Abstract
Transmembrane (TMEM)-176A and 176B proteins belong to the MS4A family of proteins whose function in the immune system remains unclear. TMEM176A transcripts were previously shown to be elevated in liver cancer or kidney tissue with proteinuria, while marked changes in TMEM176B transcripts have been found in tolerated tissue allografts and neoplastic fibroblasts. To study the functional relationship between human TMEM176A and 176B and their putative link to cancer, we used polymerase chain reaction and biochemical assays. Here, we show that TMEM176A and 176B are widely expressed in all human tissues examined. Co-immunoprecipitation of heterologously expressed TMEM176A and 176B revealed direct physical interaction. To determine the relevance of such interaction to cancer pathology, we analyzed biopsied tissue samples from a variety of normal and cancer tissues. Our data reveal that human TMEM176A and 176B protein levels are significantly elevated in lymphoma, but not in normal tissues. The protein levels of TMEM176A are also significantly increased in lung carcinoma. Finally, analysis of the protein expression ratio of TMEM176A over 176B showed significant differences between normal and cancer tissues of the breast, lymph, skin, and liver, which indicates that both TMEM proteins could be potential useful markers for certain human cancers.
Keywords: Breast cancer, Melanoma, Lymphoma, Hepatocellular carcinoma
Introduction
The transmembrane proteins TMEM176A and 176B belong to the CD20/Fc-εRIβ and membrane-spanning 4A (MS4A) family of proteins (Lurton et al., 1999; Zuccolo et al., 2010). Human TMEM176A was first identified from a screen of tumor-associated antigens in hepatocellular carcinoma (HCC) (Wang et al., 2002). However, when the author created a phage clone expressing TMEM176A and tested it in allogeneic sera (taken from different patients) with hepatitis B virus (HBV)-infected, with head and neck cancer, and healthy normal individuals, the TMEM176A protein was not reactive (Wang et al., 2002). Nevertheless, an increase in human TMEM176A mRNA transcript level has been observed in transplanted human livers that relapsed from hepatitis C virus (HCV) infection (Gehrau et al., 2011). Similarly, an elevated mouse Tmem176A transcript level has been detected in proximal tubule cells of the mouse kidney nephron (Nakajima et al., 2002). This was presumably caused by an inflammatory response to proteinuria (Nakajima et al., 2002). It is not clear if the observed upregulation of human TMEM176A transcripts, or its mouse homolog, parallels an increase in protein expression in any of these pathological states.
Human TMEM176B (also known as LR8) was first discovered in human lung fibroblasts (Lurton et al., 1999), and was recently associated in human small cell lung carcinoma (Gottschling et al., 2012). Similar to TMEM176A, the level of human TMEM176B transcripts has been shown to be markedly elevated in transplanted livers that showed recurrence of HCV infection (Gehrau et al., 2011). On the other hand, the mRNA expression of rat Tmem176B (also known as rat TORID, or mouse Clast1/LR8) is highly correlated with tolerated allografts (transplanted tissue taken from a different individual of the same species) (Louvet et al., 2005; Condamine et al., 2010). This finding suggests that Tmem176B may be involved in the regulation of immune cell activation and tolerance (Louvet et al., 2005; Condamine et al., 2010). Indeed, the tissue mRNA expression profiles of rat and mouse Tmem176A or Tmem176B point to an important role in immune function. For example, both rodent Tmem176A and Tmem176B transcripts are detected preferentially in lymphoid tissues (lymph nodes, thymus, spleen, and bone marrow), lung, kidney, as well as certain types of myeloid and lymphoid cells (Louvet et al., 2005; Condamine et al., 2010; Ghosh et al., 2010). Inflammatory stimulation using lipopolysaccharide (LPS) or poly-I:C exposure of mouse bone marrow-derived dendritic cells (BMDCs) results in down-regulation of Tmem176A and 176B transcript levels (Condamine et al., 2010). Furthermore, RNA interference (RNAi)-mediated inhibition of either mouse Tmem176A or Tmem176B has been shown to maintain the immature state of BMDCs (Condamine et al., 2010). Likewise, RNAi-induced knockdown of mouse TMEM 176A and 176B in immature BMDCs suppresses the production of co-stimulatory molecules such as Cluster of Differentiation (CD)-80, CD86, and CD40, which prevent the activation of specific T cells in culture (Condamine et al., 2010). The authors, however, did not report a significant change in the levels of major histocompatibility complex (MHC)-I and MHC-II proteins, which seemed contradictory to their previous observations that over-expression of rat Tmem176B in BMDCs results in reduction of MHC-II and CD86 proteins (Louvet et al., 2005). Nevertheless, a consistent finding for rodent TMEM proteins is its negative effects on the activation and maturation of immature conventional dendritic cells (cDCs) (Louvet et al., 2005; Condamine et al., 2010).
Bioinformatics analysis of mouse Tmem176A and 176B amino acid sequences predicts both proteins to contain four transmembrane domains with intracellular amino and carboxyl termini (Condamine et al., 2010). To elucidate further the function of mouse Tmem176B, Condamine et al. (2010) performed a genetic screening using a membrane-based (split ubiquitin) yeast two-hybrid technique. Using the bait-dependency test, the authors found that mouse Tmem176A and 176B interact with each other (Condamine et al., 2010), which possibly explains the functional relationship between the two proteins. Notwithstanding, there is a need to show biochemical evidence of the interaction between Tmem176A and 176B, in order to confirm their functional association.
