Abstract
Daily rhythms in behavior and physiology are observed in most organisms. These rhythms are controlled by internal self-sustained circadian (∼24 h) clocks, which are present in virtually all cells. The 24-h oscillations are generated by a molecular mechanism entrained by external or internal time cues and which, in turn, regulate rhythmic outputs. In mammals, the circadian system comprises a master clock located in the hypothalamus that is directly entrained by the light-dark cycle and which coordinates the phases of local clocks in the periphery in order to ensure optimal timing of the physiology. Nuclear receptors (NRs) form a large family of transcription factors that include both ligand-inducible and orphan receptors. These NRs are key regulators of major biological processes such as reproduction, development, cell growth and death, inflammation, immunity, and metabolic homeostasis. Recent observations indicate that several NR signaling pathways play a critical role in central and peripheral circadian clocks. The REV-ERB/retinoid-related orphan receptor orphan NR subfamily regulates the expression of core clock genes and contributes to the robustness of the clock mechanism. Glucocorticoid and retinoic acid receptors are involved in the resetting of peripheral clocks. Several other NRs such as peroxisome proliferator-activated receptor-α, short heterodimer partner, and constitutive androstane receptor act as molecular links between clock genes and specific rhythmic metabolic outputs. The expanding functional links between NRs and circadian clocks open novel perspectives for understanding the hormonal regulation of the mammalian circadian system as well as for exploring the role of circadian clocks in the pathogenesis of NR-related diseases such as cancer and metabolic syndrome.
IN MAMMALS, MANY aspects of behavior and physiology are subject to daily oscillations. Classical examples include sleep-wake cycles, hormone secretion, body temperature, blood pressure, renal activity, and liver metabolism (1). These rhythms are driven by self-sustained endogenous clocks located in virtually all cells of the body and forming an integrated system (2). The circadian (∼24 h) system is organized hierarchically with at the top a master clock located in the suprachiasmatic nuclei (SCN) of the hypothalamus and oscillating with an approximately 24-h period in the absence of external time cues. Every day, this central clock is directly reset by light through the retino-hypothalamic tract to keep the physiology synchronized to the external light-dark cycle. Circadian clocks present in the periphery are entrained by the SCN clock through internal synchronizers that have yet to be identified. Peripheral clocks can be synchronized by autonomic, hormonal, and metabolic time cues independently of the SCN clock (3). However, whereas they display self-sustained oscillations at the single-cell level similar to those from the SCN neurons, at the organ and systemic levels they require the SCN clock to remain in phase (4). Genetic and biochemical approaches have now comprehensively established that central and peripheral circadian clocks all use a common set of clock genes which interact together to form complex interlocked transcriptional/posttranslational feedback loops (5, 6, 7). A major autoregulatory loop in this molecular mechanism involves the two transcriptional activators CLOCK and brain and muscle ARNT-like 1 (BMAL1), which upon dimerization drive the transcription of two period (Per1 and Per2) and two cryptochrome (Cry1 and Cry2) genes. PER and CRY proteins subsequently associate and translocate to the nucleus where they repress their own transcription through inhibition of the CLOCK:BMAL1-dependent transcriptional activity. This regulatory mechanism undergoes an extensive posttranslational regulation that is essential for the maintenance of the oscillations. The resulting feedback loop generates oscillations in gene transcription that provide timing information for multiple clock-controlled genes (CCGs) regulating physiological outputs (8).
Mammalian genomes contain 48–49 nuclear receptor (NR) genes encoding a large family of proteins that function as ligand-inducible transcription factors (9). This family includes receptors for steroid and thyroid hormones, retinoic acid, vitamin D, oxysterols, and fatty acid derivatives as well as a number of orphan receptors for which no physiological ligands have been identified. NRs have been involved in all key biological processes such as development, reproduction, metabolism, cell growth, differentiation, and immunity and are consequently considered as promising therapeutic targets for the treatment of inflammatory and metabolic diseases and cancer (10). Most NRs share the same modular organization with a regulatory amino-terminal domain, a central DNA-binding domain, and a carboxy-terminal ligand-binding domain. They regulate gene transcription through binding as homo- or heterodimer to specific DNA sequences located in the promoter region of target genes. Upon ligand binding, the NR ligand-binding domain undergoes conformational changes that induce the recruitment of specific coregulators, resulting in the subsequent repression or activation of transcription. Ligand independent regulation of NR transcriptional activity has also been demonstrated for many NRs. Updated information related to NR structure and function has been extensively reviewed recently (11, 12, 13, 14, 15, 16, 17, 18).
Recent observations indicate that the NR superfamily may impinge the circadian timing system at multiple levels through the regulation of clock genes and/or CCGs. Conversely, many NRs appear to be themselves CCGs. The purpose of this article is to review the current knowledge regarding the role of all NRs that regulate, or are regulated by circadian clocks to propose an integrated view of the interactions between circadian physiology and NR signaling.
NRs WITHIN THE CORE CLOCK MECHANISM
New Roles for Old Orphans
Rev-erbα (NR1D1) is an orphan NR discovered more than 15 yr ago, which was recently reported by several groups to be a CCG in a number of peripheral tissues as well as in the SCN (19, 20, 21). REV-ERBα binds DNA as monomers to the consensus NR half-site motif flanked by a 6-bp AT-rich sequence (A/T)6 PuGGTCA, or as a homodimer to a direct repeat of the core motif separated by 2 bp and acts as a constitutive transcriptional repressor. Two such response elements are present within the promoter of the clock gene Bmal1. Consistently, disruption of the Rev-erbα gene in mice resulted in an increased of Bma1l gene expression (22). This result, together with the observation that circadian clock proteins regulate Rev-erbα, led to a model in which REV-ERBα is the molecular link between the negative and positive components of mammalian clocks (22, 23). This also explained why mice lacking PER2, a negative component of the main molecular feedback loop driving circadian clocks, were found to express elevated Bmal1 mRNA levels. Although Rev-erbα−/− mice exhibited an abnormal response to light resetting and an unstable period length of their circadian locomotor activity, they remained rhythmic (Table 1) (22). This subtle circadian phenotype could result from a redundancy with REV-ERBβ (NR1D2), a paralog of REV-ERBα also expressed rhythmically. This hypothesis was recently tested by Liu et al. (24) by using an RNA interference-based approach targeting Rev-erbβ in Rev-erbα−/− cells. They could demonstrate that whereas both REV-ERBs are indeed redundant for driving the rhythmic transcription of Bmal1, they are dispensable for the oscillation of Per2. This suggests that the main role of REV-ERBα and REV-ERBβ is to contribute to the robustness of circadian oscillations generated by the core clock mechanism and to control the transcription of rhythmic outputs (see below).
