Abstract
Organisms evolve in response to their natural environment. Consideration of natural ecological parameters are thus of key importance for our understanding of an organism’s biology. Curiously, the natural ecology of the model species Caenorhabditis elegans has long been neglected, even though this nematode has become one of the most intensively studied models in biological research. This lack of interest changed ∼10 yr ago. Since then, an increasing number of studies have focused on the nematode’s natural ecology. Yet many unknowns still remain. Here, we provide an overview of the currently available information on the natural environment of C. elegans. We focus on the biotic environment, which is usually less predictable and thus can create high selective constraints that are likely to have had a strong impact on C. elegans evolution. This nematode is particularly abundant in microbe-rich environments, especially rotting plant matter such as decomposing fruits and stems. In this environment, it is part of a complex interaction network, which is particularly shaped by a species-rich microbial community. These microbes can be food, part of a beneficial gut microbiome, parasites and pathogens, and possibly competitors. C. elegans is additionally confronted with predators; it interacts with vector organisms that facilitate dispersal to new habitats, and also with competitors for similar food environments, including competitors from congeneric and also the same species. Full appreciation of this nematode’s biology warrants further exploration of its natural environment and subsequent integration of this information into the well-established laboratory-based research approaches.
Keywords: WormBook, Caenorhabditis elegans, natural ecology, microbiome, pathogens, competition
WHY are >40% of the genes of Caenorhabditis elegans still without functional annotation and >60% without a described phenotype (Petersen et al. 2015a)? These are surprising numbers considering the enormous amount of research performed with this nematode across almost all biological disciplines. A likely reason is that the species’ natural ecology is largely neglected across these studies. These usually rely on an artificial environment consisting of agar plates supplemented with the bacterial food strain Escherichia coli OP50 and analysis of the canonical C. elegans strain N2, which shows numerous adaptations to the laboratory conditions (Sterken et al. 2015). Yet, the nematode’s ecology has not been completely ignored. During the 20th century, a handful of studies repeatedly isolated C. elegans from nature and characterized specific aspects of its ecology, for example its interaction with certain food microbes (Grewal 1991a; Grewal and Wright 1992; Venette and Ferris 1998) or variation in its reproductive system (Hodgkin and Doniach 1997). The interest in C. elegans natural populations has especially gained momentum since 2005, when several articles on its natural distribution and population genetic characteristics were published (Barrière and Félix 2005; Haber et al. 2005; Sivasundar and Hey 2005; Cutter 2006). These papers were followed by an increasing number of studies on the interaction of C. elegans with its environment and/or certain environmental components. Now, our understanding of C. elegans ecology has greatly improved since the previous review by Kiontke and Sudhaus (2006), which was published at a time when the species was only known from compost heaps and garden soil.
The biotic environment is of particular importance in this context, as biotic interactions are often a major driver of evolutionary change. The biotic environment includes interactions with competitors, parasites, predators, vectors, food, associated micro-organisms, and also interactions among C. elegans individuals, such as those among the different sexes with potentially conflicting interests. As changes in interaction characteristics are based on randomly occurring mutations in the two involved entities and are often unpredictable, they can impose high selective pressure. This has been particularly well-documented for interactions of host organisms with their coevolving parasites and pathogens (Woolhouse et al. 2002; Decaestecker et al. 2007; Schulte et al. 2010; Morran et al. 2011; Brockhurst et al. 2014; Koskella and Brockhurst 2014; King et al. 2016). Parasites/pathogens often show a high potential for evolutionary adaptation, because of comparatively shorter generation times, comparatively larger populations, or more flexible genomes (shaped by horizontal gene transfer). As parasites/pathogens by definition reduce host fitness and often depend on their hosts for survival and proliferation, their adaptation can impose continuously high selective constraints on their hosts (Woolhouse et al. 2002; Brockhurst et al. 2014), likely contributing to the evolution and maintenance of sex and recombination (Lively and Morran 2014). Sexual selection, based on diverging evolutionary interests of the sexes, can similarly cause ongoing cycles of adaptation and counteradaptation. The exceptionally high selective pressures produced by parasites or sexual interactions is reflected in the finding of significantly higher evolutionary rates in the genes associated with immunity (and thus parasite defense) and also sex-related genes (Ellegren and Parsch 2007; Fumagalli and Sironi 2014; Sironi et al. 2015; Cheng and Kirkpatrick 2016). Other biotic interactions may impose similar selective constraints, for example those involving predators or competitors (Cortez and Weitz 2014; Hiltunen and Becks 2014; Wilson 2014). Consequently, these kinds of biotic interactions are likely a key determinant in shaping the life history and underlying genome characteristics of any organism, including C. elegans. These changing selection dynamics are not only countered by single point mutations, but may account for the emergence of large gene families, when gene duplications allow a faster response to the selective challenge than point mutations or small insertions/deletions, as repeatedly documented in bacteria (Andersson and Hughes 2009; Pena-Miller et al. 2013) and suggested for some eukaryotes (Kondrashov 2012; Katju and Bergthorsson 2013; Assogba et al. 2016), including C. elegans (Farslow et al. 2015).
The aim of this review is to summarize our current understanding of the naturally occurring interactions of C. elegans with other organisms, ranging from conspecifics to interactions with other species (Figure 1). We will focus on studies that have repeatedly isolated C. elegans from nature and characterized its habitat, including locations in France and Northern Germany. We will additionally consider the increasing number of studies that have assessed naturally occurring biotic interactions under laboratory conditions, especially those with pathogens, food microbes, and the C. elegans-associated microbiome. Based on this work, we will first provide a brief overview of the characteristics of the nematode’s natural habitat (Habitats and Substrates), followed by a summary of the vectors and invertebrate hosts that are used and/or inhabited by C. elegans (Macroscopic Invertebrates as Possible Vectors or Hosts). We will discuss in detail C. elegans’ microbial environment, including potential food microbes, its microbiome, and also pathogens and parasites (The Microbial Environment and Pathogens and Parasites). We will provide an overview of the nematode’s competitors, predators, and enemies (Competitors and Predators), different types of intraspecific interactions in nature (Intraspecific Interactions), the presence of natural genetic polymorphisms as indicators for biotic interactions (Natural Genetic Polymorphisms as an Indication for Biotic Interactions), and conclude by highlighting selected topics important for future research (Perspectives).
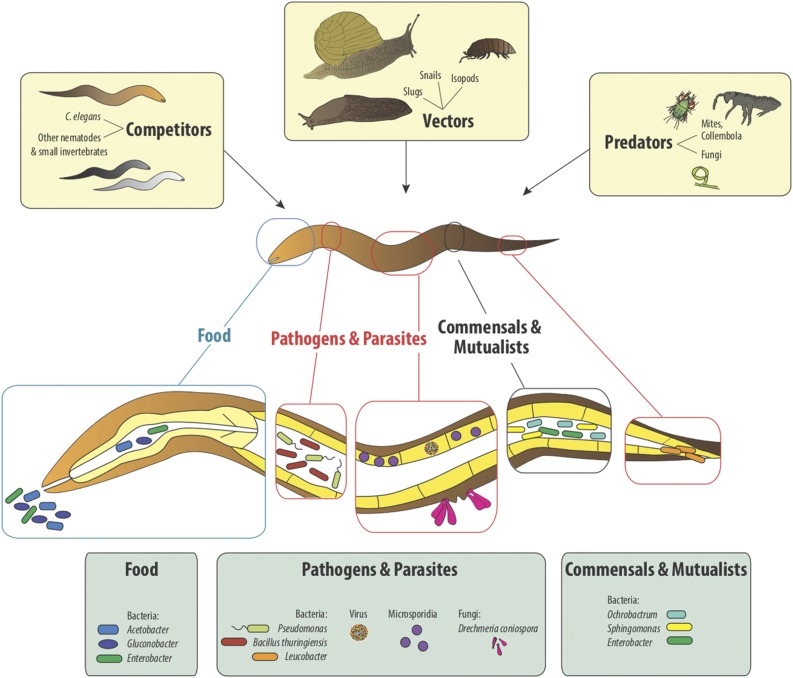
Figure 1.
Overview of biotic interactions of C. elegans. The illustration highlights examples of different types of interactions, ranging from competition with other nematodes or invertebrates over interactions with vectors (e.g., slugs, snails, or isopods), interactions with predators (e.g., mites, collembola, and nematode-trapping fungi), to the diverse interactions with microorganisms. The latter include food bacteria (as examples members of the genera Acetobacter, Gluconobacter, and Enterobacter), then pathogens and parasites (e.g., microsporidia, the Orsay virus, the fungus D. coniospora, and the bacteria P. aeruginosa, B. thuringiensis, and Leucobacter sp.), and also commensals and possibly mutualists (likely examples are members of the genera Ochrobactrum, Sphingomonas, and Enterobacter).
Habitats and Substrates
Habitat types in which C. elegans was repeatedly isolated
C. elegans appears to have a preference for humid temperate areas with a wealth of decaying vegetation. It was first sampled mostly in human-influenced habitats (compost heaps, orchards, vegetable gardens, and botanical gardens), and now also in more natural environments such as humid areas of woods and shrubland (Figure 2) (Barrière and Félix 2005, 2007; Sivasundar and Hey 2005; Kiontke et al. 2011; Félix and Duveau 2012; Petersen et al. 2014, 2015b; Frézal and Félix 2015; Cook et al. 2016). C. elegans is found on several continents (Europe, North and South Americas, Africa, Oceania, and rarely in Asia) and also on isolated islands, such as Hawaii, Madeira, Azores, and Réunion (see http://worldwideworm.banshy.fr/ for a database of C. elegans wild isolates with their location, habitat, and substrate type).
Figure 2.
Representative habitats and substrates of C. elegans. (A) A humid temperate area. (B) Rotting stems in the habitat of (A). The arrowhead indicates an isopod. (C) An orchard. (D) Rotting fruit in the habitat of (C). The arrowhead indicates collembola. The arrow indicates a slug. Bar, 1 cm. Photograph in A courtesy of Patrick Phillips; photographs in B-D by M.-A.F.
Note that sampling is biased toward substrates and landscapes where C. elegans has been previously found and also toward the geographical location of collectors. Thus, it is possible that new habitat and substrate types will be discovered in the future, especially if sampling efforts go beyond France, Germany, the UK, and the US, where most previous collections were made. Compared to other Caenorhabditis species such as C. japonica (Yoshiga et al. 2013; Okumura and Yoshiga 2014) or C. drosophilae (Kiontke 1997), the species C. elegans does not appear to have a highly specialized habitat nor a highly specialized biotic association with larger invertebrates. Although much remains to be discovered, C. elegans seems to have a more generalist lifestyle, which appears similar to that of some other Caenorhabditis species such as C. briggsae or C. remanei (see possible competition relationships in Possible competitors).
Overview of substrate types
C. elegans is most easily isolated from rotting fruits and stems, compost, and some invertebrates (see below Macroscopic Invertebrates as Possible Vectors or Hosts). In temperate areas, the large rotting fruits are chiefly found in human-associated gardens and orchards, and compost by definition is of anthropogenic origin. In more natural areas, rotting plant stems appear to be a very common substrate (Félix and Duveau 2012), as well as, occasionally, rotting flowers, fruits, and mushrooms (Kiontke et al. 2011; Félix et al. 2013; M.-A. Félix, unpublished data). The common feature of these substrates is that they consist of microbe-rich decomposing plant material. In contrast, C. elegans is rarely found in pure soil samples (except immediately adjacent to rotting fruits or stems), nor in rotting wood, leaf litter, or decomposing grass (Félix and Duveau 2012; Frézal and Félix 2015). Overall, microbe-rich rotting plant matter seems to represent the original substrate for C. elegans in nature, while it is possible that C. elegans populations have additionally adapted to human environments, such as compost or some orchards, which may provide a more stable source of nutrition across the year than found in most natural habitats. C. elegans may, thus, possess a hemerophilous (human-associated) lifestyle, as already described for various bird species or mammals (e.g., house sparrow, common pigeon, and house mouse, etc.; Marzluff et al. 2008). Finally, different C. elegans developmental stages were recently isolated from the intestines of living slugs (Petersen et al. 2015c), which may possibly be used as a completely different type of bacteria-rich substrate (see more details in Macroscopic Invertebrates as Possible Vectors or Hosts).
Life cycle of C. elegans and its biotic environment
The physical and biotic environment profoundly affects the development, physiology, and behavior of C. elegans, thereby influencing its life cycle. In the presence of food and at low population density, C. elegans develops directly from an embryo through four feeding larval stages to an adult. In contrast, in the absence of food and at high population density, C. elegans may arrest development at various stages, especially as a dauer larva, an alternative developmental stage that takes place following the second molting phase (Maupas 1915; Riddle and Albert 1997) (Figure 3). The environmental cues to enter the dauer stage are perceived during the L1 and L2 larval stages through sensory neurons, and include C. elegans density, food availability, and temperature (Bargmann and Horvitz 1991; Hu 2007; Fielenbach and Antebi 2008; Neal et al. 2015). C. elegans density operates through the secretion of ascarosides that are sensed by amphid neurons (Ludewig and Schroeder 2013). The bacteria-derived chemicals that regulate dauer entry are also sensed by amphid neurons but their chemical nature is still undefined, even for the artificial E. coli food provided to C. elegans in the laboratory. A recent study purified Nicotinamide Adenine Dinucleotide (NAD+) from a dauer exit-inducing fraction of E. coli and showed that it could induce serotonin signaling in specific sensory neurons, thus promoting dauer exit (Sze et al. 2000; Zhang 2004; Mylenko et al. 2016). Whether the same cue (i.e., its absence) acts in dauer entry is so far unclear. The effect of other more naturally encountered bacteria has hardly been studied (Jensen et al. 2010). Environmental cues regulating dauer entry vary among C. elegans wild isolates, perhaps reflecting the various encountered ecological conditions (Viney et al. 2003; Harvey et al. 2008; Diaz et al. 2014; Diaz and Viney 2015).
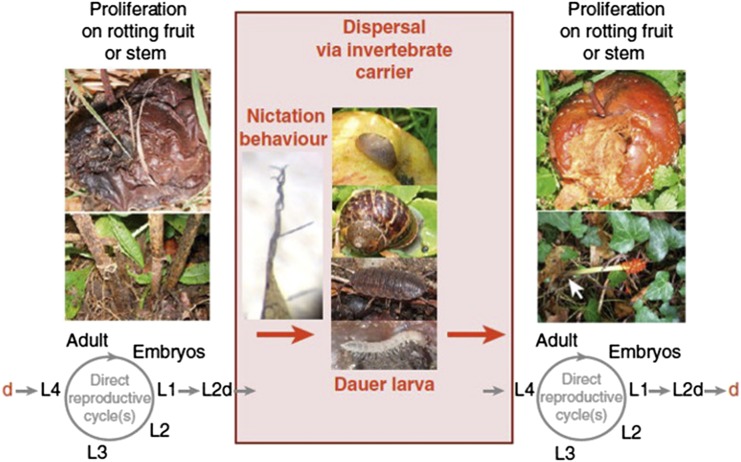
Figure 3.
The life cycle of C. elegans and the influence of biotic associations. C. elegans eats bacteria and grows in various types of bacteria-rich rotting plant material. Dauer larvae are induced by bacterial food depletion, high C. elegans density, and high temperature. Dauer larvae may actively disperse to colonize new food sources through their own locomotion. In addition, their nictation behavior may allow them to attach to carriers, such as slugs, snails, isopods, or myriapods, until a new food source is encountered, where development resumes. Reproduced from Félix and Braendle (2010) with permission from Elsevier. d, dauer larva; L1–L4, larval stages; L2d, pre-dauer larva in the L2 stage.
The dauer larva is the resistant dispersal stage that may be able to colonize new patches of food. The dauer has a closed mouth and a particularly resistant cuticle, does not feed, and yet is behaviorally active. It acts as a dispersal stage, either through its own locomotion or by hitchhiking on a larger invertebrate (isopod, gastropod, etc., see Macroscopic Invertebrates as Possible Vectors or Hosts). Dauer exit is an important decision that is also regulated by environmental cues (in particular, presence of bacteria). One or a few dauer larvae may start developing in a favorable environment, producing a proliferating population without any dauers (Félix and Duveau 2012; Figure 3). After population expansion, resource exhaustion, and density increase, the young larvae enter the dauer stage and the cycle starts again.
C. elegans, thus, adopts a boom-and-bust life cycle that is strongly dependent on its environment (Félix and Braendle 2010; Félix and Duveau 2012; Cutter 2015; Frézal and Félix 2015). Patches of rotting plant material enable fast C. elegans population growth with direct development, while resource exhaustion leads to entry into the dauer stage and migration toward a new resource patch. This life cycle is characteristic of many (but not all) members of the Rhabditidae family, which serves as an indicator of richness of soil (Yeates and Bongers 1999; Yeates 2003). Thus, through its effect on C. elegans development, the biotic environment has a key influence on population dynamics. Yet, to date, it is still unclear which exact environmental parameters are most influential. Here, the microbial environment is likely to be most important, as discussed in more detail below (The Microbial Environment).
Macroscopic Invertebrates as Possible Vectors or Hosts
Vectors or hosts: types of association
A few selected studies have assessed the association of C. elegans with other invertebrate species. Here, association is defined as the presence of C. elegans on or inside of the other animal, indicating a more intimate relationship between the two species. The focus of these studies has been on macroscopic invertebrates, especially insects, crustaceans, spiders, millipedes, chilopods, and molluscs. The common assumption is that C. elegans can use these comparatively larger invertebrates as vectors to move between locations, as do other species of rhabditid nematodes (Völk 1950; Kiontke and Sudhaus 2006). This assumption is particularly supported by the fact that dauer larvae appear to actively search for vectors for dispersal. The behavior shown has been termed nictation, whereby the dauer larvae stand on their tail, wave their body in the air, and easily attach themselves to any passing object, such as a larger animal (Lee et al. 2011). Several dauers can even jointly form a column and nictate as a group, possibly enhancing the likelihood of getting into contact with a vector (Figure 3) (Félix and Braendle 2010; Félix and Duveau 2012). C. elegans, especially when in the dauer stage, are likely able to attach to all macroscopic invertebrates that share the same habitat. Whether they may be taken away by any passing animal or whether any specificity exists is unclear.
The type of interaction with the invertebrate animals to which C. elegans physically associates is also not always clear. This is mainly due to the study approach, which is based on collecting individual invertebrates in the wild, bringing them to the laboratory, and checking for the presence of C. elegans in or on the other animal (Barrière and Félix 2007; Félix and Duveau 2012; Petersen et al. 2015c). Such associations may not only be explained by a vector-type association. Purely random relationships are conceivable, and it cannot yet be excluded that some form of parasitism or necromeny (i.e., feeding on the decomposing host after it dies) underlies the association (Kiontke and Sudhaus 2006). More specific relationships with invertebrate hosts are known, for example for the nematode Pristionchus pacificus, which in part lives in association with scarab beetles (Sommer and McGaughran 2013), or C. japonica, which shows a phoretic interaction with the bug Parastrachia japonensis (Yoshiga et al. 2013; Okumura and Yoshiga 2014). In the case of C. elegans, more details on the specificity of the interaction would now require specially designed studies, such as life history assays on the host or collection of dead hosts from the wild (beyond one anecdotal report in Barrière and Félix 2007). In addition, larger vertebrate animals interact with known C. elegans substrates such as rotting fruits, for example small rodents, certain bird species, and humans. Therefore, these could also act as hosts or vectors, but are not yet part of the available data.
Overview of vectors
Early studies found C. elegans and its relatives, such as C. briggsae and C. remanei (Baird 1999), in association with isopods, snails, slugs, and myriapods (Kiontke and Sudhaus 2006). A systematic study of snails in California revealed a high number of associations with C. elegans (Caswell-Chen 2005). A survey in Scotland isolated C. elegans from isopods such as Porcellio scaber and Po. spinicornis (Cutter 2006). In France, C. elegans was repeatedly isolated from various isopod taxa, slugs (including small slugs identified as Deroceras), and snails (including genera Helix and Pomatia) (Barrière and Félix 2005, 2007; Félix and Duveau 2012). A comprehensive survey in North Germany found C. elegans in >10% of collected slugs (mainly of the genus Arion, especially Arion lusitanicus, and occasionally Limax maximus), isopods (e.g., Po. scaber, Oniscus asellus, and Armadillidium vulgare), and also chilopods (Figure 4, A and B), while not with any of the assayed species of insects and spiders (Petersen et al. 2015c). The invertebrates were particularly prone to harbor C. elegans if collected on compost or rotting fruits, reaching a prevalence of up to or even above 30%. The association with these animals rather than insects or spiders, also present on compost or rotting plants, may be influenced by humidity (Petersen et al. 2014, 2015c). As most C. elegans stages are highly sensitive to dehydration (e.g., Erkut et al. 2011), they are likely able to better survive on animals that provide a more humid environment, such as isopods and the various molluscs.
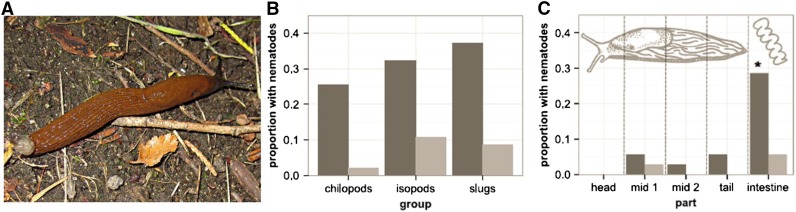
Figure 4.
Association of C. elegans with invertebrates, especially slugs. (A) Slug of the genus Arion, often found to harbor C. elegans in North Germany. (B) Proportion of different taxa associated with C. elegans (dark gray bars) or C. remanei (light gray) in North Germany, based on a screen of a total of 51 chilopods, 93 isopods, and 35 slugs carried out between July and September 2013. (C) Localization of C. elegans (dark gray) and C. remanei (light gray) in different slug body parts, based on 35 tested slugs in 2013 in North Germany, highlighting a high abundance of C. elegans in slug intestines. Photograph in A courtesy of C. Petersen; B and C are from Petersen et al. (2015c).