Several recent reports have implicated both human TMEM176A and 176B in human cancer. Abnormal DNA methylation of CpG islands in human TMEM176A and 176B has been linked to breast cancer (Strelnikov et al., 2010). A study using serial analysis of gene expression showed that both 5′ and 3′ intronic transcripts encoding the human TMEM176B gene are significantly reduced in HCC tissues (Hodo et al., 2010). It has been proposed that the oncogenic Ras protein reduces mouse TMEM176B transcription in response to hypermethylation and histone deacetylation (Ryu et al., 2010). Despite the apparent correlation between transcript levels and tumorigenesis, to date no direct evidence has been established regarding abnormal protein levels of human TMEM176A or 176B and cancer pathology. Evidence is also lacking on the functional relevance of TMEM176A and 176B interaction in human tumor formation.
In this report, we analyze the mRNA transcript and protein expression levels of human TMEM176A and 176B to shed light on its distribution pattern in tissues and organs. We also investigate the subcellular co-localization of human TMEM176A and 176B protein in cultured human cells, their putative interaction, and how such interaction could be associated with certain cancer pathology.
Materials and methods
Polymerase chain reaction (PCR): standard PCR and real-time quantitative PCR
Human tissue cDNA samples were commercially purchased from Clontech (Mountain View, CA, USA). Clontech’s multiple tissue cDNA (MTC) panels I and II include the following tissues: brain, thymus, heart, lung, liver, spleen, pancreas, small intestine, colon, kidney, skeletal muscle, prostate, ovary, testis, placenta, and peripheral blood leukocyte. According to the manufacturer, the human MTC panels have been normalized against several housekeeping genes (α-tubulin, β-actin, glyceraldehyde 3-phosphate dehydrogenase [GAPDH], phospholipase A2) and against each other to determine accurately the abundance of target mRNA. Table 1 outlines the standard and quantitative PCR primer sets used to amplify human TMEM176A, TMEM176B, GAPDH, and 18S ribosomal RNA (rRNA). The standard PCR reactions were performed using a 2× DreamTaq Green mastermix (Fermentas, Glen Burnie, MD, USA). We used GAPDH for loading control and for relative comparison of individual tissue band intensities.
Table 1.
Standard and real-time quantitative PCR primer sets.
| Gene name | Primer sequence | |
|---|---|---|
| Human TMEM176A | ||
| Standard PCR | Forward: 5′-ATGGGAACAGCCGACAGTGAT-3′ | Reverse: 5′-CTAGATTCCACTCACTTCCAA-3′ |
| Real-time QPCR | Forward: 5′-CATGGACATGCTGAAGGCCTTGTT-3′ | Reverse: 5′-ACATTCTCCAGCAGTACAGCCACA-3′ |
| Human TMEM176B | ||
| Standard PCR | Forward: 5′-ATGACGCAAAACACGGTGATT-3′ | Reverse: 5′-TCACAGGACAATGGCAGTGGA-3′ |
| Real-time QPCR | Forward: 5′-GCGAAGTCAAGAGAACCAATG-3′ | Reverse: 5′-CTACTCCCAAGGAAACCAAGG-3′ |
| Human GAPDH | ||
| Standard PCR | Forward: 5′-ATGGGGAAGGTGAAGGTCG-3′ | Reverse: 5′-AGTGGTCGTTGAGGGCAAT-3′ |
| Human 18S rRNA | ||
| Real-time QPCR | Forward: 5′-GCCCGAAGCGTTTACTTTG-3′ | Reverse: 5′-CCCTCTTAATCATGGCCTCAG-3′ |
Our QPCR assay followed the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (Bustin et al., 2009). The real-time QPCR reactions were performed using a 2× SensiMix SYBR green mastermix (Bioline, Tauton, MA, USA) in a CFX96 thermocycler (BioRad, Hercules, CA, USA). All three independent experiments included a standard curve with correlation coefficient (R2) value (means ± SD) of 0.999 ± 0.001. The average and standard deviation values of our QPCR efficiencies for each primer set were: TMEM176A = 99.5 ± 0.09%; TMEM176B = 98.6 ± 0.11%; and 18S rRNA = 94.6 ± 0.03%. The QPCR data were analyzed using the Livak method (2−ΔΔCq) to obtain the normalized expression ratio of each tissue sample’s quantitative cycle (Cq). The human 18S rRNA Cq values were used as the reference gene (normalizer) and the peripheral blood leukocyte Cq values were used as the calibrator. We chose human 18S rRNA as our internal normalization control due to its robust and stable expression levels across many tissues and cell lines (Cuajungco et al., 2003). All data were represented as means ± SEM (n = 3 independent trials).
Co-immunoprecipitation and Western blot techniques
Sequence-verified and expression-verified TruORF™ clones of human TMEM176A (HsTMEM176A-HA) and 176B (HsTMEM176A-Myc/DDK) proteins were purchased from Origene Technologies (Rockville, MD, USA). Co-immunoprecipitation (co-IP) and Western blot assays were performed as previously described with minor modifications (Cuajungco et al., 2006). In brief, heterologously expressed HsTMEM176A-HA and HsTMEM176B-Myc/DDK were co-transfected with TurboFect™ (Fermentas) into human embryonic kidney (HEK)-293 cells. The cells were lysed with standard radioimmunoprecipitation assay (RIPA) buffer plus protease inhibitor cocktail (Fermentas). The lysates were co-immunoprecipitated with anti-DDK monoclonal antibody (mAb) (OriGene Technologies, Rockville, MD, USA) and incubated with Dynabead™ protein G as per the manufacturer’s recommendation (Invitrogen, Carlsbad, CA, USA). Following a series of washes with RIPA buffer, the samples were eluted and run on 4–12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Western blotted using anti-HA mouse mAb or rabbit polyclonal (pAb) (Sigma–Aldrich, St. Louis, MO, USA), blocked with 1× LI-COR buffer, washed with Tris-buffered saline plus 0.1% Triton-X 100 (TBST), and detected with the corresponding infrared (IR)-Dye 800CW conjugated secondary antibodies using the LI-COR Odyssey Sa™ imaging scanner (LI-COR Biosciences, Lincoln, NE, USA). Single transfection experiments of HsTMEM176A-HA construct, and no primary antibody control were included in the co-IP experiments (n = 3 independent trials). The calculated molecular weight of human TMEM176A is approximately 26.11 kDa, while for human TMEM176B is approximately 29.05 kDa.
