Abstract
We present a microfluidic device, which enables single cells to be reliably trapped and cultivated while simultaneously being monitored by means of multifrequency electrical impedance spectroscopy (EIS) in the frequency range of 10 kHz–10 MHz. Polystyrene beads were employed to characterize the EIS performance inside the microfluidic device. The results demonstrate that EIS yields a low coefficient of variation in measuring the diameters of captured beads (~0.13 %). Budding yeast, Saccharomyces cerevisiae, was afterwards used as model organism. Single yeast cells were immobilized and measured by means of EIS. The bud growth was monitored through EIS at a temporal resolution of 1 min. The size increment of the bud, which is difficult to determine optically within a short time period, can be clearly detected through EIS signals. The impedance measurements also reflect the changes in position or motion of single yeast cells in the trap. By analyzing the multifrequency EIS data, cell motion could be qualitatively discerned from bud growth. The results demonstrate that single-cell EIS can be used to monitor cell growth, while also detecting potential cell motion in real-time and label-free approach, and that EIS constitutes a sensitive tool for dynamic single-cell analysis.
Keywords: Microfluidics, Single-cell analysis, Electrical impedance spectroscopy, Cell trapping, S. cerevisiae
Introduction
Cellular heterogeneity is a fundamental characteristic in any population of cells [1–4]. With the emergence of single-cell analysis methods, researchers have gained new insights into the mechanisms and kinetics of single-cell processes in cell biology, systems biology, and clinical biology [5–7]. The two most frequently used methods for single-cell analysis are flow cytometry [8] and microscopy. Flow cytometry provides high-throughput readout of cellular information based on fluorescent markers in suspended cells. However, cells of a sampled population are measured at a defined time point while moving through the detector of the instrument, and therefore, only momentary cellular information can be extracted. It is difficult to track individual cells over an extended time with high temporal resolution. Time-lapse microscopy, in contrast, is a powerful method to obtain information on dynamic cellular behavior and detailed information on intracellular components. The resulting data can be assigned to monitored individual cells. Flow cytometry and microscopy imaging require fluorescently labeled cells or cellular compartments [9, 10]. The labeling procedure, either by directly applying fluorescent stains, or by transfection of genetically encoded fluorophores into the cellular genome, may interfere with cellular functions.
The rapid development of microfabrication technologies has fueled the application of microfluidic or lab-on-a-chip systems in the fields of biology, chemistry, and medical diagnosis. Microfluidic systems, which may include multiple functions, provide new means to manipulate and study single cells [11, 12]. For instance, alternating current (AC) electrokinetics and electrical impedance spectroscopy (EIS) are electrical techniques that can be used to identify individual cells based on their size and dielectric properties [13, 14] in a noninvasive and label-free approach. AC electrokinetics, primarily electrorotation (ROT) and dielectrophoresis (DEP), have been used to study the motion of cells exposed to an external AC electric field [15–17]. The dielectric properties of cells can be extracted from the measured ROT spectrum after an analysis based on a simplified single-shell model of the biological cells [18]. However, the throughput of ROT is limited, since it usually takes seconds to measure the rotation speed of a single cell. DEP, integrated within microfluidic systems, is more applicable to the manipulation and separation of single cells than their analysis [19, 20]. In contrast, EIS enables the frequency-dependent multiparameter readout of cellular and even subcellular information in a high-throughput setting [21, 22] or dynamically [23]. EIS provides information related to the cell size at lower frequencies (from hundreds of kHz to MHz), related to the cell membrane capacitance at higher frequencies (several MHz), and information on intracellular features at even higher frequencies. By using microfabrication techniques, EIS can be integrated in microfluidic systems to detect single cells at high throughput and with good sensitivity.
Microfluidic devices with integrated EIS functions have been developed already for single-cell analysis in the past decade [13, 14, 21]. Most of those microfluidic devices, called electrical impedance cytometers, were used to characterize suspended biological samples in a flow-through setup. Holmes et al. has used impedance cytometry to identify T lymphocytes, monocytes, and neutrophils and, later, count CD4+ T cells in human whole blood for a point-of-care blood diagnostic system [24, 25]. Impedance cytometry has also been used to characterize cell disease states [26] and identify the differentiation state of single stem cells [27]. Chen et al. [28] and Zheng et al. [29] have designed a specific impedance cytometer in which single cells were deformed to pass through a constriction channel by aspiration. They used the measured impedance and transit time to classify different cell types. In order to measure single cells continuously through EIS, the function of cell seeding or cell immobilization has to be integrated into microfluidic systems by means of specifically designed electrode geometries or fluidic structures. Asphahani et al. has cultured adherent cells on metal electrodes and recorded the cellular response to drug treatment over a longer time period [30]. Ghenim et al. has monitored cell migration of single cells through EIS [31]. The variations in the measured impedance could be attributed to cell attachment, cell-substrate interaction, and cell motility. Park et al. [32] and Lan and Jang [33] have compared impedance magnitudes before and after capturing a single cell in a microfluidic trap, which enabled them to identify the presence/absence of single cells. Malleo et al. has shown the time-dependent measurements of single cell impedance in response to dynamic chemical perturbations [34].
To the best of our knowledge, EIS has not yet been used for real-time monitoring of cellular dynamics, e.g., cell growth, at single-cell resolution. In this work, we have integrated EIS into a microfluidic device for single-cell analysis. The concept of single-cell manipulation has been characterized and validated previously [35, 36] and has been adapted and extended with specifically designed electrodes. The device features reliable immobilization and the possibility to cultivate of single cells under controlled environmental conditions while performing real-time impedance measurements of the immobilized cells. Polystyrene beads have first been used to characterize the EIS function through size measurements of beads. Then, budding yeast, Saccharomyces cerevisiae, has been employed as a biological model organism in subsequent experiments. Different cell shapes during cell growth could be discriminated by using multifrequency EIS data. Moreover, cell immobilization in different orientations could be discerned. Finally, we have been able to continuously monitor the budding process and to detect potential movements of immobilized single yeast cells during measurements by extracting representative vectors from the multifrequency EIS data.
