Abstract
The regulation of expression of gluconeogenic genes including glucose-6-phosphatase (G6Pase) and phosphoenolpyruvate carboxykinase (PEPCK) in the liver plays an important role in glucose homeostasis, because aberrant expression of these genes contributes to the development of type 2 diabetes. Previous reports demonstrate that signal transducer and activator of transcription 3 (STAT3) plays a key role in regulating gluconeogenic gene expression, but the mechanism remains unclear. Herein we demonstrate that phosphorylated STAT3 is required for repression of G6Pase expression by IL-6 in both HepG2 cells and mouse liver. Interestingly, PEPCK expression is regulated by STAT3 independent of IL-6 activation. Using in vivo chromatin immunoprecipitation, we demonstrate that STAT3 binds to the promoters of the G6Pase, PEPCK, and suppressor of cytokine signaling (SOCS)3 genes, and its recruitment increases at the G6Pase and SOCS3 promoters with IL-6 treatment. Whereas persistent recruitment of RNA polymerase II is seen on the SOCS3 promoter, consistent with its induction by IL-6, a decrease in polymerase II recruitment and histone H4 acetylation is seen at the G6Pase promoter with IL-6 treatment. Thus STAT3 mediates negative regulation of hepatic gluconeogenic gene expression in vivo by interacting with regulatory regions of these genes.
STAT3 interacts with the promoter regions of gluconeogenic genes in vivo and causes transcriptional repression.
Increased hepatic glucose production (HGP) contributes to the development of diabetes, which currently affects millions worldwide. Gluconeogenesis is the major metabolic process that causes an increase in HGP in diabetic subjects (1, 2, 3); therefore, the controlled regulation of this process is critical in maintaining normal glucose homeostasis. Glucose-6-phosphatase (G6Pase) and phosphoenolpyruvate carboxykinase (PEPCK) are key enzymes involved in the rate-limiting steps of gluconeogenesis.
The expression of these gluconeogenic genes is controlled by signaling pathways and transcription factors that respond to changes in nutrient availability, and increased expression of these genes can result in increased HGP. Glucagon stimulates increased transcription of G6Pase and PEPCK through increased cAMP signaling, and this effect is mediated by several different transcription factors including cAMP-response element-binding protein (CREB), hepatocyte nuclear factor-4α, peroxisomal proliferator-activated receptor γ coactivator-1α (PGC1α), CCAAT enhancer binding protein-α, and CCAAT enhancer binding protein-β (4, 5, 6). The promoters of G6Pase and PEPCK contain cAMP response elements (CREs) to which CREB binds (5, 6), and CREB transcription involves its association with coactivators such as CREB-binding protein/p300 and transducer of regulated CREB activity (TORC2) 2 (7, 8, 9, 10). Insulin negatively regulates the transcription of gluconeogenic genes and inhibits cAMP- and glucocorticoid-induced expression of these genes through insulin responsive sites in the promoters of these genes (11, 12, 13) that recruit the transcription factor Foxo1a (14, 15, 16, 17, 18). Insulin-mediated repression of gluconeogenic genes occurs by phosphorylation and subsequent cytoplasmic redistribution of Foxo1a (19, 20, 21, 22).
Recent studies have implicated signal transducer and activator of transcription 3 (STAT3) in the regulation of gluconeogenic gene expression in mouse liver, because liver-specific STAT3 knockout mice have increased expression of these genes and are insulin resistant when placed on a high-fat diet (23). Furthermore, insulin action in the brain results in a local increase of interleukin-6 (IL-6) in the liver, phosphorylation of STAT3 (P-STAT3) and repression of gluconeogenic gene expression (24). The effect of insulin action in the brain on hepatic gluconeogenic gene expression is lost in mice lacking either STAT3 or IL-6 in the liver, demonstrating a clear role for IL- 6-activated STAT3 in transcriptional repression of these genes (24).
STAT3 is activated by all gp130-coupled cytokines, including IL-6. Cytokines bind to their cognate receptor, resulting in subsequent downstream recruitment and phosphorylation of STAT3, which then dimerizes and translocates to the nucleus, where it activates transcription of target genes. The two isoforms of STAT3, STAT3α and STAT3β, are splice variants encoded by the same structural gene (25) and appear to have unique functions (26, 27). STAT3β functions as a dominant-negative inhibitor of STAT3α function, but it may have unique transcriptional capabilities despite lacking the C-terminal activation domain (26, 27). The STAT3α isoform mediates signaling in response to IL-6, because STAT3α-knockout mouse embryonic fibroblasts (MEFs) treated with IL-6 do not show induction of STAT3 target genes such as suppressor of cytokine signaling 3 (SOCS3), but STAT3β-knockout MEFs respond in a similar manner to wild-type MEFs (26).
In this report, we have examined the mechanism by which STAT3 represses transcription of gluconeogenic genes in HepG2 cells (a model of human hepatocytes) and mouse liver. We demonstrate herein that STAT3 mediates negative regulation of G6Pase expression by IL-6 in vivo. In addition, STAT3 regulates expression levels of PEPCK independent of activation by IL-6. Importantly, STAT3 is recruited to the promoter regions of G6Pase and PEPCK, and RNA polymerase II (Pol II) recruitment decreases at the G6Pase promoter in response to IL-6 in vivo. These results implicate STAT3 as being an important integrator of different signals that result in the decreased expression of G6Pase and PEPCK, and that repression by STAT3 is achieved by direct modulation of transcriptional events.
Results
IL-6 activates STAT3 and induces SOCS3 expression, whereas forskolin induces G6Pase expression in HepG2 cells
To study the role of STAT3 activation on gluconeogenic gene expression in HepG2 cells, we first confirmed that IL-6 treatment leads to the phosphorylation of STAT3 in nuclear extracts of HepG2 cells (Fig. 1A). Consistent with this, expression of SOCS3, a known target gene of STAT3, was induced within 15 min but reached maximum levels at 30 min after treatment (60-fold increase) and remained at elevated levels at 1 h after treatment (Fig. 1B). By 2 h, SOCS3 mRNA levels were considerably decreased, which is consistent with the known function of SOCS3 in inhibiting IL-6 signaling.
Fig. 1.