Mass spectrometry (MS) and bioinformatics analysis
To validate our co-IP results, we determined the identity of the eluted protein samples using MS. We took all necessary precautions to avoid potential contamination that could confound the MS analysis. For this experiment, cell lysates co-transfected with HsTMEM176A-HA and HsTMEM176B-Myc/DDK were immunoprecipitated with anti-DDK mAb, washed, eluted, and run on SDS-PAGE to resolve the protein as described earlier. The negative control samples were run on the same gel. Subsequently, the gel was stained with Coomassie blue (SimplyBlue™ Safestain, Invitrogen, San Diego, CA, USA), and destained with distilled deionized sterile water. The detectable band corresponding to the expected size of human TMEM176A was carefully excised from the co-IP and from the control lanes. All samples were placed in a sterile tube, frozen, and sent to the Stanford University Mass Spectrometry Facility (Stanford, CA, USA) for protein analysis and identification using liquid chromatography (LC)–MS/MS technique. The data were analyzed using the Scaffold 3 software (Proteome Software, Portland, OR, USA).
For protein sequence alignment, we used Lasergene 9 software (DNAstar, Madison, WI, USA), as well as other online tools such as Clustal W (http://www.clustal.org/clustal2/) and Sequence Identities and Similarities (http://imed.med.ucm.es/Tools/sias.html).
Confocal microscopy
We subcloned both human TMEM176A and 176B into a green fluorescent protein (GFP; Clontech, Mountain View, CA, USA) tagged vector (HsTMEM176A-GFP), and a monomeric cherry (mCherry; a kind gift from Roger Tsien, UC San Diego) tagged vector (HsTMEM176B-mCherry). The integrity of both constructs was verified by sequencing prior to use. The constructs were either transfected individually, or transfected together with TurboFect™ (Fermentas, Hanover, MD, USA) in HEK-293 cells plated on glass coverslips. The cells were fixed with 4% paraformaldehyde 24–48 h post-transfection, washed 3× with phosphate-buffered saline (PBS), mounted on slides with ProLong™ Gold plus DAPI (Invitrogen, Carlsbad, CA, USA) and imaged using a Leica confocal microscope.
Immunofluorescence analysis of tissue samples
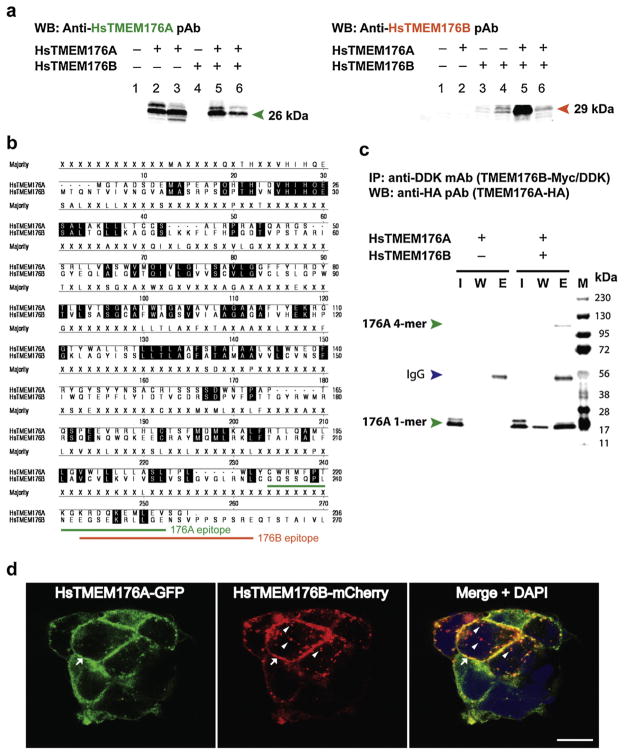
Custom rabbit polyclonal antibodies (pAb) directed at human (Hs) TMEM176A and 176B proteins were produced by Covance (Denver, PA, USA) and ProSci (Poway, CA, USA), respectively. Both anti-HsTMEM176A and anti-HsTMEM176B pAbs were affinity-purified using gel chromatography. Western blot analyses using heterologously expressed human (Hs) TMEM176A, HsTMEM176B, or co-expressed proteins provided evidence for the specificity of each corresponding antibody (Fig. 2a). The amino acid epitope corresponding to the HsTMEM176A and 176B pAbs are underlined on the sequence alignment map (Fig. 2b).
Fig. 2.
