Large granular lymphocyte (LGL) leukemia is a rare clonal disease characterized by a persistent increase in the number of CD8+ cytotoxic T cells or CD16/56+ natural killer (NK) cells. It is associated with recurrent infections, severe cytopenias and autoimmune diseases. JAK/STAT pathway activation, deregulation of pro-apoptotic pathways (sphingolipid and FAS/FAS ligand) and activation of pro-survival signaling pathways (PI3K/AKT and RAS) are known hallmarks of LGL leukemia. Activating somatic STAT3 mutations have been reported in the SH2 domain (30–70% of cases),1, 2, 3 and in the DNA-binding or coiled-coil domain (2%).4 STAT5B mutations are more rare, but typical of CD4+ T-LGL leukemia cases.5, 6, 7 The JAK/STAT pathway can also be activated by non-mutational mechanisms such as increased interleukin-6 (IL-6) secretion and epigenetic inactivation of JAK-STAT pathway inhibitors.8 Indeed, aberrant STAT signaling is observed in almost all LGL leukemia patients irrespective of the presence of JAK/STAT mutations.9
To characterize the genomic landscape of LGL leukemia, we performed whole-exome sequencing (Supplementary Methods and Supplementary Figure 1) from 19 paired tumor-control samples derived from untreated LGL leukemia patients including conventional CD8+ (n=13) T-cell cases, and more rare CD4+ or CD4+CD8+ T-cell cases (n=3), and NK LGL leukemias (n=3; Supplementary Table 1). Eleven STAT-mutation-negative patients were included for identification of new driver mutations. All sequenced samples were highly purified sorted cell populations (either CD8+ or CD4+ T cells or NK cells), and T-cell receptor Vbeta analysis confirmed monoclonal expansions in the tumor fractions of T-cell cases (see Supplementary Methods and Supplementary Table 1). The average sequencing coverage in the tumor samples was 32x (Supplementary Figure 2). Both the coverage and the number of raw called variants were similar in tumor and control samples. After selecting high confidence variants (see Supplementary Methods), and filtering out variants already described in human populations single nucleotide polymorphism database and/or with allele frequency higher than 5% in exome aggregation consortium exomes, 28 508 somatic variants in 16 518 genes were identified in the whole cohort with a high prevalence of C>T and G>A transversions (Supplementary Figure 3A). Next, among high confidence and rare variants, we selected 370 variants in 347 genes with a strong predicted functional impact (Supplementary Methods and Supplementary Table 2). The observed differences in numbers of somatic mutations (range 5–40, average 20) and genes involved (range 4–41, 19) per patient were not because of coverage differences (Supplementary Figure 3B). A slight tendency toward more mutated genes per patient in STAT-mutation-positive (22.9 in average) versus negative patients (18.4 in average) was noticed. Sanger sequencing validations of somatic variants were obtained in 14 genes (Supplementary Table 3 and Supplementary Figure 4) being recurrent or prioritized according to functional criteria and/or connections emerged by integrated pathway-derived networks. The positions of the mutations in protein domains of selected genes are shown in Supplementary Figure 5.