The peculiar relationship with slugs
The association with slugs appears peculiar. In the North German locations, slugs harbored C. elegans in habitat areas with apparently little rotting plant matter from which the nematodes could have been taken up (e.g., in some parks; Petersen et al. 2015c). This may suggest that slugs are able to harbor C. elegans for longer time periods. Detailed characterization of different body parts revealed a significantly higher preponderance of C. elegans in the slugs’ intestines, whereas they were only rarely found on the head, tail, or middle part of the body (Figure 4C; Petersen et al. 2015c). Although the majority of worms in the intestines were dauers, some non-dauers were also repeatedly isolated, suggesting survival of other stages in slug intestines and possibly their reproduction in this environment. An experimental assessment of the interaction demonstrated that some C. elegans individuals are able to pass the slug’s radula, enter the gut alive, transit through the intestine, and be released to the environment with the slug’s feces. Different C. elegans stages (including L4, adults, and dauers) succeeded in entering the gut. When slugs were exposed to nematode cultures for only 3 hr, their feces still contained worms after 30 hr (Petersen et al. 2015c). The particular association of C. elegans with slugs in North German locations is consistent with the findings from French locations, where slugs were found containing non-dauer stages, most likely present in the slug’s intestines (Félix and Duveau 2012). These results also confirmed previous reports, which focused on parasitic nematodes, yet repeatedly found C. elegans or the congeneric C. briggsae inside of slugs from Africa and Europe (Mengert 1953; Ross et al. 2010, 2012). Altogether, these findings suggest that C. elegans may use slug intestines as a microbe-rich habitat for proliferation. It is even possible that C. elegans may thereby harm slugs. The exact relationship between C. elegans and slugs clearly deserves further investigation. A similar kind of relationship is further conceivable for snail species, which in some cases were also found to harbor C. elegans feeding stages (Félix and Duveau 2012).
The Microbial Environment
Overview of the diverse interactions with microorganisms
Microorganisms are of key importance for C. elegans biology. This species proliferates on decomposing substrates that contain a high density of microorganisms, especially of bacteria (Barrière and Félix 2005, 2007; Félix and Duveau 2012; Berg et al. 2016a; Dirksen et al. 2016; Samuel et al. 2016). These bacteria in the environment can have diverse interactions with C. elegans: they may directly serve as food, they may process substrate material to make it accessible for the nematode as food, they can be part of the worm’s associated microbiome in its gut or body surface, and they may be pathogens and parasites with harmful effects (Figure 1). Microbial communities can be highly dynamic with rapidly changing compositions over space and time. Even within microbial lineages, changes can be fast, as mutations are often abundant due to usually large population sizes, frequent horizontal gene transfer, and the fact that favorable variants can spread rapidly due to the microorganism’s comparatively short generation times. As a consequence of these fast evolutionary dynamics, the microbial environment can impose continuously high selective pressures on C. elegans. The nature and type of interaction of C. elegans with possible food microbes has been described for selected microorganisms since the 1990s (e.g., Grewal 1991a; Grewal and Wright 1992; Venette and Ferris 1998), followed by additional work focused on few bacterial taxa (Avery and Shtonda 2003; Shtonda and Avery 2006; Coolon et al. 2009). The first natural pathogens and parasites of C. elegans were described in 2008 and 2011 (Troemel et al. 2008; Félix et al. 2011) (reviewed in Pathogens and Parasites). More systematic analyses of the C. elegans natural microbial environment were only published in 2016, and these addressed either the microbial composition of C. elegans substrates (Samuel et al. 2016), the native microbiome associated with wild-caught animals (Dirksen et al. 2016), or the microbiome of the canonical N2 strain exposed to soil under experimental conditions (Berg et al. 2016a). A recent meta-analysis assessed the differences and similarities among these three first systematic studies on the C. elegans microbiome (Zhang et al. 2017). Below, we summarize the findings and add information from the earlier studies, whenever appropriate. In a subsequent section, we will separately cover C. elegans’ parasites and pathogens, which are also part of its microbial environment, yet produce a specific type of relationship based on antagonistic interactions.
The general natural microbial environment of C. elegans
The bacterial environment of C. elegans, defined as the bacterial composition of the various substrates where it can be found (e.g., rotting fruit, etc.), was systematically analyzed by Samuel et al. (2016) via high-throughput sequencing of a bacterial 16S ribosomal DNA (rDNA) PCR fragment. These methods are only semiquantitative as there are numerous biases in the PCR amplification of different bacterial groups; yet, they still provide a first overview of bacterial community composition. The recent meta-analysis demonstrated that this composition differed from that associated directly with C. elegans nematodes (Zhang et al. 2017) (see also below). In the different substrate samples, sequences from the bacterial phyla Proteobacteria (mostly α- and γ-Proteobacteria), Bacteroidetes, Firmicutes, and Actinobacteria were most common. At the family and genus levels, the most common representatives were: in the γ-Proteobacteria, the families Enterobacteriaceae (e.g., genus Enterobacter), Pseudomonaceae (Pseudomonas), and Xanthomonadaceae (Stenotrophomonas); in the α-Proteobacteria: Acetobacteriaceae (Acetobacter, Gluconobacter, and Acetogluconobacter, common in fruits); in the Bacteroidetes: Flavobacteriaceae (Flavobacterium and Wautersiella) and Sphingobacteriaceae (Sphingobacterium); in the Firmicutes: Lactobacillaceae (Lactobacillus), Streptococcaceae (Lactococcus), and Leuconostocaceae (Leuconostoc); and Actinobacteria, such as Microbacteriaceae, which were commonly found, but in low amounts.
When focusing on rotting apples in a given orchard and separating the apples according to C. elegans population state (e.g., dauer vs. proliferating), a significant difference in bacterial composition was found, perhaps in part reflecting the degree of rotting of these apples (Figure 5A). C. elegans tended to be found proliferating in apples with a simpler microbiome, enriched in Acetobacteriaceae (Acetobacter and Gluconobacter) and poor in Pseudomonas, Stenotrophomonas, Flavobacterium, Chryseobacterium, Xanthomonas, and Sphingomonas. Overall, this trend matches the effect of representative genera when tested in the laboratory, as seen below. Note that Acetobacteriaceae and Lactobacillae are dominant in the gut microbiota of Drosophila fruit flies (Corby-Harris et al. 2007; Wong et al. 2013, 2015).
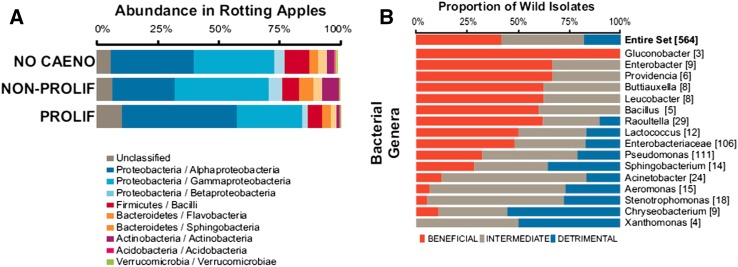
Figure 5.
The natural microbial environment of C. elegans. (A) Relative abundance of bacterial taxa (indicated by color) in rotting apples containing either no worms (indicated by NO CAENO), nonproliferating worm populations (e.g., consisting of dauer stages; NON-PROLIF), or proliferating populations of C. elegans (PROLIF). (B) Proportion of bacterial strains of a particular genus from the natural environment of C. elegans, which either have positive (orange color), intermediate (gray), or detrimental effect (blue) on worm life history. Both figures from Samuel et al. (2016).
To test the effect on C. elegans of these naturally associated bacteria, a culture collection of 565 bacteria was established from the diverse samples. Representatives of the main genera found by 16S rDNA genotyping can be cultured relatively easily in the laboratory. These isolates were tested individually for their effect on C. elegans growth and on the induction of stress and immune reporter genes (Samuel et al. 2016). Overall, some genera tended to have most representatives being beneficial (e.g., Gluconobacter and Enterobacter), while others tended to be detrimental and induce expression of the reporters (Xanthomonas, Chryseobacterium, Stenotrophomonas, and Aeromonas) (Figure 5B). Over the 111 Pseudomonas isolates, the whole spectrum of effects was found, with an overall tendency toward a detrimental effect. When C. elegans growth was tested in the presence of two bacteria (one detrimental and one beneficial), in some cases, the beneficial bacterial strain could rescue the detrimental one (better than E. coli could) and in other cases, a small proportion of the detrimental strain was already pathogenic. Finally, more complex mixtures of 18–24 bacterial strains mimicking good or bad growth environments could reconstitute good or bad C. elegans growth conditions (Samuel et al. 2016). Overall, from the pattern of natural associations and the dissection of the effect of individual bacteria or combinations of bacteria, this study provides an insight into the external bacterial environment of C. elegans.
The worm’s microbiome
We will now turn to the bacteria that are physically associated with C. elegans, mostly in the gut and some on the cuticle surface. Most C. elegans researchers only know this worm with an empty gut and a neat cuticle, devoid of any microorganisms. This is a consequence of using the N2 reference strain and culturing it monoaxenically on E. coli OP50. A routine laboratory protocol, bleaching (Stiernagle 2006), is frequently used to remove bacterial contaminants from C. elegans cultures and synchronize nematode populations. Bleaching efficiently kills all microorganisms and C. elegans stages except embryos that are protected by their eggshell. Indeed, so far there is no indication of a vertically transmitted symbiont in C. elegans (except for genome-encoded retrotransposon sequences that can assemble capsids in the germline of some isolates; Dennis et al. 2012). The bleaching protocol thus produces germ-free animals. The nematodes are then routinely combined with the E. coli OP50 strain that is used as food added on the culture plates.
In contrast, nematodes isolated from their natural substrates often contain a vast number of microorganisms in their intestines and body surface (Félix and Braendle 2010; Félix and Duveau 2012). Here, we use the word “microbiome” to refer to these microorganisms that are physically associated with C. elegans individuals. Two main studies characterized the native C. elegans-associated microbiome, using slightly different approaches. One of these focused on animals directly isolated from nature (Dirksen et al. 2016), whereas the other exposed the canonical laboratory strain N2 to defined substrates under controlled conditions, followed by microbiome characterizations (Berg et al. 2016a).
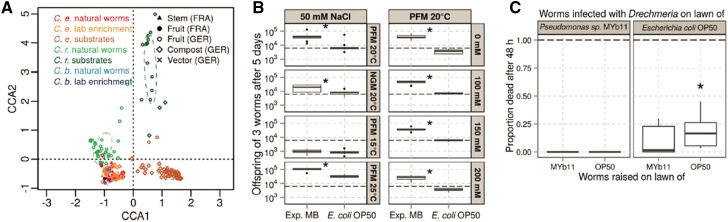
In the first case, natural strains of C. elegans and two congeneric species, C. briggsae and C. remanei, were obtained from various locations in France, one in Portugal, and two main locations in Northern Germany (Dirksen et al. 2016). Two types of samples were analyzed, both by 16S rDNA genotyping. On the one hand, the bacteria associated with single or few worms were characterized directly after their isolation from the substrates. On the other hand, Caenorhabditis populations were allowed to proliferate with their native microbiome under laboratory conditions for at least 2 weeks after original isolation (a new environmental challenge as the laboratory conditions are clearly different from the natural environment), followed by microbiome analysis. Substrate samples were also assessed as controls for many of the isolated individuals. In spite of the differences in processing protocols and collection sites of the samples from France/Portugal and those from Germany, the analysis revealed significant similarities in microbiome composition among the various C. elegans samples and, at the same time, significant differences of these to the microbiomes of the corresponding substrate and of the congeneric C. remanei (Dirksen et al. 2016) (Figure 6A). Dominant taxonomic groups that are physically associated with C. elegans are Proteobacteria, especially members of the Enterobacteriaceae and those of the genera Pseudomonas, Stenotrophomonas, Ochrobactrum, and Sphingomonas. This result strongly suggests that C. elegans possesses a microbiome that is distinct from its direct environment.
Figure 6.
The native microbiome of C. elegans. (A) Canonical correspondence analysis of the microbiome of C. elegans (C.e.), C. remanei (C.r.), C. briggsae (C.b.), and corresponding substrate samples from France (FRA) and Germany (GER). Worm samples included nematodes, which were analyzed directly after their isolation from the wild (natural worms) and after proliferation in the laboratory for ∼2 weeks (lab enrichment). The first and second axes separate the substrate samples from the worms (filled symbols in the bottom left corner), highlighting a characteristic microbiome of worms. (B) An experimental microbiome of a mixture of 14 bacterial strains (Exp. MB) enhances population growth relative to E. coli OP50 (see horizontal axes) under stress conditions such as nutrient-poor media or high temperatures (left panels), and also different salt conditions (right columns). Asterisks indicate a significant difference between Exp. MB and the E. coli OP50 control. (C) The Pseudomonas strain MYb11 protects nematodes from infection with the fungal pathogen D. coniospora, both when worms are raised on MYb11 (horizontal axis) and when infection takes place in the presence of MYb11 (left panel). The asterisk indicates significant difference in survival of worms grown under the tested conditions in the presence of the fungus versus the same conditions without fungus. All figures from Dirksen et al. (2016).
The colonization of C. elegans could be reconstituted in the laboratory after establishment of a collection of associated bacteria isolated from crushed C. elegans. Some bacteria are able to persist in the nematode gut over long time periods, especially Ochrobactrum isolates (Troemel et al. 2008; Dirksen et al. 2016), possibly indicating a more intimate relationship. In some experiments, C. elegans was cultured on plates seeded with mixes of bacteria and the C. elegans-associated microbiome was then compared to the plate microbiome by 16S rDNA genotyping. These experiments confirmed that the C. elegans microbiome is indeed distinct from that of its environment and that both C. elegans wild genotype and developmental stage have an effect on the composition of the associated microbial community. The latter result suggests that some specificity of colonization takes place. This may be a consequence of genetic adaptation of C. elegans to its microbial environment, possibly based on its behavioral choice, grinder properties, gut environment, or defecation efficacy. Alternatively, it may be determined by the ability of the various bacteria to invade and establish themselves in different C. elegans microenvironments, also determined by the genetic composition and biology of the animal. It is similarly possible that such variations result from a combination of host and bacteria properties. In this context, it is worth noting that the survey of the native microbiome did not reveal a specific bacterial species or strain (or set of strains) found across a larger number of C. elegans samples. The range of individual bacterial strains or operational taxonomic units (OTUs) varies across samples; the high similarities are only found at higher taxonomic level. This observation is inconsistent with the idea of coevolving host and microbe lineages, but it could instead be explained by host-mediated selection of favorable bacteria, which would then generate a host-specific microbiome with an important influence on host fitness (Bordenstein and Theis 2015; Douglas and Werren 2016). Our findings for C. elegans are thus consistent with the characteristics of the microbiome of other animal species, for example the house mouse or the fruitfly Drosophila melanogaster (Chandler et al. 2011; McCafferty et al. 2013; Wong et al. 2013).
The second microbiome analysis of C. elegans was based on an experimental approach (Berg et al. 2016a). Germ-free populations of the laboratory strain N2 were transferred to standardized soil samples supplemented with plant matter including various fruits. The animals were thus allowed to “collect” their preferred microbes. They were subsequently reisolated, followed by characterization of their microbial composition and that of the corresponding substrates. Consistent with the above study (Dirksen et al. 2016), the results demonstrated a worm-specific microbiome that was distinct from the corresponding substrates and that was highly similar, even if assembled from different substrates (e.g., characterized by different fruits added to the soil). Microbial composition was influenced by temperature in a host-dependent manner. Bacteria that were generally enriched in these worms again included members of the Enterobacteriaceae, Pseudomonaceae, and Xanthomonadaceae (which contains the genus Stenotrophomonas), but also other taxa such as Sphingobacteriaceae or Rhizobiaceae (Berg et al. 2016a). Representatives of the commonly identified taxonomic groups, especially the three first families, may thus be part of the core microbiome of C. elegans. However, as in the study by Dirksen et al. (2016), no single bacterial strain or OTU was systematically associated with C. elegans. In analogy with many other hosts, it is therefore likely that C. elegans is preferentially colonized by a range of bacterial taxa, whose presence is influenced by the environment, the colonization ability of individual bacterial strains, and/or selection by the host [see also Shapira (2016)].
The recent meta-analysis, which compared the first three systematic microbial characterizations, confirmed that the composition of bacterial communities from C. elegans is significantly different to that of the corresponding substrates (Zhang et al. 2017). Importantly, C. elegans-associated communities from the two very distinct study approaches are highly similar, including enrichment of eight particular families across the two studies, such as Enterobacteriaceae, Pseudomonaceae, Xanthomonadaceae, Comamonadaceae, Sphingomonadaceae, Sphingobacteriaceae, Weeksellaceae, and Flavobacteriaceae (Zhang et al. 2017). In spite of these recent advances, it is yet unclear how stable the C. elegans-associated microbiome really is, either during the lifetime of individuals or across host generations. It is possible that the microbial community can be maintained in natural C. elegans populations, at least temporarily, via some form of vertical transmission. In principle, such vertical transmission may occur through eggs (i.e., transovarial transmission, so far not seen in C. elegans), through transfer to the developing offspring in the uterus (enhanced when larvae hatch inside their mothers, an essential feature for symbiont transmission in Heterorhabditis) (Ciche et al. 2008; Griffin 2012), or via the sharing of the environment between parent and offspring (e.g., Douglas 2010). Alternatively, the worm’s microbiome is unstable across generations and determined by the changing environmental microbial community. The composition of the gut community then depends on the interaction between both host and microbe properties: on the host side on the foraging and feeding behaviors and the properties of the gut lumen; and on the microbe side, on the ability to pass the pharyngeal grinder and to persist in the gut without the population being entirely digested or expelled live by defecation. Interestingly, the recent meta-analysis highlighted that, even though substrate and nematode microbial communities are significantly different, they are still related. Taxa abundant across C. elegans samples are often abundant across the substrate samples (Zhang et al. 2017). This finding is consistent with the idea that the nematode microbiome is specifically assembled from the microbes available in the environment.
The recent studies also provided insights into the possible range of effects of the bacteria found in the environment or associated with C. elegans. Many of these bacterial strains are able to sustain C. elegans population growth, as measured as increases in population size over 5 days (Dirksen et al. 2016), developmental rate, and body size (Samuel et al. 2016), and some of them do it better than E. coli OP50. These fitness improvements were consistently expressed under different environmental conditions, including high osmolarity or various temperatures. Interestingly, on peptone-free agar media that could not sustain bacterial growth, naturally associated bacteria helped C. elegans to grow better in comparison to E. coli, likely in a mutual interaction where C. elegans provided an environment for bacterial population growth, from which C. elegans in turn obtained some food (Figure 6B) (Dirksen et al. 2016).
In contrast, some bacteria isolated from the natural environment could not sustain C. elegans growth (Samuel et al. 2016). Others induced expression of C. elegans genes usually activated by other stresses or pathogens, and at least some of these bacteria appeared pathogenic, because their effect could not be rescued by good food (Liu et al. 2014; Samuel et al. 2016). While some bacteria from the natural environment induced a mitochondrial stress response (measured with a hsp-6p::GFP reporter), other bacteria (Pseudomonas species) were able to suppress the mitochondrial stress response induced upon mitochondrial activity disruption by the Streptomyces toxin antimycin, perhaps indicating a counterdefense mechanism (Liu et al. 2014). Interestingly, certain bacterial taxa appear to have immune-protective effects, either through direct interaction with pathogens or indirectly through stimulation of host responses (Figure 6C). These immune-protective bacteria include members of the genera Pseudomonas, Gluconobacter, Providencia, and Enterobacter (Montalvo-Katz et al. 2013; Berg et al. 2016b; Dirksen et al. 2016; Samuel et al. 2016). The results complement the recent demonstration of the selective benefit of an immune-protective Enterococcus faecalis strain during experimental evolution with C. elegans and pathogenic Staphylococcus aureus (Ford et al. 2016; King et al. 2016). It is additionally possible that the associated microorganisms provide an advantage to C. elegans by shaping its natural environment, for example by eliminating harmful microbes and/or processing environmental substances and thereby making them accessible as nutrients for the nematode. These and other possible effects of the microbiome remain to be characterized and represent an exciting challenge for future research.
Food: C. elegans’ prey
Finding out what C. elegans eats in natural conditions is not a simple task. In laboratory conditions, C. elegans is fed E. coli OP50 and the N2 reference strain is able to grind E. coli efficiently through its pharyngeal grinder (Avery and You 2012). Nutrients are then imported into the intestinal cells from the gut lumen. In natural conditions, C. elegans is likely to eat mainly bacteria. Some small eukaryotes (such as yeasts) and perhaps already processed material may additionally be taken up (see below). We first focus on bacteria. From the above studies, in natural settings we know which bacteria may be found in C. elegans’ immediate environment and which are present in its gut in a form that allows 16S rDNA sequencing (Berg et al. 2016a,b; Dirksen et al. 2016; Samuel et al. 2016). Two questions arise. Is the 16S rDNA in the gut of C. elegans that of the food or mostly that of live bacteria that will not be digested? Can bacteria that survive the pharyngeal grinder of the nematode be later digested in the gut? The DNA of digested bacteria is probably rapidly degraded in the gut and metabolized, so the food may even appear as those bacterial sequences that are more abundant in the substrates than in the corresponding nematodes (Dirksen et al. 2016). One particular candidate group that is highly abundant in rotting fruits (maybe not in other substrates) but less so in the nematode are the Acetobacteriaceae, which were also a good indicator of colonization of an apple by C. elegans (Dirksen et al. 2016; Samuel et al. 2016). As mentioned above, most of the isolated strains of environmental or associated bacteria may serve as food and support growth of C. elegans. Those that do not are generally pathogens rather than bad food per se, as they affect C. elegans population growth, even in the presence of a palatable bacterial species (Samuel et al. 2016).