IL-6 treatment activates STAT3 and induces SOCS3 in HepG2 cells, and forskolin treatment induces G6Pase expression. HepG2 cells were placed in OptiMEM for 16 h and treated with 20 ng/ml IL-6 for 15 min after which cytosolic and nuclear extracts were prepared. Lysates were analyzed by Western blot for P-STAT3 levels (A). HepG2 cells were treated with IL-6 for the indicated time, and RNA was extracted. qPCR was performed for SOCS3 expression (B). HepG2 cells were treated with 10 μm forskolin for the indicated time, and RNA was extracted. qPCR was performed to detect G6Pase expression levels (C). Error bars represent ± sd of the mean.
Basal expression of gluconeogenic genes is low in HepG2 cells; therefore it was determined whether G6Pase expression could be up-regulated by the cAMP pathway similar to what is seen in vivo. As shown in Fig. 1C, forskolin treatment of HepG2 cells resulted in an increase in G6Pase mRNA by 1 h and reached maximum levels at 2 h after treatment (Fig. 1C). Therefore, HepG2 cells were determined to be a valid system in which gluconeogenic gene expression and, in particular, STAT3-mediated repression of these genes could be studied.
IL-6 treatment represses forskolin-induced expression of G6Pase in HepG2 cells
Because IL-6 and forskolin induced expression of SOCS3 and G6Pase, respectively, at 1 h, we determined whether IL-6 could repress forskolin-induced activation of G6Pase expression by cotreating HepG2 cells with IL-6 and forskolin for 1 h. As expected, IL-6 treatment alone induced expression of SOCS3 at 1 h after treatment (>150-fold), and surprisingly, cotreatment of IL-6 and forskolin further increased the expression of SOCS3 (Fig. 2A). G6Pase expression was induced by forskolin treatment at 1, 3, and 6 h after treatment, and IL-6 cotreatment resulted in 2- to 3-fold repression of forskolin-induced expression of G6Pase at all three time points (Fig. 2B).
Fig. 2.
IL-6 treatment represses forskolin-induced expression of G6Pase in HepG2 cells. HepG2 cells were placed in OptiMEM for 16 h and then treated with IL-6 (20 ng/ml) or forskolin (10 μm) or both for the indicated time, and RNA was extracted. qPCR analysis was performed for SOCS3 expression (A) and G6Pase expression (B). Error bars represent ± sd of the mean; *, P ≤ 0.05.
STAT3 is required for repression of G6Pase by IL-6 in HepG2
To determine whether STAT3 is necessary for the effect of IL-6 on G6Pase mRNA levels, we knocked down expression of STAT3 mRNA in HepG2 cells using small interfering RNA (siRNA). A specific reduction of STAT3 protein levels was achieved using this approach (Fig. 3A). Reduced levels of STAT3 resulted in a decrease in both basal and IL-6-induced levels of SOCS3 mRNA, (by 2.6-fold and 3.9-fold, respectively) consistent with the role of STAT3 as a positive regulator of SOCS3 expression (Fig. 3B). Knockdown of STAT3 caused a 4.8-fold increase in forskolin-induced expression of G6Pase (Fig. 3C) and a 2.4-fold increase in basal expression of G6Pase. Importantly, whereas IL-6 treatment repressed forskolin-induced G6Pase expression by 3.8-fold in the presence of STAT3, knockdown of STAT3 inhibits repression of G6Pase by IL-6 (1.7-fold repression, Fig. 3C). Any remaining repression is likely due to remaining STAT3. Thus, IL-6 repression of G6Pase is mediated through STAT3.
Fig. 3.
STAT3 is required for IL-6-mediated repression of G6Pase, and STAT3 represses PEPCK in the absence of IL-6. HepG2 cells were transfected with control or STAT3-specific siRNA. Whole-cell extracts were prepared and analyzed for STAT3 expression (A). siRNA-transfected cells were placed in OptiMEM for 16 h and then treated with IL-6 (20 ng/ml) or forskolin (10 μm) or both. RNA was extracted, and qPCR analysis was performed for SOCS3, G6Pase, PEPCK, PGC1α, and fatty acid synthase (FAS) (B–F). Error bars represent ± sd of the mean.
We next examined the effect of IL-6 treatment in the presence and absence of STAT3 on PEPCK. IL-6 treatment of HepG2 cells did not have an effect on PEPCK expression. However, when STAT3 protein levels were reduced, forskolin-induced expression of PEPCK was increased by 5.6-fold (Fig. 3D). Similarly, basal PEPCK expression increased 3-fold, demonstrating a role for STAT3 in repression of PEPCK expression that is independent of IL-6 activation of STAT3. In contrast to G6Pase and PEPCK, expression of PGC1α and Fasn was not altered by IL-6 treatment or by STAT3 knockdown in HepG2 cells (Fig. 3, E and F).
IL-6 induces phosphorylation of STAT3, increases SOCS3 mRNA, and represses G6Pase expression in mouse liver
Because STAT3 regulates both G6Pase and PEPCK expression in HepG2 cells, we next determined whether IL-6 administration in mice would produce similar results. Indeed, as in HepG2 cells, IL-6 treatment induced hepatic P-STAT3 within 15 min (Fig. 4A), P-STAT3 levels peaked at 30 min and decreased by 1 h after injection, whereas total STAT3 levels did not change with IL-6 treatment. Consistent with phosphorylation of STAT3, SOCS3 expression was induced by 30 min and was maximal at 1 h after injection (Fig. 4B). Importantly, IL-6 also repressed G6Pase expression within 30 min after injection, but this repression was maximal at 1 h (Fig. 4B). Because the changes in SOCS3 and G6Pase expression were greatest at 1 h after injection, we looked more closely at this time point. As shown in Fig. 4C, SOCS3 mRNA levels were induced 30-fold by IL-6, whereas G6Pase mRNA was repressed by 30% after IL-6 injection (Fig. 4D). Because deletion of STAT3 from livers of mice has been described to result in increased expression of PEPCK, FAS, PGC1α, and SREBP1c in addition to G6Pase (23), we determined the effect of IL-6 treatment on these genes. IL-6 did not affect PEPCK, FAS, or PGC1α mRNA levels but did repress SREBP1c mRNA by 28% (Fig. 4, E–H). Thus, the effects of IL-6 in HepG2 cells are paralleled in mouse liver, because only G6Pase was repressed by IL-6, whereas PEPCK, Fasn, and PGC1α were not affected (we were unable to measure SREBP1c mRNA levels in HepG2 cells). The lack of effect on PGC1α and Fasn expression is in contrast to a previously published study (23), However, neither of these proteins is required for the effects of STAT3 on gluconeogenic gene expression. Finally, blood glucose levels were examined both before injection and before euthanizing the mice (Fig. 4I), and no differences were observed between PBS- and IL-6-injected mice.