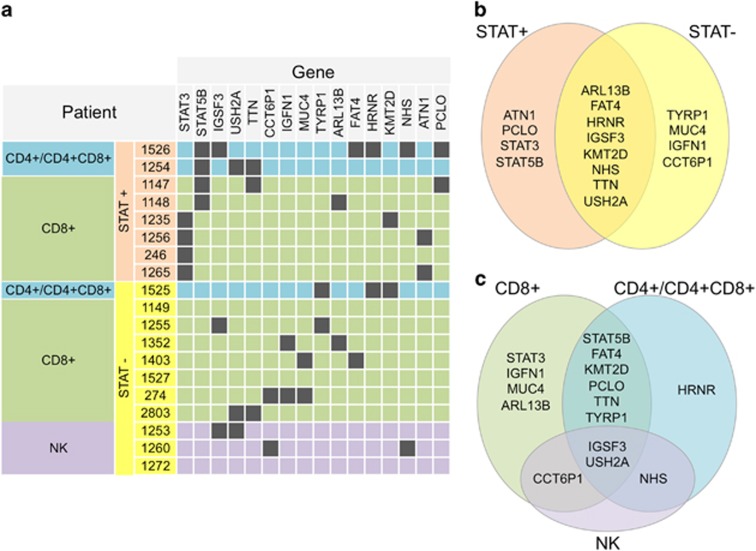
In addition to STAT3 (all in CD8+ T-LGL) and STAT5B (CD4+ and CD8+ cases) mutations (in 8/19 patients, 42%), 14 other genes had recurrent mutations including transcriptional/epigenetic regulator, tumor suppressor and cell proliferation genes (Figure 1a and 2a). KMT2D has been linked to lymphomagenesis10 and found to be frequently mutated in other cancers. Mutations of PCLO, a calcium sensor-regulating cAMP-induced exocytosis, have been previously reported in diffuse large B-cell lymphoma. FAT4 is an upstream regulator of stem cell genes both during development and cancer, functioning as a tumor growth suppressor via activation of Hippo signaling. It was previously found recurrently mutated in human cancers, including leukemias. Also the other recurrently mutated gene, ARL13B, is linked to Hippo signaling. It encodes a small GTPase of primary cilia whose role in cell cycle control has recently been recognized, and they crosstalk with several signaling pathways including Hippo. ARL13B and FAT4 genes were mutated in a mutually exclusive way. Additional non-recurrent somatic mutations of YAP1 and of its inhibitor AMOTL1 point toward an involvement of Hippo signaling deregulation in LGL leukemia.
Figure 1.
Recurrent somatic mutations in LGL leukemia patients. (a) The table indicates the genes that carry somatic variants in more than one patient, with a color code showing STAT3 and STAT5B status and classification of patients. (b) Recurrently mutated gene sets found only in STAT-mutation-negative patients (STAT−), only in STAT-mutation-positive patients (STAT+) or in both groups. (c) Recurrently mutated genes that are found only in one or are shared among patient classes (CD8+, CD4+/CD4+CD8+ and NK+).
Figure 2.
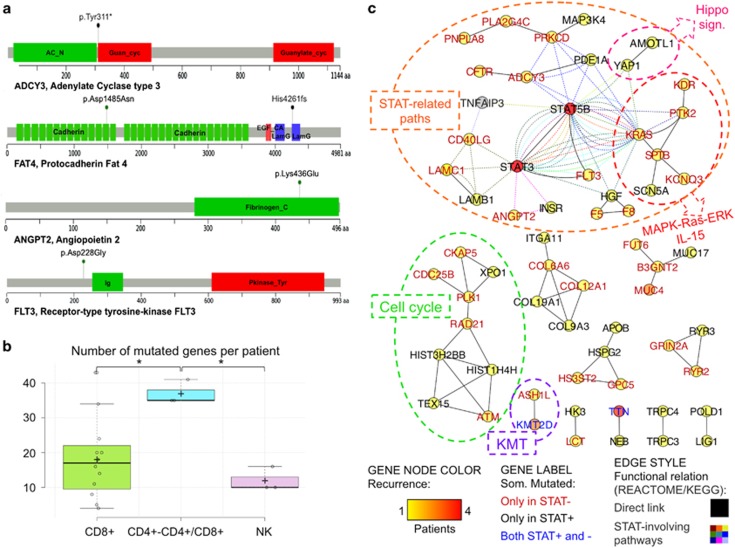
(a) Impact of selected somatic variants to protein products. Lollipop plots show the type and the position of somatic variants of four selected genes in relation to the protein sequence and domain structure (see Supplementary Figure 5 for an extended version of the figure including additional genes). The ADCY Tyr311* variant induces a very premature stop preventing the synthesis of the protein region including Guanylate cyclase, ATP and Mg2+domains; FAT4 presents two variants, the high-impact missense variant Asp1485Asn in the Cadherin 14 domain and the frameshift variant Hys4261fs inducing a stop codon before Laminin G-like domain truncating the protein before the EGF-like 6 domain and the C terminal; ANGPT2 presents the high-impact missense variant Lys463Glu in Fibrinogen C-terminal domain implicated in protein–protein interactions, and FLT3 shows a high-impact Asp228Gly variant. (b) Number of mutations per patient in each class. Normal distribution of values was confirmed with the Shapiro–Wilk test (P=0.099). Both analysis of variance (P=0.009) and pairwise Tukey s.d. post hoc tests (P-values 0.010 and 0.019 in the comparisons of CD4+/CD4+CD8+ with NK and CD8+, respectively) confirmed the statistical significance of the observed difference. (c) LGL leukemia mutation network. The network shows the functional relations of genes somatically mutated in LGL leukemia patients, according to the integration of KEGG and Reactome pathway topology (see the text and Supplementary Methods for details on the pathway-derived network reconstruction procedure); network nodes represent somatically mutated genes, node color indicates recurrence (according to the legend heat color scale) in the cohort, node label indicates the gene Symbol (different label colors indicates genes that are mutated only in STAT-mutation-positive (STAT+), only in STAT-mutation-negative (STAT−) or in both patient groups, as shown in the legend); genes are connected with black solid lines if they are directly connected in KEGG- and/or Reactome-derived networks or with colored dashed lines if they participate in pathways including STAT3 and/or with STAT5B (see Supplementary Figure 6 for a detailed version of the network).
