Abstract
The neuropeptide vasoactive intestinal peptide (VIP) strongly impacts on human pathophysiology and does so through interaction with class II G protein-coupled receptors. We characterized the C terminus-binding site of VIP in the N-terminal ectodomain (N-ted) of the human VPAC1 receptor: 1) The probe [125I-Bpa28]VIP in which the C-terminal residue (Asn28) is substituted by a photoreactive p-benzoyl-l-Phe (Bpa) was used to photolabel the receptor. After receptor cleavage and Edman sequencing, it was shown that Asn28 of VIP is in contact with Lys127 in the receptor N-ted. Taking into account previous data, it follows that the C-terminal and central parts of VIP from Asn28 to Phe6 lie in the N-ted. 2) A three-dimensional model of the N-ted was constructed, the fold being identified as a Sushi domain with two antiparallel β-sheets and three disulfide bonds. The nuclear magnetic resonance structure of VIP was then docked into this model by taking into account the constraint provided by photoaffinity experiments with [125I-Bpa28]VIP. It appeared that VIP runs parallel to the β3-β4 antiparallel sheets. 3) We performed molecular dynamic simulations over 14 nsec of the complex between VIP and receptor N-ted and the free N-ted. The structural model of the free N-ted is stable, and VIP tends to further stabilize the N-ted structure more especially in the loops connecting the β-sheets. These structural studies provide a detailed molecular understanding of the VIP-receptor interaction.
THE VASOACTIVE INTESTINAL peptide (VIP) is widely expressed in the central and peripheral nervous systems (1) as well as in immune cells (2). It strongly impacts on human pathophysiology due to its ability to control a large array of biological functions in the brain, peripheral organs (3), and the endocrine system (1). In particular, many observations highlight its great potential as an antiinflammatory agent (4) and a neuroprotector (5). VIP exerts its various biological actions by interacting with two members of the class II, also referred to as class B, family of G protein-coupled receptors (GPCRs) (6, 7). The VIP receptors are named VIP/PACAP (VPAC)1 and VPAC2 because they have two physiological ligands, VIP and another neuropeptide, the pituitary adenylate cyclase activating peptide (PACAP) (6). The class II of GPCRs comprises receptors for peptides structurally related to VIP such as glucagon, glucagon-like peptides, secretin or GH-releasing factor or peptides not related to VIP including corticotropin-releasing factor (CRF), PTH, and calcitonin (7). VPAC receptors, like other class II GPCRs, have a large N-terminal ectodomain (N-ted) that plays a pivotal role in agonist binding (7).
The molecular properties of the VPAC1 receptor have been extensively studied for many years because this receptor is responsible for many important actions of VIP (7) and also because it is prototypic of class II GPCRs (6). Site-directed mutagenesis and molecular chimerism studies (7) identified the receptor N-ted as the major site of VIP binding and suggested its role in the selectivity of the receptor toward some VIP-related peptides. The physical sites of interaction between VIP and its receptors had remained elusive until recent photoaffinity experiments showing that the central part of VIP is in direct contact with the N-ted of the VPAC1 receptor (8, 9, 10). A three-dimensional model of the N-ted of the VPAC1 receptor has been constructed (10) using the nuclear magnetic resonance (NMR) structure of the N-ted of another class II GPCR, the CRF receptor 2β (11). The fold is identified as a Sushi domain, also referred to as short consensus repeat (12). It consists of two antiparallel β-sheets and is stabilized by three disulfide bonds (11). Taking into account the constraints provided by photoaffinity, the NMR structure of VIP (10) has been docked in this structural model of the receptor N-ted, showing that the central part of VIP lies in the N-ted C-terminal part (10). However, no data are available as yet regarding the site of approximation between the C-terminal part of VIP and the human (h)VPAC1 receptor. This is an important issue because VIP has diffuse pharmacophoric domains, with the amino acids involved in the biological activity being distributed along the whole 28-amino acid peptide chain (13).
In this context, the present work has several complementary goals: 1) to develop a new affinity probe to determine the site of approximation of the C-terminal residue of VIP with the VPAC1 receptor. The probe was synthesized by substituting the C-terminal Asn28 residue by a photoreactive p-benzoyl-l-Phe (Bpa) residue. 2) To determine by radiochemical Edman degradation the pair of residues in VIP and in the receptor that ensure the covalent attachment of the [125I-Bpa-F28]VIP probe. 3) To dock VIP in the VPAC1 receptor N-ted structural model by taking into account this new constraint. 4) To perform molecular dynamic simulations over 14 nsec of the free and VIP-complexed hVPAC1 receptor N-ted, which were solvated in a box of TIP3 water molecules. The present paper describes the data.
RESULTS AND DISCUSSION
We synthesized [Bpa-F28]VIP, which consists of substitution of Asn28 residue by the photolabile residue p-benzoyl-l-phenylalanine. This probe was equipotent with native VIP in inhibiting [125I]VIP binding (Ki of 1 nm for both peptides) and in stimulating adenylyl cyclase activity (EC50 of 0.1 nm for both peptides) in Chinese hamster ovary (CHO) cells expressing the hVPAC1 receptor (Fig. 1). This is in consonance with previous alanine scanning of VIP showing that position 28 of the neuropeptide is not important for high-affinity binding to the hVPAC1 receptor (13). It follows therefore that the [Bpa-F28]VIP probe is instrumental for photoaffinity labeling of the VPAC1 receptor.
Fig. 1.
