Abstract
DNA double-strand breaks (DSBs) pose a serious threat to genomic integrity. If unrepaired, they can lead to chromosome fragmentation and cell death. If repaired incorrectly, they can cause mutations and chromosome rearrangements. DSBs are repaired using end-joining or homology-directed repair strategies, with the predominant form of homology-directed repair being synthesis-dependent strand annealing (SDSA). SDSA is the first defense against genomic rearrangements and information loss during DSB repair, making it a vital component of cell health and an attractive target for chemotherapeutic development. SDSA has also been proposed to be the primary mechanism for integration of large insertions during genome editing with CRISPR/Cas9. Despite the central role for SDSA in genome stability, little is known about the defining step: annealing. We hypothesized that annealing during SDSA is performed by the annealing helicase SMARCAL1, which can anneal RPA-coated single DNA strands during replication-associated DNA damage repair. We used unique genetic tools in Drosophila melanogaster to test whether the fly ortholog of SMARCAL1, Marcal1, mediates annealing during SDSA. Repair that requires annealing is significantly reduced in Marcal1 null mutants in both synthesis-dependent and synthesis-independent (single-strand annealing) assays. Elimination of the ATP-binding activity of Marcal1 also reduced annealing-dependent repair, suggesting that the annealing activity requires translocation along DNA. Unlike the null mutant, however, the ATP-binding defect mutant showed reduced end joining, shedding light on the interaction between SDSA and end-joining pathways.
Keywords: SDSA, DSB repair, SMARCAL1, Drosophila, homologous recombination
DNA double-strand breaks (DSBs) are deleterious to viability in somatic cells. DSBs can arise from exogenous sources such as chemical mutagens in the environment and ionizing radiation (IR) (Ciccia and Elledge 2010; Sage and Shikazono 2016). They can also occur as a byproduct of endogenous processes including replication errors, cellular metabolism resulting in oxidative stress, and repair of other types of lesions that are converted into a DSB for processing (Pfeiffer et al. 2000). Unrepaired DSBs lead to chromosome fragmentation and apoptosis; aberrant repair can cause insertions or deletions, as well as chromosomal rearrangements through inappropriate recombination (Tsai and Lieber 2010). It is vital that DSBs are not only repaired, but repaired in such a way as to restore the integrity of the genome.
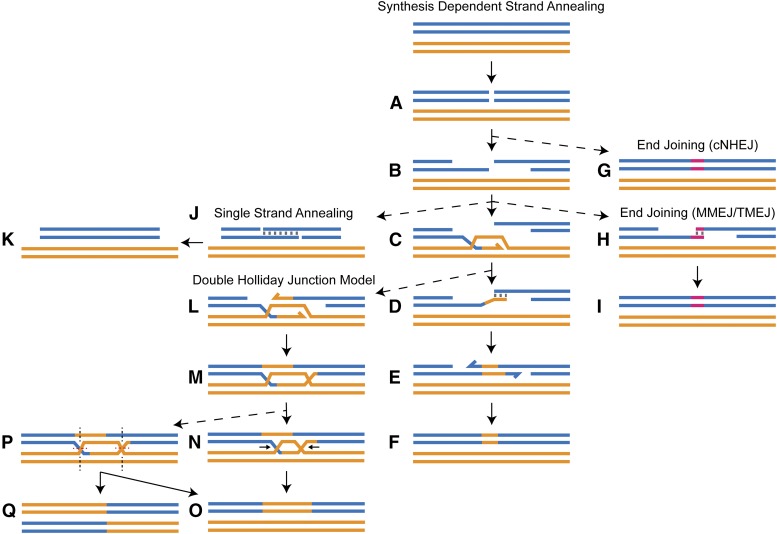
There are multiple strategies for repairing DSBs that can be separated into two main categories: strategies that do not require a template for repair, which include multiple forms of end joining (EJ); and strategies that do require a template, which are classified as homology-directed repair (HDR) (Figure 1). EJ via direct ligation or with minor processing of the ends can be employed immediately at the break, as is the case with canonical nonhomologous EJ (cNHEJ) (Figure 1G) (Mimitou and Symington 2009; Waters et al. 2014; Williams et al. 2014). EJ can also be used as an exit strategy after resection via microhomology-mediated EJ (MMEJ). In metazoans, this process requires DNA polymerase θ, and has recently been called θ-mediated EJ (TMEJ) (Figure 1, H and I) (Chan et al. 2010; Yu and McVey 2010; Garcia et al. 2011; Yousefzadeh et al. 2014; Rodgers and McVey 2016; van Schendel et al. 2016; Wyatt et al. 2016).
Figure 1.
DSB repair strategies. Blue, double-stranded DNA (dsDNA) molecule; orange, dsDNA template (sister chromatid or homologous chromosome). (A) A DSB occurs in the blue DNA molecule. (B) 5′ resection marks the first step of HDR and results in 3′ ssDNA tails. (C) Rad51-coated ssDNA tail invades a template duplex, displacing one strand to create a D-loop, and primes synthesis. (D) The D-loop is disassembled and a complementarity test between the opposing ends of the break occurs. (E) SDSA is defined by annealing between complementary sequences, followed by trimming and/or gap filling. (F) Ligation restores an intact duplex DNA molecule. Alternative strategies (dotted arrows): (G) cNHEJ can occur instead of resection, which directly ligates the ends and can generate small insertions and deletions (pink segment). (H) MMEJ (catalyzed by DNA polymerase θ in metazoans) can occur prior to strand exchange or after failure to find or anneal at complementary sequences. (I) MMEJ/TMEJ will usually generate a deletion or insertion (pink segment). (J) If the DSB occurs between two direct repeats, complementary sequences may be exposed during resection, and annealing can occur without synthesis, called SSA. (K) SSA results in deletion of one repeat. (L) Second-end capture (annealing of the opposing strand to the D-loop, allowing for extension of that strand) can occur during synthesis. (M) Ligation to the opposing 5′ ends creates a dHJ. (N) Dissolution of the dHJ involves migration of the junctions toward each other and decatenation via topoisomerase activity to (O) restore the DNA molecule. (P) Resolution involves endonucleolytic cleavage of the junctions which can be cut in either orientation, resulting in both (O) noncrossover (restoration of the DNA molecule) and (Q) crossover (recombinant) products.
HDR strategies for repair share a common set of steps that begins with resection of the 5′ ends of the break to yield 3′ single-stranded DNA (ssDNA) tails that are protected by the single-stranded binding protein RPA (Figure 1B). Studies in yeast and Drosophila melanogaster have shown that it is possible for complementary resected ends to anneal to each other in a process called single-strand annealing (SSA) (Figure 1, J and K), and SSA has been hypothesized to occur in regions of direct repeats (Rong and Golic 2003; Storici et al. 2006; Bhargava et al. 2016). In nonrepetitive regions, Brca2 and other proteins facilitate exchange of RPA for Rad51 on the resected tails, creating a filament competent to search for and invade a homologous duplex template, typically the sister chromatid or homologous chromosome (Jensen et al. 2010; Reuter et al. 2014). The invading strand anneals to the template strand, displacing its complement and generating a structure called a displacement loop (D-loop) (Figure 1C). The invading strand is then extended by synthesis, using the homologous sequence as a template.
It is at this point that the HDR strategies diverge. In synthesis-dependent strand annealing (SDSA) (central model in Figure 1), the D-loop is disassembled and a complementarity test between the nascent strand and the opposing end of the break is performed (Figure 1D). If complementarity is found, the two ends anneal. Trimming of noncomplementary overhangs, filling of gaps, and ligation restore a duplex DNA molecule (Figure 1, E and F) (Gloor et al. 1991; Nassif et al. 1994; Pâques et al. 1998). An alternative form of HDR occurs when the opposing end of the break anneals to the D-loop in a process called second-end capture (Figure 1L). Continued synthesis from both ends and subsequent ligation produces a joint molecule called a double Holliday junction (dHJ) (Figure 1M), which must be further processed to give duplex products (Figure 1, N–Q).
HDR leading to dHJ formation is often invoked as a primary form of templated repair in somatic cells, though this interpretation is largely due to studies of meiotic recombination mechanisms (reviewed in Jasin and Rothstein 2013). During meiotic recombination, the dHJ pathway is important because it can generate the crossovers that are necessary for homolog disjunction; however, crossovers can be detrimental to mitotically proliferating cells. It is thought that crossovers are avoided in somatic cells either through biased processing of dHJs or by disfavoring formation of dHJ intermediates (Ira et al. 2003; Andersen et al. 2011; Kuo et al. 2014; LaFave et al. 2014; Sarbajna et al. 2014). SDSA is gaining acceptance as a predominant form of HDR in mitotic cells due to its parsimony and a growing amount of circumstantial evidence, such as the rarity of mitotic crossovers in wild-type backgrounds (reviewed in Andersen and Sekelsky 2010). Despite this growing support, there are few assays in existence with the capacity to determine whether noncrossover gene conversion events were generated through the SDSA or the dHJ pathway. As such, little is known about the defining step of SDSA: annealing.
