Meiotic crossovers and double strand breaks (DSBs) are required for gametogenesis, but entering meiotic divisions with broken chromosomes results in...
Keywords: meiosis, Mek1, Ndt80, checkpoint, recombination
Abstract
During meiosis, homologous chromosomes are physically connected by crossovers and sister chromatid cohesion. Interhomolog crossovers are generated by the highly regulated repair of programmed double strand breaks (DSBs). The meiosis-specific kinase Mek1 is critical for this regulation. Mek1 downregulates the mitotic recombinase Rad51, indirectly promoting interhomolog strand invasion by the meiosis-specific recombinase Dmc1. Mek1 also promotes the formation of crossovers that are distributed throughout the genome by interference and is the effector kinase for a meiosis-specific checkpoint that delays entry into Meiosis I until DSBs have been repaired. The target of this checkpoint is a meiosis-specific transcription factor, Ndt80, which is necessary to express the polo-like kinase CDC5 and the cyclin CLB1 thereby allowing completion of recombination and meiotic progression. This work shows that Mek1 and Ndt80 negatively feedback on each other such that when DSB levels are high, Ndt80 is inactive due to high levels of Mek1 activity. As DSBs are repaired, chromosomes synapse and Mek1 activity is reduced below a threshold that allows activation of Ndt80. Ndt80 transcription of CDC5 results in degradation of Red1, a meiosis-specific protein required for Mek1 activation, thereby abolishing Mek1 activity completely. Elimination of Mek1 kinase activity allows Rad51-mediated repair of any remaining DSBs. In this way, cells do not enter Meiosis I until recombination is complete and all DSBs are repaired.
REPAIR of double strand breaks (DSBs) in vegetative cells occurs primarily by homologous recombination using sister chromatids as templates (Kadyk and Hartwell 1992; Bzymek et al. 2010). In contrast, during meiosis proper Meiosis I segregation is promoted by crossovers that are created by repair of programmed DSBs using homologs as templates. These DSBs are introduced preferentially into regions of the genome called “hotspots” by a highly conserved, meiosis-specific, topoisomerase-like endonuclease called Spo11 (Keeney et al. 2014). There are numerous meiosis-specific constraints on DSB repair to ensure that each homolog pair receives at least one crossover, including: (1) interhomolog bias (repair that preferentially uses the homolog instead of the sister as a template) and (2) creation of a specific class of crossovers that promotes chromosome synapsis and is distributed throughout the genome by a genetic phenomenon called interference, in which a crossover in one interval decreases the probability of crossovers nearby (Zickler and Kleckner 2015). In the budding yeast Saccharomyces cerevisiae, ∼160 DSBs are deliberately introduced into the genome in each meiosis, with potentially lethal consequences if even a single DSB goes unrepaired (pan et al. 2011). Meiotic DSB repair is therefore a highly regulated process. Meiosis-specific constraints such as interhomolog bias and interfering crossover formation are under the control of a meiosis-specific kinase called Mek1 (Kim et al. 2010; Chen et al. 2015; Hollingsworth 2016).
A unique feature of meiosis is the physical association of homologous pairs of sister chromatids by an evolutionarily conserved structure called the synaptonemal complex (SC) (Zickler and Kleckner 2015). The SC is a proteinaceous tripartite structure that plays a critical role in the regulation of recombination. SC formation begins when sister chromatids condense to form “tethered loops” along meiosis-specific protein cores called axial elements (AEs). In yeast, AEs contain the meiosis-specific proteins Hop1, Red1, and Rec8 (Hollingsworth et al. 1990; Smith and Roeder 1997; Klein et al. 1999). Hop1 is the founding member of the HORMA (Hop1-Rev7-Mad2) domain family of AE proteins required for promoting interhomolog bias during meiotic recombination in yeast, plants, nematodes, and mammals (Hollingsworth and Byers 1989; Mao-Draayer et al. 1996; Schwacha and Kleckner 1997; Couteau et al. 2004; Sanchez-Moran et al. 2007; Li et al. 2011). SC formation is completed when the AEs of each homolog pair are connected by the polymerization of a transverse filament protein (Zip1 in yeast) along the lengths of the chromosomes (Sym et al. 1993; Dong and Roeder 2000; Page and Hawley 2004). Zip1 is one of a functionally diverse set of proteins called the “ZMMs,” which are required for stabilization of interhomolog strand invasion intermediates that create stable connections between homologs that are necessary for Zip1 polymerization along the AEs (i.e., synapsis) (Dong and Roeder 2000; Börner et al. 2004). These intermediates are subsequently processed into double Holliday junctions whose resolution is biased to form crossovers (Schwacha and Kleckner 1995; Allers and Lichten 2001; Zakharyevich et al. 2012). Crossovers generated by the ZMM pathway are distributed throughout the genome by interference and constitute the bulk of the crossovers in a wild-type meiosis (Chen et al. 2008).
The G2 interval in meiosis is considerably longer than in mitosis due to Ama1, a meiosis-specific component of the Anaphase Promoting Complex that targets mitotic regulators such as the Ndd1 transcription factor and Cdc5 for degradation by the proteasome (Okaz et al. 2012). As a result, the mitotic program is abolished and exit from meiotic prophase is controlled instead by the meiosis-specific transcription factor Ndt80 (Xu et al. 1995; Chu and Herskowitz 1998). Ndt80 activates transcription of > 200 “middle” sporulation genes, including NDT80 itself, CDC5 (required for Holliday junction resolution, SC disassembly, and spindle pole body duplication), CLB1, and CLB3 (required for Meiosis I and Meiosis II, respectively) (Dahmann and Futcher 1995; Chu and Herskowitz 1998; Carlile and Amon 2008; Sourirajan and Lichten 2008; Li et al. 2015). Cells lacking NDT80 arrest at the pachytene stage of meiotic prophase, with fully synapsed chromosomes and unresolved double Holliday junctions (Xu et al. 1995; Allers and Lichten 2001).
Transcription of NDT80 occurs in two waves: the first is mediated by Ime1, a transcription factor responsible for expression of genes required for recombination and synapsis, as well as the meiosis-specific kinase IME2 [reviewed in Winter (2012)]. Ime1-mediated NDT80 transcription is delayed relative to other early genes, due to the requirement for Ime2 phosphorylation to remove the Sum1 repressor from the NDT80 promoter (Shin et al. 2010). Once sufficient active Ndt80 protein has been made, Ndt80 binds to a specific DNA sequence in its own promoter to create a positive feedback loop, as well as the promoters of its target genes (Hepworth et al. 1995; Lamoureux et al. 2002; Montano et al. 2002).
The presence of unrepaired DSBs triggers a checkpoint that arrests cells in prophase prior to Meiosis I (Lydall et al. 1996). This meiotic recombination checkpoint is a modified version of the DNA damage checkpoint in vegetative cells (Subramanian and Hochwagen 2014). The “9-1-1” clamp, comprised of Rad17-Ddc1-Mec3, binds to resected DSBs and recruits the Tel1/Mec1 checkpoint kinases (ATM/ATR in mammals), which then phosphorylate Hop1 (Carballo et al. 2008). The meiosis-specific effector kinase Mek1 binds to phospho-Hop1 via the Mek1 FHA (Forkhead associated) domain, resulting in activation of Mek1 by trans-autophosphorylation of threonine (T) 327 in the Mek1 activation loop (Niu et al. 2007; Carballo et al. 2008). Mek1 kinase activity is required for the meiotic recombination checkpoint, as evidenced by the fact that the meiotic progression delay observed for rad50S (which results in unresected DSBs that cannot be repaired) is abolished when MEK1 is deleted (Alani et al. 1990; Xu et al. 1997).
The meiotic recombination checkpoint arrest is due to inactivation of the low level of Ndt80 protein that results from Ime1 transcription (Tung et al. 2000). Under checkpoint-arrested conditions, Ndt80 is inactive due to its sequestration in the cytoplasm and the absence of the Ime2-dependent phosphorylation of Ndt80 that is necessary for transcriptional activity (Hepworth et al. 1998; Tung et al. 2000; Sopko et al. 2002; Benjamin et al. 2003; Wang et al. 2011). Abrogation of the meiotic recombination checkpoint in dmc1∆ mutants by rad17∆ results in Ndt80 activation and transcription of its target genes (Chu and Herskowitz 1998; Pak and Segall 2002). In some cases, overexpression of NDT80 can partially bypass the arrest conferred by dmc1∆ or zip1∆ mutants, suggesting that high levels of Ndt80 can partially titrate out whatever is inhibiting Ndt80 (Tung et al. 2000; Pak and Segall 2002). Importantly, the checkpoint arrest triggered by defects in recombination is thought to be an extreme case of what happens normally during meiosis, where DSBs activate the checkpoint to delay entry into Meiosis I until recombination and synapsis are complete.
Okaz et al. (2012) proposed that progression through meiosis is controlled by a switch between two stable states. In the first state, the meiotic recombination checkpoint senses the presence of DSBs and inactivates Ndt80, thereby preventing Holliday junction resolution, SC disassembly, and meiotic progression. For example, the meiosis-specific recombinase DMC1 is required for the formation of single-end invasion intermediates, which are precursors to the primary crossover pathway in yeast (Hunter and Kleckner 2001). In the absence of DMC1, DSBs are made and resected but are not repaired, thereby preventing SC formation and triggering meiotic prophase arrest (Bishop et al. 1992; Lydall et al. 1996; Xu et al. 1997). When the signal to the meiotic recombination checkpoint is eliminated, Ndt80 becomes activated, resulting in high cyclin-dependent kinase (CDK) levels (due to transcription of CLB1) that drive cells into meiosis and sporulation. But what is the signal that turns off the checkpoint to activate Ndt80? One idea is that Ime2 and CDK/Cln3 work together in a double-negative feedback loop to control NDT80 transcription (Gurevich and Kassir 2010). Another hypothesis is that synapsis of homologous chromosomes turns off Mek1 kinase activity by removing the kinase from chromosomes, thereby shutting off the checkpoint (Subramanian et al. 2016). A third model, based on the observation that ectopic expression of CDC5 bypasses the checkpoint in dmc1∆ mutants, posits a role for Cdc5 in regulating Ndt80 activity (Acosta et al. 2011).
Recently, the meiosis-specific Hed1 protein was shown to be an in vivo substrate of Mek1 (Callender et al. 2016). Hed1 binds to Rad51, thereby preventing complex formation between Rad51 and its accessory factor Rad54 (Tsubouchi and Roeder 2006; Busygina et al. 2008). Mek1 phosphorylation of Hed1 T40 inhibits Hed1 degradation, thereby contributing to the inhibition of Rad51 strand exchange activity (Callender et al. 2016). Using an antibody specific for Hed1 phospho-T40 as a sensitive marker for in vivo kinase activity, we show that Mek1 is constitutively active in ndt80-arrested cells, and that elimination of all Mek1 activity requires NDT80-mediated transcription of the polo-like kinase CDC5. We propose a model in which Mek1 kinase activity and Ndt80 negatively feedback on each other to ensure that meiotic progression does not occur until all DSBs have been repaired.
Materials and Methods
Media
Liquid and solid media were as previously described (Lo and Hollingsworth 2011) with 2% galactose used in place of glucose where indicated. Liquid sporulating culture (Spo) medium is 2% potassium acetate. To make plates containing methyl methanesulfonate (MMS), a 10% stock (Sigma Chemical, St. Louis, MO, 129925) was added to yeast peptone dextrose (YPD) or galactose (YPGal) medium at a final concentration of 0.04% immediately prior to pouring the plates. MMS plates were used the next day.
Plasmids
The plasmid pKB80 contains PGPD1-GAL4.(848) ER (hereafter GAL4-ER) in a URA3-integrating plasmid (Benjamin et al. 2003). PGPD1-GAL4.ER was subcloned into a TRP1-integrating plasmid in two steps. First, site-directed mutagenesis of pRS304 (Sikorski and Hieter 1989) was used to change T to C at base pair 529 of the TRP1 open reading frame (ORF) in pRS304 to generate pEP102. This mutation creates an NheI site but does not alter the amino acid specified by the codon (L178). Second, a fragment containing PGPD1-GAL4.ER engineered to have NsiI and KpnI ends was amplified by the polymerase chain reaction (PCR) using pKB80 as template and cloned into NsiI/KpnI-digested pEP102 to create pEP105. This plasmid can be targeted to integrate at the trp1 locus using NheI. To make PGAL1 promoter fusions using hphMX4 as the selectable marker, the PGAL1 promoter region from pFA6a-kanMX6-PGAL1 was amplified and digested with BglII and PacI and subcloned into pAG32 to make pEP104 (Longtine et al. 1998; Goldstein and McCusker 1999). pJR2 is a mek1-as URA3-integrating plasmid (Callender and Hollingsworth 2010). The plasmids pMJ830 and pMJ840 are hphMX4-integrating plasmids containing PGAL1-CDC5 and PGAL1-cdc5-N209A, respectively (generously provided by M. Lichten, National Institutes of Health). These plasmids can be targeted for integration at CDC5 by digestion with SnaBI.