Table 1.
Circadian-Related Phenotypes of NR Mutant Mice
| NR | Ligand | Tissue Distribution | Mutation | Circadian Clock Related Phenotype | Other Phenotypes | Reference Nos. |
|---|---|---|---|---|---|---|
| TRβ (NR1A2) | T3 | Retina, pituitary, periphery | Systemic KO | Loss of mid-wavelength cones, impaired photic entrainment, attenuated phase shift | Hyperthyroidism | 42 |
| REV-ERBα (NR1D1) | Orphan | SCN, periphery | Systemic KO | 0.5-h Shorter free running period, increased response to light-induced phase shift | Cerebellum defects, dyslipidemia | 22 |
| RORα (NR1F1) | Orphan | SCN, periphery | Staggerer | 0.35-h Shorter free running period | Ataxia, artherosclerosis | 25 |
| RORβ (NR1F2) | Orphan | SCN, pineal gland, retina | Systemic KO | 0.4-h Longer free running period | Vacillans phenotype, blind | 27 |
| RORγ (NR1F3) | Orphan | periphery | Systemic KO | None | Abnormal lymphoid organogenesis | 24 |
| GR (NR3C1) | Corticosterone | Ubiquitous (not in SCN) | Liver-specific KO | No resetting by glucocorticoids in liver | Hypoglycemia | 44 |
| PPAΡα (NR1C1) | Fatty acids | Liver | Systemic KO | Down-regulation of Bmal1 | Dyslipidemia | 51 |
| EAR2 (NR2F6) | Orphan | Ubiquitous | Systemic KO | Loss of Per1 rhythmic expression | Abnormal development of the locus coeruleus, Altered nociception | 40 |
KO, Knockout.
The REV-ERBα response element is also recognized by retinoid-related orphan receptors (RORs), which, in contrast to REV-ERBα, activate transcription. Using a functional genomic approach to identify new core clock components, Sato and colleagues (25) discovered that retinoid-related orphan receptor-α (RORα; NR1F1) was rhythmically expressed in the SCN and activated Bmal1 gene transcription in cotransfection assays. The RORα-dependent transactivation could be antagonized by REV-ERBα, in accordance with a model in which the oscillation of Bmal1 transcription was the result of the alternating activating and repressing activities of RORs and REV-ERBs at the same response element, respectively (26). Staggerer mice, which expressed a nonfunctional truncated form of RORα, exhibit a slightly shorter period and a reduced robustness of their circadian locomotor activity rhythm (Table 1). Interestingly, disruption of RORβ (NR1F2), a RORα paralog rhythmically expressed in the pineal gland, retina, and SCN, three key structures of the circadian system, also resulted in a measurable, albeit modest, circadian phenotype (Table 1) (27). Conversely, loss of RORγ (NR1F3) function in mice had no consequence in the SCN or in the periphery, although this NR is rhythmically expressed in multiple peripheral tissues (24). Thus it appears that in the periphery, the major drivers of rhythmic clock gene transcription are the two REV-ERBs and that circadian expression of RORγ does not play a significant role in the presence of the constitutively coexpressed RORα.
The Expanding Role of Posttranslational Regulation in the NR-Clock Connection
Recent structural and biochemical evidence suggests that cholesterol may be a RORα ligand whereas retinoids act as inverse agonists for RORβ and RORγ (28, 29). Whether these putative ROR ligands play a role in vivo and modulate circadian clock function remains unanswered. Addressing this issue may be difficult because other NR pathways are also regulated by cholesterol derivatives and retinoic acids. Based on structural data showing that their ligand-binding pocket is too small to accommodate NR ligands, REV-ERB NRs have been considered as true orphans, so far. This view was recently challenged by the discovery that the ligand-binding domain of both REV-ERBα and REV-ERBβ reversibly associates with heme, which is a prosthetic group for a wide variety of proteins (30, 31). Heme binding stimulates the recruitment of nuclear receptor corepressor and subsequent repression of Bmal1 transcription. Interestingly, heme biosynthesis is itself regulated by the clock, suggesting the existence of a metabolic feedback loop (32). It is presently not known whether heme-bound REV-ERBs are responsive to nitric oxide (NO) like their Drosophila counterpart E75 (NR1D3) (33). However, given the large number of processes regulated by NO that show daily variations such as blood pressure, metabolism, and cell cycle, it is tempting to speculate that NO may modulate specific physiological outputs of the clock via heme-bound REV-ERBs. In addition to these newly identified posttranslational regulatory mechanisms, phosphorylation plays also a significant role in the control of the clock mechanism, and at least one kinase, glycogen synthase kinase 3β (GSK3β) is acting through REV-ERBα. GSK3β was recently found to phosphorylate and stabilize the REV-ERBα protein, which is otherwise highly unstable (34). Interestingly, this effect is inhibited by lithium, a compound used in the clinic to treat bipolar disorder, a form of depression associated with altered circadian rhythms.
Although most clock components are conserved between Drosophila and vertebrates, there is no functional equivalent to the REV-ERB/ROR loop in Drosophila. Instead there exists a loop based on vrille and pdp1, two PAR transcription factors that, respectively, repress and activate the positive limb of the clock mechanism. These factors are also present in vertebrate clocks, but they serve mainly as circadian regulators of CCGs. It is tempting to speculate that the Drosophila vrille/pdp1 loop has been exchanged with the REV-ERB/ROR loop during early vertebrate evolution to build clocks able to interact with an increasingly complex endocrine system. Interestingly, the substrate for the GSK3β kinase has also shifted from TIMLESS in Drosophila to REV-ERBα in mammals, suggesting a coordinated and extensive modification of the clock-regulatory mechanisms during metazoan evolution (35, 36). A notable difference between the Drosophila and higher vertebrate clock mechanism is that peripheral clocks are photoreceptive in the fly, whereas entrainment in vertebrates in thought to occur through endocrine and autonomic cues. The REV-ERB/ROR loop may thus have been selected as an efficient molecular link between clock genes and endocrine signaling pathways. This evolutionary functional shift is supported by the presence of both peripheral photoreception and the REV-ERB loop in early vertebrates like fish.