Biochemical evidence for human TMEM176A and 176B protein-protein interaction and subcellular co-localization of heterologously expressed human TMEM176A and 176B proteins in HEK-293 cells. (a) Representative Western blot image showing the antibody specificity of anti-HsTMEM176A (left panel, green text) and anti-HsTMEM176B (right panel, orange text) polyclonal antibodies (pAbs). Expression constructs were transfected into HEK-293 cells, and the cells were lysed 24 h post-transfection. Left panel lanes 1: negative control (untransfected) lysate; 2: over-expressed HsTMEM176A lysate; 3: co-expressed HsTMEM176A and 176B lysate; 4: overexpressed HsTMEM176B lysate (control); 5: eluted HsTMEM176A from co-IP sample; 6: total protein input from co-IP sample. Right panel lanes 1: negative control (untransfected) lysate; 2: over-expressed HsTMEM176A lysate (control); 3: co-expressed HsTMEM176A and 176B lysate; 4: over-expressed HsTMEM176B lysate; 5: eluted HsTMEM176B from co-IP sample; 6: total protein input from co-IP sample. (b) Amino acid sequence alignment of HsTMEM176A and HsTMEM176B proteins using Clustal W software analysis tool. The amino acid epitopes for anti-HsTMEM176A and anti-HsTMEM176B are underlined (green and orange lines, respectively). (c) Representative Western blot image of HsTMEM176A-HA (single transfection control; left panel) and HsTMEM176A-HA co-expressed with HsTMEM176B-Myc/DDK (right panel). The samples were co-immunoprecipitated with anti-DDK mAb, while eluted samples were detected with anti-HA pAb as described in the Materials and Methods section. Lanes: I, input of total protein lysate; W, wash; E, elution. Green arrowheads indicate HsTMEM176A (monomer or tetramer); blue arrowhead represents mouse immunoglobulin (IgG). (d) Representative confocal micrographs of HsTMEM176A-GFP (left panel), HsTMEM176B-mCherry (middle panel) and composite image stained with DAPI (right panel). DAPI stains the nuclei blue. Both TMEM proteins co-localize along the plasma membrane (arrows), and appear as punctate, vesicle-like structures within the cytoplasm. Note that TMEM176B did not always co-localize in these structures (arrowheads). Scale bar = 10 μm. Hs, Homo sapiens; WB, Western blot; IP, immunoprecipitation; mAb, monoclonal antibody; pAb, polyclonal antibody; 1-mer, monomer; 4-mer, tetramer; IgG, immunoglobulin G.
The TissueFocus™ Cancer Survey Tissue Microarray (TMA) containing formalin-fixed paraffin-embedded core biopsy samples was commercially purchased from Origene Technologies (Rockville, MD, USA). The samples contained 55 normal control tissues and 110 tumors from 11 cancer types: breast, colon, lung, kidney, ovarian, endometrial, stomach, prostate, melanoma, liver, and lymphoma. To quantify the protein levels of human TMEM176A and 176B on the TMA core samples, we followed the manufacturer’s recommended standard protocol on antigen retrieval. The target proteins were detected using fluorometric technique with some modifications to suit our assay. Specifically, we conjugated both anti-TMEM176A and anti-TMEM176B primary rabbit antibodies with IRDye 800CW using a high molecular weight protein conjugation kit from LI-COR Biosciences (Lincoln, NE, USA). A working antibody concentration of 1 mg/ml was used in the assay. We used human β-actin conjugated with Acti-stain™ 670 Phalloidin (14 μM; Cytoskeleton Inc., Denver, CO, USA) as normalization control. We intentionally avoided the use of secondary antibodies for fluorescent signal detection to prevent any potential bias or background signal inflation due to non-specific binding of anti-rabbit secondary immunoglobulin G (IgGs). The tissues were simultaneously probed with anti-TMEM176A-IRDye 800CW (1:500) and Acti-stain™ 670 Phalloidin (1:1000), washed 3× with PBS, scanned, and analyzed using ImageJ (NIH, Bethesda, MD, USA) to obtain the relative fluorescence unit (RFU) intensity. The TMA was then stripped overnight with TBS plus 1% Triton-X 100 (TBST1) and β-mercaptoethanol at 50 °C. The TMA was washed with TBST1 for six times, and then washed with PBS before use. The TMA was scanned again to ensure that it was completely stripped and any background signal was subtracted out during the analysis. Subsequently, the TMA was probed with anti-TMEM176B-IRDye 800CW and Acti-stain™ 670 Phalloidin, scanned, and analyzed with ImageJ as explained earlier. The RFU intensity of each tissue core was normalized against the level of human β-actin protein. The numerical values for each sample were analyzed for significance using Student’s t-test (p-value < 0.05, two-tailed distribution) and data were represented as means ± SEM (n ≤ 10 tumor tissue samples; n ≤ 5 normal tissue samples). The ratio of protein levels was calculated by dividing the normalized TMEM176A protein values with normalized TMEM176B protein values.
Results
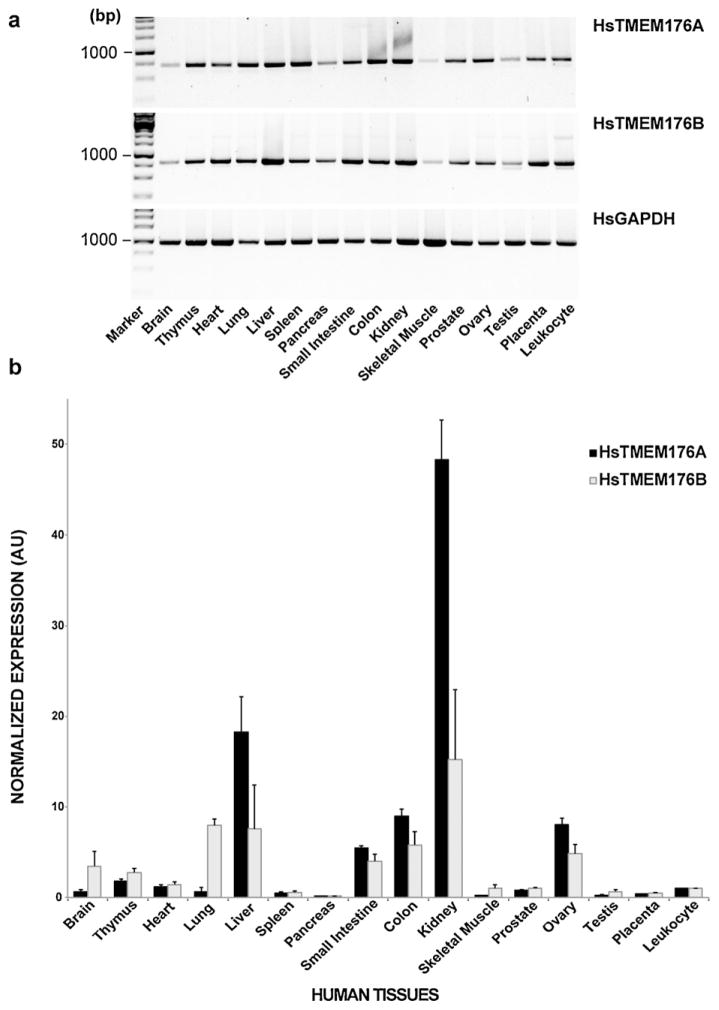
The tissue distribution of rodent Tmem176A and 176B mRNA expression has been recently published; however, the expression patterns of their human counterparts have yet to be reported. Using standard PCR, we found that human (Hs) TMEM176A and 176B mRNA expression levels to be widely distributed across multiple tissues and organs (Fig. 1a). Since a standard PCR is qualitative, we performed real-time quantitative PCR on the same tissue samples to quantitatively assess the transcript levels of both TMEM176A and 176B (Fig. 1b). Relative to peripheral blood leukocyte, we revealed that both TMEMs have a tissue-specific expression pattern. TMEM176A and 176B transcript expression levels were highest in kidney, liver, colon, small intestine, and ovary. In addition, TMEM176B transcripts were also comparably higher in lung, brain, and thymus.
