Summary
Age-related macular degeneration (AMD) affects the retinal pigment epithelium (RPE), a cell monolayer essential for photoreceptor survival, and is the leading cause of vision loss in the elderly. There are no disease-altering therapies for dry AMD, which is characterized by accumulation of subretinal drusen deposits and complement-driven inflammation. We report the derivation of human induced pluripotent stem cells (hiPSCs) from patients diagnosed with AMD, including two with the rare ARMS2/HTRA1 homozygous genotype. The hiPSC-derived RPE cells produce several AMD/drusen-related proteins, and those from AMD donors showed significantly increased complement and inflammatory factors, most exaggerated in ARMS2/HTRA1 lines. Using a panel of AMD biomarkers and candidate drug screening, combined with transcriptome analysis, we discovered that Nicotinamide (NAM) ameliorated disease-related phenotypes by inhibiting drusen proteins, inflammatory and complement factors, while upregulating nucleosome, ribosome and chromatin-modifying genes. Thus, targeting NAM-regulated pathways is a promising avenue for developing therapeutics to combat AMD.
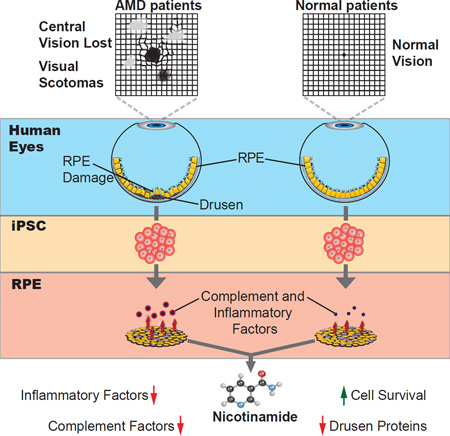
Graphical abstract
eTOC synopsis
Saini et al. show that hiPSC derived RPE from AMD patients, including ARMS2/HTRA1 homozygotes, exhibit higher complement and inflammatory factors compared to healthy controls. Nicotinamide treatment reduces these and other AMD-related molecules with no observed cytotoxicity. Pursuing Nicotinamide’s mechanism of action should reveal new therapeutic approaches for AMD.
Introduction
Age-related macular degeneration (AMD) is the leading cause of visual impairment and blindness in the elderly, affecting more than 10 million people in the US (Rein et al., 2009). In AMD, the retinal pigment epithelium (RPE), a pigmented cell monolayer lying beneath and supporting the neural retina, degenerates. Degeneration is most pronounced in the macula, the central part of the retina that is responsible for high acuity and color vision (Strauss, 2005). AMD occurs in two forms, dry and wet. The more prevalent dry form is characterized by the presence of extracellular deposits, termed drusen, which predominantly accumulate beneath the RPE layer (Jager et al., 2008; Strauss, 2005).
AMD arises through a combination of genetic as well as environmental factors (Deangelis et al., 2011). Genome wide association studies (GWAS) indicate that 65% of the total genetic contribution is accounted by approximately 20 AMD associated genetic variants, several of which have been localized to complement component genes including CFH, CFI, CFB and C3 (Deangelis et al., 2011; Fritsche et al., 2013; Fritsche et al., 2014). In addition to the complement genes, GWAS have identified a risk allele in the ARMS2 gene (SNP rs10490924) and another in the promoter region of the nearby HTRA1 gene (SNP rs11200638), which are significantly linked to AMD and to disease severity (Deangelis et al., 2011; Fritsche et al., 2013; Scholl et al., 2007). The effect of the ARMS2/HTRA1 risk SNPs, whether on RPE cells directly or some other involved cell type, and their role in pathogenesis of AMD, remain to be determined (Deangelis et al., 2011); however, a recent report indicates that ARMS2/HTRA1 results in a stress (Yang et al., 2014).
While a complete understanding of the disease process remains elusive, considerable progress has been made in elucidating the molecular composition of drusen, which consist of approximately half protein and half lipid (Crabb, 2014; Wang et al., 2010). Numerous proteins, including all the major proteins of the complement pathway, TIMP3, APOJ, Annexin, Crystallins, APOE, Vitronectin and Amyloid β (Aβ), have been identified in drusen deposits, which share several similarities to aggregates formed in Alzheimer’s disease (AD) (Ohno-Matsui, 2011). Studies have implicated dysfunction of RPE cells in AMD pathogenesis, as they can express and secrete several proteins found in drusen (An et al., 2006; De et al., 2007; Johnson et al., 2011; Rabin et al., 2013). A deeper understanding of the mechanisms underlying dry AMD and drusen formation is important in order to develop therapies that slow disease progression and inhibit transition to geographic atrophy or the more severe wet (neovascular) form of AMD, thereby preserving vision.
We previously described a rare, self-renewing multipotent stem cell from the adult human RPE layer, the RPE stem cell (RPESC) (Saini et al., 2016; Salero et al., 2012) that can model aspects of AMD in culture after treatment with an oxidative stressor (Rabin et al., 2013). However, RPESC-derived RPE have a limited ability to proliferate and maintain a polarized, cobblestone morphology (Salero et al., 2012), making a standardized disease model difficult to establish. In contrast, RPE can be readily produced at high purity from human induced pluripotent stem cells (hiPSC) reprogrammed from somatic cells. In this study, we tested whether hiPSC-derived RPE (hiPSC-RPE) from AMD and control patients can be used to model the early pathogenesis of AMD. Furthermore, we investigated the effect of the homozygous ARMS2/HTRA1 risk genotype on the production of AMD-associated proteins in RPE cells. We demonstrated that hiPSC-RPE cells express multiple biomarkers associated with AMD. Compared to healthy control lines, we found significantly higher expression of complement and inflammatory factors in the AMD hiPSC-RPE lines, more significant in the ARMS2/HTRA1 subset. We then used this in vitro model to test for drugs that would reduce AMD-associated protein production.
