To date, the existing very large genome sequence datasets of many bacterial species have not been exploited to quantify the strength and direction of...
Keywords: bacterial genomics, intergenic regions (IGRs), purifying selection, whole-genome sequencing
Abstract
Nontranslated intergenic regions (IGRs) compose 10–15% of bacterial genomes, and contain many regulatory elements with key functions. Despite this, there are few systematic studies on the strength and direction of selection operating on IGRs in bacteria using whole-genome sequence data sets. Here we exploit representative whole-genome data sets from six diverse bacterial species: Staphylococcus aureus, Streptococcus pneumoniae, Mycobacterium tuberculosis, Salmonella enterica, Klebsiella pneumoniae, and Escherichia coli. We compare patterns of selection operating on IGRs using two independent methods: the proportion of singleton mutations and the dI/dS ratio, where dI is the number of intergenic SNPs per intergenic site. We find that the strength of purifying selection operating over all intergenic sites is consistently intermediate between that operating on synonymous and nonsynonymous sites. Ribosome binding sites and noncoding RNAs tend to be under stronger selective constraint than promoters and Rho-independent terminators. Strikingly, a clear signal of purifying selection remains even when all these major categories of regulatory elements are excluded, and this constraint is highest immediately upstream of genes. While a paucity of variation means that the data for M. tuberculosis are more equivocal than for the other species, we find strong evidence for positive selection within promoters of this species. This points to a key adaptive role for regulatory changes in this important pathogen. Our study underlines the feasibility and utility of gauging the selective forces operating on bacterial IGRs from whole-genome sequence data, and suggests that our current understanding of the functionality of these sequences is far from complete.
THE ability to generate whole-genome sequence data sets from very large samples of bacterial isolates recovered from natural populations provides unprecedented power to dissect evolutionary processes. Although tests for selection are routinely carried out on the ∼85–90% of bacterial genomes corresponding to protein-coding sequences, attempts to measure the strength and direction of selection operating on nontranslated intergenic regions (IGRs) are far less common. Notable exceptions include the study by Molina and van Nimwegen (2008), who demonstrated that the number of regulatory elements per IGR is independent of genome size within bacteria, but also noted a surprising level of purifying selection operating on bacterial IGRs. However, as this study predates the advent of next-generation sequencing, very large whole-genome data sets for single species were unavailable at that time. More recently, Luo et al. (2011) found evidence for purifying selection within IGRs of a small sample (n = 13) of group A streptococcal genomes and Degnan et al. (2011) found strong evidence for sequence conservation within IGRs of eight Buchnera genomes. This observation is particularly striking in Buchnera, as it is an endosymbiont and so likely has a small effective population size (Ne). Together, these studies challenged the view held for many years that intergenic sites provide a valid proxy for neutrality (Hu et al. 2006; Wang and Chen 2013; Fu et al. 2015).
Whole-genome sequence data sets for bacteria now routinely encompass many hundreds of genomes for a single species, although currently these data have remained almost completely untapped with respect to examining selection on IGRs. In fact, despite the studies mentioned above, many commonly used pipelines and databases by default exclude IGRs altogether, with the focus instead on defining sets of “core” or “accessory” genes, upon which phylogenetic, epidemiological, or evolutionary analyses are then carried out (Jolley and Maiden 2010; Sheppard et al. 2012; Maiden et al. 2013; Feil 2015; Page et al. 2015; Maiden and Harrison 2016). One explanation for this apparently casual dismissal of IGRs is a lack of “off-the-shelf” methodology to measure selection on these sequences; the standard approach for protein-coding sequences, the dN/dS ratio, being invalid. In addition, there may be a prevailing sense that IGRs are technically challenging to work with, owing to low levels of constraint, poor annotation, and a high frequency of indels. The approach by Fu et al. (2015) is a rare exception which challenges this view. These authors generated a core genome consisting of both genes and IGRs for Salmonella enterica serovar Typhimurium, and in so doing demonstrated the feasibility of incorporating IGRs into routine analysis. Furthermore, these authors showed that IGRs contribute meaningful signal to increase discriminatory power for phylogenetic and epidemiological analyses (Fu et al. 2015).
The paucity of studies aimed at systematically measuring selection on nontranslated IGRs is strikingly at odds with the many recent examples demonstrating the phenotypic impact of mutations in riboswitches, small RNAs (sRNAs), promoters, terminators, and regulator binding sites (Waters and Storz 2009). Single nucleotide polymorphisms (SNPs) or small indels within these elements can have major phenotypic consequences. For example, in a recent genome-wide association study, 13 intergenic SNPs were found to be significantly associated with toxicity in Methicillin-resistant Staphylococcus aureus (MRSA), and 4 of these were experimentally validated (Laabei et al. 2014). In Mycobacterium tuberculosis, mutations within the eis promoter region increase expression of Eis, an enzyme which confers resistance to kanamycin and promotes intracellular survival (Casali et al. 2012). In addition to those studies focusing on naturally occurring mutations, knock-out experiments on regulatory RNAs have also confirmed their key roles in virulence and other important phenotypes such as competence. For example, the Salmonella sRNA IsrM is important for invasion of epithelial cells and replication inside macrophages (Gong et al. 2011). In S. aureus, the σB-dependent RsaA sRNA represses the global regulator MgrA; this decreases the severity of acute infection and promotes chronic infection (Romilly et al. 2014). In S. pneumoniae, the srn206 noncoding RNA is involved in competence modulation (Acebo et al. 2012).
These well-characterized regulatory elements are clearly expected to be under strong purifying selection, but there remain no broad measures of the commonality of constraint operating on IGRs at an intraspecies level. There is also currently little understanding of which intergenic regulatory elements are under strongest selection, whether a signal of selection can be detected even for those intergenic sites for which there is no known function, or to what extent positive (as well as negative) selection may be operating on IGRs. Here we use two independent approaches to address these questions: The first method is based on the established logic of site frequency spectra [the proportion of singleton mutations (PSM)], while the second is a modification of dN/dS (dI/dS; where dI is the number of intergenic SNPs per intergenic site). We apply these approaches to large, whole-genome data sets from six diverse bacterial species: Escherichia coli, S. aureus, S. enterica, Streptococcus pneumoniae, Klebsiella pneumoniae, and M. tuberculosis. With the exception of M. tuberculosis, our results demonstrate that the overall strength of selective constraint on intergenic sites in bacteria is intermediate between that operating on synonymous and nonsynonymous sites. This observation does not significantly alter even when all major regulatory IGR elements are removed from the analysis, consistent with a substantial level of cryptic functionality in these sequences. We also compare the strength and direction of selection operating on different types of regulatory element within IGRs, and note strong evidence of positive selection acting on promoters in M. tuberculosis.
Materials and Methods
Data selection
For S. aureus, S. pneumoniae, K. pneumoniae, and M. tuberculosis, isolates were selected from recently published data (Casali et al. 2014; Chewapreecha et al. 2014; Holt et al. 2015; Reuter et al. 2015). For S. enterica, isolates were selected from those whole genomes sequenced routinely by the Gastrointestinal Bacteria Reference Unit at Public Health England. Recent large-scale bacterial genome sequencing projects have been primarily motivated by efforts to understand features which are important for public health, such as disease transmission, virulence, and antibiotic resistance. Consequently, the data sets may be poorly representative of the population as a whole, with disproportionate weight given to lineages of particular clinical significance. For example, the S. aureus data were generated as part of a retrospective study of hospital-acquired MRSA in the United Kingdom (Reuter et al. 2015), and the majority of these isolates corresponded to a single clonal lineage, CC22 (EMRSA-15). We therefore subsampled the data sets to include each major lineage and a random sample from the overrepresented clonal complexes. A complete list of all isolates used in the analysis is given in Supplemental Material, Table S1.
