Abstract
Despite years of fully suppressive antiretroviral therapy (ART), HIV persists in the host and is never eradicated. One major barrier to eradication is that multiple different cell types are infected that may individually contribute to HIV persistence. Tissue macrophages are critical contributors to HIV disease (1–3); however, their specific role in HIV persistence during long-term suppressive ART has not been established (4–6). Using humanized myeloid-only mice (MoM), we demonstrate that HIV infection of tissue macrophages is rapidly suppressed by ART, as determined by a rapid drop in plasma viral load and a dramatic drop in the levels of cell-associated viral RNA and DNA. No virus rebound was observed in the plasma of 67% of the ART treated animals at seven weeks post-ART interruption, and no replication competent virus was rescued from the tissue macrophages obtained from these animals. In contrast, in a subset of animals (~33%), a significantly delayed viral rebound was observed that is consistent with the establishment of persistent infection in tissue macrophages. These observations represent the first direct evidence of HIV persistence in tissue macrophages in vivo.
Humanized Bone marrow, Liver, Thymus (BLT) mice are reconstituted with human T cells and macrophages, both of which are targets for HIV infection in vivo (7–11). Using a model reconstituted with HIV infected macrophages and T cells permits assessment of the effect of ART on tissue macrophages in the presence of T cells, allowing for the types of interactions that occur normally between the two cell types. We infected BLT mice (n=18) with macrophage-tropic viruses (HIV-1CH040 or HIV-1CH040-4013env) (7), and monitored plasma viremia longitudinally (Fig. 1a, Supplemental Table 1). Thirteen mice, at five weeks post-infection, were treated once daily with triple combination ART (12–14). Within two weeks of ART initiation, we observed a significant decrease in plasma viremia of treated (black line) vs untreated mice (dashed gray line) (Fig. 1a, Mann-Whitney test). This corresponded to an average 1.3 log reduction in plasma viral RNA after one week of treatment (Fig. 1b). We used the change in viral load after ART-initiation to estimate the half-life of productively infected cells in BLT mice which was approximately 2.4 days (Fig. 1c), similar to the previous estimate of ~2 days in humans (15–17).
Figure 1. Viral suppression, persistence and structured ART-interruption induced rebound in BLT mice.
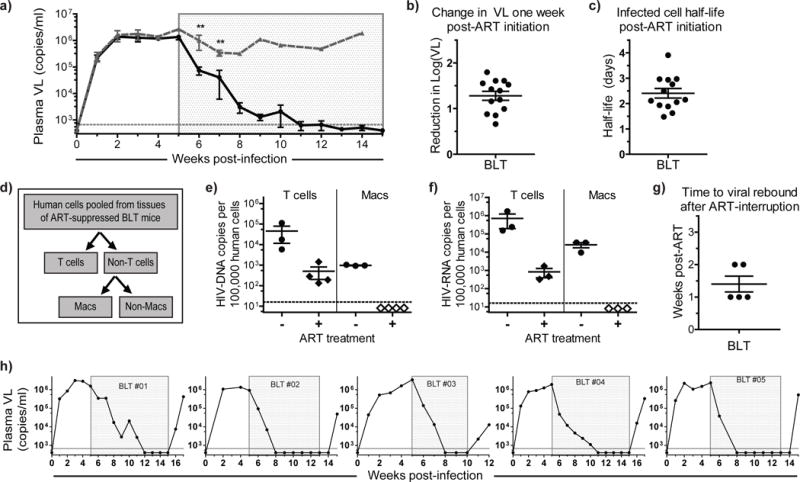
a) Plasma viral load (VL) was monitored over time in HIV-infected ART-treated (n=13, solid black line) and untreated (n=5, dashed gray line) BLT mice. Each point represents the mean ± s.e.m. The time of exposure to ART for the treated animals is indicated with a shaded gray box. A Mann-Whitney test was used to compare the plasma VL of ART-treated and untreated mice at one and two weeks post-ART initiation (p=0.0049 and p=0.0080, respectively). b) The reduction in log (base 10) plasma VL one week after ART-initiation was calculated for each treated BLT mouse (n=13). c) The half-life of productively infected cells was estimated from the change in viral load during ART (n=13). d) Schematic for the magnetic sorting and purification of human T cells and macrophages from the tissues obtained from infected animals. Human T cells (CD3+) and non-T cells were separated from pooled tissue cells of individual ART-suppressed BLT mice (n=4). Macrophages (Macs) were then isolated from the non-T cell fraction. Analysis of (e) HIV-DNA and (f) HIV-RNA levels were performed by real time PCR using purified cells isolated from ART-suppressed (n=4) and untreated (n=3) BLT mice. HIV-DNA/RNA levels are normalized and reported per 100,000 human cells. Samples with values below the level of detection are indicated by open diamonds in e & f and are shown at the average lower limit of detection. g) Time to viral rebound after ART interruption in BLT mice (n=5). h) Viral rebound was observed 1–2 weeks after ART-interruption in all BLT mice. For b, c, e, f & g, the horizontal lines represent mean ± s.e.m.
To address the effect of ART specifically on infected tissue macrophages, cells were pooled from the spleen, liver, lung, and bone marrow of individual ART-suppressed or untreated BLT mice. A step-wise magnetic cell sorting strategy was used to isolate T cells and macrophages (Fig. 1d and Supplemental Figure 1a–c) (7). We were able to detect cell-associated viral DNA (Fig. 1e) and RNA (Fig. 1f) in the purified T cell fractions of untreated mice. In ART-treated mice, the levels of viral DNA and RNA in T cells were significantly reduced but readily detectable (Fig. 1e, f). In the purified macrophage (Macs) fraction, we were able to readily detect viral DNA and RNA in untreated mice; however, the levels of viral DNA and RNA in tissue macrophages from ART-treated mice were below our level of detection using a reverse transcriptase real-time PCR assay (Fig. 1e, f). We then conducted structured treatment interruption studies to establish the presence of persistently infected cells containing replication competent virus (Fig. 1g–h) (12). In all cases, HIV rebound was observed by 2 weeks after therapy interruption, a time to rebound that is within the range observed for patients (18). Considering the absence of detectable viral RNA and DNA in macrophages isolated from ART-suppressed BLT mice, the rapid rebound observed ostensibly results from virus present in T cells.