When comparing the mutation profile between three different phenotypic LGL subgroups, qualitative and quantitative differences were observed, although the clinical characteristics of patients did not markedly differ (see details in Supplementary Results and Supplementary Table 1). Interestingly, higher mutation burden was observed in CD4+ T-LGL leukemia cases (Figure 2b). As the sequencing depth across samples did not vary significantly (Supplementary Figure 3), the differences in mutation load likely reflect a different natural history of the LGL phenotypes. Cytomegalovirus-derived stimulation and restricted usage of T-cell receptor Vβ has been associated with CD4+ T-LGL cases,11 and this could relate to the higher number of mutations. In the CD4+ group, only STAT5B and HRNR genes had recurrent mutations (Figure 1b). HRNR is a calcium-binding protein involved in hematopoietic progenitor cell differentiation, and it is mutated, amplified or overexpressed in many cancers. In NK LGL leukemias (all STAT-mutation-negative), 31 genes harbored somatic mutations including several ‘cancer genes' such as KRAS, PTK2, NOTCH2, CDC25B, HRASLS, RAB12, PTPRT and LRBA.
Next, a custom knowledge-based ‘systems genetic' approach, reminiscent of strategies recently implemented to interpret genome-wide transcriptome deregulation in cancer,12, 13 provided the functional prioritization of mutated genes. As mutations hitting different genes can drive a similar phenotype in different patients and concur to it if present in the same patient, we reconstructed a pathway-derived meta-network depicting direct interactions and functional relations between genes somatically mutated in LGL leukemias. We identified 119 KEGG and 426 Reactome pathway-derived networks, each including at least one of the 347 previously prioritized mutated genes associated to high confidence, rare and high-impact variants. The union of all path-derived networks generated a meta-network with 118 (34%) mutated genes, giving a non-redundant representation of functional relations, based on direct interactions between somatically mutated genes. Remarkably, 47 mutated genes were directly connected to at least another mutated gene in 18 multigene components (groups of genes whose products directly interact, that is, encode proteins taking part in the same molecular complex or regulating each other). Considering co-participation of mutated genes in pathways including STAT genes as additional functional link, seven multigene components connected by direct relations and three isolated genes converged into a component of 26 genes. In this reconstructed LGL leukemia network (Figure 2c and Supplementary Figure 6), 61 somatically mutated genes (occurring in many cases only in one sample) preferentially fall into a limited number of highly connected pathways, and in this manner collectively form a functional module hit by somatic mutations in LGL leukemia. The largest network component included 24 mutated genes either directly linked to STAT genes, to their neighbors and/or participating in pathways including STAT genes (Figure 2c). Beyond JAK-STAT signaling, the ‘STAT-related component' included genes intervening in several other connected paths such as acute and chronic myeloid leukemia, ErbB, HIF-1, insulin, T-cell receptor and VEGF signaling pathways. In 16 out of 19 patients, at least one gene of this group was mutated with some patients showing more than one hit in the gene group. For instance, one STAT-mutation-negative CD4+ patient presented with mutated alleles in three genes of the component (CD40LG, F8 and PLA2G4C). The similar variant allele frequency values of the validated variants support their co-presence in the dominant LGL leukemic clone (Supplementary Table 3). Altogether, 8 of 11 STAT-mutation-negative