Sequencing, mapping, and SNP calling
For each species except E. coli, reads were downloaded from the European Nucleotide Archive (ENA) (http://www.ebi.ac.uk/ena). For E. coli, completed genome sequences were downloaded from the National Center for Biotechnology Information (NCBI), and sheared into reads with ArtificialFastqGenerator (Frampton and Houlston 2012). The isolates were mapped against a single reference genome for each species (as shown in Table 1) using SMALT-0.7.6 (https://sourceforge.net/projects/smalt). SAMtools-0.1.19 (Li et al. 2009) was used to produce variant call format (VCF) files, which were filtered to call SNPs. SNPs were only called if they passed all of the following thresholds: depth ≥ 4, depth per strand ≥ 2, proportion of reads supporting the SNP ≥ 0.75, base quality ≥ 50, map quality ≥ 30, estimated site allele frequency ≥ 0.95, strand bias ≥ 0.001, map bias ≥ 0.001, and tail bias ≥ 0.001. Consensus Fasta sequences were then used to produce an alignment for each species.
Table 1. The data used in the analysis.
| Species | Data source | No. of isolates | % GC | Reference genome | RC genes | RC IGRs | IC genes | IC IGRs | SC genes | SC IGRs |
|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | NCBI complete genome | 157 | 50.8 | MG1655 | 4305 | 3647 | 3164 | 2342 | 2873 | 2060 |
| S. enterica | Public Health England | 366 | 52.2 | Typhimurium_D23580 | 4554 | 3777 | 3617 | 2830 | 3114 | 2456 |
| K. pneumoniae | Holt et al. (2015) | 208 | 57.7 | NTUH_K2044 | 4787 | 4006 | 3954 | 3150 | 2954 | 2453 |
| S. aureus | Reuter et al. (2015) | 132 | 33.2 | HO_5096_0412 | 2405 | 2084 | 2131 | 1704 | 2057 | 1609 |
| S. pneumoniae | Chewapreecha et al. (2014) | 264 | 39.5 | ATCC_700669 | 2183 | 1846 | 1574 | 1198 | 1373 | 932 |
| M. tuberculosis | Casali et al. (2014) | 144 | 65.6 | H37Rv | 4069 | 3135 | 3806 | 2940 | 3332 | 2691 |
RC, relaxed-core; IC, intermediate-core; SC, strict-core.
Validation of singleton SNPs
As singleton SNPs are potentially vulnerable to poor quality data, we performed a thorough analysis of the SNPs to validate their quality. This was based on analyzing three metrics: depth of coverage, proportion of reads supporting the variant, and the Phred quality (Q) score in both singletons and nonsingletons. Q is related to the per-base error probability according to the following equations:
In Illumina reads, the per-base Q score is ∼Q30, equating to one error every 10−3 bases. However, this error rate is substantially reduced by sequencing to high coverage, and then mapping the reads to the reference genome. For each species in our analysis (with the exception of E. coli), the sequencing depth was 50–100 times per isolate. For E. coli, we simulated reads with no errors, using the complete genome sequences, and then mapped these synthetic reads to the standard reference genome (MG1655).
To validate our SNPs, we used the mapping information in the VCF files. We focused on three metrics: the depth of coverage, the proportion of reads supporting the variant, and the Q score for the position (which takes into account the per-base-per-read error rate, and the coverage at the position). We split our SNPs into singletons and nonsingletons to check for singleton associated biases.
IGR identification and core genome definition
Each reference genome was annotated using Prokka-1.11 (Seemann 2014). This annotation was used to extract genes and IGRs (IGRs >1000 bp in length were excluded), and three core sets of genes and IGRs were defined for each species. The “relaxed-core” consisted of all genes and IGRs present in at least two genomes, the “intermediate-core” consisted of all genes and IGRs with >90% sequence present in >95% of isolates, and the “strict-core” consisted of genes and IGRs with >90% sequence present in >99% of isolates.
Calculation of dN/dS and dI/dS
Core gene and intergenic sequences were extracted from the alignments and concatenated to produce gene and intergenic alignments (reverse oriented genes were reverse complemented so all genes were in sense orientation). The codons within the gene alignment were shuffled, and the gene alignment was split into two (referred to as a and b). The YN00 program from the PAML suite (Yang 2007) was used to calculate dN/dS values by the Nei and Gojobori (1986) and Yang and Nielson (2000) methods in a pairwise manner for both gene alignments a and b. The results were almost identical, and so we used the Nei and Gojobori (1986) method as it was computationally less demanding, and enabled the results to be compared directly with the dI values. SNPs were counted between isolates in a pairwise manner from the intergenic alignment, and dI was calculated by dividing the number of SNPs by the length of the alignment, before applying a Jukes–Cantor distance correction (Jukes and Cantor 1969). For both the gene and intergenic alignments, N’s were removed from the alignment in a pairwise manner to ensure that all possible data were used. The dS values from gene alignment a were used to calculate dN/dS and dI/dS, and the dS values from gene alignment b were used as a proxy for divergence time. This ensured that when plotting dN/dS and dI/dS against dS, the dS values on each axis were calculated independently, thus controlling for statistical nonindependence.
Correcting dN/dS and dI/dS calculations for mutation biases and base composition
To confirm that the model we used to calculate dN/dS and dI/dS accurately reflects the null expected under neutrality, we simulated neutral divergence of the reference genomes based on the observed mutational spectra, then recalculated dN/dS and dI/dS from the simulated sequences. Any deviation from parity (dN/dS = 1) reflects the fact that we have not accurately incorporated mutation bias, in particular the strong AT pressure in bacterial genomes (Balbi et al. 2009; Hershberg and Petrov 2010; Hildebrand et al. 2010) and base composition, into our models. However, by calculating the magnitude of the deviation between the simulated sequences and parity we can correct for this bias.
We first calculated the per-site mutation bias for the six mutation types for each species (Figure S1 in File S1). We then simulated neutral mutations on a sequence of concatenated genes and IGRs to a divergence of 1% from the original sequence for 50 replicates. We then calculated dN/dS and dI/dS between pairwise comparisons of these 50 replicates. This gave us an expectation of dN/dS and dI/dS under neutral conditions, taking into account mutation biases and base composition. We then computed observed/expected (simulated) dN/dS and dI/dS ratios, thus providing corrected estimates. We did this for alignments of each intergenic element considered [promoters, terminators, ribosome binding sites (RBSs), noncoding RNAs, and unannotated sites] to also correct these estimates.
RBS, promoter, noncoding RNA, and terminator annotation
Promoter and terminator predictions were obtained using the PePPER web server (de Jong et al. 2012). Noncoding RNA annotations were obtained from the reference genome annotation GFF file produced by Prokka, where they were labeled as “misc_RNA.” RBS annotations were predicted using RBSfinder (Suzek et al. 2001).
Code availability and computation
All of the code used in the analysis is available at https://github.com/harry-thorpe/Intergenic_selection_paper under the GPLv3 license. The complete analysis can be reproduced by running a single script, using any alignment and annotation files as inputs. Full instructions are available in the GitHub repository. All computations were performed on the Cloud Infrastructure for Microbial Bioinformatics (CLIMB) (Connor et al. 2016). All figures were produced using the R package ggplot2 (Wickham 2009).
Data availability
The data sets supporting the conclusions of this article are available in the ENA (http://www.ebi.ac.uk/ena), except the E. coli sequences which were downloaded from the NCBI (http://www.ncbi.nlm.nih.gov/). The accession numbers of all strains used in the analysis are given in Table S1.
Results
Species and data selection
We used existing large whole-genome sequence data sets for six diverse bacterial species: E. coli, S. aureus, S. enterica, S. pneumoniae, K. pneumoniae, and M. tuberculosis. These species are diverse in terms of phylogeny (representing Gram-positive and Gram-negative taxa), in terms of population structure (ranging from the highly clonal M. tuberculosis to the freely recombining S. pneumoniae), and in terms of ecology. The K. pneumoniae and E. coli data include isolates from environmental sources and disease, the S. aureus and S. pneumoniae data includes isolates from asymptomatic carriage, and M. tuberculosis is an intracellular pathogen. The GC content of these species range from 32.9% (S. aureus) to 65.6% (M. tuberculosis) (Table 1). The diversity of these species provides a means to examine the robustness of the methods against possible confounders such as rates of recombination, demographic effects, effective population size, and population structure. In cases where very large data sets (1000s of genomes) were available, we subsampled representative strains as described in Methods. A complete list of all isolates used in the analysis is given in Table S1.