Humanized myeloid only mice (MoM) are generated by transplanting CD34+ hematopoietic stem cells into NOD/SCID mice, which are unable to support human T cell development (19). MoM are systemically reconstituted with human myeloid and B cells, with an absence of human T cells (Supplementary Fig. 2) (7). We have previously shown that MoM are susceptible to HIV infection and can efficiently sustain systemic HIV replication over several months (7). MoM were utilized to avoid the confounding complications of having an excess of T cells present during the evaluation of HIV persistence in macrophages. We infected MoM (n=14) with the same macrophage-tropic HIV isolates used for BLT mice in Figure 1 (Supplemental Table 1), and monitored infection over time in the absence (n=6) or in the presence of ART (n=8) (Supplemental Fig. 3). In all treated MoM, we noted a rapid and significant drop in plasma viremia below the limit of detection relative to untreated MoM (Fig. 2a, Mann-Whitney test). After one week of ART, this drop corresponded to an average 1.8 log reduction in plasma viral RNA levels (Fig. 2b). This reduction in plasma viral RNA levels was significantly larger than the reduction observed in BLT mice (Fig. 1b, Mann-Whitney test, p=0.039). Using change in viral load after ART-initiation we estimated the half-life of productively infected macrophages in MoM to be 1.05 days (Fig. 2c), which was notably shorter than the estimated half-life of infected T cells in humans and BLT mice (Fig. 1c, Mann-Whitney test, p=0.0002) (15–17). Although the half-life of HIV-infected macrophages in MoM is shorter than previously estimated for macrophages in SHIV infected macaques (20), our results are consistent with the more recent estimate of 1.33 days in SIV infected CD4+ T cell depleted macaques on combination ART (21).
Figure 2. ART rapidly suppresses viral replication in MoM.
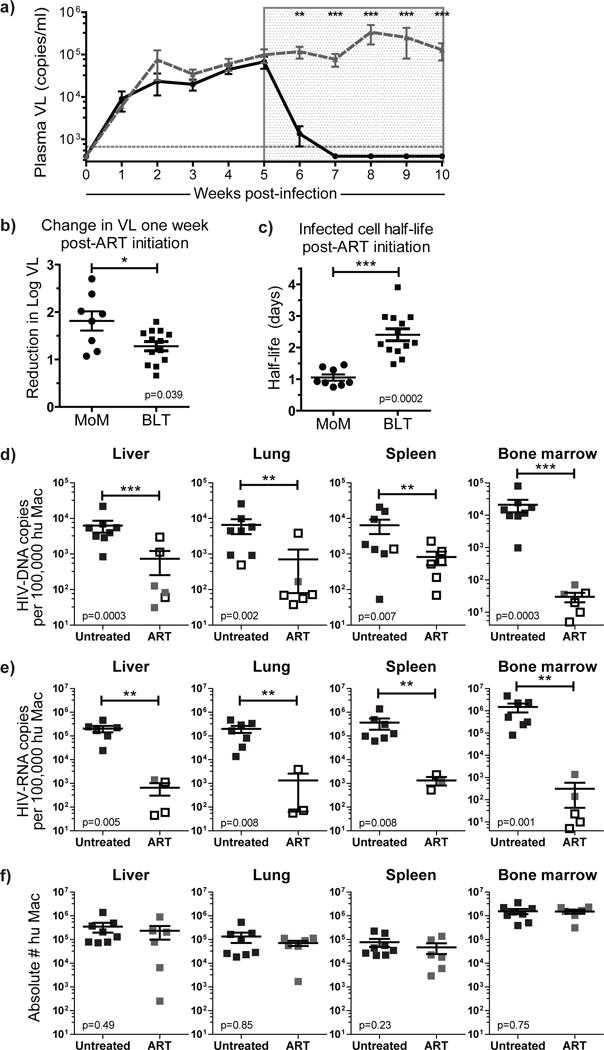
a) Longitudinal analysis of the plasma VL in HIV-infected ART-treated (n=8, solid black line) and untreated (n=6, dashed gray line) MoM. Each point represents the mean ± s.e.m.. A Mann-Whitney test was used to compare the plasma VL of treated and untreated mice at 1–5 weeks post-ART initiation (p=0.0019, p=0.0008, p=0.0008, p=0.0011 and p=0.0017, respectively). b) The reduction in log (base 10) plasma VL one week after ART-initiation was calculated for each treated MoM (n=8). c) The half-life of productively infected cells was estimated from the change in viral load during ART (n=8). A Mann-Whitney test was used to compare MoM and BLT mice in b & c. Cell-associated (d) HIV-DNA and (e) HIV-RNA levels were measured in the liver, lung, spleen and bone marrow of ART-treated (n=6, gray squares) and untreated (n=8, black squares) MoM. Undetectable samples are indicated by an empty black box shown at the limit of detection for that sample (dependent on the number of cells available for analysis). Viral DNA and RNA levels were normalized per 100,000 human macrophages and compared between treated and untreated mice. A log-rank test was used to account for censoring due to the limits of detection in d & e. f) There was no difference in the total numbers of human macrophages present in the tissues of ART-treated or untreated MoM (all p>0.05, Mann-Whitney test). In b–f, horizontal lines represent mean ± s.e.m.