Binding and Adenylyl Cyclase Activity Assays of Bpa28-VIP
Left panel, Competitive inhibition of [125I]VIP binding to CHO cell homogenates expressing hVPAC1 receptor by VIP and Bpa28-VIP. The data are expressed as percentage of initial specific binding in the absence of competitor. Right panel, Action of increasing concentrations of VIP and Bpa28-VIP on adenylyl cyclase activity in CHO cell homogenates. ○, Bpa28-VIP; •, VIP. Points are mean values from three studies; sem values are less than 4% of mean values.
Photoaffinity experiments were conducted by incubating [125I-Bpa-F28]VIP with CHO cells expressing the hVPAC1 receptor followed by UV exposure. NuPage analysis of proteins revealed a single band at approximately Mr = 95,000 (Fig. 2A). This labeled band disappeared when the labeled probe was incubated in the presence of 1 μm cold VIP (Fig. 2A). It follows that the [125I-Bpa-F28]VIP photoaffinity probe was able to specifically label the hVPAC1 receptor. The estimated Mr of the band is in good agreement with a complex between the 3-kDa probe and the 64-kDa glycosylated hVPAC1 receptor (10) fusioned at its C terminus with a 25-kDa green fluorescent protein. After CNBr cleavage of the 95-kDa complex, sodium dodecyl sulfate (SDS)-NuPAGE analysis revealed a single radiolabeled fragment of 30 kDa, which moved at 11 kDa after deglycosylation with N-glysosidase F (Fig. 2B). Considering the known glycosylation sites of the hVPAC1 receptor (14) and the theoretical receptor fragments generated by its CNBr cleavage (10), it can be concluded that [125I-Bpa-F28]VIP is attached to the Trp67-Met137 receptor fragment in the receptor N-ted. Within the native receptor, this fragment is N-glycosylated on Asn69 and Asn100 with a 9-kDa carbohydrate moiety on each glycosylation site (14). To further reduce the size of the labeled fragment obtained after CNBr treatment, the 30-kDa band was subjected to enzymatic cleavage with endopeptidase Glu-C. This treatment generated a major approximately 6-kDa band (Fig. 2B), which represented the receptor segment 109–133 considering the known cleavage sites for this enzyme (8). It should be noted that a minor labeled band at Mr 30,000 was observed (Fig. 2B), corresponding probably to incomplete digestion by endopeptidase Glu-C in our experimental conditions. Next, we created a new CNBr cleavage site within the receptor segment 109–133 by mutating Asp125 into methionine. The D125M mutant was stably expressed in CHO cells and appeared to bind VIP with an affinity similar to that of the wild-type receptor (data not shown). After incubation of the D125M mutant with the [125I-Bpa-F28]VIP probe and CNBr treatment of proteins, a 5-kDa labeled band was observed instead of a 30-kDa band for the wild-type receptor (Fig. 3A). This labeled protein represented the fragment 126–137 covalently attached to the [125I-Bpa-F28]VIP probe. To estimate which residue of the 126–137 receptor fragment was covalently linked to the [125I-Bpa-F28]VIP probe, this fragment was subjected to Edman degradation sequencing. For that purpose, the purified 5-kDa receptor fragment resulting from CNBr cleavage of the D125M mutant after labeling with [125I-Bpa-F28]VIP probe was covalently coupled to glass beads in the C-terminal position of the fragment. As shown in Fig. 3B, radiochemical sequencing of labeled Asp126-Met137 revealed that radioactivity was present in the second cycle of Edman degradation corresponding to the Lys127 residue. Altogether our results clearly indicated that Lys127 is the site of covalent attachment of the [125I-Bpa-F28]VIP affinity probe to the hVPAC1 receptor.
Fig. 2.
Photoaffinity Labeling of the hVPAC1 Receptor with the [125I-Bpa28]VIP Probe
A, Receptors expressed in CHO cells were photoaffinity-labeled with [125I-Bpa-F28]VIP in the absence (lane 1) or presence (lane 2) of an excess of cold VIP (1 μm). B, After photoaffinity labeling cell homogenates were subjected to treatment with CNBr, peptide N-glycosidase F (PNGase F), or endopeptidase Glu-C as shown in lanes 1–3, respectively. The labeled proteins were resolved on NuPAGE 4–12% Bis-Tris Gel followed by autoradiography. See Materials and Methods for details. MW, Molecular weight.
Fig. 3.
Photoaffinity Labeling of hVPAC1 Receptor and the D125M Receptor Mutant and Radiochemical Sequencing
A, CNBr-digestion products of covalent complex [125I-Bpa28]VIP/hVPAC1 receptor (lane 1) and [125I-Bpa28]VIP/D125M receptor mutant (lane 2) were resolved on SDS-NuPAGE gel. B, Radiochemical sequencing of digested peptides after photoaffinity labeling of the D125M hVPAC1 receptor mutant with the [125I-Bpa28]VIP probe. See Materials and Methods for details. MW, Molecular weight.