Materials and methods
Design and fabrication of the device
Figure 1a, b schematically shows the microfluidic single-cell EIS device and a 3D close-up of a cell trap. The fluidic network of the device consists of a cell-culturing channel (150 μm wide), a suction channel (300 μm wide), and 10 cell traps that are formed as bottleneck-like horizontal orifices with 4-μm-wide necks. The cell traps, interconnecting the cell-culturing and suction channels, are located at one sidewall of the cell-culturing channel. Only one trap is used and monitored at a time during impedance measurements. The cell suspension and medium are continuously delivered into the cell-culturing channel at a flow rate of 0.5 μl/min. To capture cells, an underpressure in the range of −30 to −5 mbar is applied to the suction channel via a precise pressure controller, and single cells are dragged towards the side of the channel by hydrodynamic forces. As soon as a single cell is captured at the monitored trap, the pressure is raised to and maintained at a value between +5 and +15 mbar to prevent the capturing of additional cells. The flow (total flow rate of 1 μl/min) generates a relatively high pressure in the cell-culturing channel, so that there is still a sufficient pressure difference across the trap to reliably retain the immobilized cell at the trap, although the pressure has been raised to positive values (overpressure). The required pressure values have to be optimized at the beginning of each set of experiments.
Fig. 1.
Microfluidic single-cell EIS device and its fabrication process. a Schematic top view of the device without PDMS cover for better visibility. b A close-up of the area for single-cell immobilization and impedance measurements. The inset shows a 3D close-up of a single-cell trap with an immobilized cell. c Fabrication sequence of the microfluidic device. Cross-sectional view along the dashed line AA′ highlighted in b (dimensions are not to scale, see text for details)
The EIS function is integrated into the microfluidic device by patterning a common electrode (20 μm wide) serving as the stimulus electrode in the cell-culturing channel and individual electrodes (20 μm wide), working as recording electrodes, at each respective trap. When a cell is immobilized at a trap, the impedance is measured by using the stimulus electrode and the corresponding recording electrode. The narrow neck of the bottleneck-like orifice has two functions. First, it prevents cells from passing into the suction channel, since the width of the neck is smaller than the diameter of the used cells. Second, the orifice constrains the electric current to flow through the small opening of the neck towards the recording electrodes. Any variation of the cross-sectional opening of the neck caused by bead/cell immobilization or cell growth will lead to a substantial change in the impedance signal and so that there is a high sensitivity of the impedance measurement to any change in the orifice. Potential electric crosstalk between adjacent electrodes is reduced with a SiNx insulation layer, which has been deposited over the whole chip surface to cover all metal tracks. This SiNx layer has been reopened only in the sensing regions close to the traps to define the electrodes and along the chip border to provide access to the electrical contact pads.
The microfluidic single-cell EIS device was fabricated by using a hybrid multilayer process as schematically shown in Fig. 1c: (1) 200-nm-thick Pt electrodes with a 20-nm-thick TiW adhesion layer underneath were patterned on the Pyrex glass wafer by a lift-off process. (2) A 500-nm SiNx insulation layer was deposited on the entire wafer by plasma-enhanced chemical vapor deposition (PECVD). (3) This SiNx layer was reopened at the sensing and contact pad regions by reactiveion etching (RIE). (4) A 30-μm-thick layer of SU-8 3025 photoresist (MicroChem, Co., USA) was spin-coated on top of the wafer and patterned to define the microfluidic channels and traps. By using a mask aligner, SU-8 patterns were precisely aligned with the Pt electrodes on the substrate. This alignment ensures accurate positioning of the cell traps between the stimulus and recording electrodes. (5) The wafer was then diced into single chips. The SU-8 surface of each chip was modified with (3-aminopropyl)triethoxysilane (APTES) (Sigma-Aldrich Co., USA) in a vapor phase silanization process. (6) In order to seal the microfluidic channels irreversibly, each chip with the modified SU-8 surface was ultimately bonded to an unstructured poly(dimethylsiloxane) (PDMS) (Sylgard® 184, Dow Corning Co., USA) cover with punched holes for fluidic inlets and outlets. The used materials, glass, SU-8, and PDMS feature excellent light transmittance, except for the 500-nm SiNx layer, which is slightly yellow. However, the SiNx has been etched away in the sensing regions, so that completely transparent regions for optical observation of cell morphology are collocated with cell-trapping sites.
Experimental setup
The assembled microfluidic device was placed on a custom-made aluminum holder, which fits onto an inverted microscope stage (Olympus IX81, Olympus Co., Japan) for imaging. The device was clamped tightly between the aluminum holder and a poly(methylmethacrylate) (PMMA) cover by using screws. A printed circuit board (PCB), comprising manual switches and spring-loaded contacts, was positioned on top of the PMMA cover. These spring-loaded pins contacted the electrode pads on the device when screwed to the aluminum holder. A commercial impedance spectroscope (HF2IS, Zurich Instruments AG, Switzerland) and a transimpedance amplifier (HF2TA, Zurich Instruments AG, Switzerland) were connected to the electrodes on the device via the PCB. For fluidic access, poly(tetrafluoroethylene) (PTFE) tubing (Bohlender GmbH, Germany) was connected through holes in the PMMA cover to the inlets and outlets of the device.
Beads, cell suspensions, and media were initially loaded into glass syringes (ILS Innovative Labor Systeme GmbH, Germany) and then delivered to the cell-culturing channel by using syringe pumps (neMESYS, Cetoni GmbH, Germany). The underpressure for capturing cells was applied to the pressure port of the suction channel by using a pressure controller (DPI 520, Druck Ltd., UK), supplied with in-house compressed air and vacuum. The instruments, including the impedance spectroscope, syringe pumps, and pressure controller, were controlled with a personal computer.
Bead and cell preparation
Commercial monodisperse polystyrene (PS) beads (Fluka, Sigma-Aldrich Production GmbH, Switzerland) with standard diameters of 8 and 10 μm (CV of the diameter calibration is 1.2 %, by manufacturer) were first employed for the EIS characterization inside the microfluidic device. Beads were mixed with 0.01 M phosphate-buffered saline (PBS) solution (Sigma-Aldrich Co., USA). Bead clusters in the suspension were mechanically separated into individual beads through ultrasonic agitation (Bioblock® Scientific 86480, Fisher Scientific GmbH, Germany). Finally, the resulting bead suspension was loaded in a syringe.