For each species, we mapped the sequence reads to a single reference genome (Table 1), and defined alternative sets of core genes and IGRs using different frequency thresholds. Defining core gene sets on the basis that each core gene is universally present, or present at a very high frequency, among all the sequenced genomes is an established first step in bacterial comparative genomics. This simplifies the analysis by removing the problem of missing data (genes), by excluding mobile elements (e.g., phage and plasmids), and it also reduces the problem of potentially conflicting signals resulting from high rates of recombination or atypical selection pressures. However, it is likely that the most frequently observed genes and IGRs that correspond to a strict-core are also the most selectively constrained, thus this approach potentially imposes a bias. To address this, we analyzed three different sets of core genes and IGRs for each species defined according to different frequency thresholds. The relaxed-core represents all genes and IGRs present in at least two genomes, the intermediate-core includes all genes and IGRs that are present in >95% isolates, and the strict-core includes all genes and IGRs present in >99% of isolates. The number of genes and IGRs included in each data set is given in Table 1. The strict-core IGR data set included at least 50% of the corresponding relaxed-core IGRs for each species, with the biggest potential bias in S. pneumoniae and E. coli, where the strict-core IGRs corresponded to 50.5 and 56.5% of the relaxed-core IGRs, respectively. For the intermediate-core data sets, 64.9 and 64.2% of the relaxed-core IGRs were included for S. pneumoniae and E. coli, respectively.
Sequence properties of genes and IGRs
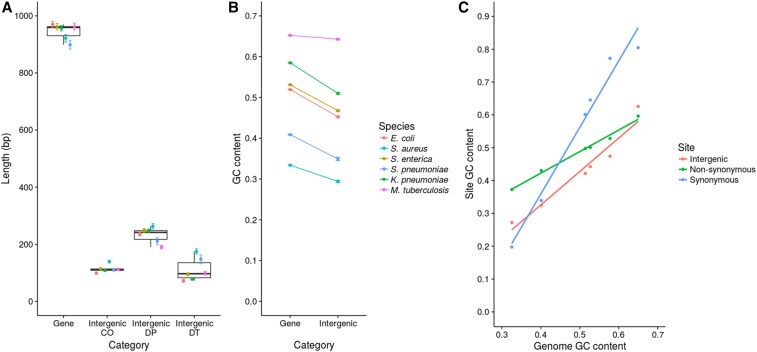
IGRs were identified based on reference genome annotations as described in Methods. The size distribution and GC content of both genes and assigned IGRs are shown in Figure 1. Figure 1A shows that IGRs with a predicted promoter at each end tend to be larger than double terminator regions and cooriented regions. This is partly explained by the fact that many cooriented regions are small spacers within operons. The GC content of IGRs is lower than in protein-coding sequences (Figure 1B); although this difference is far less marked in M. tuberculosis, it is statistically significant in all species (P < 10−16, Mann–Whitney U-test).
Figure 1.
Summary of the sequence properties of genes and IGRs. (A) Length distributions of genes and IGRs. IGRs were divided into three groups according to the orientation of the flanking genes: cooriented (CO) regions are flanked by genes in the same orientation, double promoter (DP) regions are flanked by 5′ gene starts, and double terminator (DT) regions are flanked by 3′ gene ends. The points and error bars represent mean ± SE. (B) GC contents of genes and IGRs. GC contents were calculated for each gene and IGR individually. The points and error bars represent the mean ± SE. (C) GC contents of different site classes compared to genome GC content. The GC content of synonymous, nonsynonymous, and intergenic sites was calculated and compared with the genome GC content for each species. The steepness of the slope indicates the amount of constraint on the GC content of the site class (shallower slopes indicate stronger constraint).
Muto and Osawa (1987) showed that fourfold degenerate sites exhibit the widest range of GC content across a diverse sample of genomes (that is, these sites show the most extreme values); whereas nondegenerate second codon positions exhibit the narrowest range, with first and third sites being intermediate. These authors noted that this variation in the range of GC content between different site categories mirrors the selective constraints on those sites, with second codon positions being the most constrained because they are in all cases nondegenerate (Muto and Osawa 1987; Rocha and Feil 2010). We repeated this analysis using synonymous, nonsynonymous, and intergenic sites (Figure 1C). Our data are consistent with that of Muto and Osawa: the synonymous sites exhibit the widest range of GC content (steepest slope) and the nonsynonymous sites exhibit the narrowest range of GC content (shallowest slope). However, we also note that the slope for intergenic sites is intermediate between the synonymous and nonsynonymous sites. If the original interpretation by Muto and Osawa (1987) is correct, this implies that the strength of selective constraint on intergenic sites is also intermediate between that operating on synonymous and nonsynonymous sites. Below we describe detailed analyses which examine this possibility in more detail.
The PSM is consistent with an intermediate strength of selective constraint on intergenic sites
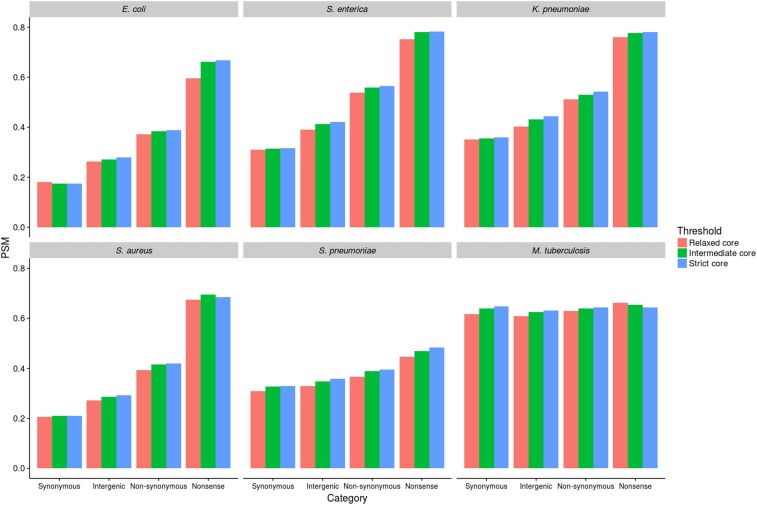
To measure the frequency of strongly deleterious intergenic mutations relative to synonymous, nonsynonymous, and nonsense mutations within coding regions, we used a simple method based on site frequency spectra. Similar methods have been used on noncoding DNA in eukaryotes (Drake et al. 2006) and in bacteria, albeit on a much smaller scale than the current study (Luo et al. 2011). Mutations affecting sites under strong selective constraint are more likely to be quickly purged by selection before they begin to rise in frequency, thus are also more likely to be very rare. Here, we define “very rare” mutations simply as those observed only once in the data set (singletons). It is thus possible to gauge the proportion of strongly deleterious SNPs for a given site category simply by computing the proportion of those SNPs that are singletons (PSM). To check to what degree the definition of core IGRs imposes a bias, we carried out the analysis using the three thresholds as defined above (relaxed-core, intermediate-core, and strict-core). We first considered four mutation categories: intergenic, synonymous, nonsynonymous, and nonsense. The PSM values for each of these mutation types, for all six species, are shown in Figure 2. An analysis of all individual genes and IGRs is given in Figure S2 in File S1 (intermediate-core only).
Figure 2.
PSM analysis of selection on different mutation categories. PSM values were calculated by dividing the number of singleton SNPs (those present in only one genome) by the total number of SNPs within that mutation category.
This analysis reveals a consistent trend across five of the six species, the exception being M. tuberculosis. Nonsense mutations had the highest PSM values, indicating the highest proportion of strongly deleterious mutations, followed by nonsynonymous mutations, intergenic mutations, and finally synonymous mutations. Thus, for five of the six species, PSM values for intergenic sites were intermediate between the synonymous and nonsynonymous PSM values. It follows that the proportion of SNPs at intergenic sites that are highly deleterious, and therefore purged rapidly by purifying selection, is intermediate between the equivalent proportions for synonymous and nonsynonymous sites.