Yeast strains
The complete genotypes of the yeast strains used in this work are given in Supplemental Material, Table S1. All strains are derivatives of the fast sporulating SK1 background. Gene deletions and PGAL1 fusions were created by PCR-based methods using kanMX6, natMX4, and hphMX4, which confer resistance to G418, nourseothricin (NAT), and Hygromycin B (HygB), respectively (Longtine et al. 1998; Goldstein and McCusker 1999; Tong and Boone 2006). In addition, some genes were deleted using the Kluyveromyces lactis URA3 gene as a selectable marker (pKlU, provided by A. Neiman, Stony Brook University). Unless stated otherwise, deletions and gene fusions were confirmed by PCR. Deletions of RAD54 were also confirmed phenotypically by sensitivity to 0.04% MMS.
The rad54∆ control strain, NH2136, was constructed by deleting RAD54 with kanMX6 in S2683 and RKY1145 and then mating to make the diploid. To make the RAD54-IN diploid, NH2319, the PGAL1 promoter was fused to the RAD54 ORF in A14154, thereby creating A14154 RAD54-IN. This haploid was then mated to SKY371 and a MATα ura3::PGPD1-GAL4-ER::URA3 trp1::hisG kanMX6-PGAL1-RAD54 segregant (NH2318-20-2) was crossed to RKY1145::pKB80 rad54∆. RKY1145::pKB80 rad54∆ was generated by first integrating pKB80 digested with NheI at the ura3 locus of RKY1145 to introduce PGPD1-GAL4-ER, followed by deletion of RAD54 with natMX4.
Estradiol (ED)-inducible alleles were created by placing the gene of interest under control of the PGAL1 promoter in the presence of GAL4-ER. These alleles are abbreviated as GENEX-IN. The NDT80-IN diploid, NH2127, was derived from mating segregants from a cross between A14154 and A28417. The NDT80-IN rad54∆ diploid, NH2126, was made by mating segregants from a cross between A14154 rad54 and A28417. The MATα parent of NH2126, NH2111-12-3, was crossed to A14154 RAD54-IN to make the NDT80-IN RAD54-IN diploid, NH2185. The NDT80-IN diploid, NH2033, was created by mating A14154 with S2683 ndt80. To create an isogenic diploid containing mek1-as, MEK1 was deleted using natMX4 in A14154 and S2683 ndt80. The S2683 ndt80 mek1 haploid was then transformed with pJR2 digested with RsrII to target integration of the plasmid downstream of the mek1∆::natMX4 and mated to A14154 mek1 to make NH2122::pJR2.
The hemizygous CDC5-IN ndt80∆ diploid, NH2296, was created by mating S3363 with RKY1145 ndt80. To create isogenic diploids homozygous for CDC5-IN and cdc5-N209A-IN, S2683 ndt80::pKB80 and RKY1145 ndt80::pKB80 were transformed with either pMJ830 or pMJ840 digested with SnaBI. The sequence of the integrated CDC5 alleles was confirmed by amplifying fragments containing PGAL1-CDC5 from the genomic DNA of the transformants followed by DNA sequencing at the Stony Brook University DNA Sequencing Facility. The haploids were then mated to create NH2398 (CDC5-IN) or NH2395 (cdc5-N209A-IN).
The NDT80-IN, NDT80-IN rad54∆, and NDT80-IN RAD54-IN genotypes were also introduced into isogenic diploids containing the HIS4-LEU2 hotspot as follows. Because the NHY1210 and NHY1215 haploids contain a deletion of URA3, thereby preventing integration of pKB80 (URA3 GAL4-ER), the TRP1 GAL4-ER-integrating plasmid pEP105 was used instead. To allow the use of TRP1 as a selectable marker, the first 222 bp of the TRP1 ORF were substituted with natMX4 to make NHY1210 trp1 and NHY1215 trp1 (this haploid was subsequently found to be disomic for chromosome III). The PGAL1 promoter was fused to NDT80 using pFA6a-kanMX6-pGAL1 (Longtine et al. 1998) in both strains, which were then transformed with pEP105 digested with NheI to introduce GAL4-ER. The resulting strains, NHY1210 trp1 NDT80-IN::pEP105 and NHY1215 trp1 NDT80-IN::pEP105, were mated to make NH2232. A rad54∆ derivative was created by deleting RAD54 with hphMX4 in the parents of NH2232 and mating the resulting haploids to create NH2240. For the NDT80-IN RAD54-IN diploid, the PGAL1 promoter was introduced upstream of RAD54 in NHY1210 trp1 NDT80-IN::pEP105 and the resulting haploid was mated to NHY1215 trp1 NDT80-IN::pEP105 to make NH2245.
The dmc1∆ NDT80-IN mek1-as diploid, NH2278, was constructed by first deleting MEK1 with hphMX4 from NHY1210 trp1 NDT80-IN::pEP105 and NHY1215 trp1 NDT80-IN::pEP105. The mek1-as URA3 plasmid pJR2 was digested with RsrII to target integration downstream of the mek1Δ deletion and transformed into each haploid (Callender and Hollingsworth 2010). The pJR2 plasmid was then popped out using 5′-fluoro-orotic acid (FOA) to select for Ura− candidates (Boeke et al. 1984). 5-FOAR colonies were screened for sensitivity to HygB to identify those popouts in which the mek1-as allele remained in the chromosome. The second exon of DMC1 was deleted using hphMX4 from NHY1210 trp1 NDT80-IN::pEP105 mek1-as and NHY1215 trp1 NDT80-IN::pEP105 mek1-as, and the haploids were mated to make NH2278. The dmc1∆ NDT80-IN mek1-as rad17∆ diploid, NH2365, was created by deleting RAD17 from NHY1210 trp1 dmc1 mek1-as NDT80-IN::pEP105 and NHY1215 trp1 dmc1 mek1-as NDT80-IN::pEP105 using K. lactis URA3 and the resulting haploids were mated.
MMS sensitivity assay
Single colonies for each strain were inoculated into 3 ml YPD and grown overnight on a roller at 30°. The optical density at wavelength 600 nm (OD600) of each culture was measured and cells were diluted to an OD600 of 0.6 in 2 ml YPD in two test tubes. The cells were incubated on a roller drum at 30° for 2 hr, at which time 5 mM ED was added to a final concentration of 1 μM to one tube for each pair, and an equal volume of 100% ethanol was added to the other tube. After a further 2 hr at 30°, the OD600 was determined, and the volume of cells equivalent to two OD600 units from each culture was transferred to 1.5-ml microcentrifuge tubes. Cells were pelleted by centrifugation at 13,000 rpm for 1 min at room temperature and then washed with 1 ml of sterile distilled water. The pellets were resuspended in 100 μl water and tenfold serial dilutions were spotted onto YPD, YPD + 0.04% MMS, and YPGal + 0.04% MMS plates. The YPD plates were incubated for 1 day at 30°, while the YPD + 0.04% MMS and YPGal+0.04%MMS plates were incubated for 2 days at 30° before scoring.
Meiotic time courses
The protocol for sporulation was described in Lo and Hollingsworth (2011). All experiments were carried out at 30°. To induce transcription of RAD54-IN, NDT80-IN, and CDC5-IN, 5 mM ED was added to a final concentration of 1 µM at the indicated times. Cells at various time points were taken and processed as follows. For meiotic progression, 0.5 ml of culture was fixed in 3.7% formaldehyde and stored at 4°. The fixed cells were subsequently spotted onto wells of 5 μl of 1 × PBS on lysine-coated slides (Carlson Scientific, #101204). The slides were left at room temperature for 5 min and the PBS solution was removed by aspiration. The cells were washed three times using 1 × PBS. After the final wash, the slides were left at room temperature for 10 min. One microliter of mounting medium containing 1.5 μg/ml 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA, #H-1200) was added to each well. A 24 × 60 mm cover slip was placed on the slides and clear nail polish was used to seal cover slips to the slides. The number of binucleate (Meiosis I) and tetranucleate (Meiosis II) cells was determined using fluorescence microscopy.
For protein analysis, 5 ml culture was transferred to a 15-ml conical tube at each time point and the cells were pelleted in a tabletop centrifuge and placed at −80°. For Southern blot analysis of DSBs, 10 ml of Spo was transferred to a 50-ml conical tube containing 10 ml 100% ethanol and 2 ml 0.5 M ethylenediaminetetraacetic acid (EDTA) and stored at −20°. Sporulation of each culture was scored after 24 hr by light microscopy using a Zeiss AxioScope microscope (Zeiss [Carl Zeiss], Thornwood, NY). Two hundred cells were counted for each time point.
For tetrad dissection, 100 μl of Spo was added to 1 ml sterile water in a 1.5-ml microcentrifuge tube. Three microliters of 10 mg/ml Zymolyase T-100 was added and the cells incubated at 37° for 15 min and then placed on ice. Tetrads were dissected using a Zeiss tetrad dissecting microscope and the spores incubated at 30° for ∼3 days.
Physical analysis of DSBs
Physical analysis was performed using the method described in Oh et al. (2009). DNA was isolated using the MasterPure Yeast DNA Purification kit (Epicentre, Cat. # MPY80200). DNA was digested with XhoI and probed after fractionation on a 0.6% agarose gel.
Western blots
For protein analyses shown in Figure 1, Figure 3, and Figure 5, 5 ml of culture were harvested using a swinging bucket rotor Beckman Coulter Allegra X-15R centrifuge (Beckman, Fullerton, CA) at 3000 rpm for 1 min at 4°. The cells were washed once with 1 ml water and the pellets resuspended in 1 ml 20% Trichloroacetic acid (TCA) (Sigma 91228). After pelleting the cells by centrifugation as before, each pellet was weighed and resuspended in 200 μl 20% TCA and stored at −80°. For Figure 2, Figure 6, and Figure S2, the extracts were prepared as described in falk et al. (2010). Protein gels were hand cast using TGX FastCast acrylamide gel solutions as described by the manufacturer (7.5 or 12%, Bio-Rad, Hercules, CA, #1610171 and #1610175, respectively). Protein samples were fractionated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using a Criterion Cell midi-format vertical electrophoresis cell (Bio-Rad, #1656001) with a PowerPac Basic power supply (Bio-Rad #1645050) at a constant voltage of 200–300 V for 40 min. Proteins were transferred to polyvinylidine fluoride (PVDF) membrane (Millipore, Bedford, CA, #IPVH00010) using a Criteron Blotter with Plate Electrodes (Bio-Rad #1704070).
Figure 1.