We determined the levels of cell-associated viral DNA and RNA in tissue macrophages isolated from liver, lung, spleen and bone marrow of untreated or ART-suppressed MoM (Supplemental Table 1). Notably, the levels of viral DNA in 18/24 samples from six ART-treated MoM were below the level of detection (indicated as empty boxes in Fig. 2d, log-rank test, p=0.0003, p=0.002, p=0.007, p=0.0003, for liver, lung, spleen and bone marrow respectively). Using an ultra-sensitive quantitative droplet PCR technique, we demonstrate that HIV was integrated into the genome in a portion of the HIV-DNA samples in the ART-treated group (total of 4 samples analyzed) and that the levels of integrated HIV DNA were significantly lower in the treated animals (Supplemental Fig. 3c). Consistent with this significant decrease in viral DNA levels, viral RNA levels of 11/15 samples analyzed were also below the level of detection in five ART-treated MoM (indicated as empty boxes in Fig. 2e, log-rank test, p=0.005, p=0.008, p=0.008, p=0.001, for liver, lung, spleen and bone marrow respectively). This difference in HIV-DNA and RNA levels between treated and untreated mice did not result from a loss of macrophages in the tissues (Fig. 2f, Mann-Whitney test, all p>0.05). The limited detection of viral DNA and RNA from ART-suppressed MoM samples is consistent with our data obtained from purified macrophages isolated from the tissues of ART-treated BLT mice (Fig. 1e, f).
To determine if HIV persists in infected tissue macrophages and can re-establish persistent infection after ART treatment, we discontinued therapy in HIV-infected MoM suppressed on ART (Supplemental Table 1). In sharp contrast from what was observed in BLT mice, where virus rebound was observed within 2 weeks after therapy discontinuation in all animals (n=5), no viral rebound was observed in 6/9 MoM (Fig. 3a). Upon further examination of tissue macrophages isolated from MoM in which no HIV rebound was observed, we detected no viral DNA or RNA (Table 1). To determine the presence of persistent, replication competent virus in tissue macrophages isolated from ART-treated MoM where no rebound was observed, we used outgrowth assays similar to those previously used in macaques to either quantify virus from infected macrophages (22) or to establish SIV eradication after vaccination with an hCMV-based vector (23, 24). For this purpose, cells obtained from MoM where rebound was not observed were directly injected into BLT humanized mice, as we have previously described (7), or co-cultured with CD8-depleted activated allogeneic CD4+ T cells (7, 25–27). Plasma or tissue culture supernatants were then monitored for the presence of virus outgrowth; however, no replication competent virus was rescued from any of the five MoM analyzed (Table 1). Fewer total numbers of cells were isolated from MoM #19, precluding additional outgrowth analysis.
Figure 3. HIV persistence in tissue macrophages during ART.
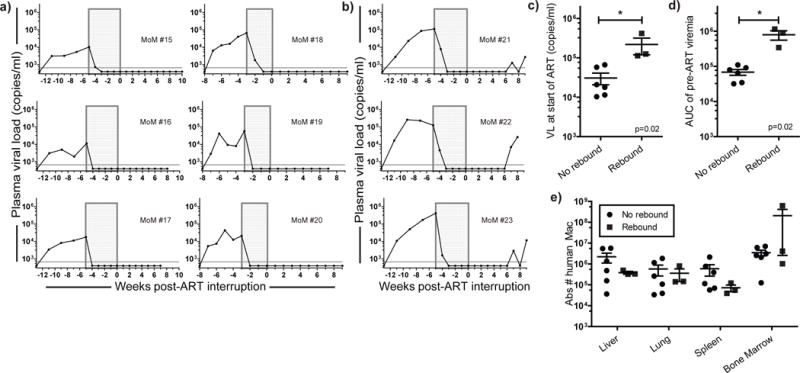
Viral rebound was absent in most infected MoM after structured ART-interruption [(a) n=6 No rebound, (b) n=3 Rebound]. c) Higher plasma VL at the start of treatment and (d) higher total viral burden [as demonstrated by area under the curve (AUC) analysis of pre-ART viremia] was associated with viral rebound after ART-interruption. e) The total numbers of human macrophages in the tissues of MoM were similar between mice where viral rebound was absent (a) or observed (b) (p>0.05). Mann-Whitney tests were used to compare mice in c–e. For c–e, the horizontal lines represent mean ± s.e.m.
Table 1. Absence of replication competent virus in tissues of MoM without plasma viral rebound after ART interruption.
HIV-DNA/RNA analysis and in vivo or in vitro viral outgrowth assays were performed with cells isolated from ART-treated MoM in which plasma viral rebound was not observed. The tissues analyzed for all mice included the spleen, liver, lung, brain and bone marrow. In MoM #18–20, the gastrointestinal and female reproductive tracts were also analyzed for presence of HIV-DNA. For the viral outgrowth assay, cells isolated from the tissues of MoM were 1) cultured with activated allogeneic CD4+ T cells and the culture supernatants tested for presence of cell-free HIV-RNA by real time PCR analysis (in vitro method) or 2) injected into BLT mice and the plasma VL monitored over time to determine the presence or absence of HIV infection in recipient mice (in vivo method). The total number of cells utilized for outgrowth analysis is indicated (# cells). LOD: limit of detection. nd: not determined.