Fig. 4.
IL-6 activates STAT3 in mouse liver, induces SOCS3 expression, and represses G6Pase expression. C57BL6 mice (10 wk of age) were fasted for approximately 16 h and injected ip with PBS or 100 ng of IL-6. Mice were euthanized at the indicated times after injection (panel B, minutes after injection; panels C–I, 1 h after injection) and livers were dissected. Lysates were prepared and analyzed by Western blot for P-STAT3 and total STAT3 levels (A). RNA was extracted and qPCR analysis for SOCS3 and G6Pase was performed (B) (n = 2). A similar experiment with n = 8 per group was performed, and qPCR analysis was performed for SOCS3, G6Pase, PEPCK, FAS, PGC1α, and SREBP1c expression (C–H). Blood glucose levels were measured before and after injections before euthanizing the mice (I). Error bars represent ± sem of the mean; *, P ≤ 0.05.
STAT3 is required in vivo for repression of G6Pase by IL-6
To determine whether STAT3 is required for repression of G6Pase and induction of SOCS3 in the liver, we overexpressed the STAT3β isoform in the liver using an adenoviral vector. Because STAT3β is a dominant-negative inhibitor of STAT3α function based on its ability to compete for DNA-binding sites, we hypothesized that STAT3β overexpression would block the observed effects of IL-6. As seen in Fig. 5A, overexpression of STAT3β caused an increase in basal P-STAT3 levels in the liver that was not seen in adenovirus-green fluorescent protein (Ad-GFP)-infected livers. IL-6 induced P-STAT3 in Ad-GFP-infected livers as well as STAT3β-overexpressing livers. SOCS3 mRNA was induced 21-fold in Ad-GFP livers (Fig. 5B), whereas it was induced only 6-fold in STAT3β-expressing livers. Importantly, G6Pase was repressed by 38% in Ad-GFP livers with IL-6 treatment, whereas significant repression was not seen in STAT3β-expressing livers (Fig. 5C). Basal G6Pase expression was not altered significantly between Ad-GFP- and STAT3β-expressing livers. In addition, whereas PEPCK expression is not repressed by IL-6, overexpression of STAT3β results in a 66% increase in basal expression of PEPCK (Fig. 5D). Therefore these results demonstrate that functional STAT3 is required in vivo to mediate the effects of IL-6 and that STAT3 regulates PEPCK expression in vivo independent of IL-6 action. The effect of STAT3 on PEPCK regulation in vivo therefore resembles regulation of PEPCK in HepG2 cells by STAT3.
Fig. 5.

Repression of G6Pase by IL-6 requires STAT3 function in vivo. C57BL/6 mice (n = 8–11 mice per group) were injected with Ad-GFP control or Ad-GFP STAT3β through the tail vein on d 1. Mice were fasted overnight on d 4, and on d 5 were injected ip with PBS or 100 ng of IL-6 and euthanized 1 h after injection, after which livers were collected. Western blot analyses for P-STAT3 and total STAT3 from a representative group of mice (A) and qPCR analysis for SOCS3, G6Pase, and PEPCK mRNA levels (B–D) are shown. Error bars represent ± sem of the mean; *, P ≤ 0.05.
STAT3 binds to the G6Pase promoter in vivo and regulates transcription
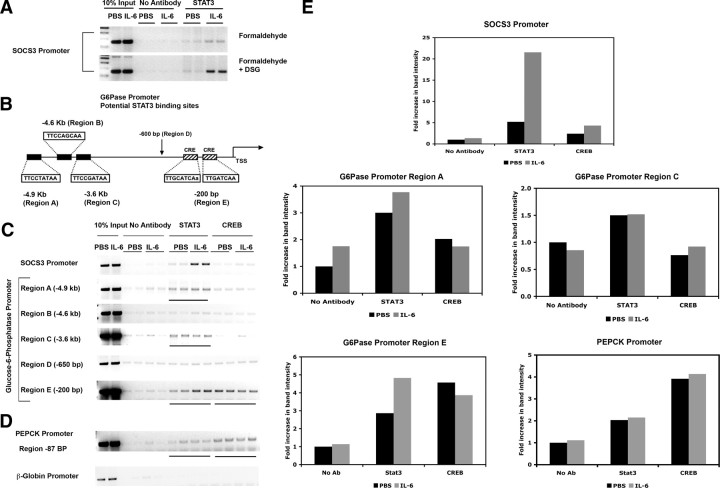
To determine how STAT3 functions as a repressor of gluconeogenic gene expression, chromatin immunoprecipitation (ChIP) assays were performed using the same livers that were used for mRNA expression analysis. It has been shown previously that phosphorylated STAT3 is recruited to the proximal SOCS3 promoter in vivo, and thus we used this target as a positive control (28). Because ChIP assays on tissue samples are not efficient, we modified our experimental protocol to include a second cross-linking step using the homo-bifunctional ester disuccinimidyl glutarate (DSG). This method has been used for ChIP assays from cells and has been shown to improve the yield of ChIP products (29). As shown in Fig. 6A, the two-step cross-linking approach employing DSG and formaldehyde results in a substantially increased yield of the SOCS3 promoter in STAT3 ChIPs without significantly increasing nonspecific binding in the control ChIPs. Therefore, for all subsequent ChIP experiments, DSG and formaldehyde cross-linking was used.
Fig. 6.
STAT3 binds to the SOCS3 and G6Pase promoters, and STAT3 recruitment is increased with IL-6 treatment. ChIP experiments were performed to detect the presence of STAT3 at the SOCS3 promoter using standard formaldehyde cross-linking vs. double cross-linking using DSG and formaldehyde (A). A schematic representation of the mouse G6Pase promoter depicting the potential STAT3 sites and known CREs is shown (B). ChIPs were then performed using the double cross-linking method to detect STAT3 and CREB at the G6Pase promoter (C) and PEPCK promoter (D). A control PCR for the β-globin promoter was also performed. Regions where STAT3 and CREB bind to the G6Pase and PEPCK promoters are underlined. Quantified band intensities from panels C and D are shown graphically (E). Ab, Antibody.