Fig. 1.
Normalized multiple tissue expression analyses of human TMEM176A and 176B transcripts. (a) Standard PCR technique. Human GAPDH was used as a positive and loading control. (b) Real-time QPCR technique. The human 18S rRNA was used as the reference gene, and the peripheral blood leukocyte was used as the calibrator (value = 1). The real-time QPCR values were calculated using the Livak method as described in the Materials and Methods section. Data are represented as means ± SEM (n = 3 independent trials). AU, arbitrary unit; Hs, Homo sapiens; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
We over-expressed HsTMEM176A alone, HsTMEM176B alone, or co-expressed both proteins in HEK-293 cells, and performed a Western blot analysis to test the specificity of the anti-HsTMEM176A and anti-TMEM176B rabbit polyclonal antibodies (pAbs). Both pAbs detected their target proteins, while the control samples did not show a band (Fig. 2a). The anti-HsTMEM176A antibody detected double bands indicative of post-translational modification. This was the case for anti-HsTMEM176B antibody. The amino acid epitope recognized by the matching antibody is located at the carboxyterminus region of the protein (Fig. 2b).
Clustal W analysis of amino acid sequence similarity between HsTMEM176A and 176B proteins revealed that the proteins were 38.6% similar, whereas their mouse homologs were 39.0% similar. Comparisons between mouse and human TMEM176A proteins showed 66.9% similarity, while the mouse and human TMEM176B proteins have 61.0% similarity. Using a yeast two-hybrid genetic screen, Condamine et al. (2010) reported that mouse Tmem176A and 176B interact with each other; however, any biochemical evidence was lacking, and it is not known if their human counterparts physically interact. To establish a biochemical proof of interaction between the two human proteins, we heterologously expressed HA peptide tagged TMEM176A (HsTMEM176A-HA) and Myc/DDK peptide tagged TMEM176B (HsTMEM176B-Myc/DDK) in HEK-293 cells. Following co-immunoprecipitation (co-IP) of cell lysates, we found that both proteins physically interacted (Fig. 2c). As expected, the single protein expression (Fig. 2c), and no primary antibody (not shown) controls were both negative. To further validate this result, we co-immunoprecipitated TMEM176B protein from cell lysates that over-expressed both TMEM176A and 176B proteins, and analyzed the eluted protein samples using mass spectrometry. Our MS data showed five hits identifying TMEM176A protein, while zero hits were obtained from control samples.
To further prove that the interaction was real, we heterologously co-expressed a green fluorescent protein tagged TMEM176A (HsTMEM176A-GFP) and an mCherry tagged TMEM176B (HsTMEM176A-GFP) in HEK-293 cells. Confocal microscopy analysis showed that both TMEM176A (green) and 176B (red) colocalize together in the plasma membrane, which appeared yellow when both images were merged (Fig. 2d, arrows). In addition, the subcellular localization of both proteins appeared punctate in distribution, and seen in vesicle-like structures (Fig. 2d). Some, but not all of these punctate structures co-stained with human LAMP-1 protein, a marker for late endocytic/lysosome structure (not shown). Noteworthy is the observation that TMEM176B-positive (red) vesicle structures did not always coincide with TMEM176A-positive vesicle structures (green) (Fig. 2d, arrowheads).
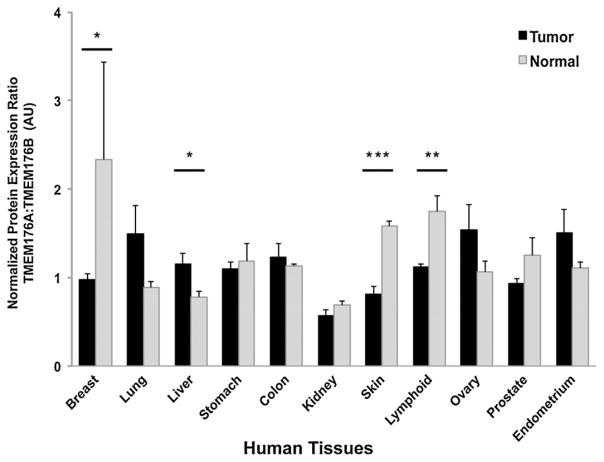
The recent reports implicating TMEM176A or TMEM176B in human cancer were primarily based on data that show abnormal mRNA transcript levels for TMEM176A and/or 176B (Wang et al., 2002; Hodo et al., 2010; Ryu et al., 2010; Strelnikov et al., 2010; Gottschling et al., 2012). One caveat about these findings is that mRNA levels do not always positively correlate with protein levels in cells or tissues due to variables such as mRNA stability, translational regulation, or proteolysis. To establish that human TMEM176A and/or 176B protein levels were abnormal and correlated with tumorigenesis, we analyzed biopsied tissue samples from normal and cancer patients (Table 2). We discovered that normalized TMEM176A protein levels were significantly elevated in lung carcinoma (p < 0.05), while both TMEM176A and 176B protein levels were significantly increased in lymphoma (p < 0.001). To assess if the functional interaction between TMEM176A and 176B is related to cancer pathology, we took the ratio of normalized TMEM176A protein levels over 176B protein levels (Fig. 3). Interestingly, we established that the normalized protein expression ratio of biopsied samples from healthy patients differed significantly from those patients with cancers of the breast (p < 0.05), liver (p < 0.05), lymphoid tissue (p < 0.001) and skin (p < 0.0001).