Standard methods were used to grow cultures of budding yeast cells (S. cerevisiae). The used yeast cells were a diploid wild-type strain. Cells were grown in a complete synthetic medium made of 0.17 % yeast nitrogen base (YNB) (Difco™, BD GmbH, Germany), 0.5 % ammonium sulfate (Sigma-Aldrich Co., USA), and 2 % glucose sulfate (Sigma-Aldrich Co., USA) at 30 °C. The prepared yeast cell suspension was diluted to reach a concentration of approximately 1×106 cells/ml in the cell-culturing medium. Before delivering the cell suspension and cell-culturing medium into the chip, the fluidic channels were flushed with 1 % bovine serum albumin (BSA) (Sigma-Aldrich Co., USA) solution in order to achieve a bubble-free channel network and a protein-coated channel surface to reduce stickiness for cells.
Electrical impedance spectroscopy
For an experiment, a bead/cell was captured at a trap, and an AC signal (Vsti, amplitude 1 V), swept over a frequency range from 10 kHz to 10 MHz and including 92 sampling frequencies, was applied to the stimulus electrode by the impedance spectroscope. The resulting signal, received by the respective recording electrode, was amplified (gain G ) and converted to a voltage signal (Vrec ) through the transimpedance amplifier, and, ultimately, recorded by the impedance spectroscope. This recorded complex signal was displayed in the format of separated magnitude (A ) and phase (θ ) signals. In parallel, a bright-field image of the immobilized bead/cell was taken by means of the inverted microscope. Afterwards, the bead/cell was released by simply increasing the pressure in the suction channel. To measure the baseline characteristics (Ae and θe ), which must be consistent during a series of measurements, the same impedance measurement was performed for the empty trap before and after particle immobilization. This measurement procedure was repeated for each immobilized bead/cell and provided a set of electrical impedance data with corresponding images for subsequent analysis. The budding process of immobilized yeast cells was monitored by using both EIS and time-lapse imaging at an interval of 1 min. The reference measurements of the empty trap were carried out before and after the cell impedance recording.
Deduced from Ohm’s law, the measured impedance can be expressed as follows:
| (1) |
| (2) |
Vsti and G are known values of the measurements. The impedance, Z, is therefore inversely proportional to the measured voltage Vrec. Hence, the magnitude (A) and phase (θ) signals recorded by the impedance spectroscope can be directly used to display impedance variations during the experiments. In order to eliminate the variations of the electrode impedance and the surrounding bulk medium resistance, differential values have been employed: The impedance changes resulting from the immobilized bead/cell were calculated relative to the values of the signals of the empty trap. The relative magnitude (Ar=A/Ae) was defined by dividing the magnitude signal when a bead/cell was trapped, through the magnitude signal of the empty trap. The relative phase (θr=θ−θe) was defined by subtracting the phase signal of the empty trap from that of the trap with an immobilized bead/cell.
Data analysis of multifrequency EIS signals
To analyze if the EIS signals alone contain enough information to determine the orientation and growth state of each cell, we represented each measurement of a cell as a vector in a 184-dimensional feature space (92 dimensions for the relative magnitude and 92 dimensions for the relative phase at 92 frequencies). Having more dimensions than data points might, however, pose problems for classification through, e.g., over fitting. Therefore, we projected all data points into a low-dimensional subspace: we used principal component analysis (PCA) [37], an established orthogonal projection method. We computed the covariance of all data points and projected each data point on its first two eigenvectors, the “principal components” (PCs). We estimated the number of necessary PCs so that more than 80 % of the variance of the original data was represented in the projected subspace. All data points in the subspace were then classified using linear discriminant analysis (LDA) [38] and leave-one-out cross validation. If the group, which a cell was assigned to, was identical to the original group, we counted it as a correct classification, otherwise as an error.
Next, we investigated if cell motion and bud growth could be discriminated according to the associated impedance signal variations. In contrast to the method detailed above, we here used temporally resolved data, i.e., multiple measurements per cell. We computed, for each cell individually, the first PC over all measurements. We averaged all first PCs of cells that were visually identified as either growing or moving and obtained two vectors, which were normalized to unit length: pgrowth and pmotion . The two vectors represent the dimensions, along which the cell motion and bud growth cause largest variations in the EIS data. For better visualization, these two vectors were orthogonalized:
| (3) |
| (4) |
Then, all recorded impedance data points were projected in the the space spanned by p̂growth and p̂motion, to recognize and qualify the effects of bud growth and cell motion.
Results and discussion
Characterization of EIS
To characterize the functionality and sensitivity of the microfluidic single-cell EIS device, we performed impedance measurements of immobilized PS beads with two different diameters, 8 and 10 μm. Figure 2 shows two images of immobilized beads as well as the measurement results of 72 beads in total, plotted as relative magnitude at 1 MHz. One megahertz was chosen as a common frequency in the bead characterization here and the cell measurements later on. The two groups of beads can be clearly discriminated, as can be seen in Fig. 2b. The mean value of the relative magnitude at 1 MHz in the case of 10-μm beads is 0.885±0.0011, which is lower than that of 8-μm beads, 0.904±0.0012. The signal difference between the two groups of beads is caused by the difference in the cross-sectional opening of the cell-trapping orifice obstructed by the immobilized beads. A 10-μm bead obstructs more of the orifice cross-sectional area than an 8-μm bead, thereby remaining a smaller cross-sectional opening, through which the electric current flows. Therefore, the measured impedance is higher when a 10-μm bead is immobilized, which leads to a lower relative signal magnitude (Ar=A/Ae) than that upon immobilization of an 8-μm bead. Moreover, the small CV (~0.13 %) within each group of beads demonstrates the high sensitivity and precision of the microfluidic single-cell EIS device in measuring the size of immobilized particles.
Fig. 2.
