello A, Cassano S, Garbi C, Musti AM, Gottesman ME, Avvedimento EV1992. v-ras and protein kinase C dedifferentiate thyroid cells by down-regulating nuclear cAMP-dependent protein kinase A. Genes Dev 6:1621–1630 [DOI] [PubMed] [Google Scholar]
- 46.Gallo A, Feliciello A, Varrone A, Cerillo R, Gottesman ME, Avvedimento VE1995. Ki-ras oncogene interferes with the expression of cyclic AMP-dependent promoters. Cell Growth Differ 6:91–95 [PubMed] [Google Scholar]
- 47.Calebiro D, de Filippis T, Lucchi S, Martinez F, Porazzi P, Trivellato R, Locati M, Beck-Peccoz P, Persani L2006. Selective modulation of protein kinase A I and II reveals distinct roles in thyroid cell gene expression and growth. Mol Endocrinol 20:3196–3211 [DOI] [PubMed] [Google Scholar]
- 48.Mayr B, Montminy M2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev 2:599–609 [DOI] [PubMed] [Google Scholar]
- 49.Carriere A, Ray H, Blenis J, Roux PP2008. The RSK factors of activating the Ras/MAPK signaling cascade. Front Biosci 13:4258–4275 [DOI] [PubMed] [Google Scholar]
- 50.Frödin M, Gammeltoft S1999. Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol Cell Endocrinol 151:65–77 [DOI] [PubMed] [Google Scholar]
- 51.García-Jiménez C, Zaballos MA, Santisteban P2005. DARPP-32 (dopamine and 3′,5′-cyclic adenosine monophosphate-regulated neuronal phosphoprotein) is essential for the maintenance of thyroid differentiation. Mol Endocrinol 19:3060–3072 [DOI] [PubMed] [Google Scholar]
- 52.Woloshin PI, Walton KM, Rehfuss RP, Goodman RH, Cone RD1992. 3′,5′-Cyclic adenosine monophosphate-regulated enhancer binding (CREB) activity is required for normal growth and differentiated phenotype in the FRTL-5 thyroid follicular cell line. Mol Endocrinol 6:1725–1733 [DOI] [PubMed] [Google Scholar]
- 53.Nguyen LQ, Kopp P, Martinson F, Stanfield K, Roth SI, Jameson JL2000. A dominant negative CREB (cAMP response element-binding protein) isoform inhibits thyrocyte growth, thyroid-specific gene expression, differentiation, and function. Mol Endocrinol 14:1448–1461 [DOI] [PubMed] [Google Scholar]
- 54.Davidson D, Chow LM, Fournel M, Veillette A1992. Differential regulation of T cell antigen responsiveness by isoforms of the src-related tyrosine protein kinase p59fyn. J Exp Med 175:1483–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Missero C, Pirro MT, Di Lauro R2000. Multiple ras downstream pathways mediate functional repression of the homeobox gene product TTF-1. Mol Cell Biol 20:2783–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rutledge RG, Côté C2003. Mathematics of quantitative kinetic PCR and the application of standard curves. Nucleic Acids Res 31:e93 [DOI] [PMC free article] [PubMed]
- 57.Sandoval J, Rodríguez JL, Tur G, Serviddio G, Pereda J, Boukaba A, Sastre J, Torres L, Franco L, López-Rodas G2004. RNAPol-ChIP: a novel application of chromatin immunoprecipitation to the analysis of real-time gene transcription. Nucleic Acids Res 32:e88 [DOI] [PMC free article] [PubMed]
- 58.Civitareale D, Lonigro R, Sinclair AJ, Di Lauro R1989. A thyroid-specific nuclear protein essential for tissue-specific expression of the thyroglobulin promoter. EMBO J 8:2537–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]