patients carried validated somatic mutations in at least one of the ‘STAT-related component' genes, such as in FLT3, KRAS, ADCY3, ANGPT2 and PTK2. These mutated genes also connect the STAT component to the MAPK-Ras-ERK (Figure 2c) pathway and to the IL-15, all known to be deregulated in LGL leukemia.14 For example, PTK2 is a non-receptor protein-tyrosine kinase, which is highly expressed in T cells and it regulates several processes, including cell cycle progression, cell proliferation and apoptosis, activation of numerous pathways such as PI3K/AKT signaling MAPK/ERK and MAP kinase signaling cascades. Also the mutated ANGPT2 is linked to PI3K-AKT and RAS signaling pathways that it antagonizes. ANGPT2 is expressed in lymphocytes and controls T-cell proliferation. ANGPT2 and other angiogenic factors are reportedly involved in chronic lymphocytic leukemia where they exert pro-survival effects. Other STAT-connected genes were receptors such as CD40LG (modulates B-cell function, regulates immune system and participates in STAT3 as well as in IL and NFAT signaling pathways) and FLT3 (a class III receptor tyrosine kinase that promotes the phosphorylation of various proteins and kinases in the PI3K/AKT/mTOR, RAS and JAK/STAT signaling pathways). Interestingly, CD40LG was annotated in the same KEGG pathways as TNFAIP3 (Figure 2c), which is a negative regulator of NF-κB signaling and known tumor suppressor gene, and was recently found to be mutated in 8% of T-LGL leukemia patients.15 Other relevant variants confirmed in STAT-mutation-negative patients and connected to the STAT pathway were KRAS and the kinase KDR/VEGFR2.
Other components (and pathways) not directly linked to the main lesions were also of interest. Nine genes were linked to cell cycle regulation, and include the CDC25b gene and ATM, which is involved in apoptosis and P53 signaling (Figure 2c). Furthermore, the epigenetic nodule included the recurrently mutated KMT2D, which is connected to ASH1L. Both are histone methyltransferases involved in epigenetic regulation of gene expression programs and are part of the ASCOM complex, involved in transcriptional co-activation. The networks of genes mutated in individual CD8+ and CD4+ or NK LGL leukemia patients and in each patient subgroup are presented in the Supplementary Figures 7–9.
To conclude, with the systems genetic approach, we were able to map individual mutations found in LGL leukemia patients in novel functional modules. The central role of JAK-STAT network was further highlighted, and our data provide important new insights of the activation of this pathway in those LGL leukemias that do not carry STAT mutations.
Acknowledgments
We would like to thank the personnel at the Hematology Research Unit Helsinki and FIMM for their expert clinical and technical assistance. We thank also Dr Geertruy te Kronnie and Silvia Bresolin for useful discussion during development of the network-based method. This work was supported by the European Research Council (project M-IMM), Academy of Finland, the Finnish Cancer Institute, the Finnish Cancer Societies, the Signe and Ane Gyllenberg Foundation, Sigrid Juselius Foundation, Instrumentarium Science Foundation, State funding for university-level health research in Finland, Swedish Cultural Foundation, Blood Disease Foundation, the Finnish Cultural Foundation. SB received funding from Fondazione Cassa di Risparmio di Padova e Rovigo Progetti di Eccellenza 2011/2012 (http://www.fondazionecariparo.net/english-version/), Ministero dell'Istruzione, dell'Università e della Ricerca (http://www.istruzione.it/), PRIN 2010/115 (2010NYKNS7_002) and FIRB 2011 (RBAP11CZLK) and from ‘Special Program Molecular Clinical Oncology 5 × 1000' to Associazione Italiana per la Ricerca sul Cancro (http://www.airc.it/english/) and from the University of Padova. MH received funding from the German Research Foundation (FOR1961; HE3552/4-2).
