Abstract
We address the task of genotype imputation to a dense reference panel given genotype likelihoods computed from ultralow coverage sequencing as inputs. In this setting, the data have a high-level of missingness or uncertainty, and are thus more amenable to a probabilistic representation. Most existing imputation algorithms are not well suited for this situation, as they rely on prephasing for computational efficiency, and, without definite genotype calls, the prephasing task becomes computationally expensive. We describe GeneImp, a program for genotype imputation that does not require prephasing and is computationally tractable for whole-genome imputation. GeneImp does not explicitly model recombination, instead it capitalizes on the existence of large reference panels—comprising thousands of reference haplotypes—and assumes that the reference haplotypes can adequately represent the target haplotypes over short regions unaltered. We validate GeneImp based on data from ultralow coverage sequencing (0.5×), and compare its performance to the most recent version of BEAGLE that can perform this task. We show that GeneImp achieves imputation quality very close to that of BEAGLE, using one to two orders of magnitude less time, without an increase in memory complexity. Therefore, GeneImp is the first practical choice for whole-genome imputation to a dense reference panel when prephasing cannot be applied, for instance, in datasets produced via ultralow coverage sequencing. A related future application for GeneImp is whole-genome imputation based on the off-target reads from deep whole-exome sequencing.
Keywords: genotype imputation, genotype likelihood, imputation from genotype likelihoods, GeneImp, phasing, no prephasing
THE cost of next-generation sequencing has fallen remarkably since its first adoption by sequencing centers in 2008 (van Dijk et al. 2014; Wetterstrand 2016)—from $1,352,982 per genome in April 2008 to $1245 in October 2015. This profound cost reduction, and the ongoing improvement of experimental and computational pipelines, have made next generation sequencing a competing technology to the historically more established micro-array platforms (Hurd and Nelson 2009; Baker 2013). In a proof-of-concept study, Pasaniuc et al. (2012) demonstrated that ultralow coverage DNA-sequencing (sequencing at 0.1–0.5×), followed by imputation to a dense reference panel, captures almost as much of the common (minor allele frequency >5%) and low-frequency (1–5%) variation as single-nucleotide polymorphism (SNP) arrays, and argued that this paradigm could become cost-effective for genome-wide association studies (GWAS) as sample preparation and sequencing costs would continue to fall.
The cost-efficiency of ultralow coverage sequencing predicted by Pasaniuc et al. (2012) is not yet realized, primarily due to the concurrent reduction in the cost of whole-genome SNP arrays. Nevertheless, their proof-of-concept has important implications not only for the design of new genomic studies, where one needs to consider the trade-off between sample size and sequencing read depth (Sims et al. 2014) before comparing to SNP-array alternatives, but also for existing whole-exome sequencing datasets, where off-target reads can be used to acquire whole-genome imputation data. In recent years, whole-exome sequencing has been used extensively in translational research studies (Majewski et al. 2011; Rabbani et al. 2014; van Dijk et al. 2014), owing to its reduced cost per sample and easier interpretation of findings compared to whole-genome sequencing. For many exome capture systems, a 0.2–0.6× coverage of the whole genome can be acquired from off-target reads (Chilamakuri et al. 2014), if we assume a recommended 80× average coverage of the exome (Sims et al. 2014).
Genotype imputation methods use a reference panel comprising haplotypes of individuals at a large set of genetic variants to infer the untyped genotypes in target samples that have been assayed for only a subset of the variants. Accurate imputation of untyped variants allows for joint analysis of individual-level data (or summary statistics) from samples typed at a different set of genetic loci; a detailed account of imputation uses in the context of GWAS is given in Marchini and Howie (2010). This has greatly facilitated the meta-analysis of GWAS [Franke et al. 2010; Berndt et al. 2013; Global Lipids Genetics Consortium 2013; Al Olama et al. 2014; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium et al. 2014], and has led to the robust identification of hundreds of genetic variants associated with various phenotypes.
In this work, we address the computational challenge of scaling up the imputation task to the whole genome and to large reference panels when the data come from ultralow coverage sequencing. In Pasaniuc et al. (2012), the experimental feasibility of this procedure was demonstrated by imputing 10 distinct 5 Mb regions (∼0.015% of the genome) using BEAGLE [algorithm v.4.0 (Browning and Browning 2007, 2009)] and a relatively small reference panel, comprising 381 haplotypes. However, the computational tractability of extending this task to genome-wide scale was not examined.
Genotype imputation is a computationally expensive task, and this cost is exacerbated when the data have a probabilistic representation, due to a high-level of missingness or uncertainty. The complexity of genotype inference is driven by the computation of the hidden state, which is typically defined in terms of the underlying pair of haplotypes. For data with a deterministic representation, a key methodological advancement that made whole-genome imputation computationally efficient was to split the task of genotype imputation into two steps: phasing, followed by haploid imputation (Howie et al. 2009, 2012). Refinement of the original algorithms and computational speed-ups have since been developed in software tools implementing these two steps.
The common procedure for genotype imputation using definite genotype calls with a low level of missing data is to prephase the genotypes using a program such as SHAPEIT2 (Delaneau et al. 2013), which infers phase from the target genotypes with or without a reference panel, and then use the phased genotypes with an imputation program such as IMPUTE2 (Howie et al. 2009, 2011), Minimac (Fuchsberger et al. 2015), or, more recently, BEAGLE v.4.1 (Browning and Browning 2016). Imputation without prephasing is usually substantially slower. We summarize the algorithms and list key properties of the most established imputation software packages in Table 1 and Table 2.
Table 1. Summary of imputation methods.
| Name | Description | Hidden State Approximation |
|---|---|---|
| BEAGLE v.4.1 (Browning and Browning 2016) | Li and Stephens (2003) haplotype frequency model with parsimonious state-space | HMM calculations restricted to clusters of markers (aggregates) within small regions |
| BEAGLE v.4.0 (Browning and Browning 2007, 2009) | Iterates between building a suffix tree from current-guess haplotypes and updating haplotypes based on probabilities from the tree. The suffix tree resembles a “cluster-haplotype model,” but has a variable number of clusters depending on LD in each region | Suffix tree is pruned to produce parsimonious representation of the data |
| fastPHASE (Scheet and Stephens 2006) | EM algorithm that iterates between fitting hyper-parameters of a “haplotype-cluster HMM” and running forward-backward algorithm in fitted HMM to get imputed genotypes | Hidden state modeled by haplotype clusters (20 clusters good empirically) |
| IMPUTE (v.1) (Marchini et al. 2007) | HMM similar to Li-Stephens model. Forward-backward algorithm to compute hidden state probabilities, then analytically integrate over all hidden states | Can restrict computation to reference panel haplotypes |
| IMPUTE2 (v.2) (Howie et al. 2009, 2011) | MCMC iterating between phasing and imputing. Phasing done with IMPUTE v.1 HMM (HMM forward path sampling). Imputation done by haploid HMM (HMM forward-backward) | Only subset of haplotypes with smallest Hamming distance to current-guess haplotypes in phasing step |
| MaCH (Li et al. 2010) | HMM similar to Li-Stephens model. Iteratively update phase of each individual based on haplotypes of other individuals (HMM forward-backward). Additionally update HMM hyper-parameters at every iteration | Only a subset of haplotypes randomly selected |
| MiniMac (Howie et al. 2012) | Fast implementation of MaCH model using prephased data | NA (phasing precomputed) |
| MiniMac2 (Fuchsberger et al. 2015) | Computational speed-ups to the MiniMac software | NA |
| SNPTools (Wang et al. 2013) | “Constrained” Li-Stephens method | Only four parental haplotypes, selected based on metropolis-hastings MCMC sampling |
| GeneImp | Haplotype-pair sequence within window is exact copy of reference haplotype-pair. Reference haplotype-pairs compatible with genotype likelihoods are analytically integrated over | Only a subset of “filtered” reference haplotypes |
Table 2. Key properties of imputation methods.