Scanning of the initial 5 kb of the G6Pase promoter identified three potential STAT3 binding sites (Fig. 6B) that conform to the known consensus site TT(N5)AA for STAT3 (marked region A, potential site at −4.9 kb, region B, potential site at −4.6 kb, and region C, potential site at −3.6 kb). In addition we identified region D (−650 bp) where there were no potential STAT3 sites seen. Region E at −200 bp has been previously characterized to contain two functional CRE sites to which CREB has been shown to bind in EMSAs. ChIP assays were performed using STAT3 and CREB antibodies to detect the presence of these transcription factors on the G6Pase promoter (Fig. 6C). Interestingly, STAT3 was detected at region A, region C, and region E of the G6Pase promoter in the basal state, but its recruitment to the proximal region E is enhanced by IL-6. In addition, region E of the G6Pase promoter recruits CREB but this is unaffected by IL-6 treatment. As expected, STAT3 binds only minimally to the SOCS3 promoter in the basal state, whereas its recruitment is maximally enhanced by IL-6 treatment. Importantly, no binding of STAT3 or CREB was seen to region D of the G6Pase promoter or to the control β-globin promoter (Fig. 6C), demonstrating specificity of STAT3 binding.
STAT3 and CREB binding was also assessed at the PEPCK promoter using ChIP assays. CREB bound to the PEPCK promoter at the CRE site located −85 bp upstream of the transcription start site (Fig. 6D). STAT3 also bound weakly to the PEPCK promoter, but there was no significant change in binding of either STAT3 or CREB with IL-6 treatment.
Acetylation of histone H4 and recruitment of RNA Pol II are decreased at the G6Pase promoter but not PEPCK promoter in the presence of IL-6
To determine whether the recruitment of STAT3 to the SOCS3 and G6Pase promoters leads to changes in RNA Pol II recruitment and histone acetylation, ChIP assays were again performed. In the presence of IL-6, there was a slight increase in Pol II recruitment to the SOCS3 promoter (Fig. 7A), which is consistent with increased transcription of SOCS3. However, although there were no changes in histone acetylation at the proximal SOCS3 promoter, histone H4 is deacetylated at several distal regions of the G6Pase promoter (Fig. 7B), which is consistent with reduced transcription of this gene. In addition, RNA Pol II recruitment to the proximal G6Pase promoter was reduced in the presence of IL-6, again consistent with its reduced transcription (Fig. 7B). In contrast to the effects of IL-6 on histone acetylation and RNA Pol II recruitment to the G6Pase promoter, there was no change in either of these parameters at the PEPCK promoter in the presence of IL-6 (Fig. 7C), which correlates with the apparent lack of effect of IL-6 on PEPCK mRNA levels. These results suggest that STAT3 recruitment to the G6Pase promoter results in chromatin remodeling, which then leads to a block in transcription.
Fig. 7.
Decrease in acetylated histone H4 and RNA Pol II recruitment is seen at the G6Pase promoter in response to IL-6 treatment. ChIP assays were performed to detect the presence of acetylated histone H4, acetylated histone H3, and RNA Pol II at the SOCS3 (A), G6Pase (B), and PEPCK (C) promoters. A control PCR for the β-globin promoter was also performed. Regions where changes in acetylated histone H4 and RNA Pol II were seen at the G6Pase promoter are underlined. Quantified band intensities from panels A, B, and C are shown graphically (D). Ab, Antibody; TSS, transcription start site.
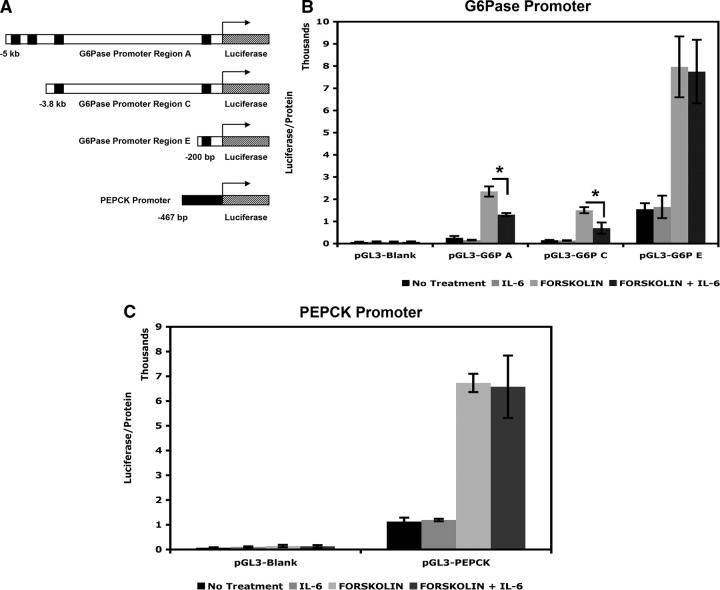
Distal elements of the G6Pase promoter are required for transcriptional repression by IL-6
To determine whether the actions of STAT3 on G6Pase expression require binding at all three regions identified in ChIP assays, we studied the transcriptional response of three reporter constructs that contain −5 kb (pGL3-G6P A), −3.9 kb (pGL3-G6P C), and −200 bp (pGL3-G6P E) of the G6Pase promoter upstream of luciferase (depicted in Fig. 8A). HepG2 cells were transfected with these reporter constructs and then treated with IL-6, forskolin, or both in a paradigm identical to experiments that studied expression of endogenous G6Pase mRNA in these cells. As expected, forskolin induced transcription of all three promoter constructs via the CREs present at −200 bp. Remarkably, whereas region E alone does not confer the ability to be repressed by IL-6, the reporter constructs containing regions up to −3.9 kb (regions C and E) and −5 kb (regions A, C, and E) are significantly repressed by IL-6 (Fig. 8B). Thus, although there is increased recruitment of STAT3 to the −200 bp region of G6Pase with IL-6, the distal elements are required for repression. In addition, we also examined the transcriptional response of a reporter construct that contained a 470-bp region of the rat PEPCK promoter (Fig. 8A) that is 95% identical to the mouse G6Pase promoter in this region. Expression of luciferase is increased with forskolin treatment, but IL-6 has no effect on basal or induced expression of this construct (Fig. 8C).
Fig. 8.
Distal elements of the G6Pase promoter are required for transcriptional repression by IL-6. Schematic diagram of the different luciferase constructs is shown (A). HepG2 cells were transfected with empty pGL3 vector, pGL3-G6P A, pGL3-G6P C or pGL3-G6P E constructs and treated with IL-6, forskolin, or both and luciferase expression was measured (B). HepG2 cells were transfected with pGL3 empty vector or pGL3-PEPCK and treated as in panel B, and luciferase expression was measured (C). Error bars represent ± sd of the mean; *, P ≤ 0.05.