| Mouse # | % Human | Virus | HIV-DNA | HIV-RNA | Viral outgrowth analysis performed on tissue cells | ||
|---|---|---|---|---|---|---|---|
| Observation | Method | # cells | |||||
| MoM #15 | 42 | CH040 | below LOD | nd | no outgrowth | in vitro | 63×106 |
| MoM #16 | 63 | CH040 | below LOD | nd | no outgrowth | in vitro | 44×106 |
| MoM #17 | 49 | CH040 | below LOD | nd | no outgrowth | in vitro | 30×106 |
| MoM #18 | 28 | CH040 | below LOD | below LOD | no outgrowth | in vivo | 4×106 |
| MoM #19 | 8 | CH040 | below LOD | nd | nd | nd | nd |
| MoM #20 | 16 | CH040 | below LOD | below LOD | no outgrowth | in vivo | 4×106 |
We did observe viral rebound in the plasma from three MoM, seven weeks post-ART interruption (Fig. 3b). We confirmed the absence of human T cells in the tissues of “Rebound” MoM (Supplemental Fig. 4). We demonstrated the replication competence of the rebound virus (from MoM #23) to infect both T cells and macrophages using the outgrowth assays performed in the “No rebound” MoM (Supplemental Fig. 5, Supplemental Table 2). Additionally, HIV env was sequenced from the plasma of “Rebound” MoM and compared to the original virus used for infection. No mutations were found in MoM #22. In MoM #21 and #23, a point mutation was present resulting in single amino acid changes (I 262 L and I 107 S, respectively) in the env sequence (GenBank accession numbers KX825880 and KX825881). In MoM #23, this mutation was present in ~50% of the bulk sequence. A post-hoc comparison of mice with or without HIV rebound demonstrated a higher pre-ART viral load (Fig. 3c) and larger total pre-ART viral burden (as demonstrated by the area under the curve for pre-ART viremia, Fig. 3d) in the “Rebound” mice (Mann-Whitney test, p=0.02). No differences were noted in the overall levels of human cells in the blood (Mann-Whitney test, p=0.381) or in the number of macrophages in the tissues of mice that did or did not rebound (Fig. 3e, p>0.05, Mann-Whitney test). This suggests that under our experimental conditions, the lack of rebound observed in 6/9 mice was not simply due to insufficient numbers of human tissue macrophages but rather to the absence of productively infected cells capable of restarting the infection. We then used these data to model the likely persistence of HIV infected macrophages under ART in view of their half-life. Our estimate of approximately 1 day for the half-life of infected macrophages indicates that half of all infected macrophages are lost every day. Therefore, the number of productively infected macrophages under ART would be expected to decay to about 1% after one week and to less than 0.00001% of their initial value during the 5 weeks of treatment. The fact that viral rebound was observed seven weeks after therapy interruption (and was absent in most treated MoM) is consistent with the existence of a small population of infected tissue macrophages that persist despite ART, especially when pre-treatment viral burden is greatest.
As in any study involving the use of a model system, there are some caveats to this analysis. First, the strain of immunodeficient mice used to generate MoM (NOD/SCID mice) develop spontaneous thymic lymphomas resulting in a relatively short life-span (9–14 months) (28). This limitation prevented us from extending our observations of infected MoM after ART-interruption for longer periods of time. Therefore, outgrowth assays were used to confirm the absence of cells containing replication competent virus. Second, interactions between T cells and macrophages may be critical for persistent HIV infection; therefore, our results in MoM may underestimate the frequency of viral rebound from macrophages. However, this is unlikely to be the case since the disappearance of infected macrophages was noted in both ART-treated HIV infected BLT and MoM, suggesting that the presence of human T cells is not critical for HIV persistence in human macrophages.
Antiretroviral therapy alone is not sufficient to eradicate HIV from the body. Hill et al. indicate that reductions of 2–3 logs in the number of latently infected cells are needed to delay rebound for the time reported herein for MoM (29). The delay in rebound observed in MoM is greater than that reported for patients initiating ART during the chronic phase of infection (30) and from patients with an ultralow HIV reservoir (18). Our results indicate that HIV-infected tissue macrophages have a short half-life and that ART alone can significantly reduce the levels of infected tissue macrophages, further demonstrating the critical contribution of T cells to the viral reservoir. It is possible that under extended ART treatment, no viral rebound from macrophages would be observed in MoM. In patients where viral rebound occurs after a long period post ART-interruption, the cellular identity and location within the tissue of the reservoir is unknown (31, 32). However, the fact that viral rebound can be observed in 33% of the infected/suppressed animals suggests that macrophages may represent a persistent viral reservoir. Specialized long-lived macrophages, such as microglia, are thought to persist in the CNS long-term despite infection (33, 34), and could therefore contribute to viral persistence over time. Therefore our results show that productively infected tissue macrophages can persist despite effective treatment and re-establish productive infection after structured treatment interruption.
Online Methods
Generation of humanized mice
Mice were created and monitored as previously described (7, 12, 13). BLT mice were generated by implanting human thymus and liver tissue under the kidney capsule of sub-lethally irradiated NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG, The Jackson Laboratory) mice. BLT mice received 3.5×105 autologous CD34+ hematopoietic stem cells. Humanized MoM were created by transplanting sub-lethally irradiated NOD.CB17-Prkdcscid/J mice (NOD/SCID, The Jackson Laboratory) with approximately 1×106 cord blood or liver-derived CD34+ hematopoietic stem cells. All animals were irradiated prior to stem cell transplantation (BLT mice: 200 rad, MoM: 250 rad). Mice were maintained in a specific pathogen-free facility by the Division of Laboratory Animal Medicine at the University of North Carolina at Chapel Hill (UNC-CH) according to protocols approved by the Institutional Animal Care and Use Committee.
HIV exposures
Stocks of CH040 and the chimeric virus, CH040-4013 env, were prepared and titered as previously described (7). Briefly, virus supernatants were prepared via transient transfection of 293T cells, and were titered using TZM-bl cells (an indicator cell line). The chimeric virus was created by replacing env in CH040 (Accession #jn944905, 6396-8863) with the corresponding restriction fragment from 4013 (Accession #jn562796). For the ART-treatment experiments, humanized MoM and BLT mice were injected intravenously with 3.6 × 105 TCIU of HIV. Untreated BLT mice in Figure 1A were exposed vaginally to 3.6 × 105 TCIU of HIV. BLT mice were exposed 8–36 weeks post-humanization (mean 18.3 weeks) and MoM were exposed 8–26 weeks post-humanization (mean 16.7 weeks); there was no difference in the weeks post-humanization between MoM and BLT mice (Mann-Whitney test, p=0.59).