Although comparisons of PSM values between species are not valid, as species-specific factors (e.g., the rate of recombination) will also affect PSM, it is reasonable to assume that these potential confounders are at least consistent between different mutation types within a single species. This is strongly evidenced by the consistency of the relative strengths of selective constraint operating on each mutation type (nonsense > nonsynonymous > intergenic > synonymous). The same trend is observed when individual genes and IGRs were analyzed separately (Figure S2 in File S1). Moreover, the pattern is highly robust to the definition of core genes and IGRs. Although the strict-core gene and IGR sets within each species correspond to marginally higher PSM values (as expected and consistent with a higher selective constraint), again the relative trends within each species remain robust. We are therefore confident that this analysis is not confounded by biases resulting from species-specific factors, or from selecting unrepresentative genes and IGRs. In M. tuberculosis, the exceptional species, there was very little difference between all mutation categories, and PSM scores were high in all cases. Multiple interpretations of the apparent patterns of selection and the high frequency of rare variants in M. tuberculosis have been discussed in the literature. These include very weak purifying selection, short coalescent time, linkage (background selection), a combination of purifying and positive selection, rapid demographic expansion combined with bottlenecks (leading to a reduction in the effective population size and increased drift), selective sweeps, and diversifying selection (Hershberg et al. 2008; Namouchi et al. 2012; Pepperell et al. 2013). We consider some of these possibilities in the context of our results in more detail below.
We recognize that this analysis is potentially vulnerable to sequencing errors, as these are most likely to generate singleton SNPs. The consistency of the results across five diverse species is reassuring, as this is very difficult to reconcile with a high error rate without assuming this systematically affects some site categories more than others. Nevertheless, to gauge whether our analysis has been affected by a high frequency of error-derived singleton SNPs, we repeated the analysis by first removing all singleton SNPs and instead computing, for each site category, the proportion of doubleton mutations (PDM). These are SNPs present in exactly two genomes within each sample; although still rare, these are a priori far less likely to have been generated by random sequencing error than singleton SNPs. PDM values were ordered as nonsense > nonsynonymous > intergenic > synonymous for all species except M. tuberculosis and S. pneumoniae (Figure S3 in File S1). Thus, the only discrepancy between the PSM and PDM results was S. pneumoniae, where intergenic < synonymous. However, we note this discrepancy is marginal, and the distinction between site categories is less robust for this species even when considering PSM, probably reflecting very high rates of recombination in this species.
To further examine to what extent singleton SNPs may have been generated by sequencing error, we carried out a detailed comparative analysis of the quality scores of singleton and nonsingleton SNPs (Figure S4 in File S1). We analyzed three metrics: depth of coverage, proportion of reads supporting each variant, and the Q score in both singletons and nonsingletons. The analysis revealed that the vast majority (>99%) of all SNPs were of extremely high quality, and that there are negligible differences in the quality scores between singleton and nonsingleton SNPs. For example, across all species 99.5% of singleton SNPs and 99.8% of nonsingleton SNPs had a Q score of >100. This quality score corresponds to an error rate of 10−10, or equivalently 1 erroneous SNP every 2000 genomes (given a 5-Mb genome). Given these combined checks, we are highly confident that errors in the singleton SNPs have not confounded our analysis.
The signal of purifying selection on intergenic sites is time dependent
To further examine selective constraint on intergenic sites, we extended the logic of dN/dS by computing dI/dS, where dI = intergenic SNPs per intergenic site. dI has previously been used in M. tuberculosis as a neutral reference by calculating dI/dS for individual IGRs using neighboring genes as a source of synonymous sites (Wang and Chen 2013). In contrast, we drew pairwise comparisons by pooling sites across the whole genome, and used the genome-wide dI as the numerator and the genome-wide dS as the denominator. We computed genome-wide dN/dS in the same way to draw valid comparisons between the strength of selection on intergenic sites and nonsynonymous sites, both relative to synonymous sites.
Previous work has shown that dN/dS decreases with divergence time due to a lag in purifying selection, which operates much more strongly on nonsynonymous than synonymous sites as the former are more likely to be slightly deleterious (Rocha et al. 2006; Namouchi et al. 2012). We tested for the same time dependence in dI/dS by comparing pairs of very closely related genomes [within “clonal complexes” (CCs); where dS < 0.003] with those representing more distantly related genomes (“between CCs”; dS > 0.003). This analysis was also carried out for all three alternative gene sets (relaxed, intermediate, and strict-core; Figure 3). All genome comparisons within M. tuberculosis were defined as “within CC” due to the very low level of variation in this species. We also plotted, for each pair of isolates and for each species, dN/dS and dI/dS against dS to further explore the impact of divergence time on dI/dS (intermediate-core only; Figure S5 in File S1).
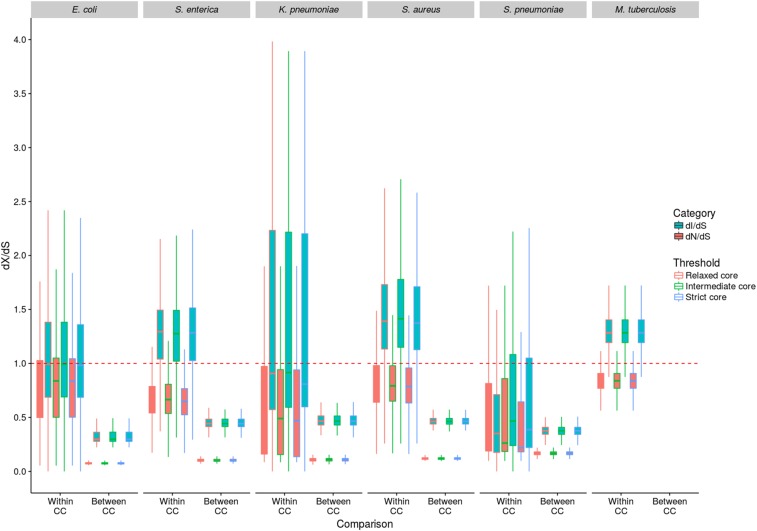
Figure 3.
dN/dS and dI/dS analysis of selection. dN/dS and dI/dS were calculated between isolates in a pairwise manner. The results were categorized into within-CC (dS < 0.003) and between-CC (dS > 0.003) comparisons, to account for the effect of divergence time on the observed levels of selection. All comparisons between M. tuberculosis isolates were classified as within CC due to the extremely low level of diversity in this species. The dashed red line shows where dN/dS and dI/dS = 1, and therefore indicates neutrality.
Figure 3 shows that for each species, dI/dS is consistently greater than dN/dS for both within- and between-CC comparisons. The between-CC dN/dS and dI/dS values are universally <1; but the within-CC values are more equivocal, with the dN/dS values being mostly <1; and the dI/dS values being <1 in E. coli, K. pneumoniae, and S. pneumoniae, and >1 in S. enterica, S. aureus, and M. tuberculosis. This trend is consistent across all three core gene and IGR sets, with very little difference in dI/dS and dN/dS values between the sets. Low dN/dS and dI/dS values (particularly in the between-CC comparisons) are strong evidence of purifying selection on nonsynonymous and intergenic sites, and lower dN/dS values compared to dI/dS values indicate stronger constraint on nonsynonymous sites than intergenic sites. It is worth noting that this observation (dI < dS) has previously been interpreted as evidence for positive selection on synonymous sites (Wang and Chen 2013). Given previous work and the results of the PSM analysis, we argue instead that it confirms greater selective constraint on intergenic sites than on synonymous sites. Moreover, the difference between within- and between-CC comparisons is evidence of time dependence consistent with ongoing purifying selection operating over increasing divergence, as noted previously for nonsynonymous sites (Rocha et al. 2006). For four of the five species for which such a comparison was possible (E. coli, S. aureus, S. enterica, and K. pneumoniae), dN/dS and dI/dS were both significantly higher for within-CC comparisons than between-CC comparisons (P < 10−16, Mann–Whitney U-test). These differences are expected if nonsynonymous and intergenic SNPs are preferentially purged (relative to synonymous SNPs) over divergence time, although we recognize that our pairwise methodology might lead to an amplification of these differences due to the oversampling of long internal branches in the between-CC comparisons. In S. pneumoniae, dN/dS was significantly higher for within-CC comparisons compared to between-CC comparisons (P < 10−16, Mann–Whitney U-test) but dI/dS was not (P = 0.19). It is possible that the signal of time dependence is weaker in this species owing to high rates of recombination (Chaguza et al. 2016). The statistical analysis described above was carried out based on the intermediate-core gene set. As there was negligible difference between the three core gene and IGR sets in both the PSM and dI/dS analyses presented thus far, we performed all subsequent analyses on the intermediate-core sets (where at least 90% of genes and IGRs are present in at least 95% of isolates).
