Figure 3. Pre-assembled Pol III* complexes do not exchange Pol III core.
(A) Red and green Pol III* are separately pre-assembled by treatment at 37°C for 15 min (30 nM Pol III core and 10 nM τ3-CLC). These are then mixed in equal ratios and kept at 37°C for 1 hr prior to dilution to 6 pM Pol III* for imaging. (B) Red Pol III* complexes and green Pol III* complexes do not co-localize to produce any white spots as seen in Figure 3—figure supplement 1, demonstrating the α–τ interaction within the Pol III* complex remains intact for the duration of the DNA replication assays. (C) Pre-assembled red and green Pol III* complexes that have participated in DNA replication (at 3.3 nM of each) do not co-localize, demonstrating that the Pol III cores within a Pol III* do not exchange with cores from other Pol III*s at the replication fork during active DNA synthesis. White scale bars represent 5 μm.
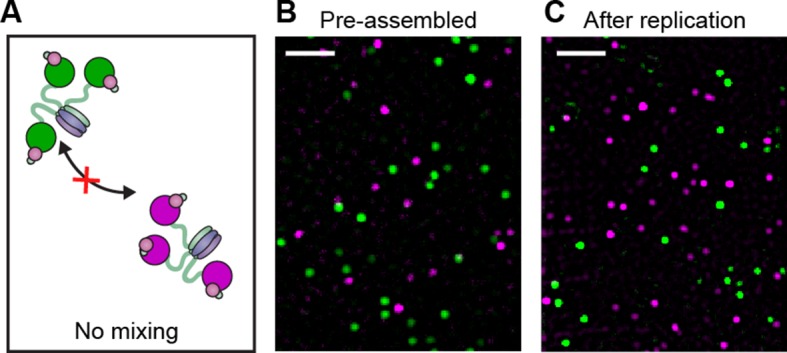
Figure 3—figure supplement 1. Pol III* complexes of mixed Pol III core composition (1:1) show co-localization.
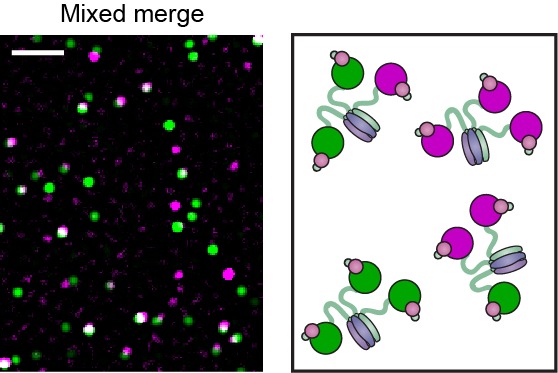
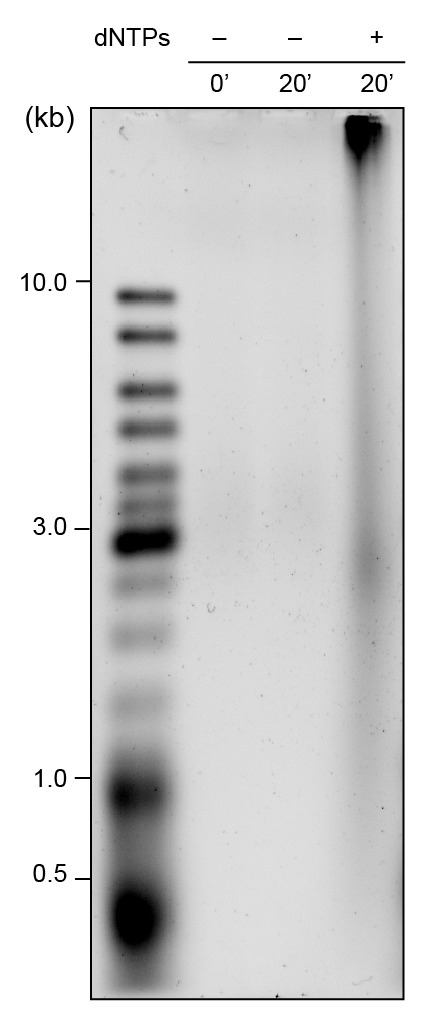
Figure 3—figure supplement 2. Alkaline gel showing leading- and lagging-strand products using pre-assembled red and green Pol III*s.