Discussion
The controlled expression of gluconeogenic genes is essential to maintain normal glucose homeostasis, because an increase in expression of these genes results in increased hepatic glucose output. In this report, we have determined that STAT3 directly targets the regulatory regions of both G6Pase and PEPCK in vivo to regulate their expression. Indeed, P-STAT3 is induced by IL-6 and mediates repression of G6Pase, whereas PEPCK responds to basal levels of STAT3 demonstrating unique mechanisms of negative regulation by STAT3.
Although STAT3 regulates transcription of most genes by increasing mRNA synthesis, a few examples of STAT3 functioning as a negative regulator of transcription are known. For example, STAT3 has been shown to repress the expression of p53, and this contributes to cellular transformation in oncogenesis (30). STAT3 also represses endothelial nitric oxide synthase expression in aortic endothelial cells (31). STAT3 is therefore capable of activating and repressing target genes in response to certain signals, and the molecular mechanisms by which this differential regulation occurs is yet to be determined. The majority of STAT3-regulated genes contain STAT3 binding sites in the promoter, suggesting that a direct binding event precedes transcriptional changes. Previous work has demonstrated that STAT3 plays a putative role in suppressing expression of gluconeogenic genes, because the loss of STAT3 in the liver resulted in increased expression of these genes (23, 24). However, it was not clear from these studies whether STAT3 phosphorylation leads to direct recruitment to and repression of gluconeogenic genes.
Several signaling pathways are known to activate STAT3, including cytokines such as IL-6, interferons, and leptin. The beneficial role of IL-6 in glucose homeostasis remains controversial because some studies have shown increased circulating IL-6 levels in obese and diabetic animals (32, 33), whereas IL-6 knockout mice develop mature onset obesity and impaired glucose tolerance and fail to respond to insulin action in the brain (24, 34). For the purpose of this study, IL-6 was used as a tool to activate STAT3 to study transcriptional regulation of gluconeogenic genes in HepG2 cells and in vivo. We demonstrate that IL-6 potently activates STAT3 in HepG2 cells and in mouse liver and increases expression of SOCS3. Furthermore, IL-6-activated STAT3 also strongly represses G6Pase expression in HepG2 cells and in mouse liver.
This study demonstrates, for the first time, that STAT3 is present at the G6Pase and PEPCK promoters and is required for their regulation in vivo. There is increased recruitment of STAT3 to the proximal G6Pase promoter in response to IL-6. In addition, STAT3 was detected at two upstream regions of the G6Pase promoter. Using truncated promoter constructs, we determined that the distal regions at −3.9 kb and −5 kb are necessary for IL-6-mediated repression of the G6Pase promoter in cell culture. The recruitment of STAT3 is necessary for repression of G6Pase by IL-6 because knockdown of STAT3 in HepG2 cells or expression of STAT3β in vivo blocks repression. In contrast, STAT3 binds to the PEPCK promoter in an IL-6-independent manner and modulates basal activity of PEPCK expression because both knockdown of STAT3 in HepG2 cells and expression of STAT3β in vivo strongly stimulate PEPCK expression.
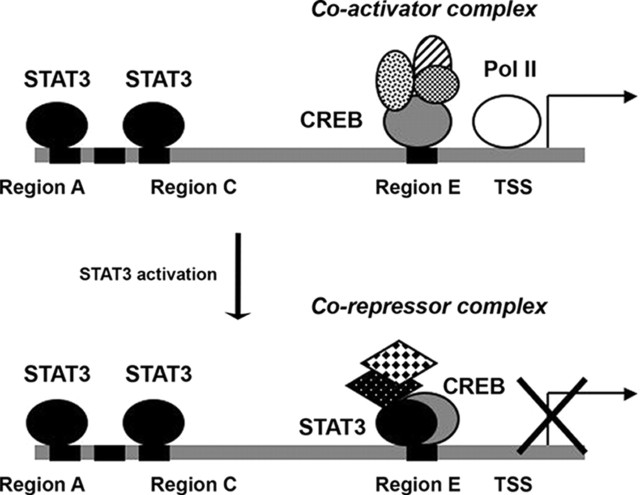
Although the mechanism by which STAT3 mediates different transcriptional events at the SOCS3 and gluconeogenic gene promoters has not been entirely discerned, we have shown that STAT3 recruitment leads to distinct transcriptional events. IL-6 treatment leads to persistent or enhanced RNA Pol II recruitment to the SOCS3 promoter, consistent with transcriptional activation. In contrast, at the G6Pase promoter there is reduced recruitment of RNA Pol II with IL-6 treatment, which correlates with decreased transcription. In addition, IL-6 treatment resulted in decreased histone H4 acetylation at the G6Pase locus, implying that when STAT3 is recruited a unique cofactor complex is recruited that then allows for transcriptional repression via chromatin remodeling. Given that STAT3 is recruited to the proximal G6Pase promoter in the region of CREB binding, it is tempting to speculate that STAT3 interferes with CREB-mediated activation. Based on the fact that the distal regions of the G6Pase promoter are required for repression by IL-6 from reporter experiments, and the fact that STAT3 increases at the proximal region of the G6Pase promoter in response to IL-6 treatment (ChIP assays), we propose a model in which STAT3 is bound to the distal sites of the G6Pase promoter to maintain basal transcription (Fig. 9). Upon activation, STAT3 is recruited to the proximal promoter, chromatin remodeling occurs at several regions of the promoter, and RNA Pol II decreases at the transcriptional start site, leading to transcriptional repression of G6Pase.
Fig. 9.
Model of regulation of the G6Pase promoter by STAT3. In the basal state, STAT3 is present at regions A and C (−5 kb and −3.9 kb). Upon activation, STAT3 is also recruited to the proximal region to result in repression of expression. Chromatin remodeling occurs, and RNA Pol II recruitment is decreased. TSS, Transcription start site.
In summary, we demonstrate that the transcription factor STAT3 regulates gluconeogenic gene expression by interacting with their promoter regions and further demonstrates that the mechanisms by which this repression is achieved are likely to be different for individual genes. It is very likely that STAT3 has several additional uncharacterized targets that could play a role in glucose and lipid homeostasis, and a more global approach to identifying these targets needs to be addressed. Although these data throw light on the function of STAT3 as a repressor of gluconeogenic genes, it remains to be determined whether hepatic STAT3 signaling is compromised in diabetic obese animals. Indeed, hepatic STAT3 may serve as a key integrator of nutrient signals that leads to changes in hepatic glucose production; thus, disrupting this signaling pathway could contribute to the onset and progression of diabetes.