| Name | Can Use Reference Panel | Can Perform Phasing | Can Use Genotype Likelihood Inputs | Requires Prephasing | Can use Prephasing to Speed up Computation | Computational Complexity |
|---|---|---|---|---|---|---|
| BEAGLE v.4.1a (Browning and Browning 2016) | Yes | No | No | Yes | Yes | Linear in number of haplotypes |
| BEAGLE v.4.0 (Browning and Browning 2007, 2009) | Yes | Yes | Yes | No | No | Quadratic in number of haplotypes |
| fastPHASE (Scheet and Stephens 2006) | Yes | Yes | No | No | No | M-step linear in number of haplotypes, quadratic in number of clusters |
| IMPUTE (v.1) (Marchini et al. 2007) | Yes | No | No | No | No | Quadratic in number of haplotypes |
| IMPUTE2 (v.2) (Howie et al. 2009, 2011) | Yes | Yes | Yes | No | Yes | Phasing quadratic in number of haplotypes, imputing linear in number of haplotypes |
| MaCH (Li et al. 2010) | Yes | Yes | No | No | No | Quadratic in number of haplotypes |
| MiniMac (Howie et al. 2012) | Yes | No | No | Yes | Yes | Linear in number of haplotypes (phasing precomputed) |
| MiniMac2 (Fuchsberger et al. 2015) | Yes | No | No | Yes | Yes | NA |
| SNPTools (Wang et al. 2013) | No | Yes | Yes | No | No | Constant in number of haplotypes |
| GeneImp | Yes | No (but can be added) | Yes | No | No | Subquadratic in number of reference haplotypesb |
Attributes for the options that invoke the v.4.1 algorithm in the BEAGLE software package.
GeneImp hidden state considers reference haplotypes only (i.e., ignores haplotypes of other target individuals).
When the data are produced via ultralow coverage whole-genome sequencing, a probabilistic representation such as genotype likelihoods is more suitable. Making definite genotype calls in this setting would result in either a high level of missingness or high genotype misspecification. Without definite genotype calls, phasing has a substantially higher computational cost, as there is no single haplotype reconstruction that contains all the information in the original genotype data. This makes the two-step pipeline computationally inefficient for this setting. In addition to the algorithmic complexity, genotype likelihoods are also not supported at the software level, with most existing methods for prephasing requiring a low proportion of missing data (Browning and Browning 2011).
An exception to this is the BEAGLE v.4.0 algorithm, which performs phasing and imputation simultaneously. Nevertheless, BEAGLE’s v.4.0 algorithm scales quadratically with the number of samples in the reference panel (Browning and Browning 2016), and it is thus computationally too expensive to apply on a genome-wide scale. We note that BEAGLE’s v.4.1 algorithm, which is substantially faster than v.4.0, cannot be used with genotype likelihoods, and recedes to v.4.0 when genotype likelihoods are specified as inputs. IMPUTE2 can also accept genotype likelihoods as inputs, but, without prephasing, it is substantially slower, as it implements a Markov Chain Monte Carlo scheme that iterates between phasing and haploid imputation, which is also computationally intractable for whole-genome imputation.
SNPTools (Wang et al. 2013) is another software package that accepts genotype likelihoods as inputs. However, SNPTools’ imputation algorithm is designed to improve the task of variant-calling in next-generation sequencing data rather than to impute completely untyped loci. The hidden state is represented by only four parental haplotypes at each region, selected using a Markov Chain Monte Carlo sampling scheme. It is unclear whether the algorithm could efficiently impute untyped loci based on a reference panel. Currently this cannot be evaluated, as the software package does not support usage of a reference panel.
MarViN (Arthur et al. 2015) was recently proposed for genotype imputation of low-coverage sequencing data, where the average coverage is ∼7×. MarViN’s recommended usage is for low and intermediate coverage data, where some reads are recorded in the majority of sites, rather than ultralow coverage data, where many sites have no reads. In the latter case, MarViN’s Expectation-Maximization algorithm is likely to take considerably more iterations to converge. Moreover, as reported in Arthur et al. (2015), its homogeneity assumption for Linkage Disequilibrium (LD) is likely to break when the reference panel consists of a heterogeneous mix of populations with finer LD substructure, such as the 1000 Genomes panel (1000 Genomes Project Consortium 2015).
Finally, imputation methods from the animal breeding community, such as FINDHAP (VanRaden et al. 2011), typically exploit long-range Identity-By-Descent, and thus are not well suited when the target cohort comprises unrelated samples, as is often the case in human genetics.
In this paper, we describe and validate GeneImp, a program that imputes genotypes at reference sites from ultralow coverage sequencing, or any other platform that generates genotype likelihoods, and has subquadratic complexity in the number of individuals in the reference panel.
Our algorithm is motivated by a second pivotal advancement in genotype imputation, i.e., the availability of large reference haplotype panels. Most imputation programs were originally developed at a time when reference panels were small compared with the typical size of a GWAS cohort. The original HapMap panel (International HapMap Consortium 2007) consisted of only 120 gametes in each of three continental groups. Therefore, imputation quality depended heavily on information coming from the target individuals for estimating the hidden state-space of true underlying haplotypes and performing phasing.
Larger reference panels with wider coverage of local genetic variation are now available. The most widely used reference panel is the one produced by the 1000 Genomes Project (1000 Genomes Project Consortium 2015), which contains haplotypes for 2504 individuals. Recently, the Haplotype Reference Consortium (McCarthy et al. 2015) has produced a panel comprising >32,000 individuals from 20 cohorts. In these situations, the extra information about haplotype frequencies contributed by the target cohort is likely to be small compared with the information in the reference panel. Therefore, we can phase or impute one target individual at a time without losing too much information. In GeneImp, we exploit this independence assumption, and parallelize imputation of target individuals.
We hypothesize that, given a large reference panel from the same ancestral population as the target cohort, the reference haplotypes represent haplotypes in the target individuals over short regions adequately, without an explicit model of recombination. We infer genotype probabilities over short regions based on how probable each reference haplotype-pair is given the genotype likelihoods in the region. In the following sections, we outline the GeneImp algorithm and its tuning parameters, and evaluate its performance on imputing ultralow coverage sequence data.
Materials and Methods
The GeneImp algorithm
GeneImp imputation is based on a sliding window—where each window corresponds to a short region of the genome—and is performed for one target individual at a time. Our main hypothesis is that the reference haplotypes adequately represent target haplotypes within each window.
Let index a sequence of W consecutive, nonoverlapping windows across the genome; let denote the set of haplotypes in the reference panel, and let K be the number of haplotypes in the set Then, the two haplotype sequences of a target individual can be represented by two W-dimensional vectors and with each element being a categorical variable, mapping the haplotype of the target sample in window w to K available haplotypes in the reference panel,
The joint probability distribution under GeneImp is given by:
| (1) |
where and are the hidden haplotype sequences for individual i, is a vector of genotypes with elements j indexes sites across the genome, and is a data-derived vector representing observed sequence reads across the genome, so that is the data likelihood at site j for genotype A glossary of technical terms we use to describe sequencing data is given in Table 3.