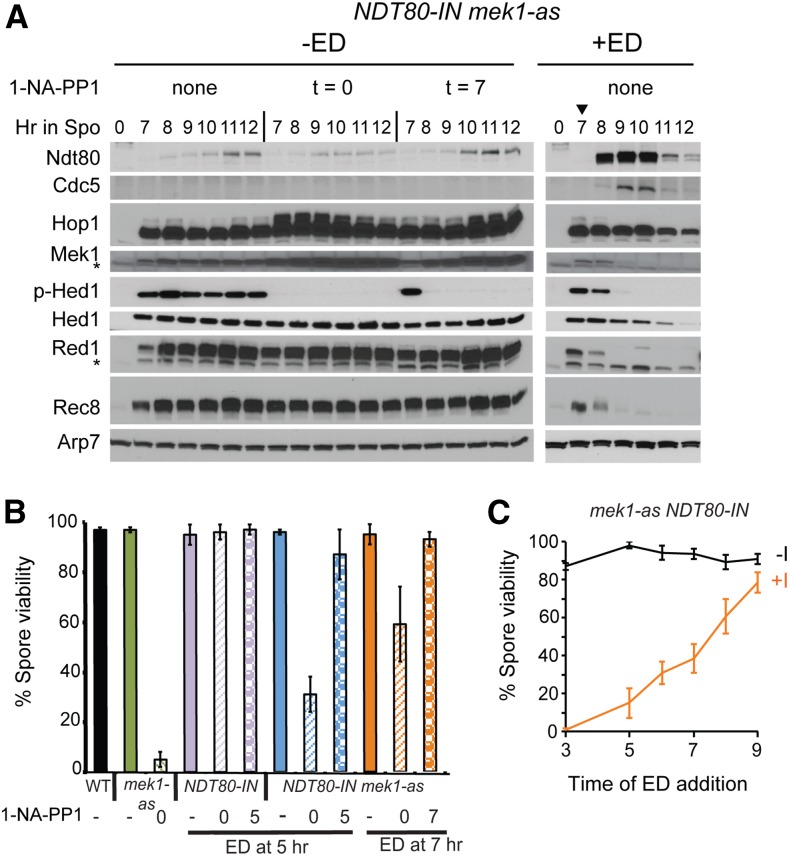
Mek1 inactivation resulting from NDT80 induction correlates with the presence of Cdc5. (A) The NDT80-IN mek1-as diploid, NH2122::pJR2, was transferred to Spo medium. The Mek1-as inhibitor, 1-NA-PP1, was added at a final concentration of 1 μM at the indicated times. Either DMSO (−ED) or a final concentration of 1 μM ED (+ED) was added at 7 hr (indicated by arrowhead). Protein extracts were then probed with antibodies against Ndt80, Cdc5, Hop1, Mek1, phospho-Hed1 threonine 40 (p-Hed1), Hed1, Red1, Rec8, and Arp7 as a loading control. Asterisks indicate nonspecific bands. “p-Hop1” indicates the Hop1 mobility shift due to phosphorylation. Three biological replicates of this experiment gave similar results. (B) Spore viability in NDT80-IN diploids in which Mek1-as was inactivated either before or after NDT80 induction. WT (NH144), mek1-as (NH729::pJR2), NDT80-IN (NH2033), and NDT80-IN mek1-as (NH2122::pJR2) were transferred to Spo medium. NDT80 was induced using 1 μM ED and Mek1-as was inhibited using 1 μM 1-NA-PP1 at the indicated times. The resulting tetrads were dissected to determine the percent viable spores. The averages of at least three biological replicates are shown with error bars indicating the standard deviations (SDs). (C) Spore viability in the absence of Mek1 activity as a function of time of NDT80 induction. The NDT80-IN mek1-as diploid, NH2122::pJR2, was transferred to Spo medium and divided between two flasks. One flask had no 1-NA-PP1 (−I) while the other contained 1 μM 1-NA-PP1 added at t = 0 (+ I). Cells were removed at the indicated timepoints, ED was added (+ED), and the cells were placed back on the shaker for > 24 hr to complete sporulation. “−ED” indicates that no ED was added. Error bars represent the SD from the dissections performed on different plates. A minimum of 77 and 103 tetrads were dissected for the no inhibitor and plus inhibitor time points, respectively. ED, estradiol; Spo, sporulation medium; WT, wild-type.
Figure 3.
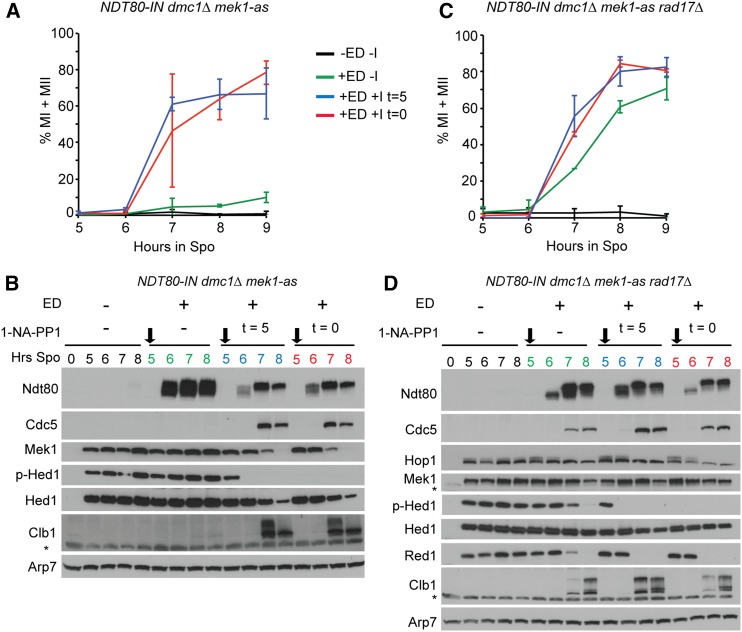
NDT80-dependent inactivation of Mek1 is independent of DMC1-mediated strand invasion. Meiotic progression. (A) The NDT80-IN dmc1∆ mek1-as diploid (NH2278) was transferred to Spo medium for 5 hr and a final concentration of 1 μM 1-NA-PP1 and/or ED were added at the indicated times. Meiotic progression was assayed using fluorescent microscopy of fixed DAPI-stained nuclei to determine the percentage of MI and MII cells. Two hundred cells were counted for each strain at each time point. The average values from two experiments were plotted with error bars indicating the range. (B) Meiotic progression in the NDT80-IN dmc1∆ mek1-as rad17∆ diploid (NH2365) was analyzed as in (A). (C) Immunoblot analysis of various proteins from extracts of cells taken from one of the NDT80-IN dmc1∆ mek1-as time courses shown in (A). Asterisks indicate nonspecific bands. Black arrows indicate the time of ED addition. (D) Immunoblot analysis using extracts from one of the NDT80-IN dmc1∆ mek1-as rad17∆ time courses shown in (B). Two biological replicates of each strain gave similar results. ED, estradiol; MI, binucleate; MII, tetranucleate; Spo, sporulation medium.
Figure 5.
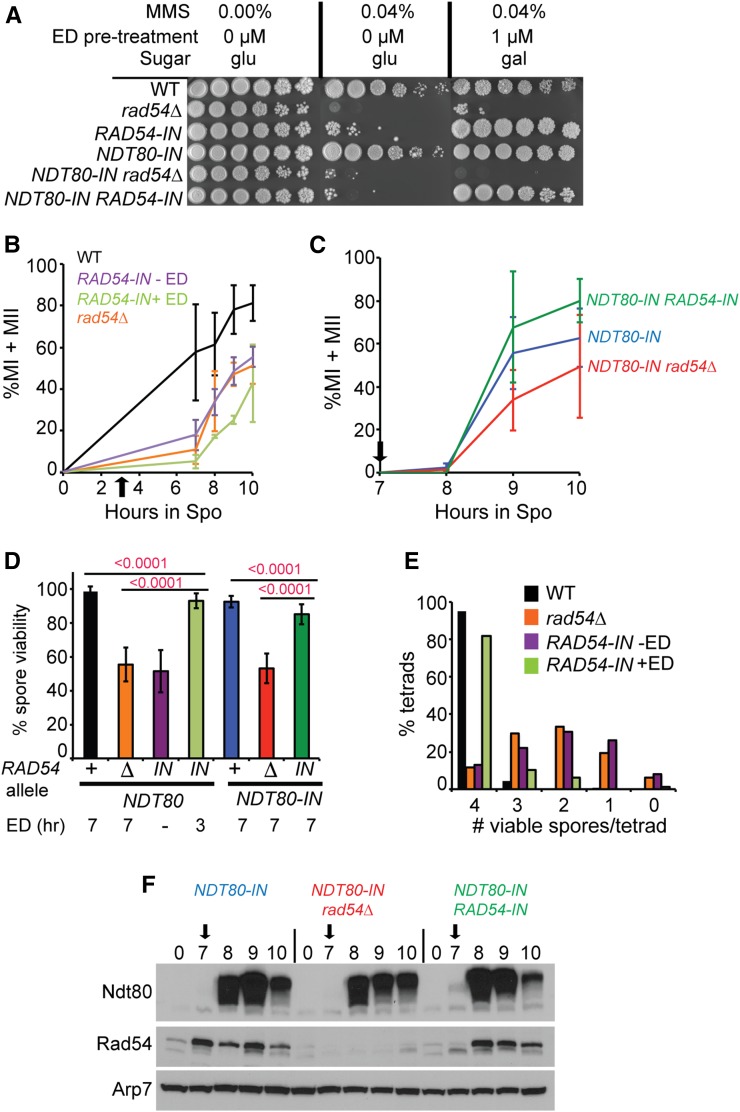
RAD54 functions after NDT80 induction to promote meiotic progression and the formation of viable spores. (A) MMS sensitivity. WT (NH144), rad54Δ (NH2136), RAD54-IN (NH2319), NDT80-IN (NH2127), NDT80-IN rad54Δ (NH2126), and NDT80-IN RAD54-IN (NH2185) diploids were grown to log phase in YPD. For one set of strains, ED was added to a final concentration of 1 μM for 2 hr prior to plating. Tenfold serial dilutions were spotted onto YP plates containing either 2% glu or gal with or without 0.04% MMS as indicated. (B) Meiotic progression in NDT80 diploids. WT, rad54∆, and RAD54-IN diploids were transferred to Spo medium and incubated at 30°. After 3 hr, the RAD54-IN culture was split with a final concentration of 1 μM ED added to one of the RAD54-IN cultures (indicated by black arrow). Meiotic progression was assayed using fluorescent microscopy of fixed DAPI-stained nuclei to determine the percentage of MI and MII cells. Two hundred cells were counted for each strain at each time point. The average values from three biological replicates are shown. Error bars indicate the SDs. (C) Meiotic progression in NDT80-IN diploids. ED was added at 7 hr (black arrow) and progression was monitored as in (B). (D) Spore viability. ED was added to 1 μM at the indicated timepoints. Lines indicate the P-values using a χ2 test. Average values are shown with error bars indicating the SDs. For the NDT80-IN strains, two independently constructed diploids using different SK1 parents exhibited similar results and therefore the data were combined. For each strain, the number of biological replicates: number of tetrads is: NH144 (3:120), (4:162), NH2319 −ED (7:185), NH2319 +ED at 3 hr (7:172), NH2127 + NH2232 (14:524), NH2126 + NH2240 (10:423), and NH2185 + NH2245 (10:318). (E) Distribution of viable spores in tetrads for WT, rad54∆, and RAD54-IN without and with ED from the dissections presented in (D). (F) Induction of Rad54 and Ndt80 in NDT80-IN diploids. The black arrows indicate addition of ED at 7 hr after transfer to Spo medium. This experiment was performed three times with similar results. ED, β-estradiol; gal, galactose; glu, glucose; MI, binucleate; MII, tetranucleate; MMS, methyl methanesulfonate; Spo, sporulation medium; WT, wild type.
Figure 2.
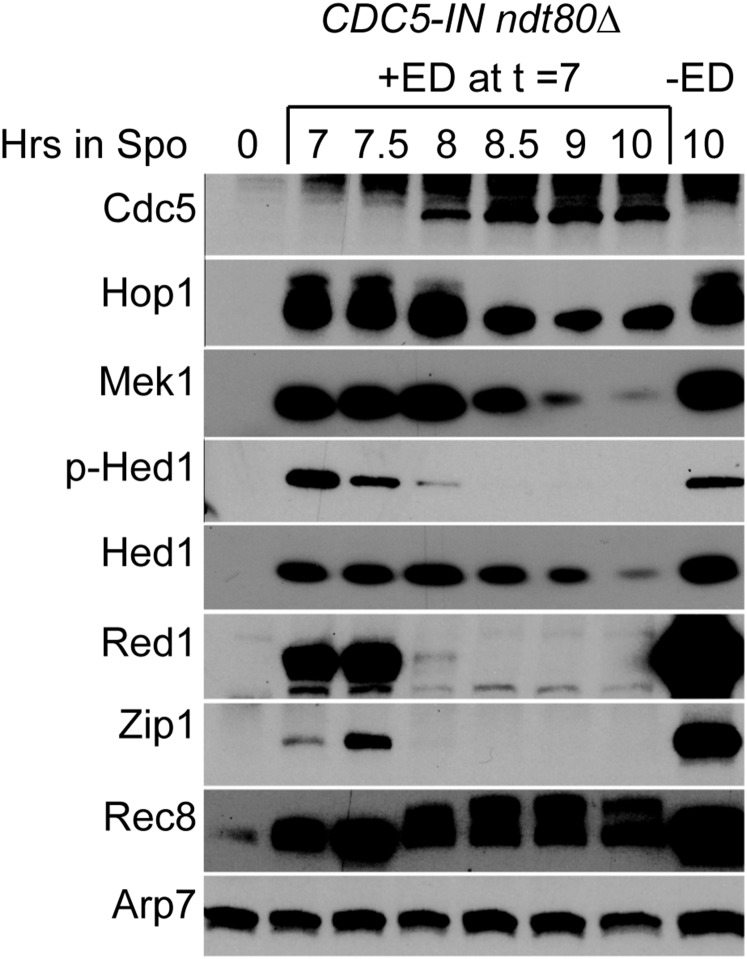
CDC5 is sufficient to inactivate Mek1. The CDC5-IN ndt80Δ diploid, NH2296, was transferred to Spo medium and incubated for 7 hr, at which time the culture was split in half and either DMSO (−ED) or a final concentration of 1 μM ED (+ED) was added. Protein extracts were then probed with antibodies against, Cdc5, Hop1, Mek1, phospho-Hed1 T40 (p-Hed1), Hed1, Red1, Zip1, and Rec8. Arp7 was used as a loading control. This experiment was repeated five times with similar results. ED, estradiol; Spo, sporulation medium.