Monitoring of HIV infection in humanized mice
Peripheral blood was collected and plasma was isolated by centrifugation. HIV infection was monitored in peripheral blood plasma with a one-step reverse transcriptase real-time PCR assay (ABI custom TaqMan Assays-by-design) according to the manufacturer’s instructions (with primers 5′-CATGTTTTCAGCATTATCAGAAGGA-3′ and 5′-TGCTTGATGTCCCCCCACT-3′; assay sensitivity of 668 RNA copies per mL). At necropsy, mononuclear cells (MNCs) were isolated from tissues and analyzed for the presence of viral RNA and DNA. For viral outgrowth analysis (Table 1), an in vitro or ex vivo outgrowth analysis was performed as done previously in MoM (7). For the in vitro analysis, macrophages were plated and allowed to adhere. Within 24 hours of plating, approximately one million allogenic, PHA-stimulated feeder cells (CD8-depleted PBMCs from healthy human donors) were added to the cultures as targets for viral outgrowth (kindly provided by Nancie Archin at UNC-CH). After 10 days, culture supernatants were analyzed for the presence of viral RNA by real-time PCR. For the ex vivo outgrowth analysis, cells were isolated from the tissues of ART-suppressed MoM and ~4 million cells were injected intravenously into naïve BLT mice. The plasma viral load was monitored for 4–8 weeks post-injection and tissues harvested from BLT mice were analyzed for the presence of viral DNA at necropsy.
Integrated HIV-DNA analysis
High molecular weight (15kb in length or greater) DNA was separated by Pulse-field Gel Electrophoresis with the automated BluePippin cassette system [presented by Lada et al at the 2016 Conference on Retroviruses and Opportunistic Infections (CROI)]. 30μL of total DNA and 10μL of loading buffer were pipetted onto one lane of a 0.75% agarose cassette. Using a 15kb cutoff high-pass protocol and BluePippin S1 marker, high molecular weight DNA was purified and collected in 80μL total volume. Afterwards HIV Gag, HIV 2-LTR junction and host cell RPP30 were assayed by ddPCR. Digital droplet PCR analysis was performed as previously described (35).
Antiretroviral treatment of mice
For HIV treatment, we used a previously described triple combination of drugs that we have shown to be effective at suppressing viral load in T cell-only mice (ToM) and BLT mice (12, 13, 36). Mice were administered daily intraperitoneal injections of emtricitabine (FTC; 211 mg/kg), tenofovir disoproxil fumarate (TDF; 205 mg/kg) and raltegravir (RAL; 56 mg/kg) for three to eleven weeks as indicated on individual graphs. HIV infection was monitored throughout ART-treatment via plasma viral load analysis as described above.
Tissue harvesting of humanized mice
To minimize the numbers of blood-derived monocytes in tissues harvested from HIV-infected MoM and BLT mice, at necropsy animals were transcardially perfused with ~20 ml of room temperature PBS. MNCs were isolated from the bone marrow, spleen, lung, and liver as previously described. Tissues were minced and/or digested and filtered through a 70 μm strainer. Liver, lung and brain samples were purified by Percoll density centrifugation. Red blood cells were lysed as needed (namely for the spleen, bone marrow and liver tissues). MNCs were washed and counted via trypan blue exclusion.
Macrophage and T cell sorting
Total MNCs were isolated from the spleen, liver, lung, and bone marrow of HIV-infected BLT mice. Mouse cells were depleted using the EasySep Mouse Human Chimera Isolation kit (Stem Cell Technologies, Cat. 19849) followed by positive magnetic selection for human CD3+ T cells (Miltenyi, CD3 MicroBeads, Cat. 130-050-101). Next, negative magnetic selection for human monocytes and macrophages (Miltenyi, Pan Monocyte Isolation Kit, Cat. 130-096-537) was performed on the non-CD3 cell population to yield a purified macrophage population (<1% T cell contamination). Cell purity was verified using flow cytometric staining.
Flow cytometric analysis of humanized mice
All flow antibodies were purchased from BD Pharmingen. The antibody panel used to analyze cells isolated from humanized mice included antibodies directed against hCD45 (APC, Cat. 555485 or APC-Cy7, Cat. 557833), hCD3 (FITC, Cat. 555339), hCD19 (PE-Cy7, Cat. 557835), hCD33 (PE, Cat. 340679), hCD14 (FITC, Cat. 555397), hCD16 (PE-Cy7, Cat. 557744), hCCR5 (APC, Cat. 550856) and hCD11b (PE, Cat. 555388). Live cells were distinguished by their forward and side scatter profiles. Flow cytometry data was collected on a BD FACSCanto and analyzed with BD FACSDiva software (v.6.1.3).
Immunohistochemical analysis
Tissues for IHC were harvested from MoM and fixed in 4% paraformaldehyde for 24 hour at 4°C, embedded in paraffin, cut into 5-μm sections and mounted onto poly-L-lysine coated glass slides. Following paraffin removal, antigen retrieval (DIVA Decloaker, Biocare Medical, Cat. DV2004.) and blocking of non-specific Ig-binding sites (Background Sniper, Biocare Medical), tissue sections were stained with primary antibodies overnight at 4°C and developed with a biotin-free horseradish peroxidase (HRP)-polymer system (MACH3 Mouse HRP-Polymer Detection, Biocare Medical). All tissue sections were then counterstained with hematoxylin. Primary antibodies directed against CD45 LCA (2B11&PD7/26, Dako), CD3 (SP7, Thermo Scientific), CD20 (L26, Biocare Medical) and CD68 (KP1, Dako) were used to identify human cells in the spleen, liver and lung. For comparison, tissue sections were stained with mouse IgG1k or IgG2a isotype controls. Light microscopy images were taken with a Nikon H550S microscope at 40×.