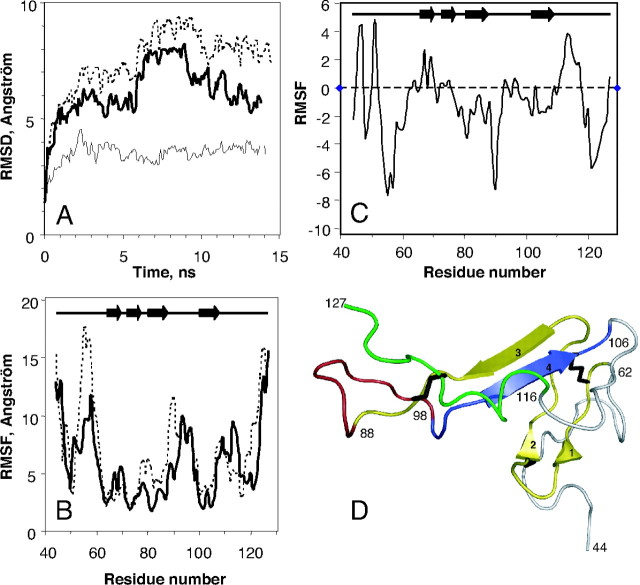
We developed a three-dimensional model of the hVPAC1 receptor N-ted (10) and docked the NMR structure of VIP, in this model using the new constraint provided by photoaffinity experiments described above. This model is shown in Fig. 4 and nicely accommodated VIP. Indeed, after docking the distances between residues involved in photolabeling are lower than 8 Å and compatible with the use of benzophenone probes that exhibit an extended distance of about 7 Å (15). This included the contacts between Phe6, Tyr22, Asn24 (10), and Asn28 (this paper) of VIP with Asp107, Gly116, Cys122, and Lys127 in the N-ted of the hVPAC1 receptor, respectively (Fig. 4). In the model the entire α-helix of VIP from Phe6 to the C-terminal Asn28 runs parallel to the β3–β4 antiparallel sheets of the Sushi domain of the receptor N-ted. To further validate this model, we performed molecular dynamic simulations over 14 nsec in explicit solvent (see Materials and Methods). To gain insight into the possible role of VIP in stabilizing the receptor-binding domain, both the complex between VIP and hVAPC1 receptor N-ted and the free hVPAC1 receptor N-ted were submitted to molecular dynamic simulations. In both cases, after a 5 nsec equilibration period, some fluctuations of the root mean square deviations (RMSDs) were observed but RMSDs remained overall stable (Fig. 5, A). It could be also noticed that the RMSDs were constantly higher for hVPAC1 receptor N-ted alone than when it is complexed with VIP. Averaged RMSDs were 8 and 6.5 Å for hVPAC1-Nted alone and in the presence of VIP, respectively. This observation suggests that the receptor N-ted is more stable in the presence than in the absence of VIP. Figure 5A also showed that after the equilibration step, the RMSDs of VIP in the presence of hVPAC1 receptor N-ted were stable up to 14 nsec in the range of 3–4 Å. This low RMSD suggests that VIP remains highly structured when associated with the receptor N-ted, even in our conditions of molecular dynamic simulations in a water box. Previous circular dichroism experiments indicated that VIP has a poorly defined structure in aqueous solution (16). It follows that the interaction of VIP with the receptor N-ted is probably sufficient to stabilize the structured form of the peptide.
Fig. 4.
Three-Dimensional Molecular Model of VIP Docked into the hVPAC1 Receptor N-ted
Magenta, VIP; cyan, main chain of hVPAC1 N-ted (sequence 44–127); yellow, β-strand of hVPAC1 N-ted; orange, disulfide bonds C50-C72, C63-C105, and C86-C122. Residues involved in the photoaffinity labeling are shown in red (F6, Y22, N24, and N28) and blue (D107, G116, C122 and K127) for VIP and hVPAC1 N-ted, respectively. Figure was produced using Pymol software (http://www.pymol.org).
Fig. 5.
Molecular Dynamic Simulations of hVPAC1 N-ted in the Presence or the Absence of VIP
A, Conformational drift from the initial structure during the simulations, measured as backbone RMSD for the hVPAC1 N-ted in the presence (dashed line) and in the absence (bolded line) of VIP. The same data are also shown for VIP itself (thin line) docked in the receptor N-ted. Plots were drawn using a 0.1-nsec sampling. B, Conformational flexibility of hVPAC1 N-ted measured as RMSF of the backbone atoms during the 14-nsec molecular dynamic simulation as a function of residue number. Data are shown for hVPAC1 N-ted in the presence (bolded line), and the absence (dashed line) of VIP. C, Conformational flexibility of hVPAC1 N-ted due to the presence of VIP measured as ΔRMSF, which represents difference between the RMSFs of hVPAC1 N-ted in the presence and in the absence of VIP. D, Several segments of the receptor N-ted were colored to more easily follow the text in Results and Discussion. Yellow, 63–88 segment; red, 89–98 segment; blue, 99–106 segment; green, 116–127 segment. The three disulfide bridges (C50-C72, C63-C105, and C86-C122) were represented in black. Panel D was produced using Pymol software (http://www.pymol.org). ns, Nanosecond.
The root mean square fluctuation (RMSFs) were calculated over 14 nsec for the backbone atoms of the hVPAC1 receptor N-ted, complexed or not with VIP, at the level of each residue along the N-ted primary sequence (Fig. 5B). In both conditions, the structured core of the receptor N-ted (residues 62–88 and 98–106), which includes the two antiparallel β-sheets, presented a small fluctuation (∼3–5 Å) compared with the rest of the protein. The differences between the RMSFs calculated in the absence of VIP and in the presence of VIP, referred to as ΔRMSF (Fig. 5C), were very small in the segments comprising residues 62–88 and 98–106, suggesting that VIP does not further stabilize this already highly structured fold of the receptor N-ted, i.e. Sushi domain (10). An exception is the proximal part of the segment 62–88 (Fig. 5D), which appears to be stabilized by VIP (Fig. 5C). The remaining regions of the receptor N-ted exhibited much larger RMSF (Fig. 5B). However, it is clear that the structure of the hVPAC1 N-ted is overall stabilized by VIP as evidenced by the negative ΔRMSF which largely exceeded the positive ΔRMSF (Fig. 5C). Analysis of the ΔRMSF (Fig. 5C) reveals that two main regions are stabilized by VIP. First, VIP may stabilize the N-ted region around residue 120 (Fig. 5C). This region is localized at the end of the β4-sheet and at proximity of the C86–C122 disulfide bridge (Fig. 5D). Furthermore, this region (sequence 116–127) of the N-ted is in close contact with VIP (Fig. 4). VIP also stabilizes the loop comprising residues 89–97 (Fig. 5B), which connects the β3- to the β4-sheet (Fig. 5D).