NR SIGNALING AND CLOCK SYNCHRONIZATION
Photic Resetting Pathways
Light is a major and universal external resetting signal (or zeitgeber) of the circadian system in all species. In mammals, the photic information is conveyed to the SCN via the retino-hypothalamic tract and resets the central clock. When animals are kept under constant darkness, light resetting operates only when light is normally not expected. This occurs independently of the image formation pathway because mice or humans devoid of light perception still respond to light synchronization. At the level of the retina, an important role in light-dependent entrainment of the circadian clock is played by a group of photosensitive retinal ganglion cells that contain a recently discovered nonvisual pigment termed melanopsin (37). How the SCN perceives the light information at the molecular level has been the subject of numerous studies which collectively established that Per1 and Per2 play a critical role in the light response and in the entrainment of the SCN clock, through dynamic chromatin remodeling and cAMP regulation of their promoters (38, 39).
Several lines of evidence suggest that NRs are involved in the photic resetting mechanism. 1) As briefly mentioned above, Rev-erbα−/− mice kept in constant darkness and exposed to a light pulse at a time when Rev-erbα is normally peaking display a dramatic phase advance response (>5 h) as compared with wild-type animals (∼1 h) (22); 2) EAR2 (NR2F6) is an orphan NR required for the development of the locus coeruleus norepinephrine-producing neurons. Mice lacking EAR2 display an unstable circadian locomotor activity rhythm and abnormal light resetting (40). Additional circadian defects include a loss of Per1 circadian expression, a dampening of Per2 circadian expression in the frontal cortex as well as an altered resynchronization after a light-dark cycle phase shift. 3) Ventrolateral SCN neurons express the androgen receptor AR (NR3C4), and androgens are required for a normal light response of FOS in the SCN (41).
The developmental role of some NRs can also be critical for the integrity of the photic resetting pathway. For instance, mice lacking the thyroid hormone receptor-β (NR1A2) are devoid of mid-wavelength cones and consequently have an abnormal light entrainment and phase-shifting response (Table 1) (42).
Light regulation of clock gene expression is not restricted to the SCN because photic induction of mouse Per1 and Per2 gene expression was recently demonstrated to occur in the adrenal glands via the SCN-sympathetic pathway (43). This effect is independent of the hypothalamo-pituitary-adrenal axis. Nevertheless, it is associated with a large increase of plasma and cerebrospinal fluid corticosterone levels and the concomitant induction of three NRs including Nurr77 (NR4A1), Nurr1 (NR4A2), and steroidogenic factor 1 (NR5A1), a critical regulator of steroidogenesis. These data show that specific NRs directly participate in the conversion of the environmental light information to endogenous glucocorticoid signaling that may, in turn, entrain peripheral clocks as suggested by previous studies (see below).
Clearly, these data demonstrate that NRs may play a prominent role in the resetting mechanism of circadian clocks. Given the fact that many additional NRs including photoreceptor-specific NR (NR2E3), RORα, estrogen receptor-β (ERβ; NR3A2), and retinoid X receptors (RXRs; NR2B1–3) are expressed in retinal ganglion cells, it would be important to analyze more systematically the light-induced phase-shifting response of null mutants or agonist-treated animals.
Nonphotic Resetting Pathways
Whereas light is unambiguously the major synchronizer of the SCN clock, accumulating evidence indicates that nonphotic time cues including hormonal, metabolic, and autonomic signals are important synchronizers of peripheral clocks. Glucocorticoid hormones are secreted rhythmically with maximum plasma levels at the end of the sleep phase in mammals and have been therefore considered as ideal candidates for such a role. Indeed, glucocorticoids reset peripheral clocks in rodents, and this effect is mediated by the glucocorticoid receptor (NR3C1) and antagonized by RU486 (Table 1) (44, 45). Importantly, the clock-resetting activity of glucocorticoids is independent of the SCN, which does not express GR. In contrast to light resetting, which can occur only during the dark period in rodents, glucocorticoids could phase shift peripheral clocks at any time of the light-dark cycle. How glucocorticoids regulate circadian clocks is not understood at the molecular level, but this may implicate the regulation of Per genes because treating cells with the glucocorticoid agonist dexamethasone acutely induces the expression of Per1, a gene already known to participate in the resetting of the SCN clock (46). Consistently, a functional glucocorticoid response element was identified in the mouse Per1 gene promoter and acute stress, a condition that elevates endogenous corticosterone secretion, was shown to up-regulate Per1 expression in peripheral tissues (47).
More surprising was the finding that retinoids also act as putative synchronizers of circadian clocks in the vascular system. The developmental and physiological effects of retinoids are mediated by retinoic acid receptors (RARα/β/γ, NR1B1–3) and RXRs (RXRα/β/γ, NR2B1–3). Whereas RARs bind all-trans retinoic acid (tRA) and 9-cis retinoic acid (9-cis-RA), RXRs bind only 9-cis-RA. Based on a two-hybrid approach, McNamara and colleagues showed that RARα and RXRα could physically interact in a ligand-dependent manner with CLOCK and MOP4/NPAS2, a CLOCK paralog expressed in some extra hypothalamic brain areas and in peripheral tissues (48). This interaction prevents DNA binding and subsequent transcriptional activation by the CLOCK:BMAL1 heterodimer. In human vascular smooth muscle cells, tRA but no other NR ligands could reset the clock. In contrast to glucocorticoids this tRA-mediated phase-shifting effect occurred only at specific times.
Restricting feeding to the light period is a very potent synchronizer of peripheral clocks independently of the SCN in nocturnal rodents (3, 49). Although the underlying mechanisms remain unresolved, one might expect that some plasma metabolites may operate as metabolic time cues for peripheral clocks. Additionally, NRs that are key regulators of energy metabolism such as the peroxisome proliferator-activated receptors (PPARs; NR1C1–3), liver X receptors (NR1H2–3), and estrogen-related receptors (ERRs; NR3B1–3) are also potential mediators of the entraining effect of the feeding schedule (50). This hypothesis is supported by the recent identification of PPARα (NR1C1) as an upstream regulator of the clock gene Bmal1 (Table 1) (51). The PPARα agonist fenofibrate was further shown to synchronize quiescent fibroblasts (51). Interestingly, long-chain unsaturated fatty acids and their derivatives are natural PPAR ligands, and their plasma levels undergo changes with the nutritional status, notably with an increase during the unfed state. It is therefore conceivable that part of the feeding schedule-dependent resetting of peripheral clocks involves fatty acid-activated PPARα. Because the three PPAR paralogs exhibit distinct expression patterns, if this hypothesis is valid, then one might expect that this role is achieved by PPARβ (NR1C2) an PPARγ (NR1C–3) in skeletal muscle and adipose tissue, respectively.