Sequencing of HIV-1 in MoM plasma
For sequence analysis of HIV-1 envelope, viral RNA was isolated from plasma from infected MoM using the QIAamp viral RNA Mini kit (Qiagen). Viral cDNA was generated using Superscript III Reverse Transcriptase (Invitrogen) and amplified by nested PCR using the Expand High Fidelity PCR System (Roche). The amplified region was a 1.2 kb region of HIV env (CH040 6426-7702; CH040-4013env 6426-7702). The primers used for CH040 env amplification include env outer forward primer (CACCACTCTATTTTGTGCATCAGATG), env outer reverse primer (GCCCATAGTGCTTCCTGCTGC), env inner forward primer (CACACATGCCTGTGTACCCACAGAC) and env inner reverse primer (CTCTTCTCTTTGCCTTGGTAGGTGC). The primers used for CH040-4013env env amplification include env outer forward primer (CACCACTCTATTTTGTGCATCAGATG), env outer reverse primer (GCCCATAGTGCTTCCTGCTGC), env inner forward primer (CACACATGCCTGTGTACCCACAGAC) and env inner reverse primer (CTCTTCTCTTTGCCTTGGTGGGTGC). Amplified DNA was sequenced and aligned to HIV-1 CH040 or CH040-4013 env to determine if nucleotide changes had occurred. FinchTV (Geospiza Inc., Seattle, WA) was used for the sequence analysis of the chromatograms and ClustalW (http://www.genome.jp/tools/clustalw/) was used to compare the MoM envelope sequences to the reference CH040 or CH040-4013env sequences.
Experimental groups
The total number of experiments performed was determined based on total number of tissue cohorts (representing different human donors) and viral exposures utilized in each figure. For Figure 1, 12 human donors and 8 exposure groups are represented. For Figure 2, 13 human donors and 13 exposure groups are represented. For Figure 3, 5 human donors and 5 exposure groups are represented.
Statistical analyses
All data was graphed using GraphPad Prism (version 5.04). The statistical tests used are indicated in the figure legends and/or text for each comparison performed. All statistical tests were exact. A mixed-effects approach using Monolix (Lixoft, France), allowing for censored values, was utilized to model the decay slope for viremia after ART initiation, leading to an estimate for the half-life of productively infected cells in MoM and BLT mice. For half-life analyses, we assumed a near-perfect efficacy of the ART regimen to prevent new infections (>99%), yielding upper bound estimates for the half-lives. For comparisons in Fig. 2d/2e & Supplemental Fig. 3c, a log-rank test was utilized, thus accounting for the variation in the lower limit of detection between samples similar to what has been done for other assays (37–39). For all Mann-Whitney comparisons, a two-tailed test was used. For all comparisons, a p-value <0.05 was considered statistically significant. No adjustment was made for multiple hypothesis testing. In all figures, the following representations were used to indicate the p-values: * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. No statistical methods were used to pre-determine sample size. No randomization was used. Blinding of the statisticians (RMM and YC) was used when accessing and modeling the infected cell half-life post-ART initiation (Fig. 1c, 2c). Investigators were given viral loads for lettered groups of mice (A, B, C), but were not privy to the identity of the groups (MoM, ToM or BLT mice) until all half-lives were calculated. No blinding was used for comparisons in the vDNA and vRNA levels of treated or untreated MoM (Fig. 2d, 2e).
Data availability
The sequencing data demonstrating point mutations in env for MoM #21 and MoM #23 were deposited in GenBank (accession numbers KX825880 & KX825881).
Supplementary Material
Acknowledgments
JVG was supported by grants from the National Institute of Mental Health (MH-108179) and the National Institute of Allergy and Infectious Diseases (NIAID) (AI-111899) and the UNC CFAR (P30 AI050410). Research reported in this publication was also supported by CARE, a Martin Delaney Collaboratory, the National Institute of Allergy and Infectious Diseases, National Institute of Neurological Disorders and Stroke (NINDS), National Institute On Drug Abuse (NIDA) and the National Institute of Mental Health (NIMH) of the National Institutes of Health, grant number 1UM1AI126619-01 (JVG and DDR). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. JBH was supported in part by the Virology Training Grant (T32 AI-007419). RMR was supported by the NIAID (R01 AI-104373). The funders had no part in study design, data collection/analysis, decision to publish, or preparation of the manuscript. We thank Nancie Archin for providing the CD8-depleted feeder cells for in vitro viral outgrowth analysis. We thank J. Kappes and C. Ochsenbauer for providing CH040 (cat#11740) via the AIDS Research and Reference Reagent Program. We thank Ron Swanstrom for providing the 4013-env for use in our studies. We thank Janice Clements for her intellectual input in these studies. We thank Myron Cohen, Bart Haynes, Stan Lemon, Angela Wahl and Martina Kovarova for their comments and suggestions regarding this manuscript. We also thank former and current members of the Garcia laboratory, the husbandry technicians at the UNC Division of Laboratory Animal Medicine for their assistance.
Footnotes
Author contributions: JBH provided the experimental design, collected and processed blood and tissues samples from mice, prepared viral stocks, performed viral inoculations, administered ART and performed flow cytometric analyses. WOT processed blood and tissue samples from mice and prepared/administered ART. CEB provided RNA/DNA analysis, prepared ART and processed tissue samples. RAC performed immunohistochemical analysis of tissues. SML/DDR performed the integrated DNA analysis and analyzed the results. JBH, RMR, YC and MGH analyzed the data and wrote the manuscript. JVG conceived of the study, designed and coordinated the study, and wrote the manuscript.