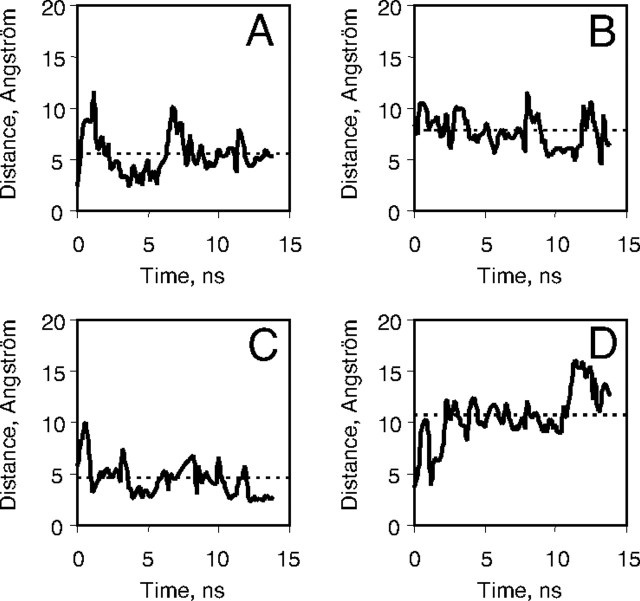
To get insights into the flexibility of the interaction between VIP and the hVPAC1 receptor N-ted, we consider the distances between residues of VIP and receptor N-ted, which have been experimentally shown by photoaffinity experiments to be in close proximity in the VIP-receptor complex (Fig. 6). Over the 14 nsec molecular dynamic simulations, the distances between Phe6 (VIP) and Asp107 (receptor), Tyr22 (VIP) and Gly116 (receptor), and Asn24 (VIP) and Cys122 (receptor) (see Fig. 4) were relatively stable and close to each other with averaged values of 5.4, 7.7, and 4.5 Å, respectively. In contrast the distance between Asn28 (VIP) and Lys127 (receptor) exhibited higher fluctuations (averaged value of 10.6 Å), as compared with the initial value of 3.85 Å at the beginning of molecular dynamic simulation. A possible explanation for such a large distance is that Lys127 is located in the last loop of our model of hVPAC1 receptor N-ted, and it is quite common to observe high fluctuation in free extremities of a protein. As mentioned above, this may be artifactual because beyond Lys127 there is a connecting peptide in the native hVPAC1 receptor that links the N-ted to the first transmembrane helix.
Fig. 6.
Distance Drifts of Residues Involved in Photoaffinity Labeling throughout the 14-nsec Molecular Dynamic Simulation
A–D, F6 (VIP)-D107 (N-ted), Y22 (VIP)-G116 (N-ted), N24 (VIP)-C122 (N-ted), and N28 (VIP)-K127 (N-ted), respectively. Dashed lines correspond to distance averages. Plots were drawn using a 0.2-nsec sampling. ns, Nanosecond.
In this work, we show that VIP, a natural peptide agonist, nicely lies in the N-ted of the hVPAC1 receptor, a class II GPCR. The entire C-terminal segment of VIP from Phe6 to C-terminal Asn28 is in close contact with the receptor N-ted as demonstrated by photoaffinity experiments. The molecular dynamic simulations in a water box show that the VPAC1 receptor N-ted structure alone is stable. In particular, the Sushi domain made of two antiparallel β-sheets with three disulfide bonds has a low RMSD whereas the loops connecting the β sheets are more flexible. The presence of VIP does not further stabilize the highly structured fold of the Sushi domain but stabilizes some loops (see above) resulting in an overall stabilization of the receptor N-ted. The fine positioning of the VIP 6–28 segment of VIP in its hVPAC1 receptor N-ted structure shown in this paper raises three major questions.
1) The first question is related to the positioning of VIP in the hVPAC1 receptor N-ted as compared with what is known in the literature regarding the site of interaction of class II GPCR with peptide ligands. Although several studies of photoaffinity labeling have been described for a few class II GPCRs, including secretin receptor (17) and PTH1 receptor (18), structural studies are scarce. The three-dimensional NMR structure of N-ted of CRF-2β receptor in complex with the peptide antagonist, astressin, has been reported (19). Astressin is a peptide analog of the hCRF fragment 12–41, which has been shown clearly to interact with the CRF-2 β receptor N-ted (19). The data reported by Grace et al. (19) described the positioning of the 27–41 segment of astressin in the CRF receptor N-ted in a region that is different from that described for the 6–28 segment of VIP in the VPAC1 receptor N-ted. In particular, the loop 86–98 in the VPAC1 receptor (Fig. 6) does not interact with VIP whereas the equivalent loop 84–98 in CRF-2β is important for astressin interaction (18). It could be argued that the major reason for such difference is the fact that astressin is an antagonist whereas VIP is an agonist. However, astressin is a peptide antagonist analog based on human CRF with high sequence homology between the two peptides. The antagonistic properties of astressin are mainly due to the deletion of the 1–12 N-terminal segment, which plays a crucial role in receptor activation. In this context, it has been speculated that CRF and astressin bind in a similar manner to the CRF N-ted (19). A much more relevant explanation of the difference of positioning of VIP 6–28 and astressin 27–41 in their respective class II GPCR, is directly related to the length of the peptides. Whereas CRF is a long 41-amino acid peptide, VIP is much shorter with only 28 amino acids, a property shared with other peptides of its structural family such as glucagon (29 amino acids) or secretin (27 amino acids). In that respect, the segment 27–41 of astressin, which has been positioned in the CRF-2 β receptor, simply has no equivalent in VIP or in short peptides acting at other class II GPCRs. In this context, it can be speculated that long peptides acting at class II GPCRs, such as CRF or astressin, have an additional site of interaction with the receptor N-ted as compared with short peptides such as VIP. Of course, it would be very interesting to know the site of interaction of the N-terminal part of CRF, up to residue 26, with the CRF2 β-receptor, but such data are not yet available. Very recently, the solution structure of the N-ted of a PAC1 receptor variant complexed to the peptide antagonist PACAP 6–38 has been determined by NMR (20). Quite interestingly the data are consistent with our model of VIP binding to VPAC1 receptor N-ted with an interesting difference. Whereas in our model VIP runs parallel with the β3-β4 antiparallel sheets (Fig. 4), in the PAC1 N-ted structure, residues 6–10 of PACAP run across β3-β4 (20). This may be tentatively related to the fact that the PAC1 receptor is complexed with a truncated PACAP lacking the 1–5 N-terminal segment, which behaves as an antagonist, whereas we deal with the entire VIP peptide 1–28, which behaves as an agonist.