Annealing is the step at which the cell commits to SDSA (Figure 1D), so understanding this step is critical to understanding DSB repair. Studies in budding yeast have identified Rad52 as an important mediator of annealing during SSA (Ivanov et al. 1996; Storici et al. 2006; Jensen 2013). Mammalian Rad52 mutations do not result in strong HDR defects (Rijkers et al. 1998), although human RAD52 has been found to be important for second-end capture through interactions with RPA and Rad51 (McIlwraith and West 2008; Nimonkar et al. 2009; Khade and Sugiyama 2016). This suggests that Rad52 functions in animals may be confined to steps of HDR where Rad51 is active, such as strand invasion, making it unlikely that RAD52 is the mediator of annealing during SDSA.
Recent studies in mammalian systems have uncovered a class of helicases with ATP-driven annealing activity called annealing helicases (Yuan et al. 2012). Members of this family can anneal two RPA-coated, complementary single DNA strands, making these enzymes ideal candidates for annealing during SDSA. The first member of this family to be identified, SMARCAL1, has a C-terminal Swi/Snf2-family helicase domain and two N-terminal HepA-related protein (HARP) domains (SMARCAL1 was originally called HARP) (Coleman et al. 2000; Yusufzai and Kadonaga 2008). SMARCAL1 plays roles in replication-associated DNA damage repair and has been shown to regress model replication forks by annealing the parental strands (Bétous et al. 2012). SMARCAL1 is activated by ATR-dependent phosphorylation and recruited to troubled forks via an interaction with RPA (Ciccia et al. 2009). Biallelic mutations in SMARCAL1 cause the autosomal, recessive disorder Schimke immuno-osseous dysplasia (SIOD). SIOD patients have multiple clinical features, some of which are similar to those of DNA damage-repair disorders such as Bloom syndrome and Fanconi anemia. These include poor growth, immune deficiencies, and premature aging symptoms (Lou et al. 2002; Baradaran-Heravi et al. 2012b; Morimoto et al. 2016).
Orthologs of SMARCAL1 are found throughout metazoans, as well as in plants and some protists, though are notably absent from yeasts. D. melanogaster Marcal1 is 41% identical and 60% similar to human SMARCAL1 across the helicase domain (based on BLAST alignment of residues 154–679 of Marcal1 to residues 337–869 of SMARCAL1). Marcal1 has a single HARP domain vs. the two in SMARCAL1; the presence of two HARP domains appears to be unique to chordates. The distance between the helicase ATPase domain and the proximal HARP domain is critical for the annealing function of SMARCAL1 in vitro (Ghosal et al. 2011), and that distance is conserved in Marcal1. In vitro studies comparing the activity of Marcal1 to SMARCAL1 showed that both proteins have robust annealing activity (Kassavetis and Kadonaga 2014). Further support for a role of Marcal1 in annealing during SDSA is suggested by a previous study showing that mutations in mei-41, which encodes the Drosophila ortholog of ATR, significantly reduce annealing during both SDSA and SSA (LaRocque et al. 2007); suggesting that an ATR-activated protein, such as SMARCAL1/Marcal1, catalyzes annealing during DSB repair.
Drosophila is one of the few model organisms with genetic tools available to assay SDSA, making it an ideal system to test whether SMARCAL1 plays a role in annealing during SDSA in vivo. We show here that Drosophila Marcal1 mutants have elevated lethality when exposed to DSB-inducing agents, indicating Marcal1 has a role in HDR. We used well-characterized assays to demonstrate that annealing during both SDSA and SSA is severely reduced in Marcal1 mutants. Abrogating Marcal1 ATP binding reduces EJ as well as annealing, suggesting that Marcal1 activity is epistatic to TMEJ. Altogether, these data uncover new information about HDR that further our understanding of DSB repair, which will aid in improving the efficiency of chemotherapeutics as well as laboratory technologies such as CRISPR/Cas9.
Materials and Methods
Drosophila stocks
Fly stocks were maintained at 25° on standard cornmeal medium. All Marcal1 null assays were performed using the heteroallelic null mutations Marcal1del and Marcal1kh1. Marcal1del is a 679-bp deletion of part of the first exon and second intron generated via imprecise P-element excision as described in Baradaran-Heravi et al. (2012a). Marcal1kh1 was generated using CRISPR/Cas9 technology (Gratz et al. 2013; Bassett and Liu 2014). Oligonucleotides (Integrated DNA Technologies) used for guide RNA (gRNA) were cloned into pU6 BbsI chimeric RNA (chiRNA) vector and then injected into Bloomington stock number 51323 (y1 M{vas::Cas9}ZH-2A w1118/FM7c) (BestGene). Marcal1kh1 deletes 2L: 4,955,554–4,956,729 (Drosophila annotated genome r6.13), which encompasses exon 1 through part of exon 2. The Brca243 null mutation was used in trans to the Brca247 null mutation in all assays. Both are large deletions produced via imprecise P-element excision, described in Thomas et al. (2013).
Marcal1K275M mutation was generated using CRISPR/Cas9. The gRNA vector was prepared as described for the Marcal1kh1 allele. A repair template vector was generated by amplifying 1234-bp upstream and 643-bp downstream of the conserved lysine codon from BACPAC genomic DNA clone library (identifier: BACR13M11) and inserted into the pSL1180 vector. The QuickChange Site-Directed Mutagenesis Kit (Agilent Technologies) was used to introduce a mutation in the PAM recognition site as well as two single base pair changes to alter the lysine codon to methionine (primer sequence: 5′-GAAATGGGCCTGGGCATGACCTATCAGGCCTTGGC CGTAGCCG-3′). The repair vector was injected with the BbsI chiRNA vector into the same stock used for generating the Marcal1kh1 allele.
Sensitivity assays
Vials of five heterozygous Marcal1del females and three heterozygous Marcal1kh1 males each were incubated for 3 days (brood one) before being transferred to fresh vials of food for 2 additional days (brood two), after which the adult flies were discarded. Then, 1 day later, 250 μl of aqueous mutagenic solution was added directly to the food in brood-two vials and the larvae were allowed to develop to adulthood (dosages listed in Supplemental Material, Table S1 in File S1). For IR experiments, larvae were exposed to 137Cs for the required time to reach the desired dosage. Surviving adults in both broods were quantified by genotype (heterozygous or heteroallelic null) and the ratio of heterozygous to heteroallelic was calculated per vial. Exposed vials were normalized to unexposed paired brood-one ratios. Paired t-tests were performed between unexposed and exposed ratios. Each round of biological replicates had 10–20 vials and each mutagen had two or more rounds.
P{wa} assay
The P{wa} assay was performed as previously described (Adams et al. 2003). Briefly, single males of the genotype y w P{wa}; Marcal1del/Marcal1kh1; Sb P{∆2-3, ry+}/TM6B were crossed to four females homozygous for y w P{wa} and Sb+ female progeny were scored for red, yellow, or apricot eyes. Representative samples of red- and yellow-eyed females were collected and crossed to FM7w males to recover the repair product in subsequent males. DNA was extracted from a single adult male progeny with the repair product for PCR analysis. Multiple repair events were recovered per male, however, only repair events that could be confirmed unique were analyzed and reported. The same methods and transposase source were used for all genotypes.
Each vial of progeny from a single male was scored independently and a ratio of either red- or yellow-eyed progeny to total progeny was calculated per vial. Outliers were identified using the ROUT method and removed from the data set. No outliers were removed from the Brca2 data set; one was removed from Marcal1 Brca2; and three were removed from Marcal1, wild type, and Marcal1K275M data sets. To determine and compare mean frequency of repair events, the red-eye (SDSA) ratios were compared between genotypes using parametric ANOVA and the mean ratios of each genotype were compared to the mean ratios of all other genotypes; the analysis was repeated for yellow-eye (EJ) ratios. Rate of lack of excision was calculated by subtracting the EJ rate of Brca2 mutants from the total scored progeny, as Brca2 mutants are incapable of strand invasion. Rate of complete restoration of P{wa} was calculated by summing the frequency of all repair events in wild-type males and subtracting that rate from the EJ rate of Brca2 mutants, using the assumption that all excisions were repaired by EJ in these mutants.
To assess synthesis capability, each repair event was analyzed with a series of seven PCRs (primers are listed in Table S2 in File S1). Amplified products using one primer set were calculated as a percentage of total events analyzed and repeated for all primer sets (intervals). Percentage of events with positive PCRs were compared between genotypes using parametric ANOVA. When compared as individual PCRs, P < 0.05 was considered to be statistically significant; however, when asking how much difference exists between genotypes across all PCRs, P-values for each interval were multiplied by the number of intervals (seven) to correct for multiple comparisons.