NRs LINKING CLOCK GENES TO PHYSIOLOGICAL OUTPUTS
Our understanding of the mammalian circadian system at the molecular and integrated levels remains limited, in part, because the pathways that link clock genes to other homeostatic processes have not been identified. Although a nearly unique set of clock genes seems to be responsible for the generation of circadian rhythms throughout the body, accumulating evidence from genomic studies indicate that the transcriptional program regulated by these clock genes is highly tissue specific. Identifying the genetic circuitry that clock genes use to regulate physiological outputs in specific organs is key for understanding the tissue specificity of the circadian regulation. In that context, NRs, a number of which exhibit a restricted expression pattern and are clock controlled, appear to be good candidates to transmit the rhythmic information generated by clock genes to physiological output in specific tissues or cells.
Oscillations in Plasma Levels of NR Ligands
It has been known for decades that plasma levels of many hormones from the pituitary and endocrine tissues, including adrenal and sex steroids, fluctuate daily in rodents and humans. Consistently, circadian expression profiling of the mouse adrenal gland transcriptome has recently revealed that 33 genes involved in the corticosterone biosynthesis pathway are clock controlled (52). Additionally, the adrenal clock exerts a gating of the glucocorticoid biosynthetic response to ACTH, possibly through the transcriptional control of specific steps between cholesterol intake and corticosterone secretion (53). Glucocorticoids regulate glucose metabolism in the liver and several enzymes in this pathway, including glucokinase and glucose-6-phosphatase, are clock controlled presumably through the rhythmic activation of the glucocorticoid receptor (54). Another example is the circadian expression and glucocorticoid regulation of topoisomerase 1, a DNA replication enzyme targeted by camptothecin derivative drugs such as irinotecan in the treatment of colorectal cancer (55). Such a regulation may explain the influence of the administration time on the efficacy and toxicity of irinotecan (56).
GnRH and gonadotropins are released predominantly during the night in humans. Data obtained with clock/clock mutant mice, which exhibit irregular estrous cycles (57), indicate that CLOCK may even be required for the pulsatile secretion pattern of GnRH (58). Interestingly, clock gene expression has been reported in the ovary (59). Thus it appears that the whole female gonadal axis is subjected to a clock control that is likely to interfere with estrogen signaling. The situation is somewhat different in the male because no functional clock has been identified in the testis (60). However, an important level at which sex steroid levels may be regulated by the clock is their metabolism, which occurs primarily in the liver. Accordingly, it has been demonstrated that the steroid 15α-hydroxylase, which catalyzes one of the hydroxylation steps in the metabolism of estrogens and testosterone in the liver, is controlled by bZIP proline acidic amino acid-rich basic leucine zipper albumin site D box-binding protein, which is itself a direct clock target (61, 62). Daily variation in the plasma levels of these hormones is therefore likely to result, at least in part, from their rhythmic catabolism.
Rhythmic Transcriptional and Posttranslational Regulation of NRs
Several independent lines of evidence suggest that indirect rather than direct pathways are involved in the regulation of most clock outputs. Indeed, many CCGs lack in their promoter region the canonical E-box response elements that mediate CLOCK:BMAL1 transcriptional activation, and initial work in Drosophila has shown that the protein CLOCK may directly regulate less than 10% of the rhythmic transcriptome in this model system (63, 64). Importantly, clock-controlled expression of numerous transcriptional regulators has been demonstrated in the SCN and in peripheral organs (7). For instance, transcriptional cascades involving the DEC1 and D box-binding protein transcription factors have been identified (8, 65, 66). Among these transcriptional clock outputs, NRs may play a prominent role because more than 50% of NRs display a diurnal expression pattern (67).
Liver metabolism is one of the biological processes most extensively influenced by the circadian system. An increasing number of studies suggest that NRs are critical in transmitting the timing information to specific metabolic targets in the liver. PPARα is a liver-enriched fatty acid-activated NR the expression of which is rhythmically regulated at the systemic level by glucocorticoids but also by clock genes in liver cells (20, 51, 68, 69, 70, 71). This is thought to influence, in turn, the responsiveness of metabolic targets, such as the pyruvate dehydrogenase kinase 4, to fatty acids (51, 68). PPARα regulates the expression of many other genes of lipid metabolism in the liver, a number of which are clock controlled, such as those encoding lipoprotein lipase and carnitine palmitoyl transferase. PPARα appears as both an input (see above) and an output of the clock mechanism in the liver. Its role as a critical metabolic sensor in this organ is thus likely to integrate an overlooked potentially important circadian component. Lipid metabolism in liver is also regulated by small heterodimer partner (SHP, NR0B2), which is a tissue-specific and inducible orphan NR. SHP is devoid of DNA-binding activity and represses the transcriptional activity of multiple metabolic NRs including liver receptor homolog 1 (LRH-1; NR5A2), farnesoid X receptor-α (NR1H4), hepatocyte nuclear receptor 4 (NR2A1), ERα (NR3A1), PPARs, pregnane X receptor (NR1I2) and constitutive androstane receptor-α (CARα; NR1I3) through heterodimerization (72). This unusual NR is expressed with a robust circadian expression pattern in the liver, and its promoter is synergistically activated by the CLOCK:BMAL1 heterodimer and LRH-1, a mechanism that involves a physical interaction between CLOCK and LRH-1 (73). This finding suggests a posttranslational mechanism by which nonrhythmic metabolic NRs may regulate bile acid, triglyceride, and glucose homeostasis rhythmically through time-dependent interaction with SHP.
In addition to its role in the transcriptional regulation of clock genes, REV-ERBα has also been implicated in lipid metabolism in the liver as well as in the adipocyte differentiation process (74, 75). In the liver, REV-ERBα and most likely REV-ERBβ drive the circadian expression of the plasminogen activator inhibitor type 1, a critical regulator of the fibrinolytic system increasingly recognized as a regulator of the inflammatory response in the vascular wall (76). Recent data also indicate that REV-ERBα and REV-ERBβ are circadian regulators of Cdkn1a expression, providing evidence for a molecular link between clock genes and cell proliferation (77).
Another important liver metabolic function is the detoxification of xenobiotic compounds, a process in which two NRs, CAR (NR1I3) and pregnane X receptor, play a central role as transcriptional regulators of xenobiotic/steroid and drug-metabolizing enzymes (17). CAR is expressed rhythmically in the liver, and phenobarbital induction of Cyp2B p450 cytochrome, a CAR target, is higher at the time of maximal CAR expression (78).