Characterization results of the microfluidic single-cell EIS device by using PS beads. a Images of immobilized single 10- and 8-μm beads at a trap. Scale bar is 5 μm. b Histograms of impedance measurements of 10-μm (n=31) and 8-μm (n=41) beads plotted as the relative magnitude Ar=A/Ae
Measuring cell growth states of budding yeast through EIS
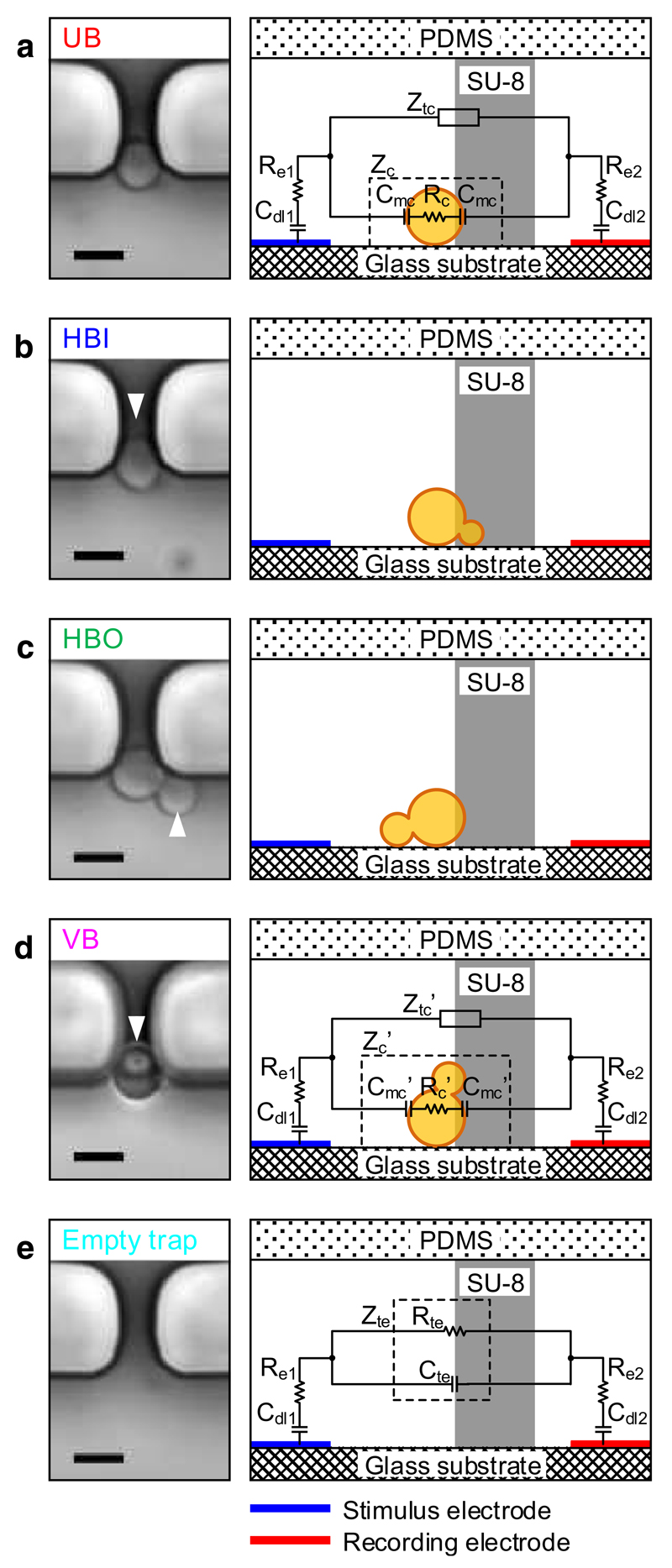
Cell growth of budding yeast is accompanied with corresponding morphological changes. Cells can be classified into unbudded and budded cells in a first simple approach. We observed that cells with small buds tend to lie down flat on the horizontal substrate with the bud pointing inside or outside the trapping orifice as a consequence of hydrodynamic forces. In contrast, cells at a more advanced growth state with larger buds, i.e., with buds of a diameter larger than the width of the cell-trapping orifice, can be retained at the trap in a vertical position. Figure 3 shows representative microscopy images of typical orientations of immobilized single yeast cells and their corresponding cross-sectional views in schematics, including an unbudded cell (UB, Fig. 3a), a horizontally immobilized cell with the bud inside the trap (HBI, Fig. 3b), a horizontally immobilized cell with the bud outside the trap (HBO, Fig. 3c), and a vertically immobilized cell with the bud and mother cell stacked vertically (VB, Fig. 3d).
Fig. 3.
Images of immobilized single yeast cells in all observed orientations and their corresponding cross-sectional views in schematics with the impedance components of the equivalent circuits. a UB, unbudded cell. b HBI, horizontally immobilized cell with bud inside the trap. c HBO, horizontally immobilized cell with bud outside the trap. d VB, vertically immobilized cell with mother cell and bud stacked vertically. e Empty trap. Buds are marked with arrowheads. Scale bar is 5 μm
For an empty trap (Fig. 3e), the measured impedance includes the electrical double-layer capacitances (Cdl1, Cdl2) and the resistances (Re1, Re2) of both electrodes, and the impedance (Zte ) of the bulk medium across the empty trap, which consists of a resistance (Rte) and a capacitance (Cte) in parallel. Thus, the impedance across the stimulus and recording electrodes is as follows:
| (5) |
For a trap with an immobilized cell as shown in Fig. 3a–d, an additional element, the impedance (Zc or Zc′) of the immobilized cell, is added in parallel to the impedance (Ztc or Ztc′) of the bulk medium across the trap. The cell impedance is composed of the cell membrane capacitance (Cmc or Cmc′) and the Ohmic cell resistance (Rc or Rc′). Hence, the modeled impedance of a trap with an immobilized unbudded cell in Fig. 3a is the following:
| (6) |
The modeled impedance of a trap with a budded cell in Fig. 3d can be expressed as follows:
| (7) |
From the three formulas above, we can see that several factors collectively influence the measured impedance of a trap with an immobilized cell: The impedance of the electrodes, the impedance of the bulk medium across the trap (Zte, Ztc, or Ztc′), and the impedance of the immobilized cell (Zc or Zc′). Generally, the impedance of electrodes, including the resistances (Re1 and Re2) and the double-layer capacitances (Cdl1 and Cdl2), are present in measurements at low frequencies, typically below 100 kHz. This portion of the impedance stays relatively constant whether there is a cell immobilized or not, because it depends on the characteristics of the electrode and the surrounding medium. When a cell is immobilized, the impedance of the bulk medium across the trap, Ztc or Ztc′, increases, since the yeast cell with its volume blocks a significant portion of the trapping orifice and forces the electric current to flow around the obstacle through a reduced opening. The impedance, therefore, is dominated by the parallel resistance of the cell and the trap, which will consequently show a variation in the magnitude spectrum of the EIS signals at frequencies typically below 1 MHz. At higher frequencies, from hundreds of kilohertz on, the electric current starts to penetrate the cell wall and plasma membrane partially. Thus, the capacitance of the cell membrane will lead to a variation in the phase spectrum especially around several megahertz, in clear contrast to the phase spectrum of an empty trapping site. This variation is most pronounced for a VB cell (Fig. 3d), which blocks the largest fraction of space of the trapping orifice and features more cell membrane areas in comparison to a UB cell. Therefore, it is expected that UB and VB cells are distinguishable according to their impedance magnitude and phase signals.