2) A second question which can be raised following the present study is related to the site of interaction of the proximal N-terminal 1–5 segment of VIP with its VPAC1 receptor. Does this segment also interact with the receptor N-ted and/or does it rather position in close proximity to the serpentine region of the receptor to initiate signaling? To answer these questions, new photoaffinity probes consisting of substitution of amino acids in the N-terminal segment of VIP by photolabile residues are certainly needed. Such probes have not yet been developed for VIP due to the fact that changes in the VIP N-terminal part drastically decrease affinity for receptors. The interaction of peptide agonist N terminus with class II GPCR serpentine region is suggested by the two-step model of activation of class II GPCR (6, 21) in which the receptor N-ted binds the C-terminal part of the ligand and then positions the N-terminal portion of the peptide agonist in proximity of the serpentine receptor core. This would be consistent with previous mutagenesis experiments of the hVPAC1 receptor providing indirect evidence that the VIP Asp3 side chain fitted inside the transmembrane helix bundle of the receptor (22) and demonstrating that a few amino acid residues in the juxtamembrane region of the receptor play a significant role in VIP binding (7). Further experiments are clearly needed to delineate the site(s) of interaction of the N-terminal portion of VIP with its receptor and more generally of natural peptide agonists with class II GPCRs.
3) A third question raised by the studies described herein is related to the role of the N-terminal 1–5 segment of VIP in the positioning of VIP in the VPAC1 receptor N-ted. It is well known that deletion of the first residues of VIP drastically decreases affinity for receptors and potency in stimulating adenylyl cyclase activity (23, 24). As discussed above, a likely explanation is that the VIP N-terminal segment has its own site of interaction with the VPAC1 receptor. However, an alternative explanation could be that deletion of the VIP N terminus alters the positioning of the 6–28 helical segment in the VPAC1 receptor N-ted. Unfortunately, such deletion results in a decrease of affinity for receptors (23, 24) so important that it is not possible to determine experimentally if it would change the photoaffinity labeling pattern of the receptor.
In summary, this study shows clearly that the entire α helix of VIP from Phe6 to C-terminal Asn28 lies in the VPAC1 receptor N-ted. This is the first characterization of the site of interaction of a natural peptide agonist with a class II GPCR. The next step will be to delineate the receptor domains that physically interact with the N-terminal end of VIP. This is a crucial issue because the VIP N-terminal domain, as well as the N-terminal domain of all peptides interacting with class II GPCRs, plays a crucial role in the activation of such receptors.
MATERIALS AND METHODS
Materials
The hVPAC1 receptor was cloned and fused in the C-terminal position with green fluorescent protein as described elsewhere (25). This receptor construct was stably transfected in CHO cells (15) and displayed the same phenotype as compared with the wild-type receptor (wt) in term of VIP binding and adenylyl cyclase activation by VIP (15). The [Bpa28]-VIP photoaffinity probe in which the C-terminal residue (Asn28) is substituted by a photoreactive p-benzoyl-l-Phe residue (Bpa), was obtained by custom solid-phase synthesis from GL Biochem (Shangaï, China). [125I]VIP and [125I-Bpa28]VIP were prepared and purified as described (13). Addition of CNBr cleavage site in the hVPAC1 receptor was obtained by site-directed mutagenesis by methionine substitution of 120I as reported (9). All chemical compounds used were from Sigma (Saint-Quentin-Fallavier, France).
Cell Culture and Cell Homogenate Preparation
The CHO cell clones expressing the hVPAC1 receptor or the I120M receptor mutant were grown in F12-HAM medium supplemented with 10% decomplemented fetal calf serum, 100 U/ml penicillin G, and 100 μg/ml streptomycin in a humidified atmosphere containing 95% air and 5% CO2 at 37 C. Cell membrane preparation was performed as described (26). Briefly, after removing the culture medium, cells were washed two times with PBS, and then harvested with rubber policeman and centrifuged for 5 min at 3000 × g. Cell pellets were resuspended in 5 mm HEPES buffer, pH 7.5, and incubated for 30 min at 4 C. Cell suspension was homogenized with a Dounce homogenizer and centrifuged for 20 min at 11,000 × g, and pellets were stored at −80 C until use.