The relationship of C. elegans to food around it has been studied in various ways, but so far not using the recently isolated bacteria naturally found with C. elegans. The type of bacteria that was offered as food was found to influence the growth rate of C. elegans populations (Grewal 1991a; Venette and Ferris 1998). Food availability affects behavior, including progeny production (Goranson et al. 2005), egg-laying, and bagging (Chen and Caswell-Chen 2004). Using a set of soil bacteria, Darby and Herman (2014) showed that in a mixture of bacteria, C. elegans grows as fast as the best available prey allows. Smaller size bacteria tend to be better food (Avery and Shtonda 2003). When given the choice between different soil bacteria (Avery and Shtonda 2003), C. elegans is able to learn to discriminate the food on which it grows best (Shtonda and Avery 2006): in a behavioral assay with patches of different bacteria, C. elegans tends to dwell on patches of good food and leave patches of worse food. In natural conditions, this is likely to be relevant, because the substrate is not a well-stirred bacterial mix and instead is structured with bacterial patches, i.e., colonies forming from single bacterial cells.
Nutritional requirements of C. elegans have been studied in efforts to devise a chemically defined medium on which C. elegans can grow (Lu and Goetsch 1993; Perelman and Lu 2000; Szewczyk et al. 2003; Balachandar and Lu 2005; Xiong and Lu 2008; Zhao and Lu 2011). In addition to standard requirements for salts, amino acids, sugar, nucleotides, and various specific compounds/vitamins, C. elegans is defective in heme synthesis and must import it from bacteria (Hieb et al. 1970; Rao et al. 2005). In modern standard culture conditions, the heme is provided by E. coli, but in axenic conditions, heme must be provided together with a carrier protein (Buecher et al. 1970; Vanfleteren 1974). The rate of C. elegans growth and reproduction seems to be dependent on metabolically active bacteria or possibly a heat-labile nonsoluble component of live bacteria (Lenaerts et al. 2008). In the absence of such components, the nematodes reproduce more slowly and display an increased life span that is mediated by elevated activity of the Foxo transcription factor DAF-16, as in the dietary restriction response (Szewczyk et al. 2006; Lenaerts et al. 2008). This may suggest that C. elegans is able to express different healthy life histories in response to bacterial availability (Szewczyk et al. 2006). The growth conditions and physiological state of bacteria thus, in turn, affect C. elegans development and physiology.
Different bacteria vary in the nutritional supplies they provide (Watson and Walhout 2014; Yilmaz and Walhout 2014). Vitamin B12, a coenzyme required for breakdown of the short-chain fatty acid propionate (CH3CH2CO) and the methionine/S-adenosylmethionine cycle, is only synthesized by some species of bacteria. E. coli OP50 is a poor provider of vitamin B12 and C. elegans individuals were found to grow faster on a better provider such as Comamonas aquatica DA1877 (MacNeil et al. 2013; Watson et al. 2014). When grown on E. coli OP50, C. elegans activates the transcription of metabolic enzymes in a shunt pathway of propionate breakdown (Watson et al. 2016). Whether E. coli uses a fermentative or a respiratory metabolism further appears to matter for C. elegans longevity (Saiki et al. 2008). Other examples of nutrients that differ between bacteria concern the amount of dietary folate (Virk et al. 2012, 2016), tryptophan (Gracida and Eckmann 2013), or nitric oxide (Gusarov et al. 2013).
Bacteria do not provide all nutrients required for C. elegans, and specifically this nematode requires an external sterol source (Hieb and Rothstein 1968; Lu et al. 1977). In the standard laboratory medium, cholesterol is added as the sterol source. Where does C. elegans get its sterols in natural settings? One possible source of sterols is fungi, another being the degraded plant tissue itself. Fungal cells or cell walls are sometimes observed in the gut of wild-caught C. elegans (Félix and Duveau 2012). Therefore, it is possible that this nematode specifically takes up fungal cells (mostly unicellular yeast forms) and/or plant material and/or material from other animals (e.g., dead and decomposing animals found in rotting fruits or compost) to satisfy its nutritional needs.
In this context, it may be speculated that access to these environmental nutrients may be mediated by bacterial members of the nematode’s microbiome. In analogy to the gut microbiomes of termites (Brune 2014; Peterson and Scharf 2016) or ruminants (Krause et al. 2013), these bacteria may express the relevant enzymes to process environmental substances, which the worm itself cannot digest directly. The associated bacteria may thus fulfill a twofold function by directly serving as food and by indirectly providing access to environmental nutrients through metabolizing and further processing the available components. The latter ability and the relevance for nematode viability and fitness clearly deserve further study in the future.
C. elegans as a disperser
Besides potentially providing a substrate for bacterial growth, C. elegans may also disperse micro-organisms. It is so far unclear whether dauer larvae may carry bacteria, but feeding stages certainly do. Studies of dispersal by C. elegans have been performed using different experimental approaches. C. elegans can be found in the rich compost of mushroom farms and in this context may affect growth of the commercial mushroom Agaricus bisporus (Grewal 1991b), in part but not only because it spreads bacteria, such as Pseudomonas tolaasii (Grewal 1991c). Moreover, in microcosm experiments, C. elegans was found to mediate the spread of different bacteria (e.g., specific strains of E. coli and Salmonella newport) to new substrates such as compost or plant material (Kenney et al. 2005, 2006; Anderson et al. 2006). Similar laboratory-based experiments demonstrated that C. elegans can enhance the dispersal of pathogenic P. aeruginosa (Diaz and Restif 2014) and of a bacteriophage of P. syringae (possibly with its host bacterium) (Dennehy et al. 2006). A recent study demonstrated nematode-mediated dispersal of E. coli, which resulted in a growth advantage for the bacteria on the new substrate patches and, most impressively, a subsequent increase in population growth of C. elegans (Thutupalli et al. 2017). In this experiment, the bacteria likely dispersed mostly by attaching to the animal’s cuticle, as srf-3 cuticle mutants did not disperse E. coli. Because of the fitness advantage for the nematode, the phenomenon was termed “farming” (Thutupalli et al. 2017). However, this term is misleading because it implies specific adaptations that allow C. elegans to disperse and initiate new E. coli colonies. To date, it cannot be excluded that bacterial dispersal is simply a by-product of normal C. elegans foraging behavior instead of the result of past adaptive evolution specific for this trait.
Another dispersal system uses the artificial pairing of two model organisms, Dictyostelium discoideum and C. elegans. These interactions may possibly occur in nature, as various slime molds are found in the same samples as C. elegans. C. elegans adults (although not necessarily the larvae; H. Schulenburg, unpublished data) can feed on the amoebae but not on the aggregated stages of Dictyostelium. Instead, on the aggregated stage, C. elegans helps by dispersing spores that survive the animal’s gut (Kessin et al. 1996). Finally, dauers crawl up the fruiting bodies and nictate, helping in their own dispersal. This constitutes an interesting example of the web of relationships that C. elegans could have with associated organisms.
Pathogens and Parasites
Overview of the diversity of microbial antagonists
We will now turn to the microbes that entertain an antagonistic relationship with C. elegans. Pathogens are defined as harming C. elegans, while parasites are defined as taking advantage of it. Both types of relationship usually coexist for a given interaction. Moreover, a given organism can be pathogenic in one condition and beneficial in another. Note that, in immunity studies, longevity assays are often used, yet they are not necessarily informative concerning pathogenicity. For example, a particular microorganism may shorten the C. elegans postreproductive life span, but may not really matter to the nematode (or its evolution) if it does not affect the animal’s fitness in a given environment. The consequences of pathogens are thus not always visible in reduced life span, but rather reduced offspring production. Indeed, in exponentially growing C. elegans populations, a slight slowing down of progeny production and/or brood size decrease should have a much stronger effect than decreased longevity (Hodgkin and Barnes 1991) [see below in Viruses and the competition experiment with a C. elegans virus in Ashe et al. (2013)].
Harmful effects of pathogens may be caused by toxic substances, which are known to be produced by various C. elegans pathogens, including both bacteria and fungi (Griffitts and Aroian 2005; Cezairliyan et al. 2013; Kirienko et al. 2013; Li and Zhang 2016). The pathogen may induce harm by disrupting cellular integrity or cellular and physiological homeostasis. For a parasite, it is additionally necessary that it can benefit from the interaction, generally by invading and replicating in the nematode body. Below, we provide an overview of naturally associated pathogens and parasites, ranging from fungal taxa, microsporidia, viruses, and oomycetes to bacterial pathogens.
Fungal pathogens
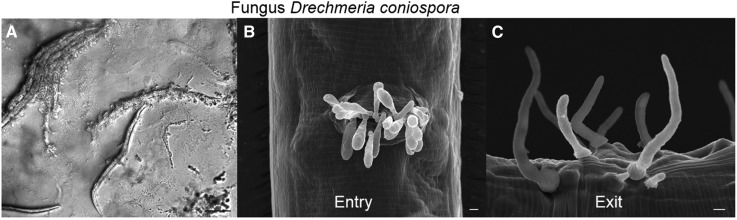
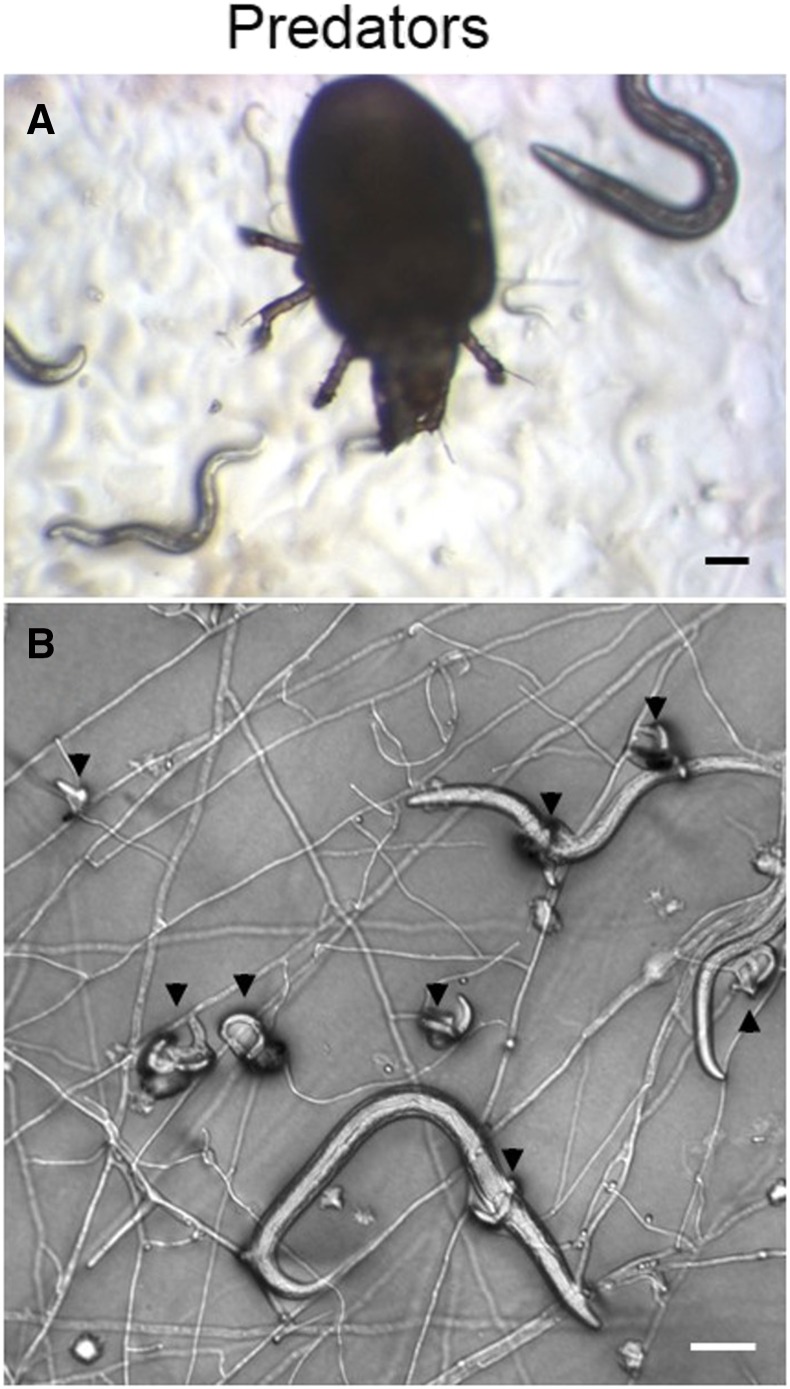
For nematode-harming fungi, a rather arbitrary frontier between predation and pathogens/parasites is usually set depending on the size of the relevant fungal life stage relative to the worm: hyphae for predators (Competitors and Predators) and spores for pathogens/parasites (here). In the latter case, spores attack worms and subsequently grow into hyphae inside the nematode. To date, two types of fungal pathogens have been found to infect C. elegans and other rhabditids in nature: Drechmeria and Harposporium (Félix and Duveau 2012). Most extensively studied is the interaction between C. elegans and Drechmeria coniospora. Spores of this fungus attach to the nematode cuticle, mostly around the mouth and the vulva, pierce it, and hyphae start invading the nematode, killing it within 2–4 days (Jansson 1994; Pujol et al. 2001). The hyphae then pierce out again and, once emerged, start budding spores along their axis (Figure 7). A single infected animal can yield thousands of spores and an infected C. elegans culture can be completely killed under laboratory culture conditions. C. elegans responds to Drechmeria infection by the secretion of antimicrobial peptides and other cellular responses that ameliorate its survival and reproduction capacity. The signaling pathways and effectors for the response have been extensively studied using C. elegans genetics and have become a model for the complexity of invertebrate immune responses [reviewed in Kim and Ewbank (2015)]. The genome of D. coniospora has been recently assembled and annotated (Lebrigand et al. 2016), providing a valuable resource for studying the genetics of interacting pathogen and host molecules.
Figure 7.
Infection of C. elegans by the fungus D. coniospora. (A) Infected C. elegans larvae on an agar plate. In these last stages of infection, fungal hyphae bearing spores exit from the dead nematodes. Bar, 100 μm. (B and C) Scanning electron microscopy. Bar, 2 μm. (B) Adhesion of D. coniospora spores to the vulva (they also adhere preferentially to the mouth periphery). (C) Exit of the fungus after infection. Pictures by M.-A.F.
In contrast to spores of Drechmeria, those of Harposporium enter through ingestion (at least for most species of this genus) (Esser and El-Gholl 1992). The intestinal eptihelium is thus the first tissue to be attacked by the growing fungus. The transcription/RNA turnover response to infections of various pathogens depends greatly on the infected tissue rather than on the type of pathogen (Engelmann et al. 2011).
Microsporidia
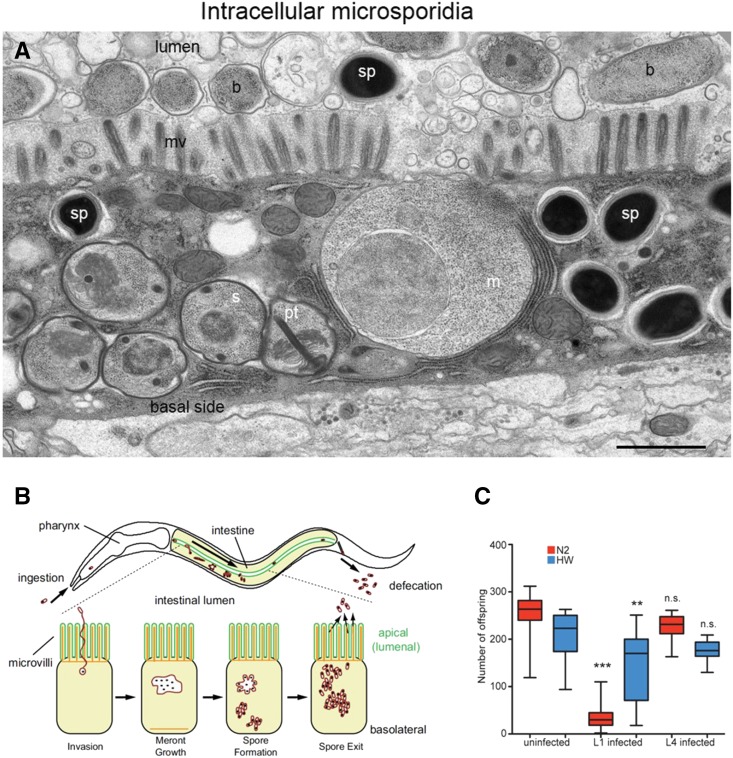
Microsporidia were the first parasites found in wild-caught C. elegans (Troemel et al. 2008). These close relatives of fungi are obligate intracellular parasites. They can be visualized inside the host cells by Nomarski microscopy. The most visible stage is the spore stage, when they resemble rod-shaped bacteria such as E. coli [indeed, they were mistakenly dubbed intracellular bacteria in Barrière and Félix (2005)]. Four species of microsporidia have been found in C. elegans so far, all placed in a new genus called Nematocida (Troemel et al. 2008; Luallen et al. 2016; G. Zhang et al. 2016; Reinke et al. 2017). The first and most common one, Nematocida parisii, and its close relatives N. ironsii and N. ausubeli, infect C. elegans’ intestinal cells and are horizontally transmitted through ingestion and defecation (Figure 8). Microsporidia spores contain a characteristic “polar tube,” which is discharged in some specific environments such as the C. elegans gut and injects the microsporidian DNA into the host cell. DNA replication and nuclear divisions ensue in a syncytial meront stage inside the host cell. The microsporidia can spread laterally from intestinal cell to intestinal cell (Balla et al. 2016). The meronts then secrete an envelope, cellularize (sporont stage), and start maturing in spores with a polar tube and its anchoring disc. The mature spores are surrounded by an additional membrane and routed through a vesicular pathway, recruiting RAB-11 and an actin coat, until they exit to the gut lumen (Szumowski et al. 2014, 2016). One infected animal may yield over 1000 spores.
Figure 8.
Microsporidian infections of C. elegans. (A) Transmission electron microscopy of N. ausubeli infecting C. elegans. The different developmental stages of the microsporidia are visible within a host intestinal cell: m, meront; s, sporont; sp, spore; pt, polar tube. Also visible are b, bacteria; mv, microvilli. Bar, 1 μm. (B) Schematic depiction of the life cycle of N. parisii in C. elegans. (C) Effect of Nematocida infection on C. elegans brood size. Red bars: N2 strain. Blue bars: CB4856 strain, from Hawaii (HW). The CB4856 strain is able to actively clear infection when infected early in the L1 stage. (A) Courtesy of M. Sachse and G. Zhang. (B) Reproduced from Szumowski et al. (2014). (C) Reproduced from Balla et al. (2015).
N. parisii slows down progeny production and severely reduces C. elegans progeny number. If the animals are infected at the L1 stage, death of the animal may occur after ∼3–5 days (Troemel et al. 2008; Balla et al. 2015). As with many intestinal pathogens, an obvious consequence of microsporidian infection is a severe shrinkage of intestinal cells, including of their storage granules and microvilli. Thus, the animals become pale under the dissecting microscope and likely display a reduced input of nutrients and metabolic activity. N. parisii infection results in upregulation of SCF (Skp/Cullin/F-box containing) ubiquitin-ligase subunits and ubiquitinylation of the pathogen, which may target the microsporidia to degradation by autophagy (Bakowski et al. 2014b). Individuals of some wild isolates of C. elegans are able to fully clear the infection: if inoculated with spores as young larvae, they can be infected intracellularly by N. parisii and then clear it entirely from their body. The genetic basis for natural variation in this process is being studied (Balla et al. 2015). Interestingly, the microsporidian proteins that are exposed to the host cytoplasm or nucleus tend to bear signal peptides or transmembrane domains and belong to large and/or fast-evolving, species-specific gene families (Reinke et al. 2017). Further details on the etiology and genetics of C. elegans–microsporidia interactions have been nicely summarized elsewhere (Balla and Troemel 2013; Bakowski et al. 2014a; Szumowski and Troemel 2015).
The ability to infect other nematode species varies among the microsporidia species. N. parisii and N. ausubeli both appear to infect C. elegans, C. briggsae, and other wild-caught Caenorhabditis species of the Elegans group. N. parisii is not able to infect Oscheius tipulae in the laboratory (G. Zhang et al. 2016). Conversely, microsporidia infecting O. tipulae, a very common rhabditid nematode species, cannot infect C. elegans. Individual microsporidia taxa may thus be able to infect closely related species, but not necessarily all rhabditids. A third Nematocida species with a different tissue tropism has been found recently in a wild-caught C. elegans, and named N. displodere (Luallen et al. 2016). This species proliferates mostly in the epidermis, as well as muscles and neurons. The polar tube in the spore is long and may directly reach the epidermis from the gut lumen. Spores are then released from the epidermis and other tissues via bursting of the animal at the vulva, thus also killing the host.