Table 3. Glossary of terms for sequencing data.
| Term | Description |
|---|---|
| Reads | Data recording nucleotide signal strengths for short DNA fragments. These data are used for base calling |
| Base calling | The process of assigning nucleotide bases to reads. Encompasses a measure for the uncertainty and quality of assigning a nucleotide at each site |
| Genotype likelihoods | Probability of observing the reads given each possible genotype. Uses the uncertainty and quality scores for all reads at a given site for a given individual. For a diallelic SNPs with reference allele denoted by r and alternate allele denoted by a, the genotype likelihood for an individual i at site j is the three-dimensional vector where and denote reads and genotype at site j for individual i, respectively |
We make two assumptions regarding data dependencies when we formulate the probabilistic model of GeneImp. The joint probability distribution of observed sequencing reads and latent haplotype states factorizes over samples. This ignores information about the phase coming from other target samples, and allows us to impute each target individual independently. Second, the genotype likelihoods factorize over sites given the hidden haplotype sequences. This is equivalent to assuming that sequencing errors are independent at different sites of the sequencing read, an assumption commonly made by widely adopted variant calling algorithms (Li et al. 2008). A graphical model depicting the conditional independence assumptions under GeneImp is given in Supplemental Material, Figure S1.
Inference:
In the following, we describe the inference procedure for one target sample. To simplify the notation, we drop the index i for individuals.
For every pair of reference haplotypes, we first calculate the posterior probability of being the underlying haplotype-pair in a target window given the sequencing reads. This is proportional to the product of genotype likelihoods for genotypes consistent with the haplotype-pair at sites within the window, and two flanking regions left and right of the window. Then, at each site within the window, we calculate the probability distribution over the three possible genotypes by summing the posterior probabilities of reference haplotype-pairs giving rise to each of the three genotypes.
Specifically, we are interested in the posterior probability distribution over the sequence of genotypes given a set of K reference haplotypes, and the data vector representing sequencing reads:
| (2) |
The first factor is the probability of the genotype sequence given the sequences of latent state assignments for the two haplotypes. Given the haplotype-pair assignment, we define the genotype deterministically, so that this term factorizes over windows and is given by:
| (3) |
where denotes the vector with the genotypes at all sites belonging to window w, and are the haplotype sequences of latent states and at window w, with for a reference haplotype k and a window w with sites, and is an indicator function evaluating to one if the condition holds and to zero otherwise. The condition is satisfied for the vector of genotypes arising by the reference haplotypes corresponding to latent states and within window w.
The second factor in Equation 2 is the posterior distribution of the sequence of latent state assignments given the data. This term is computationally expensive to compute exactly, as conditioning on the data vector, introduces dependencies between hidden state assignments in consecutive windows.
To perform inference we approximate the full joint distribution over hidden state assignments with a simpler distribution that factorizes over windows:
| (4) |
This approximation allows us to decompose the summation over the whole sequence of hidden states in Equation 2 into summations of the hidden states in each window:
| (5) |
Finally, the distribution over the hidden state within a window can be computed using Bayes rule:
| (6) |
where is the set of sites whose sequence reads depend on the haplotype assignment in window w (sequence reads within the window and the two flanking regions), Z is a normalizing constant summing over the hidden states for the haplotype-pair assignment, is the genotype likelihood at site j for the genotype given by adding haplotypes and at site j, and we have assumed, a priori, that the haplotype assignments are independent, and that all haplotypes in the reference set have equal prior probability.
Overall, inference is performed in a single pass over windows, with the joint distribution over hidden state assignments being approximated by the product of distributions over hidden states in each window (Equation 4). This resembles a cluster-based mean-field approximation (Jordan et al. 1999), where a complex graph is divided into small clusters, each of which can be inferred by exact inference.
Time complexity:
The complexity of the inference procedure is quadratic in the number of reference haplotypes, and linear in the number of target individuals and in the number of windows. In the section Filtering reference haplotypes, we reduce the state-space over hidden haplotype-pairs by considering only a subset of haplotypes in each window. This is a second approximation to the posterior distribution, and keeps the complexity of GeneImp subquadratic in the number of reference haplotypes, as the summation over haplotype-pairs in Equation 5 is now over pairs instead of
Splitting into windows:
Our objective is to keep windows short, so that the underlying haplotypes in each window can be matched to haplotypes in the reference panel, while ensuring that each window contains enough sequencing reads (i.e., observations) to perform inference reliably. To mitigate this trade-off, we adopt a data-driven approach, where the informativeness of the observed reads is used to split each individual’s genome into the smallest windows that contain sufficient information.
We use the following heuristic approach to quantify the information content of sequencing reads. Consider a simple model where the genotype at a site j is independent of genotypes at other sites. A prior distribution for can be defined in terms of allele frequencies in the reference panel, where and are the frequencies of the reference and the alternate allele, respectively. Using genotype likelihoods derived from the sequencing data, we can apply Bayes rule to get the posterior probability for under this fully factorized model.
We define the information content, at site j, as the Kullback–Leibler divergence between the posterior and the prior distributions of this simple factorized model:
| (7) |
where a indexes the three possible genotypes (ref/ref, ref/alt, alt/alt), and is a normalizing constant.
To divide each chromosome into windows, we require that each subsequent window is the smallest possible, while the total information content of reads in each window is above a fixed threshold. The threshold for the total information content in a window is a tuning parameter of GeneImp. Smaller values result in smaller average windows, as fewer sequence reads are needed to reach the required level.
Flanking regions:
The posterior distribution over the haplotype-pair assignment in a window considers sequencing reads from two flanking regions left and right of the window—in addition to reads within the window (Equation 6). The flanking regions allow us to use more observations for inferring the underlying haplotype-pair, while keeping the imputation window short. Furthermore, they smooth out boundary effects that would occur if each window was processed independently of its two bordering regions.
In this work, we set the size of each flanking region to be roughly half the size of the window, which we empirically found to work well in preliminary experiments. This means that, when computing the posterior distribution over haplotype-pairs in a window (Equation 6), ∼25% of sequencing reads in the set come from the preceding window, ∼50% come from within the window, and ∼25% come from the following window.
Averaging over window splits:
Due to LD, we expect that the sequence of hidden haplotypes will have a block dependency structure. If the blocks were known, we could group sequencing reads from each block in a single window, and perform inference in each window in isolation. However, the “correct” grouping into windows is unknown. Therefore any arbitrary grouping based on heuristic approaches will misrepresent some of the dependency structure in the target haplotypes.
To alleviate this problem, we employ a simple averaging scheme. We run GeneImp a number of times, each time splitting the genome into different windows by choosing a different value for the window-size parameter. This results in different subsets of loci being grouped together in each run. Then, we compute the posterior distribution over haplotype-pairs at a single locus j by taking a flat average of the corresponding distributions from each run:
| (8) |
where s indexes different splits into windows, and S is the total number of different splits, is the w-th window of the s-th split, and denotes the haplotype assignment in the -th window, where is the window the j-th locus belongs to.
Averaging over a large number of approximations is often used for the optimization of nonconvex functions. Our approach is motivated by structured mean-field approximations of loopy graphs (distributions), where we approximate a complex structured distribution by an average over multiple simpler approximations such as trees or chains (Jordan et al. 1999; Xing et al. 2003).
Filtering reference haplotypes:
Similarly to most imputation algorithms, the computational complexity of GeneImp is reduced by introducing an approximation to the state space over the hidden haplotype-pair assignment. Specifically, in Equation 5, instead of summing over all possible haplotype-pair combinations from the reference panel, we select a subset of haplotypes, and sum over the possible haplotype-pair combinations.
We perform the filtering step at each window (and for each individual). Since we are directly imputing diploid sequences (genotypes), our objective is to choose haplotypes that give rise to “good” haplotype-pairs, i.e., haplotype-pairs that are compatible with the genotype likelihoods for the window we are imputing. However, we also want the selected haplotypes to cover the state space adequately, i.e., to also assign probability mass to hidden configurations that are less likely, but still compatible with the sequencing reads. Therefore, we also want the selected haplotypes to give rise to diverse haplotype-pairs.