Nicotinamide (NAM), a vitamin B3 derivative with anti-oxidant and anti-inflammatory properties (Maiese et al., 2009), can restore cognition in AD mice (Green et al., 2008) and is in clinical trial for AD (NCT00580931). Here we show that NAM can significantly inhibit the production of AMD-related proteins including drusen proteins, and markedly repress key genes in the complement and inflammatory pathways as well as improve RPE survival. In contrast, inhibition of complement only, currently a major therapeutic approach being developed for AMD treatment, did not reduce inflammatory markers nor improve RPE cell health and survival. This study demonstrates the utility of hiPSC-RPE to model AMD and compare therapeutic strategies, and illuminates potential new pathways for treatment development.
Results
Derivation of hiPSC-RPE from AMD and control adult RPE and fibroblasts
Adult RPE or corneal fibroblasts were cultured from donated globes from patients with an AMD diagnosis (age range 73–93 years) and from unaffected controls (age range 71–91 years). The resulting cell cultures were reprogrammed using non-integrating strategies (Figure 1A and Table S1), generating five hiPSC lines from four AMD patients, and three hiPSC lines from three unaffected, age-matched individuals. All hiPSC lines showed typical iPSC morphology (tightly packed, flat colonies with prominent nucleoli), expressed the pluripotency markers (Figure S1A and Figure S1B), and had normal karyotype (Figure S1C). Staining for markers of the three embryonic germ layers, demonstrated that each hiPSC line was pluripotent (Figure S1D). Since certain hiPSCs originated from adult RPE tissue and were differentiated back into the same cell type as the source tissue, the absence of RPE contamination was demonstrated for these lines (Figure S1E). We genotyped each donor for the ARMS2/HTRA1 susceptibility SNPs and found that all the controls harbored protective alleles at both loci, while two out of four AMD donors were homozygous for the risk alleles and one AMD donor was heterozygous (Table S1). As expected, there was a 100% linkage disequilibrium between the rs10490924 locus in ARMS2 and the rs11200638 locus in HTRA1 genes (Table S1). Additionally, we genotyped for high AMD risk SNPs in CFH (rs1061170) and C3 (rs2230199) genes. While these were present in some donors in the heterozygous condition, the AMD odds ratio (OR) is much lower (C3 risk allele OR: 1.42; CFH risk allele OR: 2.43) compared to that of the ARMS2/HTRA1 risk allele in the homozygous condition (OR: 7–8) (Table S1) (Fritsche et al., 2013; Scholl et al., 2007).
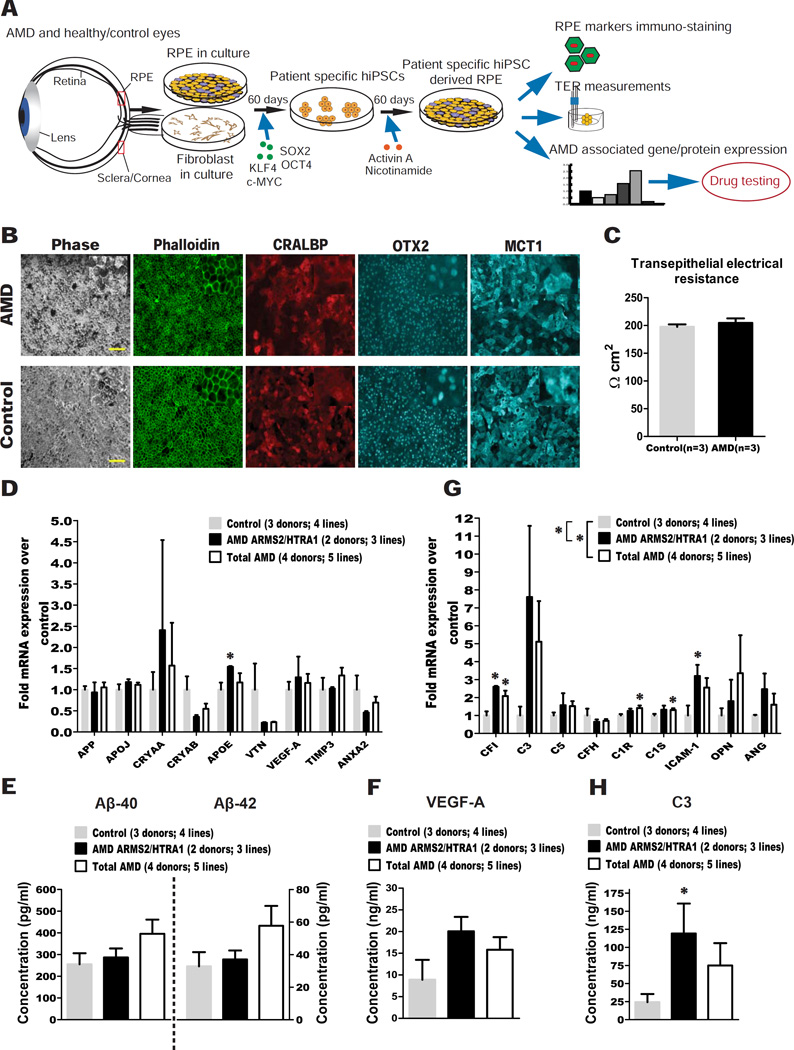
Figure 1. hiPSC-RPE from total AMD and AMD ARMS2/HTRA1 donors express higher levels of disease-related markers compared to controls.
(A) Experiment schematic.