VIP Binding and Adenylyl Cyclase Activity Assays
Ligand binding assay was performed as reported (13). Briefly, cell homogenates were incubated for 1 h at 30 C with 0.05 nm [125I]VIP in the presence of increasing concentrations of VIP or [Bpa28]VIP, in 20 mm HEPES buffer, pH 7.5, containing 2% (wt/vol) BSA. Reaction was stopped by addition of ice-cold HEPES buffer, pH 7.5, and centrifugation for 20 min at 11,000 × g. Specific binding was calculated as the difference between the amount of [125I]VIP bound in the absence (total binding) and the presence (nonspecific binding) of 1 μm unlabeled VIP. The concentrations of peptides that elicited half-maximal inhibition of specific [125I]VIP binding (Ki) were determined by computer analysis. Adenylyl cyclase activity was determined in presence of increasing concentration of VIP or [Bpa28]VIP as described (26). Dose-response curves were fitted, and concentrations of peptides giving half-maximal responses (EC50) were calculated using the Prism software suite (GraphPad Software, San Diego, CA).
Receptor Photoaffinity Labeling
Transfected CHO cells were incubated in the dark for 1 h at 25 C in the presence of 10 nm [125I-Bpa28]VIP in 20 mm HEPES buffer, pH 7.5, containing 2% (wt/vol) BSA, 1 mm phenylmethylsulfonyl fluoride, and 50 μg/ml tosyl lysyl chloromethylketone as reported (10). Reaction was stopped by adding ice-cold 20 mm HEPES buffer. After centrifugation for 5 min at 3000 × g, cell pellets were suspended in 1 ml of ice-cold 20 mm HEPES buffer and exposed to UV irradiation (λ = 365 nm) for 30 min. After UV exposure, cells were collected and washed two times with 10 mm HEPES containing 25 mm glycine, 75 mm NaCl buffer, pH 7.5, and one time with 20 mm HEPES as described (10). Cell pellets were solubilized in SDS sample buffer containing 20 mm dithiothreitol and then subjected to SDS-PAGE separation on 12% polyacrylamide gel for subsequent detection of labeled protein by autoradiography.
CNBr and Enzyme Cleavages
Radiolabeled receptor bands were cut out from polyacrylamide gel and incubated overnight in 1 ml of pure water at room temperature under agitation, and water solution was lyophilized under vacuum. Purified labeled receptor was subjected to CNBr or enzyme cleavages as described (9). Briefly, labeled receptor was resuspended in 80% formic acid containing 10 mg/ml CNBr and incubated overnight at room temperature. CNBr cleavage products were separated by SDS-PAGE electrophoresis. Radiolabeled band was excised, eluted, lyophilized, and then submitted to enzymatic cleavage. Endopeptidase Glu-C and endoglycosidase F were used separately or in sequence to cleave radiolabeled hVPAC1 receptor or mutant as previously reported (10). Digested materials were analyzed on NuPAGE Bis-Tris Gel using 2-(N-morpholino)ethanesulfonic acid running buffer (Invitrogen, Carlsbad, CA). The apparent molecular masses of radiolabeled receptor fragments were determined by interpolation on a plot of the mobility of the Rainbow-colored protein molecular weight markers from Amersham Pharmacia Biotech (Piscataway, NJ) or Benchmark-prestained protein ladder from Invitrogen vs. the log values of their masses.
Edman Degradation Sequencing
In addition to peptide mapping by chemical or enzymatic cleavages, determination of hVPAC1 receptor fragment affinity-labeled with [125I-Bpa28]VIP probe was carried out by Edman degradation sequencing as previously described (10). Briefly, CHO cells expressing the I120M receptor mutant were incubated with [125I-Bpa28]VIP probe, washed, and UV exposed. Cells were washed and centrifuged, and pellets were analyzed by NuPage electrophoresis. Radiolabeled receptor was purified from gel and cleaved by CNBr as described above. CNBr fragments were then dissolved in a buffer containing 10 mg/ml of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and immobilized in C-terminal position by coupling to 20 mg of N-(2-aminoethyl-1)-3-aminopropyl glass beads for 2 h at room temperature. It should be noted that the carboxy group of [125I-Bpa28]VIP was amidated and did not interfere with the coupling to the beads. Immobilized fragments were subjected to manual Edman degradation sequencing up to six cycles as follows: beads were washed with 50 μl triethylamine for three times and dried. Triethylamine/methanol/phenylisothiocy-anate mixture (60 μl; 1:7:1) was added to beads and incubated at 50 C for 5 min. Beads were washed three times and dried. Dried beads were incubated with 50 μl of trifluoroacetic acid for 5 min at 25 C and washed three times with 200 μl of methanol. Washing methanol effluents were pooled and evaporated, and the radioactivity released was quantified in a γ-spectrometer. The beads were dried and ready to use for the next cycle.
Molecular Modeling
The three-dimensional NMR structure of the amino-terminal domain of the mouse CRF 2β receptor (11) was used to model the N-ted of the hVPAC1 receptor by homology (10). Sequence of the N-ted (1–137) of the hVPAC1 receptor was aligned with that of the N-ted of the CRF 2β receptor (PDB code: 1U34) using FASTA algorithm (27). Based on sequence alignment, 100 homology models were built using Modeler v6.0, and objective function was calculated for each model to select the best score (28). Energy minimization calculation was performed with the AMBER force field. The model has 77% of the Φ, Ψ angle pairs in the allowed region of Ramachandran plot indicating a correct stereochemistry. It must be noticed that RMSD analysis of the 20 NMR structures for the CRF receptor (11) gave a value of 0.81 Å for the structured core, but a value of 17.7 Å for the entire structure, which includes largely variable structures for N- and C-terminal regions. We used only a part of the CRF2β receptor N-ted (residue 39–124) to construct our model of the hVPAC1 receptor N-ted, and RMSD of this domain of CRF2β receptor N-ted shows a value of 4.5 Å close to RMSD observed in molecular dynamic simulations (see below).