We also plotted, for each pair of isolates for each species, dN/dS and dI/dS against dS (based on the intermediate-core only; Figure S5 in File S1). In the case of E. coli, S. aureus, S. enterica, and K. pneumoniae, a large number of points are evident at very low values of dS; these reflect the presence of clusters of closely related genomes in these species (i.e., CCs). The absence of significant clonal clustering in S. pneumoniae reflects high rates of recombination, and can help to explain the lack of significant difference within and between CCs in this species as noted above. However, for all species except S. enterica and M. tuberculosis, there is a significant decrease of both dN/dS and dI/dS against dS (P < 10−16, Spearman’s correlation). The time dependence of dN/dS potentially poses a problem for comparing between species, as those species with longest time to most recent common ancestor will appear to be under stronger selection (as dN/dS decreases with divergence time). However, Figure S5 in File S1 shows that dN/dS decreases very quickly initially and then begins to plateau very early, suggesting that this is not a major problem in our analysis.
As noted above, the genetic diversity within M. tuberculosis is so low that between-CC comparisons were not possible, as the dS for all pairwise comparisons was <0.003. Values of dN/dS are ∼0.8 in this species, which is comparable to the within-CC values for all the other species, and is slightly higher than that reported previously for this species (Hershberg et al. 2008; Pepperell et al. 2013). Both the observation of high dN/dS in M. tuberculosis and the PSM analysis described above are consistent with, though not demonstrative of, weak purifying selection in this species. It has been argued that weak purifying selection in M. tuberculosis reflects its lifestyle as an obligate pathogen subject to frequent bottlenecks, and thus a reduction in effective population size (Hershberg et al. 2008). Contrary to this view, Namouchi et al. (2012) highlighted the absence of the classic footprints of genome degradation expected to result from increased drift, and that, in contrast to Hershberg et al. (2008), nonsynonymous SNPs are more common (compared to synonymous SNPs) in terminal branches of the tree (as predicted if purifying selection is operating). Moreover, other authors have reported evidence of both positive and purifying selection (Farhat et al. 2013; Pepperell et al. 2013), and even diversifying selection in M. tuberculosis (Osório et al. 2013). Given this complex picture, it is possible that our results represent a mixture of contrasting forces acting over short coalescence times, converging on a signal that is indistinguishable from very weak purifying selection. In favor of this argument, the diversity in M. tuberculosis is so low that only a small number of mutations would need to be positively selected to have a large impact on the patterns observed, and our M. tuberculosis sample is enriched for antibiotic resistance which is known to be positively selected (Farhat et al. 2013; Pepperell et al. 2013; Casali et al. 2014).
Purifying selection on intergenic sites is strongest near gene borders
Although values of dI/dS < 1 are consistent with stronger selective constraint on intergenic sites than on synonymous sites, this could also arise due to slower mutation rates within IGRs than in coding regions. This might be expected if a nonnegligible fraction of mutations arose during transcription, which would also affect intergenic sites near to the gene border (Chen and Zhang 2013). The demonstration of the time dependence of dI/dS, specifically the difference between within- and between-CC comparisons, acts to mitigate these concerns; but as a further check we calculated dI/dS values from intergenic sites immediately upstream of genes (30 bp upstream from the start codon). If intergenic sites immediately upstream of genes are transcribed, and transcription-derived mutation significantly elevates dS, then dI/dS should approach 1 in these regions. However, for each species (except M. tuberculosis) we noted the opposite: dI/dS immediately upstream of genes was in fact lower than dI/dS for intergenic sites in general (P < 10−16, Mann–Whitney U-test), suggesting that transcription-derived mutation is not confounding our analysis (Figure S6 in File S1). This suggests that intergenic sites close to the start of genes are under particularly strong purifying selection, which may be due to the presence of regulatory elements upstream of genes, or selection for messenger RNA stability to enable efficient translation (Molina and van Nimwegen 2008).
The strength of purifying selection on different classes of intergenic regulatory element
Above we demonstrate that intergenic sites in the majority of bacterial species are likely to be under selective constraint. However, we have not yet considered to what extent the strength of purifying selection may vary within a given IGR according to the presence or absence of different regulatory elements. It would be expected that sites associated with known or predicted regulatory elements should be under stronger selective constraint than sites with no known function, and it may be the case that certain classes of regulatory element are under stronger selective constraint than others. To test this possibility, we identified all RBSs, noncoding RNAs, predicted promoters, and Rho-independent terminators for each species (see Methods). We then applied both methods (PSM and dI/dS) to compare the strength of selective constraint on these different elements, as well as on all the remaining intergenic sites that do not correspond to any of these elements (“unannotated sites”).
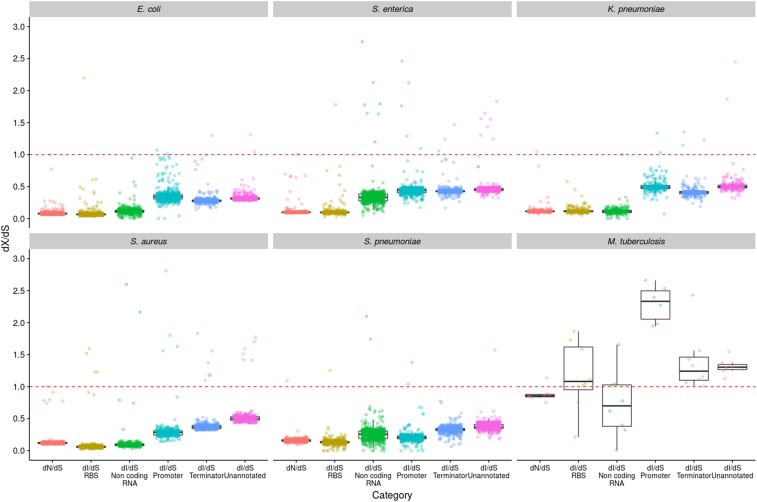
With the exception of M. tuberculosis, we note that the PSM values for the RBSs tend to be higher than for other regulatory elements and unannotated sites (Figure S7 in File S1), suggesting that these elements are particularly strongly constrained. In E. coli, S. aureus, and K. pneumoniae, noncoding RNAs also appear to be strongly constrained. In contrast, promoters and terminators tend to exhibit similar PSM values to the unannotated sites. We next drew the same comparisons using dI/dS values (Figure 4). This confirmed the observation from the PSM analysis of particularly strong purifying selection on RBSs in all species except M. tuberculosis, and in noncoding RNAs of E. coli, S. aureus, and K. pneumoniae. Indeed, the dI/dS values for RBSs and noncoding RNAs in these species are similar to the dN/dS values, suggesting that the strength of purifying selection on these elements is similar to that operating on nonsynonymous sites (Figure 4). This analysis also reveals that predicted promoters and terminators tend to be under more similar levels of selective constraint to unannotated sites. The two analyses (PSM and dI/dS) are highly concordant, with both showing a clear signal of strong purifying selection on RBSs and noncoding RNAs in the same species, and showing that promoters and terminators are under similar levels of purifying selection to unannotated sites. Importantly, however, it is clear that (with the exception of M. tuberculosis) dI/dS is <1 in all cases, including unannotated sites, which suggests a high level of constraint even when excluding major regulatory elements.