(B) AMD and control hiPSC-RPE stained for Phalloidin, and the RPE markers CRALBP, OTX2 and MCT1 (Scale bar: 100µM). Insets show digitally zoomed high magnification images.
(C) AMD and control hiPSC-RPE showed similar TER.
(D) qPCR analysis of AMD ARMS2/HTRA1 hiPSC-RPE and total AMD hiPSC-RPE versus control hiPSC-RPE for AMD/drusen transcripts.
(E-F) ELISA testing of culture supernatant from AMD ARMS2/HTRA1, total AMD and control hiPSC-RPE obtained 60–72 hours after the last medium change for secretion of Aβ peptides (E) and VEGF-A (F).
(G) qPCR analysis of AMD ARMS2/HTRA1 hiPSC-RPE and total AMD hiPSC-RPE versus control hiPSC-RPE for complement/inflammatory transcripts.
(H) ELISA testing of culture supernatant from AMD ARMS2/HTRA1, total AMD and control hiPSC-RPE obtained 60–72 hours after the last medium change for secretion of C3.
Data are expressed as mean± SEM. Unpaired Student’s t test (one-tailed) (D–H) and two-way ANOVA (D, G) were used for statistical analysis (*= p<0.05).
The hiPSC lines were differentiated to produce RPE cells (Figure 1A), using a slight modification of our previously published protocol (Ferrer et al., 2014) (see STAR Methods), the RPE colonies were then manually picked and passaged 1–7 times in order to obtain sufficient pure RPE cells. The resulting monolayers of RPE cells had a typical cobblestone morphology as shown by Phalloidin staining, and expressed markers of mature RPE including CRALBP, OTX2 and MCT1 (Figure 1B). Transepithelial resistance (TER) measurement, an indicator of epithelial tightness and RPE health, on cells growing in transwells confirmed that hiPSCs had been successfully differentiated into a tight RPE monolayer (Figure 1C). Immunostaining and TER analysis did not reveal any difference in the expression and functional status between AMD patient-specific hiPSC-RPE and age-matched healthy control hiPSC-RPE.
AMD hiPSC-RPE show higher levels of AMD-related markers compared to healthy control hiPSC-RPE
hiPSC-RPE from each line were then passaged, plated at the same density and cultured for the same amount of time (3–4 weeks) to form stable, pure monolayers of RPE cells, prior to analysis. The hiPSC-RPE lines were considered as three groups: total healthy control hiPSC-RPE (homozygous for the ARMS2/HTRA1 protective allele), total AMD patient hiPSC-RPE (including those homozygous for the ARMS2/HTRA1 risk allele), and the AMD ARMS2/HTRA1 hiPSC-RPE (homozygous for the ARMS2/HTRA1 risk allele) subset. Since two RPE lines were derived from donors 2 (AMD 2 and AMD 2’ in Table S1) and 5 (Control 1 and Control 1’ in Table S1), the mean of the readouts from the two RPE lines for each donor was used for the analysis.
We first assessed the expression of a panel of nine AMD and drusen-associated protein transcripts (Rabin et al., 2013) to determine whether AMD hiPSC-RPE showed higher production. Overall, the expression of the panel was not significantly different between total AMD or the AMD ARMS2/HTRA1 hiPSC-RPE compared to control hiPSC-RPE (ANOVA p>0.05), although a t-test suggested a significant increase in expression of APOE in the AMD ARMS2/HTRA1 hiPSC-RPE (Figure 1D). Given the importance of Aβ for AMD progression (Ohno-Matsui, 2011), we examined the secretion of Aβ-40 and Aβ-42 from the RPE monolayers, and found no significant difference between the groups (Figure 1E). VEGF-A, which is involved in neo-vascularization in wet AMD (Jager et al., 2008), was secreted at a marginally but not significantly higher level (P=0.09) by AMD ARMS2/HTRA1 hiPSC-RPE compared to control hiPSC-RPE (Figure 1F).
Dysregulation of the alternative complement system leading to local and systemic inflammation is a major driver of AMD pathogenesis (Bradley et al., 2011). HTRA1 encodes a serine peptidase, which has been hypothesized to regulate the complement pathway, leading to increased susceptibility to AMD (An et al., 2010). To test the hypothesis that the AMD hiPSC-RPE, and especially those with the ARMS2/HTRA1 risk genotype, model susceptibility to AMD pathogenesis by exacerbating these disease-associated pathways, we assessed the expression of a second panel of nine transcripts comprising a subset of complement and inflammatory proteins, including C3, CFI and CFH, which have been reported in drusen deposits. There was a significantly increased overall expression of these transcripts in total AMD hiPSC-RPE (ANOVA p<0.01), more significant in the AMD ARMS2/HTRA1 hiPSC-RPE subset (ANOVA p<0.001) compared to controls (Figure 1G). By analyzing individual transcripts, we determined that CFI and ICAM1 in the AMD ARMS2/HTRA1 hiPSC-RPE, and CFI, C1R and C1S in total AMD hiPSC-RPE, were expressed at significantly higher levels compared to control hiPSC-RPE. Complement component 3 (C3) protein (detected by ELISA) was secreted significantly more by AMD ARMS2/HTRA1 hiPSC-RPE compared to controls (Figure 1H). In contrast, total AMD hiPSC-RPE and control hiPSC-RPE showed similar levels of C3 secretion, indicating that the higher level was mostly due to the AMD ARMS2/HTRA1 lines (Figure 1H). Further, C3 protein secretion from lines heterozygous for the CFH risk allele (all AMD and Control 3 in Table S1) was not significantly different from lines without the risk allele (Figure S1F).