Materials and Methods
Cell culture
HepG2 cells were maintained in DMEM containing 4.5 g/liter glucose supplemented with 10% fetal bovine serum, 2 mm l-glutamine, and 100 U/ml penicillin, 100 μg/ml streptomycin sulfate, 0.025 μg/ml amphotericin B at 37 C in 5% CO2. Cells were routinely passaged using 0.05% trypsin-EDTA and passed through a 23.5-gauge needle three times before cells were seeded on a fresh plate to avoid cells forming aggregates. All cell culture reagents were obtained from Life Technologies (Carlsbad, CA) (GIBCO). In cell culture-based experiments, n = 3 per treatment group.
IL-6 and forskolin experiments
HepG2 cells were seeded in six-well plates and allowed to reach 80% confluency. The medium was replaced with OptiMEM approximately 16 h before treatment with IL-6 or forskolin. Cells were then treated with 20 ng/ml IL-6 (Sigma Chemical Co., St. Louis, MO) or 10 μm forskolin (Sigma) or both for the time indicated in each experiment.
siRNA experiments
HepG2 cells were seeded in DMEM supplemented with fetal bovine serum without antibiotics and allowed to reach 30% confluency. Cells were transfected with100 nm control (nonspecific) siRNA or siRNA against STAT3 (HSS11280 obtained from Invitrogen) using LipofectAMINE 2000 according to the manufacturer’s protocol. Cells were monitored for 48 h and inspected visually for toxicity/cell death. Medium was replaced with OptiMEM 48 h after transfection, for 16 h before treatment with IL-6 or forskolin. Cells were treated for 1 h, followed by RNA extractions and gene expression analysis. For protein lysates, cells were lysed without treatment and analyzed by Western blot. A representative experiment of three separate experiments is shown.
Plasmid constructs, transfections, and luciferase assays
pGL3-G6P A (−5 kb to +21 of the G6Pase promoter) and pGL3-G6P C (−3.9 kb to +21) were generated by PCR amplification of the G6Pase promoter using a G6P BAC clone (CHORI RP23-429G15) as a template, followed by cloning using a TOPO-TA cloning kit (Invitrogen). The promoter regions were then subcloned into the pGL3 vector using KpnI-XhoI restriction sites. The constructs were sequenced and used for transfection experiments. pGL3-G6P E (−231 to + 66 of the mouse G6Pase promoter) was obtained from Dr. Richard O'Brien (Vanderbilt University, Memphis, TN). pGL3-PEPCK (−467 bp of the rat PEPCK promoter) was obtained from Dr. Daryl Granner (Vanderbilt University). HepG2 cells were seeded in 12-well plates and transfected with 50 ng of the appropriate luciferase construct and 450 ng of empty vector pKCR2 (to bring the total amount of transfected DNA to 500 ng) using LipofectAMINE 2000 according to the manufacturer’s protocol. Media was changed to fresh OptiMEM 6 h after transfection. After 16 h, cells were treated with IL-6, forskolin, or both for 6 h and lysed. Luciferase expression was measured using a Promega luciferase assay kit and luminometer (Promega Corp., Madison, WI), and expression was normalized to total protein concentration in the sample. Protein concentrations were measured by BCA assay (Pierce Chemical Co., Rockford, IL). This experiment was repeated three to five times, and a representative experiment performed in triplicate is shown.
Mouse liver experiments
All animal experiments were approved by the Institutional Animal Care and Use Committee at BIDMC. C57BL/6 mice (9 wk of age) were obtained from Jackson Labs for all experiments. The mice were housed in single cages for 1 wk at the BIDMC animal facility and maintained on a 14-h light, 10-h dark cycle and were fed a rodent chow diet (Harlan Teklad F6 Rodent Diet, Harlan Laboratories). Mice were handled daily for a week to acclimate them before the experiment and were fasted overnight for approximately 16 h before the start of experiments. After the fast, animals were injected ip with PBS (control) or IL-6 (100 ng per mouse; Sigma). Blood glucose was measured using a glucometer (One-touch Ultra; Lifescan, Inc., Milpitas, CA) before injection and before the mice were killed. All mice were killed at the indicated time point by CO2 inhalation. Tissues were dissected and frozen immediately in liquid nitrogen.
Adenovirus experiments
A construct expressing STAT3β cDNA (Origene Technologies, Inc., Rockville, MD) was generated in an adenoviral vector containing a bidirectional promoter for expression of GFP in one direction and STAT3β in the other. The Ad-GFP-STAT3β virus and a control virus expressing only GFP (Ad-GFP) were obtained from the Harvard Gene Therapy Initiative. C57BL/6 mice (10 wk of age) were injected with adenovirus (1 × 109 pfu/mouse) through the tail vein. On d 4, mice were fasted overnight in clean cages, and on d 5, IL-6 experiments were performed as described above. Livers that had comparable STAT3β and GFP expression (for Ad-GFP-STAT3β) or only GFP (for Ad-GFP) as determined by Western blotting were used for further analysis of gene expression.
Western blotting
For HepG2 cells, cells were treated with IL-6 for 15 min, and nuclear extracts were prepared based on a previously described protocol (35). For siRNA experiments, HepG2 cells were lysed in 1× RIPA buffer [10 mm Tris-Cl (pH 8.0), 1 mm EDTA, 0.5 mm EGTA, 140 mm NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), and protease inhibitors]. For mouse liver, approximately 50 mg of tissue was homogenized in 1 ml lysis buffer (Cell Signaling Technology, Beverly, MA) using a TissueLyser (QIAGEN, Chatsworth, CA) for 1 min at 30 Hz. Proteins were resolved on 10% NuPAGE Bis-Tris gels (Invitrogen) or 8% Tris-tricine gels and transferred to nitrocellulose membrane, and blots were probed for P-STAT3 (anti-Phospho STAT3-Tyr705, Cell Signaling Technology). Proteins were visualized using an ECL-Plus kit (Pierce). Blots were then stripped and reprobed for total STAT3 (Santa Cruz K15, Santa Cruz Biotechnology, Santa Cruz, CA) or RNA Pol II (antibody clone 8WG16; Upstate Biotechnology, Lake Placid, NY) as a loading control.