Figure 6.
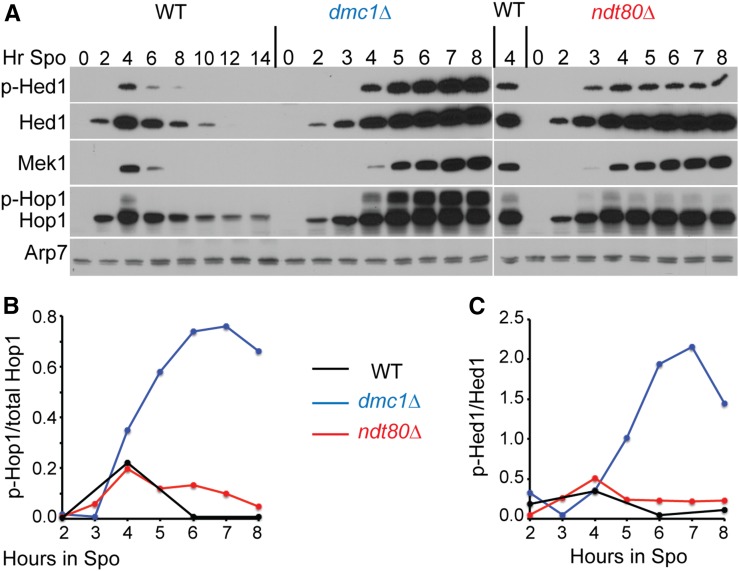
Mek1 kinase activity correlates with the amount of phosphorylated Hop1. (A) WT (NH716), dmc1∆ (NH792), and ndt80∆ (NH2188) diploids were transferred to Spo medium and assayed at different time points for the presence of Hed1, Hed1 phospho-T40 (p-Hed1), Hop1, Mek1, and Arp7. (B) Fraction of phosphorylated Hop1 as a function of time. The Hop1 and p-Hop1 bands from the same lanes were quantified and normalized to Arp7. The ratio of p-Hop1/(p-Hop1 + Hop1) was plotted. (C) Fraction of p-Hed1 as a function of time. Hed1 and p-Hed1 were normalized using Arp7 from the same lane for each timepoint and the p-Hed1/Hed1 ratio was plotted. This experiment was repeated twice with similar results. Spo, sporulating culture; WT, wild type.
Antibodies
Primary antibodies were incubated overnight between 16 and 20 hr at 4°, except α-Ndt80 and α-Hop1, which were incubated at room temperature for 2 hr. Secondary antibodies were incubated at room temperature for 1 hr. Antibody dilutions and sources are listed in Table S2.
Mek1 and Rec8 antibodies were generated by Covance Research Products in guinea pigs using the peptides CISQAIPKKKKVLE and QKDGNDFKFNYQDEC, respectively. The Rec8 antibodies were affinity purified from the serum by Covance using the peptide. The specificity of the antibodies was confirmed by probing yeast extracts from wild-type, mek1∆, or rec8∆ diploids isolated either at 0 or 4 hr after transfer to Spo medium. For Mek1, a protein of the correct molecular weight was observed only in the wild-type diploid undergoing meiosis, as expected given that MEK1 is a meiosis-specific gene (Rockmill and Roeder 1991; Leem and Ogawa 1992) (Figure S1A). There is a nonspecific, NDT80-induced band that runs above Mek1, so it is important to include a zero time point or mek1∆ as a control. For Rec8, a highly induced protein band was observed between 95 and 130 kDa after 4 hr, consistent with Rec8 being meiosis-specific (Figure S1B) (Klein et al. 1999). The detected protein ran slower in the gel than the predicted molecular weight of 77 kDa, likely due to extensive phosphorylation of Rec8 (Brar et al. 2006; Katis et al. 2010). A weak band of the same molecular weight was observed both in the 0 hr time point from wild-type and the 4 hr time point from rec8∆, indicating a nonspecific band (Figure S1B, data not shown). This band can be eliminated by using large dilutions of the antibody (1:100,000).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. All strains, plasmids, and antibodies are available upon request.
Results
Mek1 is constitutively active at the ndt80 arrest
To analyze Mek1 kinase activity in the absence or presence of NDT80 in the same culture, a diploid containing a conditional allele of NDT80 (NDT80-IN) was used. Transcription of NDT80-IN can be induced by addition of ED to the Spo medium (Benjamin et al. 2003; Carlile and Amon 2008). In addition, the diploid contained the mek1-as analog-sensitive mutant, which encodes a kinase that can be inactivated by addition of the purine analog 1-NA-PP1 to the medium (Wan et al. 2004).
An NDT80-IN mek1-as diploid was transferred to Spo medium and separated into three flasks. No Mek1-as inhibitor was added to one flask, while 1-NA-PP1 was added either at 0 or 7 hr to the other flasks, respectively. Cells were taken at various time points and protein extracts probed with antibodies to detect various proteins. In the absence of ED, low levels of Ndt80 protein were observed, but these amounts were not sufficient to induce expression of CDC5, as no Cdc5 protein was detected (Figure 1A, −ED). Hop1 phosphorylation is an indirect indicator of meiotic DSB formation, as well as being required for Mek1 activation (Niu et al. 2005; Carballo et al. 2008). Phosphorylated Hop1 results in slower migrating species that can be detected on SDS polyacrylamide gels (Niu et al. 2005). In the absence of ED, phosphorylated Hop1 persisted, regardless of whether Mek1 was inhibited (Figure 1A). This observation is consistent with the fact that DSBs continue to occur at the ndt80 arrest and that Hop1 phosphorylation occurs prior to Mek1 activation (Allers and Lichten 2001; Carballo et al. 2008; Goldfarb and Lichten 2010; Thacker et al. 2014; Subramanian et al. 2016). Inactivation of Mek1-as, either 0 or 7 hr after transfer to Spo medium, resulted in an increase in total Hop1 and Mek1 levels, as well as Hop1 phosphorylation (Figure 1A). Similar increases were not observed for other meiosis-specific proteins such as Hed1, Red1, or Rec8. Why the combination of prophase arrest and inactivation of Mek1 specifically increases the amount of Hop1 and Mek1 is not clear.
Total Hed1 levels were constant for up to 12 hr in the absence of ED (Figure 1A, −ED). Probing with an antibody specific for the phosphorylated form of Hed1 T40 revealed Hed1 phosphorylation similarly persists until at least 12 hr. No Hed1 T40 phosphorylation was observed when Mek1-as was inactivated immediately after transfer to Spo medium, consistent with previous experiments demonstrating that Mek1 directly phosphorylates Hed1 T40 (Callender et al. 2016). Inactivation of Mek1-as after 7 hr in Spo medium resulted in the disappearance of Hed1 T40 phosphorylation within 1 hr, proving that constitutive Mek1 activity is needed to maintain Hed1 in the phosphorylated state in pachytene-arrested cells (Figure 1A, −ED).
Induction of NDT80 results in Red1 degradation and inactivation of Mek1
Induction of NDT80 by the addition of ED resulted in the production of Cdc5 and a loss of Hop1 phosphorylation, suggesting that DSBs were repaired (Figure 1A, +ED). The disappearance of Hed1 T40 phosphorylation (an indirect indicator of Mek1 activity) similarly correlated with the presence of Cdc5. Hed1 T40 phosphorylation was lost more quickly than total Hed1 protein, indicating that dephosphorylation precedes Hed1 degradation, consistent with previous work showing that a negative charge at Hed1 T40 promotes protein stability (Figure 1A, +ED) (Callender et al. 2016). However, dephosphorylation is not sufficient for Hed1 degradation, since the Hed1 protein was stable in the absence of ED and Mek1 kinase activity. Therefore, induction of NDT80 is also necessary for proteolysis of Hed1.
After induction of NDT80, Hop1 levels slowly decreased, while both Red1 and Rec8 proteins were degraded within 2 hr (Figure 1A, +ED). Red1 and Rec8 degradation was not due to the loss of Mek1 kinase activity, because steady-state levels of both proteins were unchanged in the ndt80-arrested cells in which Mek1-as was inactivated (Figure 1A, −ED). Induction of NDT80 resulted in progression through the meiotic divisions and Rec8 degradation was likely due to separase-mediated proteolytic cleavage of Rec8 (Buonomo et al. 2000).
Mek1 kinase activity is required before, but not after, NDT80 induction for the formation of viable spores
Because MEK1 is required to suppress intersister recombination during meiosis, diploids lacking Mek1 activity make dead spores due to chromosome missegregation at Meiosis I (Rockmill and Roeder 1991; Wan et al. 2004; Kim et al. 2010) (Figure 1B). Execution point experiments using mek1-as demonstrated that Mek1 kinase activity is required only in prophase to produce viable spores (Wan et al. 2004). To further resolve the timing of Mek1 function relative to Ndt80, an NDT80-IN mek1-as diploid was incubated in Spo medium for either 5 or 7 hr. NDT80 was induced by the addition of ED at the same time that Mek1 inhibitor was added and the resulting tetrads dissected to determine spore viability. The efficacy of the inhibitor was confirmed by showing that a mek1-as NDT80 diploid sporulated in the absence of 1-NA-PP1 exhibited 97.8% viable spores, compared to 5.1% in the presence of inhibitor (Figure 1B). The inhibitor only affects Mek1-as, as 1-NA-PP1 had no effect on the spore viability of a MEK1 NDT80-IN strain (Figure 1B). Inhibiting Mek1-as immediately after transfer to Spo medium significantly reduced spore viability when NDT80-IN was induced at either 5 or 7 hr compared to the no inhibitor control (χ2 test, P < 0.0001) (Figure 1B). The spore viability was not as low as the mek1-as + 1-NA-PP1 control, however, and increased when cells were arrested longer prior to NDT80 induction (Figure 1, B and C). These results indicate that prolonged time at the ndt80 arrest allows increasing amounts of interhomolog recombination in the absence of Mek1 kinase activity and are consistent with physical analyses of mek1∆ ndt80∆ diploids showing a switch from intersister to interhomolog joint molecules around the time that NDT80 would normally be induced (Goldfarb and Lichten 2010).
Inactivation of Mek1-as at the same time as NDT80 induction resulted in wild-type levels of viable spores after the 7-hr arrest (χ2 test, P < 0.142) (Figure 1B). A similar result was observed for the 5-hr arrest, although spore viability was slightly reduced compared to the no inhibitor control (χ2 test, P < 0.0001) (Figure 1B). These data support the idea that Mek1-as kinase activity is required prior to NDT80 induction while the bulk of interhomolog recombination is occurring, but is not required after NDT80 is transcribed, as expected given that the kinase is inactivated by Ndt80.