These findings show that hiPSC-RPE express a broad panel of drusen and AMD-associated molecules, enabling studies to define factors that mitigate their expression. Further, they show that hiPSC-RPE from AMD patients show an increased expression of inflammatory and complement factors compared to controls. Finally, we found evidence of aggravation of these pathways, and in particular complement-associated factors, in AMD patient lines carrying the homozygous ARMS2/HTRA1 risk alleles. Together, these findings indicate the utility of hiPSC-RPE as a disease model to shed light on the mechanism of AMD progression and for drug screening.
NAM inhibits AMD-related biomarkers in hiPSC-RPE
NAM has multiple actions that could be beneficial for AMD, including preserving the mitochondrial membrane potential, protecting against oxidative damage, and preventing inflammation (Maiese et al., 2009). Given the established role of oxidative stress and inflammation in AMD, we tested whether NAM can reduce the production of AMD- and drusen-associated transcripts and proteins in our hiPSC-RPE based model.
AMD (including the homozygous AMD ARMS2/HTRA1 hiPSC-RPE from 2 donors) and control hiPSC-RPE (passage 2–8) were cultured for 3–4 weeks in NAM until they formed confluent monolayers (see STAR Methods). The cells were then cultured in either 10mM NAM or vehicle for three additional weeks, after which we assessed the expression of the first panel of transcripts by qPCR. Remarkably, NAM significantly reduced the expression of most of the AMD-related transcripts examined, in AMD as well as control hiPSC-RPE (Figure 2A). The effect of NAM was dose-dependent, as 1mM NAM was much less effective, only significantly inhibiting the expression of the VEGF-A transcript (Figure S2A).
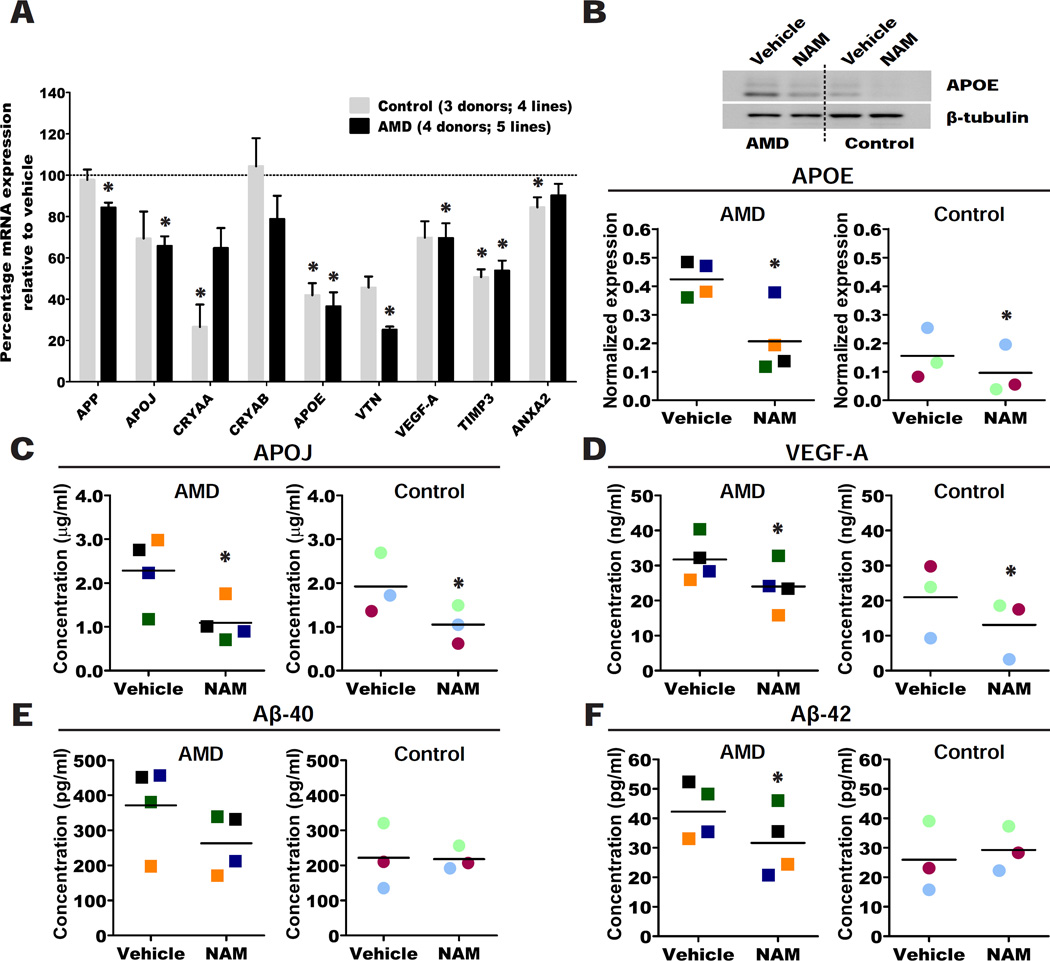
Figure 2. NAM suppresses production of AMD-related disease markers in AMD and control hiPSC-RPE.
(A) Percentage mRNA expression for AMD/drusen transcripts analyzed by qPCR in AMD and control hiPSC-RPE treated with 10mM NAM relative to vehicle (baseline defined as 100%). Data are expressed as mean± SEM.
(B) Representative western blot of APOE protein in AMD and control hiPSC-RPE treated with 10mM NAM or vehicle; (quantification of western blot in Figure S2B, lower panel).
(C-F) Secretion of APOJ (C), VEGF-A (D), Aβ-40 (E) and Aβ-42 (F) measured by ELISA into the culture supernatant 60–72 hours after the last medium change in AMD and control hiPSC-RPE treated with 10mM NAM or vehicle.