Genetic and biochemical evidence for the role of several additional NRs as circadian clock outputs regulating rhythmic physiological outputs have also recently been obtained for the thyroid hormone receptor α1 (NR1A1), ERRα (NR3B1), and ERβ (NR3A2) (79, 80, 81). Given the fact that ER and ERR cross talk at multiple levels (coregulators, ligands, target genes), these observations suggest that estrogen signaling is a potentially important target of the circadian system (82).
The present survey of the literature dealing with the involvement of NR signaling pathways in the clock control of physiological outputs leads to several comments. First, some NR ligands or activators are produced locally through the tissue-specific expression of biosynthetic enzymes. Although some data show, for instance, the rhythmic expression of the 5′-deiodinase in liver, this remains an unexplored mechanism that may play a significant role in the cell or tissue-specific control of circadian outputs. Second, when compared with other transcription factor families, NRs appear to be overrepresented among CCGs. In the liver for instance, 20% of the NRs are rhythmic whereas it is generally assumed that 10% of the transcriptome is clock controlled in this organ. This extensive involvement of NRs in the circadian control suggests that the molecular integration of timing and hormonal cues at a single target protein represents an advantage for the control of body homeostasis. Third, an emerging principle is that many NRs seem to be components of both the input and output pathways of circadian clocks. The existence of such multiple secondary interlocked NR-dependent feedback loops suggests that the clock mechanism is constantly adjusting and adjusted by the metabolic and endocrine environment, most likely in a tissue-specific manner (Fig. 1). Indeed, NRs like Rev-erbs, RORs, and PPARs are all metabolic sensors and/or metabolic regulators. Given the importance of NRs in health and disease, this implicates that a trade-off may exist between an increased robustness of circadian clocks and metabolic homeostasis under the normal physiological situation and increased sensitivity to metabolic and endocrine perturbations.
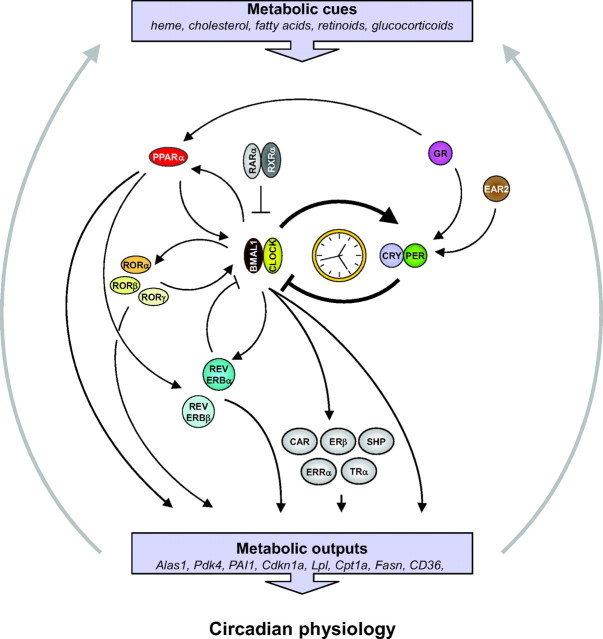
Fig. 1.
An Integrated View of the Links between NRs and the Mammalian Circadian System
The positive (CLOCK and BMAL1) and negative (PER, CRY) limbs that constitute the main feedback loop of circadian clocks are shown at the center. This core clock mechanism is regulated by multiple secondary loops formed by PPARα, REV-ERBs, and RORs. Additional NRs also regulate core clock genes. These NR-dependent input pathways may contribute to the tissue-specific synchronization of peripheral clocks by integrating specific metabolic signals. Simultaneously, the same secondary loops, together with core clock genes and additional clock-controlled NRs, regulate the expression of rhythmic metabolic outputs that, in turn, may contribute to generation of metabolic time cues. Only NRs for which a mechanistic or genetic evidence for their circadian regulation in addition to their rhythmic expression has been provided, are mentioned. FXR, Farnesoid X receptor; GR, glucocorticoid receptor; TR, thyroid hormone receptor; CRY, crytochrome; PER, period.
CONCLUSION
One third of the NR family has already been involved in the circadian system through biochemical and genetic studies, and, collectively, more than half undergo a circadian regulation in specific tissues. The resulting increasingly recognized importance of NRs in circadian physiology suggests that it may be possible to improve the therapeutic index of NR ligands by modulating the time of delivery. This concept has proven to be valid for many anticancer drugs and awaits further development in the endocrinology field (83). This also opens new pharmacological perspectives for disorders associated with circadian dysfunction because it may be possible to treat them by targeting specific NRs. Importantly, as a potential NR target, the circadian system should become systematically evaluated for the absence of unwanted effects during the development of new NR ligands.
NURSA Molecule Pages:
Coregulators: NCOR;
Nuclear Receptors: AR | CAR | ERα | ERβ | ERRα | ERRβ | ERRγ | Eip75 | FXRα | GR | HNF4α | LRH-1 | LXRα | LXRβ | NGFIB | NURR1 | PNR | PPARα | PPARγ | PPARδ | PXR | RARα | RARβ | RARγ | REV-ERBα | REV-ERBβ | RORα | RORβ | RORγ | RXRα | RXRβ | RXRγ | SF-1 | SHP | TRα | TRβ.
Footnotes
This work was supported by the University of Nice, Centre National de la Recherche Scientifique, Association pour la Recherche sur le Cancer Grant 3729, European Commission, LSHM-CT-2005-01865, and LSHG-CT-2006-037543.
Author Disclosure: The authors have nothing to disclose.
First Published Online July 24, 2008
Abbreviations: BMAL1, Brain and muscle ARNT-like 1; CAR, constitutive androstane receptor; CCG, clock-controlled gene; ER, estrogen receptor; ERR, estrogen-related receptor; GSK3β, glycogen synthase kinase 3β; LRH-1, liver receptor homolog 1; NR, nuclear receptor; PPAR, peroxisome proliferator-activated receptor; RAR, retinoic acid receptor; ROR, retinoid-related orphan receptor; RXR, retinoid X receptor; SCN, suprachiasmatic nuclei; SHP, small heterodimer partner; tRA, all-trans retinoic acid.