Figure 4.
dI/dS analysis of selection on different regulatory elements. dI/dS was calculated between isolates in a pairwise manner, and the results were binned by dS (bin width = 0.0001) to control for oversampling of very closely related isolates (such as those belonging to the same CC). The genome-wide dN/dS values are included to enable comparisons to be made between nonsynonymous sites and the different regulatory intergenic sites. The dashed red line shows where dN/dS and dI/dS = 1, and therefore indicates neutrality.
We then further examined the strength of selective constraint on transcriptional terminators which appear to be under only marginally stronger selective constraint than unannotated sites. Transcriptional terminators consist of a stem-loop structure, and it seemed likely that the stem should be under stronger constraint than the loop, due to requirement of the stem sequence to maintain complementary base pairing. To test this, we calculated dI/dS for the terminator stems and loops separately (Figure S8 in File S1). As expected, the stem dI/dS values are substantially lower than those for the loop, confirming that the stem is more constrained than the loop, and providing additional validation of our methodology. However, we also note that the dI/dS values for the loops are clearly <1 in S. aureus, E. coli, and S. pneumoniae, indicating they are not completely free to change in these species.
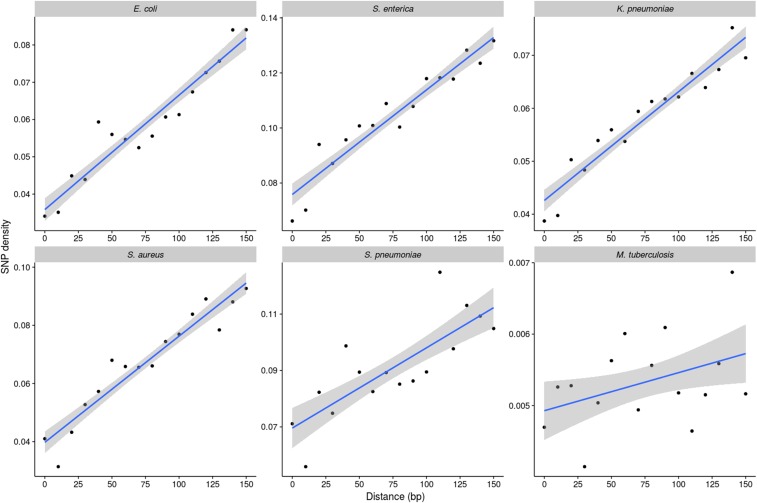
As discussed, our analysis points to considerable selective constraint (relative to synonymous sites) on intergenic sites even when the major regulatory elements are excluded. We noted earlier (Figure S6 in File S1) that the strength of selective constraint appears to be particularly high within 30 bp of the gene borders. To examine to what extent this trend reflects the presence of known regulatory elements, we first excluded these elements and then investigated SNP density as a function of the distance from gene start codons in cooriented IGRs (where the genes flanking these regions are in the same orientation). In each species (except M. tuberculosis), SNP densities increased with distance from the 5′ gene starts (P < 10−4, Spearman’s correlation) (Figure 5), demonstrating that the relatively high level of selective constraint on intergenic sites near gene borders (noted earlier) remains even when excluding promoters, terminators, RBSs, and noncoding RNAs.
Figure 5.
Analysis of SNP densities within cooriented IGRs (those flanked by genes in the same orientation as each other). SNP densities were calculated in 10-bp windows moving away from the gene start codon by dividing the number of SNPs by the number of IGRs of that length or greater (to normalize for the unequal lengths of IGRs). Only unannotated intergenic sites were considered in the analysis.
Evidence for positive selection within IGRs of M. tuberculosis
Throughout this analysis, M. tuberculosis has repeatedly proved the exception as it exhibits very little evidence of purifying selection on protein-coding sequences, and even some evidence of positive selection on IGRs. Considering different intergenic regulatory elements separately reveals that positive selection is strongly associated with predicted promoter regions. The mean dI/dS for M. tuberculosis promoters was 2.8 (Figure 4), and the vast majority (97%) of comparisons exhibit a dI/dS of >1 (Figure S9 in File S1). This result is not solely a consequence of the approach we have used to calculate dI/dS, which corrects for mutation biases and base composition, as even without this correction the mean dI/dS for promoters is 1.9. The evidence for positive selection is highly statistically significant. Of the 10,513 promoter sites, 99 have experienced a SNP, compared with 6330 of the 954,745 synonymous sites (P < 0.0001 by a Fisher’s exact test). We further confirmed significance by resampling the predicted promoter and synonymous sites 1000 times and comparing the distributions with a z-test (P < 10−16). Figure 3 shows that the within-CC values of dI/dS for S. enterica and S. aureus are >1, thus are also indicative of positive selection. We therefore also calculated dI/dS separately for the different intergenic elements for all the other five species, but restricting the analysis to within-CC comparisons. This did not reveal any evidence of positive selection on promoters or any other IGR elements (Figure S10 in File S1).
To further investigate the potential functional relevance of promoter SNPs in the M. tuberculosis data set, we identified genes downstream of predicted promoters harboring SNPs (Table S2). The 71 promoter SNPs identified corresponded to 58 genes; 11 genes were identified for which the corresponding promoter harbored 2 SNPs, and 1 gene where the promoter harbored 3 SNPs. Many of the downstream genes are known to play a key role in virulence, resistance, or global regulation. For example, eight genes were transcriptional regulators, and promoters in four of these experienced two independent SNPs: MT0026 (a putative HTH-type regulator), CmtR (a cadmium sensing repressor; Chauhan et al. 2009), and WhiB2 and WhiB4 (transcription factors; Larsson et al. 2012; Smith et al. 2012; Ma et al. 2015). Six genes were members of the PE/PPE protein family that are recognized virulence factors (Fishbein et al. 2015). Genes known to play a role in resistance were also identified, including one mutation in the ethA promoter; mutations in this promoter have previously been implicated in resistance to ethionamide (Casali et al. 2014). The promoter for the alanine dehydrogenase gene ald is the only example harboring three independent SNPs, loss of function of this gene has recently been shown to confer resistance to D-cycloserine (Desjardins et al. 2016). Other notable genes include ctpJ, which encodes an ATPase that controls cytoplasmic metal levels (Raimunda et al. 2014); and psk2, which plays a critical role in the synthesis of cell wall lipids (Sirakova et al. 2001). In addition, 15 hypothetical genes residing downstream of mutated promoters were identified, and in five of these cases the promoter experienced two independent SNPs.
Discussion
Here we demonstrate consistent evidence for purifying selection on intergenic sites in five diverse species (excluding M. tuberculosis), even when major regulatory elements are excluded. This further challenges the view that IGRs can be used as mostly neutral markers to estimate neutral mutation rates or profiles (Wang and Chen 2013). Rather, our results suggest these regions are rich with functional elements, many of which are yet to be characterized, and are selectively conserved and maintained (Molina and van Nimwegen 2008; Degnan et al. 2011; Luo et al. 2011). Although consistent with previous work, this observation is pertinent with respect to the default exclusion of IGRs from bacterial databases based on the core genome multilocus sequence typing model (Jolley and Maiden 2010; Sheppard et al. 2012; Maiden et al. 2013; Maiden and Harrison 2016). Our analysis suggests the exclusion of IGRs from these databases is not warranted either for biological or technical reasons.
We have used two fast and efficient approaches to measuring selection on nonprotein coding sequences based on established principles of population genetics and suited for large whole-genome data sets. According to the nearly neutral theory, slightly deleterious mutations are not eliminated immediately from a population, but can persist for a period of time determined by the selection coefficient (s) and the effective population size (Ne) (Kimura and Ohta 1971; Ohta 1973). Highly deleterious mutations will be lost more quickly while they are still very rare. The rarest SNPs are those that are observed in only one genome (singletons), thus the PSM reflects the frequency of highly deleterious mutations (Hershberg et al. 2008). The weaker effect mutations will tend to be lost more gradually over time (Rocha et al. 2006). Whereas the PSM approach provides a measure of how many SNPs are purged very quickly due to highly deleterious effects, dI/dS provides a measure of how many deleterious mutations have been purged relative to the coalescence time of the genomes under consideration. Thus, these two methods are not only independent but also provide complementary comparisons encompassing both strongly and more weakly deleterious mutations.