In order to choose “good” haplotype-pairs, we perform filtering in two steps. In the first step, we select haplotypes at random. In the second step, we select the remaining haplotypes that give rise to haplotype-pairs with high posterior probability (Equation 6) when paired with the haplotypes already selected.
Data description and preprocessing
Target sample:
Our target sample comprises 16 individuals of Scottish ancestry who were included in the Scottish Early Rheumatoid Arthritis (SERA) cohort (Kronisch et al. 2016). The SERA study was reviewed and approved by the West of Scotland Research Ethics Committee Number 4, REC reference number 10/S0704/20. Written informed consent was obtained from all participants. Ultralow coverage sequencing for study participants was available through an industry collaboration, but chip genotyping was not available for the majority of samples. Therefore, imputation to a dense reference panel was necessary for performing downstream analyses with the genetic data.
The raw sequencing reads were aligned to the HG19 reference genome using the Torrent Mapping Alignment Program for Ion Torrent Data (TMAP) software program. Aligned reads were stored as BAM files. We removed duplicate reads using the MarkDuplicates tool from the Picard suite (Data availability). After removing duplicate reads the average sequencing coverage in the 16 individuals was 0.62, ranging from 0.45 to 0.76.
To evaluate the quality of the sequencing and alignment, we considered the aligned reads from chromosome 22, and compared the variant alleles inferred at loci in dbSNP with the variant alleles given in dbSNP. The agreement was >99%, compared with 33% that would be expected by chance. Specifically, only 43 out of 36,846 SNPs with an alternate (ALT) allele called in chromosome 22 had different alternate allele to that reported in dbSNP.
We used the UnifiedGenotyper tool of the GATK suite (Data availability) to compute genotype likelihoods at sites with sequencing reads. We restricted genotype calling to diallelic SNP sites from the 1000 Genomes reference panel, since calling of indels and multiallelic variants from ultralow coverage sequencing can be unreliable, and sites not typed in the reference panel do not contribute any information to the imputation model.
To tune and evaluate the quality of imputation from the sequencing data, the 16 pilot individuals in our target sample were also typed with the Illumina HumanOmniExpressExome-8-v1-2-B chip.
Reference panels:
We evaluated two reference panels: the 1000 Genomes Phase 3 (1000 Genomes Project Consortium 2015) panel of 2504 individuals, and a larger reference panel formed by combining the 1000 Genomes Phase 3 panel with 2432 individuals from the UK10K panel (UK10K Consortium 2015), and 398 individuals from the ORCADES study (McQuillan et al. 2008). We refer to the former reference panel as “1000G,” and to the latter as “Combined.” We note that the representation of mainland Scottish population in the two added cohorts is low, individuals in the UK10K panel were primarily recruited in England, while the ORCADES population is a genetic isolate.
Measuring imputation quality
Multiple evaluation metrics have been proposed in the literature for measuring imputation quality. Here, we used two of the most established metrics, (a) the allelic mean which is the squared Pearson’s correlation coefficient between true genotypes and imputed dosages at each site, averaged over sites; and (b) the calibration of the posterior genotype probabilities, which compares the probability of predicted genotypes to the concordance rate between predicted and true genotypes (Browning and Browning 2009). We assume that the true genotypes are the ones recorded by the SNP-chip platform. We describe these imputation metrics in more detail below.
Mean-R2:
We measure imputation quality using the squared correlation between the expected value for the genotype dosage under the posterior probability distribution inferred by the imputation algorithm, and the genotype recorded in the SNP-chip array. Specifically, for individual i and for a diallelic locus j with alleles r and a, let denote the genotypic dosage for the alternate allele a, i.e., the number of a alleles that individual i carries at locus j. Then, the expected dosage under the imputation model is defined as:
| (9) |
We then compute the squared correlation coefficient between the expected dosage and the true genotype at each locus and average across loci, where we assume that the true genotype is the one recorded by the SNP-chip. We refer to this measure as the mean- The mean- is a standard measure for imputation quality. It can be interpreted as the reduction in effective sample size for a GWAS due to imperfect imputation (Browning and Browning 2009).
Calibration:
To assess whether the imputation probabilities are well-calibrated, we first compute the concordance rate between the most likely genotype and the true genotype. For well-calibrated probabilities, we expect that predicted genotypes with posterior probability α will have concordance rate of approximately α.
BEAGLE settings
To run BEAGLE, we used the following parameter values:
Xmx64G
nthreads = 32 (default = 1)
windowsize = 25,000 (default = 50,000)
The smaller window-size was used in order to reduce memory usage. We used the default settings for all other parameters.
Data availability
The GeneImp code is available as an R package that can be downloaded from https://pm2.phs.ed.ac.uk/geneimp/. The target samples used in this work are from the SERA study. The data custodian is the Stratified Medicine Scotland Innovation Centre (SMS-IC). Managed access to the raw sequence data and the chip genotypes can be negotiated with SMS-IC, who may be contacted at Admin@stratmed.co.uk. The software packages and reference datasets we used in the analyses reported in this manuscript, including data preprocessing steps, can be accessed as follows.
1000 Genomes project, June 2013 phase 3 release downloaded from the ftp server at ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/.
UK10K project: ALSPAC cohort accession number EGAD00001000195, TWINSUK cohort accession number EGAD00001000194.
ORCADES study: http://www.orcades.ed.ac.uk/orcades.
BEAGLE v.4.0 software (beagle.r1399.jar): https://faculty.washington.edu/browning/beagle/b4_0.html.
Picard Tools (v 1.128): http://broadinstitute.github.io/picard/.
Genome Analysis ToolKit (GATK) suite (McKenna et al. 2010) v3.3-0-g37228af: http://www.broadinstitute.org/gatk.
Results
In this section, we present results based on (a) imputation of the whole genome; and (b) imputation of chromosome 22, which is ∼1% of the human genome. In both cases, we assessed imputation quality (mean- and calibration) by considering the subset of SNPs that are included in the combined reference panel, and are also typed in the SNP-chip genotyping platform. This was a total of 810,219 SNPs across the whole genome, and 8911 SNPs on chromosome 22.
GeneImp imputation quality
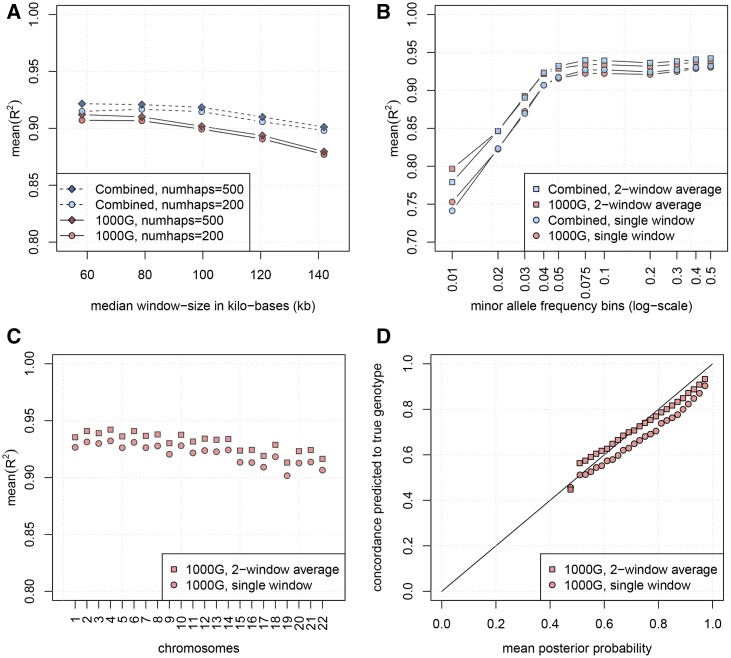
We first evaluated a number of settings for the window-size parameter, and the number of filtered haplotypes, using either the 1000G or the Combined reference panel, by comparing imputation quality achieved by GeneImp on chromosome 22 (Figure 1A). We then selected a smaller subset of settings, and performed imputation of the whole genome (Figure 1, B–D).