Viruses
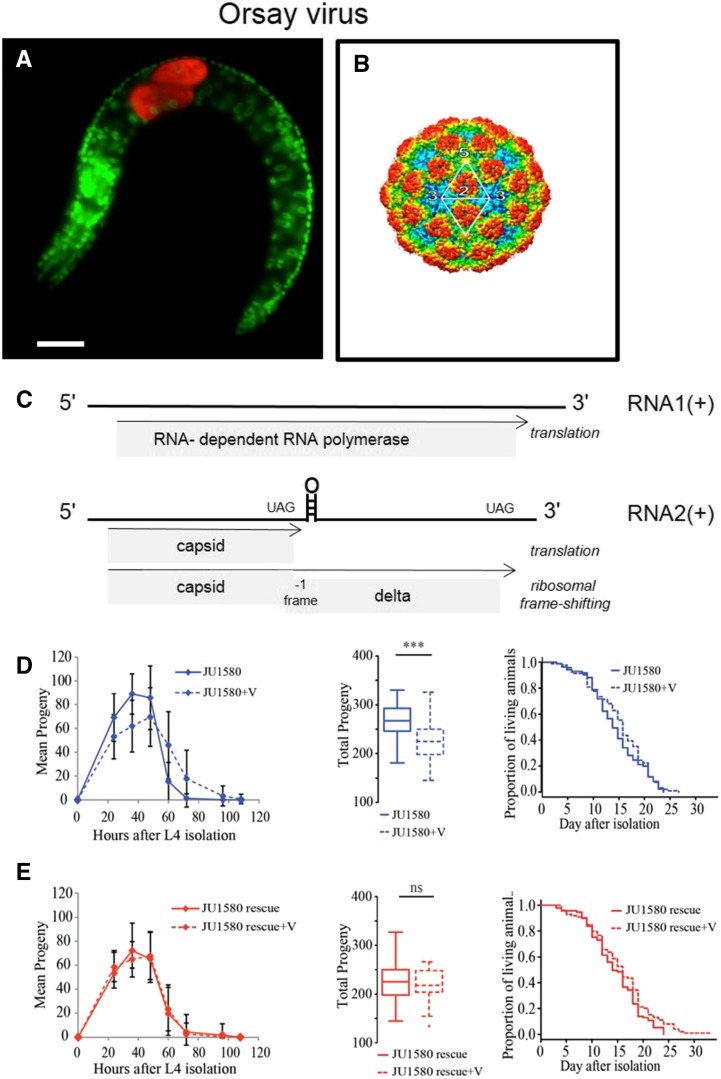
The discovery of viruses infecting C. elegans came with the observation of wild-caught C. elegans and C. briggsae animals with intestinal cell abnormalities, yet without visible pathogens, by Nomarski microscopy. These symptoms could be cured by bleaching and reinfection experiments after 0.2 μm filtration yielded the same cellular symptoms. These experiments led to the discovery of a group of nematode viruses related to fish and arthropod nodaviruses (Félix et al. 2011; Franz et al. 2012). These three viruses are transmitted horizontally and all infect intestinal cells (Félix et al. 2011; Franz et al. 2014) (Figure 9). Their genome is composed of two positive-strand RNA molecules: RNA1 encoding a RNA-dependent RNA polymerase and RNA2, a capsid with a facultative N-terminal δ domain translated by facultative ribosomal frameshifting at the first stop codon (Félix et al. 2011; Jiang et al. 2014) (Figure 9C). These viruses are species-specific, with the Orsay virus able to infect C. elegans but not C. briggsae, and conversely for the Santeuil and Le Blanc viruses (Félix et al. 2011; M.-A. Félix, T. Bélicard, G. Brésard and L. Frézal, unpublished data). The Orsay virus has, so far, only been found on rare occasions in C. elegans isolates of the region around Paris (L. Frézal and M.-A. Félix, unpublished data). These viruses all cause the same symptoms in the host intestinal cells, which progressively lose internal structures such as cytoplasmic granule content and nuclei, and fuse to each other. At the level of the organism, these viral infections are detrimental to population growth by slowing down progeny production and slightly lowering the brood size, without a detectable effect on worm longevity (Ashe et al. 2013) (Figure 9, D and E).
Figure 9.
Orsay virus infection in C. elegans. (A) Fluorescent in situ hybridization of the Orsay virus in C. elegans JU1580 using a probe against RNA2 (red). DAPI is in green. Bar, 20 μm. Picture courtesy of L. Frézal. (B) Cryo-electron microscopy structure of capsid assembly (Guo et al. 2014). (C) Structure of the Orsay virus, with the facultative ribosomal frameshifting in the translation of RNA2 (Jiang et al. 2014). (D) Infection by the Orsay virus has an effect on progeny production but not longevity. (E) Rescue of the wild isolate JU1580 (carrying a drh-1 deletion) with a drh-1(N2) transgene. Panels from left to right: dynamics of progeny production, total brood size, and survival curves (n = 40 animals for brood size, n =130 for longevity). ***P < 0.001. (D and E) Reprinted from Ashe et al. (2013).
The replication cycle of RNA viruses includes a double-stranded stage, to which C. elegans may respond by initiating a small RNA cascade that degrades the viral RNAs (Félix et al. 2011; Ashe et al. 2013). However, wild isolates of C. elegans differ widely in their sensitivity toward the Orsay virus: after laboratory infection, a wide range of viral load is observed, with some isolates being fully unable to sustain viral replication. A genome-wide association study pointed to a single main locus explaining most of the species’ phenotypic variation in viral replication, identified as a deletion polymorphism in the drh-1 resistance gene. Figure 9, panels D and E, shows the rescue of the wild isolate JU1580 by an intact drh-1(N2) transgene. The DRH-1 protein is a homolog of the vertebrate RIG-I family, which starts the transcriptional interferon response in mammals. In C. elegans, DRH-1 instead starts the small RNA antiviral response (Ashe et al. 2013). That the derived allele is a deletion in a resistance gene is odd in terms of evolution of host–pathogen interactions. One possible explanation is that this conditional deleterious variant hitchhiked on another variant due to the low level of outcrossing and the high level of linkage disequilibrium in C. elegans (Ashe et al. 2013).
Oomycetes
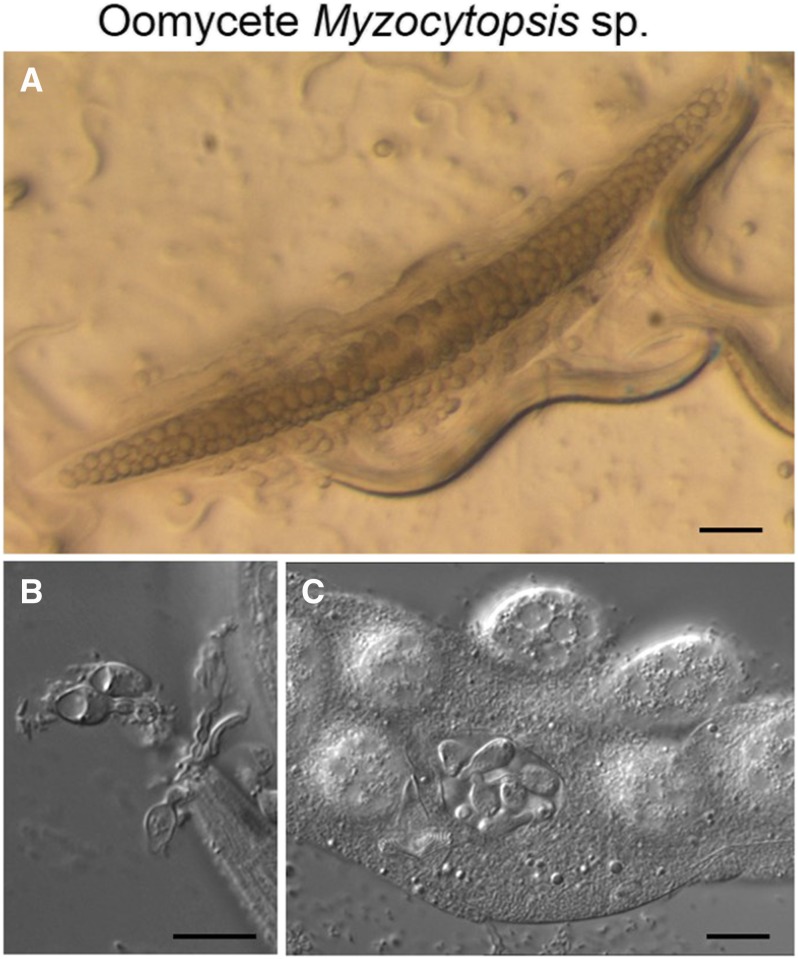
Oomycetes were previously classified with Fungi because of similarities in their life cycle, but are now known to belong to the clade of Heterokonts, together with brown algae and diatoms (Beakes et al. 2012). Oomycetes include mostly plant-infecting species, such as Phytophthora infestans, which causes the potato blight disease. Oomycetes were previously found infecting other rhabditid nematodes (Maupas 1915). Their life cycle was studied morphologically (Beakes et al. 2012), but as for trapping fungi the authors did not pay much attention to the nematode species being infected, presumably because of the low host specificity. C. elegans was recently found infected with an oomycete of the genus Myzocytiopsis (M.-A. Félix, unpublished results), and this now allows the study of oomycete–C. elegans molecular interactions (M. Barkoulas, personal communication) (Figure 10).
Figure 10.
Infection of C. elegans by the oomycete Myzocytopsis sp. (A) Infected C. elegans adult on an agar plate. In this last stage of infection, the oomycete forms in the host body spherical multinucleate structures called sporangia. Bar, 100 μm. (B and C) Nomarski microscopy. Bar, 10 μm. (B) Adhesion of Myzocytopsis spores to the C. elegans mouth (they may adhere elsewhere on the cuticle as well). (C) Formation of spores in the sporangia. Only the cuticle and grinder of C. elegans remain visible. Pictures by M.-A.F.
Bacterial antagonists
Overview of bacterial pathogens:
Despite the fact that C. elegans immune defenses must have evolved in the context of naturally encountered bacteria, the first focus has been to study C. elegans interaction with human pathogenic bacteria rather than natural pathogens of C. elegans itself (Darby 2005; Kim and Ewbank 2015). Several of these bacteria are nevertheless likely to be relevant in the natural context, for example P. aeruginosa, Bacillus thuringiensis, and Serratia marcescens, for which there is some indication for coexistence with C. elegans and which we describe in more detail below. We left out some of the other previously studied opportunistic human pathogens, for which it is possible, yet currently less clear, that they interact with C. elegans in the field, for example Burkholderia pseudomallei (e.g., Day and Sifri 2012; Lee et al. 2013; Lim et al. 2016).
Note that the natural coexistence of pathogens and C. elegans is difficult to uncover, in contrast to the natural interaction with mutualists or commensals. Because of the antagonistic nature of the interaction, the animals are often paralyzed or even dissolved before they can be isolated and identified using the common C. elegans isolation protocols. Therefore, it is likely that the taxa that have so far been found are biased toward those mildly pathogenic for C. elegans. Many more pathogens are likely to be discovered in the future. Nevertheless, there is little doubt that pathogens have had a strong impact on C. elegans evolution. Although the species possesses a comparatively simple immune system that lacks specialized immune cells, it is still based on complex, interconnected immunity pathways and comprises defenses in the form of behavior, physical barrier, and physiology. Thus, C. elegans has become an important model system for studying the genetics of the innate immune system [reviewed in Irazoqui et al. (2010), Pukkila-Worley and Ausubel (2012), and Kim and Ewbank (2015)] as well as behavioral responses to pathogens. The latter include direct avoidance behaviors, reduced oral uptake of pathogens, and a learning response upon first encounter of pathogens, leading to enhanced avoidance upon secondary encounters [reviewed in Schulenburg and Ewbank (2007), Zhang (2008), and Meisel and Kim (2014)].
The naturally coexisting pathogens may infect C. elegans either through the cuticle or the gut. Below, we will start with several naturally associated bacteria that are likely to infect the worm through the gut, followed by cuticle-attaching pathogens, and then an overview of the nematode’s interaction with P. aeruginosa, B. thuringiensis and S. marcescens.
Bacterial pathogens naturally associated with C. elegans.
Live bacteria in the C. elegans gut may be beneficial in some cases, when the bacteria do not overproliferate. However, microscopic observations of wild-caught C. elegans indicate that naturally associated bacteria may proliferate so much that the gut lumen is filled with a large number of bacteria, leading to a substantial enlargement of the lumen at the expense of the worm’s intestinal cells. In this case, the nematode appears compromised in its ability to feed and process nutrients, leading to reduced progeny production (Félix and Duveau 2012). Many bacteria that were isolated in the microbiota studies are likely to accumulate in the gut. Of those tested for their effect on C. elegans, some strains strongly diminish brood size and are thus potential pathogens, for example some (not all) members of Pseudomonas (MYb193), Microbacterium (MYb45 and MYb50), Bacillus (MYb78 and MYb56), Chryseobacterium (MYb7 and MYb120), Arthrobacter (MYb27), Rhodococcus (MYb53), Leuconostoc (MYb83), and Sphingobacterium (MYb181 and MYb210) (Dirksen et al. 2016). Moreover, specific strains of Chryseobacterium (JUb44), Serratia (JUb9), and Pseudomonas (GRb427) slowed down the growth of C. elegans individuals considerably (Samuel et al. 2016). Another pathogenic bacterium that was isolated with C. elegans is Chryseobacterium (or Elizabethkingia) sp. JUb129, a member of family Flavobacteriaceae in the Bacteroidetes phylum (Félix and Duveau 2012). This bacterium is able to kill C. elegans within a day and seems to consume it completely, including the cuticle (Félix and Duveau 2012). It is so far unclear how this infection starts, whether through the gut or through the cuticle. More work is required to determine its interaction with C. elegans.
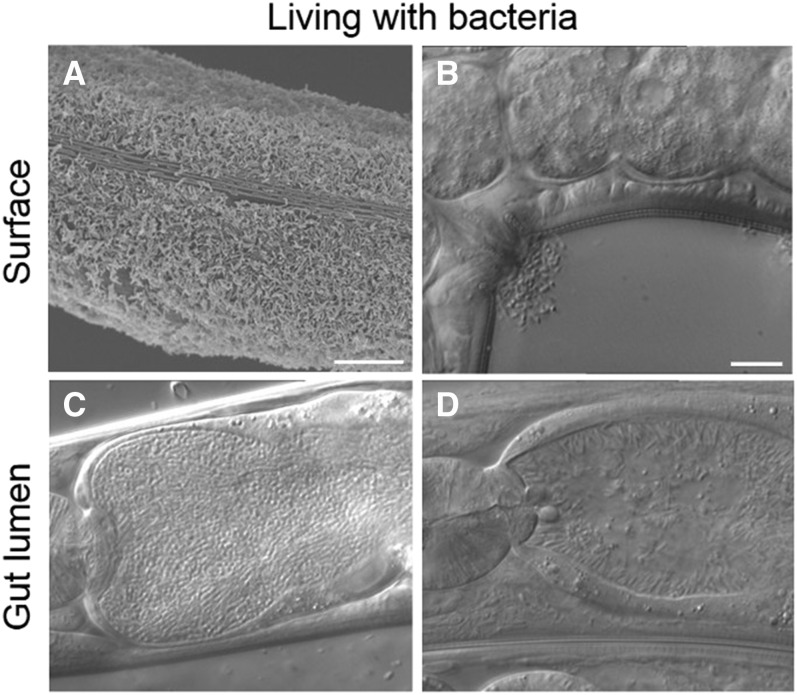
Cuticle-attaching bacteria were first studied by J. Hodgkin’s laboratory, making use of an infection occurring repeatedly in laboratory cultures. Microbacterium nematophilum bacteria attach to the cuticle next to the rectum (potentially a good place to get food) and proliferate (Hodgkin et al. 2000). They induce a local swelling of the anal region and harm the animal. They show some specificity among rhabditids; in C. briggsae they also adhere to the vulval region, following the expression pattern of the bus-1 gene encoding a membrane O-acyltransferase (Gravato-Nobre and Hodgkin 2008). Genetic screens for mutants with an altered response to infection uncovered a number of genes involved in cuticle composition (Gravato-Nobre 2005; Yook and Hodgkin 2007; Gravato-Nobre et al. 2011). Although M. nematophilum was not found associated with C. elegans in natural settings so far, other bacteria of the same genus are common in the worm’s microbiome (although their effect is yet unclear; H. Schulenburg, unpublished data; Dirksen et al. 2016), and bacteria attaching to the cuticle and sometimes affecting locomotion are often found on wild-caught C. elegans animals (Figure 11; M.-A. Félix and H. Schulenburg, unpublished data).
Figure 11.
Types of physical interaction of C. elegans with bacteria (other than food). (A) Scanning electron microscopy picture of C. elegans covered with Leucobacter celer CBX151. Bar, 10 μm. (B–D) Nomarski pictures of unidentified bacteria with their wild C. elegans associate: adhering to the vulva of C. elegans (B), proliferating in the intestinal lumen of a L4 larva (C), and adhering to the apical intestinal border (D). Bar, 10 µm. Pictures by M.-A.F.
Three other bacteria found associated with C. elegans or C. tropicalis in nature were identified as Leucobacter spp., in the same family Microbacteriaceae (bacterial phylum Actinobacteria) as M. nematophilum (Hodgkin et al. 2013). Two of them, Leucobacter musarum CBX152/Verde 2 and CBX130, induce rectal swelling similar to M. nematophilum (Hodgkin et al. 2013; Clark and Hodgkin 2015). The third one, L. celer CBX151/Verde 1, coats the whole surface of the nematode (Figure 11A). Remarkably, in liquid culture, it induces the aggregation of C. elegans individuals through their tail (worm star formation) and rapidly kills them within a day (Hodgkin et al. 2013; Clark and Hodgkin 2014). The pathogenic mechanism appears to result from a physical injury of the animal’s tail and subsequent invasion. The nematodes may escape via autotomy of the tail, which can heal, resulting in a sometimes partially fertile worm with a posterior body truncation. Strikingly, this lethal bacterium in liquid culture rescues on agar plate surfaces the lethality caused by L. musarum (CBX152/Verde 2 and CBX130) (Hodgkin et al. 2013). Mutant animals that are resistant to M. nematophilum and L. musarum due to a change in cuticular composition are hypersensitive to L. celer, suggesting a tradeoff in resistance between the two bacteria (Hodgkin et al. 2013). Members of genera Microbacterium and Leucobacter have been found in studies of the microbiota associated with C. elegans (see above), which suggests that they are relevant for C. elegans in the wild. However, it is not entirely clear which of the medium-dependent effects of these bacteria are most relevant in natural C. elegans populations. Newly infested plant matter or compost may still provide a rather intact substrate surface, thus resembling solid agar plates. Increased decomposition of plant material, especially fruits, may yield a liquid environment similar to the liquid lab medium, although usually with higher viscosity. Simulation of the most relevant natural medium conditions under laboratory conditions still represents a particular challenge for future research.
P. aeruginosa as a likely natural pathogen:
Another likely natural pathogen of C. elegans is the Gram-negative bacterium P. aeruginosa. This bacterium was reported to coexist with C. elegans in compost collected from a mushroom farm in the UK (Grewal 1991a). Subsequent experimental analysis revealed that the bacterial isolate led to reduced worm population growth (Grewal 1991a). Almost a decade later (Darby et al. 1999; Mahajan-Miklos et al. 1999; Tan et al. 1999a,b), C. elegans was exposed to clinical isolates of this pathogen species, demonstrating the bacteria’s ability for infecting and killing the worms. Three main types of killing dynamics were identified, which were dependent on the exact composition of the medium: (i) fast toxin-mediated killing on high osmolarity agar plates (Darby et al. 1999; Mahajan-Miklos et al. 1999; Cezairliyan et al. 2013); (ii) slow killing through bacterial accumulation in the gut and expression of various virulence factors on low osmolarity agar plates (Tan et al. 1999a,b; Feinbaum et al. 2012), and (iii) iron-dependent, hypoxia-mediated killing in liquid medium (Kirienko et al. 2013, 2015). This model infection system was explored in numerous studies to dissect the molecular basis of the nematode’s immune defense system. It revealed the ability of C. elegans to behaviorally avoid this pathogen, either upon direct contact, mediated through pathogen secondary metabolites and a neuroendocrine host response based on G protein-coupled chemoreceptor and TGF-β signaling (Meisel and Kim 2014; Meisel et al. 2014), through the neuropeptide receptor gene npr-1 (Styer et al. 2008; Reddy et al. 2009; Chang et al. 2011; Nakad et al. 2016), or upon a learned avoidance response mediated by serotonin and TGF-β signaling (Zhang et al. 2005; Zhang 2008; Zhang and Zhang 2012). Moreover, C. elegans can respond by activating its immune system, especially through the p38 MAPK pathway, the GATA transcription factor ELT-2, the insulin-like signaling cascade, and the hypoxia response (Kim et al. 2002; Shapira et al. 2006; Evans et al. 2008b; Kawli and Tan 2008; Kirienko et al. 2013). In-depth genetic analysis of the response to this pathogen permitted characterization of the complex signaling network underlying invertebrate immunity (Irazoqui et al. 2010; Pukkila-Worley and Ausubel 2012; Kim and Ewbank 2015).
The commonly used P. aeruginosa strain PA14 is able to interfere with the nematode’s defense mechanisms, in particular with the insulin-like signaling cascade, thereby increasing its ability to infect (Evans et al. 2008a). Secreted Pseudomonas compounds, such as acylated homoserine lactones, also attract C. elegans (Beale et al. 2006). Such an attraction response has been found toward other pathogens. For example, C. elegans is initially attracted to pathogenic S. marcescens, which it subsequently avoids (Pradel et al. 2007) (see also below S. marcescens as a likely natural pathogen). The nematode also responds positively to specific volatile organic compounds, such as 2-heptanone, of B. nematocida (Niu et al. 2010; C. Zhang et al. 2016). These observations may suggest a manipulative behavior of the bacteria, a “trojan horse” mechanism (Niu et al. 2010), that could facilitate successful infection of the host. In general, they support the idea that pathogen-mediated manipulation of the host has evolved repeatedly and should subsequently lead to increased selection on the host to counteradapt (Schmid-Hempel 2008).
Another study was based on an evolution experiment, which favored the emergence of infectious P. aeruginosa varieties that supported proliferating C. elegans populations (Jansen et al. 2015). This selection regime caused the evolving bacteria to lose virulence while maintaining their ability to accumulate inside worms. Pathogens thus evolved a commensal and perhaps mutualistic phenotype within few generations, highlighting that changes along the parasite–mutualist continuum can happen fast in response to appropriate selective constraints (Jansen et al. 2015). None of the recent studies used P. aeruginosa strains that coexist with C. elegans in nature and are thus likely shaping worm life history. Similarly, only a single study has yet assessed the response of a range of natural C. elegans isolates to this pathogen (using one of the clinical isolates of P. aeruginosa). QTL analysis of C. elegans strains in this study revealed that variation in avoidance behavior mapped to a genomic region in the middle of chromosome IV (Andersen et al. 2012).