Docking Calculations
Using HADDOCK 1.2 software (29) the NMR lowest-energy conformers of VIP (10) were docked into the three-dimensional model (44–137) of the hVPAC1 receptor N-ted under interaction restraints between the residues Phe6, Tyr22, Asn24 (8, 9, 10), and Asn24 (this paper) of VIP and the residues Asp107, Gly116, Cys122, and Lys127 of the receptor N-ted, respectively. For the first iteration, 2000 rigid docking structures were generated, and then in the second iteration the 200 best structures were subjected to a semiflexible simulated annealing.
Molecular Dynamic Simulations
Docked structures of hVPAC1 receptor N-ted complexed with VIP as well as hVPAC1 receptor N-ted alone were solvated using a cubic box of TIP3 water molecules extended 15 Å from protein atoms using the Visual Molecular Dynamics 1.8.5 package (30). Minimization of 4000 steps was then performed using the standard procedure of Nanoscale Molecular Dynamics package (31). Molecular dynamic simulations were calculated using the Charmm 27 parameters into Nanoscale Molecular Dynamics code (32). A time step of 1 fsec was used, and all bonds involving hydrogen were constrained using the SHAKE algorithm. A cutoff of 12 Å combined with a pair list distance of 13.5 Å was applied with the switching method. Periodic boundary conditions and Particle Mesh Ewald method were employed. In a first step, the simulation system was slowly brought from 0 to 310 K over 75 psec, and then the temperature was equilibrated over 4 nsec; finally pressure and temperature were both equilibrated for 1 nsec using the Langevin Piston Method. During the production step conducted for 9 nsec, only the pressure was equilibrated. Distance measurements between two amino acids were performed by the following procedure: distances between all atoms of these two residues were calculated and the minimal distance was retained. The RMSD of the selected backbone atoms was calculated from the trajectory at 25-ps intervals with the initial structure as reference. The RMSF of each backbone atom within a given residue of the hVPAC1 N-ted was calculated similarly.
Footnotes
This work was supported by the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique and the Université Paris 7. The research is also supported by Association pour la Recherche sur la Polyarthrite (Grant 2007 to A.C.) and Association Charles Debray.
Disclosure Statement: The authors have nothing to disclose.
First Published Online September 20, 2007
Abbreviations: Bpa, p-Benzoyl-l-phenylalanine; CHO cells, Chinese hamster ovary cells; CRF, corticotropin-releasing factor; GPCR, G protein-coupled receptor; h, human; NMR, nuclear magnetic resonance; N-ted, N-terminal ectodomain; PACAP, pituitary adenylate cyclase-activating peptide; RMSD, root mean square deviation; RMSF, root mean square fluctuation; SDS, sodium dodecyl sulfate; VIP, vasoactive intestinal peptide; VPAC, VIP/ PACAP receptor.
References
- 1.Sherwood NM, Krueckl SL, McRory, JE 2000. The origin and function of the pituitary adenylate cyclase-activating polypeptide (PACAP)/glucagon superfamily. Endocr Rev 21:619–670 [DOI] [PubMed] [Google Scholar]
- 2.Delgado MD, Pozo D, Ganea D 2004. The significance of vasoactive intestinal peptide in immunomodulation. Pharmacol Rev 56:249–290 [DOI] [PubMed] [Google Scholar]
- 3.Vaudry H, Laburthe M 2006. VIP, PACAP and related peptides: from gene to therapy. Ann NY Acad Sci 1070:1–633 [PubMed] [Google Scholar]
- 4.Gomariz RP, Juarranz Y, Abad C, Arranz A, Leceta J, Martinez C 2006. VIP-PACAP system in immunity: new insights for multitarget therapy. Ann NY Acad Sci 1070:51–74 [DOI] [PubMed] [Google Scholar]
- 5.Gozes I., Fridkinb M, Hill JM, Brenneman DE 1999. Pharmaceutical VIP: prospects and problems. Curr Med Chem 6:1019–1034 [PubMed] [Google Scholar]
- 6.Laburthe M, Couvineau A, Tan V 2007. Class II G protein-coupled receptors for VIP and PACAP: structure, models of activation and pharmacology. Peptides 28:1631–1639 [DOI] [PubMed] [Google Scholar]
- 7.Laburthe M, Couvineau A, Marie JC 2002. VPAC receptors for VIP and PACAP. Receptors Channels 8:137–153 [PubMed] [Google Scholar]
- 8.Tan YV, Couvineau A, Van Rampelbergh J, Laburthe M 2003. Photoaffinity labeling demonstrates physical contact between vasoactive intestinal peptide and the N-terminal ectodomain of the human VPAC1 receptor. J Biol Chem 278:36531–36536 [DOI] [PubMed] [Google Scholar]
- 9.Tan YV, Couvineau A, Laburthe M 2004. Diffuse pharmacophoric domains of vasoactive intestinal peptide (VIP) and further insights into the interaction of VIP with the N-terminal ectodomain of human VPAC1 receptor by photoaffinity labeling with [Bpa6]-VIP. J Biol Chem 279:38889–38894 [DOI] [PubMed] [Google Scholar]
- 10.Tan YV, Couvineau A, Murail S, Ceraudo E, Neumann JM, Lacapère JJ, Laburthe M 2006. Peptide agonist docking in the N-terminal ectodomain of a class II G protein-coupled receptor, the VPAC1 receptor. Photoaffinity, NMR, and molecular modeling. J Biol Chem 281:12792–12798 [DOI] [PubMed] [Google Scholar]