P{wIw} assay
The P{wIw} assay was performed as previously described (Mukherjee et al. 2009). Briefly, four Marcal1del/CyO; P{wIw}/TM6B females were crossed to three Marcal1kh1/CyO; Sb P{70I-SceI}/TM6B males for 3 days and the parents were discarded. Then, 1 day later, the first-instar larval progeny were heat-shocked at 37° for 1 hr. Heat shock was repeated on the following day to ensure all first-instar larvae received treatment. Larvae were allowed to develop to adulthood. All Sb+ progeny were scored for red or white eyes. All red-eyed flies were collected (up to a maximum of 10) from each vial; 10 white-eyed flies were collected per vial. DNA was extracted from each collected fly for analysis.
Red-eye events: The linker region was amplified (primer sets in Table S3 in File S1) and subjected to digestion with I-SceI. Events that failed to amplify or were not cut by I-SceI were categorized as EJ events. Events that were successfully cut by I-SceI could not be distinguished between EJ and uncut, and were removed from the dataset.
White-eye events: The 5′ region of the upstream mini-white (mini-w) gene is 480 bp whereas the 5′ region of the downstream mini-w is 369 bp using the same primer set (Table S3 in File S1). The presence of both products indicates SSA did not occur; these events were categorized as EJ events. A small number of events had only the 369-bp product, these were verified to be in the upstream location via a primer anchored in the P element and categorized as SSA events. PCRs with no product were categorized as EJ events. The percentage of SSA events in the white class was calculated per vial. SSA and EJ events were then adjusted by this number.
The adjusted percentages of SSA events per vial were compared via unpaired t-tests between control and mutant genotypes per experiment. Marcal1K275M heterozygotes and homozygotes were compared to each other, wild type, and Marcal1 using parametric ANOVA.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Drosophila stocks are available upon request.
Results
Marcal1 mutants show elevated lethality when exposed to DSB-inducing agents
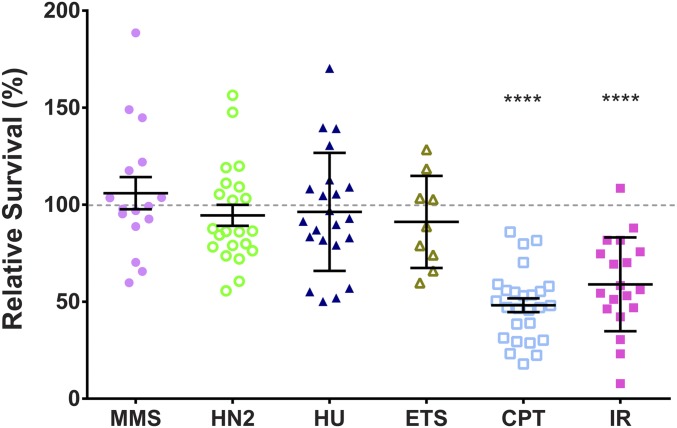
We tested whether Marcal1 has a role in DSB repair by exposing mutant larvae to DNA damaging agents and measuring survival to adulthood relative to unexposed siblings. Marcal1 mutant survival was not affected when exposed to methyl methanesulfonate (MMS), an alkylating agent (Lundin et al. 2005); or nitrogen mustard (HN2), which generates both mono-adducts and interstrand cross-links (Povirk and Shuker 1994) (Figure 2). Studies in mice have shown that SMARCAL1 null mutations confer sensitivity to killing by hydroxyurea (HU) (Baradaran-Heravi et al. 2012b), a ribonucleotide reductase inhibitor thought to result in stalled replication forks (Hammond et al. 2003); however, Marcal1 mutant larvae showed no decrease in survival when exposed to HU. HU treatment is most detrimental in cells sensitive to perturbations in replication, which is consistent with published in vitro evidence that Marcal1 cannot regress a four-stranded model replication fork (Kassavetis and Kadonaga 2014) and suggests Marcal1 may not have a significant role in protecting stalled forks in flies.
Figure 2.
Marcal1 mutants are sensitive to killing by DSB-inducing agents. Flies heterozygous for null mutations in Marcal1 were mated in two broods of at least 10 vials, with each vial representing a biological replicate. Brood one was unexposed; brood two received a dose of MMS, HN2, HU, ETS, CPT, or IR during larval feeding. Relative survival was calculated as the ratio of homozygous mutant to heterozygous control adults in treated vials, normalized to the same ratio in the corresponding unexposed vial. Dotted line represents 100% relative survival. Dosage, number of replicates, and total progeny counts are in Table S1 in File S1. **** P < 0.0001 in paired t-tests between unexposed and exposed vials.
We found a significant reduction in survival of Marcal1 mutant larvae exposed to IR, an established DSB-inducing agent (Radford 1985). Marcal1 mutant flies were also sensitive to killing by camptothecin (CPT), similar to both mouse and human cell studies (Baradaran-Heravi et al. 2012b). CPT prevents topoisomerase I from religating DNA after it has nicked a strand and become covalently bound to the end (Pommier et al. 2010). CPT is thought to be most lethal during replication, where the nick can become a DSB. Previous studies have suggested that CPT lesions are repaired via HDR in Drosophila (Andersen et al. 2011) and in chicken DT40 cell lines (Maede et al. 2014). Interestingly, Marcal1 mutant flies did not have elevated lethality when exposed to etoposide (ETS), a topoisomerase II poison that generates DSBs (Pommier et al. 2010). A genetic screen of DT40 cells found that mutations in EJ genes conferred sensitivity to ETS and resistance to CPT, whereas mutations in HR genes resulted in higher sensitivity to CPT than to ETS (Maede et al. 2014). It is possible that Marcal1 mutants are sensitive to ETS at higher doses than those tested here, however the data from CPT and IR treatments sufficiently support our hypothesis that Marcal1 is involved in DSB repair.
Marcal1 mutants have reduced annealing capacity during gap repair
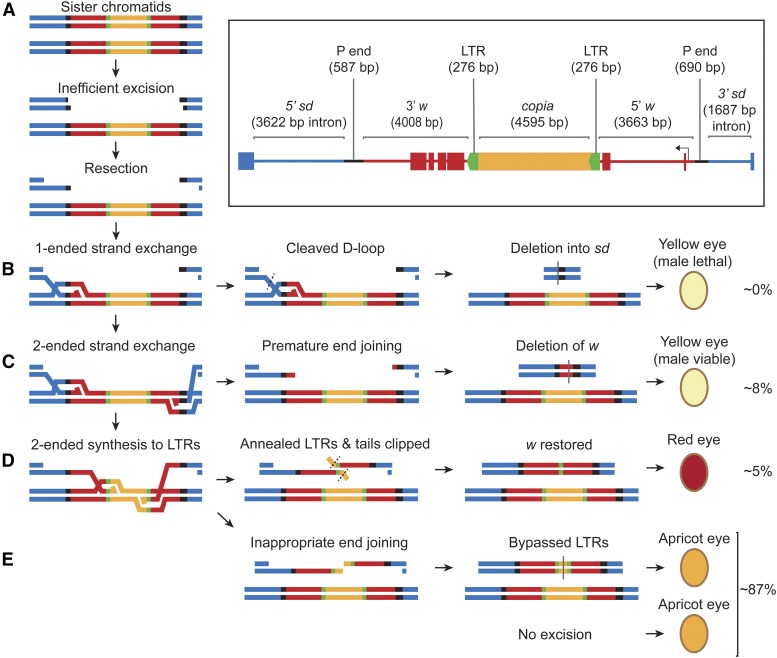
Because Marcal1 mutants are sensitive to agents that cause DSBs, we tested the ability of these mutants to repair a double-stranded gap by SDSA. We used the well-characterized P{wa} assay in the germline of male Drosophila (Figure 3) (Kurkulos et al. 1994; McVey et al. 2007). P{wa} is a 14-kb P element that is a nonlethal insertion into the first intron of the essential gene scalloped (sd) on the X chromosome (Figure 3, inset). The P element contains a white (w) gene, the product of which loads red pigment into the eye, interrupted by an intronic copia retrotransposon flanked by two 276-bp long terminal repeats (LTRs). The copia insertion alters w splicing and results in an apricot-colored eye in hemizygous males or homozygous females. When exposed to an inefficient source of P-element transposase, the P{wa} element is excised from one chromatid; the intact sister chromatid serves as an efficient template for HDR. Excision generates 17-nt, noncomplementary overhangs on both sides of the break, which are structurally similar to short resected ends and are poor substrates for EJ via cNHEJ (Symington and Gautier 2011). Repair events from single males are recovered in female progeny by crossing to females homozygous for P{wa}.