Figure 4 shows the results of impedance measurements of immobilized cells in all four orientations that have been mentioned above and of the empty trap as reference. Among the four groups of cells, one can observe differences in the magnitude spectra of EIS signals, especially at low frequencies from 100 kHz to a few megahertz, while the differences in the phase spectra are more pronounced at high frequencies above 1 MHz (Fig. 4a). The relative magnitude and phase signals in Fig. 4b, c give a better representation of those signal differences. Selected frequencies at which the relative magnitude and phase vary most between the four groups have, therefore, been chosen to classify the orientations of immobilized cells. The largest differences can be observed in the relative magnitude spectra at around 1 MHz and in the relative phase spectra at around 4 MHz. By using the corresponding data at those two frequencies, VB cells can be clearly separated from the other cells, as shown in Fig. 4d. The differences between the EIS signals of the VB cells and the other groups of cells mainly arise from the additional volume of the bud. Compared to horizontally immobilized cells, the vertical stack of bud and mother cell of a VB cell obstructs a larger cross-sectional area of the cell-trapping orifice, which is reflected in the EIS signals. In some cases, VB-classified cells do not feature a perfectly vertical stack of bud and mother cell. An example is inserted as a picture in Fig. 4d. The values of the EIS signals are, in this case, closer to those of horizontally immobilized cells.
Fig. 4.
Multifrequency EIS of immobilized single yeast cells: UB (n= 34), HBI (n=10), HBO (n=13), and VB (n=11), with reference measurements of the empty traps. a Magnitude and phase spectra of raw signals over the swept frequency range from 10 kHz to 10 MHz. b, c Relative magnitude and phase spectra. The color curves and shaded regions in a, b, and c represent the mean values and standard deviations in the measurements, respectively. d Separation of VB cells by using the relative magnitude at 1 MHz versus the relative phase at 4 MHz shown in a scatter plot. e Separation of HBI cells by using the relative magnitude at 100 kHz versus the relative phase at 900 kHz shown in a scatter plot. Inserts show images of cell samples. Buds are marked with arrowheads. Scale bar is 5 μm. f Discrimination of UB, HBI, and HBO cells by means of LDA on the full multifrequency data set (projections on first two PCs shown). Colored areas depict the regions in which a cell sample is assigned to the respective group, and dashed lines show classification boundaries
HBI cells can be discriminated from the data clusters of UB, HBI, and HBO cells by means of the relative magnitude and phase signals at other frequencies, as shown in Fig. 4e. Distinct signal differences of immobilized HBI cells occur at around 100 kHz in the relative magnitude spectra (Fig. 4b) and at around 900 kHz in the relative phase spectra (Fig. 4c), compared to those of VB, HBO, and UB cells. Plotting the signals of all measured cells at these two frequencies allows for separating three clusters of cell orientations. The buds of HBI cells are oriented horizontally towards the inside of the trap. This orientation of buds, in contrast to the vertically stacked buds, obstructs less of the cross-sectional opening of the cell-trapping orifice and consequently leads to a smaller impedance change. Further, compared to UB cells, the buds of HBI cells are still situated in the opening of the orifice, thereby influencing the impedance values. However, some cells with small buds, as for example, the cell shown in the left image inserted in Fig. 4e, are difficult to distinguish from UB and HBO cells solely on the basis of EIS data.
It is difficult to discriminate HBO cells from UB cells by simply looking at the spectra of the relative signals over the whole frequency range, since the buds of HBO cells are often dragged to the downstream side of the trap by flow-induced hydrodynamic forces (see Fig. 3c). In such a scenario, a bud, even though it may feature a big volume, does not produce any effective contribution to cross-sectional obstruction of the cell-trapping orifice, so that no distinctive feature or change in the EIS signal is observed. The multifrequency EIS data were, therefore, analyzed by means of PCA. Since VB cells can be directly discriminated through the EIS data (Fig. 4d), only UB, HBI, and HBO cells have been classified by LDA based on the first and second PCs (accounting for 89 % of the variance), through PCA, as shown in Fig. 4f. The three orientations of immobilized cells can be classified into 3 groups with 14 out of 57 misclassifications (20 % average error rate per class, leave-one-out cross-validation, 67 % chance error rate). It can be seen that some HBO and UB cells are not located with their class boundaries, which shows the limitation of the current system. Some of the misclassifications can be explained by the small size of the bud, which, for an HBO-classified cell, lies outside of the trap and only minimally contributes to an impedance variation. Therefore, HBO cells with small buds show, over all frequencies, EIS signal values that are similar to those of UB cells.
Overall, all four orientations of immobilized single yeast cells can be discriminated either by directly using the relative impedance signals at specific frequencies or by using statistical analysis of the multifrequency EIS data. VB cells display more distinct features that can be used for classification in comparison to other cell orientations. Therefore, the vertical immobilization of single yeast cells yields the highest sensitivity for monitoring cell growth and the budding process by means of EIS and will be used in the next sections.