Figure 1.
Mean- and calibration for imputation based on GeneImp. (A) Mean- as a function of window-size. Results are from chromosome 22. A smaller window and the Combined panel lead to higher mean- while more filtered haplotypes lead to very small gains. (B) Mean- as a function of MAF. Results are from the whole genome using filtered haplotypes. Single window-split corresponds to median window-size of 58.2 kb, average of two window-splits is taken over results with median window-sizes of 58.2 and 78.9 kb. Mean- increases as a function of the MAF, leveling-off around Averaging posterior probabilities from two window-splits leads to higher mean- especially for rarer SNPs. (C) Mean- in different chromosomes. Results are based on filtered haplotypes. Single window-split corresponds to median window-size of 58.2 kb, average of two window-splits is taken over results with median window-sizes of 58.2 and 78.9 kb. Imputation is marginally worse in shorter chromosomes. (D) Calibration of posterior probabilities from a single window-split corresponding to median window-size of 58.2 kb, and an average of two window-splits taken over results from median window-sizes of 58.2 and 78.9 kb. To evaluate calibration we split imputed genotypes into bins according to their posterior probability distribution. We plot the mean posterior probability in each bin (x-axis) against the percentage of correctly predicted genotypes in each bin (y-axis). Averaging across window-splits leads to well calibrated posterior probabilities (most points lie close to the diagonal), while imputation probabilities based on a single window-split are over-confident (points lie below the diagonal).
In the following sections, we examine imputation quality with respect to (a) GeneImp tuning parameters, (b) minor allele frequency, (c) chromosome, and (d) choice of reference panel.
Window size:
GeneImp performance is relatively robust across different settings, with the mean- ranging from 0.870 to 0.922 on chromosome 22 (Figure 1A). Table 4 shows the median and the 10 and 90% quantiles of window lengths in kilobases corresponding to the five window-size values displayed in Figure 1A. The maximum mean- is achieved with a median length of 58.2 kb. Imputation quality is similar for a median length of 78.9 kb, and starts decreasing when we further increase the window size.
Table 4. Median, 10 and 90% quantiles of window length (kb) for different settings of the window-size parameter.
| 10% Quantile | Median | 90% Quantile |
|---|---|---|
| 23.2 | 58.2 | 131.3 |
| 33.0 | 78.9 | 173.8 |
| 42.4 | 99.6 | 216.8 |
| 53.0 | 120.6 | 258.2 |
| 63.7 | 141.9 | 302.3 |
We report the length of the total region that is used to compute the posterior probabilities over haplotype-pair assignments, i.e., the length of the imputation window, plus the lengths of the two flanking regions left and right of the window. We only impute the middle part of this region, which corresponds to imputation blocks with median length of 29–39 kb. The haplotype blocks shared between unrelated individuals in European populations are typically of length 20–100 kb (Daly et al. 2001; De La Vega et al. 2005), suggesting that smaller windows could lead to better imputation quality if the information content of the sequencing reads was increased (e.g., through deeper sequencing).
In the whole-genome experiments, we used the window-size settings corresponding to median length of 58.2 and 78.9 kb. Generally, the optimal window size will depend on the size of the reference panel, on how closely the target individual is related to the individuals in the reference panel, and on the depth of sequencing coverage. With deeper sequencing, a smaller window size may be optimal. With a reference panel containing many individuals closely related to the target individual, a larger window size may be optimal.
Number of filtered haplotypes:
The number of filtered haplotypes, controls the trade-off between computational complexity and the degree of the approximation in the posterior distribution over genotypes when we reduce the state-space from to where K is the total number of reference haplotypes (Equation 5). We assessed imputation quality using and filtered haplotypes, depicted by circles and diamonds in Figure 1A. Using more haplotypes leads to an increase in the mean- however, this increase is always marginal. Therefore, if the computational cost is a key consideration, setting haplotypes is a good baseline for imputation of European populations to the 1000 Genomes reference panel. In the whole-genome experiments, we used haplotypes.
Evaluation per minor allele frequency:
Figure 1B plots imputation mean- as a function of minor allele frequency (MAF), where we have divided the SNPs from the whole genome into bins according to their MAF in the Combined panel. The first bin shows imputation quality for SNPs with MAF ≤ 1%. The mean- initially increases sharply as we consider bins with higher MAF, and levels out when we reach MAF > 5%. For rare SNPs (MAF ≤ 1%), imputation quality is still at a reasonable level, especially if we consider results from the average over two window-splits (square markers).
Evaluation per chromosome:
Figure 1C shows imputation mean- for different chromosomes. Performance is relatively robust across chromosomes, with the mean- ranging from 0.90 to 0.93 for imputation based on a single window-split, and from 0.91 to 0.94 for imputation based on the average of two window-splits. Imputation quality is marginally worse for shorter chromosomes.
Averaging over window splits:
Any single split into windows will misrepresent some dependencies in the target haplotypes, as the “correct” grouping into windows is unknown. To alleviate this problem, we compute imputation probabilities by taking a flat average over genotype probabilities from different splits into windows. Results from taking an average over two window-splits, corresponding to median window-sizes of 58.2 and 78.9 kb, are presented in Figure 1, B and C in terms of mean- and in Figure 1D in terms of calibration (circles vs. squares). Results are based on SNPs from the whole genome.
The mean- achieved by averaging over the two window-splits is always higher compared to the mean- from the single window-split. The improvement is only marginal for common SNPs (MAF > 5%), but becomes substantial for rarer SNPs (Figure 1B). Averaging over window-splits is also important for calibration, with imputation probabilities giving overconfident predictions when we use genotype posterior probabilities from a single window-split, while being well-calibrated when we take the average over two window-splits (Figure 1D).
Choice of reference panel:
We have used two reference panels denoted by 1000G (1000 Genomes Phase 3) and Combined (1000 Genomes Phase 3 + UK10K + ORCADES). These are depicted by red and blue colors, respectively, in Figure 1. For common SNPs, imputation based on the combined panel results in higher mean- compared to the 1000G panel (Figure 1, A and B). However, the improvement is generally marginal. We believe that this is due to the fact that the added samples from the UK10K and the ORCADES panels are roughly as representative of the samples in our target cohort as samples from the 1000 Genomes panel. Therefore, our approximation of the state-space of haplotype-pair assignments for the target samples, which is based on the available reference haplotypes, does not improve substantially by adding haplotypes from the UK10K and the ORCADES panels.
Interestingly, for rare SNPs (MAF ≤ 1%), imputation based on the 1000G panel results in slightly higher mean- compared to the combined panel (Figure 1B). A possible explanation is that, in the 1000G Phase 3 panel, all individuals were sequenced using both whole-genome sequencing with a mean depth of 7.4×, and targeted exome sequencing with a mean depth of 65.7×, while the UK10K and ORCADES cohorts were only whole-genome sequenced at a mean depth of 7×. Therefore, SNPs with low MAF are likely to be better tagged in the 1000G Phase 3 panel. It is worth noting that Huang et al. (2015) proposed a method for rephasing the UK10K panel, and showed that this rephasing improved imputation of low MAF variants. In this work, we have used a version of the UK10K panel without the proposed rephasing.