Author contributions
AC, EIA, SM and StB designed the study, coordinated the project, analyzed the data and wrote the paper. AC, AB and StB designed and performed the bioinformatics analysis. EIA, VRG and SaB validated mutations. SM and EA provided sequence data. MC, JM and MH provided patient samples and clinical data. All authors read and approved the final manuscript.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
SM has received honoraria and research funding from Novartis, Pfizer and Bristol-Myers Squibb and research funding from Ariad (none of these related to this project). The remaining authors declare no conflict of interest.
Supplementary Material
References
- Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmaki H, Andersson EI et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med 2012; 366: 1905–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasan A, Kern W, Grossmann V, Haferlach C, Haferlach T, Schnittger S. STAT3 mutations are highly specific for large granular lymphocytic leukemia. Leukemia 2013; 27: 1598–1600. [DOI] [PubMed] [Google Scholar]
- Jerez A, Clemente MJ, Makishima H, Koskela H, Leblanc F, Peng NgK et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood 2012; 120: 3048–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson E, Kuusanmaki H, Bortoluzzi S, Lagstrom S, Parsons A, Rajala H et al. Activating somatic mutations outside the SH2-domain of STAT3 in LGL leukemia. Leukemia 2016; 30: 1204–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson E, Tanahashi T, Sekiguchi N, Gasparini VR, Bortoluzzi S, Kawakami T et al. High incidence of activating STAT5B mutations in CD4-positive T-cell large granular lymphocyte leukemia. Blood 2016; 128: 2465–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajala HL, Eldfors S, Kuusanmaki H, van Adrichem AJ, Olson T, Lagstrom S et al. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood 2013; 121: 4541–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajala HL, Porkka K, Maciejewski JP, Loughran Jr TP, Mustjoki S. Uncovering the pathogenesis of large granular lymphocytic leukemia-novel STAT3 and STAT5b mutations. Ann Med 2014; 46: 114–122. [DOI] [PubMed] [Google Scholar]
- Teramo A, Gattazzo C, Passeri F, Lico A, Tasca G, Cabrelle A et al. Intrinsic and extrinsic mechanisms contribute to maintain the JAK/STAT pathway aberrantly activated in T-type large granular lymphocyte leukemia. Blood 2013; 121: 3843–3854, S1. [DOI] [PubMed] [Google Scholar]
- Epling-Burnette PK, Liu JH, Catlett-Falcone R, Turkson J, Oshiro M, Kothapalli R et al. Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. J Clin Invest 2001; 107: 351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dominguez-Sola D, Hussein S, Lee JE, Holmes AB, Bansal M et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med 2015; 21: 1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Caballero A, Garcia-Montero AC, Barcena P, Almeida J, Ruiz-Cabello F, Tabernero MD et al. Expanded cells in monoclonal TCR-alphabeta+/CD4+/NKa+/CD8−/+dim T-LGL lymphocytosis recognize hCMV antigens. Blood 2008; 112: 4609–4616. [DOI] [PubMed] [Google Scholar]
- Calura E, Pizzini S, Bisognin A, Coppe A, Sales G, Gaffo E et al. A data-driven network model of primary myelofibrosis: transcriptional and post-transcriptional alterations in CD34+ cells. Blood Cancer J 2016; 6: e439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calura E, Bisognin A, Manzoni M, Todoerti K, Taiana E, Sales G et al. Disentangling the microRNA regulatory milieu in multiple myeloma: integrative genomics analysis outlines mixed miRNA-TF circuits and pathway-derived networks modulated in t(4;14) patients. Oncotarget 2016; 7: 2367–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc F, Zhang D, Liu X, Loughran TP. Large granular lymphocyte leukemia: from dysregulated pathways to therapeutic targets. Future Oncol 2012; 8: 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson P, Bergmann A, Rahmann S, Wohlers I, Scholtysik R, Przekopowitz M et al. Recurrent alterations of TNFAIP3 (A20) in T-cell large granular lymphocytic leukemia. Int J Cancer 2016; 138: 121–124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.