We demonstrate for the first time that, like dN/dS (Rocha et al. 2006; Castillo-Ramírez et al. 2011), dI/dS also decreases with divergence time, as the ratio is lower when considering between- (rather than within-) CC comparisons. This confirms that the lower prevalence of segregating sites in IGRs when compared to synonymous sites (i.e., dI/dS < 1) does not simply reflect differences in mutation rate, and moreover our analysis of IGR sequences near gene borders reveals that dS has not been significantly inflated by transcription-derived mutation. We also note that our analyses are likely to be conservative. The comparator (dS) is not a perfect neutral benchmark; selection at synonymous sites operates on codon usage bias (Sharp et al. 2005), secondary RNA structure (Molina and van Nimwegen 2008), and possibly GC content (Balbi et al. 2009; Hildebrand et al. 2010; Rocha and Feil 2010; Namouchi et al. 2012). Moreover, dI/dS and dN/dS will continue to decrease with divergence time until the synonymous sites are saturated, and there is no reason to suppose that the available data corresponds to the minima for a given species.
Our analyses provides a novel comparison of the strength and direction of selection on different classes of regulatory element within IGRs. This reveals that RBSs and noncoding RNAs tend to be under relatively strong constraint, broadly comparable to nonsynonymous sites. We have shown that the average selection operating on terminator regions reflects strong selection on the stem, combined with much weaker selective constraint on the loop. Our results also demonstrate that purifying selection is operating on IGRs (relative to synonymous sites) even when predicted promoters, terminators, RBSs, and noncoding RNAs are excluded, and that this constraint is strongest close to gene starts. This suggests that many functional elements in IGRs remain uncharacterized, and unannotated intergenic sites close to gene borders may have particular functional significance.
The power of our approach is underscored by novel evidence for positive selection in predicted promoter regions in M. tuberculosis. This result is highly statistically significant, meaning that the signal of positive selection must be strong enough not to be confounded by any background purifying selection in our global comparisons. To gauge the functional relevance of these promoter SNPs, we identified all downstream genes, and noted a number global regulators, transcription factors, and genes implicated in virulence or resistance. A large number of hypothetical proteins were also identified, which could form targets for future studies (Table S2). This observation thus points to a key role of subtle changes within promoters for short-term adaptation through regulatory rewiring in this species, which may also help to account for the paucity of variation within coding regions. A recent report by McNally et al. (2016) is consistent with this view as it implicated a key role for changes in promoters within a single E. coli clone (ST131) as an adaptive response coinciding with the gain and loss of accessory elements.
Conclusion
Here we have applied two tests to quantify the strength and direction of selection acting on IGRs in bacteria. We demonstrate consistent evidence of strong purifying selection on IGRs in five species, even when major regulatory elements are excluded. We also note the strength and direction of selection varies with the class of intergenic regulatory element, the species under consideration, and distance from gene border. We show that the signal of purifying selection increases with divergence time for intergenic sites, just as it does for nonsynonymous sites, and consistent with expectations under the nearly neutral model of evolution. Although our analysis is consistent with previous reports of very weak purifying selection in M. tuberculosis (Hershberg et al. 2008), we are cognizant that this evidence is equivocal and that our data may in fact reflect a complex combination of purifying, positive, and possibly diversifying selection operating over short coalescence times (Farhat et al. 2013; Osório et al. 2013; Pepperell et al. 2013; Casali et al. 2014). In support of this, we note strong evidence for positive selection within M. tuberculosis promoters, and argue that regulatory rewiring represents a major adaptive mechanism in this species.
We conclude that our current understanding of the functions encoded in IGRs is fragmented, and we would therefore urge utmost caution before excluding these regions from bacterial databases or core genome analyses. Our results call for the routine analysis of the selection pressure operating on, and hence functional relevance of, IGRs similar to those carried out on protein-coding regions.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.195784/-/DC1.
Acknowledgments
We are very grateful to Kathie Grant and Tim Dallman of the Gastrointestinal Bacteria Reference Unit at Public Health England for permission to use the Salmonella enterica data. We also thank Adam Eyre-Walker for critical reading of the manuscript and invaluable discussions and acknowledge the anonymous referees for insightful and constructive comments. We are also very grateful to the Medical Research Council-funded Cloud Infrastructure for Microbial Bioinformatics consortium for granting access and support. The Staphylococcus aureus genome sequences were generated as part of a study supported by a grant from the United Kingdom Clinical Research Collaboration Translational Infection Research Initiative and the Medical Research Council (grant number G1000803, held by Sharon Peacock) with contributions from the Biotechnology and Biological Sciences Research Council; the National Institute for Health Research on behalf of the Department of Health; and the Chief Scientist Office of the Scottish Government Health Directorate, on which E.J.F. was a principal investigator and S.C.B. is a postdoctoral researcher. H.A.T. is funded by a University of Bath research studentship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare that they have no competing interests.
Author contributions: Basic study design, data analysis, and early manuscript drafts: H.A.T. and E.J.F. Additional analysis and suggestions: S.C.B. and L.D.H. Final preparation of manuscript: H.A.T., E.J.F., S.C.B., and L.D.H.
Footnotes
Communicating editor: J. Lawrence
Literature Cited
- Acebo P., Martin-Galiano A. J., Navarro S., Zaballos A., Amblar M., 2012. Identification of 88 regulatory small RNAs in the TIGR4 strain of the human pathogen Streptococcus pneumoniae. RNA 18: 530–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbi K. J., Rocha E. P. C., Feil E. J., 2009. The temporal dynamics of slightly deleterious mutations in Escherichia coli and Shigella spp. Mol. Biol. Evol. 26: 345–355. [DOI] [PubMed] [Google Scholar]
- Casali N., Nikolayevskyy V., Balabanova Y., Ignatyeva O., Kontsevaya I., et al. , 2012. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res. 22: 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali N., Nikolayevskyy V., Balabanova Y., Harris S. R., Ignatyeva O., et al. , 2014. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat. Genet. 46: 279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Ramírez S., Harris S. R., Holden M. T. G., He M., Parkhill J., et al. , 2011. The impact of recombination on dN/dS within recently emerged bacterial clones. PLoS Pathog. 7: e1002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaguza C., Andam C. P., Harris S. R., Cornick J. E., Yang M., et al. , 2016. Recombination in Streptococcus pneumoniae lineages increase with carriage duration and size of the polysaccharide capsule. MBio 7: e01053-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S., Kumar A., Singhal A., Tyagi J. S., Krishna Prasad H., 2009. CmtR, a cadmium-sensing ArsR-SmtB repressor, cooperatively interacts with multiple operator sites to autorepress its transcription in Mycobacterium tuberculosis. FEBS J. 276: 3428–3439. [DOI] [PubMed] [Google Scholar]
- Chen X., Zhang J., 2013. No gene-specific optimization of mutation rate in Escherichia coli. Mol. Biol. Evol. 30: 1559–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chewapreecha C., Harris S. R., Croucher N. J., Turner C., Marttinen P., et al. , 2014. Dense genomic sampling identifies highways of pneumococcal recombination. Nat. Genet. 46: 305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor, T. R., N. J. Loman, S. Thompson, A. Smith, J. Southgate et al., 2016 CLIMB (the Cloud Infrastructure for Microbial Bioinformatics): an online resource for the medical microbiology community. Microbial Genomics 2 Available at: http://mgen.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.000086. [DOI] [PMC free article] [PubMed]
- Degnan P. H., Ochman H., Moran N. A., 2011. Sequence conservation and functional constraint on intergenic spacers in reduced genomes of the obligate symbiont Buchnera. PLoS Genet. 7: e1002252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong A., Pietersma H., Cordes M., Kuipers O. P., Kok J., 2012. PePPER: a webserver for prediction of prokaryote promoter elements and regulons. BMC Genomics 13: 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins C. A., Cohen K. A., Munsamy V., Abeel T., Maharaj K., et al. , 2016. Genomic and functional analyses of Mycobacterium tuberculosis strains implicate ald in D-cycloserine resistance. Nat. Genet. 48: 544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. A., Bird C., Nemesh J., Thomas D. J., Newton-Cheh C., et al. , 2006. Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nat. Genet. 38: 223–227. [DOI] [PubMed] [Google Scholar]