Overall, if the interest is in common variation (MAF > 1%), we recommend using the 1000 Genomes Phase 3 reference panel for imputing samples of European ancestry, since the computational burden increases with a larger reference panel. If the interest is in rare variation, a combination of the rephased UK10K panel (Huang et al. 2015) with the 1000 Genomes Phase 3 panel would be more suitable. Adding the current-guess haplotypes of target individuals to the reference panel and rerunning GeneImp is unlikely to increase accuracy substantially, unless the target sample contains related individuals.
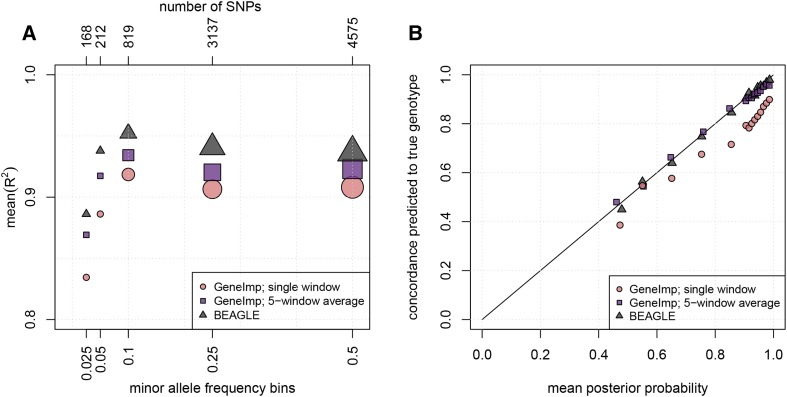
Comparison with BEAGLE
Figure 2 compares imputation quality of GeneImp to BEAGLE v.4.0 in terms of mean- and calibration. Results are based on imputation of chromosome 22 using the 1000G reference panel. For GeneImp, we used filtered haplotypes. The single window-split corresponds to median window-size of 58.2 kb, and the average of five window-splits is taken over results corresponding to the median window-sizes reported in Table 4. BEAGLE has the highest mean- at all MAF bins, followed closely by GeneImp, when averaging posterior probabilities over five window-splits. The difference in mean- between BEAGLE and GeneImp-5-window-average is ∼0.02 at all MAF bins. GeneImp based on a single window-split has the lowest mean- and the difference to the other two methods is higher for rarer SNPs (Figure 2A). As there is only a small number of rare SNPs in chromosome 22, we cannot estimate the mean- for smaller MAF bins reliably in this case.
Figure 2.
Comparison of GeneImp with BEAGLE. (A) Imputation mean- as a function of MAF. Each point is computed by averaging the values for SNPs within a MAF bin. The size of the marker reflects the number of SNPs in the MAF-bin. BEAGLE has the highest mean- followed closely by GeneImp-5-window-average. (B) Calibration of genotype posterior probabilities. Imputed genotypes are split into bins according to their posterior probability distribution. We plot the mean posterior probability in each bin (x-axis) against the percentage of correctly predicted genotypes in each bin (y-axis). BEAGLE and GeneImp-5-window-average are well-calibrated, while GeneImp-single-window is overconfident.
The imputation probabilities of BEAGLE and GeneImp-5-window-average are well-calibrated with most points lying on the diagonal, while the imputation probabilities of GeneImp-single-window are over-confident, i.e., the mean posterior probability is higher than the corresponding percentage of correctly predicted genotypes (Figure 2B).
Overall, combining results from different window splits improves imputation quality both in terms of mean- and calibration, but has diminishing returns. The mean- for GeneImp imputation of chromosome 22 using a single split into windows, and an average over 2, 3, 4, and 5 windows is respectively. Therefore, averaging over two window splits is a good trade-off between quality and efficiency.
We report the mean- averaged over all SNPs, together with the running time and allocated CPUs for BEAGLE and the two GeneImp experiments in Table 5. GeneImp is 15, and up to 90, times faster than BEAGLE, with only a small drop in imputation quality, which makes it a practical choice for real-world applications. For this comparison, we ran BEAGLE with twice as many allocated CPUs (specified through the nthreads option) as GeneImp. In terms of memory complexity, BEAGLE needed at least 64 GB of pre-allocated memory (through the -Xmx Java option) and gave out-of-memory errors when we used smaller values (see also BEAGLE settings). On the other hand, GeneImp is implemented using the bigmemory R package to create, store, and access the reference panel. This allows the program to use RAM, when there is sufficient RAM available, but can also create file-backed data structures, which are accessed in a fast manner when not enough RAM is available. The timings reported here are based on file-backed versions of the reference panels.
Table 5. Performance and running time for GeneImp and BEAGLE imputation of chromosome 22 (16 target samples).
| Method | Mean- | Time in hra | Allocated CPUsb |
|---|---|---|---|
| GeneImp; single-window | 0.907 | 1.5 | 16 parallel processes (doMC R package) |
| GeneImp; five-window-average | 0.922 | 9.0 | 16 parallel processes (doMC R package) |
| BEAGLE | 0.939 | 135.6 | 32 parallel threads (nthreads option) |
We report the elapsed time between start and finish of the experiment. This was recorded using the time unix command for BEAGLE and the proc.time() R function for GeneImp. The server was solely used for this job during this time.
Experiments performed on a server with 4× Intel Xeon E7-4870 processors, with 2.40 GHz and 10 cores (20 threads) each and 256 GB RAM.
Finally, we also compared the running time of BEAGLE to that of GeneImp as we increase the number of target individuals. For this comparison, we used an additional 112 samples with ultralow coverage sequencing from the SERA cohort. These samples could not be used to assess imputation quality, as they had not been typed with the SNP-chip. Table 6 shows the time BEAGLE and GeneImp-single-window took to impute a chunk from chromosome 22 comprising 100,000 variants in the 1000G reference panel (∼1/7th of chromosome 22). Again, we ran BEAGLE with twice as many allocated CPUs as GeneImp, and with other settings as specified in section BEAGLE settings. For GeneImp-single-window, we used the same settings as in the previous comparison (median window-size kb, filtered haplotypes, and file-backed data structure for the reference panel). The GeneImp single-window was 90 times faster than BEAGLE when imputing 16 target individuals, which is consistent with the previous comparison based on imputation of the whole chromosome 22. The GeneImp single-window was 108 times faster than BEAGLE when imputing 128 target individuals. Thus the scaling of computational time with respect to target sample size is slightly favorable for GeneImp over BEAGLE.
Table 6. Running times for GeneImp and BEAGLE imputation with different target sample sizes (100 kb from chromosome 22).
| Method | Time in Minutes for 16 Target Samplesa | Time in Minutes for 128 Target Samples | Allocated CPUsb |
|---|---|---|---|
| GeneImp; single-window | 6.9 | 24.4 | 16 parallel processes (doMC R package) |
| BEAGLE | 615 | 2646 | 32 parallel threads (nthreads option) |
We report the elapsed time between start and finish of the experiment. This was recorded using the time unix command for BEAGLE and the proc.time() R function for GeneImp. The server was solely used for this job during this time.
Experiments performed on a server with 4× Intel Xeon E5-4650 processors, with 2.70 GHz and eight cores (16 threads) each and 1 TB RAM.