Figure 3.
The P{wa} assay for SDSA. (Inset, top right) The construct is a 14-kb P element inserted into the second intron of the essential gene sd (blue) in reverse orientation (diagram is relative to genome coordinates on X chromosome). Black segments represent P-element sequences needed for excision. Red segments are exons (boxes) and introns (lines) of a w gene, the product of which loads eye pigments when functional. This copy of w is interrupted by a copia retrotransposon (orange central box) which is flanked by two 276-bp LTRs (green directional boxes), resulting in partial loss of w function and an apricot-eyed phenotype. (A) Line representations of the construct on two sister chromatids in the male germline. Exposure to inefficient P transposase results in excision of the construct from one sister, leaving 17-nt noncomplementary overhangs on each side, and the ends are resected. (B) One of the 3′ ssDNA tails invades the intact sister to initiate synthesis. If D-loop dissociation is defective, the D-loop is cleaved, creating a deletion into an sd exon on one or both sides of the construct. When mated to a homozygous P{wa} female, the progeny with flanking deletions will have a yellow eye (due to the full copy of the construct from the mother) but the event will be male lethal in subsequent generations (∼0% of progeny from wild-type males have this phenotype). (C) Premature EJ after two-ended strand exchange and some synthesis results in complete loss of w function; progeny will have yellow eyes and viable males in subsequent generations (∼8% of progeny from wild-type males). (D) Synthesis of the LTRs followed by annealing, tail clipping, and gap filling restores w function and progeny have a red eye (∼5% of progeny from wild-type males). (E) Synthesis to the LTRs followed by inappropriate EJ in copia results in an apricot eye in the progeny and is indistinguishable from nonexcision or full gene conversion events via dHJ intermediates (∼87% of progeny from wild-type males). These progeny are scored but not counted as repair events.
For SDSA to occur, both sides of the break must be extended (via synthesis) beyond the first region of complementarity, the copia LTRs, and these must be annealed correctly (Figure 3). The resultant product deletes copia except for a single LTR, resulting in restoration of w splicing, observable as red eyes in progeny inheriting this product. If EJ occurs either without synthesis or after incomplete synthesis, w function is lost, resulting in yellow-eyed progeny (due to the maternally inherited complete P{wa} copy). Complete restoration of P{wa} could occur via a dHJ intermediate or through SDSA or EJ that synthesizes past the LTRs. These are not scored as repair events because they cannot be differentiated from a lack of excision; however, lack of excision is the most frequent class (>85% of progeny), whereas complete restoration is estimated to be <2% in wild-type males. SDSA and EJ events are quantified as a percentage of total scorable progeny (daughters that do not inherit the transposase source) from each male, including apricot-eyed progeny. We also measured the distribution of events per male.
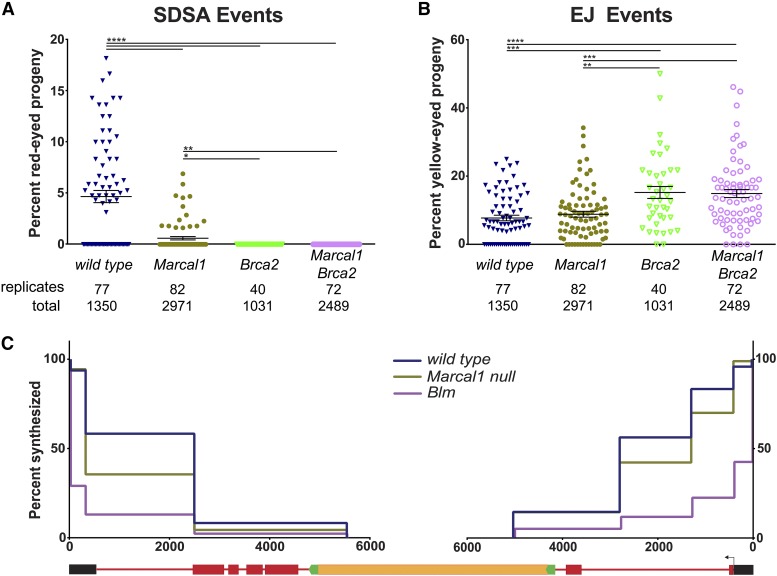
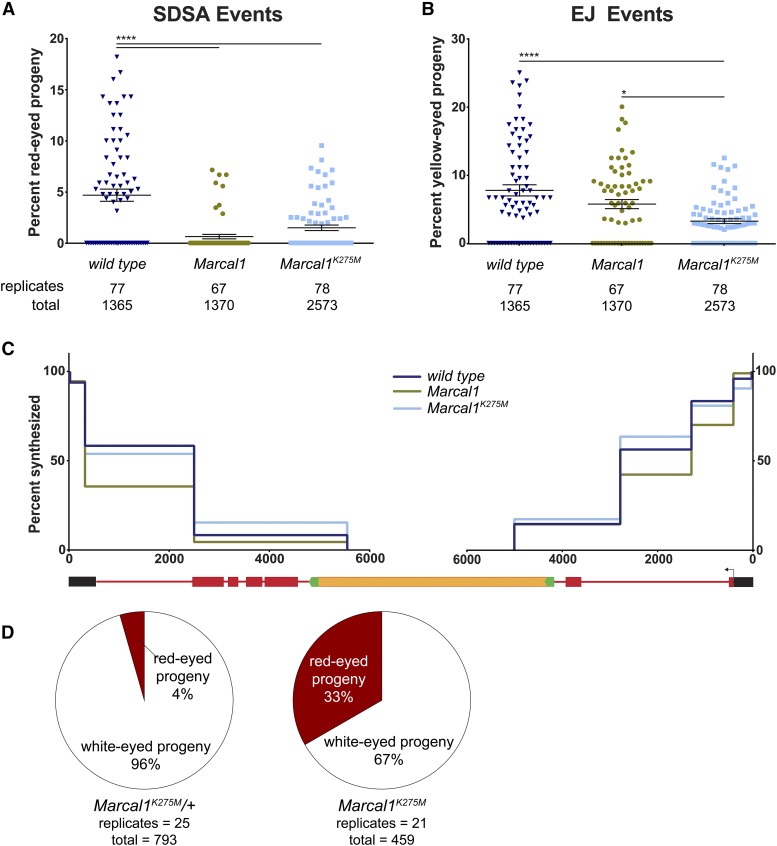
We found that Marcal1 mutants had significantly reduced SDSA (red-eyed progeny) compared to wild type, both in percentage of total progeny scored (Figure 4A) and in number of males with observable SDSA events in the progeny (Figure S1 in File S1). EJ (yellow-eyed progeny) was not significantly changed in Marcal1 mutants (Figure 4B). SDSA could be reduced if P-element excision is reduced or strand exchange is impaired. To test this, we performed the assay in a Brca2 mutant. Drosophila Brca2 has a strand-exchange function similar to that of human BRCA2 (Brough et al. 2008); so we expected strand exchange to be defective in Brca2 mutants and for all repair to be the result of EJ events prior to strand invasion, which would be observable as yellow-eyed progeny. As expected, we observed no red-eyed progeny in Brca2 single mutants and a compensatory increase in yellow-eyed progeny compared to wild type (Figure 4, A and B). If Marcal1 mutations reduce P-element excision or affect the pathway upstream of Brca2 function in DSB repair, we would expect Marcal1 Brca2 double mutants to have reduced EJ compared to Brca2 single mutants due to an overall reduction in observable repair products. However, we found Marcal1 Brca2 double mutants to have a repair phenotype that was not significantly different from Brca2 single mutants (Figure 4, A and B). We therefore conclude that the decreased SDSA in Marcal1 mutants results from a loss of function downstream of strand invasion.
Figure 4.
Marcal1 mutants have reduced SDSA capacity in the P{wa} assay. (A) SDSA events are measured as the percentage of scored progeny with red eyes. Mean and SEM are indicated. Marcal1 null mutant, Brca2 mutant, and Marcal1 Brca2 double mutant frequencies were all significantly reduced compared to wild type. The numbers of single males (biological replicates) and total progeny scored are listed below the graph. (B) EJ events were measured as the percentage of scored progeny with yellow eyes. Brca2 and Marcal1 Brca2 mutants had significantly elevated EJ compared to wild type and Marcal1 single mutants. P-values: **** P < 0.0001, ** P < 0.002, * P < 0.05, based on parametric ANOVA. (C) Synthesis tracts in repair events recovered in yellow-eyed progeny were measured using a series of PCRs. Each interval was measured independently and quantified as a percentage of total independent events analyzed. x-axis denotes distance (in nucleotides) from each end of the gap, on the same scale as the schematic of P{wa} below. y-axis is percent of events analyzed that had a positive PCR and therefore synthesized at least as far as the most internal primer. Marcal1 (n = 90) was not significantly different from wild type (n = 48) when corrected for multiple intervals (Materials and Methods). Blm (n = 75) mutants were significantly different (P < 0.0001) from both wild type and Marcal1.