Data are expressed as mean of all samples (line), and individual sample values are plotted: AMD hiPSC-RPE are represented by squares (n=4 AMD donors; 5 lines) and control hiPSC-RPE by circles (n=3 control donors; 4 lines). Colors correspond to hiPSC-RPE samples in Table S1; each sample is color matched across NAM and vehicle treatment.
Paired Student’s t test (one-tailed) was used for statistical analysis (*= p<0.05).
To determine whether NAM can also inhibit the synthesis of AMD- and drusen-associated proteins, we quantified the expression of APOE in cell lysates using western blotting, and the secretion of Aβ, APOJ and VEGFA in culture supernatants using ELISA, following treatment of hiPSC-RPE with vehicle or 10mM NAM. NAM significantly inhibited the levels of APOE, APOJ and VEGF-A protein produced from both AMD and control hiPSC-RPE lines (Figure 2B–D; Figure S2B–B’), while secretion of only Aβ-42 was inhibited in AMD hiPSC-RPE lines but not in all control hiPSC-RPE lines (Figure 2E–F). In order to exclude the possibility that transcript and protein production was reduced because 10mM NAM was toxic to the hiPSC-RPE cells, we analyzed the release of lactate dehydrogenase (LDH), an indicator of cytotoxicity. Notably, NAM at 10mM was beneficial for the RPE cells, significantly reducing LDH release (Figure S2C) and preserving expression of RPE markers (Figure S2D). The similar expression of RPE markers in NAM and vehicle treated hiPSC-RPE also suggests that RPE were stable without NAM over the 5–6 weeks time course.
To test the effect of treatment duration, a subset of AMD and control hiPSC-RPE lines was cultured with NAM or vehicle for 12 weeks. Similar to a three-week treatment, long-term treatment with NAM resulted in decreased AMD-related transcripts (Figure S2E) and protein expression (Figure S2F–G), indicating that 10mM NAM was not toxic to the cells and that it can effectively dampen AMD-associated pathways over the long term. We also found that the effect of NAM can be observed as early as one week in hiPSC-RPE cells (Figure S2H). Further, we show that the increase in the expression of AMD associated proteins in hiPSC-RPE cultured in vehicle can be reversed upon re-treatment with NAM (Figure S2I–J).
In order to determine whether these findings made on hiPSC-derived RPE are relevant to primary adult human RPE, we tested the effect of NAM on five RPESC-derived human RPE lines from two AMD and three control donors (Table S2), and found that NAM significantly inhibited APOJ and VEGF-A but not Aβ-40 (Figure S2K–M), while Aβ-42 was below the detection limit.
Overall, our results showed that AMD, including the homozygous ARMS2/HTR1A subset, and control hiPSC-RPE exhibit similar responses to NAM, albeit to different extents, with several markers showing the most inhibition in AMD lines. This indicates that AMD hiPSC-RPE may be the most sensitive to identify potential therapeutics. Nevertheless, given the common response of both control and AMD lines to NAM, we studied the responses to NAM of both groups together to increase power, in order to dissect its mechanism of action.
Gene expression analysis after NAM treatment highlights AMD-associated pathways
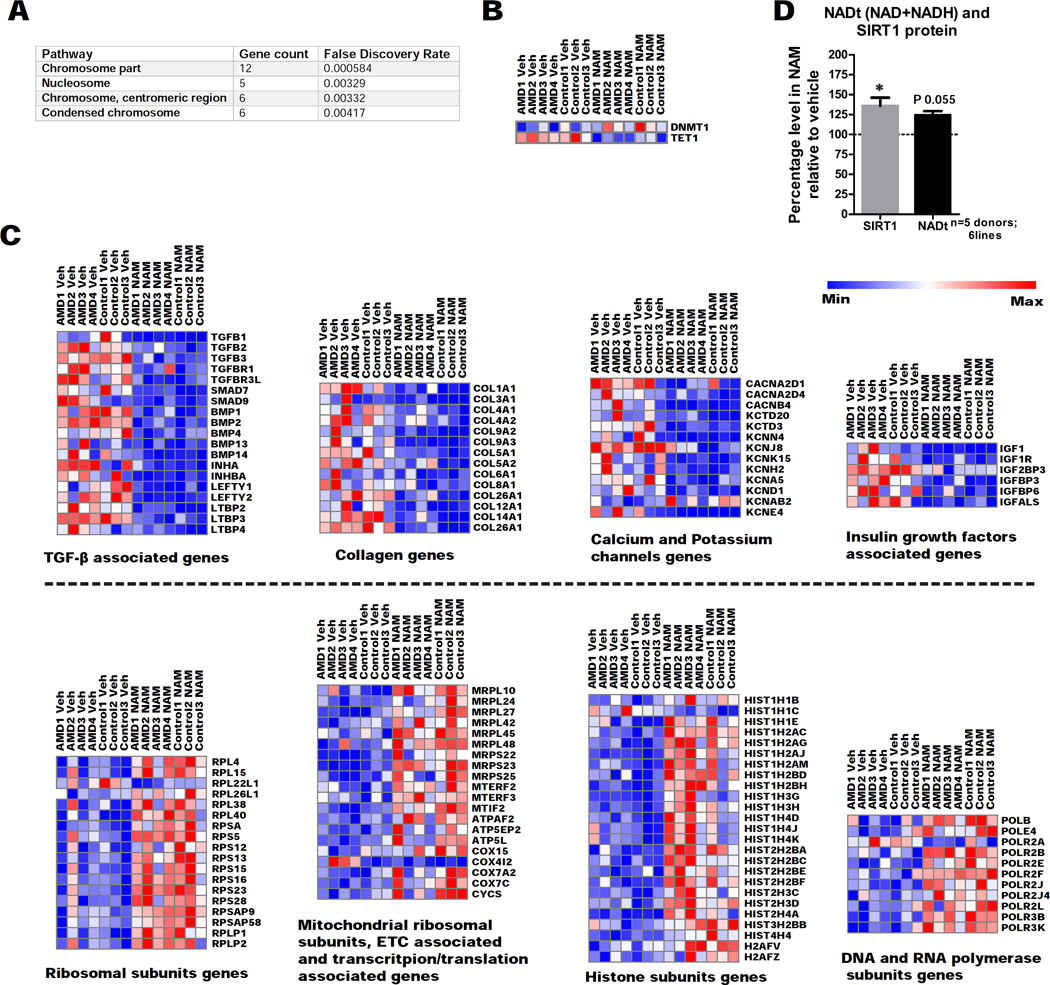
In order to better understand the mechanism of action of NAM, we examined the global impact on the RPE transcriptome using hiPSC-RPE lines from all seven donors under study (AMD 1, AMD 2, AMD 3, AMD 4, Control 1, Control 2 and Control 3 in Table S1), cultured with 10mM NAM or vehicle for three weeks. The RNA-seq analysis identified 2025 differentially expressed genes (FDR p≤0.1) in NAM-treated versus vehicle-treated hiPSC-RPE, with 713 genes showing higher expression and 1312 genes showing lower expression. In order to validate the RNA-seq data, several differentially-expressed genes were confirmed by qPCR (Figure S3A–B).