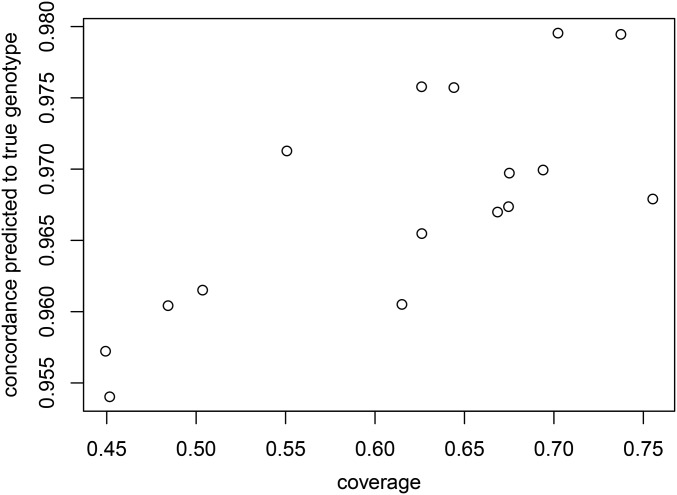
Minimum sequencing coverage
Imputation quality is proportional to the sequencing coverage of each sample. Figure 3 plots the concordance between true and predicted genotypes against the average coverage for each of the 16 samples. As expected, imputation quality improves with higher sequencing coverage.
Figure 3.
Concordance against sequencing coverage. Imputation quality improves with higher sequencing coverage.
Discussion
We described GeneImp, an algorithm that performs genotype imputation to a dense reference panel using genotype likelihoods as inputs, and evaluated its performance on data produced via ultralow coverage sequencing. Although imputation from ultralow coverage sequencing has been shown to be experimentally feasible (Pasaniuc et al. 2012), this is the first study to demonstrate a computational method that can scale up to whole-genome imputation in this setting.
We compared the imputation quality of GeneImp to that of BEAGLE v.4.0, the algorithm used in Pasaniuc et al. (2012) to demonstrate the proof-of-concept that imputation from ultralow coverage sequencing data is possible. Our results show that, although BEAGLE v.4.0 remains state-of-the-art for imputation quality, GeneImp achieves imputation quality very close to that of BEAGLE, using one to two orders of magnitude less time, and without an increase in memory complexity.
Recently, Davies et al. (2016) developed STITCH, a method for imputing genotypes from sequencing data without the use of a reference panel. Their method is motivated by the need for genotype imputation in nonhuman species where SNP genotyping arrays have not been developed, or do not work well, and large reference panels are not available. They showed that STITCH was able to impute genotypes from ultralow coverage sequencing (0.15×) in outbred mice with high accuracy (mean- between 0.948 and 0.972). Furthermore, they showed that in a human cohort of 11,670 Han Chinese sequenced to coverage, STITCH outperformed BEAGLE without a reference panel (mean- of 0.922 and 0.886, respectively), and was slightly outperformed by BEAGLE with a reference panel (mean- of 0.943), though in the latter case BEAGLE took 7.3 times longer to run. When the sequencing depth was reduced to the coverage examined in this work, the performance of STITCH dropped (mean- between 0.8 and 0.82 for coverage, and between 0.58 and 0.7 for coverage for target sample sizes ranging from 2000 to 12,000 individuals). Therefore, in the case of ultralow coverage sequencing (<1×), and an available reference panel, GeneImp would give the best trade-off between imputation quality and efficiency, with a mean- above 0.9.
Similar to many imputation algorithms, GeneImp’s inference complexity is driven by the computation of the hidden state space. Our effective reduction of the diploid hidden state-space introduced by the haplotype filtering step keeps GeneImp’s time complexity subquadratic in the number of reference haplotypes, making it a practical choice when large reference panels are available. In addition to its comparably low algorithmic complexity, GeneImp is also highly parallelizable, since we can perform inference independently for each sample, each window and each window-split, and we only need to combine results from different window-splits in the end. This is in contrast to many iterative schemes, for instance based on Markov-Chain Monte Carlo or Expectation-Maximization, where results typically need to be combined after each iteration, and convergence of the iterative process must be evaluated. Therefore, additional scalability of GeneImp can be achieved by taking advantage of existing computing infrastructures that offer large, shared-memory supercomputers, while further computational speed-ups can be developed through a GPU implementation, which can take advantage of this type of parallelizable computation.
The settings for the GeneImp tuning parameters reported here should work well for imputation of European individuals with ultralow coverage sequencing (0.5×). Guidelines on how to set the window-size parameter when deeper sequencing is available are given in the GeneImp R package (Data availability). We recommend running GeneImp two or three times, each time with a different value for the window-size parameter, and computing the imputation probabilities as a flat average over runs. To evaluate the quality of imputation in the absence of SNP-chip data, we suggest “masking” a number of loci with confident genotype calls, i.e., setting their genotype likelihoods to missing, and assessing how well the imputed genotypes match the original calls. This only needs to be performed in a small region of the genome.
A useful extension for GeneImp, which we plan to implement in the future, is an option for phasing. The program already finds the most likely haplotype pair within each window, and thus we only need to add a method for stitching together the most likely haplotype pairs based on the overlapping segments of adjacent windows.
Our evaluation of GeneImp was based on imputation of ultralow coverage sequencing data. In contrast to SNP arrays, the cost of sequencing depends on the required read depth, which leads to a trade-off between sample size and read depth for a fixed amount of resources (Sims et al. 2014). Although the cost of SNP arrays has fallen more rapidly than the cost of sequencing in recent years, it is not clear how these costs will compare in the future. Therefore, the capability offered by GeneImp to impute ultralow coverage sequencing data to a dense reference panel accurately and efficiently on a genome-wide scale is an important consideration when examining the cost-effectiveness of different technologies for the design of new genomic studies.
The successful application of GeneImp to ultralow coverage sequencing data motivates an interesting future usage: acquiring whole-genome imputation in existing deep whole-exome sequencing datasets based on their off-target reads. In recent years, whole-exome sequencing has been used extensively to explore rare variation (Norton et al. 2012; Tennessen et al. 2012; Lek et al. 2016), since it is more cost-effective, and findings are easier to interpret compared to whole-genome sequencing. According to a recent review (Sims et al. 2014), whole-exome sequencing would require an 80× average read depth of targeted regions to cover ∼90% of targeted bases by at least 10-fold. At this level of average coverage for the exome, we would expect a 0.2–0.6× average coverage of the rest of the genome from off-target reads, depending on the exome capture system, with Illumina’s Nextera and TrueSeq technologies giving the highest expected whole-genome coverage (Chilamakuri et al. 2014). A 0.5× average read depth was used for the ultralow coverage whole-genome sequencing data analyzed in this work.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.200063/-/DC1.
Acknowledgments
We thank all the co-investigators in the PROMISERA project, including Duncan Porter, Iain B. McInnes, and Caron Paterson, and our colleagues at the Stratified Medicine Scotland Innovation Centre, including Hannah Child and Carolyn Low. We thank Scott Howell for drawing the GeneImp logo. Pharmatics acknowledges support of European Union (EU) FP7 MIMOmics.
Footnotes
Communicating editor: J. Shendure
Literature Cited
- 1000 Genomes Project Consortium, Auton A., Brooks L. D., Durbin R. M., Garrison E. P., et al. , 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Olama A. A., Kote-Jarai Z., Berndt S. I., Conti D. V., Schumacher F., et al. , 2014. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat. Genet. 46: 1103–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur R., O’Connell J., Schulz-Trieglaff O., Cox A. J., 2015. Rapid genotype refinement for whole-genome sequencing data using multi-variate normal distributions. Bioinformatics 32: 2306–2312. [DOI] [PubMed] [Google Scholar]
- Baker, S. C., 2013 Next-generation sequencing vs. microarrays: is it time to switch? GEN BioPerspectives. Available at: http://www.genengnews.com/gen-articles/next-generation-sequencing-vs-microarrays/4689.