References
- 1.Brown SA, Schibler U 1999. The ins and outs of circadian timekeeping. Curr Opin Genet Dev 9:588–594 [DOI] [PubMed] [Google Scholar]
- 2.Ko CH, Takahashi JS 2006. Molecular components of the mammalian circadian clock. Hum Mol Genet 15(Spec No 2):R271–R277 [DOI] [PubMed]
- 3.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U 2000. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14:2950–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS 2004. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA 101:5339–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reppert SM, Weaver DR 2002. Coordination of circadian timing in mammals. Nature 418:935–941 [DOI] [PubMed] [Google Scholar]
- 6.Lowrey PL, Takahashi JS 2004. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet 5:407–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panda S, Hogenesch JB, Kay SA 2002. Circadian rhythms from flies to human. Nature 417:329–335 [DOI] [PubMed] [Google Scholar]
- 8.Delaunay F, Laudet V 2002. Circadian clock and microarrays: mammalian genome gets rhythm. Trends Genet 18:595–597 [DOI] [PubMed] [Google Scholar]
- 9.Robinson-Rechavi M, Carpentier AS, Duffraisse M, Laudet V 2001. How many nuclear hormone receptors are there in the human genome? Trends Genet 17:554–556 [DOI] [PubMed] [Google Scholar]
- 10.Gronemeyer H, Gustafsson JA, Laudet V 2004. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov 3:950–964 [DOI] [PubMed] [Google Scholar]
- 11.Benoit G, Cooney A, Giguere V, Ingraham H, Lazar M, Muscat G, Perlmann T, Renaud JP, Schwabe J, Sladek F, Tsai MJ, Laudet V 2006. International Union of Pharmacology. LXVI. Orphan nuclear receptors. Pharmacol Rev 58:798–836 [DOI] [PubMed] [Google Scholar]
- 12.Flamant F, Baxter JD, Forrest D, Refetoff S, Samuels H, Scanlan TS, Vennstrom B, Samarut J 2006. International Union of Pharmacology. LIX. The pharmacology and classification of the nuclear receptor superfamily: thyroid hormone receptors. Pharmacol Rev 58:705–711 [DOI] [PubMed] [Google Scholar]
- 13.Germain P, Chambon P, Eichele G, Evans RM, Lazar MA, Leid M, De Lera AR, Lotan R, Mangelsdorf DJ, Gronemeyer H 2006. International Union of Pharmacology. LX. Retinoic acid receptors. Pharmacol Rev 58:712–725 [DOI] [PubMed] [Google Scholar]
- 14.Germain P, Chambon P, Eichele G, Evans RM, Lazar MA, Leid M, De Lera AR, Lotan R, Mangelsdorf DJ, Gronemeyer H 2006. International Union of Pharmacology. LXIII. Retinoid X receptors. Pharmacol Rev 58:760–772 [DOI] [PubMed] [Google Scholar]
- 15.Lu NZ, Wardell SE, Burnstein KL, Defranco D, Fuller PJ, Giguere V, Hochberg RB, McKay L, Renoir JM, Weigel NL, Wilson EM, McDonnell DP, Cidlowski JA 2006. International Union of Pharmacology. LXV. The pharmacology and classification of the nuclear receptor superfamily: glucocorticoid, mineralocorticoid, progesterone, and androgen receptors. Pharmacol Rev 58:782–797 [DOI] [PubMed] [Google Scholar]
- 16.Michalik L, Auwerx J, Berger JP, Chatterjee VK, Glass CK, Gonzalez FJ, Grimaldi PA, Kadowaki T, Lazar MA, O'Rahilly S, Palmer CN, Plutzky J, Reddy JK, Spiegelman BM, Staels B, Wahli W 2006. International Union of Pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacol Rev 58:726–741 [DOI] [PubMed] [Google Scholar]
- 17.Moore DD, Kato S, Xie W, Mangelsdorf DJ, Schmidt DR, Xiao R, Kliewer SA 2006. International Union of Pharmacology. LXII. The NR1H and NR1I receptors: constitutive androstane receptor, pregnene X receptor, farnesoid X receptor α, farnesoid X receptor β, liver X receptor α, liver X receptor β, and vitamin D receptor. Pharmacol Rev 58:742–759 [DOI] [PubMed] [Google Scholar]
- 18.Dahlman-Wright K, Cavailles V, Fuqua SA, Jordan VC, Katzenellenbogen JA, Korach KS, Maggi A, Muramatsu M, Parker MG, Gustafsson JA 2006. International Union of Pharmacology. LXIV. Estrogen receptors. Pharmacol Rev 58:773–781 [DOI] [PubMed] [Google Scholar]
- 19.Lazar MA, Hodin RA, Darling DS, Chin WW 1989. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA α transcriptional unit. Mol Cell Biol 9:1128–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torra IP, Tsibulsky V, Delaunay F, Saladin R, Laudet V, Fruchart JC, Kosykh V, Staels B 2000. Circadian and glucocorticoid regulation of Rev-erbα expression in liver. Endocrinology 141:3799–3806 [DOI] [PubMed] [Google Scholar]
- 21.Balsalobre A, Damiola F, Schibler U 1998. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93:929–937 [DOI] [PubMed] [Google Scholar]
- 22.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U 2002. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110:251–260 [DOI] [PubMed] [Google Scholar]
- 23.Triqueneaux G, Thenot S, Kakizawa T, Antoch MP, Safi R, Takahashi JS, Delaunay F, Laudet V 2004. The orphan receptor Rev-erbα gene is a target of the circadian clock pacemaker. J Mol Endocrinol 33:585–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu A, Tran H, Zhang E, Priest A, Welsh D, Kay SA 2008. Redundant function of REV-ERBα and β and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet 4:e1000023 [DOI] [PMC free article] [PubMed]
- 25.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB 2004. A functional genomics strategy reveals RORa as a component of the mammalian circadian clock. Neuron 43:527–537 [DOI] [PubMed] [Google Scholar]
- 26.Guillaumond F, Dardente H, Giguere V, Cermakian N 2005. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms 20:391–403 [DOI] [PubMed] [Google Scholar]
- 27.Andre E, Conquet F, Steinmayr M, Stratton SC, Porciatti V, Becker-Andre M 1998. Disruption of retinoid-related orphan receptor β changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. EMBO J 17:3867–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kallen J, Schlaeppi JM, Bitsch F, Delhon I, Fournier B 2004. Crystal structure of the human RORα ligand binding domain in complex with cholesterol sulfate at 2.2 A. J Biol Chem 279:14033–14038 [DOI] [PubMed] [Google Scholar]