CDC5 is sufficient to inactivate Mek1
Ndt80 activates the transcription of > 200 genes, one of which encodes the polo-like kinase Cdc5, which is required for Holliday junction resolution and SC disassembly (Clyne et al. 2003). CDC5 is the sole Ndt80 target required for these processes as ectopic expression of CDC5-IN in the ndt80∆ background is sufficient for Holliday junction resolution and SC disassembly (Sourirajan and Lichten 2008). To see whether CDC5 is the sole Ndt80 target responsible for inactivation of Mek1, a diploid homozygous for ndt80∆ and containing one copy of an inducible allele of CDC5 (CDC5-IN) was constructed. In the absence of ED, Hed1 was still phosphorylated after 10 hr in Spo medium (Figure 2). The SC proteins, Hop1, Red1, and Zip1, were stable and the meiosis-specific cohesin subunit, Rec8, was unphosphorylated. Addition of ED after 7 hr in Spo medium resulted in the appearance of Cdc5 an hour later (Figure 2). The timing of Cdc5 production correlated with the disappearance of Red1 and Zip1. Mek1 was inactivated, as phosphorylated Hed1 was greatly reduced by 8 hr and eliminated by 9 hr. Mek1 protein was still present at 8 hr, albeit at a lower level, suggesting that the kinase may be inactivated by dephosphorylation of T327 in the activation loop of Mek1, as opposed to simply undergoing protein degradation (Figure 2) (Niu et al. 2007). Rec8 mobility decreased at 8 hr, likely due to CDC5-dependent phosphorylation (Brar et al. 2006; Attner et al. 2013). In contrast to the NDT80-IN diploid, the Rec8 protein was not degraded because the absence of the Ndt80 target, CLB1, prevents entry to the meiotic divisions (Chu and Herskowitz 1998).
Previously, it was reported that induction of NDT80, but not CDC5, results in Zip1 degradation, in contrast to our results (Figure 2 and Figure S2) (Sourirajan and Lichten 2008). One difference between the two studies was the allele of CDC5 that was used. The published work using a hemizygous N-terminally 3HA-tagged version of CDC5 failed to see degradation of Zip1, while Zip1 was degraded in our experiments using a hemizygous untagged CDC5 allele, indicating that the presence of the HA tag decreases CDC5 function (Figure 2 and Figure S2). To test whether Cdc5 kinase activity is necessary for Red1 degradation and Mek1 inactivation, diploids homozygous for either CDC5-IN or the kinase dead cdc5-N209A mutant (cdc5-N209A-IN) were compared (Hardy and Pautz 1996). The Zip1 protein was stable in the mutant, but the level of untagged Cdc5-N209A protein was reduced, making it unclear whether the phenotype was due to catalytic inactivation of the kinase or reduced protein levels (Figure S2). The N209A mutation and the 3HA tag appear to independently destabilize Cdc5, as the 3HA-Cdc5-N209A protein was more unstable than either Cdc5-N209A or 3HA-Cdc5 alone (Figure S2). Therefore, caution should be used when interpreting results obtained from either the 3HA-CDC5 or cdc5-N209A alleles. We conclude that Cdc5 is sufficient to induce degradation of both Red1 and Zip1.
Mek1 kinase inactivation by NDT80 does not require CDC5-mediated Holliday junction resolution
In addition to Red1 and Zip1 degradation, another function of CDC5 is Holliday junction resolution to generate crossovers (Sourirajan and Lichten 2008). One possibility is Holliday junction resolution is the signal to inactivate Mek1. This idea was tested by examining Mek1 activity in dmc1∆ mek1-as NDT80-IN diploids. If Holliday resolution is a prerequisite for Mek1 inactivation by Ndt80, then Hed1 phosphorylation should persist when NDT80 is induced with ED.
Previous work using the BR strain background showed that overexpression of NDT80 partially suppresses the dmc1∆ checkpoint arrest (Tung et al. 2000), suggesting that induction of NDT80-IN by ED might also bypass this arrest. However, this was not the case in our SK1 strains. Although high levels of Ndt80 protein were observed in the NDT80-IN dmc1∆ mek1-as diploid after addition of ED, the cells remained arrested in meiotic prophase (Figure 3, A and B). Furthermore, Cdc5 and Clb1 were not detected, indicating that the Ndt80 generated during the checkpoint arrest was unable to activate transcription of either CDC5 or CLB1 (Figure 3B) (Chu and Herskowitz 1998; Chu et al. 1998; Clyne et al. 2003). This result is consistent with the meiotic recombination checkpoint inactivating Ndt80 by sequestration of the Ndt80 protein in the cytoplasm (Wang et al. 2011). Hed1 T40 phosphorylation and Mek1 protein persisted, as expected given the lack of Cdc5 (Figure 3B).
Inactivation of Mek1-as in dmc1∆ diploids results in intersister repair of DSBs, thereby allowing meiotic progression by eliminating the unrepaired DSBs that trigger the checkpoint (Wan et al. 2004; Niu et al. 2005). In fact, inhibition of Mek1-as either immediately after transfer to Spo medium or at the time of NDT80 induction resulted in production of both Cdc5 and Clb1, inactivation of Mek1 (i.e., disappearance of Hed1 T40 phosphorylation), and meiotic progression (Figure 3, A and B). The robust checkpoint inhibition of Ndt80 activity precluded testing whether NDT80 induction inactivates Mek1 in the absence of DMC1 in this diploid.
To eliminate the checkpoint inhibition of Ndt80 in the dmc1∆ background, RAD17 was deleted from the NDT80-IN dmc1∆ mek1-as strain (Chu and Herskowitz 1998; Pak and Segall 2002). In this strain, addition of ED resulted in the production of transcriptionally active Ndt80 and the cells progressed through the meiotic divisions (Figure 3C). Both Cdc5 and Clb1 proteins appeared within 1 hr after Ndt80 protein was observed and this timing correlated with the disappearance of Red1 and phosphorylated Hed1 (Figure 3D). Therefore, Dmc1-mediated interhomolog recombination is not required for Ndt80-mediated inactivation of Mek1, nor is the Holliday resolution function of Cdc5.
RAD17 is required for one of two pathways necessary to activate Mek1 (Ho and Burgess 2011). In addition, deletion of another component of the checkpoint, RAD24, exhibits reduced Mek1 phosphorylation, which is indicative of activation (Gray et al. 2013). A reduction in Mek1 kinase activity could explain why Ndt80 was active when induced in the dmc1∆ rad17 strain, but this reduction was not low enough to affect Mek1’s ability to phosphorylate Hed1 (compare p-Hed and Hed1 in the uninduced strains in Figure 3, B and D). The elimination of p-Hed1 in the rad17∆ strain required the presence of Ndt80 (Figure 3D, +ED no 1-Na-PP1). CDC5 is the sole target of Ndt80 necessary to inactivate Mek1 in the dmc1∆ background, as induction of CDC5 alone in dmc1∆-arrested cells results in Red1 degradation and meiotic progression (Okaz et al. 2012). Induction of CDC5 bypasses the prophase checkpoint as Red1 degradation was also observed in the dmc1∆ ndt80∆ background (Okaz et al. 2012). A simple interpretation of these results is that Ndt80 induction of Cdc5 results in disassembly of the AEs by degradation of Red1, thereby eliminating all remaining Mek1 activity.
NDT80-dependent inactivation of Mek1 promotes DSB repair in dmc1∆ mutants
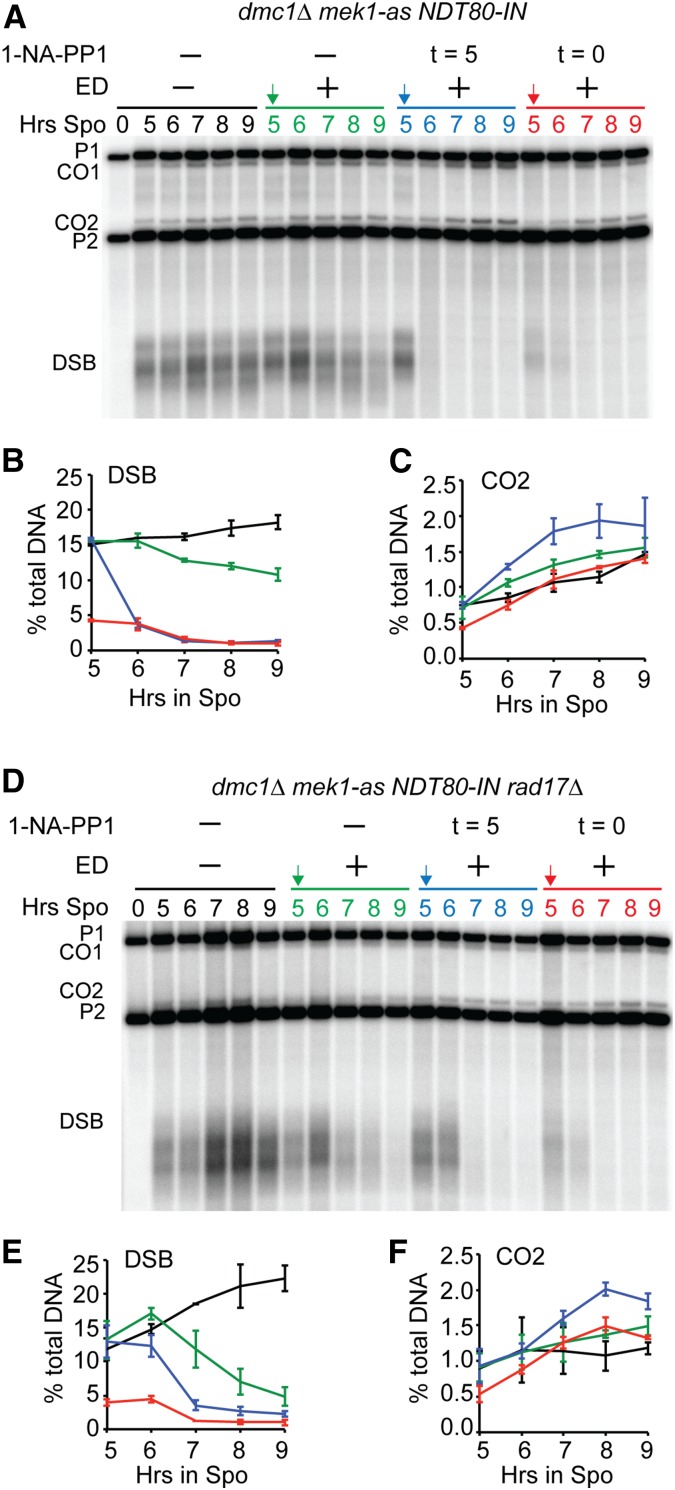
Mek1 downregulates Rad51 during meiosis by inhibiting Rad51-Rad54 complex formation through phosphorylation of Rad54 and Hed1 (Niu et al. 2009; Callender et al. 2016). Therefore, inactivation of Mek1 by Ndt80 should allow DSB repair in the dmc1∆ rad17∆ diploid. To test this idea, Southern blot analysis was performed to look at repair of DSBs at the HIS4-LEU2 hotspot on chromosome III (Hunter and Kleckner 2001). The Spo11 DSB site at this hotspot is flanked by XhoI restriction sites, allowing the detection of DSBs and crossovers using XhoI-digested genomic DNA (Hunter and Kleckner 2001; Oh et al. 2009).
Induction of NDT80 in the dmc1∆ background resulted in a slow decrease in DSBs, suggesting that some cells escaped the checkpoint (Figure 4, A and B). A band corresponding to the CO2 fragment was observed in all of the dmc1∆ diploids, even in the absence of NDT80 and Cdc5, suggesting that it was not generated by Holliday junction resolution (Figure 3, B and D and Figure 4, A and D). One explanation for this band is that the extensive resection that occurs in dmc1∆ mutants resulted in noncrossover gene conversion of the diagnostic XhoI polymorphism closest to the DSB site. Artificially inactivating Mek1-as by addition of inhibitor, either at 0 or 5 hr after transfer to Spo medium, resulted in repair of the breaks as expected (Figure 4, A and B). In contrast, in the dmc1 mek1-as NDT80-IN rad17∆ strain, DSBs persisted in the absence of ED but disappeared more quickly upon expression of NDT80 compared to the RAD17 strain (Figure 4, D and E). The kinetics of repair was slower than that observed for the culture in which Mek1-as was directly inactivated by inhibitor. Two possible explanations for this are: (1) since there are two independent pathways for activating Mek1, one that requires RAD17 and the other PCH2, some Mek1 is still activated in the rad17∆ (Ho and Burgess 2011); and (2) addition of 1-NA-PP1 rapidly inhibits the kinase, whereas inactivation of Mek1 via Ndt80 requires time to transcribe and translate both NDT80 and CDC5 and for Red1 to be degraded.
Figure 4.