The authors have no competing interests as defined by Springer Nature, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
References
- 1.Kumar A, Herbein G. The macrophage: a therapeutic target in HIV-1 infection. Mol Cell Ther. 2014;2:10. doi: 10.1186/2052-8426-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koppensteiner H, Brack-Werner R, Schindler M. Macrophages and their relevance in Human Immunodeficiency Virus Type I infection. Retrovirology. 2012;9:82. doi: 10.1186/1742-4690-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell JH, Hearps AC, Martin GE, Williams KC, Crowe SM. The importance of monocytes and macrophages in HIV pathogenesis, treatment, and cure. AIDS. 2014;28:2175–2187. doi: 10.1097/QAD.0000000000000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sattentau QJ, Stevenson M. Macrophages and HIV-1: An Unhealthy Constellation. Cell Host Microbe. 2016;19:304–310. doi: 10.1016/j.chom.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collman RG, Perno CF, Crowe SM, Stevenson M, Montaner LJ. HIV and cells of macrophage/dendritic lineage and other non-T cell reservoirs: new answers yield new questions. J Leukoc Biol. 2003;74:631–634. doi: 10.1189/jlb.0703357. [DOI] [PubMed] [Google Scholar]
- 6.Gavegnano C, Schinazi RF. Antiretroviral therapy in macrophages: implication for HIV eradication. Antivir Chem Chemother. 2009;20:63–78. doi: 10.3851/IMP1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honeycutt JB, Wahl A, Baker C, Spagnuolo RA, Foster J, Zakharova O, Wietgrefe S, Caro-Vegas C, Madden V, Sharpe G, Haase AT, Eron JJ, Garcia JV. Macrophages sustain HIV replication in vivo independently of T cells. J Clin Invest. 2016;126:1353–1366. doi: 10.1172/JCI84456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denton PW, Krisko JF, Powell DA, Mathias M, Kwak YT, Martinez-Torres F, Zou W, Payne DA, Estes JD, Garcia JV. Systemic administration of antiretrovirals prior to exposure prevents rectal and intravenous HIV-1 transmission in humanized BLT mice. PLoS One. 2010;5:e8829. doi: 10.1371/journal.pone.0008829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wahl A, Swanson MD, Nochi T, Olesen R, Denton PW, Chateau M, Garcia JV. Human breast milk and antiretrovirals dramatically reduce oral HIV-1 transmission in BLT humanized mice. PLoS Pathog. 2012;8:e1002732. doi: 10.1371/journal.ppat.1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olesen R, Wahl A, Denton PW, Garcia JV. Immune reconstitution of the female reproductive tract of humanized BLT mice and their susceptibility to human immunodeficiency virus infection. J Reprod Immunol. 2011;88:195–203. doi: 10.1016/j.jri.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melkus MW, Estes JD, Padgett-Thomas A, Gatlin J, Denton PW, Othieno FA, Wege AK, Haase AT, Garcia JV. Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-1. Nat Med. 2006;12:1316–1322. doi: 10.1038/nm1431. [DOI] [PubMed] [Google Scholar]
- 12.Denton PW, Olesen R, Choudhary SK, Archin NM, Wahl A, Swanson MD, Chateau M, Nochi T, Krisko JF, Spagnuolo RA, Margolis DM, Garcia JV. Generation of HIV latency in humanized BLT mice. J Virol. 2012;86:630–634. doi: 10.1128/JVI.06120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denton PW, Long JM, Wietgrefe SW, Sykes C, Spagnuolo RA, Snyder OD, Perkey K, Archin NM, Choudhary SK, Yang K, Hudgens MG, Pastan I, Haase AT, Kashuba AD, Berger EA, Margolis DM, Garcia JV. Targeted cytotoxic therapy kills persisting HIV infected cells during ART. PLoS Pathog. 2014;10:e1003872. doi: 10.1371/journal.ppat.1003872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson MA, Aberg JA, Hoy JF, Telenti A, Benson C, Cahn P, Eron JJ, Gunthard HF, Hammer SM, Reiss P, Richman DD, Rizzardini G, Thomas DL, Jacobsen DM, Volberding PA. Antiretroviral treatment of adult HIV infection: 2012 recommendations of the International Antiviral Society-USA panel. JAMA. 2012;308:387–402. doi: 10.1001/jama.2012.7961. [DOI] [PubMed] [Google Scholar]
- 15.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 16.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 17.Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 18.Calin R, Hamimi C, Lambert-Niclot S, Carcelain G, Bellet J, Assoumou L, Tubiana R, Calvez V, Dudoit Y, Costagliola D, Autran B, Katlama C, U. S. Group Treatment interruption in chronically HIV-infected patients with an ultralow HIV reservoir. AIDS. 2016;30:761–769. doi: 10.1097/QAD.0000000000000987. [DOI] [PubMed] [Google Scholar]
- 19.Shultz LD, Ishikawa F, Greiner DL. Humanized mice in translational biomedical research. Nat Rev Immunol. 2007;7:118–130. doi: 10.1038/nri2017. [DOI] [PubMed] [Google Scholar]