B. thuringiensis as a likely natural pathogen:
Another possibly natural pathogen of C. elegans is the Gram-positive spore-producing B. thuringiensis. This species is defined by plasmids that encode crystal toxins. Single strains usually contain only one to few toxin genes, which in turn determine specificity of the pathogen toward various insect and nematode hosts, including C. elegans (Vilas-Boas et al. 2007). Nematode infection begins with the oral uptake of the spores and their associated crystallized toxins. The toxins are solubilized in the nematode gut, proteolytically activated, and then specifically bind to sugars on intestinal cells, which leads to formation of pores in these host cells. Intestinal damage through the pore-forming toxins leads to a currently not yet well-understood change in milieu in the gut, which causes germination of the spores, followed by proliferation of vegetative cells. This process ultimately destroys all worm tissues. The animal is filled more or less completely by bacterial cells, which then start producing new spores and associated crystal toxins (Griffitts and Aroian 2005; Nielsen-LeRoux et al. 2012) (Figure 12). A nonpathogenic strain of this species was co-isolated with C. elegans from nature (M.-A. Félix and H. Schulenburg, unpublished data). The interaction of C. elegans with a variety of B. thuringiensis strains and specific toxins was analyzed in detail. The animals can behaviorally avoid this bacterium, a behavior mediated at least to some extent through the insulin-like signaling cascade and the neuropeptide receptor gene npr-1 (Hasshoff et al. 2007; Nakad et al. 2016). Glycolipids on intestinal cells act as receptors for the crystal toxin Cry5B (Griffitts et al. 2005; Barrows et al. 2007), while ASP-1-mediated necrosis enhances susceptibility to the toxin Cry6Aa (F. Zhang et al. 2016). The physiological immune response is influenced by p38 and Jun N-terminal kinase (JNK) MAPK signaling, the hypoxia pathway, and insulin-like signaling (Huffman et al. 2004; Hasshoff et al. 2007; Bellier et al. 2009; Kao et al. 2011; Yang et al. 2015). Immune effectors include several lysozymes and caenopores (Boehnisch et al. 2011; Hoeckendorf and Leippe 2012; Hoeckendorf et al. 2012). Some micro RNAs additionally influence the response to the highly virulent B. thuringiensis strain DB27 (Iatsenko et al. 2013).
Figure 12.
Infection of C. elegans infected with B. thuringiensis. (A) C. elegans killed by an infection with B. thuringiensis on an agar plate; the nematode is already disintegrating and only spores of the pathogen remain (picture courtesy of Rebecca Schulte). (B) Vegetative cells of B. thuringiensis inside a killed worm (picture courtesy of A. Papkou). (C) and (D) Electron micrograph of a killed infected nematode, containing mainly vegetative cells of the pathogen inside of its body. Picture in (C) from Schulte et al. (2010), and picture in (D) by H.S.
The interaction between C. elegans and B. thuringiensis was established as a model for experimental evolution, to study the dynamics of host–pathogen coevolution. Using genetically diverse host populations, C. elegans was shown in three fully independent evolution experiments to be able to adapt rapidly and in a highly specific manner to the continuously evolving pathogen challenge (Schulte et al. 2010, 2011, 2012, Masri et al. 2013, 2015; H. Schulenburg, unpublished data). At the same time, B. thuringiensis adapted to the host in a similarly specific manner (Schulte et al. 2010, 2011; Masri et al. 2015), apparently through changes in the copy number of a toxin-encoding plasmid and possibly also additional virulence factors (Masri et al. 2015). Taken together, these studies highlight that the interaction with B. thuringiensis has the potential to impose high selection pressure on C. elegans, possibly also in nature. However, such dynamics have not yet been studied in a more natural context.
S. marcescens as a likely natural pathogen:
The Gram-negative opportunistic pathogen S. marcescens is common in diverse environments, and an isolate showing the ability to swarm was previously found together with C. elegans in a compost sample in France (M.-A. Félix, unpublished results) (Pradel et al. 2007). Moreover, natural C. elegans strains produce highly genotype-specific interactions with natural S. marcescens isolates (Schulenburg and Ewbank 2004), possibly suggesting reciprocal coadaptations between these two antagonists. Therefore, it is likely that C. elegans interacts with this pathogen in nature and is able to respond to it in a highly specific form. This idea is supported by several evolution experiments under controlled conditions in the laboratory, under which the two antagonists were allowed to coadapt to each other (i.e., both host and pathogen could reciprocally adapt to the other) or only one was allowed to adapt to a nonchanging partner (e.g., C. elegans was allowed to adapt, while the same S. marcescens strain was newly added at each transfer step from a frozen stock culture, and vice versa). These experiments highlighted that both are able to specifically adapt to the antagonist and that the high selective constraints imposed by the continuous need to coadapt favor increased outcrossing rates in C. elegans (Morran et al. 2009, 2011, 2013, 2014; Slowinski et al. 2016).
The interaction between C. elegans and a specific S. marcescens isolate was further analyzed at the genetic and genomic level. These studies revealed that the inducible immune response is mediated by a TGF-β pathway and possibly a GATA transcription factor (Mallo et al. 2002; W. Yang et al. 2016), while resistance is influenced by lysozymes and C type lectins (Mallo et al. 2002; Miltsch et al. 2014). Most impressively, C. elegans is able to specifically recognize a surfactant from S. marcescens and then respond through an avoidance behavior, mediated by G protein signaling and the Toll-like receptor TOL-1 (Pujol et al. 2001; Pradel et al. 2007). TOL-1 acts by influencing the development and functioning of specific chemosensory neurons, the BAG neurons, which are involved in the surveillance of bacterial metabolism and the initiation of a pathogen avoidance response (Brandt and Ringstad 2015). C. elegans isolates show variation in their behavioral response to S. marcescens, which can be mapped to three main QTL, although the exact underlying genetic changes and molecular processes are yet unknown (Glater et al. 2014). It is of particular interest to find out to what extent such behavioral and physiological responses are expressed in natural C. elegans isolates toward coexisting S. marcescens varieties.
Competitors and Predators
Possible competitors
The competitors of C. elegans have been little studied. Other bacteriovorous nematodes can be found in the same samples, including other species of Caenorhabditis. C. briggsae was the Caenorhabditis species found most often in the same fruit or stem in surveys in France (Félix and Duveau 2012). In an apple orchard (Orsay), C. briggsae and C. elegans have overlapping but distinct seasonal distributions; while C. briggsae dominates in summer and early fall, C. elegans thrives in late fall. This seasonal distribution fits the species temperature preference as tested through competitions in the laboratory (Félix and Duveau 2012). However, an overlap occurred in the fall, and many samples in rotting stems in woods also contained both species in feeding stages, indicating possible competition (Félix and Duveau 2012). In contrast, in the surveys in Germany C. briggsae was rare, while C. remanei was most abundant in fruits and C. elegans in compost (Petersen et al. 2014). For the two latter species, humidity was a much stronger predictor for nematode presence than temperature (Petersen et al. 2014), opposite to the findings from France. To date, it is unclear why. It is similarly unknown why C. briggsae is apparently rare in German locations, while C. remanei shows low abundance in France. Moreover, it is also unclear whether the differential distribution of C. elegans and C. remanei in the German locations (compost vs. apples, respectively) is due to competitive exclusion. The feeding stages of other bacteriovorous nematode genera are also well-represented in the same samples containing C. elegans, such as members of the genera Oscheius, Pristionchus, Panagrellus (in fruits only), Panagrolaimus, Mesorhabditis, and various other rhabditids. Competition for live bacteria as a food source may also come from amoebas, slime molds, ciliates, and other small invertebrates (Félix and Duveau 2012).
A curious aspect of the competition with congeners is that Caenorhabditis nematodes readily mate with the opposite sex of closely related species under controlled laboratory conditions. These interspecies matings come with two types of cost. On the one hand, energetic resources that are invested in mating behavior, gamete production, and embryo development are wasted because hybrid offspring is usually not viable (Baird et al. 1992; Baird and Yen 2000; Hill and L’Hernault 2001). On the other hand, mating with a closely related species has a sterilizing effect on the production of self-fertilized offspring, resulting in significantly reduced progeny numbers in the case of C. elegans (Ting et al. 2014). The sterilization effect may result from the absence of coadapted antagonist gene(s) in the opposite sex, which is/are present in intraspecific combinations, thus preventing sterilization. As a nonexclusive alternative explanation, it is possible that the sterilization effect evolved to control population size of interspecific competitors (Ting et al. 2014). Whatever the cause, the effect is likely relevant in natural populations of C. elegans, which was found to coexist with congeners in single pieces of rotting plant material, fruit, and compost (Félix and Duveau 2012; Petersen et al. 2014).
Overview of possible predators
Predators feeding on C. elegans in natural settings have similarly been poorly studied. C. elegans moves forward rapidly when touched on the tail and backward when touched on the head (Croll 1975; Chalfie and Sulston 1981; Goodman 2006). This behavior could be an adaptive avoidance response to any of these predators (Pirri and Alkema 2012) and may indicate frequent interactions of C. elegans with such predatory antagonists. While observing predation of C. elegans in the wild context is a challenge, testing whether an organism preys on C. elegans under artificial laboratory conditions is not necessarily relevant. A first requirement for relevance of an interaction is the cooccurrence of predator and prey in the same samples. One approach, not implemented so far for putative C. elegans predators, is to analyze the gut content of wild-caught predators (Read et al. 2006). The closest situation to a natural setting where preying on C. elegans was observed is in freshly collected substrate samples placed on a Petri dish with nutrient agar and brought under a dissecting microscope. In this context, the most common and obvious predators are trapping fungi. Various mites are also found in the same samples and at least one species (tentatively identified as Sancassania sp.; Félix and Braendle 2010) has been observed to eat C. elegans, swallowing it like spaghetti (Figure 13A). Nematode-eating collembola have also been observed in samples with C. elegans. Whether there is any specificity in the nematode species that these small arthropods eat is unclear. Note that predators can also potentially act as vectors if C. elegans avoids being eaten.
Figure 13.
Predators of C. elegans. (A) Sancassania mite. (B) C. elegans larvae trapped by a fungus. Arrowheads designate traps. Bar, 100 μm in (A) and 20 μm in (B). Pictures by M.-A.F.
Besides studies in wild-derived microcosms, predation has been studied in the laboratory by exposing C. elegans or another small nematode to a candidate predator, although this approach does not test whether the interaction is ecologically relevant. Under these artificial conditions, C. elegans can be eaten by the collembola Folsomia candida (Lee and Widden 1996) or the nematode P. pacificus (Serobyan et al. 2014). In Pristionchus species, different adult mouth forms develop depending on the environment (Serobyan et al. 2014). In dire conditions, the mouth develops with strong teeth that enable the adults to prey on other nematodes. Feeding stages of C. elegans and Pristionchus species (other than P. pacificus) share the rotting vegetal matter environment (Félix and Duveau 2012), but the mouth form of the Pristionchus animals has not been assayed. Other nematodes such as mononchs (e.g., Prionchulus spp.) are specialized predators (e.g., Mikola and Sulkova 2001), but have not been noted so far in samples with Caenorhabditis.
Nematode-trapping fungi as predators of C. elegans
Nematode-trapping fungi are diverse and overall the best studied predators of small nematodes like C. elegans (Drechsler 1941; Gray 1987; Zhang and Hyde 2016). Nematode-trapping fungi have been frequently observed in samples with C. elegans (M.-A. Félix, unpublished results, see Figure 13B), but have not been characterized nor their impact on C. elegans populations assessed. Most of the work on nematode-trapping fungi has been carried out using other bacteriovorous nematodes or parasitic nematodes, against which they serve as biological control agents. Most nematode-trapping fungi belong to a clade in Orbiliomycetes (Li et al. 2005). These fungi are facultative predators and may also live as saprophages. Nematode traps are specialized derivatives of fungal hyphae, in the form of adhesive knobs (e.g., Monacrosporium haptotylum; Ahren 2005), adhesive loops or networks (e.g., Arthrobotrys oligospora), or circular rings that constrict upon entry of a nematode (e.g., Drechslerella doedycoides; Drechsler 1941; Zhang and Hyde 2016). The fungi secrete proteases that are able to digest the nematode cuticle (J. Yang et al. 2016). Due to the size of its traps, D. doedycoides mostly traps the young larval stages of C. elegans. Some C. elegans larvae escape using active mechanosensation and suppression of lateral head movements while backing (Maguire et al. 2011). The fungal traps can develop constitutively or be induced by the presence of nematodes (Drechsler 1941; Zhang and Hyde 2016). For example, the traps of A. oligospora are induced upon sensing of ascarosides (Xie et al. 2010; Hsueh et al. 2013), chemicals produced by C. elegans that also act in dauer induction (as mentioned above) and male attraction behavior (Ludewig and Schroeder 2013). Thus, the fungus is capable of using the intraspecific communication signals of C. elegans to induce trap formation, where and when worms appear abundant.
Intraspecific Interactions
Overview of different types of intraspecific interactions and their importance
The biotic environment of C. elegans individuals is not only shaped by interactions with other species, but also by interaction with members of its species. Indeed, these interactions occur frequently and may be associated with highly selective dynamics. Two types of conspecific interaction can be particularly important. On the one hand, competition occurs within growing populations, especially when density increases and nutrient availability decreases. On the other hand, interactions among hermaphrodites and males occur during mating. These interactions are likely in part shaped by conflicting evolutionary interests. Fitness in females and possibly C. elegans hermaphrodites is enhanced by the availability and choice of high-quality mates, whereas that of males is driven by the number of matings (i.e., Bateman’s principle; Arnold 1994). To date, we lack data on the relative importance of these two types of interaction for C. elegans in nature. Yet, as explained below, several lines of evidence suggest them to be influential.
Intraspecific competition and its resolution through C. elegans pheromones
As detailed above (Life cycle of C. elegans and its biotic environment), C. elegans in nature is likely subject to a boom-and-bust life cycle (Frézal and Félix 2015). One or a few dauer larvae are assumed to colonize a new substrate. If conditions are favorable, rapid population growth ensues, until food depletion, crowding, and possibly other factors induce entry of the young larvae into the dauer stage. The increase in population density can be very dramatic, likely reaching up to >100,000 worms/g of substrate (Félix and Duveau 2012; Frézal and Félix 2015). During population growth and especially once higher densities are reached, individuals are likely to compete with each other for resources, particularly food microbes, but also for space and possibly mating opportunities (if males are available). Intraspecific competition can occur either within the same genotype or between distinct genotypes, as substrates were reported to host either a single or several genetically distinct lineages (Barrière and Félix 2007; Frézal and Félix 2015; Petersen et al. 2015b). In principle, coexistence of several genotypes may lead to expression of more aggressive behaviors, whereas competition within the same genotype may be shaped by kin selection and could result in cooperative behaviors (Hamilton 1963; Bourke 2014).
The dynamics of intraspecific interactions remain to be characterized in natural C. elegans populations. Substantial variation can be detected among natural C. elegans isolates in their propensity to enter the dauer stage under fixed pheromone and population size conditions and in growing populations (Viney et al. 2003; Green et al. 2013; Diaz et al. 2014). Moreover, different natural C. elegans strains vary both in the composition of the dauer pheromone they produce and in their response to the dauer pheromone of conspecifics. The result is in an intricate matrix of dauer-promoting and dauer-repressing effects of the pheromones produced and perceived by the various natural isolates (Diaz et al. 2014). This finding may suggest the presence of differences among strains in resolving competition through this alternate life stage. It is even conceivable that the dauer pheromone is used to induce dauer formation in competing genotypes and to remove competitors from the shared environment (Diaz et al. 2014), in agreement with the recent observations made for strains of the nematode species P. pacificus (Bose et al. 2014; Mayer et al. 2015; Sommer and Mayer 2015). As the dauer stage neither requires food nor mates, competition at these two levels is automatically removed in a population of dauers. The low levels of dauer formation that are seen in some C. elegans isolates might have evolved as a defense against manipulation by competitors. Further work is needed to distinguish this possibility from other ecological and evolutionary scenarios.
The C. elegans pheromone does not only influence dauer formation, it can also affect foraging behavior. One component of the pheromone, the ascaroside icas#9, can suppress foraging activity (Greene et al. 2016b). The response to icas#9 varies among natural C. elegans isolates due to polymorphisms in two interacting G protein-coupled chemoreceptor genes, srx-43 and srx-44 (Greene et al. 2016a,b). These two genes are subject to balancing selection in natural C. elegans populations (Greene et al. 2016a,b), possibly indicating the presence of alternative foraging strategies in response to food availability and population density. Thus, these alternative strategies may also contribute to resolve intraspecific competition. It is yet unclear to what extent natural food bacteria similarly influence alternative foraging behaviors in this context.
Male–hermaphrodite interactions
Another influential type of intraspecific interaction is found among the two sexes. C. elegans has an androdioecious reproductive system, consisting of sequential XX hermaphrodites and X0 males. Hermaphrodites can reproduce either by selfing or through mating with the males. Males can be produced by hermaphrodites as a consequence of X chromosome nondisjunction or as crossprogeny of a male and a hermaphrodite. Male frequencies in natural C. elegans strains were shown to be influenced by the rate of X chromosome nondisjunction and the mating efficiency of the males (Hodgkin and Doniach 1997; Wegewitz et al. 2008; Anderson et al. 2010), leading to significant variation in male abundance among C. elegans isolates and environments, ranging from 0.1% up to >30% males stably maintained in the populations (Hodgkin and Doniach 1997; Teotónio et al. 2006; Wegewitz et al. 2008; Anderson et al. 2010). Additional factors such as hermaphrodite receptivity may further contribute to variation in the proportion of males. Surprisingly, males are rare in nature, showing an abundance that is compatible with the X chromosome nondisjunction rate (Barrière and Félix 2005, 2007; Teotónio et al. 2006; Félix and Duveau 2012; Petersen et al. 2014). Nevertheless, matings do occur in the wild, leading to the occasional presence of heterozygote individuals and genetic exchange (Barrière and Félix 2005, 2007; Haber et al. 2005; Sivasundar and Hey 2005; Cutter et al. 2009; Andersen et al. 2012; Félix and Duveau 2012; Petersen et al. 2015b).
Laboratory-based evolution experiments demonstrated that mating and outcrossing can provide a fitness advantage in the presence of coevolving pathogens, such as S. marcescens or B. thuringiensis (Morran et al. 2009, 2011; Schulte et al. 2010; Masri et al. 2013), and other novel environments (Teotónio et al. 2012; Carvalho et al. 2014). Moreover, analysis of small molecular metabolites produced by C. elegans suggests a modular and flexible assembly of molecular signals that not only function as regulators of development, life span, and dauer formation (see above), but also aggregation and male–hermaphrodite interactions (i.e., sex pheromone; Ludewig and Schroeder 2013; Schroeder 2015; Dong et al. 2016). Laboratory evolution experiments additionally showed that male–male competition can lead to strong sexual selection and cause changes in the competitiveness of male sperm (Figure 14) (LaMunyon and Ward 2002; Anderson et al. 2010; Palopoli et al. 2015). Interactions of hermaphrodites with males can lead to reduced life span and offspring production of hermaphrodites (Gems and Riddle 1996; Wegewitz et al. 2008; Shi and Murphy 2014; Palopoli et al. 2015), apparently through mechanical damage of the cuticle (Woodruff et al. 2014) or via secreted harmful compounds (Maures et al. 2014) that can decrease the number of germline progenitor cells, fat storage, or somatic stress resistance (Shi and Murphy 2014; Aprison and Ruvinsky 2016). These costs of mating or contact with males are possibly a consequence of sexual selection, which can favor the production of manipulative and damaging substances by males to increase male reproductive rate (Arnqvist and Rowe 2005). Such sexual selection is likely to act more strongly in gonochoristic Caenorhabditis species, as indicated for example in C. remanei male–female interactions (Garcia et al. 2007; Palopoli et al. 2015) and through comparison of sex-linked gene expression patterns between gonochoristic and androdioecious nematodes (Thomas et al. 2012). Yet, even for C. elegans, it remains an exciting challenge to find out how frequent potentially conflicting male–hermaphrodite interactions really occur in nature and to what extent they have shaped C. elegans life history evolution.
Figure 14.
Male–male sexual competition enhances male competitiveness and male-induced harm. (A) Scanning electron micrograph of male C. elegans (Bar, 50 µm; picture by H.S. and A. Thomas). Sixty generations of experimental evolution under high male–male competition (indicated by High Comp., with three replicate lines called ABC) leads to increased sperm size (B) and thus male sperm competitiveness; to longer spicule insertion time (D) and thus mating duration; and also to increased killing of mated hermaphrodites (C). Graphs from Palopoli et al. (2015).
Natural Genetic Polymorphisms as an Indication for Biotic Interactions
Many biotic interactions impose high selection on the involved organisms. Such selective dynamics are likely to leave traces in the organism’s genomes. In particular, selective sweeps result in reduced variation in the locus/loci under selection. As they are usually caused by locally restricted interactions (e.g., a pathogen highly abundant at a specific site), they should additionally cause geographic variation. Overdominant selection and other forms of balancing selection can produce variation locally and possibly globally. These types of selective dynamics are likely associated with host–parasite coevolutionary interactions (Woolhouse et al. 2002; Schulenburg et al. 2009) and may also result from other types of interaction, especially if antagonistic or unpredictable (Brockhurst et al. 2014). Natural genetic polymorphisms and the functional characterization of the underlying loci may thus point to important biotic interactions.