- Berndt S. I., Gustafsson S., Mägi R., Ganna A., Wheeler E., et al. , 2013. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet. 45: 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning B. L., Browning S. R., 2009. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84: 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning B. L., Browning S. R., 2016. Genotype imputation with millions of reference samples. Am. J. Hum. Genet. 98: 116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S. R., Browning B. L., 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81: 1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S. R., Browning B. L., 2011. Haplotype phasing: existing methods and new developments. Nat. Rev. Genet. 12: 703–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilamakuri C. S. R., Lorenz S., Madoui M.-A., Vodák D., Sun J., et al. , 2014. Performance comparison of four exome capture systems for deep sequencing. BMC Genomics 15: 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly M. J., Rioux J. D., Schaffner S. F., Hudson T. J., Lander E. S., 2001. High-resolution haplotype structure in the human genome. Nat. Genet. 29: 229–232. [DOI] [PubMed] [Google Scholar]
- Davies R. W., Flint J., Myers S., Mott R., 2016. Rapid genotype imputation from sequence without reference panels. Nat. Genet. 48: 965–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O., Zagury J.-F., Marchini J., 2013. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 10: 5–6. [DOI] [PubMed] [Google Scholar]
- De La Vega F. M., Isaac H., Collins A., Scafe C. R., Halldórsson B. V., et al. , 2005. The linkage disequilibrium maps of three human chromosomes across four populations reflect their demographic history and a common underlying recombination pattern. Genome Res. 15: 454–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium, South Asian Type 2 Diabetes (SAT2D) Consortium, Mexican American Type 2 Diabetes (MAT2D) Consortium, Type 2 Diabetes Genetic Exploration by Next-generation sequencing in multi-Ethnic Samples (T2D-GENES) Consortium et al. , 2014. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 46: 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A., McGovern D. P. B., Barrett J. C., Wang K., Radford-Smith G. L., et al. , 2010. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 42: 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsberger C., Abecasis G. R., Hinds D. A., 2015. minimac2: faster genotype imputation. Bioinformatics 31: 782–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Lipids Genetics Consortium, Willer C. J., Schmidt E. M., Sengupta S., Peloso G. M., et al. 2013. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45: 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B., Marchini J., Stephens M., 2011. Genotype imputation with thousands of genomes. G3 1: 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G. R., 2012. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44: 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B. N., Donnelly P., Marchini J., 2009. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Howie B., McCarthy S., Memari Y., Walter K., et al. , 2015. Improved imputation of low-frequency and rare variants using the UK10k haplotype reference panel. Nat. Commun. 6: 8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd P. J., Nelson C. J., 2009. Advantages of next-generation sequencing vs. the microarray in epigenetic research. Brief. Funct. Genomics 8: 174–183. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium , 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan M. I., Ghahramani Z., Jaakkola T. S., Saul L. K., 1999. An introduction to variational methods for graphical models. Mach. Learn. 37: 183–233. [Google Scholar]
- Kronisch C., McLernon D. J., Dale J., Paterson C., Ralston S. H., et al. , 2016. Brief report: predicting functional disability: one year results from the Scottish early rheumatoid arthritis inception cohort. Arthritis Rheumatol. 68: 1596–1602. [DOI] [PubMed] [Google Scholar]
- Lek M., Karczewski K. J., Minikel E. V., Samocha K. E., Banks E., et al. , 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Stephens M., 2003. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165: 2213–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ruan J., Durbin R., 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18: 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Willer C. J., Ding J., Scheet P., Abecasis G. R., 2010. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 34: 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J., Schwartzentruber J., Lalonde E., Montpetit A., Jabado N., 2011. What can exome sequencing do for you? J. Med. Genet. 48: 580–589. [DOI] [PubMed] [Google Scholar]
- Marchini J., Howie B., 2010. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 11: 499–511. [DOI] [PubMed] [Google Scholar]
- Marchini J., Howie B., Myers S., McVean G., Donnelly P., 2007. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39: 906–913. [DOI] [PubMed] [Google Scholar]
- McCarthy S., Das S., Kretzschmar W., Durbin R., Abecasis G., et al. , 2015. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 48: 1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillan R., Leutenegger A.-L., Abdel-Rahman R., Franklin C. S., Pericic M., et al. , 2008. Runs of homozygosity in European populations. Am. J. Hum. Genet. 83: 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton N., Robertson P. D., Rieder M. J., Züchner S., Rampersaud E., et al. , 2012. Evaluating pathogenicity of rare variants from dilated cardiomyopathy in the exome era. Circ. Cardiovasc. Genet. 5: 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasaniuc B., Rohland N., McLaren P. J., Garimella K., Zaitlen N., et al. , 2012. Extremely low-coverage sequencing and imputation increases power for genome-wide association studies. Nat. Genet. 44: 631–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani B., Tekin M., Mahdieh N., 2014. The promise of whole-exome sequencing in medical genetics. J. Hum. Genet. 59: 5–15. [DOI] [PubMed] [Google Scholar]
- Scheet P., Stephens M., 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78: 629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims D., Sudbery I., Ilott N. E., Heger A., Ponting C. P., 2014. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 15: 121–132. [DOI] [PubMed] [Google Scholar]
- Tennessen J. A., Bigham A. W., O’Connor T. D., Fu W., Kenny E. E., et al. , 2012. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337: 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UK10K Consortium, Walter K., Min J. L., Huang J., Crooks L., et al. , 2015. The UK10K project identifies rare variants in health and disease. Nature 526: 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E. L., Auger H., Jaszczyszyn Y., Thermes C., 2014. Ten years of next-generation sequencing technology. Trends Genet. 30: 418–426. [DOI] [PubMed] [Google Scholar]
- VanRaden P. M., O’Connell J. R., Wiggans G. R., Weigel K. A., 2011. Genomic evaluations with many more genotypes. Genet. Sel. Evol. 43: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Lu J., Yu J., Gibbs R. A., Yu F., 2013. An integrative variant analysis pipeline for accurate genotype/haplotype inference in population NGS data. Genome Res. 23: 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterstrand, K. A., 2016 DNA sequencing costs: data from the NHGRI genome sequencing program (GSP). Available at: www.genome.gov/sequencingcosts. Accessed: March 31, 2016.
- Xing E. P., Jordan M. I., Russell S., 2003. A generalized mean field algorithm for variational inference in exponential families. Proceedings of UAI, San Francisco, CA, pp. 583–591. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GeneImp code is available as an R package that can be downloaded from https://pm2.phs.ed.ac.uk/geneimp/. The target samples used in this work are from the SERA study. The data custodian is the Stratified Medicine Scotland Innovation Centre (SMS-IC). Managed access to the raw sequence data and the chip genotypes can be negotiated with SMS-IC, who may be contacted at Admin@stratmed.co.uk. The software packages and reference datasets we used in the analyses reported in this manuscript, including data preprocessing steps, can be accessed as follows.
1000 Genomes project, June 2013 phase 3 release downloaded from the ftp server at ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/.
UK10K project: ALSPAC cohort accession number EGAD00001000195, TWINSUK cohort accession number EGAD00001000194.
ORCADES study: http://www.orcades.ed.ac.uk/orcades.
BEAGLE v.4.0 software (beagle.r1399.jar): https://faculty.washington.edu/browning/beagle/b4_0.html.
Picard Tools (v 1.128): http://broadinstitute.github.io/picard/.
Genome Analysis ToolKit (GATK) suite (McKenna et al. 2010) v3.3-0-g37228af: http://www.broadinstitute.org/gatk.