RNA extraction, cDNA synthesis, and quantitative PCR (qPCR)
HepG2 cells were treated with IL-6, forskolin, or both for the time indicated in each experiment and the media were aspirated. Cells were lysed in RNA-STAT60 (Tel-Test, Inc., Friendswood, TX), and RNA was extracted according to the manufacturer’s protocol. For liver, approximately 30 mg of tissue was homogenized in RNA-STAT60 using a TissueLyser for 2 min at 30 Hz, and RNA was extracted. cDNA was synthesized from 1 μg of RNA for each sample using the Advantage RT for PCR kit (BD Biosciences, Franklin Lakes, NJ). cDNA was diluted as necessary, and quantitative PCR was performed using SYBR green reaction chemistry (Dynamo SYBR green qPCR reagent; Finnzymes, Espoo, Finland) or Taqman assays (Applied Biosystems) and a Stratagene Mx 3000 thermal cycler. Primer sequences are available upon request. Gene expression was normalized to glyceraldehyde-3-phosphate dehydrogenase or cyclophilin mRNA levels.
ChIP assays
Approximately 70 mg of tissue was minced in 5 ml of ice-cold PBS and cross-linked using 2 mm DSG at room temperature for 45 min. The tissue was then washed twice in PBS and cross-linked using 1% formaldehyde at room temperature for 10 min. The cross-linked tissue was washed in PBS and resuspended in 1.6 ml of ChIP lysis buffer (50 mm Tris-Cl, pH 8.0; 10 mm EDTA; 1% SDS; and protease inhibitors). Chromatin was sonicated in 800-μl aliquots to an average shear size of 500–1000 bp and centrifuged to remove debris. Chromatin was pooled and precleared by incubating 100 μl of chromatin diluted in 1 ml of IP buffer (0.5× RIPA buffer, protease inhibitors, and 10 μg of salmon sperm DNA) with 40 μl (packed volume) of Protein A agarose for 90 min at 4 C. The precleared chromatin was then immunoprecipitated overnight using 4 μg of antibody. Antibodies used were α-STAT3 (C-20) and α-CREB (C-21) (Santa Cruz), α-acetyl histone H4, and α-RNA Pol II 8WG16 (Upstate). Immunoprecipitated complexes were captured using Protein A agarose for 90 min at 4 C and washed three times with 0.5× RIPA buffer. Cross-links were reversed by incubating the complexes with digesting buffer (50 mm Tris-Cl, pH 8.0; 1 mm EDTA; 100 mm NaCl; 0.5% SDS) at 65 C for 4 h, DNA was extracted, and PCR was performed. Primer sequences are available upon request. ChIP results are representative of three independent experiments.
Statistics
Statistical significance (P value) was determined by Student’s t test. P ≤ 0.05 was considered to be significant.
Acknowledgments
We thank Dr. Jeng-Shin Lee (Harvard Gene Therapy Initiative) for adenovirus generation, and Kaila Holtz for technical assistance. We thank Dr. Richard O'Brien for providing the pGL3-G6P E construct, and Dr. Daryl Granner for providing the pGL3-PEPCK construct.
Footnotes
This work was supported by the Smith Family Pinnacle Award from the American Diabetes Association (to A.N.H.).
Disclosure Summary: The authors have nothing to disclose.
First Published Online March 5, 2009
Abbreviations: Ad-GFP, Adenovirus-green fluorescent protein; ChIP, chromatin immunoprecipitation; CRE, cAMP response element; CREB, CRE-binding protein; DSG, disuccinimidyl glutarate; G6Pase, glucose-6-phosphatase; HGP, hepatic glucose production; MEF, mouse embryonic fibroblast; PEPCK, phosphoenolpyruvate carboxykinase; PGC1α, peroxisomal proliferator-activated receptor-γ coactivator-1α; Pol II, polymerase II; P-STAT3, phosphorylation of STAT3; qPCR, quantitative PCR; SDS, sodium dodecyl sulfate; siRNA, small interfering RNA; SOCS, suppressor of cytokine signaling; STAT3, signal transducer and activator of transcription 3.
References
- 1.Cline GW, Rothman DL, Magnusson I, Katz LD, Shulman GI1994. 13C-nuclear magnetic resonance spectroscopy studies of hepatic glucose metabolism in normal subjects and subjects with insulin-dependent diabetes mellitus. J Clin Invest 94:2369–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Consoli A1992. Role of liver in pathophysiology of NIDDM. Diabetes Care 15:430–441 [DOI] [PubMed] [Google Scholar]
- 3.Consoli A, Nurjhan N, Capani F, Gerich J1989. Predominant role of gluconeogenesis in increased hepatic glucose production in NIDDM. Diabetes 38:550–557 [DOI] [PubMed] [Google Scholar]
- 4.Gautier-Stein A, Mithieux G, Rajas F2005. A distal region involving hepatocyte nuclear factor 4α and CAAT/enhancer binding protein markedly potentiates the protein kinase A stimulation of the glucose-6-phosphatase promoter. Mol Endocrinol 19:163–174 [DOI] [PubMed] [Google Scholar]
- 5.Lin B, Morris DW, Chou JY1997. The role of HNF1α, HNF3γ, and cyclic AMP in glucose-6-phosphatase gene activation. Biochemistry 36:14096–14106 [DOI] [PubMed] [Google Scholar]
- 6.Liu JS, Park EA, Gurney AL, Roesler WJ, Hanson RW1991. Cyclic AMP induction of phosphoenolpyruvate carboxykinase (GTP) gene transcription is mediated by multiple promoter elements. J Biol Chem 266:19095–19102 [PubMed] [Google Scholar]
- 7.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365:855–859 [DOI] [PubMed] [Google Scholar]
- 8.Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M2003. TORCs: transducers of regulated CREB activity. Mol Cell 12:413–423 [DOI] [PubMed] [Google Scholar]
- 9.Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bächinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH1994. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370:223–226 [DOI] [PubMed] [Google Scholar]
- 10.Ravnskjaer K, Kester H, Liu Y, Zhang X, Lee D, Yates 3rd JR, Montminy M2007. Cooperative interactions between CBP and TORC2 confer selectivity to CREB target gene expression. EMBO J 26:2880–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forest CD, O'Brien RM, Lucas PC, Magnuson MA, Granner DK1990. Regulation of phosphoenolpyruvate carboxykinase gene expression by insulin. Use of the stable transfection approach to locate an insulin responsive sequence. Mol Endocrinol 4:1302–1310 [DOI] [PubMed] [Google Scholar]