Induction of NDT80 allows Rad51-mediated DSB repair in the dmc1∆ rad17∆ background. Diploids containing either dmc1∆ mek1-as NDT80-IN (NH2278) or dmc1∆ mek1-as NDT80-IN rad17∆ (NH2365) were transferred to Spo medium for 5 hr, and a final concentration of 1 μM 1-NA-PP1 and/or ED was added at the indicated times. Arrows indicate ED addition. DSBs and COs at the HIS4::LEU2 hotspot were detected by XhoI digestion of genomic DNA probed with a 0.6-kb AgeI/BglII fragment from pNH90 (hunter and kleckner 2001). P1 and P2 = parental bands; CO1 and CO2 = CO bands; DSB = DSB fragments. “t” indicates the time that the Mek1-as inhibitor, 1-NA-PP1, was added. (A) Southern blot of NDT80-IN dmc1∆ mek1-as from one of two independent time courses. (B) DSB fragments as a percent of the total DNA is plotted for each timepoint. The averages between two independent experiments are shown with error bars indicating the range. (C) COs for the experiment shown in (A) plotted as in (B). (D) Southern blot of NDT80-IN dmc1∆ mek1-as rad17∆ from one of two independent time courses. (E and F) DSBs and COs, respectively, for the experiment shown in (D). CO, crossover; DSB, double strand break; ED, estradiol; Spo, sporulation medium.
RAD54 functions after the bulk of meiotic interhomolog recombination to promote spore viability
During wild-type meiosis, the bulk of Mek1 activity is eliminated by chromosome synapsis (Subramanian et al. 2016). Depletion of RAD54 from ndt80∆-arrested cells results in increased Rad51 foci, suggesting that the reduction in Mek1 activity has resulted in Rad51-Rad54 complexes repairing some DSBs at the ndt80∆ arrest. However, the persistence of Hed1 phosphorylation at the ndt80 arrest, as well as the fact that some hotspots still exhibit interhomolog bias, indicates that synapsis is not sufficient to eliminate all Mek1 activity, nor to completely activate Rad51. Furthermore, the depletion experiment does not rule out an early function for RAD54, nor does it address whether the Rad54-mediated DSB repair that occurs after the bulk of interhomolog recombination is necessary for spore viability. To address these questions, a complementary approach was used to induce RAD54 at the same time as NDT80 to see if addition of RAD54 after interhomolog recombination is sufficient to restore the sporulation and spore viability defects observed for rad54 mutants (Shinohara et al. 1997; Schmuckli-Maurer and Heyer 2000).
A conditional allele of RAD54 (RAD54-IN) was created by putting RAD54 under control of the GAL1 promoter in a strain containing GAL4-ER. To confirm that RAD54-IN is inducible with ED, several phenotypes were examined. In vegetative cells, rad54∆ is sensitive to the alkylating agent MMS (Game and Mortimer 1974; Shinohara et al. 1997). Serial dilutions of RAD54, rad54∆, and RAD54-IN in the presence of either NDT80 or NDT80-IN were plated onto 0.04% MMS with either glucose or galactose as the carbon source. In the absence of ED, the RAD54-IN strains were killed by MMS on the glucose medium, similar to the rad54∆ controls (Figure 5A). In contrast, addition of 1 μM ED to liquid cultures 2 hr prior to plating on galactose medium rescued the MMS sensitivity of RAD54-IN, but not rad54∆, confirming that RAD54-IN is a conditional allele.
In meiotic cells, RAD54-IN was examined for rescue of the rad54∆ spore viability defect. Wild-type, rad54∆, and RAD54-IN diploids were transferred to Spo medium and the RAD54-IN culture divided in half. After 3 hr, ethanol alone was added to one half and ED to the other half. The uninduced RAD54-IN culture was functionally null in meiosis, exhibiting a similar delay in meiotic progression and reduction in spore viability as the rad54∆ (Figure 5, B and D). For reasons that are not clear, addition of ED to RAD54-IN exacerbated the meiotic progression delay, as opposed to rescuing it. However, induction of RAD54-IN improved sporulation and spore viability compared to the uninduced strain and the rad54∆, but was not to the level of wild-type (wild-type, 78.8 ± 1.6; rad54∆ 53.3 ± 26.0; RAD54-IN − ED 56.9 ± 11.6; and RAD54-IN + ED 65.6 ± 4.5) (Figure 5D). The pattern of spore lethality for both rad54∆ and RAD54-IN suggests that the spore inviability is not due to Meiosis I nondisjunction, which results in decreased numbers of tetrads with four viable spores and increased numbers of tetrads with two and zero viable spores (Hollingsworth et al. 1995) (Figure 5E). Instead, the increase in tetrads with one, two, or three viable spores suggests that the spore lethality was due to unrepaired DSBs, similar to what is observed in mms4∆ diploids (de los Santos et al. 2001). Finally, while RAD54 is naturally induced in meiotic cells (Figure 5F, compare the 0- and 7-hr time points for NDT80-IN), for RAD54-IN the increase in Rad54 protein was dependent upon ED (Figure 5F, compare 0-, 7-, and 8-hr time points for NDT80-IN RAD54-IN). Therefore, RAD54-IN is an ED-inducible allele that is null in mitotic and meiotic cells in the absence of ED.
To test whether co-induction of RAD54 with NDT80 rescues the defects in sporulation and spore viability that occur in the absence of RAD54 function, NDT80-IN, NDT80-IN rad54∆, and NDT80-IN RAD54-IN cells were arrested in prophase by incubating cells in Spo medium for 7 hr, prior to induction using 1 μM ED. Ndt80 protein was observed in all three diploids 1 hr after induction (Figure 5F), after which time cells proceeded through the meiotic divisions (Figure 5C) (Benjamin et al. 2003). The NDT80-IN rad54∆ diploid exhibited a similar decrease in spore viability as NDT80 rad54∆ (Figure 5D). Meiotic progression was delayed in NDT80-IN rad54∆, despite similar timing and levels of Ndt80 protein compared to NDT80-IN (Figure 5, C and F). This delay may be due to a DNA damage checkpoint that is triggered by DSBs that persist after Meiosis I (Cartagena-Lirola et al. 2008). Co-induction of NDT80-IN RAD54-IN rescued the delay in meiotic progression and restored sporulation and spore viability to nearly wild-type levels (Figure 5, C and D) [sporulation:NDT80-IN 58.5 ± 25.3 (n = 13), NDT80-IN rad54∆ 20.3 ± 14.8 (n = 8); and NDT80-IN RAD54-IN 50.5 ± 17.4 (n = 8)]. These results indicate that RAD54 functions after interhomolog recombination to generate viable spores, most likely in the repair of residual DSBs.
Mek1 kinase activity is correlated with the number of DSBs
Synapsis results in Mek1 removal from chromosomes and the weakening of interhomolog bias at some hotspots (Subramanian et al. 2016), yet it is clear that some Mek1 remains active in ndt80-arrested cells. One explanation for this apparent contradiction is that there are two sequential stages to Mek1 inactivation, with the first stage occurring due to repair of the bulk of DSBs that result in synapsis and the second stage occurring due to Cdc5-dependent disassembly of AEs. This hypothesis is based on the assumption that the amount of Mek1 activity reflects the number of DSBs in the cell. This idea was tested by comparing the amount of Hed1 phosphorylation (an indicator of Mek1 activity) (Callender et al. 2016) and Hop1 phosphorylation (an indicator of DSBs) (Niu et al. 2005; Carballo et al. 2008) in dmc1∆- and ndt80∆-arrested cells. These two mutants were chosen because Hed1 and Hop1 are both stable in the arrested cells, in contrast to wild-type cells where the proteins are degraded as meiosis progresses (Figure 6A). In addition, the levels of DSBs are very different; dmc1∆ strains accumulate unrepaired DSBs while ndt80∆ diploids contain only ∼2–10 DSBs/cell (Bishop et al. 1992; Subramanian et al. 2016). This difference was reflected in the amount of phosphorylated Hop1 in the two mutants. At 4 hr, the level of phosphorylated Hop1 was similar in all three strains (Figure 6, A and B). In the wild-type diploid, phosphorylated Hop1 disappeared by 6 hr, while it increased and accumulated in the dmc1∆ mutant. A low level of phospho-Hop1 persisted in the ndt80∆ strain, consistent with the low number of DSBs. Mek1 kinase activity mirrored the results with phospho-Hop1, including the persistence of a low level phospho-Hed1 in the ndt80∆ for up to 8 hr of meiotic prophase arrest. Therefore, Mek1 kinase activity is correlated with the number of DSBs in the cell.
Discussion
Meiotic progression and repair of residual DSBs are coordinated through Mek1 kinase activity and Ndt80
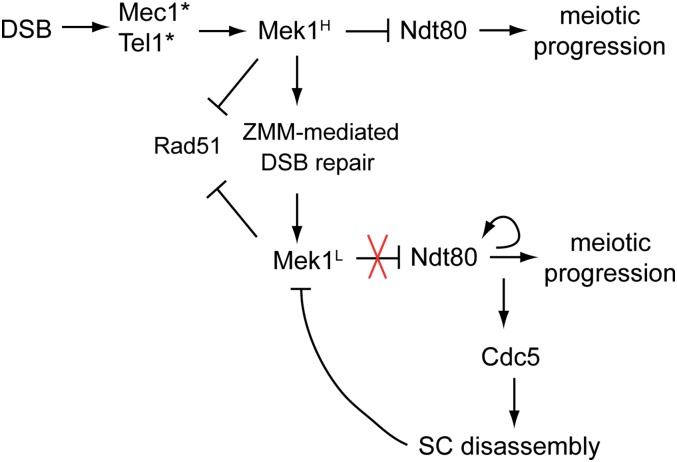
A critical issue for a meiotic yeast cell is determining when to exit prophase and enter the meiotic divisions. Doing so prematurely would be deleterious to the cell, as breaks may remain unrepaired and trigger a DNA damage response prior to Meiosis II (Cartagena-Lirola et al. 2008; Gurevich and Kassir 2010). We propose that coordination between the completion of DSB repair and meiotic progression is controlled by the interplay between Mek1 kinase activity and the Ndt80 transcription factor. Our hypothesis is that Ndt80 is a sensor that responds to the global level of Mek1 kinase activity, which is directly related to the number of DSBs. Early in meiosis, when there are relatively few DSBs, breaks are repaired without interhomolog bias, indicating that insufficient Mek1 has been activated (Joshi et al. 2015). Therefore, meiotic DSB repair requires a threshold number of breaks to activate sufficient Mek1 for the imposition of interhomolog bias. We propose that a similar threshold exists for the amount of Mek1 kinase activity (Mek1H) needed to inactivate the Ndt80 protein generated by Ime1, perhaps by sequestration of Ndt80 to the cytoplasm (Figure 7) (Wang et al. 2011). The idea that there is a DSB threshold required for activation of the meiotic recombination checkpoint has been previously proposed by Gray et al. (2013), based on the observation that synergistic reductions in activated Mek1 and spore viability were observed when spo11 hypomorphs (which reduce DSB formation) were combined with deletions of checkpoint components such as RAD24. The reduced spore viability was rescued by delaying NDT80 expression (similar to what we observed in the mek1∆ NDT80-IN). Mek1H inhibits Rad51 by phosphorylation of Rad54 and Hed1, and promotes Dmc1-mediated strand invasion of homologs, as well as the ZMM pathway of interfering crossover formation that is required for chromosome synapsis (Börner et al. 2004; Niu et al. 2009; Chen et al. 2015; Callender et al. 2016). As DSBs are repaired, Mek1 is removed from chromosomes and the global level of Mek1 kinase activity decreases (Mek1L), resulting in a weakening of interhomolog bias at some hotspots (Subramanian et al. 2016) (Figure 7). While this reduced level of Mek1 activity is still sufficient to phosphorylate Hed1, it is below the amount necessary to inactivate Ndt80. As a result, Ndt80 transcribes the NDT80 gene in a positive feedback loop, as well as expressing the other Ndt80 targets, including CDC5 (Figure 7). Cdc5 activity results in double Holliday junction resolution, disassembly of the SC through degradation of Red1 and Zip1, and inactivation of Mek1 (Sourirajan and Lichten 2008; Okaz et al. 2012) (Figure 2 and Figure 7). With Mek1 activity abolished, Rad51 can bind to Rad54 and mediate repair of any residual DSBs. In this way, cells do not begin the meiotic divisions until all DSBs have been repaired.
Figure 7.
Model indicating how Mek1 and Ndt80 negatively feedback on each other. DSBs activate the Mec1/Tel1 checkpoint kinases, which in turn activates Mek1. Mek1H indicates a level of kinase activity above the threshold necessary to inactivate Ndt80. Mek1 inhibits Rad51-mediated DSB repair by phosphorylation of Rad54 and Hed1 and promotes the ZMM pathway of interhomolog DSB repair leading to interfering crossovers and synapsis. As a result of this repair, Mek1 kinase activity is lowered below the threshold necessary to inactivate Ndt80 (Mek1L). Induction of NDT80 results in transcription of CDC5, leading to Holliday junction resolution and SC disassembly via Red1 and Zip1 degradation. As a result, the remaining Mek1 is inactivated, allowing Rad51-mediated repair of any residual DSBs. In addition, Ndt80-mediated transcription of CLB1 allows meiotic progression. DSB, double strand break; SC, synaptonemal complex.