While the Marcal1 mutant phenotype could result from defective LTR annealing, it could also be due to compromised D-loop disassembly or reduced capacity to synthesize past the LTRs. In Blm mutants, which are believed to be defective in D-loop disassembly, the synthesis length in repair products is significantly shorter than in wild-type males (McVey et al. 2007). In addition, many repair events have deletions from the break site into the flanking sd gene; these are hypothesized to arise from nucleolytic cleavage of D-loops that cannot be disassembled (Adams et al. 2003; McVey et al. 2004a). We analyzed the aggregated amount of synthesis from each end of the break in all observed EJ events to determine the overall synthesis pattern within the population of EJ events (Figure 4C). Synthesis tracts in Marcal1 mutants were similar in length to those of wild-type flies, but there were significantly longer tracts in Blm mutants (Figure 4C). Repeated rounds of strand exchange, synthesis, and D-loop dissociation have been observed in previous assays of gap repair (McVey et al. 2004b), and our finding that aggregated synthesis-tract lengths in Marcal1 mutants is similar to wild type suggests that the length of synthesis per cycle, while likely having some stochastic component, is unchanged by loss of Marcal1. Also, we did not observe any flanking deletions among EJ repair products from Marcal1 mutants, whereas 48% of EJ repair in Blm mutants was associated with deletion. Altogether, these data suggest that the reduction in red-eyed progeny in Marcal1 mutants is not due to defects in synthesis or inability to disassemble D-loops. These data support the hypothesis that Marcal1 mutants have a defect in LTR annealing.
Marcal1 mediates annealing independent of synthesis
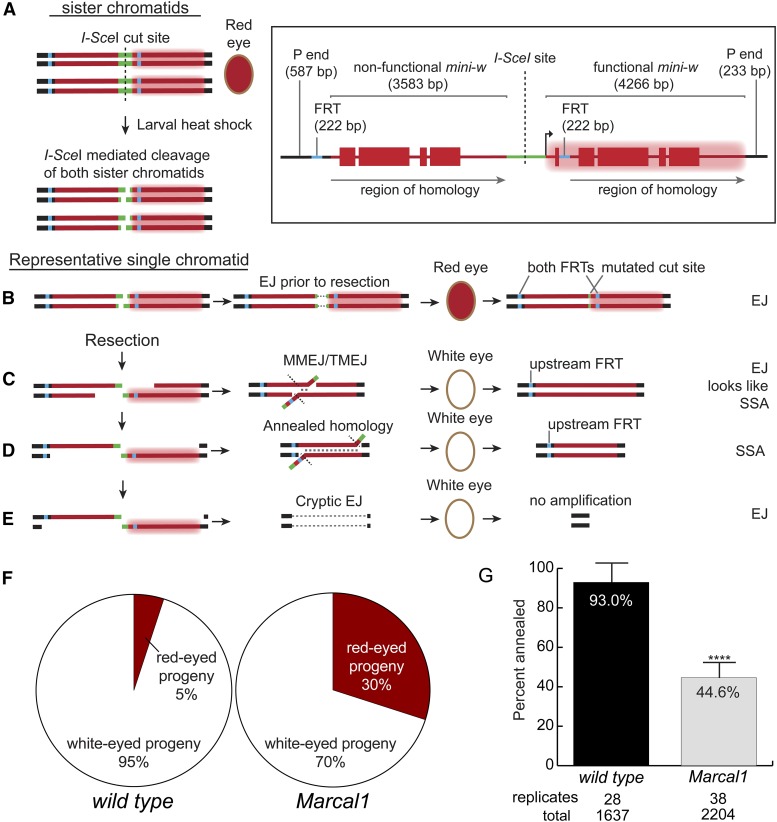
SMARCAL1 annealing studies have been restricted to replication-associated roles and we observed reduced annealing in Marcal1 mutants using the P{wa} assay, which requires synthesis for annealing. We wanted to know if Marcal1 mediates annealing in contexts that do not require synthesis, so we used an SSA assay called P{wIw} (Rong and Golic 2003).
P{wIw} is a P element that has two mini-w genes in tandem (Figure 5, inset). The downstream copy (∼4.3 kb) is functional, while the upstream copy (∼3.6 kb) is nonfunctional due to a deletion of the promoter and first exon. The two are separated by a linker region containing an I-SceI recognition site and can be differentiated by amplifying FRT sites present in the 5′ region of each copy (Figure 5, inset). I-SceI is expressed in the germlines of male larvae heterozygous for an insertion of P{wIw} on chromosome 3, and repair events are recovered in the progeny (Figure 5A). Previous studies have shown that I-SceI cuts at >90% efficiency in this context, thus the most common outcome is cutting of both sister chromatids (Rong and Golic 2003). Since the homologous chromosome does not have a P{wIw} insertion, strand invasion is a rare event; however, full resection of both sides can reveal the 3.6 kb of complementarity between the upstream and downstream copies of mini-w. If these sequences are annealed, completion of SSA repair gives a product that retains only a nonfunctional copy of mini-w; progeny that inherit this repair event will have white eyes and the upstream FRT site (Figure 5D). Red-eyed progeny can result from EJ with little or no resection or from an uncut construct; which will have both FRT sites and a mutated I-SceI cut site (Figure 5B). White eyes can also result from EJ repair with deletion into the promoter of the downstream mini-w, which will have both FRT sites; however, EJ events with deletions that extend into the downstream FRT site can be indistinguishable from SSA events (Figure 5C).
Figure 5.
Marcal1 mutants have reduced annealing capacity in the P{wIw} assay. (Inset, top right) The construct is on chromosome 3 and consists of two copies of the mini-w gene which are tandemly arrayed and separated by a linker containing an I-SceI recognition site. The upstream copy is missing exon 1 and intron 1, rendering it nonfunctional, while the downstream copy is functional (represented by a red glow in the schematic) and produces a wild-type red eye in a w null background. The upstream mini-w has a 5′ FRT site and the downstream copy has an FRT insertion in intron 1. PCR amplification of the FRT anchored in mini-w yields different size products for each gene, which is used to identify the presence or absence of each copy. (A) The assay is performed in the male germline with P{wIw} heterozygous. I-SceI is expressed via heat shock during larval development, resulting in DSBs with 4-nt overhangs on both sister chromatids. (B) Mutational cNHEJ can occur at the cut site, yielding red-eyed progeny and the presence of both FRTs but a mutated I-SceI cut site. (C) If resection is insufficient to expose complementary sequences, MMEJ/TMEJ can give white-eyed progeny with a deletion that includes the downstream FRT. These events cannot be differentiated from products of SSA unless the deletion is sufficiently large (>1000 bp); if no difference in size was observed, these events were classified as SSA. (D) Full resection of at least 3.6 kb reveals complementarity between the ssDNA ends that can be annealed. This results in white-eyed progeny with only the upstream FRT site; such events were categorized as SSA. (E) Excessive resection can result in deletion of the entire construct, resulting in white-eyed progeny with no amplification products. (F) Percentage of total progeny with white or red eyes for wild type and Marcal1 mutants. P < 0.0001 using χ2 test. (G) Percentage of repair products that involved annealing, after molecular analysis correction (Figure S2 in File S1). Marcal1 null mutants had significantly reduced annealing compared to wild type, **** P < 0.0001. Error bars are SD. Biological replicates (single males) and total progeny scored are denoted below each genotype.
We observed a significant difference in the distribution of eye color between wild type and Marcal1 mutants (Figure 5F). Every Marcal1 mutant male had progeny with both eye colors, whereas almost 40% of wild-type males did not produce any red-eyed progeny. We collected 10 (or all if <10) red-eyed progeny from each male for molecular analyses. These analyses showed no difference in cutting efficiency of I-SceI between Marcal1 and wild type (Figure S2 in File S1), suggesting that the increase in red-eyed progeny in Marcal1 mutants is not due to reduced induction of DSBs. Furthermore, the distribution of EJ events in red-eyed flies was strikingly similar between Marcal1 and wild type, which suggests that the type of EJ used is not affected by Marcal1. This is consistent with a role for Marcal1 after resection, since the type of EJ is dictated by the structure of the DNA ends.
We also collected 10 white-eyed progeny for analysis. A total of 98% of the white-eyed males from wild-type flies were consistent with annealing by SSA (Figure S2 in File S1). In stark contrast, 34% of analyzed white-eyed progeny from Marcal1 mutant males were confirmed as EJ. It is likely that this number is underreported because the large regions of homology in the construct obscures identification of EJ events that are similar in size to the predicted annealed length. Altogether, these data show significantly reduced SSA capacity in Marcal1 null mutants (44.6%) compared to wild type (93.0%) (Figure 5G), indicating Marcal1 is important for annealing in both synthesis-dependent (SDSA) and synthesis-independent (SSA) repair strategies.