Table 2.
Normalized human TMEM176A and 176B protein expression levels from biopsied tissues of healthy normal and cancer patients. Human β-actin protein was used as normalization control. The protein levels were quantified and analyzed as described in the Materials and Methods section. Data are represented as means ± SEM. Statistical significance was calculated using Student’s t-test (p < 0.05, two-tailed distribution).
| Tissue samples | HsTMEM176A normalized protein levels (AU) | HsTMEM176B normalized protein levels (AU) | Sample size (n) |
|---|---|---|---|
| Breast cancer | 4.53 ± 0.48 | 4.64 ± 0.52 | 9 |
| Breast normal | 4.65 ± 1.66 | 2.93 ± 1.57 | 4 |
| Lung cancer | 3.60 ± 0.43* | 3.26 ± 0.56 | 10 |
| Lung normal | 1.71 ± 0.52 | 2.14 ± 0.88 | 5 |
| Liver cancer | 2.30 ± 0.54 | 3.23 ± 1.00 | 9 |
| Liver normal | 1.25 ± 0.12 | 1.11 ± 0.07 | 5 |
| Stomach cancer | 4.58 ± 0.49 | 4.12 ± 0.35 | 10 |
| Stomach normal | 4.14 ± 0.63 | 3.62 ± 0.59 | 3 |
| Colon cancer | 4.51 ± 0.50 | 3.96 ± 0.55 | 10 |
| Colon normal | 3.48 ± 0.97 | 3.06 ± 0.83 | 5 |
| Kidney cancer | 2.17 ± 0.43 | 4.00 ± 0.90 | 9 |
| Kidney normal | 1.89 ± 0.34 | 2.70 ± 0.42 | 5 |
| Skin cancer (melanoma) | 2.84 ± 0.57# | 4.14 ± 1.12 | 9 |
| Skin normal | 5.26 ± 1.63 | 3.42 ± 1.11 | 3 |
| Lymphoid cancer (lymphoma) | 5.14 ± 0.53* | 4.58 ± 0.46** | 10 |
| Lymphoid normal | 2.89 ± 0.64 | 1.68 ± 0.33 | 5 |
| Ovary cancer | 4.18 ± 0.51 | 3.11 ± 0.42 | 10 |
| Ovary normal | 3.05 ± 0.69 | 3.20 ± 0.98 | 4 |
| Prostate cancer | 5.03 ± 0.36 | 5.45 ± 0.27# | 8 |
| Prostate normal | 4.66 ± 0.79 | 4.05 ± 0.82 | 5 |
| Endometrial cancer | 4.15 ± 0.42 | 3.26 ± 0.50 | 10 |
| Endometrium normal | 4.02 ± 0.80 | 3.63 ± 0.64 | 5 |
p < 0.05.
p < 0.001.
p = 0.08 (barely significant value).
Fig. 3.
Normalized protein expression ratio between human TMEM176A and TMEM176B across biopsied samples from normal and cancer patients. The ratio was calculated by dividing normalized protein levels of HsTMEM176A over HsTMEM176B values. Human β-actin was used as normalization control as described in the Materials and Methods section. Data are represented as means ± SEM. Significant p-values: *p < 0.05, **p < 0.001, ***p < 0.0001, Student’s t-test (two-tailed distribution). AU, arbitrary unit.
Discussion
Defining the tissue distribution of human TMEM176A and 176B is vital to increasing our understanding of their cellular function and possible connection to cancer pathology. Here, we show for the first time that human TMEM176A and 176B mRNA transcripts exhibit a tissue-specific expression pattern. We also found that both proteins interact, co-localize in plasma membrane and intracellular compartments, and that their protein expression levels are altered in several cancer tissues.
To explore the tissue distribution pattern of human TMEM176A and 176B, we assessed their transcript levels from an array of normalized tissue cDNA samples. We determined that TMEM176A transcripts are highest in liver and kidney – two tissues where the gene was first identified and associated with hepatocellular carcinoma (Wang et al., 2002) and kidney proteinuria (Nakajima et al., 2002), respectively. Similarly, TMEM176B transcripts are also relatively higher in liver and kidney, as well as in lung, which happens to be the tissue where this gene was first identified (Lurton et al., 1999). In addition, the detected levels of TMEM176B transcripts in the human brain are consistent with a previous report in rats (Louvet et al., 2005), suggesting that this protein may contribute to brain function. A case in point, knocking out Tmem176B in mice produces ataxia (loss of motor coordination) due to abnormal development of granule cell neurons in the cerebellum (Maeda et al., 2006).
While the tissue expression pattern of human TMEM176A and 176B transcripts coincided with their mouse (Condamine et al., 2010) and rat (Louvet et al., 2005) homologs, their relative amounts, particularly in certain tissues markedly differed. For example, we detected low levels of human TMEM176A and 176B transcripts in spleen (<1 arbitrary units [AU]), while transcript levels of their rat (Louvet et al., 2005) or mouse (Condamine et al., 2010) counterparts showed higher levels in spleen (>5 AU). Human brain transcript levels of TMEM176B parallel that of its rat homolog (Louvet et al., 2005), but not the transcript levels reported for its mouse homolog (Condamine et al., 2010). Such transcript level variations are likely due to methodological differences and/or the salient nature of species-specific gene regulation and expression.
