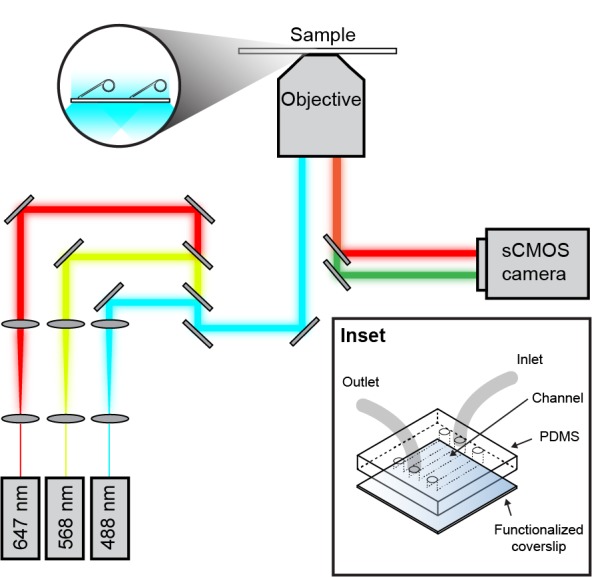
Figure 1. Single-molecule rolling-circle replication assay.
(A) Canonical view of the organization of the E. coli replication fork. The DnaB helicase encircles the lagging strand, facilitates unwinding of dsDNA through ATP hydrolysis, and recruits DnaG primase for synthesis of RNA primers that initiate synthesis of 1–2 kb Okazaki fragments on the lagging strand. The extruded single-stranded (ss) DNA is protected by ssDNA-binding protein, SSB. The Pol III holoenzyme (HE) uses the ssDNA of both strands as a template for coupled, simultaneous synthesis of a pair of new DNA duplex molecules. The β2 sliding clamp confers high processivity on the Pol III HE by tethering the αεθ Pol III core onto the DNA. The clamp loader complex (CLC) assembles the β2 clamp onto RNA primer junctions. Up to three Pol III cores interact with the CLC through its τ subunits to form the Pol III* complex, and the τ subunits also interact with DnaB, thus coupling the Pol III HE to the helicase. (B) Schematic representation of the experimental design. 5'-Biotinylated M13 DNA is coupled to the passivated surface of a microfluidic flow cell through a streptavidin linkage. Addition of the E. coli replication proteins and nucleotides initiates DNA synthesis. The DNA products are elongated hydrodynamically by flow, labeled with intercalating DNA stain, and visualized using fluorescence microscopy (Figure 1—figure supplement 1).
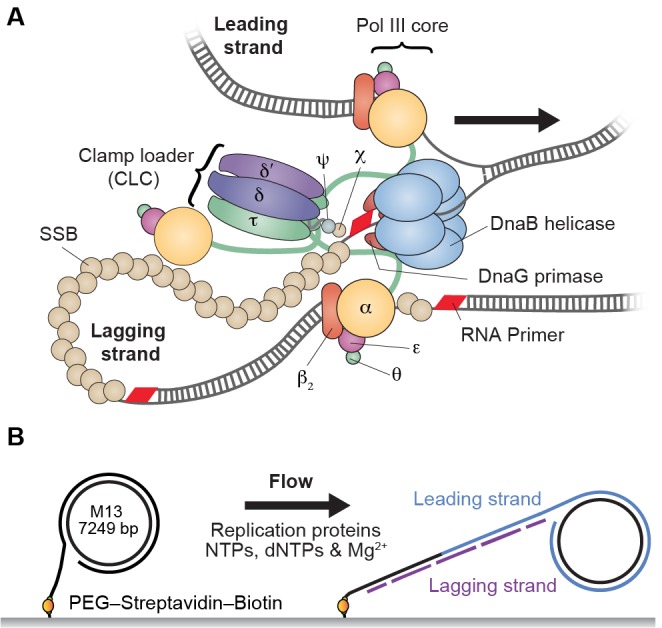
Figure 1—figure supplement 1. Schematic overview of the single-molecule fluorescence microscope.