- 29.Stehlin-Gaon C, Willmann D, Zeyer D, Sanglier S, Van Dorsselaer A, Renaud JP, Moras D, Schule R 2003. All-trans retinoic acid is a ligand for the orphan nuclear receptor RORβ. Nat Struct Biol 10:820–825 [DOI] [PubMed] [Google Scholar]
- 30.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA 2007. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science 318:1786–1789 [DOI] [PubMed] [Google Scholar]
- 31.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F 2007. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nat Struct Mol Biol 14:1207–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaasik K, Lee CC 2004. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 430:467–471 [DOI] [PubMed] [Google Scholar]
- 33.Reinking J, Lam MM, Pardee K, Sampson HM, Liu S, Yang P, Williams S, White W, Lajoie G, Edwards A, Krause HM 2005. The Drosophila nuclear receptor e75 contains heme and is gas responsive. Cell 122:195–207 [DOI] [PubMed] [Google Scholar]
- 34.Yin L, Wang J, Klein PS, Lazar MA 2006. Nuclear receptor Rev-erbα is a critical lithium-sensitive component of the circadian clock. Science 311:1002–1005 [DOI] [PubMed] [Google Scholar]
- 35.Martinek S, Inonog S, Manoukian AS, Young MW 2001. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105:769–779 [DOI] [PubMed] [Google Scholar]
- 36.Iitaka C, Miyazaki K, Akaike T, Ishida N 2005. A role for glycogen synthase kinase-3β in the mammalian circadian clock. J Biol Chem 280:29397–29402 [DOI] [PubMed] [Google Scholar]
- 37.Panda S, Sato TK, Castrucci AM, Rollag MD, DeGrip WJ, Hogenesch JB, Provencio I, Kay SA 2002. Melanopsin (Opn4) requirement for normal light-induced circadian phase shifting. Science 298:2213–2216 [DOI] [PubMed] [Google Scholar]
- 38.Crosio C, Cermakian N, Allis CD, Sassone-Corsi P 2000. Light induces chromatin modification in cells of the mammalian circadian clock. Nat Neurosci 3:1241–1247 [DOI] [PubMed] [Google Scholar]
- 39.Albrecht U, Zheng B, Larkin D, Sun ZS, Lee CC 2001. MPer1 and mper2 are essential for normal resetting of the circadian clock. J Biol Rhythms 16:100–104 [DOI] [PubMed] [Google Scholar]
- 40.Warnecke M, Oster H, Revelli JP, Alvarez-Bolado G, Eichele G 2005. Abnormal development of the locus coeruleus in Ear2(Nr2f6)-deficient mice impairs the functionality of the forebrain clock and affects nociception. Genes Dev 19:614–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karatsoreos IN, Wang A, Sasanian J, Silver R 2007. A role for androgens in regulating circadian behavior and the suprachiasmatic nucleus. Endocrinology 148:5487–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dkhissi-Benyahya O, Gronfier C, De Vanssay W, Flamant F, Cooper HM 2007. Modeling the role of mid-wavelength cones in circadian responses to light. Neuron 53:677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishida A, Mutoh T, Ueyama T, Bando H, Masubuchi S, Nakahara D, Tsujimoto G, Okamura H 2005. Light activates the adrenal gland: timing of gene expression and glucocorticoid release. Cell Metab 2:297–307 [DOI] [PubMed] [Google Scholar]
- 44.Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U 2000. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289:2344–2347 [DOI] [PubMed] [Google Scholar]
- 45.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H 2000. Resetting central and peripheral circadian oscillators in transgenic rats. Science 288:682–685 [DOI] [PubMed] [Google Scholar]
- 46.Balsalobre A, Marcacci L, Schibler U 2000. Multiple signaling pathways elicit circadian gene expression in cultured rat-1 fibroblasts. Curr Biol 10:1291–1294 [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto T, Nakahata Y, Tanaka M, Yoshida M, Soma H, Shinohara K, Yasuda A, Mamine T, Takumi T 2005. Acute physical stress elevates mouse period1 mRNA expression in mouse peripheral tissues via a glucocorticoid-responsive element. J Biol Chem 280:42036–42043 [DOI] [PubMed] [Google Scholar]
- 48.McNamara P, Seo SP, Rudic RD, Sehgal A, Chakravarti D, FitzGerald GA 2001. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell 105:877–889 [DOI] [PubMed] [Google Scholar]
- 49.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M 2001. Entrainment of the circadian clock in the liver by feeding. Science 291:490–493 [DOI] [PubMed] [Google Scholar]
- 50.Desvergne B, Michalik L, Wahli W 2006. Transcriptional regulation of metabolism. Physiol Rev 86:465–514 [DOI] [PubMed] [Google Scholar]
- 51.Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, Michalik L, Delaunay F, Wahli W, Laudet V 2006. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor α defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol 20:1715–1727 [DOI] [PubMed] [Google Scholar]
- 52.Oster H, Damerow S, Hut RA, Eichele G 2006. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J Biol Rhythms 21:350–361 [DOI] [PubMed] [Google Scholar]
- 53.Oster H, Damerow S, Kiessling S, Jakubcakova V, Abraham D, Tian J, Hoffmann MW, Eichele G 2006. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab 4:163–173 [DOI] [PubMed] [Google Scholar]
- 54.Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL, Antoch MP, Walker JR, Esser KA, Hogenesch JB, Takahashi JS 2007. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci USA 104:3342–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuramoto Y, Hata K, Koyanagi S, Ohdo S, Shimeno H, Soeda S 2006. Circadian regulation of mouse topoisomerase I gene expression by glucocorticoid hormones. Biochem Pharmacol 71:1155–1161 [DOI] [PubMed] [Google Scholar]
- 56.Filipski E, Lemaigre G, Liu XH, Mery-Mignard D, Mahjoubi M, Levi F 2004. Circadian rhythm of irinotecan tolerability in mice. Chronobiol Int 21:613–630 [DOI] [PubMed] [Google Scholar]