In C. elegans, such natural polymorphisms have been studied for a variety of traits. As expected, phenotypic polymorphisms were found for the interaction with different microbes, including microsporidian parasites (see Microsporidia; Figure 8C; Balla et al. 2015), pathogenic bacteria such as P. aeruginosa (P. aeruginosa as a likely natural pathogen; Styer et al. 2008; Reddy et al. 2009; Chang et al. 2011; Andersen et al. 2012), S. marcescens (S. marcescens as a likely natural pathogen; Glater et al. 2014), B. thuringiensis (B. thuringiensis as a likely natural pathogen; Volkers et al. 2013; Nakad et al. 2016), and also food bacteria (Bendesky et al. 2011; Volkers et al. 2013). Importantly, the genetic dissection of such polymorphisms can lead to the discovery of novel molecular mechanisms, as demonstrated and explained above for the interaction with the Orsay virus (e.g., discovery of the drh-1 resistance gene; Viruses; Ashe et al. 2013). It may also indicate a yet unrecognized interaction with a pathogen. The study by Ghosh et al. (2012) identified a long-term polymorphism in the chloride channel subunit gene glc-1 that explains resistance to the antihelminthic avermectin. Such antihelminthic compounds are produced by bacteria of the genus Streptomyces (Burg et al. 1979), which may coexist with C. elegans in nature. Thus, the observed balancing selection on this gene may result from repeated exposure of the nematode to avermectin (e.g., applied in agriculture) or from an ongoing interaction between C. elegans and Streptomyces pathogens. Another polymorphism was detected toward a different Streptomyces antihelminthic compound in Andersen et al. (2012).
Other genetic polymorphisms could be linked to C. elegans reproductive behavior and may emphasize the particular selective dynamics associated with intersexual interactions. Intriguingly, a natural polymorphism was found at two interacting genes, peel-1 and zeel-1, which determine mating compatibility of C. elegans in the wild (Seidel et al. 2008, 2011). These two genes define a toxin–antitoxin system at the same genetic locus. The gene peel-1 encodes the toxin, a transmembrane protein expressed in sperm and delivered to the embryo, which it kills at the twofold stage in the absence of an antidote (Figure 15). The gene zeel-1 is transiently expressed in the embryo. Its transmembrane domain can suppress the killing effect of PEEL-1 (Seidel et al. 2011). The polymorphism concerns the absence or presence of these two linked genes. It is yet unclear which selective dynamics determine maintenance of the polymorphism in nature. Another case is the variation in genes involved in copulatory plug formation, such as plg-1 (Palopoli et al. 2008) and plep-1 (Noble et al. 2015), possibly as a consequence of male–male competition in this predominantly selfing species.
Figure 15.
Natural genetic polymorphism for mating incompatibility. (A) Overview of the compatibilities defined by presence of the paternally expressed toxin PEEL-1 in the sperm (indicated by purple color) and rescue through zygotic expression of the antidote ZEEL-1 (indicated in yellow), which may be obtained through the father and/or mother genome. Absence of PEEL-1 (not shown) always results in zygote survival. (B) Worldwide distribution of the two compatibility types defined through activity (blue) or inactivity (orange) of the two genes. In one case (green dot), zeel-1 is active, but there is not paternal killing through peel-1. (A) From Petersen et al. (2015a) with permission from Elsevier and (B) from Seidel et al. (2008), reprinted with permission from AAAS.
Yet other natural polymorphisms were linked to pheromone perception (Greene et al. 2016a,b) and, thus, possibly to competition within the species C. elegans, and also between C. elegans and closely related species (see also above Intraspecific competition and its resolution through C. elegans pheromones). Interestingly, the characterized QTL consisted of polymorphic G protein-coupled chemoreceptors (Greene et al. 2016a,b). These chemoreceptors are likely to perceive and subsequently mediate the response to environmental signals, for example from con-specifics (above) or also pathogens (Zugasti et al. 2014). They are part of the largest gene superfamily in C. elegans, containing >1300 genes (Thomas and Robertson 2008). Several chemoreceptor families are subject to adaptive sequence evolution (Thomas et al. 2005) and substantial copy number variations (Volkers et al. 2013). These polymorphisms may be a consequence of past natural selection on these genes as central mediators of the interaction with the environment.
Perspectives
We are only scratching the surface of the complex biotic interactions between C. elegans and other organisms in nature. Since these interactions have shaped C. elegans along its evolutionary history, they are pivotal for an in-depth characterization of C. elegans biology and for understanding the evolutionary maintenance of its many genes. Thus, we are in dire need of additional studies on the natural ecology of C. elegans. Especially neglected fields are the interactions with enemies, competitors, and also hosts and vectors. Is C. elegans really exclusively free-living or has it evolved a semiparasitic and/or necromenic lifestyle, at least temporarily, e.g., as indicated with slugs (see above)? Do the well-characterized phenotypes and molecular processes in development, neurobiology, or cell biology, all intensively studied under artificial laboratory conditions, behave similarly when examined under more natural conditions? And are such processes and phenotypes then still influenced by mutations in the same set of genes? How do we best analyze such environmental contributions under laboratory conditions? Are the natural conditions best approximated with the help of two-dimensional solid agar media, three-dimensional liquid cultures, or some type of viscous medium?
The microbial environment should be of central importance, because it is highly dynamic in nature and is a required interaction of C. elegans with its environment through feeding. The recent studies provided a glimpse at the possible diversity of microbial interactors, ranging from possible mutualists to commensals and pathogens. Yet, to date, it is not entirely clear to what extent C. elegans is associated with specific taxa over longer time periods, allowing for repeated reciprocal coevolutionary adaptations. Alternatively, the animal may be inhabited by specific microbes only for short time periods, whereby the presence of these short-term visitors may be a consequence of selection by C. elegans, or the colonization ability of the microbes, or a combination thereof. Moreover, it is not clear which of the taxa only act as food or indeed represent commensals or even mutualists. Are some bacteria both food and also mutualists, because C. elegans carries them along to new habitats, where they process nutrients available in rotten fruits and then allow the worms to use them by feeding on them? Are pathogens exclusively detrimental or can they also serve as food or commensals under specific conditions? What is the influence of the large variety of microbes and their combination on nematode biology, particularly regarding dauer entry and exit? What is the diversity of viruses and phages associated with the worm’s microbiome, and what are their effect on C. elegans life history? Beyond the example of the viruses, is there a strong specificity in the biotic interactions of C. elegans compared to those of congeners such as C. briggsae and C. remanei, and among C. elegans genotypes? These are some of the many questions that need to be addressed in future research.
Intraspecific interactions can be highly antagonistic (e.g., sexual conflict or competition for resources) and could have resulted in selection dynamics that have shaped C. elegans’ biology, but how common are such competitive interactions among C. elegans genotypes in nature? Does the cooccurrence of different genotypes result in competitive interactions, for example mediated by the dauer pheromone? To what extent is this influenced by resource availability or the level of relatedness among genotypes, as predicted by theory? How common are sexual interactions and do these associate with antagonistic sexual conflict? Are these intraspecific interactions and conflicts mainly mediated by ascarosides or similar small molecules? What is the role of the many sperm-associated proteins of unknown function? These represent highly promising foci for future research programs on C. elegans ecology.
Acknowledgments
We thank Christian Braendle and the reviewers for helpful suggestions on the manuscript. We also thank Susanne Landis (http://www.scienstration.com/) for creating Figure 1. M.-A.F. acknowledges funding from the Centre National de la Recherche Scientifique, the École Normale Supérieure, and through grants of the Fondation pour la Recherche Médicale “Equipe FRM DEQ20150331704” and the Agence Nationale pour la Recherche “ANR-14-CE10-0003-02”. H.S. is supported by grants from the German Science Foundation within the Collaborative Research Center 1182 on the origin and function of metaorganisms (projects A1.1 and A4.3).
Footnotes
Communicating editor: A. D. Cutter
Literature Cited
- Ahrén D., Tholander M., Fekete C., Rajashekar B., Friman E., et al. , 2005. Comparison of gene expression in trap cells and vegetative hyphae of the nematophagous fungus Monacrosporium haptotylum. Microbiology 151: 789–803. [DOI] [PubMed] [Google Scholar]
- Andersen E. C., Gerke J. P., Shapiro J. A., Crissman J. R., Ghosh R., et al. , 2012. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat. Genet. 44: 285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson G. L., Kenney S. J., Millner P. D., Beuchat L. R., Williams P. L., 2006. Shedding of foodborne pathogens by Caenorhabditis elegans in compost-amended and unamended soil. Food Microbiol. 23: 146–153. [DOI] [PubMed] [Google Scholar]
- Anderson J. L., Morran L. T., Phillips P. C., 2010. Outcrossing and the maintenance of males within C. elegans populations. J. Hered. 101: S62–S74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson D. I., Hughes D., 2009. Gene amplification and adaptive evolution in bacteria. Annu. Rev. Genet. 43: 167–195. [DOI] [PubMed] [Google Scholar]
- Aprison E. Z., Ruvinsky I., 2016. Sexually antagonistic male signals manipulate germline and soma of C. elegans hermaphrodites. Curr. Biol. 26: 2827–2833. [DOI] [PubMed] [Google Scholar]
- Arnold S. J., 1994. Bateman’s principles and the measurement of sexual selection in plants and animals. Am. Nat. 144: S126–S149. [Google Scholar]
- Arnqvist G., Rowe L., 2005. Sexual Conflict. Princeton University Press, NJ. [Google Scholar]
- Ashe A., Bélicard T., Le Pen J., Sarkies P., Frézal L., et al. , 2013. A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. Elife 2: e00994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assogba B. S., Milesi P., Djogbénou L. S., Berthomieu A., Makoundou P., et al. , 2016. The ace-1 locus is amplified in all resistant Anopheles gambiae mosquitoes: fitness consequences of homogeneous and heterogeneous duplications. PLoS Biol. 14: e2000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L., Shtonda B. B., 2003. Food transport in the C. elegans pharynx. J. Exp. Biol. 206: 2441–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L., and You Y.-J., 2012 C. elegans feeding (May 21, 2012), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.150.1, http://www.wormbook.org.
- Baird S. E., 1999. Natural and experimental associations of Caenorhabditis remanei with Trachelipus rathkii and other terrestrial isopods. Nematology 1: 471–475. [Google Scholar]
- Baird S. E., Yen W. C., 2000. Reproductive isolation in Caenorhabditis: terminal phenotypes of hybrid embryos. Evol. Dev. 2: 9–15. [DOI] [PubMed] [Google Scholar]
- Baird S. E., Sutherlin M. E., Emmons S. W., 1992. Reproductive isolation in Rhabditidae (Nematoda: Secernentea): mechanisms that isolate six species of three genera. Evolution 46: 585–594. [DOI] [PubMed] [Google Scholar]
- Bakowski M. A., Luallen R. J., Troemel E. R., 2014a Microsporidia infections in Caenorhabditis elegans and other nematodes, pp. 341–356 in Microsporidia - Pathogens of Opportunity, edited by Weiss L. M., Becnel J. J. John Wiley & Sons, Chichester, UK. [Google Scholar]
- Bakowski M. A., Desjardins C. A., Smelkinson M. G., Dunbar T. A., Lopez-Moyado I. F., et al. , 2014b Ubiquitin-mediated response to microsporidia and virus infection in C. elegans. PLoS Pathog. 10: e1004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandar R., Lu N. C., 2005. Nutritional requirements for pantothenate, pantethine or coenzyme A in the free-living nematode Caenorhabditis elegans. Nematology 7: 761–766. [Google Scholar]
- Balla K. M., Troemel E. R., 2013. Caenorhabditis elegans as a model for intracellular pathogen infection. Cell. Microbiol. 15: 1313–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla K. M., Andersen E. C., Kruglyak L., Troemel E. R., 2015. A wild C. elegans strain has enhanced epithelial immunity to a natural microsporidian parasite. PLoS Pathog. 11: e1004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla K. M., Luallen R. J., Bakowski M. A., Troemel E. R., 2016. Cell-to-cell spread of microsporidia causes Caenorhabditis elegans organs to form syncytia. Nat. Microbiol. 1: 16144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann C. I., Horvitz H. R., 1991. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7: 729–742. [DOI] [PubMed] [Google Scholar]
- Barrière A., Félix M.-A., 2005. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr. Biol. 15: 1176–1184. [DOI] [PubMed] [Google Scholar]
- Barrière A., Félix M.-A., 2007. Temporal dynamics and linkage disequilibrium in natural Caenorhabditis elegans populations. Genetics 176: 999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrows B. D., Haslam S. M., Bischof L. J., Morris H. R., Dell A., et al. , 2007. Resistance to Bacillus thuringiensis toxin in Caenorhabditis elegans from loss of fucose. J. Biol. Chem. 282: 3302–3311. [DOI] [PubMed] [Google Scholar]
- Beakes G. W., Glockling S. L., Sekimoto S., 2012. The evolutionary phylogeny of the oomycete “fungi.” Protoplasma 249: 3–19. [DOI] [PubMed] [Google Scholar]
- Beale E., Li G., Tan M.-W., Rumbaugh K. P., 2006. Caenorhabditis elegans senses bacterial autoinducers. Appl. Environ. Microbiol. 72: 5135–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellier A., Chen C.-S., Kao C.-Y., Cinar H. N., Aroian R. V., 2009. Hypoxia and the hypoxic response pathway protect against pore-forming toxins in C. elegans. PLoS Pathog. 5: e1000689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendesky A., Tsunozaki M., Rockman M. V., Kruglyak L., Bargmann C. I., 2011. Catecholamine receptor polymorphisms affect decision-making in C. elegans. Nature 472: 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg M., Stenuit B., Ho J., Wang A., Parke C., et al. , 2016a Assembly of the Caenorhabditis elegans gut microbiota from diverse soil microbial environments. ISME J. 10: 1998–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg M., Zhou X. Y., Shapira M., 2016b Host-specific functional significance of Caenorhabditis gut commensals. Front. Microbiol. 7: 1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehnisch C., Wong D., Habig M., Isermann K., Michiels N. K., et al. , 2011. Protist-type lysozymes of the nematode Caenorhabditis elegans contribute to resistance against pathogenic Bacillus thuringiensis. PLoS One 6: e24619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein S. R., Theis K. R., 2015. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 13: e1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose N., Meyer J. M., Yim J. J., Mayer M. G., Markov G. V., et al. , 2014. Natural variation in dauer pheromone production and sensing supports intraspecific competition in nematodes. Curr. Biol. 24: 1536–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourke A. F., 2014. Hamilton’s rule and the causes of social evolution. Philos. Trans. R. Soc. B Biol. Sci. 369: 20130362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt J. P., Ringstad N., 2015. Toll-like receptor signaling promotes development and function of sensory neurons required for a C. elegans pathogen-avoidance behavior. Curr. Biol. 25: 2228–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst M. A., Chapman T., King K. C., Mank J. E., Paterson S., et al. , 2014. Running with the Red Queen: the role of biotic conflicts in evolution. Proc. Biol. Sci. 281: 20141382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune A., 2014. Symbiotic digestion of lignocellulose in termite guts. Nat. Rev. Microbiol. 12: 168–180. [DOI] [PubMed] [Google Scholar]
- Buecher E. J., Hansen E. L., Yarwood E. A., 1970. Growth of nematodes in defined medium containing hemin and supplemented with commercially available proteins. Nematologica 16: 403–409. [Google Scholar]
- Burg R. W., Miller B. M., Baker E. E., Birnbaum J., Currie S. A., et al. , 1979. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob. Agents Chemother. 15: 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho S., Chelo I. M., Goy C., Teotónio H., 2014. The role of hermaphrodites in the experimental evolution of increased outcrossing rates in Caenorhabditis elegans. BMC Evol. Biol. 14: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell-Chen E. P., Chen J., Lewis E. E., Douhan G. W., Nadler S. A., et al. , 2005. Revising the standard wisdom of C. elegans natural history: ecology of longevity. Sci. Aging Knowledge Environ. 2005: pe30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cezairliyan B., Vinayavekhin N., Grenfell-Lee D., Yuen G. J., Saghatelian A., et al. , 2013. Identification of Pseudomonas aeruginosa phenazines that kill Caenorhabditis elegans. PLoS Pathog. 9: e1003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M., Sulston J., 1981. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev. Biol. 82: 358–370. [DOI] [PubMed] [Google Scholar]
- Chandler J. A., Lang J. M., Bhatnagar S., Eisen J. A., Kopp A., 2011. Bacterial communities of diverse Drosophila species: ecological context of a host–microbe model system. PLoS Genet. 7: e1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. C., Paek J., Kim D. H., 2011. Natural polymorphisms in C. elegans HECW-1 E3 ligase affect pathogen avoidance behaviour. Nature 480: 525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Caswell-Chen E. P., 2004. Facultative vivipary is a life-history trait in Caenorhabditis elegans. J. Nematol. 36: 107. [PMC free article] [PubMed] [Google Scholar]
- Cheng C., Kirkpatrick M., 2016. Sex-specific selection and sex-biased gene expression in humans and flies. PLoS Genet. 12: e1006170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciche T. A., Kim K. S., Kaufmann-Daszczuk B., Nguyen K. C., Hall D. H., 2008. Cell invasion and matricide during Photorhabdus luminescens transmission by Heterorhabditis bacteriophora nematodes. Appl. Environ. Microbiol. 74: 2275–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark L. C., Hodgkin J., 2014. Commensals, probiotics and pathogens in the Caenorhabditis elegans model. Cell. Microbiol. 16: 27–38. [DOI] [PubMed] [Google Scholar]
- Clark L. C., Hodgkin J., 2015. Leucobacter musarum subsp. musarum sp. nov., subsp. nov., Leucobacter musarum subsp. japonicus subsp. nov., and Leucobacter celer subsp. astrifaciens subsp. nov., three nematopathogenic bacteria isolated from Caenorhabditis, with an emended description of Leucobacter celer. Int. J. Syst. Evol. Microbiol. 65: 3977–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D. E., Zdraljevic S., Tanny R. E., Seo B., Riccardi D. D., et al. , 2016. The genetic basis of natural variation in Caenorhabditis elegans telomere length. Genetics 204: 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolon J. D., Jones K. L., Todd T. C., Carr B. C., Herman M. A., 2009. Caenorhabditis elegans genomic response to soil bacteria predicts environment-specific genetic effects on life history traits. PLoS Genet. 5: e1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corby-Harris V., Pontaroli A. C., Shimkets L. J., Bennetzen J. L., Habel K. E., et al. , 2007. Geographical distribution and diversity of bacteria associated with natural populations of Drosophila melanogaster. Appl. Environ. Microbiol. 73: 3470–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez M. H., Weitz J. S., 2014. Coevolution can reverse predator-prey cycles. Proc. Natl. Acad. Sci. USA 111: 7486–7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll N. A., 1975. Components and patterns in the behavior of the nematode Caenorhabditis elegans. J. Zool. 176: 159–176. [Google Scholar]
- Cutter A. D., 2006. Nucleotide polymorphism and linkage disequilibrium in wild populations of the partial selfer Caenorhabditis elegans. Genetics 172: 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter A. D., 2015. Caenorhabditis evolution in the wild. Bioessays 37: 983–995. [DOI] [PubMed] [Google Scholar]
- Cutter A. D., Dey A., Murray R. L., 2009. Evolution of the Caenorhabditis elegans genome. Mol. Biol. Evol. 26: 1199–1234. [DOI] [PubMed] [Google Scholar]
- Darby B. J., Herman M. A., 2014. Effect of prey richness on a consumer’s intrinsic growth rate. Oecologia 175: 243–250. [DOI] [PubMed] [Google Scholar]
- Darby C., 2005. Interactions with microbial pathogens (September 6, 2005), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.21.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Darby C., Cosma C. L., Thomas J. H., Manoil C., 1999. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96: 15202–15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day S. R., Sifri C. D., 2012. Intoxication vs. infection: a decade of studying Burkholderia pseudomallei virulence in a simple infection model. Virulence 3: 471–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaestecker E., Gaba S., Raeymaekers J. A. M., Stoks R., Van Kerckhoven L., et al. , 2007. Host–parasite “Red Queen” dynamics archived in pond sediment. Nature 450: 870–873. [DOI] [PubMed] [Google Scholar]
- Dennehy J. J., Friedenberg N. A., Yang Y. W., Turner P. E., 2006. Bacteriophage migration via nematode vectors: host-parasite-consumer interactions in laboratory microcosms. Appl. Environ. Microbiol. 72: 1974–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis S., Sheth U., Feldman J. L., English K. A., Priess J. R., 2012. C. elegans germ cells show temperature and age-dependent expression of Cer1, a Gypsy/Ty3-related retrotransposon. PLoS Pathog. 8: e1002591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz S. A., Restif O., 2014. Spread and transmission of bacterial pathogens in experimental populations of the nematode Caenorhabditis elegans. Appl. Environ. Microbiol. 80: 5411–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz S. A., Viney M., 2015. The evolution of plasticity of dauer larva developmental arrest in the nematode Caenorhabditis elegans. Ecol. Evol. 5: 1343–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz S. A., Brunet V., Lloyd-Jones G. C., Spinner W., Wharam B., et al. , 2014. Diverse and potentially manipulative signalling with ascarosides in the model nematode C. elegans. BMC Evol. Biol. 14: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen P., Marsh S. A., Braker I., Heitland N., Wagner S., et al. , 2016. The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol. 14: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Dolke F., von Reuss S. H., 2016. Selective MS screening reveals a sex pheromone in Caenorhabditis briggsae and species-specificity in indole ascaroside signalling. Org. Biomol. Chem. 14: 7217–7225. [DOI] [PubMed] [Google Scholar]
- Douglas A. E., 2010. The Symbiotic Habit. Princeton University Press, New Jersey. [Google Scholar]
- Douglas A. E., Werren J. H., 2016. Holes in the hologenome: why host-microbe symbioses are not holobionts. MBio 7: e02099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler C., 1941. Predaceous fungi. Biol. Rev. Camb. Philos. Soc. 16: 265–290. [Google Scholar]
- Ellegren H., Parsch J., 2007. The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 8: 689–698. [DOI] [PubMed] [Google Scholar]
- Engelmann I., Griffon A., Tichit L., Montañana-Sanchis F., Wang G., et al. , 2011. A comprehensive analysis of gene expression changes provoked by bacterial and fungal infection in C. elegans. PLoS One 6: e19055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkut C., Penkov S., Khesbak H., Vorkel D., Verbavatz J. M., et al. , 2011. Trehalose renders the dauer larva of Caenorhabditis elegans resistant to extreme desiccation. Curr. Biol. 21: 1331–1336. [DOI] [PubMed] [Google Scholar]
- Esser R. P., El-Gholl N. E., 1992. Harposporium, a fungus that parasitizes and kills nematodes utilizing conidia swallowed by or sticking to its prey. Nematol. Circ. 200: 6. [Google Scholar]
- Evans E. A., Kawli T., Tan M. W., 2008a Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signaling pathway in Caenorhabditis elegans. PLoS Pathog. 4: e1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E. A., Chen W. C., Tan M.-W., 2008b The DAF-2 insulin-like signaling pathway independently regulates aging and immunity in C. elegans. Aging Cell 7: 879–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farslow J. C., Lipinski K. J., Packard L. B., Edgley M. L., Taylor J., et al. , 2015. Rapid increase in frequency of gene copy-number variants during experimental evolution in Caenorhabditis elegans. BMC Genomics 16: 1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinbaum R. L., Urbach J. M., Liberati N. T., Djonovic S., Adonizio A., et al. , 2012. Genome-wide identification of Pseudomonas aeruginosa virulence-related genes using a Caenorhabditis elegans infection model. PLoS Pathog. 8: e1002813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M.-A., Braendle C., 2010. The natural history of Caenorhabditis elegans. Curr. Biol. 20: R965–R969. [DOI] [PubMed] [Google Scholar]
- Félix M.-A., Duveau F., 2012. Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol. 10: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M.-A., Ashe A., Piffaretti J., Wu G., Nuez I., et al. , 2011. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 9: e1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M.-A., Jovelin R., Ferrari C., Han S., Cho Y. R., et al. , 2013. Species richness, distribution and genetic diversity of Caenorhabditis nematodes in a remote tropical rainforest. BMC Evol. Biol. 13: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N., Antebi A., 2008. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 22: 2149–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford S. A., Kao D., Williams D., King K. C., 2016. Microbe-mediated host defence drives the evolution of reduced pathogen virulence. Nat. Commun. 7: 13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C. J., Zhao G., Félix M.-A., Wang D., 2012. Complete genome sequence of Le Blanc virus, a third Caenorhabditis nematode-infecting virus. J. Virol. 86: 11940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C. J., Renshaw H., Frezal L., Jiang Y., Félix M.-A., et al. , 2014. Orsay, Santeuil and Le Blanc viruses primarily infect intestinal cells in Caenorhabditis nematodes. Virology 448: 255–264. [DOI] [PubMed] [Google Scholar]
- Frézal L., Félix M.-A., 2015. C. elegans outside the Petri dish. Elife 4: e05849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M., Sironi M., 2014. Human genome variability, natural selection and infectious diseases. Curr. Opin. Immunol. 30: 9–16. [DOI] [PubMed] [Google Scholar]
- Garcia L. R., LeBoeuf B., Koo P., 2007. Diversity in mating behavior of hermaphroditic and male-female Caenorhabditis nematodes. Genetics 175: 1761–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D., Riddle D. L., 1996. Longevity in Caenorhabditis elegans reduced by mating but not gamete production. Nature 379: 723–725. [DOI] [PubMed] [Google Scholar]
- Ghosh R., Andersen E. C., Shapiro J. A., Gerke J. P., Kruglyak L., 2012. Natural variation in a chloride channel subunit confers avermectin resistance in C. elegans. Science 335: 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glater E. E., Rockman M. V., Bargmann C. I., 2014. Multigenic natural variation underlies Caenorhabditis elegans olfactory preference for the bacterial pathogen Serratia marcescens. G3 (Bethesda) 4: 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M. B., 2006 Mechanosensation (January 6, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.62.1, http://www.wormbook.org.