Although both Red1 and Zip1 disappear upon CDC5 induction, it is the removal of Red1 that is most likely to be responsible for inactivation of Mek1. When Zip1 is artificially removed from ndt80∆-arrested cells, Hop1 phosphorylation increases and Mek1 relocalizes to chromosomes, suggesting that the absence of the transverse filament protein indirectly activates Mek1, rather than abolishing Mek1 activity (Subramanian et al. 2016). In contrast, Red1 forms a complex with Hop1 in the AE and is required for Mec1/Tel1 phosphorylation of Hop1 in response to DSBs and Mek1 activation (Niu et al. 2007; Lo et al. 2014). Therefore, degrading Red1 removes the chromosomal infrastructure that Mek1 requires for activation.
Our model is consistent with several published observations. First, since DSBs are necessary to activate Mek1, mutants defective in DSB formation (e.g., spo11∆) or Mek1 activation (hop1∆, red1∆) should prematurely activate Ndt80 and enter more rapidly into the meiotic divisions. This is, indeed, the case (Malone et al. 2004). Second, in haploid cells, DSBs remain unrepaired, due to the lack of a homolog for Dmc1-mediated repair and the inhibition of Rad51 activity by Mek1 (De Massy et al. 1994; Callender and Hollingsworth 2010). In disomic cells, DSBs are repaired by Dmc1 on the disomic chromosomes, but remain unrepaired on the haploid chromosomes, and meiotic progression is very delayed (Callender and Hollingsworth 2010). Therefore, synapsis of a single pair of homologs is insufficient to inactivate the checkpoint. In contrast, robust intersister DSB repair occurs at DSBs located on one arm of a chromosome that is hemizygous in an otherwise diploid cell (Goldfarb and Lichten 2010). We propose that the DSB repair resulting from synapsis of the other homologs in the latter case is sufficient to lower the level of Mek1 activity below the threshold needed to activate Ndt80, thereby enabling Cdc5 to eliminate Mek1 activity altogether to allow Rad51-mediated repair using sister chromatids. Third, double Holliday junctions are recombination intermediates that require CDC5 for their resolution and therefore accumulate in ndt80∆-arrested cells (Clyne et al. 2003; Sourirajan and Lichten 2008). Unresolved double Holliday junctions do not trigger the meiotic recombination checkpoint (Zakharyevich et al. 2012), indicating that it is not the absence of crossovers that is being monitored by the checkpoint, but the presence of a threshold number of DSBs. This result also shows that the level of Mek1 kinase activity in pachytene cells is not high enough to inactivate Ndt80. Finally, our model explains why ectopic expression of CDC5 is able to suppress the meiotic checkpoint arrest/delay conferred by dmc1∆ and zip1∆ (Acosta et al. 2011; Okaz et al. 2012). The artificial production of Cdc5 prior to Ndt80 activation bypasses the need for DSB repair to decrease Mek1 activity so that Ndt80 can be activated. Instead, Cdc5-promoted degradation of Red1 eliminates Mek1 activity, thereby allowing Ndt80 to transcribe its target genes (Figure 7).
This model also reconciles what initially appeared to be two contradictory results. Subramanian et al. (2016) showed that Mek1 activity is reduced by synapsis and that depletion of RAD54 at the ndt80∆ arrest increases the number of DSBs. This result suggests that Rad51-Rad54-mediated repair is occurring prior to NDT80 induction. In contrast, our data show that elimination of Hed1, the major repressor of Rad51, requires Ndt80 to inactivate Mek1-mediated phosphorylation of Hed1 and subsequent Hed1 degradation. A possible explanation for this conundrum comes from the interesting result that the reduced level of Mek1 activity observed in ndt80∆-arrested cells produces variable effects on interhomolog bias depending upon the hotspot (Subramanian et al. 2016). Chromosomes contain domains rich in Hop1 and Red1, which most likely produce increased amounts of localized Mek1 activity relative to Hop1/Red1-poor domains (Panizza et al. 2011; Sun et al. 2015). A recent study has shown that crossovers formed in Hop1 rich domains are more likely to occur via the ZMM pathway, while crossovers formed in domains with reduced levels of Hop1 are generated by structure-specific nucleases used in mitotic recombination (Medhi et al. 2016). We propose that Rad51-mediated intersister recombination occurs in ndt80∆-arrested cells at hotspots with low levels of local Mek1 activity, perhaps in Hop1 poor domains. Other DSB sites, such as the HIS4-LEU2 hotspot, in which interhomolog bias is maintained after synapsis (Subramanian et al. 2016), may be exposed to higher local levels of Mek1 and require the elimination of all Mek1 activity by Cdc5-mediated disassembly of the AEs to be repaired.
The meiotic prophase checkpoint in yeast likely monitors unrepaired DSBs, not synapsis
In nematodes, SC formation occurs independently of recombination, with homologs brought together by pairing sites (Dernburg et al. 1998; MacQueen et al. 2005). In this organism, two separate checkpoints monitor synapsis and DSB repair (Bhalla and Dernburg 2005; Kim et al. 2015). The synapsis checkpoint in nematodes is dependent upon a conserved, meiosis-specific AAA ATPase called PCH2 (Bhalla and Dernburg 2005). PCH2 was originally identified in budding yeast by a mutant that partially bypassed the meiotic progression delay exhibited by zip1∆ and dmc1∆ strains (San-Segundo and Roeder 1999). Variation in the ability of pch2∆ and rad17∆ to suppress the meiotic prophase delay triggered by different mutants led to the suggestion that a synapsis checkpoint operates in S. cerevisiae as well (Wu and Burgess 2006). However, subsequent work has shown that PCH2 and RAD17 independently promote activation of Mek1 (Ho and Burgess 2011), raising the possibility that the variable suppression of the checkpoint delays/arrest observed for pch2∆ and rad17∆ reflects different requirements for the levels of Mek1 kinase activity to trigger the checkpoint. In fact, pch2∆ rad17∆ diploids enter Meiosis I faster than wild-type cells, similar to spo11∆, consistent with our model that failure to activate Mek1 results in premature activation of Ndt80 (Wu and Burgess 2006).
We propose that, during meiotic prophase in yeast, a single checkpoint monitoring DSB repair determines when cells exit from pachytene and begin Meiosis I. In yeast, unlike nematodes, synapsis is not required for meiotic progression, as spo11∆ diploids progress efficiently through the meiotic divisions (Malone et al. 2004). While zmm mutants are defective in synapsis and exhibit delayed meiotic progression, they are also defective in the stable interhomolog interactions necessary to inhibit Spo11, resulting in increased numbers of DSBs (Börner et al. 2004; Thacker et al. 2014). We propose that the meiotic progression delay in zip1∆ mutants is related to inefficient DSB repair, rather than its synapsis defect. For example, phosphorylation of a conserved region in the C terminus of Zip1 is required for synapsis and the ZMM pathway of interfering crossover formation (Chen et al. 2015). Despite similar defects in synapsis, the nonphosphorylatable zip1-4A mutant exhibits significantly more DSBs than the zip1∆ mutant and a greater delay in meiotic progression, supporting the idea that the checkpoint is monitoring DSB repair as opposed to synapsis.
Ndt80 marks the transition between two phases of recombination in yeast
In nematodes, there are changes in chromosome structure at mid to late pachytene that result in a transition from interhomolog to intersister recombination (Hayashi et al. 2007; Couteau and Zetka 2011). In mice, nonhomologous chromosomal regions that undergo heterosynapsis late in zygotene are associated with the loss of DSB-associated marker γ-H2AX, leading to the proposal that the constraints on intersister recombination are relaxed (Mahadevaiah et al. 2001). Our results suggest that a similar transition occurs during pachytene in budding yeast and that activation of Ndt80 is the switch. Leading up to pachytene, Mek1 suppresses intersister recombination and promotes the ZMM-pathway of DSB repair that leads to the formation of interhomolog double Holliday junctions and synapsis (börner et al. 2004; goldfarb and Lichten 2010; kim et al. 2010; chen et al. 2015). In addition, Mek1 directly or indirectly inhibits Ndt80. Using Mek1 kinase activity to control Ndt80 activity ensures that Ndt80 will not be activated until sufficient DSB repair has occurred to reduce Mek1 below a threshold level. Having both double Holliday junction resolution and Mek1 inactivation under control of Cdc5 allows the repair of residual DSBs to occur during or immediately after crossovers have been formed, so that the completion of recombination and onset of the meiotic divisions are coordinated.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.199703/-/DC1.
Acknowledgments
We thank Bruce Futcher, Andreas Hochwagen, Neil Hunter, Scott Keeney, Michael Lichten, and Aaron Neiman for helpful discussions and/or comments on the manuscript. We are grateful to Angelika Amon, Wolf Heyer, Michael Lichten, Patrick Sung, and Linda Wang for providing helpful reagents. Thanks to Saif Laljee and David Chen for their help with some of the experiments. This work was supported by National Institutes of Health grant R01 GM-50717 to N.M.H.