ATP binding is required for Marcal1 activity during SDSA
We next asked if Marcal1 translocation is required for annealing during SDSA. Helicases use the conserved Walker-A and Walker-B motifs to bind and hydrolyze ATP for translocation. SMARCAL1 can bind DNA in the absence of ATP (Yusufzai and Kadonaga 2008), so we abolished the ATP-binding site in the Walker-A motif by mutating the conserved lysine to a methionine (K275M) in the endogenous Marcal1 gene via CRISPR/Cas9. We then tested the ability of Marcal1K275M ATP-binding defective mutants to repair a gap using the P{wa} assay.
Marcal1K275M mutants had a reduction in SDSA similar to that of Marcal1 null mutants (Figure 6A), suggesting that translocation is required for its function during SDSA. Surprisingly, EJ events were significantly reduced compared to wild type and Marcal1 null mutants (Figure 6B), revealing a genetic interaction between Marcal1 and EJ pathways. We compared synthesis patterns in Marcal1K275M mutants to wild type and Marcal1 null mutants. We found that Marcal1K275M mutants did not have significantly different synthesis length compared to either genotype (Figure 6C).
Figure 6.
Marcal1K275M mutants have reduced SDSA and EJ capacity. (A) SDSA and (B) EJ events were measured in Marcal1K275M mutants as described in Figure 4. SDSA was similar between Marcal1K275M and Marcal1 null, but EJ events were significantly reduced in Marcal1K275M compared to both wild type and Marcal1 null mutants. (C) Synthesis-tract lengths measured as described in Figure 4C. No significant differences were found between Marcal1K275M (n = 52) and wild type (n = 48) or Marcal1 null mutants (n = 90). (D) Marcal1K275M heterozygotes and Marcal1K275M mutants were tested in the P{wIw} SSA assay as described in Figure 5. Heterozygotes had 96% white-eyed progeny, which is not statistically different from wild type (Figure 5F) based on a parametric ANOVA test. Marcal1K275M mutants had 67% white-eyed progeny, which was significantly reduced compared to heterozygotes but not significantly different from Marcal1 null mutants (Figure 5F). * P = 0.0185; **** P < 0.0001.
Lastly, we tested the ability of Marcal1K275M mutants and Marcal1K275M heterozygotes to anneal complementary sequences during SSA using the P{wIw} assay. White-eyed flies were significantly decreased in Marcal1K275M mutants (66.88%) compared to Marcal1K275M heterozygotes (95.59%) (Figure 6D). The white-eyed frequency in heterozygotes was not statistically different from wild type (wild-type data from Figure 5B), and like wild type, 40% of Marcal1K275M heterozygous males had no red-eyed progeny (all Marcal1K275M homozygous males had red-eyed progeny). The percentage of red- and white-eyed progeny in Marcal1K275M mutants was not significantly different from Marcal1 null mutants (null data from Figure 5B), suggesting that ATP binding does not affect EJ in the absence of strand exchange, synthesis, and D-loop dissociation. These data also show that ATP binding is required for annealing during SSA, supporting our findings from P{wa} and establishing a requirement for ATP hydrolysis in annealing activities of Marcal1.
Discussion
DSB-repair strategies can be thought of as a series of choices that are made by weighing a combination of complex factors, including availability of a repair template, cell-cycle timing, nuclear architecture, and the chromatin context of the break itself. The first key decision point is resection, which dictates a commitment to HDR strategies (sequestering the ends of the break from resection results in direct ligation via cNHEJ (reviewed in Mimitou and Symington 2009). Brca2 acts downstream of resection to facilitate strand exchange with the template and the phenotype of Brca2 Marcal1 double mutants shows Brca2 is epistatic to Marcal1, indicating that Marcal1 acts later in the HDR pathway and is unlikely to be involved in resection.
Resection almost invariably leads to strand exchange and synthesis. SSA is an exception, but this can occur only when there are direct repeats flanking the break and perhaps primarily when there is no available template for HDR; it is unclear how much SSA is used outside of specialized assays such as P{wIw}. Regardless, SSA is efficiently used in specific situations and the data presented here provide evidence that it shares a common annealing mediator, Marcal1, with other HDR strategies.
The second key decision point in HDR repair is disassembly of the D-loop, which dictates the choice between SDSA and the dHJ pathway. Disassembly favors SDSA by promoting complementarity tests and annealing, whereas continued synthesis increases the likelihood of second-end capture and dHJ formation. In Drosophila, Blm helicase has been identified as a key mediator of D-loop disassembly (Adams et al. 2003; McVey et al. 2004b) and recent studies suggest Fancm may play a minor role in this step as well (Kuo et al. 2014; Romero et al. 2016), though studies in human cells do not show a role for FANCM in SDSA (Zapotoczny and Sekelsky 2017). We did not observe phenotypes suggesting defects in D-loop dissociation in Marcal1 mutants, suggesting that the role of Marcal1 is downstream of D-loop dissociation.
The third key decision point is annealing, which we propose impacts the choice between SDSA, EJ (primarily TMEJ), and reinvasion. Prior to the studies reported here, little was known about annealing during HDR in animals or how the decision between these three options is regulated. Our data here provide in vivo evidence that Marcal1 mediates annealing during SDSA and SSA in Drosophila; and suggest that Marcal1 acts directly to anneal complementary strands, as abrogating Marcal1 translocation activity via a Walker-A mutation recapitulates the null phenotype.
The Marcal1K275M mutation reduces EJ as well as annealing during SDSA, which suggests that Marcal1K275M antagonizes EJ in contexts where EJ follows strand exchange, synthesis, and D-loop dissociation. EJ in Marcal1 null mutants is unaffected, which further suggests the EJ phenotype in Marcal1K275M mutants is likely due to defective annealing activity leading to aberrant interactions with the DNA rather than a nonannealing protein-protein interaction. We propose the Marcal1K275M phenotype is a consequence of localization to the DNA without translocation activity, which may indicate that Marcal1 precedes recruitment of EJ factors after D-loop dissociation. Further studies are needed to test this hypothesis.
Interestingly, we observed that Marcal1K275M is completely recessive in the P{wIw} assay (Figure 6D). It is possible that the Marcal1K275M mutant protein may bind DNA less tightly than the wild-type protein. While ATP binding is not required for DNA binding by SMARCAL1, it can cause a conformational change that influences the DNA-binding constant (Gupta et al. 2015). It is also possible that multiple Marcal1 molecules bind the nascent DNA and the presence of any wild-type protein is sufficient to rescue the Marcal1K275M phenotype. Recent work on the RPA-SMARCAL1 interface has shown that SMARCAL1 binds to the C-terminal region of RPA32 with 1:1 stoichiometry (Bhat et al. 2014; Xie et al. 2014) and it is likely that many molecules of Marcal1 bind the RPA-coated nascent DNA, allowing for multiple complementarity searches to occur simultaneously. Studies have revealed a similar mechanism for homology searching by Rad51-coated ssDNA (Wright and Heyer 2014; Qi and Greene 2016). Mutation of the Walker-B motif to allow ATP binding (thus preserving DNA-binding kinetics) but prevent ATP hydrolysis (translocation), as well as in vitro studies of Marcal1 interactions with long RPA-bound filaments, may help to clarify the mechanistic basis of the Marcal1K275M phenotype.
An intriguing finding from our study is that synthesis-tract length is not elevated in Marcal1 mutants even though annealing is defective. Early studies of gap repair in Drosophila have shown that continuous synthesis averages 1379 bp, and complete restoration decreases as template length increases (Gloor et al. 1991); additional studies with the P{wa} assay suggest that gap filling involves multiple cycles of strand exchange, synthesis, and D-loop dissociation (McVey et al. 2004b). These attributes appear unaffected in Marcal1 mutants, suggesting that the choice to strand reinvade or EJ during a given round is not solely dependent on the outcome of complementarity tests, even though the reduction in EJ in Marcal1K275M mutants argues that Marcal1 is recruited to the nascent ends prior to recruitment of EJ factors. It is possible that rounds of strand invasion, synthesis, and D-loop disassembly are simply stochastic in nature, and that after each a complementarity test is performed by Marcal1. When Marcal1 is present but defective, EJ factors may be excluded after D-loop dissociation; whereas when Marcal1 is completely absent, EJ factors would have access to the DNA, but EJ would still be reliant on the synthesis machinery and unknown regulatory signals. These data support a model where complementarity tests are upstream of the EJ vs. strand-reinvade decision but have no effect on that decision if annealing is unsuccessful. Further studies of genetic interactions between Marcal1 and EJ factors may help to clarify this interaction.