- Goranson N. C., Ebersole J. P., Brault S., 2005. Resolving an adaptive conundrum: reproduction in Caenorhabditis elegans is not sperm-limited when food is scarce. Evol. Ecol. Res. 7: 325–333. [Google Scholar]
- Gracida X., Eckmann C. R., 2013. Fertility and germline stem cell maintenance under different diets requires nhr-114/HNF4 in C. elegans. Curr. Biol. 23: 607–613. [DOI] [PubMed] [Google Scholar]
- Gravato-Nobre M. J., 2005. Multiple genes affect sensitivity of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics 171: 1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravato-Nobre M. J., Hodgkin J., 2008. The acyltransferase gene bus-1 exhibits conserved and specific expression in nematode rectal cells and reveals pathogen-induced cell swelling. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 237: 3762–3776. [DOI] [PubMed] [Google Scholar]
- Gravato-Nobre M. J., Stroud D., O’Rourke D., Darby C., Hodgkin J., 2011. Glycosylation genes expressed in seam cells determine complex surface properties and bacterial adhesion to the cuticle of Caenorhabditis elegans. Genetics 187: 141–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N. F., 1987. Nematophagous fungi with particular reference to their ecology. Biol. Rev. Camb. Philos. Soc. 62: 245–304. [Google Scholar]
- Green J. W. M., Snoek L. B., Kammenga J. E., Harvey S. C., 2013. Genetic mapping of variation in dauer larvae development in growing populations of Caenorhabditis elegans. Heredity 111: 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene J. S., Dobosiewicz M., Butcher R. A., McGrath P. T., Bargmann C. I., 2016a Regulatory changes in two chemoreceptor genes contribute to a Caenorhabditis elegans QTL for foraging behavior. Elife 5: e21454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene J. S., Brown M., Dobosiewicz M., Ishida I. G., Macosko E. Z., et al. , 2016b Balancing selection shapes density-dependent foraging behaviour. Nature 539: 254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal P. S., 1991a Influence of bacteria and temperature on the reproduction of Caenorhabditis elegans (Nematoda: Rhabditidae) infesting mushrooms (Agaricus bisporus). Nematologica 37: 72–82. [Google Scholar]
- Grewal P. S., 1991b Relative contribution of nematodes (Caenorhabditis elegans) and bacteria towards the disruption of flushing patterns and losses in yield and quality of mushrooms (Agaricus bisporus). Ann. Appl. Biol. 119: 483–499. [Google Scholar]
- Grewal P. S., 1991c Effects of Caenorhabditis elegans (Nematoda: Rhabditidae) on the spread of the bacterium Pseudomonas tolaasii in mushrooms (Agaricus bisporus). Ann. Appl. Biol. 118: 47–55. [Google Scholar]
- Grewal P. S., Wright D. J., 1992. Migration of Caenorhabditis elegans (Nematoda: Rhabditidae) larvae towards bacteria and the nature of the bacterial stimulus. Fundam. Appl. Nematol. 15: 159–166. [Google Scholar]
- Griffin C. T., 2012. Perspectives on the behavior of entomopathogenic nematodes from dispersal to reproduction: traits contributing to nematode fitness and biocontrol efficacy. J. Nematol. 44: 177–184. [PMC free article] [PubMed] [Google Scholar]
- Griffitts J. S., Aroian R. V., 2005. Many roads to resistance: how invertebrates adapt to Bt toxins. Bioessays 27: 614–624. [DOI] [PubMed] [Google Scholar]
- Griffitts J. S., Haslam S. M., Yang T., Garczynski S. F., Mulloy B., et al. , 2005. Glycolipids as receptors for Bacillus thuringiensis crystal toxin. Science 307: 922–925. [DOI] [PubMed] [Google Scholar]
- Guo Y. R., Hryc C. F., Jakana J., Jiang H., Wang D., et al. , 2014. Crystal structure of a nematode-infecting virus. Proc. Natl. Acad. Sci. USA 111: 12781–12786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov I., Gautier L., Smolentseva O., Shamovsky I., Eremina S., et al. , 2013. Bacterial nitric oxide extends the lifespan of C. elegans. Cell 152: 818–830. [DOI] [PubMed] [Google Scholar]
- Haber M., Schüngel M., Putz A., Müller S., Hasert B., et al. , 2005. Evolutionary history of Caenorhabditis elegans inferred from microsatellites: evidence for spatial and temporal genetic differentiation and the occurrence of outbreeding. Mol. Biol. Evol. 22: 160–173. [DOI] [PubMed] [Google Scholar]
- Hamilton W. D., 1963. The evolution of altruistic behavior. Am. Nat. 97: 354–356. [Google Scholar]
- Harvey S. C., Shorto A., Viney M. E., 2008. Quantitative genetic analysis of life-history traits of Caenorhabditis elegans in stressful environments. BMC Evol. Biol. 8: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasshoff M., Bohnisch C., Tonn D., Hasert B., Schulenburg H., 2007. The role of Caenorhabditis elegans insulin-like signaling in the behavioral avoidance of pathogenic Bacillus thuringiensis. FASEB J. 21: 1801–1812. [DOI] [PubMed] [Google Scholar]
- Hieb W. F., Rothstein M., 1968. Sterol requirement for reproduction of a free-living nematode. Science 160: 778–780. [DOI] [PubMed] [Google Scholar]
- Hieb W. F., Stokstad E. L., Rothstein M., 1970. Heme requirement for reproduction of a free-living nematode. Science 168: 143–144. [DOI] [PubMed] [Google Scholar]
- Hill K. L., L’Hernault S. W., 2001. Analyses of reproductive interactions that occur after heterospecific matings within the genus Caenorhabditis. Dev. Biol. 232: 105–114. [DOI] [PubMed] [Google Scholar]
- Hiltunen T., Becks L., 2014. Consumer co-evolution as an important component of the eco-evolutionary feedback. Nat. Commun. 5: 5226. [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Barnes T. M., 1991. More is not better: brood size and population growth in a self-fertilizing nematode. Proc. Biol. Sci. 246: 19–24. [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Doniach T., 1997. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics 146: 149–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., Kuwabara P. E., Corneliussen B., 2000. A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr. Biol. CB 10: 1615–1618. [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Félix M.-A., Clark L. C., Stroud D., Gravato-Nobre M. J., 2013. Two Leucobacter strains exert complementary virulence on Caenorhabditis including death by worm-star formation. Curr. Biol. 23: 2157–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeckendorf A., Leippe M., 2012. SPP-3, a saposin-like protein of Caenorhabditis elegans, displays antimicrobial and pore-forming activity and is located in the intestine and in one head neuron. Dev. Comp. Immunol. 38: 181–186. [DOI] [PubMed] [Google Scholar]
- Hoeckendorf A., Stanisak M., Leippe M., 2012. The saposin-like protein SPP-12 is an antimicrobial polypeptide in the pharyngeal neurons of Caenorhabditis elegans and participates in defence against a natural bacterial pathogen. Biochem. J. 445: 205–212. [DOI] [PubMed] [Google Scholar]
- Hsueh Y.-P., Mahanti P., Schroeder F. C., Sternberg P. W., 2013. Nematode-trapping fungi eavesdrop on nematode pheromones. Curr. Biol. 23: 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P. J., 2007 Dauer (August 8, 2007), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.144.1, http://www.wormbook.org.
- Huffman D. L., Abrami L., Sasik R., Corbeil J., van der Goot F. G., et al. , 2004. Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proc. Natl. Acad. Sci. USA 101: 10995–11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iatsenko I., Sinha A., Rodelsperger C., Sommer R. J., 2013. New role for DCR-1/Dicer in Caenorhabditis elegans innate immunity against the highly virulent bacterium Bacillus thuringiensis DB27. Infect. Immun. 81: 3942–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui J. E., Urbach J. M., Ausubel F. M., 2010. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat. Rev. Immunol. 10: 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G., Crummenerl L. L., Gilbert F., Mohr T., Pfefferkorn R., et al. , 2015. Evolutionary transition from pathogenicity to commensalism: global regulator mutations mediate fitness gains through virulence attenuation. Mol. Biol. Evol. 32: 2883–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson H.-B., 1994. Adhesion of conidia of Drechmeria coniospora to Caenorhabditis elegans wild type and mutants. J. Nematol. 26: 430. [PMC free article] [PubMed] [Google Scholar]
- Jensen V. L., Simonsen K. T., Lee Y.-H., Park D., Riddle D. L., 2010. RNAi screen of DAF-16/FOXO target genes in C. elegans links pathogenesis and dauer formation. PLoS One 5: e15902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Franz C. J., Wu G., Renshaw H., Zhao G., et al. , 2014. Orsay virus utilizes ribosomal frameshifting to express a novel protein that is incorporated into virions. Virology 450–451: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C.-Y., Los F. C. O., Huffman D. L., Wachi S., Kloft N., et al. , 2011. Global functional analyses of cellular responses to pore-forming toxins. PLoS Pathog. 7: e1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katju V., Bergthorsson U., 2013. Copy-number changes in evolution: rates, fitness effects and adaptive significance. Front. Genet. 4: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawli T., Tan M.-W., 2008. Neuroendocrine signals modulate the innate immunity of Caenorhabditis elegans through insulin signaling. Nat. Immunol. 9: 1415–1424. [DOI] [PubMed] [Google Scholar]
- Kenney S. J., Anderson G. L., Williams P. L., Millner P. D., Beuchat L. R., 2005. Persistence of Escherichia coli O157:H7, Salmonella newport, and Salmonella Poona in the gut of a free-living nematode, Caenorhabditis elegans, and transmission to progeny and uninfected nematodes. Int. J. Food Microbiol. 101: 227–236. [DOI] [PubMed] [Google Scholar]
- Kenney S. J., Anderson G. L., Williams P. L., Millner P. D., Beuchat L. R., 2006. Migration of Caenorhabditis elegans to manure and manure compost and potential for transport of Salmonella newport to fruits and vegetables. Int. J. Food Microbiol. 106: 61–68. [DOI] [PubMed] [Google Scholar]
- Kessin R. H., Gundersen G. G., Zaydfudim V., Grimson M., 1996. How cellular slime molds evade nematodes. Proc. Natl. Acad. Sci. USA 93: 4857–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H., Ewbank J. J., 2015. Signaling in the innate immune response (September 16, 2015), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.83.2, http://www.wormbook.org.
- Kim D. H., Feinbaum R., Alloing G., Emerson F. E., Garsin D. A., et al. , 2002. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297: 623–626. [DOI] [PubMed] [Google Scholar]
- King K. C., Brockhurst M. A., Vasieva O., Paterson S., Betts A., et al. , 2016. Rapid evolution of microbe-mediated protection against pathogens in a worm host. ISME J. 10: 1915–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K., 1997. Description of Rhabditis (Caenorhabditis) drosophilae n. sp. and R.(C.) sonorae n. sp. (Nematoda: Rhabditida) from saguaro cactus rot in Arizona. Fundam. Appl. Nematol. 20: 305–315. [Google Scholar]
- Kiontke K., Sudhaus W., 2006. Ecology of Caenorhabditis species (January 9, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.37.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Kiontke K. C., Félix M.-A., Ailion M., Rockman M. V., Braendle C., et al. , 2011. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol. Biol. 11: 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirienko N. V., Kirienko D. R., Larkins-Ford J., Wählby C., Ruvkun G., et al. , 2013. Pseudomonas aeruginosa disrupts Caenorhabditis elegans iron homeostasis, causing a hypoxic response and death. Cell Host Microbe 13: 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirienko N. V., Ausubel F. M., Ruvkun G., 2015. Mitophagy confers resistance to siderophore-mediated killing by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 112: 1821–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov F. A., 2012. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. Biol. Sci. 279: 5048–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella B., Brockhurst M. A., 2014. Bacteria–phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 38: 916–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause D. O., Nagaraja T. G., Wright A. D. G., Callaway T. R., 2013. Rumen microbiology: leading the way in microbial ecology. J. Anim. Sci. 91: 331–341. [DOI] [PubMed] [Google Scholar]
- LaMunyon C. W., Ward S., 2002. Evolution of larger sperm in response to experimentally increased sperm competition in Caenorhabditis elegans. Proc. R. Soc. Lond. B Biol. Sci. 269: 1125–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrigand K., He L. D., Thakur N., Arguel M.-J., Polanowska J., et al. , 2016. Comparative genomic analysis of Drechmeria coniospora reveals core and specific genetic requirements for fungal endoparasitism of nematodes. PLoS Genet. 12: e1006017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Choi M., Lee D., Kim H., Hwang H., et al. , 2011. Nictation, a dispersal behavior of the nematode Caenorhabditis elegans, is regulated by IL2 neurons. Nat. Neurosci. 15: 107–112. [DOI] [PubMed] [Google Scholar]
- Lee Q., Widden P., 1996. Folsomia candida, a “fungivorous” collembolan, feeds preferentially on nematodes rather than soil fungi. Soil Biol. Biochem. 28: 689–690. [Google Scholar]
- Lee S.-H., Wong R.-R., Chin C.-Y., Lim T.-Y., Eng S.-A., et al. , 2013. Burkholderia pseudomallei suppresses Caenorhabditis elegans immunity by specific degradation of a GATA transcription factor. Proc. Natl. Acad. Sci. USA 110: 15067–15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaerts I., Walker G. A., Van Hoorebeke L., Gems D., Vanfleteren J. R., 2008. Dietary restriction of Caenorhabditis elegans by axenic culture reflects nutritional requirement for constituents provided by metabolically active microbes. J. Gerontol. A Biol. Sci. Med. Sci. 63: 242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.-H., Zhang K.-Q., 2016. Nematode-toxic fungi and their nematicidal metabolites, pp. 313–375 in Nematode-Trapping Fungi, Fungal Diversity Research, edited by Zhang K.-Q., Hyde K. D. Springer, New York. [Google Scholar]
- Li Y., Hyde K. D., Jeewon R., Cai L., Vijaykrishna D., et al. , 2005. Phylogenetics and evolution of nematode-trapping fungi (Orbiliales) estimated from nuclear and protein coding genes. Mycologia 97: 1034–1046. [DOI] [PubMed] [Google Scholar]
- Lim M.-P., Firdaus-Raih M., Nathan S., 2016. Nematode peptides with host-directed anti-inflammatory activity rescue Caenorhabditis elegans from a Burkholderia pseudomallei infection. Front. Microbiol. 7: 1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Samuel B. S., Breen P. C., Ruvkun G., 2014. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature 508: 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lively C. M., Morran L. T., 2014. The ecology of sexual reproduction. J. Evol. Biol. 27: 1292–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N. C., Goetsch K. M., 1993. Carbohydrate requirement of Caenorhabditis elegans and the final development of a chemically defined medium. Nematologica 39: 303–311. [Google Scholar]
- Lu N. C., Newton C., Stokstad E. L. R., 1977. Requirement of sterol and various sterol precursors in free-living nematodes. Nematologica 23: 57–61. [Google Scholar]
- Luallen R. J., Reinke A. W., Tong L., Botts M. R., Félix M.-A., et al. , 2016. Discovery of a natural microsporidian pathogen with a broad tissue tropism in Caenorhabditis elegans. PLoS Pathog. 12: e1005724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig A. H, Schroeder F. C. 2013. Ascaroside signaling in C. elegans (January 8, 2013), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.155.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- MacNeil L. T., Watson E., Arda H. E., Zhu L. J., Walhout A. J. M., 2013. Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell 153: 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire S. M., Clark C. M., Nunnari J., Pirri J. K., Alkema M. J., 2011. The C. elegans touch response facilitates escape from predacious fungi. Curr. Biol. 21: 1326–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S., Tan M.-W., Rahme L. G., Ausubel F. M., 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa–Caenorhabditis elegans pathogenesis model. Cell 96: 47–56. [DOI] [PubMed] [Google Scholar]
- Mallo G. V., Kurz C. L., Couillault C., Pujol N., Granjeaud S., et al. , 2002. Inducible antibacterial defense system in C. elegans. Curr. Biol. 12: 1209–1214. [DOI] [PubMed] [Google Scholar]
- Marzluff J. M., Shulenberger E., Endlicher W., Alberti M., Bradley G. et al. (Editors), 2008. Urban Ecology. Springer, New York. [Google Scholar]
- Masri L., Schulte R. D., Timmermeyer N., Thanisch S., Crummenerl L. L., et al. , 2013. Sex differences in host defence interfere with parasite-mediated selection for outcrossing during host-parasite coevolution. Ecol. Lett. 16: 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri L., Branca A., Sheppard A. E., Papkou A., Laehnemann D., et al. , 2015. Host-pathogen coevolution: the selective advantage of Bacillus thuringiensis virulence and its cry toxin genes. PLoS Biol. 13: e1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupas E., 1915. Sur un champignon parasite des Rhabditis. Bull Soc Hist Nat Afr Nord 7: 34–49. [Google Scholar]
- Maures T. J., Booth L. N., Benayoun B. A., Izrayelit Y., Schroeder F. C., et al. , 2014. Males shorten the life span of C. elegans hermaphrodites via secreted compounds. Science 343: 541–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M. G., Rödelsperger C., Witte H., Riebesell M., Sommer R. J., 2015. The orphan gene dauerless regulates dauer development and intraspecific competition in nematodes by copy number variation . PLoS Genet. 11: e1005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCafferty J., Mühlbauer M., Gharaibeh R. Z., Arthur J. C., Perez-Chanona E., et al. , 2013. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J. 7: 2116–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel J. D., Kim D. H., 2014. Behavioral avoidance of pathogenic bacteria by Caenorhabditis elegans. Trends Immunol. 35: 465–470. [DOI] [PubMed] [Google Scholar]
- Meisel J. D., Panda O., Mahanti P., Schroeder F. C., Kim D. H., 2014. Chemosensation of bacterial secondary metabolites modulates neuroendocrine signaling and behavior of C. elegans. Cell 159: 267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengert H., 1953. Nematoden und schnecken. Zoomorphology 41: 311–349. [Google Scholar]
- Mikola J., Sulkova P., 2001. Responses of microbial-feeding nematodes to organic matter distribution and predation in experimental soil habitat. Soil Biol. Biochem. 33: 811–817. [Google Scholar]
- Miltsch S. M., Seeberger P. H., Lepenies B., 2014. The C-type lectin-like domain containing proteins Clec-39 and Clec-49 are crucial for Caenorhabditis elegans immunity against Serratia marcescens infection. Dev. Comp. Immunol. 45: 67–73. [DOI] [PubMed] [Google Scholar]
- Montalvo-Katz S., Huang H., Appel M. D., Berg M., Shapira M., 2013. Association with soil bacteria enhances p38-dependent infection resistance in Caenorhabditis elegans. Infect. Immun. 81: 514–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran L. T., Parmenter M. D., Phillips P. C., 2009. Mutation load and rapid adaptation favour outcrossing over self-fertilization. Nature 462: 350–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran L. T., Schmidt O. G., Gelarden I. A., Parrish R. C., Lively C. M., 2011. Running with the Red Queen: host-parasite coevolution selects for biparental sex. Science 333: 216–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran L. T., Parrish R. C., Gelarden I. A., Lively C. M., 2013. Temporal dynamics of outcrossing and host mortality rates in host-pathogen experimental coevolution. Evolution 67: 1860–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran L. T., Parrish R. C., Gelarden I. A., Allen M. B., Lively C. M., 2014. Experimental coevolution: rapid local adaptation by parasites depends on host mating system. Am. Nat. 184: S91–S100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylenko M., Boland S., Penkov S., Sampaio J. L., Lombardot B., et al. , 2016. NAD+ is a food component that promotes exit from dauer diapause in Caenorhabditis elegans. PLoS One 11: e0167208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakad R., Snoek L. B., Yang W., Ellendt S., Schneider F., et al. , 2016. Contrasting invertebrate immune defense behaviors caused by a single gene, the Caenorhabditis elegans neuropeptide receptor gene npr-1. BMC Genomics 17: 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal S. J., Takeishi A., O’Donnell M. P., Park J., Hong M., et al. , 2015. Feeding state-dependent regulation of developmental plasticity via CaMKI and neuroendocrine signaling. Elife 4: e10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen-LeRoux C., Gaudriault S., Ramarao N., Lereclus D., Givaudan A., 2012. How the insect pathogen bacteria Bacillus thuringiensis and Xenorhabdus/Photorhabdus occupy their hosts. Curr. Opin. Microbiol. 15: 220–231. [DOI] [PubMed] [Google Scholar]