Real-time monitoring of bud growth through EIS
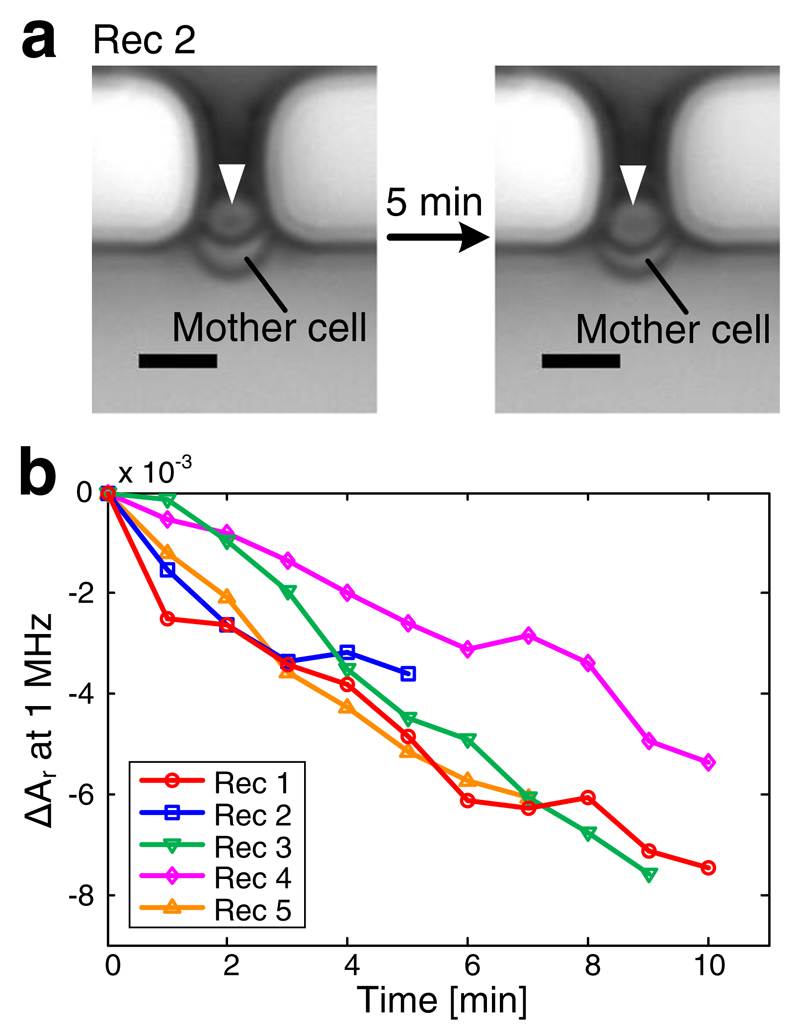
Budding yeast, S. cerevisiae, proliferates in that the new cell (daughter) develops from a bud on the old cell (mother) at one particular site. The dynamic budding process of vertically immobilized budding cells was recorded in real-time by using simultaneously EIS and time-lapse imaging. Figure 5a shows the microscopy images of an immobilized yeast cell at the beginning and end of a recording. Figure 5b shows five independent EIS-based recordings of the budding process of five different yeast cells plotted as the variation of relative magnitude at 1 MHz versus time. It can be clearly seen that the relative magnitude signals of all five cells decrease over the recording period. The magnitude decrease is attributed to the growth of the bud, which obstructs more and more cross-sectional area of the cell-trapping orifice and, consequently, induces an increase in the measured impedance. The bud growth can be clearly detected through the signal variations in the impedance measurements, even within a comparably short time period (Rec 2). In contrast, the size increase of a bud during 5 min is hardly measurable by optical images (Fig. 5a). The relative magnitude variation between the empty trap before and after the recordings at 1 MHz in Figs. 5 and 6 is below 1×10−3, a value, which is much smaller than the signal variations derived from the cellular dynamics. The measured signals can, therefore, be clearly assigned to bud growth and demonstrate sufficient stability, sensitivity, and temporal resolution of the EIS method for real-time monitoring of vertically immobilized yeast cells.
Fig. 5.
Real-time EIS recordings of the budding process of five immobilized single yeast cells. a Images of an immobilized budding yeast cell at the beginning and the end of Rec 2. Buds are marked with arrowheads. Scale bar is 5 μm. b Growth curves displayed as variations in relative magnitude signals at 1 MHz versus time
Fig. 6.
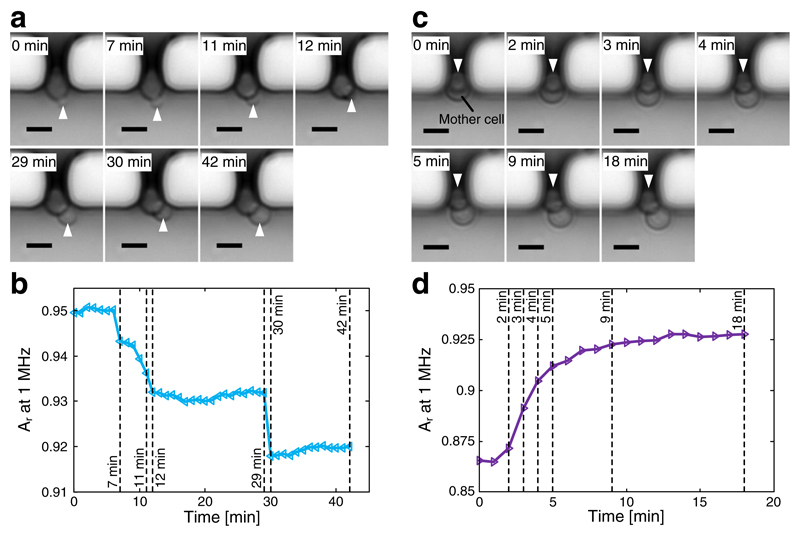
Real-time EIS recordings of cell motion of immobilized budding yeast cells. a Images at defined time points showing an immobilized cell with its bud growing and moving around in the trap. b Recorded bud growth and movement, displayed as relative magnitude signal at 1 MHz versus time. c Images at specific time points showing the bud of an immobilized cell at the trap with the mother cell moving towards the outside of the trap. d Recorded cell movement displayed as relative magnitude signal at 1 MHz versus time. In a and c, buds are marked with arrowheads, and the scale bar is 5 μm
Real-time monitoring of cell motion/growth through EIS
In order to monitor the entire budding process of S. cerevisiae by using EIS, a mother cell with a tiny bud has to be captured, held in place, and will then be recorded. The continuous medium flow along the traps may, however, cause a movement of the immobilized cell, since the bud is initially too small to be clamped in the cell-trapping orifice. Figure 6a shows time-lapse images of a cell starting with a tiny bud and then growing the bud size during 42 min. During the recording, we can observe several movements. At 7 min, the bud together with the mother cell starts to rotate into a more upright position so that the contours of the bud in the optical image partially overlap with that of the mother at around 12 min. This rotation leads to a sudden drop in the magnitude signal of EIS recording (Fig. 6b), as the cell with the bud in a more vertical orientation obstructs more of the orifice opening. The medium flow then pushes the growing bud towards the right side of the orifice, where the further growth of the bud becomes almost invisible in the impedance measurement. Between 29 and 30 min, the immobilized cell again undergoes an upward rotation, which results in a stronger overlap of the bud and mother cell contours in the optical image. This rotation also triggers a steep decrease in the magnitude signal.