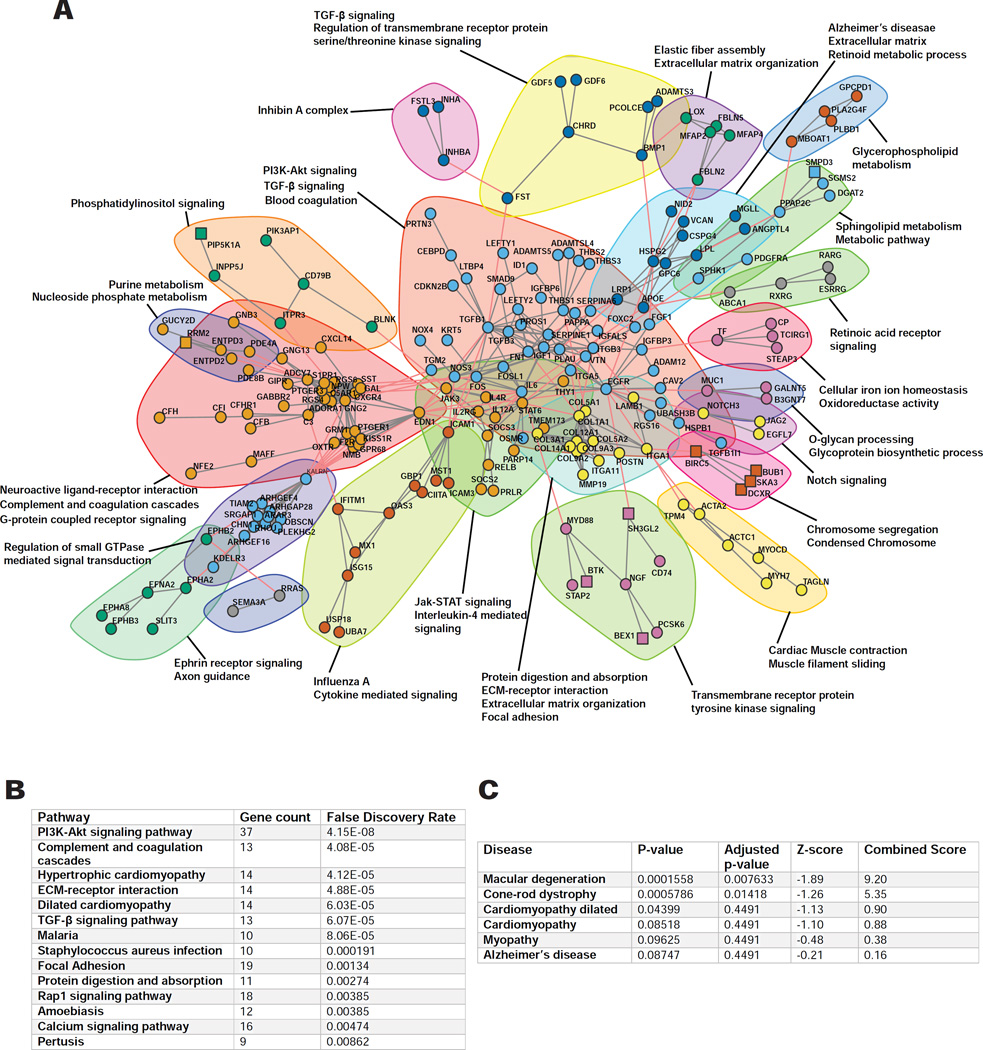
Of the total differentially expressed genes (FDR p≤0.1), a subset demonstrated greater than a two-fold change in expression (719 total); the vast majority of these genes (647) were found to be suppressed by NAM. We performed bioinformatic analysis using the STRING database to predict the associations between this 719 gene subset and extracted subnetworks or neighborhoods based on functional annotations (Figure 3A). Pathway enrichment analysis showed that NAM primarily affects the PI3K-Akt signaling pathway, followed by complement cascades, extracellular matrix (ECM) receptor interactions, TGF-β signaling, and to a lesser extent the calcium signaling pathway (Figure 3A–B). Remarkably, the most significant disease-associated Gene Ontology (GO) terms, analyzed using ENRICHR gene enrichment tool, were those involved in macular degeneration (Figure 3C), due to impact on several AMD-associated genes such as C3, CFB, ABCA4, THBS1, HMCN1, APOE and FBLN5, re-enforcing the applicability of the model and NAM as a preventative small molecule.