Unlike P{wa}, the P{wIw} assay did not reveal any differences between Marcal1K275M and null mutants. P{wa} requires multiple iterations of the anneal vs. EJ vs. reinvade decision (McVey et al. 2004b), providing increased opportunities to observe moderate defects in that decision among repair products. On the other hand, in the P{wIw} assay there is usually no intact homologous template for repair (in wild-type flies <1% of the products were uncut or restored to the original sequence). While it is not possible to determine which repair strategy is initially favored at the break, the efficiency of I-SceI cutting may ultimately select for HDR, since direct ligation would often restore the cut site, perhaps to be cut again. Once resected, annealing becomes the strongly favored strategy since long ssDNA tails are not ideal substrates for TMEJ or cNHEJ (Waters et al. 2014; Wyatt et al. 2016) and strand invasion is not possible. SSA represents >93% of all repair products in wild type and Marcal1K275M heterozygotes, supporting this interpretation. Because the anneal vs. EJ vs. strand-reinvade decision point is highly altered and potentially deregulated by the assay design, it is likely that EJ events in P{wIw} are facilitated through alternative pathways that are less influenced by Marcal1. Our data support this interpretation as we see a large (more than sevenfold) increase in EJ events in both Marcal1K275M and null mutants. These EJ events are predominantly joined near the break site without large deletions, suggesting an alternative form of EJ is used on long resected ends when annealing is compromised. EJ was reported by Chan et al. (2010) in mutants defective in both cNHEJ and TMEJ, though the key mediator of this form of EJ remains unknown.
We observed phenotypes indicative of residual annealing activity in Marcal1 mutants. It is possible that another protein is capable of some annealing, though Drosophila lacks orthologs of other candidate annealases such as Rad52 and ZRANB3 (an annealing helicase paralog of SMARCAL1). It is more likely that our observation reveals key limitations in the ability to differentiate annealed events vs. MMEJ/TMEJ using both the P{wa} and the P{wIw} assay. In P{wa}, restoration of w gene function is interpreted as LTR annealing; however, it is possible that EJ near the LTR could also result in restoration of w, and the size of insertion tolerated before w function is compromised remains unknown. It is possible that synthesis and MMEJ/TMEJ that occurs past the LTRs can result in red-eyed progeny. Extensive synthesis past the LTRs may result in apricot eyes that would not be recovered for analysis, which may also explain why we did not observe a compensatory increase in EJ events in Marcal1 mutants as well.
The limitations of P{wIw} are predominantly due to extensive homology to either side of the break, which makes it difficult to definitively identify annealed events vs. MMEJ/TMEJ of approximately the same size. Molecular analysis revealed that 22% of white-eyed progeny in Marcal1 mutants had this type of event (Figure S2 in File S1) and it is highly probable that the true percentage is even higher, but sequence similarity is an impediment to fine-scale amplification. Amplification of the entire construct followed by single-molecule sequencing, as done in the Saccharomyces cerevisiae gap-repair assay of Guo et al. (2015), might allow sampling of a larger number of repair events from multiple tissues of a single individual and it could be further adapted to recover events in different progeny from a single male germline, as in our strategy. Our work has highlighted the need for fine-scale assays capable of distinguishing true annealing events from microhomology-mediated events.
The findings reported here provide evidence that Marcal1 mediates annealing in both SDSA and SSA. We have further shown that Marcal1 affects EJ pathways (most likely TMEJ) during HDR, providing new information on the regulation of the anneal–EJ–strand-reinvade decision point. Understanding this decision has broad implications for multiple applications, including chemotherapeutics and genome-editing technologies. Many cancer drugs generate DSBs as a primary mechanism; the role in DSB repair discovered here suggests SMARCAL1 is important for multiple repair mechanisms during S/G2, making it an attractive target for drug development, as has been proposed by Zhang et al. (2012). Additionally, insertion of long fragments during CRISPR/Cas9 genome editing has been proposed to occur via SDSA (Byrne et al. 2015); understanding the regulation of SDSA will improve the efficiency of this technology. Lastly, understanding the interplay of multiple repair strategies, as well as gaining insight into which strategies are used in different contexts, will enhance our understanding of both the basis of genome-stability disorders and their progression.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.200238/-/DC1.
Acknowledgments
We thank E. Buzhardt and A. Cook for preliminary Marcal1 null P{wa} data acquisition. This work was supported by grants from the National Institute of General Medical Sciences to J.S. (1-R01 GM-061252 and 1-R35 GM-118127). J.K.H. was supported in part by National Institute of General Medical Sciences grants 5-T32 GM-007092 and F31 GM-113550.
Footnotes
Communicating editor: J. A. Birchler
Literature Cited
- Adams M. D., McVey M., Sekelsky J. J., 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299: 265–267. [DOI] [PubMed] [Google Scholar]
- Andersen S. L., Sekelsky J., 2010. Meiotic vs. mitotic recombination: two different routes for double-strand break repair. BioEssays 32: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S. L., Kuo H. K., Savukoski D., Brodsky M. H., Sekelsky J., 2011. Three structure-selective endonucleases are essential in the absence of BLM helicase in Drosophila. PLoS Genet. 7: e1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baradaran-Heravi A., Cho K. S., Tolhuis B., Sanyal M., Morozova O., et al. , 2012a Penetrance of biallelic SMARCAL1 mutations is associated with environmental and genetic disturbances of gene expression. Hum. Mol. Genet. 21: 2572–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baradaran-Heravi A., Raams A., Lubieniecka J., Cho K. S., DeHaai K. A., et al. , 2012b SMARCAL1 deficiency predisposes to non-Hodgkin lymphoma and hypersensitivity to genotoxic agents in vivo. Am. J. Med. Genet. A. 158A: 2204–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett A. R., Liu J. L., 2014. CRISPR/Cas9 and genome editing in Drosophila. J. Genet. Genomics 41: 7–19. [DOI] [PubMed] [Google Scholar]
- Bétous R., Mason A. C., Rambo R. P., Bansbach C. E., Badu-Nkansah A., et al. , 2012. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 26: 151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava R., Onyango D. O., Stark J. M., 2016. Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 32: 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat K. P., Betous R., Cortez D., 2014. High-affinity DNA binding domains of replication protein A (RPA) direct SMARCAL1-dependent replication fork remodeling. J. Biol. Chem. 290: 4110–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brough R., Wei D., Leulier S., Lord C. J., Rong Y. S., et al. , 2008. Functional analysis of Drosophila melanogaster BRCA2 in DNA repair. DNA Repair (Amst.) 7: 10–19. [DOI] [PubMed] [Google Scholar]
- Byrne S. M., Ortiz L., Mali P., Aach J., Church G. M., 2015. Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res. 43: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. H., Yu A. M., McVey M., 2010. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 6: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., Elledge S. J., 2010. The DNA damage response: making it safe to play with knives. Mol. Cell 40: 179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., Bredemeyer A. L., Sowa M. E., Terret M. E., Jallepalli P. V., et al. , 2009. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 23: 2415–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M. A., Eisen J. A., Mohrenweiser H. W., 2000. Cloning and characterization of HARP/SMARCAL1: a prokaryotic HepA-related SNF2 helicase protein from human and mouse. Genomics 65: 274–282. [DOI] [PubMed] [Google Scholar]
- Garcia A., Salomon R. N., Witsell A., Liepkalns J., Calder R. B., et al. , 2011. Loss of the bloom syndrome helicase increases DNA ligase 4-independent genome rearrangements and tumorigenesis in aging Drosophila. Genome Biol. 12: R121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal G., Yuan J., Chen J., 2011. The HARP domain dictates the annealing helicase activity of HARP/SMARCAL1. EMBO Rep. 12: 574–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor G. B., Nassif N. A., Johnson-schlitz D. M., Preston C. R., Engels W. R., 1991. Targeted gene replacement in Drosophila via P element-induced gap repair. Science 253: 1110–1117. [DOI] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., et al. , 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Lehner K., O’Connell K., Zhang J., Dave S. S., et al. , 2015. SMRT sequencing for parallel analysis of multiple targets and accurate SNP phasing. G3 (Bethesda) 5: 2801–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M., Mazumder M., Dhatchinamoorthy K., Nongkhlaw M., Haokip D. T., et al. , 2015. Ligand-induced conformation changes drive ATP hydrolysis and function in SMARCAL1. FEBS J. 282: 3841–3859. [DOI] [PubMed] [Google Scholar]
- Hammond E. M., Green S. L., Giaccia A. J., 2003. Comparison of hypoxia-induced replication arrest with hydroxyurea and aphidicolin-induced arrest. Mutat. Res. Fundam. Mol. Mech. Mutagen. 532: 205–213. [DOI] [PubMed] [Google Scholar]