Although mouse TMEM176A and 176B proteins were recently shown to interact (Condamine et al., 2010), it was not known if their human counterparts also functionally interacted. We set out to determine if this was the case and we show here that human TMEM176A and 176B do bind each other (Fig. 2c). The physical association between the two proteins was validated by both mass spectrometry and subcellular co-localization studies. Confocal microscopy revealed that both proteins are found in the plasma membrane, and within intracellular compartments that exhibit a punctate distribution (Fig. 2d). The appearance of two very close bands for TMEM176A or TMEM176B upon Western blot analyses indicates post-translational modification likely involved in membrane localization. Indeed, our confocal microscopy observation supports this interpretation. A closer look at cells co-expressing both proteins demonstrates that the TMEM176A protein does not always co-localize in the same intracellular vesicle-like compartments as TMEM176B protein. This outcome suggests that the functional TMEM176A/176B protein complex may exist as both homomeric and heteromeric structures. It would be interesting to identify in future studies the subunit composition of the complex and whether homomeric TMEM176A or 176B proteins serve a completely different function in cells in comparison to their heteromeric equivalent. In addition, future investigations of specific post-translational modifications conferred to both TMEM176A and 176B could provide additional information on the proteins’ function in cells.
Several reports have associated distinct types of human cancer with changes in tissue mRNA expression level or transcriptional regulation of TMEM176A and/or 176B genes (Wang et al., 2002; Hodo et al., 2010; Strelnikov et al., 2010; Gottschling et al., 2012) however, evidence that clearly shows abnormality of TMEM176A or 176B protein levels in these cancer tissues was lacking. In this study, we show that protein levels of both TMEM176A and 176B are significantly increased in lymphoma tissues, while TMEM176A protein alone is significantly elevated in lung carcinoma tissues (Table 2). A caveat in our findings is that the population of cells in any given biopsied tumor tissue samples is very heterogeneous, since infiltrating myeloid and lymphoid immune cells as well as vascular and stromal cells are typically found in tumors (Motz and Coukos, 2011).
There are several possible reasons why TMEM176A and/or 176B protein levels are elevated in tumor tissues. First, it is possible that tumor cells over-express both TMEM176A and 176B proteins to evade the immune system and to bestow protection and tolerance from attacks by the immune system (Louvet et al., 2005). Second, tumor cells may induce infiltrating immune cells to over-express both proteins to negatively impact their detection by the immune system, since the expression of these proteins has been reported to inhibit both the maturation and activation of cDCs (Condamine et al., 2010). These explanations are in line with a previous report that over-expression of rat Tmem176B maintains long-term tolerance of kidney allografts, reduced expression of molecules critical to antigen presentation such as MHC-II, and decreased levels of co-stimulatory molecules involved in T-cell activation such as CD86 proteins (Louvet et al., 2005). Likewise, immune cells associated with tumors are known to influence carcinogenesis (Zamarron and Chen, 2011). On the other hand, the substantial abundance of TMEM176A protein levels in lung carcinoma tissues is highly suspect and anomalous, since our findings indicate that TMEM176A transcript levels in human lung is very low (<1 AU, Fig. 1b). Although transcript levels do not necessarily equate to protein levels, it would be interesting to discover if this is causal to, and not as a consequence of, tumor formation. With regard to liver cancer tissues, the level of TMEM176A protein was higher compared to non-cancer tissues (Table 1), which supports the previous study implicating the protein with HCC (Wang et al., 2002), but the comparison between the samples failed to reach statistical significance. Notwithstanding, we believe that the statistical power of the effect could increase by increasing the sample size, which would eventually make the values significant. Intriguingly, a significant association between liver carcinoma and TMEM176A proteins is observed when the level of TMEM176B is taken into consideration in the analysis (Fig. 3, see below for details).
To further assess if both the interaction between TMEM176A and 176B and their protein levels are associated with cancer, we compared the normalized protein expression ratio (TMEM176A:TMEM176B) of normal tissues with cancer tissues. The TMEM176A:TMEM176B ratio uncovered significant differences between normal tissues and cancer tissues of the breast, liver, skin, and lymphoid tissues. All three TMEM176A:TMEM176B ratios analyzed from breast cancer, melanoma, and lymphoma are significantly lower in comparison with non-cancer samples. The data suggest that tissues with higher TMEM176B protein levels with respect to TMEM176A protein are quite atypical and only found in these types of cancer tissues. In contrast, the TMEM176A:TMEM176B ratio in liver cancer tissues is significantly higher compared with non-cancer tissues. This result indicates that a reduction in the amount of TMEM176B protein in relation to TMEM176A protein is likely to be detected only in liver cancer tissues. It is not clear if the protein expression level of TMEM176B influences the level of TMEM17A proteins, since the transcript levels of both TMEM proteins have been reported to be stable and independent of each other (Condamine et al., 2010). It is evident, however, that the current observed variations in human TMEM176B protein levels with respect to TMEM176A protein levels are highly predictive of tumor pathology.
In conclusion, we established that human TMEM176A and 176B transcripts are differentially expressed in various tissues examined. We found that both TMEM proteins physically interact, form homomeric and heteromeric structures, as well as co-localize in the plasma membrane and vesicular compartments. Finally, we showed for the first time that the protein expression levels of both TMEM proteins are closely linked to distinct types of cancer in humans. Specifically, the amount of TMEM176B protein appears intimately associated with TMEM176A, which could be used as a potential biomarker for lymphoma, melanoma, breast carcinoma, or liver carcinoma (HCC). Future research should aim to discover if the abnormal quantities of both TMEM proteins is primary (causal) or secondary (consequential) to tumorigenesis in humans.