- Farhat M. R., Shapiro B. J., Kieser K. J., Sultana R., Jacobson K. R., et al. , 2013. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat. Genet. 45: 1183–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil E. J., 2015. Toward a synthesis of genotypic typing and phenotypic inference in the genomics era. Future Microbiol. 10: 1897–1899. [DOI] [PubMed] [Google Scholar]
- Fishbein S., van Wyk N., Warren R. M., Sampson S. L., 2015. Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Mol. Microbiol. 96: 901–916. [DOI] [PubMed] [Google Scholar]
- Frampton M., Houlston R., 2012. Generation of artificial FASTQ files to evaluate the performance of next-generation sequencing pipelines. PLoS One 7: e49110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S., Octavia S., Tanaka M. M., Sintchenko V., Lan R., 2015. Defining the core genome of Salmonella enterica serovar typhimurium for genomic surveillance and epidemiological typing. J. Clin. Microbiol. 53: 2530–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H., Vu G. P., Bai Y., Chan E., Wu R., et al. , 2011. A Salmonella small non-coding RNA facilitates bacterial invasion and intracellular replication by modulating the expression of virulence factors. PLoS Pathog. 7: e1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R., Petrov D. A., 2010. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 6: e1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R., Lipatov M., Small P. M., Sheffer H., Niemann S., et al. , 2008. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 6: e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand F., Meyer A., Eyre-Walker A., 2010. Evidence of selection upon genomic GC-content in bacteria. PLoS Genet. 6: e1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt K. E., Wertheim H., Zadoks R. N., Baker S., Whitehouse C. A., et al. , 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA 112: E3574–E3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Lan R., Reeves P. R., 2006. Adaptation of multilocus sequencing for studying variation within a major clone: evolutionary relationships of Salmonella enterica serovar Typhimurium. Genetics 172: 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley K. A., Maiden M. C. J., 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes T. H., Cantor C. R., 1969. Evolution of protein molecules, pp. 21–132 in Mammalian Protein Metabolism, edited by Munro H. N. Academic Press, New York. [Google Scholar]
- Kimura M., Ohta T., 1971. Protein polymorphism as a phase of molecular evolution. Nature 229: 467–469. [DOI] [PubMed] [Google Scholar]
- Laabei M., Recker M., Rudkin J. K., Aldeljawi M., Gulay Z., et al. , 2014. Predicting the virulence of MRSA from its genome sequence. Genome Res. 24: 839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson C., Luna B., Ammerman N. C., Maiga M., Agarwal N., et al. , 2012. Gene expression of Mycobacterium tuberculosis putative transcription factors whiB1-7 in redox environments. PLoS One 7: e37516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Tang J., Friedman R., Hughes A. L., 2011. Ongoing purifying selection on intergenic spacers in group A streptococcus. Infect. Genet.Evol. 11: 343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Minch K. J., Rustad T. R., Hobbs S., Zhou S.-L., et al. , 2015. Integrated modeling of gene regulatory and metabolic networks in Mycobacterium tuberculosis. PLoS Comput. Biol. 11: e1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden M. C. J., Harrison O. B., 2016. The population and functional genomics of the Neisseria revealed with gene-by-gene approaches. J. Clin. Microbiol. 54: 1949–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden M. C. J., Jansen van Rensburg M. J., Bray J. E., Earle S. G., Ford S. A., et al. , 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat. Rev. Microbiol. 11: 728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally A., Oren Y., Kelly D., Ben Pascoe S. D., Sreecharan T., et al. , 2016. Combined analysis of variation in core, accessory and regulatory genome regions provides a super-resolution view into the evolution of bacterial populations. PLoS Genet. 12: e1006280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina N., van Nimwegen E., 2008. Universal patterns of purifying selection at noncoding positions in bacteria. Genome Res. 18: 148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Osawa S., 1987. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc. Natl. Acad. Sci. USA 84: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namouchi A., Didelot X., Schöck U., Gicquel B., Rocha E. P. C., 2012. After the bottleneck: genome-wide diversification of the Mycobacterium tuberculosis complex by mutation, recombination, and natural selection. Genome Res. 22: 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Gojobori T., 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Ohta T., 1973. Slightly deleterious mutant substitutions in evolution. Nature 246: 96–98. [DOI] [PubMed] [Google Scholar]
- Osório N. S., Rodrigues F., Gagneux S., Pedrosa J., Pinto-Carbó M., et al. , 2013. Evidence for diversifying selection in a set of Mycobacterium tuberculosis genes in response to antibiotic- and nonantibiotic-related pressure. Mol. Biol. Evol. 30: 1326–1336. [DOI] [PubMed] [Google Scholar]
- Page A. J., Cummins C. A., Hunt M., Wong V. K., Reuter S., et al. , 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31: 3691–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepperell C. S., Casto A. M., Kitchen A., Granka J. M., Cornejo O. E., et al. , 2013. The role of selection in shaping diversity of natural M. tuberculosis populations. PLoS Pathog. 9: e1003543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimunda D., Long J. E., Padilla-Benavides T., Sassetti C. M., Argüello J. M., 2014. Differential roles for the Co2+ /Ni2+ transporting ATPases, CtpD and CtpJ, in Mycobacterium tuberculosis virulence. Mol. Microbiol. 91: 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter S., Török E. M., Holden M. T. G., Reynolds R., Raven K. E., et al. , 2015. Building a genomic framework for prospective MRSA surveillance in the United Kingdom and the republic of Ireland. Genome Res. 26: 263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha E. P. C., Feil E. J., 2010. Mutational patterns cannot explain genome composition: are there any neutral sites in the genomes of bacteria? PLoS Genet. 6: e1001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha E. P. C., Smith J. M., Hurst L. D., Holden M. T. G., Cooper J. E., et al. , 2006. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J. Theor. Biol. 239: 226–235. [DOI] [PubMed] [Google Scholar]
- Romilly C., Lays C., Tomasini A., Caldelari I., Benito Y., et al. , 2014. A non-coding RNA promotes bacterial persistence and decreases virulence by regulating a regulator in Staphylococcus aureus. PLoS Pathog. 10: e1003979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T., 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30: 2068–2069. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Bailes E., Grocock R. J., Peden J. F., Elizabeth Sockett R., 2005. Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 33: 1141–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard S. K., Jolley K. A., Maiden M. C. J., 2012. A gene-by-gene approach to bacterial population genomics: whole genome MLST of Campylobacter. Genes (Basel) 3: 261–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirakova T. D., Thirumala A. K., Dubey V. S., Sprecher H., Kolattukudy P. E., 2001. The Mycobacterium tuberculosis pks2 gene encodes the synthase for the hepta- and octamethyl-branched fatty acids required for sulfolipid synthesis. J. Biol. Chem. 276: 16833–16839. [DOI] [PubMed] [Google Scholar]
- Smith L. J., Stapleton M. R., Buxton R. S., Green J., 2012. Structure-function relationships of the Mycobacterium tuberculosis transcription factor WhiB1. PLoS One 7: e40407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzek B. E., Ermolaeva M. D., Schreiber M., Salzberg S. L., 2001. A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics 17: 1123–1130. [DOI] [PubMed] [Google Scholar]
- Wang T.-C., Chen F.-C., 2013. The evolutionary landscape of the Mycobacterium tuberculosis genome. Gene 518: 187–193. [DOI] [PubMed] [Google Scholar]
- Waters L. S., Storz G., 2009. Regulatory RNAs in bacteria. Cell 136: 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H., 2009. Ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York. [Google Scholar]
- Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R., 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17: 32–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets supporting the conclusions of this article are available in the ENA (http://www.ebi.ac.uk/ena), except the E. coli sequences which were downloaded from the NCBI (http://www.ncbi.nlm.nih.gov/). The accession numbers of all strains used in the analysis are given in Table S1.