Figure 3. RNA-seq analysis of action of NAM on hiPSC-RPE.
(A) Gene interaction network (confidence level=0.9) of the differentially expressed genes in NAM treated hiPSC-RPE compared to vehicle from RNA-seq analysis, using the STRING database (n=7 donors; 4 AMD and 3 control; 7 lines). Subnetworks (Neighborhoods) are colored and annotated with enriched functional categories. Gray lines: connections within a neighborhood; Red lines: connections between neighborhoods; Squares: Upregulated genes; Circles: Downregulated genes.
(B-C) GO enrichment for KEGG pathways (B) and disease associations (C) of the differentially expressed genes (n=7 donors; 4 AMD and 3 control; 7 lines).
See also Figure S3.
NAM decreases inflammatory cytokine production in hiPSC-RPE
Although AMD is a complex multifactorial disorder, inflammation has been consistently associated with AMD pathogenesis, with complement factors being a principal source of inflammation (Bradley et al., 2011). NAM is suggested to act on inflammation via the Akt pathway (Maiese et al., 2009), which is significantly affected by NAM treatment in hiPSC-RPE. In order to explore the effect of NAM on inflammation more fully, as well as to investigate the capability of hiPSC-RPE to secrete inflammatory cytokines, we tested the supernatant collected individually from four hiPSC-RPE lines (AMD 1, AMD 4, Control 1 and Control 2 in Table S1) for 102 cytokines (Table S3 and Figure S4A–B), several of which have been associated with AMD, comparing three weeks of NAM treatment versus vehicle.
On quantifying the pixel intensity of the cytokine array and upon discounting the cytokines showing substantial variation between the paired technical replicates (see STAR Methods), the secretion of 29 cytokines was inhibited by NAM (ANOVA p<0.0001). After correcting for multiple comparisons, nine cytokines (FDR p≤0.1) were reduced by NAM (Figure 4A–B; Figure S4C), with CXCL12, THBS1 and ICAM1 being the most significantly inhibited. Of these nine, MCP1, CXCL12, MCP3, ICAM1 and IL16 have been shown to be involved in migration and recruitment of immune cells (Ambati et al., 2013; Griffith et al., 2014), which could potentially promote progression to advanced forms of AMD (Ambati et al., 2013). THBS1, MCP1, EMMPRIN and ICAM1 have been documented in drusen deposits (Alcazar et al., 2009; He et al., 2006) or in the aqueous humor (Jonas et al., 2012) from AMD patients. However, notably, we additionally report modulation of IL16 and IGFBP3, which have not previously been associated with AMD.
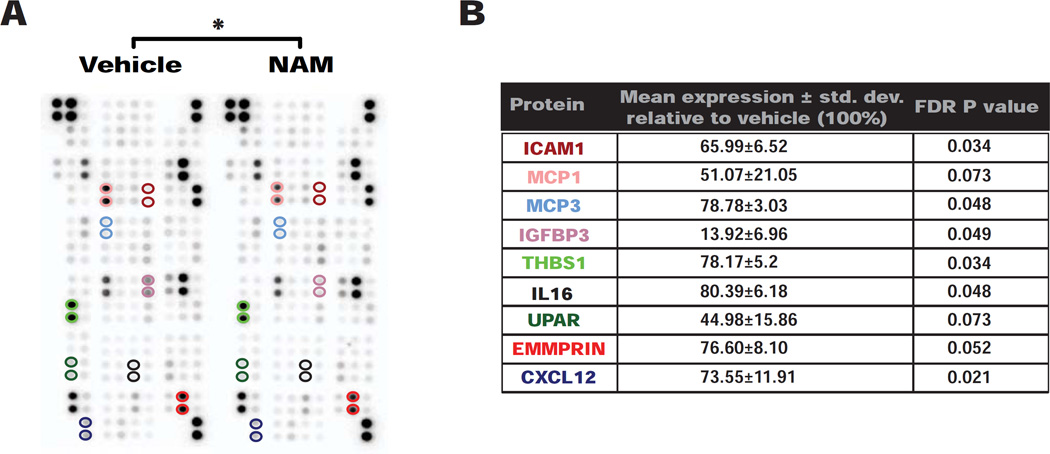
Figure 4. NAM reduces production of pro-inflammatory cytokines from hiPSC-RPE cells.
(A) Representative image of an array analyzed for 102 human cytokines in supernatant collected 60–72 hours after the last medium change from AMD and control hiPSC-RPE lines treated with 10mM NAM or vehicle (Figure S4B) (n=4 donors; 2 AMD and 2 control; 4 lines). Colored circles represent proteins that showed a significant change, also listed in Figure 4B.
(B) Nine cytokines showed significantly differential secretion in hiPSC-RPE treated with 10mM NAM relative to vehicle, their mean expression ± std. dev. relative to vehicle (baseline defined as 100%) (Figure S4C) and P values. Protein names are color-matched to the circles highlighting the cytokine localization on the array in Figure 4A.
Paired Student’s t test (two-tailed) with FDR adjustment and two-way ANOVA were used for statistical analysis (*= p<0.05).
In examining the expression of these nine inhibited cytokines in the RNA-seq analysis, five showed reduced transcript levels after NAM treatment, three (EMMPRIN, CXCL12 and MCP3) showed no significant difference, and one transcript (IL16) was upregulated. Hence, NAM appears to impact cytokine secretion at both transcriptional and post-transcriptional levels.