Footnotes
Communicating editor: N. Hunter
Literature Cited
- Acosta I., Ontoso D., San-Segundo P. A., 2011. The budding yeast polo-like kinase Cdc5 regulates the Ndt80 branch of the meiotic recombination checkpoint pathway. Mol. Biol. Cell 22: 3478–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E., Padmore R., Kleckner N., 1990. Analysis of wildtype and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61: 419–436. [DOI] [PubMed] [Google Scholar]
- Allers T., Lichten M., 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. [DOI] [PubMed] [Google Scholar]
- Attner M. A., Miller M. P., Ee L. S., Elkin S. K., Amon A., 2013. Polo kinase Cdc5 is a central regulator of meiosis I. Proc. Natl. Acad. Sci. USA 110: 14278–14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin K. R., Zhang C., Shokat K. M., Herskowitz I., 2003. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 17: 1524–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N., Dernburg A. F., 2005. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science 310: 1683–1686. [DOI] [PubMed] [Google Scholar]
- Bishop D. K., Park D., Xu L., Kleckner N., 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation and cell cycle progression. Cell 69: 439–456. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., La Croute F., Fink G. R., 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197: 345–346. [DOI] [PubMed] [Google Scholar]
- Börner G. V., Kleckner N., Hunter N., 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. [DOI] [PubMed] [Google Scholar]
- Brar G. A., Kiburz B. M., Zhang Y., Kim J. E., White F., et al. , 2006. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature 441: 532–536. [DOI] [PubMed] [Google Scholar]
- Buonomo S. B., Clyne R. K., Fuchs J., Loidl J., Uhlmann F., et al. , 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103: 387–398. [DOI] [PubMed] [Google Scholar]
- Busygina V., Sehorn M. G., Shi I. Y., Tsubouchi H., Roeder G. S., et al. , 2008. Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev. 22: 786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek M., Thayer N. H., Oh S. D., Kleckner N., Hunter N., 2010. Double Holliday junctions are intermediates of DNA break repair. Nature 464: 937–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callender T. L., Hollingsworth N. M., 2010. Mek1 suppression of meiotic double-strand break repair is specific to sister chromatids, chromosome autonomous and independent of Rec8 cohesin complexes. Genetics 185: 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callender T. L., Laureau R., Wan L., Chen X., Sandhu R., et al. , 2016. Mek1 down regulates Rad51 activity during yeast meiosis by phosphorylation of Hed1. PLoS Genet. 12: e1006226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo J. A., Johnson A. L., Sedgwick S. G., Cha R. S., 2008. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 132: 758–770. [DOI] [PubMed] [Google Scholar]
- Carlile T. M., Amon A., 2008. Meiosis I is established through division-specific translational control of a cyclin. Cell 133: 280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartagena-Lirola H., Guerini I., Manfrini N., Lucchini G., Longhese M. P., 2008. Role of the Saccharomyces cerevisiae Rad53 checkpoint kinase in signaling double-strand breaks during the meiotic cell cycle. Mol. Cell. Biol. 28: 4480–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. Y., Tsubouchi T., Rockmill B., Sandler J. S., Richards D. R., et al. , 2008. Global analysis of the meiotic crossover landscape. Dev. Cell 15: 401–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Suhandynata R. T., Sandhu R., Rockmill B., Mohibullah N., et al. , 2015. Phosphorylation of the synaptonemal complex protein Zip1 regulates the crossover/noncrossover decision during yeast meiosis. PLoS Biol. 13: e1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S., Herskowitz I., 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1: 685–696. [DOI] [PubMed] [Google Scholar]
- Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., et al. , 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705 (erratum: Science 282: 1421). [DOI] [PubMed] [Google Scholar]
- Clyne R. K., Katis V. L., Jessop L., Benjamin K. R., Herskowitz I., et al. , 2003. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 5: 480–485. [DOI] [PubMed] [Google Scholar]
- Couteau F., Zetka M., 2011. DNA damage during meiosis induces chromatin remodeling and synaptonemal complex disassembly. Dev. Cell 20: 353–363. [DOI] [PubMed] [Google Scholar]
- Couteau F., Nabeshima K., Villeneuve A., Zetka M., 2004. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 14: 585–592. [DOI] [PubMed] [Google Scholar]
- Dahmann C., Futcher B., 1995. Specialization of B-type cyclins for mitosis or meiosis in S. cerevisiae. Genetics 140: 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T., Loidl J., Larkin B., Hollingsworth N. M., 2001. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics 159: 1511–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Massy B., Baudat F., Nicolas A., 1994. Initiation of recombination in Saccharomyces cerevisiae haploid meiosis. Proc. Natl. Acad. Sci. USA 91: 11929–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A. F., McDonald K., Moulder G., Barstead R., Dresser M., et al. , 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. [DOI] [PubMed] [Google Scholar]
- Dong H., Roeder G. S., 2000. Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. J. Cell Biol. 148: 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk J. E., Chan A. C., Hoffmann E., Hochwagen A., 2010. A Mec1- and PP4-dependent checkpoint couples centromere pairing to meiotic recombination. Dev. Cell 19: 599–611. [DOI] [PubMed] [Google Scholar]
- Game J. C., Mortimer R. K., 1974. A genetic study of x-ray sensitive mutants in yeast. Mutat. Res. 24: 281–292. [DOI] [PubMed] [Google Scholar]
- Goldfarb T., Lichten M., 2010. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 8: e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H., 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Gray S., Allison R. M., Garcia V., Goldman A. S., Neale M. J., 2013. Positive regulation of meiotic DNA double-strand break formation by activation of the DNA damage checkpoint kinase Mec1(ATR). Open Biol. 3: 130019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich V., Kassir Y., 2010. A switch from a gradient to a threshold mode in the regulation of a transcriptional cascade promotes robust execution of meiosis in budding yeast. PLoS One 5: e11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy C. F. J., Pautz A., 1996. A novel role for Cdc5p in DNA replication. Mol. Cell. Biol. 16: 6775–6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Chin G. M., Villeneuve A. M., 2007. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 3: e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth S. R., Ebisuzaki L. K., Segall J., 1995. A 15-base-pair element activates the SPS4 gene midway through sporulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 3934–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth S. R., Friesen H., Segall J., 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 5750–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho H. C., Burgess S. M., 2011. Pch2 acts through Xrs2 and Tel1/ATM to modulate interhomolog bias and checkpoint function during meiosis. PLoS Genet. 7: e1002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N. M., 2016. Mek1/Mre4 is a master regulator of meiotic recombination in budding yeast. Microb. Cell 3: 129–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N. M., Byers B., 1989. HOP1: a yeast meiotic pairing gene. Genetics 121: 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N. M., Goetsch L., Byers B., 1990. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell 61: 73–84. [DOI] [PubMed] [Google Scholar]
- Hollingsworth N. M., Ponte L., Halsey C., 1995. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 9: 1728–1739. [DOI] [PubMed] [Google Scholar]
- Hunter N., Kleckner N., 2001. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106: 59–70. [DOI] [PubMed] [Google Scholar]
- Joshi N., Brown M. S., Bishop D. K., Borner G. V., 2015. Gradual implementation of the meiotic recombination program via checkpoint pathways controlled by global DSB levels. Mol. Cell 57: 797–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk L. C., Hartwell L. H., 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132: 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis V. L., Lipp J. J., Imre R., Bogdanova A., Okaz E., et al. , 2010. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev. Cell 18: 397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., Lange J., Mohibullah N., 2014. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu. Rev. Genet. 48: 187–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. P., Weiner B. M., Zhang L., Jordan A., Dekker J., et al. , 2010. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143: 924–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Kostow N., Dernburg A. F., 2015. The chromosome axis mediates feedback control of CHK-2 to ensure crossover formation in C. elegans. Dev. Cell 35: 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F., Mahr P., Galova M., Buonomo S. B. C., Michaelis C., et al. , 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements and recombination during meiosis. Cell 98: 91–103. [DOI] [PubMed] [Google Scholar]
- Lamoureux J. S., Stuart D., Tsang R., Wu C., Glover J. N., 2002. Structure of the sporulation-specific transcription factor Ndt80 bound to DNA. EMBO J. 21: 5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leem S.-H., Ogawa H., 1992. The MRE4 gene encodes a novel protein kinase homologue required for meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 20: 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Shao Y., Jin H., Yu H. G., 2015. Ndj1, a telomere-associated protein, regulates centrosome separation in budding yeast meiosis. J. Cell Biol. 209: 247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. C., Bolcun-Filas E., Schimenti J. C., 2011. Genetic evidence that synaptonemal complex axial elements govern recombination pathway choice in mice. Genetics 189: 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H. C., Hollingsworth N. M., 2011. Using the semi-synthetic epitope system to identify direct substrates of the meiosis-specific budding yeast kinase, Mek1. Methods Mol. Biol. 745: 135–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y. H., Chuang C. N., Wang T. F., 2014. Pch2 prevents Mec1/Tel1-mediated Hop1 phosphorylation occurring independently of Red1 in budding yeast meiosis. PLoS One 9: e85687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lydall D., Nikolsky Y., Bishop D. K., Weinert T., 1996. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383: 840–843. [DOI] [PubMed] [Google Scholar]
- MacQueen A. J., Phillips C. M., Bhalla N., Weiser P., Villeneuve A. M., et al. , 2005. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 123: 1037–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah S. K., Turner J. M., Baudat F., Rogakou E. P., de Boer P., et al. , 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27: 271–276. [DOI] [PubMed] [Google Scholar]
- Malone R. E., Haring S. J., Foreman K. E., Pansegrau M. L., Smith S. M., et al. , 2004. The signal from the initiation of meiotic recombination to the first division of meiosis. Eukaryot. Cell 3: 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao-Draayer Y., Galbraith A. M., Pittman D. L., Cool M., Malone R. E., 1996. Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics 144: 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medhi D., Goldman A. S., Lichten M., 2016. Local chromosome context is a major determinant of crossover pathway biochemistry during budding yeast meiosis. eLife 5: e19669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montano S. P., Cote M. L., Fingerman I., Pierce M., Vershon A. K., et al. , 2002. Crystal structure of the DNA-binding domain from Ndt80, a transcriptional activator required for meiosis in yeast. Proc. Natl. Acad. Sci. USA 99: 14041–14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Wan L., Baumgartner B., Schaefer D., Loidl J., et al. , 2005. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell 16: 5804–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Li X., Job E., Park C., Moazed D., et al. , 2007. Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol. Cell. Biol. 27: 5456–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Wan L., Busygina V., Kwon Y., Allen J. A., et al. , 2009. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol. Cell 36: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. D., Jessop L., Lao J. P., Allers T., Lichten M., et al. , 2009. Stabilization and electrophoretic analysis of meiotic recombination intermediates in Saccharomyces cerevisiae. Methods Mol. Biol. 557: 209–234. [DOI] [PubMed] [Google Scholar]
- Okaz E., Arguello-Miranda O., Bogdanova A., Vinod P. K., Lipp J. J., et al. , 2012. Meiotic prophase requires proteolysis of M phase regulators mediated by the meiosis-specific APC/CAma1. Cell 151: 603–618. [DOI] [PubMed] [Google Scholar]
- Page S. L., Hawley R. S., 2004. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20: 525–558. [DOI] [PubMed] [Google Scholar]
- Pak J., Segall J., 2002. Role of Ndt80, Sum1, and Swe1 as targets of the meiotic recombination checkpoint that control exit from pachytene and spore formation in Saccharomyces cerevisiae. Mol. Cell. Biol. 22: 6430–6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Sasaki M., Kniewel R., Murakami H., Blitzblau H. G., et al. , 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144: 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza S., Mendoza M. A., Berlinger M., Huang L., Nicolas A., et al. , 2011. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146: 372–383. [DOI] [PubMed] [Google Scholar]
- Rockmill B., Roeder G. S., 1991. A meiosis-specific protein kinase homologue required for chromosome synapsis and recombination. Genes Dev. 5: 2392–2404. [DOI] [PubMed] [Google Scholar]
- San-Segundo P. A., Roeder G. S., 1999. Pch2 links chromatin silencing to meiotic checkpoint control. Cell 97: 313–324. [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E., Santos J. L., Jones G. H., Franklin F. C., 2007. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 21: 2220–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmuckli-Maurer J., Heyer W. D., 2000. Meiotic recombination in RAD54 mutants of Saccharomyces cerevisiae. Chromosoma 109: 86–93. [DOI] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N., 1995. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83: 783–791. [DOI] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N., 1997. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90: 1123–1135. [DOI] [PubMed] [Google Scholar]
- Shin M. E., Skokotas A., Winter E., 2010. The Cdk1 and Ime2 protein kinases trigger exit from meiotic prophase in Saccharomyces cerevisiae by inhibiting the Sum1 transcriptional repressor. Mol. Cell. Biol. 30: 2996–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M., Shita-Yamaguchi E., Buerstedde J.-M., Shinagawa H., Ogawa H., et al. , 1997. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homolog of RAD54, RDH54/TID1 in mitosis and meiosis. Genetics 147: 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. V., Roeder G. S., 1997. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol. 136: 957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R., Raithatha S., Stuart D., 2002. Phosphorylation and maximal activity of Saccharomyces cerevisiae meiosis-specific transcription factor Ndt80 is dependent on Ime2. Mol. Cell. Biol. 22: 7024–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourirajan A., Lichten M., 2008. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev. 22: 2627–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V. V., Hochwagen A., 2014. The meiotic checkpoint network: step-by-step through meiotic prophase. Cold Spring Harb. Perspect. Biol. 6: a016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V. V., MacQueen A. J., Vader G., Shinohara M., Sanchez A., et al. , 2016. Chromosome synapsis alleviates Mek1-dependent suppression of meiotic DNA repair. PLoS Biol. 14: e1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Huang L., Markowitz T. E., Blitzblau H. G., Chen D., et al. , 2015. Transcription dynamically patterns the meiotic chromosome-axis interface. eLife 4: e07424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M., Engebrecht J., Roeder G. S., 1993. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72: 365–378. [DOI] [PubMed] [Google Scholar]
- Thacker D., Mohibullah N., Zhu X., Keeney S., 2014. Homologue engagement controls meiotic DNA break number and distribution. Nature 510: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., Boone C., 2006. Synthetic genetic array (SGA) analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 313: 171–192. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H., Roeder G. S., 2006. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 20: 1766–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung K. S., Hong E. J., Roeder G. S., 2000. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc. Natl. Acad. Sci. USA 97: 12187–12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L., de los Santos T., Zhang C., Shokat K., Hollingsworth N. M., 2004. Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic double strand break repair in budding yeast. Mol. Biol. Cell 15: 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chang C. Y., Wu J. F., Tung K. S., 2011. Nuclear localization of the meiosis-specific transcription factor Ndt80 is regulated by the pachytene checkpoint. Mol. Biol. Cell 22: 1878–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter E., 2012. The Sum1/Ndt80 transcriptional switch and commitment to meiosis in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 76: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Y., Burgess S. M., 2006. Two distinct surveillance mechanisms monitor meiotic chromosome metabolism in budding yeast. Curr. Biol. 16: 2473–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Ajimura M., Padmore R., Klein C., Kleckner N., 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 6572–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Weiner B. M., Kleckner N., 1997. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 11: 106–118. [DOI] [PubMed] [Google Scholar]
- Zakharyevich K., Tang S., Ma Y., Hunter N., 2012. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149: 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D., Kleckner N., 2015. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7: a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. All strains, plasmids, and antibodies are available upon request.