- 57.Miller BH, Olson SL, Turek FW, Levine JE, Horton TH, Takahashi JS 2004. Circadian clock mutation disrupts estrous cyclicity and maintenance of pregnancy. Curr Biol 14:1367–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chappell PE, White RS, Mellon PL 2003. Circadian gene expression regulates pulsatile gonadotropin-releasing hormone (GnRH) secretory patterns in the hypothalamic GnRH-secreting GT1–7 cell line. J Neurosci 23:11202–11213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fahrenkrug J, Georg B, Hannibal J, Hindersson P, Gras S 2006. Diurnal rhythmicity of the clock genes Per1 and Per2 in the rat ovary. Endocrinology 147:3769–3776 [DOI] [PubMed] [Google Scholar]
- 60.Morse D, Cermakian N, Brancorsini S, Parvinen M, Sassone-Corsi P 2003. No circadian rhythms in testis: period1 expression is clock independent and developmentally regulated in the mouse. Mol Endocrinol 17:141–151 [DOI] [PubMed] [Google Scholar]
- 61.Lavery DJ, Lopez-Molina L, Margueron R, Fleury-Olela F, Conquet F, Schibler U, Bonfils C 1999. Circadian expression of the steroid 15 α-hydroxylase (Cyp2a4) and coumarin 7-hydroxylase (Cyp2a5) genes in mouse liver is regulated by the PAR leucine zipper transcription factor DBP. Mol Cell Biol 19:6488–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ripperger JA, Shearman LP, Reppert SM, Schibler U 2000. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev 14:679–689 [PMC free article] [PubMed] [Google Scholar]
- 63.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB 2002. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109:307–320 [DOI] [PubMed] [Google Scholar]
- 64.McDonald MJ, Rosbash M 2001. Microarray analysis and organization of circadian gene expression in Drosophila Cell 107:567–578 [DOI] [PubMed] [Google Scholar]
- 65.Gachon F, Fonjallaz P, Damiola F, Gos P, Kodama T, Zakany J, Duboule D, Petit B, Tafti M, Schibler U 2004. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev 18:1397–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grechez-Cassiau A, Panda S, Lacoche S, Teboul M, Azmi S, Laudet V, Hogenesch JB, Taneja R, Delaunay F 2004. The transcriptional repressor STRA13 regulates a subset of peripheral circadian outputs. J Biol Chem 279:1141–1150 [DOI] [PubMed] [Google Scholar]
- 67.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM 2006. Nuclear receptor expression links the circadian clock to metabolism. Cell 126:801–810 [DOI] [PubMed] [Google Scholar]
- 68.Lemberger T, Saladin R, Vazquez M, Assimacopoulos F, Staels B, Desvergne B, Wahli W, Auwerx J 1996. Expression of the peroxisome proliferator-activated receptor α gene is stimulated by stress and follows a diurnal rhythm. J Biol Chem 271:1764–1769 [DOI] [PubMed] [Google Scholar]
- 69.Inoue I, Shinoda Y, Ikeda M, Hayashi K, Kanazawa K, Nomura M, Matsunaga T, Xu H, Kawai S, Awata T, Komoda T, Katayama S 2005. CLOCK/BMAL1 is involved in lipid metabolism via transactivation of the peroxisome proliferator-activated receptor (PPAR) response element. J Atheroscler Thromb 12:169–174 [DOI] [PubMed] [Google Scholar]
- 70.Oishi K, Shirai H, Ishida N 2005. CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor α (PPARα) in mice. Biochem J 386:575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stavinoha MA, Rayspellicy JW, Hart-Sailors ML, Mersmann HJ, Bray MS, Young ME 2004. Diurnal variations in the responsiveness of cardiac and skeletal muscle to fatty acids. Am J Physiol Endocrinol Metab 287:E878–E887 [DOI] [PubMed]
- 72.Bavner A, Sanyal S, Gustafsson JA, Treuter E 2005. Transcriptional corepression by SHP: molecular mechanisms and physiological consequences. Trends Endocrinol Metab 16:478–488 [DOI] [PubMed] [Google Scholar]
- 73.Oiwa A, Kakizawa T, Miyamoto T, Yamashita K, Jiang W, Takeda T, Suzuki S, Hashizume K 2007. Synergistic regulation of the mouse orphan nuclear receptor SHP gene promoter by CLOCK-BMAL1 and LRH-1. Biochem Biophys Res Commun 353:895–901 [DOI] [PubMed] [Google Scholar]
- 74.Raspe E, Duez H, Mansen A, Fontaine C, Fievet C, Fruchart JC, Vennstrom B, Staels B 2002. Identification of Rev-erbα as a physiological repressor of apoC-III gene transcription. J Lipid Res 43:2172–2179 [DOI] [PubMed] [Google Scholar]
- 75.Fontaine C, Dubois G, Duguay Y, Helledie T, Vu-Dac N, Gervois P, Soncin F, Mandrup S, Fruchart JC, Fruchart-Najib J, Staels B 2003. The orphan nuclear receptor Rev-Erbα is a peroxisome proliferator-activated receptor (PPAR) γ target gene and promotes PPARγ-induced adipocyte differentiation. J Biol Chem 278:37672–37680 [DOI] [PubMed] [Google Scholar]
- 76.Wang J, Yin L, Lazar MA 2006. The orphan nuclear receptor Rev-erb α regulates circadian expression of plasminogen activator inhibitor type 1. J Biol Chem 281:33842–33848 [DOI] [PubMed] [Google Scholar]
- 77.Grechez-Cassiau A, Rayet B, Guillaumond F, Teboul M, Delaunay F 2008. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J Biol Chem 283:4535–4542 [DOI] [PubMed] [Google Scholar]
- 78.Kanno Y, Otsuka S, Hiromasa T, Nakahama T, Inouye Y 2004. Diurnal difference in CAR mRNA expression. Nucl Recept 2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zandieh Doulabi B, Platvoet-Ter Schiphorst M, Kalsbeek A, Fliers E, Bakker O, Wiersinga WM 2004. Diurnal variation in rat liver thyroid hormone receptor (TR)-α messenger ribonucleic acid (mRNA) is dependent on the biological clock in the suprachiasmatic nucleus, whereas diurnal variation of TR β 1 mRNA is modified by food intake. Endocrinology 145:1284–1289 [DOI] [PubMed] [Google Scholar]
- 80.Horard B, Rayet B, Triqueneaux G, Laudet V, Delaunay F, Vanacker JM 2004. Expression of the orphan nuclear receptor ERRα is under circadian regulation in estrogen-responsive tissues. J Mol Endocrinol 33:87–97 [DOI] [PubMed] [Google Scholar]
- 81.Cai W, Rambaud J, Teboul M, Masse I, Benoit G, Gustafsson JA, Delaunay F, Laudet V, Pongratz I 2008. Expression levels of estrogen receptor β are modulated by components of the molecular clock. Mol Cell Biol 28:784–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Giguere V 2002. To ERR in the estrogen pathway. Trends Endocrinol Metab 13:220–225 [DOI] [PubMed] [Google Scholar]
- 83.Levi F, Schibler U 2007. Circadian rhythms: mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol 47:593–628 [DOI] [PubMed] [Google Scholar]