- Ira G., Malkova A., Liberi G., Foiani M., Haber J. E., 2003. Srs2 and Sgs1 – Top3 suppress crossovers during double-strand break repair in yeast. Cell 115: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov E. L., Sugawara N., Fishman-Lobell J., Haber J. E., 1996. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics 142: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M., Rothstein R., 2013. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 5: a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. B., 2013. BRCA2: one small step for DNA repair, one giant protein purified. Yale J. Biol. Med. 86: 479–489. [PMC free article] [PubMed] [Google Scholar]
- Jensen R. B., Carreira A., Kowalczykowski S. C., 2010. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467: 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Kadonaga J. T., 2014. The annealing helicase and branch migration activities of Drosophila HARP. PLoS One 9: e98173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khade N. V., Sugiyama T., 2016. Roles of C-terminal region of yeast and human rad52 in rad51-nucleoprotein filament formation and ssDNA annealing. PLoS One 11: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H. K., McMahan S., Rota C. M., Kohl K. P., Sekelsky J., 2014. Drosophila FANCM helicase prevents spontaneous mitotic crossovers generated by the MUS81 and SLX1 nucleases. Genetics 198: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkulos M., Weinberg J. M., Roy D., Mount S. M., 1994. P element-mediated in vivo deletion analysis of white-apricot: deletions between direct repeats are strongly favored. Genetics 136: 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFave M. C., Andersen S. L., Stoffregen E. P., Holsclaw J. K., Kohl K. P., et al. , 2014. Sources and structures of mitotic crossovers that arise when BLM helicase is absent in Drosophila. Genetics 196: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRocque J. R., Jaklevic B., Su T. T., Sekelsky J., 2007. Drosophila ATR in double-strand break repair. Genetics 175: 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou S., Lamfers P., McGuire N., Boerkoel C. F., 2002. Longevity in Schimke immuno-osseous dysplasia. J. Med. Genet. 39: 922–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin C., North M., Erixon K., Walters K., Jenssen D., et al. , 2005. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 33: 3799–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maede Y., Shimizu H., Fukushima T., Kogame T., Nakamura T., et al. , 2014. Differential and common DNA repair pathways for topoisomerase I- and II-targeted drugs in a genetic DT40 repair cell screen panel. Mol. Cancer Ther. 13: 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwraith M. J., West S. C., 2008. DNA repair synthesis facilitates RAD52-mediated second-end capture during DSB repair. Mol. Cell 29: 510–516. [DOI] [PubMed] [Google Scholar]
- McVey M., Larocque J. R., Adams M. D., Sekelsky J. J., 2004a Formation of deletions during double-strand break repair in Drosophila DmBlm mutants occurs after strand invasion. Proc. Natl. Acad. Sci. USA 101: 15694–15699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Adams M., Staeva-Vieira E., Sekelsky J. J., 2004b Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics 167: 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Andersen S. L., Broze Y., Sekelsky J., 2007. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics 176: 1979–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E. P., Symington L. S., 2009. DNA end resection: many nucleases make light work. DNA Repair (Amst.) 8: 983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto, M., D. B. Lewis, T. Lücke, and C. F. Boerkoel, 2016 Schimke immunoosseous dysplasia, in GeneReviews [Internet], edited by R. A. Pagon, M. P. Adam, H. H. Ardinger et al. University of Washington, Seattle. [PubMed] [Google Scholar]
- Mukherjee S., LaFave M. C., Sekelsky J., 2009. DNA damage responses in Drosophila nbs mutants with reduced or altered NBS function. DNA Repair (Amst.) 8: 803–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif N., Penney J., Pal S., Engels W. R., Gloor G. B., 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14: 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar A. V., Sica R. A., Kowalczykowski S. C., 2009. Rad52 promotes second-end DNA capture in double-stranded break repair to form complement-stabilized joint molecules. Proc. Natl. Acad. Sci. USA 106: 3077–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F., Leung W. Y., Haber J. E., 1998. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol. 18: 2045–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer P., Goedecke W., Obe G., 2000. Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Mutagenesis 15: 289–302. [DOI] [PubMed] [Google Scholar]
- Pommier Y., Leo E., Zhang H., Marchand C., 2010. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17: 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk L. F., Shuker D. E., 1994. DNA damage and mutagenesis induced by nitrogen mustards. Mutat. Res. Genet. Toxicol. 318: 205–226. [DOI] [PubMed] [Google Scholar]
- Qi Z., Greene E. C., 2016. Visualizing recombination intermediates with single-stranded DNA curtains. Methods 105: 62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford I. R., 1985. The level of induced DNA double-strand breakage correlates with cell killing after x–irradiation. Int. J. Radiat. Biol. 48: 45–54. [DOI] [PubMed] [Google Scholar]
- Reuter M., Zelensky A., Smal I., Meijering E., van Cappellen W. A., et al. , 2014. BRCA2 diffuses as oligomeric clusters with RAD51 and changes mobility after DNA damage in live cells. J. Cell Biol. 207: 599–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijkers T., van den Ouweland J., Morolli B., Rolink A. G., Baarends W. M., et al. , 1998. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 18: 6423–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers K., McVey M., 2016. Error-prone repair of DNA double-strand breaks. J. Cell. Physiol. 231: 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero N. E., Matson S. W., Sekelsky J., 2016. Biochemical activities and genetic functions of the Drosophila melanogaster fancm helicase in DNA repair. Genetics 204: 531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G., 2003. The Homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics 165: 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage E., Shikazono N., 2016. Radiation-induced clustered DNA lesions: repair and mutagenesis. Free Radic. Biol. Med. DOI: 10.1016/j.freeradbiomed.2016.12.008. [DOI] [PubMed] [Google Scholar]
- Sarbajna S., Davies D., West S. C., 2014. Roles of SLX1–SLX4, MUS81–EME1, and GEN1 in avoiding genome instability and mitotic catastrophe. Genes Dev. 28: 1124–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F., Snipe J. R., Chan G. K., Gordenin D. A., Resnick M. A., 2006. Conservative repair of a chromosomal double-strand break by single-strand DNA through two steps of annealing. Mol. Cell. Biol. 26: 7645–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., Gautier J., 2011. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45: 247–271. [DOI] [PubMed] [Google Scholar]
- Thomas A. M., Hui C., South A., McVey M., 2013. Common variants of Drosophila melanogaster Cyp6d2 cause camptothecin sensitivity and synergize with loss of Brca2. G3 (Bethesda) 3: 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai A. G., Lieber M. R., 2010. Mechanisms of chromosomal rearrangement in the human genome. BMC Genomics 11: S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schendel R., van Heteren J., Welten R., Tijsterman M., 2016. Genomic scars generated by polymerase theta reveal the versatile mechanism of alternative end-joining. PLoS Genet. 12: e1006368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters C. A., Strande N. T., Wyatt D. W., Pryor J. M., Ramsden D. A., 2014. Nonhomologous end joining: a good solution for bad ends. DNA Repair (Amst.) 17: 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G. J., Hammel M., Radhakrishnan S. K., Ramsden D., Lees-Miller S. P., et al. , 2014. Structural insights into NHEJ: building up an integrated picture of the dynamic DSB repair super complex, one component and interaction at a time. DNA Repair (Amst.) 17: 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright W. D., Heyer W. D., 2014. Rad54 functions as a heteroduplex DNA pump modulated by its DNA substrates and Rad51 during D Loop formation. Mol. Cell 53: 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt D. W., Feng W., Conlin M. P., Yousefzadeh M. J., Roberts S. A., et al. , 2016. Essential roles for polymerase θ-mediated end joining in the repair of chromosome breaks. Mol. Cell 63: 662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S., Lu Y., Jakoncic J., Sun H., Xia J., et al. , 2014. Structure of RPA32 bound to the N-terminus of SMARCAL1 redefines the binding interface between RPA32 and its interacting proteins. FEBS J. 281: 3382–3396. [DOI] [PubMed] [Google Scholar]
- Yousefzadeh M. J., Wyatt D. W., Takata K., Mu Y., Hensley S. C., et al. , 2014. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 10: e1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A. M., McVey M., 2010. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 38: 5706–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Ghosal G., Chen J., 2012. The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol. Cell 47: 410–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T., Kadonaga J. T., 2008. HARP is an ATP-driven annealing helicase. Science 322: 748–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapotoczny G., Sekelsky J., 2017. Human cell assays for synthesis-dependent strand annealing and crossing over during double-strand break repair. G3 (Bethesda) DOI: 10.1534/g3.116.037390. [DOI] [PMC free article] [PubMed]
- Zhang L., Fan S., Liu H., Huang C., 2012. Targeting SMARCAL1 as a novel strategy for cancer therapy. Biochem. Biophys. Res. Commun. 427: 232–235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Drosophila stocks are available upon request.