Acknowledgments
This project was funded by grants from the NSF (MCB 920127), NIH (R15-NS070774), HHMI Undergraduate Research Program, and CSUF Office of Grants and Contracts to MPC. We are very grateful to Dr. Roger Y. Tsien (University of California San Diego) for the generous gift of fluorescent-tagged vectors. We thank the following individuals for their technical contribution in this project: Steve Karl for assistance in confocal microscopy and Chris Adams of the Stanford University Mass Spectrometry Facility for protein sample processing and analysis. We are also grateful to Sean Murray for critically reading this manuscript.
References
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Condamine T, Le Texier L, Howie D, Lavault A, Hill M, Halary F, et al. Tmem176B and Tmem176A are associated with the immature state of dendritic cells. J Leukoc Biol. 2010;88:507–15. doi: 10.1189/jlb.1109738. [DOI] [PubMed] [Google Scholar]
- Cuajungco MP, Grimm C, Oshima K, D’Hoedt D, Nilius B, Mensenkamp AR, et al. PACSINs bind to the TRPV4 cation channel. PACSIN 3 modulates the subcellular localization of TRPV4. J Biol Chem. 2006;281:18753–62. doi: 10.1074/jbc.M602452200. [DOI] [PubMed] [Google Scholar]
- Cuajungco MP, Leyne M, Mull J, Gill SP, Lu W, Zagzag D, et al. Tissue-specific reduction in splicing efficiency of IKBKAP due to the major mutation associated with familial dysautonomia. Am J Hum Genet. 2003;72:749–58. doi: 10.1086/368263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrau R, Maluf D, Archer K, Stravitz R, Suh J, Le N, et al. Molecular pathways differentiate hepatitis C virus (HCV) recurrence from acute cellular rejection in HCV liver recipients. Mol Med. 2011;17:824–33. doi: 10.2119/molmed.2011.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh HS, Cisse B, Bunin A, Lewis KL, Reizis B. Continuous expression of the transcription factor e2-2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity. 2010;33:905–16. doi: 10.1016/j.immuni.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling S, Jauch A, Kuner R, Herpel E, Mueller-Decker K, Schnabel PA, et al. Establishment and comparative characterization of novel squamous cell non-small cell lung cancer cell lines and their corresponding tumor tissue. Lung Cancer. 2012;75:45–57. doi: 10.1016/j.lungcan.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Hodo Y, Hashimoto S, Honda M, Yamashita T, Suzuki Y, Sugano S, et al. Comprehensive gene expression analysis of 5′-end of mRNA identified novel intronic transcripts associated with hepatocellular carcinoma. Genomics. 2010;95:217–23. doi: 10.1016/j.ygeno.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Louvet C, Chiffoleau E, Heslan M, Tesson L, Heslan JM, Brion R, et al. Identification of a new member of the CD20/FcepsilonRIbeta family overexpressed in tolerated allografts. Am J Transplant. 2005;5:2143–53. doi: 10.1111/j.1600-6143.2005.01007.x. [DOI] [PubMed] [Google Scholar]
- Lurton J, Rose TM, Raghu G, Narayanan AS. Isolation of a gene product expressed by a subpopulation of human lung fibroblasts by differential display. Am J Respir Cell Mol Biol. 1999;20:327–31. doi: 10.1165/ajrcmb.20.2.3368. [DOI] [PubMed] [Google Scholar]
- Maeda Y, Fujimura L, O-Wang J, Hatano M, Sakamoto A, Arima M, et al. Role of Clast1 in development of cerebellar granule cells. Brain Res. 2006;1104:18–26. doi: 10.1016/j.brainres.2006.05.068. [DOI] [PubMed] [Google Scholar]
- Motz GT, Coukos G. The parallel lives of angiogenesis and immunosuppression: cancer and other tales. Nat Rev Immunol. 2011;11:702–11. doi: 10.1038/nri3064. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Takenaka M, Kaimori JY, Nagasawa Y, Kosugi A, Kawamoto S, et al. Gene expression profile of renal proximal tubules regulated by proteinuria. Kidney Int. 2002;61:1577–87. doi: 10.1046/j.1523-1755.2002.00300.x. [DOI] [PubMed] [Google Scholar]
- Ryu SH, Kim KH, Kim HB, Kim MH, Kim NH, Kang Y, et al. Oncogenic Ras-mediated downregulation of Clast1/LR8 is involved in Ras-mediated neoplastic transformation and tumorigenesis in NIH3T3 cells. Cancer Sci. 2010;101:1990–6. doi: 10.1111/j.1349-7006.2010.01626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelnikov V, Tanas A, Shkarupo V, Kuznetsova E, Gorban N, Zaletaev D. Non-microarray DNA differential methylation screening in breast cancer. Cancer Genet Cytogenet. 2010;203:93. [Google Scholar]
- Wang Y, Han KJ, Pang XW, Vaughan HA, Qu W, Dong XY, et al. Large scale identification of human hepatocellular carcinoma-associated antigens by autoantibodies. J Immunol. 2002;169:1102–9. doi: 10.4049/jimmunol.169.2.1102. [DOI] [PubMed] [Google Scholar]
- Zamarron BF, Chen WF. Dual roles of immune cells and their factors in cancer development and progression. Int J Biol Sci. 2011;7:651–8. doi: 10.7150/ijbs.7.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccolo J, Bau J, Childs SJ, Goss GG, Sensen CW, Deans JP. Phylogenetic analysis of the MS4A and TMEM176 gene families. PLoS One. 2010;5:e9369. doi: 10.1371/journal.pone.0009369. [DOI] [PMC free article] [PubMed] [Google Scholar]