- 11.Grace CR, Perrin MH, DiGruccio MR, Miller CL, Rivier JE, Vale WW, Riek R 2004. NMR structure and peptide hormone binding site of the first extracellular domain of a type B1 G protein-coupled receptor. Proc Natl Acad Sci USA 101:12836–12841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarzenbacher R, Zeth K, Diederichs K, Gries A, Kostner GM, Laggner P, Prassl R 1999. Crystal structure of human β2-glycoprotein I: implications for phospholipid binding and the antiphospholipid syndrome. EMBO J 18:6228–6239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicole P, Lins L, Rouyer-Fessard C, Drouot C, Fulcrand P, Thomas A, Couvineau A, Martinez J, Brasseur R, Laburthe M 2000. Identification of key residues for interaction of vasoactive intestinal peptide with human VPAC1 and VPAC2 receptors and development of a highly selective VPAC1 receptor agonist. Alanine scanning and molecular modeling of the peptide. J Biol Chem 275:24003–24012 [DOI] [PubMed] [Google Scholar]
- 14.Couvineau A, Fabre C, Gaudin P, Maoret J, Laburthe M 1996. Mutagenesis of N-glycosylation sites in the human vasoactive intestinal peptide 1 receptor. Evidence that asparagine 58 or 69 is crucial for correct delivery of the receptor to plasma membrane. Biochemistry 35:1745–1752 [DOI] [PubMed] [Google Scholar]
- 15.Sankaranarayanan K, Ramasamy P 2006. Growth of benzophenone single crystals from solution: a novel approach with 100% solute-crystal conversion efficiency. Cryst Res Technol 41:225–230 [Google Scholar]
- 16.Wray V, Nokihara K, Naruse S, Ando E, Kakoschke C, Wei M 1995. Synthesis, solution structure and biological action of PACAP-related peptide. Biomed Pept Proteins Nucleic Acids 1:77–82 [PubMed] [Google Scholar]
- 17.Dong M, Miller LJ 2006. Use of photoaffinity labeling to understand the molecular basis of ligand binding to the secretin receptor. Ann NY Acad Sci 1070:248–264 [DOI] [PubMed] [Google Scholar]
- 18.Gensure RC, Shimizu N, Tsang J, Gardella TJ 2003. Identification of a contact site for residue 19 of parathyroid hormone (PTH) and PTH-related protein analogs in transmembrane domain two of the type 1 PTH receptor. Mol Endocrinol 17:2647–2658 [DOI] [PubMed] [Google Scholar]
- 19.Grace CR, Perrin MH, Gulyas J, Digruccio MR, Cantle JP, Rivier JE, Vale WW, Riek R 2007. Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand. Proc Natl Acad Sci USA 104:4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun C, Song D, Davis-Taber RA, Barrett LW, Scott VE, Richardson PL, Pereda-Lopez A, Uchic ME, Solomon LR, Lake MR, Walter KA, Hajduk PJ, Olejniczak ET 2007. Solution structure and mutational analysis of pituitary adenylate cyclase-activating polypeptide binding to the extracellular domain of PAC1-RS. Proc Natl Acad Sci USA 104:7875–7880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoare SRJ 2005. Mechanisms of peptide and nonpeptide ligand binding to class B G-protein-coupled receptors. Drug Discov Today 10:417–427 [DOI] [PubMed] [Google Scholar]
- 22.Couvineau A, Rouyer-Fessard C, Fournier A, St Pierre S, Pipkorn R, Laburthe M 1984. Structural requirements for VIP interaction with specific receptors in human and rat intestinal membranes: effect of nine partial sequences. Biochem Biophys Res Commun 121:493–498 [DOI] [PubMed] [Google Scholar]
- 23.Langer I, Vertongen P, Perret J, Cnudde J, Gregoire F, De Neef P, Robberecht P, Waelbroeck M 2002. VPAC(1) receptors have different agonist efficacy profiles on membrane and intact cells. Cell Signal 14:689–694 [DOI] [PubMed] [Google Scholar]
- 24.Solano RM, Langer I, Perret J, Vertongen P, Juarranz MG, Robberecht P, Waelbroeck M 2001. Two basic residues of the h-VPAC1 receptor second transmembrane helix are essential for ligand binding and signal transduction. J Biol Chem 276:1084–1088 [DOI] [PubMed] [Google Scholar]
- 25.Gaudin P, Maoret J, Couvineau A, Rouyer-Fessard C, Laburthe M 1998. Constitutive activation of the human vasoactive intestinal peptide 1 receptor, a member of the new class II family of G protein-coupled receptors. J Biol Chem 273:4990–4996 [DOI] [PubMed] [Google Scholar]
- 26.Couvineau A, Lacapère JJ, Tan YV, Rouyer-Fessard C, Nicole P, Laburthe M 2003. Identification of cytoplasmic domains of hVPAC1 receptor required for activation of adenylyl cyclase. Crucial role of two charged amino acids strictly conserved in class II G protein-coupled receptors. J Biol Chem 278:24759–24766 [DOI] [PubMed] [Google Scholar]
- 27.Pearson WR, Lipman DJ 1988. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85:2444–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lins L, Couvineau A, Rouyer-Fessard C, Nicole P, Maoret J, Benhamed M, Brasseur R, Thomas A, Laburthe M 2001. The human VPAC1 receptor: three-dimensional model and mutagenesis of the N-terminal domain. J Biol Chem 276:10153–10160 [DOI] [PubMed] [Google Scholar]
- 29.Dominguez C, Boelens R, Bonvin AMJJ 2003. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125:1731–1737 [DOI] [PubMed] [Google Scholar]
- 30.Humphrey W, Dalke A, Schulten K 1996. VMD: visual molecular dynamics. J Mol Graphics 14:33–38 [DOI] [PubMed] [Google Scholar]
- 31.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid V, Villa E, Chipot C, Skeel R D, Kale L, Schulten K 2005. Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 4:187–217 [Google Scholar]