Moreover, some cells with an already large bud, which had been initially trapped vertically (VB), also moved and changed their orientations during EIS recording. This motion and reorientation can be attributed to the growth of buds, which squeezed the mother cells out of the trap. Figure 6c shows the time-lapse images of such an event. At 0 min, a budding yeast cell has been immobilized with its bud and mother cell stacked vertically. As the bud grows within the first 5 min, the mother cell is squeezed out of the trap and then remains in the resulting position until the end of the recording (18 min). This cell movement is analogous to a class transition from VB to HBO. Accordingly, the relative magnitude in the real-time EIS recording increases during the movement of the mother cell (Fig. 6d). Once in the horizontal position, the signal remains relatively constant. Compared to the growth-only recordings in Fig. 5b, the curve in Fig. 6d shows different characteristics: the move of the mother cell into a more horizontal position decreases the fraction of obstructed cross-sectional opening area of the cell-trapping orifice, thereby decreasing the measured impedance, which, consequently, gives rise to an increase of the relative magnitude signal.
As is evident from the results above, any motion of the immobilized budding yeast cell, of the mother cell or of the bud, affects the EIS signals to a much larger extent than the potential signal variation during growth or upon a size increase of the bud. However, this issue can, to some extent, be addressed by using the complete multifrequency EIS data set and conducting a statistical analysis as described in the “Data analysis of multi-frequency EIS signals” section. Two vectors, which represent cell growth and cell motion, have been extracted from all data sets and the associated image information (cell relative position with respect to orifice). Then the recordings of Figs. 5 and 6 have been projected with respect to these two vectors. The results are shown in Fig. 7. We cans see that the data points of the bud-growth recordings are aligned with the growth vector, whereas the data points of the recordings that include cell motion are prevailingly aligned along the motion vector. Since the cell in Mot 1 moves from a horizontal position to a more vertical position, whereas the cell in Mot 2 transits from a VB to a HBO orientation, the data points of the two recordings align along opposite directions in Fig. 7. Moreover, the more horizontal sections of Mot 1 reflect the bud growth of the cell, which can be correlated to the respective periods in Fig. 6b (12–29 and 30–42 min). The multifrequency EIS recording data can be used to qualitatively correlate the variation in impedance signals to cell activities, i.e., cell growth and cell motion for the given device and immobilization scenario.
Fig. 7.
Projection of multifrequency EIS data with respect to cell growth and cell motion vectors, which show the possibility to discriminate cell activities during the overall recording duration. Rec 1 to Rec 5 are from the five bud-growth recordings in Fig. 5. Mot 1 and Mot 2 represent the two cell motion/growth recordings in Fig. 6. For each recording, the temporally first data point was set to (0, 0) for better illustration
Conclusion
A microfluidic device that combines immobilization and localized multifrequency electrical impedance measurements of single cells has been presented in this work. Monodisperse polystyrene beads have been used to characterize the device and yielded results that demonstrate the high precision of measuring the size of particles through EIS.
The experiments using budding yeast cells, S. cerevisiae, have validated the functionality and sensitivity of the EIS-integrated microfluidic device. The yeast cells have been captured in different orientations as a consequence of the specific configuration of the cell traps and of the cell morphologies (with buds or without buds). VB and HBI cells have been discriminated from other immobilized single yeast cells directly by using the raw data of the impedance measurements. By analyzing the multifrequency EIS data with PCA, HBO and UB cells have been classified with 20 % misclassification. Among the four potential orientations of immobilized cells, VB cells, where the bud and the mother cell are stacked vertically, can be most sensitively monitored by impedance measurements. The signal contribution of the buds is largest in VB orientation so that this orientation has been chosen to perform real-time monitoring of bud growth. The budding process of vertically immobilized yeast cells has been successfully monitored by using EIS. Only cells with a large enough pre-developed bud can be vertically immobilized and reliably measured as a consequence of the geometry of the cell traps, and the accessible recording periods are relatively short (10 min). Moreover, the immobilized yeast cells are subject to movements induced by either the medium flow or the growth of bud. These movements significantly influence the EIS signals and may completely mask a bud-growth-induced signal variation. By extracting vectors, which represent cell growth and cell motion, from the multifrequency EIS data, a potential movement of immobilized single cells can be identified during the cell growth process.
In summary, the results demonstrate that multifrequency EIS enables real-time monitoring of cell growth and cell motion at immobilization sites. We believe that EIS provides a sensitive approach for dynamic analysis of single-cell proliferation in real-time and label-free monitoring. The microfluidic device and the immobilization geometry (defined cell orientation and stable cell positioning) will have to be optimized so as to allow for long-term impedance recordings without cell movement.
Acknowledgments
The yeast cells used in this study were kindly supplied by Diana Ottoz and Dr. Fabian Rudolf, ETH Zurich, D-BSSE, CSB Group. This work was financially supported through the Swiss SystemX.ch program within the RTD project “CINA”, the Commission for Technology and Innovation (CTI project) in Switzerland, and the ERC Advanced Grant NeuroCMOS (AdG 267351). Zhen Zhu received individual funding from the Chinese Scholarship Council.