- Niu Q., Huang X., Zhang L., Xu J., Yang D., et al. , 2010. A Trojan horse mechanism of bacterial pathogenesis against nematodes. Proc. Natl. Acad. Sci. USA 107: 16631–16636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble L. M., Chang A. S., McNelis D., Kramer M., Yen M., et al. , 2015. Natural variation in plep-1 causes male-male copulatory behavior in C. elegans. Curr. Biol. 25: 2730–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura E., Yoshiga T., 2014. Host orientation using volatiles in the phoretic nematode Caenorhabditis japonica. J. Exp. Biol. 217: 3197–3199. [DOI] [PubMed] [Google Scholar]
- Palopoli M. F., Rockman M. V., TinMaung A., Ramsay C., Curwen S., et al. , 2008. Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature 454: 1019–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palopoli M. F., Peden C., Woo C., Akiha K., Ary M., et al. , 2015. Natural and experimental evolution of sexual conflict within Caenorhabditis nematodes. BMC Evol. Biol. 15: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Miller R., Laehnemann D., Jansen G., Fuentes-Hernandez A., Rosenstiel P., et al. , 2013. When the most potent combination of antibiotics selects for the greatest bacterial load: the smile-frown transition. PLoS Biol. 11: e1001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman D., Lu N. C., 2000. Requirements for branched chain amino acids and their interactions in the nematode Caenorhabditis elegans. Nematology 2: 501–506. [Google Scholar]
- Petersen C., Dirksen P., Prahl S., Strathmann E. A., Schulenburg H., 2014. The prevalence of Caenorhabditis elegans across 1.5 years in selected North German locations: the importance of substrate type, abiotic parameters, and Caenorhabditis competitors. BMC Ecol. 14: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C., Dirksen P., Schulenburg H., 2015a Why we need more ecology for genetic models such as C. elegans. Trends Genet. 31: 120–127. [DOI] [PubMed] [Google Scholar]
- Petersen C., Saebelfeld M., Barbosa C., Pees B., Hermann R. J., et al. , 2015b Ten years of life in compost: temporal and spatial variation of North German Caenorhabditis elegans populations. Ecol. Evol. 5: 3250–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C., Hermann R. J., Barg M.-C., Schalkowski R., Dirksen P., et al. , 2015c Travelling at a slug’s pace: possible invertebrate vectors of Caenorhabditis nematodes. BMC Ecol. 15: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson B. F., Scharf M. E., 2016. Lower termite associations with microbes: synergy, protection, and interplay. Front. Microbiol. 7: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirri J. K., Alkema M. J., 2012. The neuroethology of C. elegans escape. Curr. Opin. Neurobiol. 22: 187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E., Zhang Y., Pujol N., Matsuyama T., Bargmann C. I., et al. , 2007. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 104: 2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol N., Link E. M., Liu L. X., Kurz C. L., Alloing G., et al. , 2001. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr. Biol. 11: 809–821. [DOI] [PubMed] [Google Scholar]
- Pukkila-Worley R., Ausubel F. M., 2012. Immune defense mechanisms in the Caenorhabditis elegans intestinal epithelium. Curr. Opin. Immunol. 24: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A. U., Carta L. K., Lesuisse E., Hamza I., 2005. Lack of heme synthesis in a free-living eukaryote. Proc. Natl. Acad. Sci. USA 102: 4270–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read D. S., Sheppard S. K., Bruford M. W., Glen D. M., Symondson W. O. C., 2006. Molecular detection of predation by soil micro-arthropods on nematodes. Mol. Ecol. 15: 1963–1972. [DOI] [PubMed] [Google Scholar]
- Reddy K. C., Andersen E. C., Kruglyak L., Kim D. H., 2009. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science 323: 382–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke A. W., Balla K. M., Bennett E. J., Troemel E. R., 2017. Identification of microsporidia host-exposed proteins reveals a repertoire of rapidly evolving proteins. Nat. Commun. 8: 14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle D. L., Albert P. S., 1997. Genetic and environmental regulation of dauer larva development, in C. elegans II, edited by Riddle D. L., Blumenthal T., Priess J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- Ross J. L., Ivanova E. S., Severns P. M., Wilson M. J., 2010. The role of parasite release in invasion of the USA by European slugs. Biol. Invasions 12: 603–610. [Google Scholar]
- Ross J. L., Ivanova E. S., Sirgel W. F., Malan A. P., Wilson M. J., 2012. Diversity and distribution of nematodes associated with terrestrial slugs in the Western Cape Province of South Africa. J. Helminthol. 86: 215–221. [DOI] [PubMed] [Google Scholar]
- Saiki R., Lunceford A. L., Bixler T., Dang P., Lee W., et al. , 2008. Altered bacterial metabolism, not coenzyme Q content, is responsible for the lifespan extension in Caenorhabditis elegans fed an Escherichia coli diet lacking coenzyme Q. Aging Cell 7: 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel B. S., Rowedder H., Braendle C., Félix M.-A., Ruvkun G., 2016. Caenorhabditis elegans responses to bacteria from its natural habitats. Proc. Natl. Acad. Sci. USA 113: E3941–E3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Hempel P., 2008. Parasite immune evasion: a momentous molecular war. Trends Ecol. Evol. 23: 318–326. [DOI] [PubMed] [Google Scholar]
- Schroeder F. C., 2015. Modular assembly of primary metabolic building blocks: a chemical language in C. elegans. Chem. Biol. 22: 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenburg H., Ewbank J. J., 2004. Diversity and specificity in the interaction between Caenorhabditis elegans and the pathogen Serratia marcescens. BMC Evol. Biol. 4: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenburg H., Ewbank J. J., 2007. The genetics of pathogen avoidance in Caenorhabditis elegans. Mol. Microbiol. 66: 563–570. [DOI] [PubMed] [Google Scholar]
- Schulenburg H., Kurtz J., Moret Y., Siva-Jothy M. T., 2009. Introduction. Ecological immunology. Philos. Trans. R. Soc. B Biol. Sci. 364: 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte R. D., Makus C., Hasert B., Michiels N. K., Schulenburg H., 2010. Multiple reciprocal adaptations and rapid genetic change upon experimental coevolution of an animal host and its microbial parasite. Proc. Natl. Acad. Sci. USA 107: 7359–7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte R. D., Makus C., Hasert B., Michiels N. K., Schulenburg H., 2011. Host-parasite local adaptation after experimental coevolution of Caenorhabditis elegans and its microparasite Bacillus thuringiensis. Proc. Biol. Sci. 278: 2832–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte R. D., Hasert B., Makus C., Michiels N. K., Schulenburg H., 2012. Increased responsiveness in feeding behaviour of Caenorhabditis elegans after experimental coevolution with its microparasite Bacillus thuringiensis. Biol. Lett. 8: 234–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., Rockman M. V., Kruglyak L., 2008. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science 319: 589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., Ailion M., Li J., van Oudenaarden A., Rockman M. V., et al. , 2011. A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. PLoS Biol. 9: e1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serobyan V., Ragsdale E. J., Sommer R. J., 2014. Adaptive value of a predatory mouth-form in a dimorphic nematode. Proc. Biol. Sci. 281: 20141334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira M., 2016. Gut microbiotas and host evolution: scaling up symbiosis. Trends Ecol. Evol. 31: 539–549. [DOI] [PubMed] [Google Scholar]
- Shapira M., Hamlin B. J., Rong J., Chen K., Ronen M., et al. , 2006. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc. Natl. Acad. Sci. USA 103: 14086–14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Murphy C. T., 2014. Mating induces shrinking and death in Caenorhabditis mothers. Science 343: 536–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtonda B. B., Avery L., 2006. Dietary choice behavior in Caenorhabditis elegans. J. Exp. Biol. 209: 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi M., Cagliani R., Forni D., Clerici M., 2015. Evolutionary insights into host-pathogen interactions from mammalian sequence data. Nat. Rev. Genet. 16: 224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasundar A., Hey J., 2005. Sampling from natural populations with RNAi reveals high outcrossing and population structure in Caenorhabditis elegans. Curr. Biol. 15: 1598–1602. [DOI] [PubMed] [Google Scholar]
- Slowinski S. P., Morran L. T., Parrish R. C., II, Cui E. R., Bhattacharya A., et al. , 2016. Coevolutionary interactions with parasites constrain the spread of self-fertilization into outcrossing host populations. Evolution 70: 2632–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer R. J., Mayer M. G., 2015. Toward a synthesis of developmental biology with evolutionary theory and ecology. Annu. Rev. Cell Dev. Biol. 31: 453–471. [DOI] [PubMed] [Google Scholar]
- Sommer R. J., McGaughran A., 2013. The nematode Pristionchus pacificus as a model system for integrative studies in evolutionary biology. Mol. Ecol. 22: 2380–2393. [DOI] [PubMed] [Google Scholar]
- Sterken M. G., Snoek L. B., Kammenga J. E., Andersen E. C., 2015. The laboratory domestication of Caenorhabditis elegans. Trends Genet. 31: 224–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle, T., 2006 Maintenance of C. elegans (February 11, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.101.1, http://www.wormbook.org.
- Styer K. L., Singh V., Macosko E., Steele S. E., Bargmann C. I., et al. , 2008. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science 322: 460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze J. Y., Victor M., Loer C., Shi Y., Ruvkun G., 2000. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 403: 560–564. [DOI] [PubMed] [Google Scholar]
- Szewczyk N. J., Kozak E., Conley C. A., 2003. Chemically defined medium and Caenorhabditis elegans. BMC Biotechnol. 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk N. J., Udranszky I. A., Kozak E., Sunga J., Kim S. K., et al. , 2006. Delayed development and lifespan extension as features of metabolic lifestyle alteration in C. elegans under dietary restriction. J. Exp. Biol. 209: 4129–4139. [DOI] [PubMed] [Google Scholar]
- Szumowski S. C., Troemel E. R., 2015. Microsporidia–host interactions. Curr. Opin. Microbiol. 26: 10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szumowski S. C., Botts M. R., Popovich J. J., Smelkinson M. G., Troemel E. R., 2014. The small GTPase RAB-11 directs polarized exocytosis of the intracellular pathogen N. parisii for fecal-oral transmission from C. elegans. Proc. Natl. Acad. Sci. USA 111: 8215–8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szumowski S. C., Estes K. A., Popovich J. J., Botts M. R., Sek G., et al. , 2016. Small GTPases promote actin coat formation on microsporidian pathogens traversing the apical membrane of Caenorhabditis elegans intestinal cells: actin coats on microsporidia-containing vesicles. Cell. Microbiol. 18: 30–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M.-W., Mahajan-Miklos S., Ausubel F. M., 1999a Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 96: 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M.-W., Rahme L. G., Sternberg J. A., Tompkins R. G., Ausubel F. M., 1999b Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 96: 2408–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotónio H., Manoel D., Phillips P. C., 2006. Genetic variation for outcrossing among Caenorhabditis elegans isolates. Evolution 60: 1300–1305. [PubMed] [Google Scholar]
- Teotónio H., Carvalho S., Manoel D., Roque M., Chelo I. M., 2012. Evolution of outcrossing in experimental populations of Caenorhabditis elegans. PLoS One 7: e35811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. G., Li R., Smith H. E., Woodruff G. C., Oliver B., et al. , 2012. Simplification and desexualization of gene expression in self-fertile nematodes. Curr. Biol. CB 22: 2167–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. H., Robertson H. M., 2008. The Caenorhabditis chemoreceptor gene families. BMC Biol. 6: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. H., Kelley J. L., Robertson H. M., Ly K., Swanson W. J., 2005. Adaptive evolution in the SRZ chemoreceptor families of Caenorhabditis elegans and Caenorhabditis briggsae. Proc. Natl. Acad. Sci. USA 102: 4476–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thutupalli S., Uppaluri S., Constable G. W., Levin S. A., Stone H. A., et al. , 2017. Farming and public goods production in Caenorhabditis elegans populations. Proc. Natl. Acad. Sci. USA 114: 2289–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting J. J., Woodruff G. C., Leung G., Shin N.-R., Cutter A. D., et al. , 2014. Intense sperm-mediated sexual conflict promotes reproductive isolation in Caenorhabditis nematodes. PLoS Biol. 12: e1001915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel E. R., Félix M.-A., Whiteman N. K., Barrière A., Ausubel F. M., 2008. Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans. PLoS Biol. 6: 2736–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanfleteren J. R., 1974. Nematode growth factor. Nature 248: 255–257. [DOI] [PubMed] [Google Scholar]
- Venette R. C., Ferris H., 1998. Influence of bacterial type and density on population growth of bacterial-feeding nematodes. Soil Biol. Biochem. 30: 949–960. [Google Scholar]
- Vilas-Boas G. T., Peruca A. P. S., Arantes O. M. N., 2007. Biology and taxonomy of Bacillus cereus, Bacillus anthracis, and Bacillus thuringiensis. Can. J. Microbiol. 53: 673–687. [DOI] [PubMed] [Google Scholar]
- Viney M. E., Gardner M. P., Jackson J. A., 2003. Variation in Caenorhabditis elegans dauer larva formation. Dev. Growth Differ. 45: 389–396. [DOI] [PubMed] [Google Scholar]
- Virk B., Correia G., Dixon D. P., Feyst I., Jia J., et al. , 2012. Excessive folate synthesis limits lifespan in the C. elegans: E. coli aging model. BMC Biol. 10: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virk B., Jia J., Maynard C. A., Raimundo A., Lefebvre J., et al. , 2016. Folate acts in E. coli to accelerate C. elegans aging independently of bacterial biosynthesis. Cell Rep. 14: 1611–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völk J., 1950. Die Nematoden der Regenwürmer und aasbesuchenden Käfer. Zool Jahrb. Abt. Syst. 79: 1–70. [Google Scholar]
- Volkers R. J., Snoek L. B., van Hellenberg Hubar C. J., Coopman R., Chen W., et al. , 2013. Gene-environment and protein-degradation signatures characterize genomic and phenotypic diversity in wild Caenorhabditis elegans populations. BMC Biol. 11: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E., Walhout A. J. M., 2014. Caenorhabditis elegans metabolic gene regulatory networks govern the cellular economy. Trends Endocrinol. Metab. 25: 502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E., MacNeil L. T., Ritter A. D., Yilmaz L. S., Rosebrock A. P., et al. , 2014. Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits. Cell 156: 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E., Olin-Sandoval V., Hoy M. J., Li C.-H., Louisse T., et al. , 2016. Metabolic network rewiring of propionate flux compensates vitamin B12 deficiency in C. elegans. Elife 5: e17670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegewitz V., Schulenburg H., Streit A., 2008. Experimental insight into the proximate causes of male persistence variation among two strains of the androdioecious Caenorhabditis elegans (Nematoda). BMC Ecol. 8: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A. J., 2014. Competition as a source of constraint on life history evolution in natural populations. Heredity 112: 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. C., Chaston J. M., Douglas A. E., 2013. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 7: 1922–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. C., Luo Y., Jing X., Franzenburg S., Bost A., et al. , 2015. The host as the driver of the microbiota in the gut and external environment of Drosophila melanogaster. Appl. Environ. Microbiol. 81: 6232–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff G. C., Knauss C. M., Maugel T. K., Haag E. S., 2014. Mating damages the cuticle of C. elegans hermaphrodites. PLoS One 9: e104456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M. E., Webster J. P., Domingo E., Charlesworth B., Levin B. R., 2002. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet. 32: 569–577. [DOI] [PubMed] [Google Scholar]
- Xie H., Aminuzzaman F. M., Xu L., Lai Y., Li F., et al. , 2010. Trap induction and trapping in eight nematode-trapping fungi (Orbiliaceae) as affected by juvenile stage of Caenorhabditis elegans. Mycopathologia 169: 467–473. [DOI] [PubMed] [Google Scholar]
- Xiong X., Lu N. C., 2008. Optimal levels of the essential amino acids histidine, lysine and threonine in Caenorhabditis elegans maintenance medium. Nematology 10: 539–544. [Google Scholar]
- Yang J., Liang L., Zhou C., Zhang K.-Q., 2016. Molecular mechanism of nematophagous fungi infection of nematodes, pp. 263–311 in Nematode-Trapping Fungi, Fungal Diversity Research, edited by Zhang K.-Q., Hyde K. D. Springer, New York. [Google Scholar]
- Yang W., Dierking K., Esser D., Tholey A., Leippe M., et al. , 2015. Overlapping and unique signatures in the proteomic and transcriptomic responses of the nematode Caenorhabditis elegans toward pathogenic Bacillus thuringiensis. Dev. Comp. Immunol. 51: 1–9. [DOI] [PubMed] [Google Scholar]
- Yang W., Dierking K., Rosenstiel P. C., Schulenburg H., 2016. GATA transcription factor as a likely key regulator of the Caenorhabditis elegans innate immune response against gut pathogens. Zoology 119: 244–253. [DOI] [PubMed] [Google Scholar]
- Yeates G. W., 2003. Nematodes as soil indicators: functional and biodiversity aspects. Biol. Fertil. Soils 37: 199–210. [Google Scholar]
- Yeates G. W., Bongers T., 1999. Nematode diversity in agroecosystems. Agric. Ecosyst. Environ. 74: 113–135. [Google Scholar]
- Yilmaz L. S., Walhout A. J. M., 2014. Worms, bacteria, and micronutrients: an elegant model of our diet. Trends Genet. 30: 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook K., Hodgkin J., 2007. Mos1 mutagenesis reveals a diversity of mechanisms affecting response of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics 175: 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiga T., Ishikawa Y., Tanaka R., Hironaka M., Okumura E., 2013. Species-specific and female host-biased ectophoresy in the roundworm Caenorhabditis japonica. Naturwissenschaften 100: 205–208. [DOI] [PubMed] [Google Scholar]
- Zhang C., Zhao N., Chen Y., Zhang D., Yan J., et al. , 2016. The signaling pathway of Caenorhabditis elegans mediates chemotaxis response to the attractant 2-heptanone in a Trojan Horse-like pathogenesis. J. Biol. Chem. 291: 23618–23627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Peng D., Cheng C., Zhou W., Ju S., et al. , 2016. Bacillus thuringiensis crystal protein Cry6Aa triggers Caenorhabditis elegans necrosis pathway mediated by aspartic protease (ASP-1). PLoS Pathog. 12: e1005389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Berg M., Dierking K., Félix M.-A., Shapira M., et al. , 2017. Caenorhabditis elegans as a model for microbiome research. Front. Microbiol. 8: 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Sachse M., Prevost M.-C., Luallen R. J., Troemel E. R., et al. , 2016. A large collection of novel nematode-infecting microsporidia and their diverse interactions with Caenorhabditis. elegans and other related nematodes. PLoS Pathog. 12: e1006093 (erratum: PLoS Pathog 3: e1006204). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K.-Q., Hyde K. D. (Editors), 2016. Nematode-Trapping Fungi. Springer; New York. [Google Scholar]
- Zhang S., 2004. Caenorhabditis elegans TRPV ion channel regulates 5HT biosynthesis in chemosensory neurons. Development 131: 1629–1638. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhang Y., 2012. DBL-1, a TGF-β, is essential for Caenorhabditis elegans aversive olfactory learning. Proc. Natl. Acad. Sci. USA 109: 17081–17086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., 2008. Neuronal mechanisms of Caenorhabditis elegans and pathogenic bacteria interactions. Curr. Opin. Microbiol. 11: 257–261. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Lu H., Bargmann C. I., 2005. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438: 179–184. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Lu N. C., 2011. Requirements of non-haem iron and haem iron in the nematode Caenorhabditis elegans. Nematology 13: 853–858. [Google Scholar]
- Zugasti O., Bose N., Squiban B., Belougne J., Kurz C. L., et al. , 2014. Activation of a G protein–coupled receptor by its endogenous ligand triggers the innate immune response of Caenorhabditis elegans. Nat. Immunol. 15: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]