- 20.Igarashi T, Brown CR, Endo Y, Buckler-White A, Plishka R, Bischofberger N, Hirsch V, Martin MA. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): Implications for HIV-1 infections of humans. Proc Natl Acad Sci U S A. 2001;98:658–663. doi: 10.1073/pnas.021551798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Micci L, Alvarez X, Iriele RI, Ortiz AM, Ryan ES, McGary CS, Deleage C, McAtee BB, He T, Apetrei C, Easley K, Pahwa S, Collman RG, Derdeyn CA, Davenport MP, Estes JD, Silvestri G, Lackner AA, Paiardini M. CD4 depletion in SIV-infected macaques results in macrophage and microglia infection with rapid turnover of infected cells. PLoS Pathog. 2014;10:e1004467. doi: 10.1371/journal.ppat.1004467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avalos CR, Price SL, Forsyth ER, Pin JN, Shirk EN, Bullock BT, Queen SE, Li M, Gellerup D, O’Connor SL, Zink MC, Mankowski JL, Gama L, Clements JE. Quantitation of Productively Infected Monocytes and Macrophages of Simian Immunodeficiency Virus-Infected Macaques. J Virol. 2016;90:5643–5656. doi: 10.1128/JVI.00290-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, Legasse AW, Chiuchiolo MJ, Parks CL, Axthelm MK, Nelson JA, Jarvis MA, Piatak M, Jr, Lifson JD, Picker LJ. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473:523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen SG, Piatak M, Jr, Ventura AB, Hughes CM, Gilbride RM, Ford JC, Oswald K, Shoemaker R, Li Y, Lewis MS, Gilliam AN, Xu G, Whizin N, Burwitz BJ, Planer SL, Turner JM, Legasse AW, Axthelm MK, Nelson JA, Fruh K, Sacha JB, Estes JD, Keele BF, Edlefsen PT, Lifson JD, Picker LJ. Immune clearance of highly pathogenic SIV infection. Nature. 2013;502:100–104. doi: 10.1038/nature12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sung JA, Lam S, Garrido C, Archin N, Rooney CM, Bollard CM, Margolis DM. Expanded cytotoxic T-cell lymphocytes target the latent HIV reservoir. J Infect Dis. 2015;212:258–263. doi: 10.1093/infdis/jiv022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Archin NM, Eron JJ, Palmer S, Hartmann-Duff A, Martinson JA, Wiegand A, Bandarenko N, Schmitz JL, Bosch RJ, Landay AL, Coffin JM, Margolis DM. Valproic acid without intensified antiviral therapy has limited impact on persistent HIV infection of resting CD4+ T cells. AIDS. 2008;22:1131–1135. doi: 10.1097/QAD.0b013e3282fd6df4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spina CA, Anderson J, Archin NM, Bosque A, Chan J, Famiglietti M, Greene WC, Kashuba A, Lewin SR, Margolis DM, Mau M, Ruelas D, Saleh S, Shirakawa K, Siliciano RF, Singhania A, Soto PC, Terry VH, Verdin E, Woelk C, Wooden S, Xing S, Planelles V. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS Pathog. 2013;9:e1003834. doi: 10.1371/journal.ppat.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prochazka M, Gaskins HR, Shultz LD, Leiter EH. The nonobese diabetic scid mouse: model for spontaneous thymomagenesis associated with immunodeficiency. Proc Natl Acad Sci U S A. 1992;89:3290–3294. doi: 10.1073/pnas.89.8.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill AL, Rosenbloom DI, Goldstein E, Hanhauser E, Kuritzkes DR, Siliciano RF, Henrich TJ. Real-Time Predictions of Reservoir Size and Rebound Time during Antiretroviral Therapy Interruption Trials for HIV. PLoS Pathog. 2016;12:e1005535. doi: 10.1371/journal.ppat.1005535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steingrover R, Pogany K, Fernandez Garcia E, Jurriaans S, Brinkman K, Schuitemaker H, Miedema F, Lange JM, Prins JM. HIV-1 viral rebound dynamics after a single treatment interruption depends on time of initiation of highly active antiretroviral therapy. AIDS. 2008;22:1583–1588. doi: 10.1097/QAD.0b013e328305bd77. [DOI] [PubMed] [Google Scholar]
- 31.Persaud D, Gay H, Ziemniak C, Chen YH, Piatak M, Jr, Chun TW, Strain M, Richman D, Luzuriaga K. Absence of detectable HIV-1 viremia after treatment cessation in an infant. N Engl J Med. 2013;369:1828–1835. doi: 10.1056/NEJMoa1302976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henrich TJ, Hu Z, Li JZ, Sciaranghella G, Busch MP, Keating SM, Gallien S, Lin NH, Giguel FF, Lavoie L, Ho VT, Armand P, Soiffer RJ, Sagar M, Lacasce AS, Kuritzkes DR. Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J Infect Dis. 2013;207:1694–1702. doi: 10.1093/infdis/jit086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nath A, Clements JE. Eradication of HIV from the brain: reasons for pause. AIDS. 2011;25:577–580. doi: 10.1097/QAD.0b013e3283437d2f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joseph SB, Arrildt KT, Sturdevant CB, Swanstrom R. HIV-1 target cells in the CNS. J Neurovirol. 2015;21:276–289. doi: 10.1007/s13365-014-0287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH, Spina CA, Woelk CH, Richman DD. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One. 2013;8:e55943. doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Honeycutt JB, Wahl A, Archin N, Choudhary S, Margolis D, Garcia JV. HIV-1 infection, response to treatment and establishment of viral latency in a novel humanized T cell-only mouse (TOM) model. Retrovirology. 2013;10:121. doi: 10.1186/1742-4690-10-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang D, Fan C, Zhang J, Zhang CH. Nonparametric methods for measurements below detection limit. Stat Med. 2009;28:700–715. doi: 10.1002/sim.3488. [DOI] [PubMed] [Google Scholar]
- 38.Lafleur B, Lee W, Billhiemer D, Lockhart C, Liu J, Merchant N. Statistical methods for assays with limits of detection: Serum bile acid as a differentiator between patients with normal colons, adenomas, and colorectal cancer. J Carcinog. 2011;10:12. doi: 10.4103/1477-3163.79681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gillespie BW, Chen Q, Reichert H, Franzblau A, Hedgeman E, Lepkowski J, Adriaens P, Demond A, Luksemburg W, Garabrant DH. Estimating population distributions when some data are below a limit of detection by using a reverse Kaplan-Meier estimator. Epidemiology. 2010;21(Suppl 4):S64–70. doi: 10.1097/EDE.0b013e3181ce9f08. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data demonstrating point mutations in env for MoM #21 and MoM #23 were deposited in GenBank (accession numbers KX825880 & KX825881).