- 12.Magnuson MA, Quinn PG, Granner DK1987. Multihormonal regulation of phosphoenolpyruvate carboxykinase-chloramphenicol acetyltransferase fusion genes. Insulin’s effects oppose those of cAMP and dexamethasone. J Biol Chem 262:14917–14920 [PubMed] [Google Scholar]
- 13.O'Brien RM, Lucas PC, Forest CD, Magnuson MA, Granner DK1990. Identification of a sequence in the PEPCK gene that mediates a negative effect of insulin on transcription. Science 249:533–537 [DOI] [PubMed] [Google Scholar]
- 14.Altomonte J, Richter A, Harbaran S, Suriawinata J, Nakae J, Thung SN, Meseck M, Accili D, Dong H2003. Inhibition of Foxo1 function is associated with improved fasting glycemia in diabetic mice. Am J Physiol Endocrinol Metab 285:E718–E728 [DOI] [PubMed]
- 15.Barthel A, Schmoll D, Krüger KD, Bahrenberg G, Walther R, Roth RA, Joost HG2001. Differential regulation of endogenous glucose-6-phosphatase and phosphoenolpyruvate carboxykinase gene expression by the forkhead transcription factor FKHR in H4IIE-hepatoma cells. Biochem Biophys Res Commun 285:897–902 [DOI] [PubMed] [Google Scholar]
- 16.Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, Spiegelman BM2003. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature 423:550–555 [DOI] [PubMed] [Google Scholar]
- 17.Hall RK, Yamasaki T, Kucera T, Waltner-Law M, O'Brien R, Granner DK2000. Regulation of phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein-1 gene expression by insulin. The role of winged helix/forkhead proteins. J Biol Chem 275:30169–30175 [DOI] [PubMed] [Google Scholar]
- 18.Vander Kooi BT, Streeper RS, Svitek CA, Oeser JK, Powell DR, O'Brien RM2003. The three insulin response sequences in the glucose-6-phosphatase catalytic subunit gene promoter are functionally distinct. J Biol Chem 278:11782–11793 [DOI] [PubMed] [Google Scholar]
- 19.Biggs 3rdWH, Meisenhelder J, Hunter T, Cavenee WK, Arden KC1999. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci USA 96:7421–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo S, Rena G, Cichy S, He X, Cohen P, Unterman T1999. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J Biol Chem 274:17184–17192 [DOI] [PubMed] [Google Scholar]
- 21.Rena G, Guo S, Cichy SC, Unterman TG, Cohen P1999. Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J Biol Chem 274:17179–17183 [DOI] [PubMed] [Google Scholar]
- 22.Nakae J, Kitamura T, Ogawa W, Kasuga M, Accili D2001. Insulin regulation of gene expression through the forkhead transcription factor Foxo1 (Fkhr) requires kinases distinct from Akt. Biochemistry 40:11768–11776 [DOI] [PubMed] [Google Scholar]
- 23.Inoue H, Ogawa W, Ozaki M, Haga S, Matsumoto M, Furukawa K, Hashimoto N, Kido Y, Mori T, Sakaue H, Teshigawara K, Jin S, Iguchi H, Hiramatsu R, LeRoith D, Takeda K, Akira S, Kasuga M2004. Role of STAT-3 in regulation of hepatic gluconeogenic genes and carbohydrate metabolism in vivo. Nat Med 10:168–174 [DOI] [PubMed] [Google Scholar]
- 24.Inoue H, Ogawa W, Asakawa A, Okamoto Y, Nishizawa A, Matsumoto M, Teshigawara K, Matsuki Y, Watanabe E, Hiramatsu R, Notohara K, Katayose K, Okamura H, Kahn CR, Noda T, Takeda K, Akira S, Inui A, Kasuga M2006. Role of hepatic STAT3 in brain-insulin action on hepatic glucose production. Cell Metab 3:267–275 [DOI] [PubMed] [Google Scholar]
- 25.Caldenhoven E, van Dijk TB, Solari R, Armstrong J, Raaijmakers JA, Lammers JW, Koenderman L, de Groot RP1996. STAT3β, a splice variant of transcription factor STAT3, is a dominant negative regulator of transcription. J Biol Chem 271:13221–13227 [DOI] [PubMed] [Google Scholar]
- 26.Maritano D, Sugrue ML, Tininini S, Dewilde S, Strobl B, Fu X, Murray-Tait V, Chiarle R, Poli V2004. The STAT3 isoforms α and β have unique and specific functions. Nat Immunol 5:401–409 [DOI] [PubMed] [Google Scholar]
- 27.Yoo JY, Huso DL, Nathans D, Desiderio S2002. Specific ablation of Stat3β distorts the pattern of Stat3-responsive gene expression and impairs recovery from endotoxic shock. Cell 108:331–344 [DOI] [PubMed] [Google Scholar]
- 28.Guo F, Bakal K, Minokoshi Y, Hollenberg AN2004. Leptin signaling targets the thyrotropin-releasing hormone gene promoter in vivo Endocrinology 145:2221–2227 [DOI] [PubMed] [Google Scholar]
- 29.Nowak DE, Tian B, Brasier AR2005. Two-step cross-linking method for identification of NF-κB gene network by chromatin immunoprecipitation. Biotechniques 39:715–725 [DOI] [PubMed] [Google Scholar]
- 30.Niu G, Wright KL, Ma Y, Wright GM, Huang M, Irby R, Briggs J, Karras J, Cress WD, Pardoll D, Jove R, Chen J, Yu H2005. Role of Stat3 in regulating p53 expression and function. Mol Cell Biol 25:7432–7440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saura M, Zaragoza C, Bao C, Herranz B, Rodriguez-Puyol M, Lowenstein CJ2006. Stat3 mediates interleukin-6 [correction of interelukin-6] inhibition of human endothelial nitric-oxide synthase expression. J Biol Chem 281:30057–30062 [DOI] [PubMed] [Google Scholar]
- 32.Klover PJ, Clementi AH, Mooney RA2005. Interleukin-6 depletion selectively improves hepatic insulin action in obesity. Endocrinology 146:3417–3427 [DOI] [PubMed] [Google Scholar]
- 33.Klover PJ, Zimmers TA, Koniaris LG, Mooney RA2003. Chronic exposure to interleukin-6 causes hepatic insulin resistance in mice. Diabetes 52:2784–2789 [DOI] [PubMed] [Google Scholar]
- 34.Wallenius V, Wallenius K, Ahrén B, Rudling M, Carlsten H, Dickson SL, Ohlsson C, Jansson JO2002. Interleukin-6-deficient mice develop mature-onset obesity. Nat Med 8:75–79 [DOI] [PubMed] [Google Scholar]
- 35.Schreiber E, Matthias P, Müller MM, Schaffner W1989. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]