References
- 1.Altschuler SJ, Wu LF. Cell. 2010;141:559–563. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dexter DL, Kowalski HM, Blazar BA, Fligiel Z, Vogel R, Heppner GH. Cancer Res. 1978;38(10):3174–3181. [PubMed] [Google Scholar]
- 3.Dexter DL, Spremulli EN, Fligiel Z, Barbosa JA, Vogel R, VanVoorhees A, Calabresi P. Am J Med. 1981;71(6):949–956. doi: 10.1016/0002-9343(81)90312-0. [DOI] [PubMed] [Google Scholar]
- 4.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Science. 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 5.Fritzsch FSO, Dusny C, Frick O, Schmid A. Annu Rev Chem Biomol Eng. 2012;3(1):129–155. doi: 10.1146/annurev-chembioeng-062011-081056. [DOI] [PubMed] [Google Scholar]
- 6.Schubert C. Nature. 2011;480(7375):133–137. doi: 10.1038/480133a. [DOI] [PubMed] [Google Scholar]
- 7.Di Carlo D, Lee LP. Anal Chem. 2006;78(23):7918–7925. doi: 10.1021/ac069490p. [DOI] [PubMed] [Google Scholar]
- 8.Macey MG. Flow cytometry: principles and applications. Humana; Totowa: 2007. [Google Scholar]
- 9.Nolan JP, Sklar LA. Nat Biotech. 1998;16(7):633–638. doi: 10.1038/nbt0798-633. [DOI] [PubMed] [Google Scholar]
- 10.Krutzik PO, Nolan GP. Nat Methods. 2006;3(5):361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- 11.Lecault V, White AK, Singhal A, Hansen CL. Curr Opin Chem Biol. 2012;16(3–4):381–390. doi: 10.1016/j.cbpa.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 12.Yin H, Marshall D. Curr Opin Biotech. 2012;23(1):110–119. doi: 10.1016/j.copbio.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Morgan H, Sun T, Holmes D, Gawad S, Green NG. J Phys D: Appl Phys. 2007;40(1):61–70. [Google Scholar]
- 14.Heileman K, Daoud J, Tabrizian M. Biosens Bioelectron. 2013;49:348–359. doi: 10.1016/j.bios.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 15.Holzapfel C, Vienken J, Zimmermann U. J Membr Biol. 1982;67(1):13–26. doi: 10.1007/BF01868644. [DOI] [PubMed] [Google Scholar]
- 16.Jones TB. IEEE Eng Med Biol. 2003;22(6):33–42. doi: 10.1109/memb.2003.1304999. [DOI] [PubMed] [Google Scholar]
- 17.Gawad S, Cheung K, Seger U, Bertsch A, Renaud P. Lab Chip. 2004;4(3):241–251. doi: 10.1039/b313761a. [DOI] [PubMed] [Google Scholar]
- 18.Han SI, Joo YD, Han KH. Analyst (Cambridge, U K) 2013;138(5):1529–1537. doi: 10.1039/c3an36261b. [DOI] [PubMed] [Google Scholar]
- 19.Valero A, Braschler T, Rauch A, Demierre N, Barral Y, Renaud P. Lab Chip. 2011;11(10):1754–1760. doi: 10.1039/c1lc00007a. [DOI] [PubMed] [Google Scholar]
- 20.Vahey MD, Pesudo LQ, Svensson JP, Samson LD, Voldman J. Lab Chip. 2013;13(14):2754–2763. doi: 10.1039/c3lc50162k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun T, Morgan H. Microfluid Nanofluid. 2010;8(4):423–443. [Google Scholar]
- 22.Haandbaek N, Burgel SC, Heer F, Hierlemann A. Lab Chip. 2014;14(2):369–377. doi: 10.1039/c3lc50866h. [DOI] [PubMed] [Google Scholar]
- 23.Yang L, Arias LR, Lane T, Yancey M, Mamouni J. Anal Bioanal Chem. 2011;399(5):1823–1833. doi: 10.1007/s00216-010-4584-9. [DOI] [PubMed] [Google Scholar]
- 24.Holmes D, Pettigrew D, Reccius CH, Gwyer JD, van Berkel C, Holloway J, Davies DE, Morgan H. Lab Chip. 2009;9(20):2881–2889. doi: 10.1039/b910053a. [DOI] [PubMed] [Google Scholar]
- 25.Holmes D, Morgan H. Anal Chem. 2010;82(4):1455–1461. doi: 10.1021/ac902568p. [DOI] [PubMed] [Google Scholar]
- 26.Du E, Ha S, Diez-Silva M, Dao M, Suresh S, Chandrakasan AP. Lab Chip. 2013;13(19):3903–3909. doi: 10.1039/c3lc50540e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song H, Wang Y, Rosano JM, Prabhakarpandian B, Garson C, Pant K, Lai E. Lab Chip. 2013;13(12):2300–2310. doi: 10.1039/c3lc41321g. [DOI] [PubMed] [Google Scholar]
- 28.Chen J, Zheng Y, Tan Q, Shojaei-Baghini E, Zhang YL, Li J, Prasad P, You L, Wu XY, Sun Y. Lab Chip. 2011;11(18):3174–3181. doi: 10.1039/c1lc20473d. [DOI] [PubMed] [Google Scholar]
- 29.Zheng Y, Shojaei-Baghini E, Azad A, Wang C, Sun Y. Lab Chip. 2012;12(14):2560–2567. doi: 10.1039/c2lc21210b. [DOI] [PubMed] [Google Scholar]
- 30.Asphahani F, Wang K, Thein M, Veiseh O, Yung S, Xu J, Zhang MQ. Phys Biol. 2011;8(1):15006. doi: 10.1088/1478-3975/8/1/015006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghenim L, Kaji H, Hoshino Y, Ishibashi T, Haguet V, Gidrol X, Nishizawa M. Lab Chip. 2010;10(19):2546–2550. doi: 10.1039/c004115g. [DOI] [PubMed] [Google Scholar]
- 32.Park H, Kim D, Yun KS. Sens Act B. 2010;150(1):167–173. [Google Scholar]
- 33.Lan KC, Jang LS. Biosens Bioelectron. 2011;26(5):2025–2031. doi: 10.1016/j.bios.2010.08.080. [DOI] [PubMed] [Google Scholar]
- 34.Malleo D, Nevill JT, Lee LP, Morgan H. Microfluid Nanofluid. 2010;9(2–3):191–198. doi: 10.1007/s10404-009-0534-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Z, Frey O, Ottoz DS, Rudolf F, Hierlemann A. Lab Chip. 2012;12(5):906–915. doi: 10.1039/c2lc20911j. [DOI] [PubMed] [Google Scholar]
- 36.Zhu Z, Frey O, Haandbaek N, Ottoz DS, Rudolf F, Hierlemann A. Proceedings of the international conference on solid state sensors and actuators, transducers ’13; Barcelona, Spain. 2013. pp. 1527–1530. [Google Scholar]
- 37.Jolliffe IT. Principle component analysis. 2nd edn. Springer; New York: 2002. [Google Scholar]
- 38.McLachlan GJ. Discriminant analysis and statistical pattern recognition. Wiley; Hoboken: 2004. [Google Scholar]