Contribution of the complement pathway
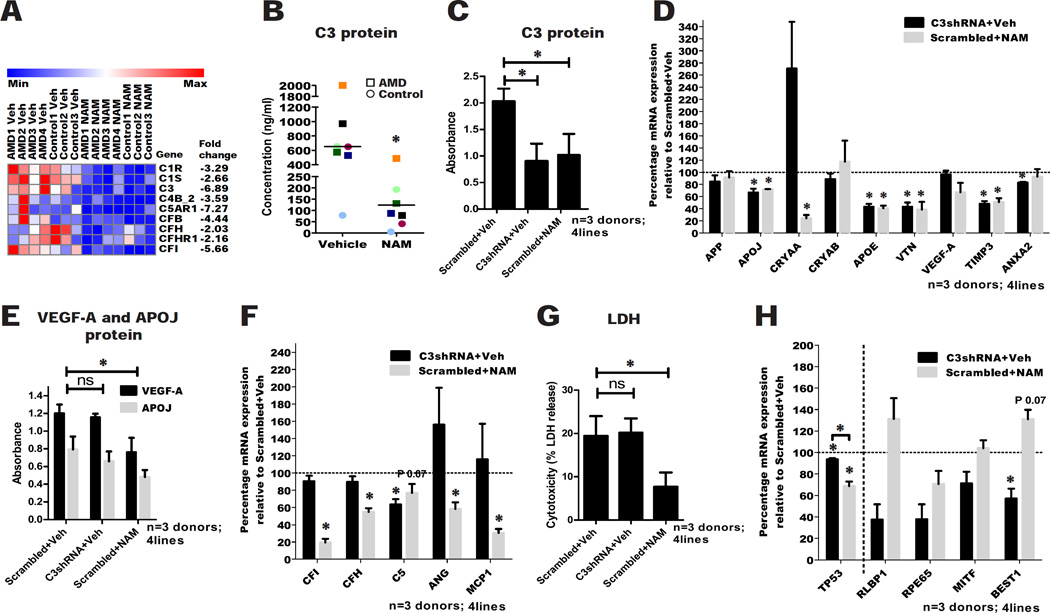
From the RNA-seq analysis, NAM inhibited nine genes (FDR p≤0.1) involved in the complement cascade (Figure 5A), and we validated a subset of these by qPCR (Figure S5A), revealing a dramatic (approximately 90%) reduction in the C3 transcript level with NAM. Using ELISA, we found an average 6–7 fold inhibition of C3 protein in the culture supernatant from hiPSC-RPE by NAM treatment (Figure 5B). NAM also inhibited the complement regulatory factor CFH, but notably, the ratio of CFH to C3 was consistently increased across the hiPSC-RPE lines (Figure S5B), suggesting that relatively more CFH is available in the presence of NAM, revealing a potentially important protective action of NAM on complement pathway inhibition. Interestingly, lower CFH/C3 ratios segregated more closely with the AMD status of the donors (Figure S5B), supporting evidence of higher complement activity in AMD donor lines.
Figure 5. NAM effectively reduces production of complement factors.
(A) Altered expression of several genes in the complement pathway detected by RNA-seq analysis of NAM treatment (n=7 donors; 4 AMD and 3 control; 7 lines).
(B) Secretion of C3 into the culture supernatant 60–72 hours after the last medium change in AMD and control hiPSC-RPE treated with 10mM NAM or vehicle. Data are expressed as mean of all samples (line), and individual sample values are plotted: AMD hiPSC-RPE are represented by squares and control hiPSC-RPE by circles (n=7 donors; 4 AMD and 3 control; 9 lines). Colors correspond to hiPSC-RPE samples in Table S1; each sample is color matched across NAM and vehicle treatment.
(C) ELISA measurement of secretion of C3 into the culture supernatant 60–72 hours after the last medium change in vehicle, 10mM NAM and C3shRNA treated hiPSC-RPE. Data are expressed as mean± SEM of absorbance from ELISA assay (n=3 donors; 2 AMD and 1 control; 4 lines).
(D) qPCR analysis of AMD/drusen associated protein transcripts in C3shRNA and 10mM NAM treated hiPSC-RPE relative to vehicle (baseline defined as 100%). Data are expressed as mean± SEM (n=3 donors; 2 AMD and 1 control; 4 lines).
(E) ELISA measurement of secretion of VEGF-A and APOJ into the culture supernatant 60–72 hours after the last medium change in vehicle, 10mM NAM and C3shRNA treated hiPSC-RPE. Data are expressed as mean± SEM of absorbance from ELISA assay (n=3 donors; 2 AMD and 1 control; 4 lines).
(F) qPCR analysis of complement and inflammation associated protein transcripts in C3shRNA and 10mM NAM treated hiPSC-RPE relative to vehicle (baseline defined as 100%). Data are expressed as mean± SEM (n=3 donors; 2 AMD and 1 control; 4 lines).
(G) LDH release into the culture supernatant of vehicle, 10mM NAM and C3shRNA treated hiPSC-RPE. Data are expressed as mean± SEM (n=3 donors; 2 AMD and 1 control; 4 lines).
(H) qPCR analysis of TP53 and RPE genes expression in C3shRNA and 10mM NAM treated hiPSC-RPE relative to vehicle (baseline defined as 100%). Data are expressed as mean± SEM (n=3 donors; 2 AMD and 1 control; 4 lines). Paired Student’s t test (two-tailed) was used for statistical analysis.
Paired Student’s t test (one-tailed) was used for statistical analysis, unless stated otherwise above (*= p<0.05). ns: not significant; veh: vehicle.
Given the strong involvement of complement factors in AMD, we tested whether the effect of NAM was largely explained via this pathway. We generated a lentivirus to knock down C3 mRNA (C3shRNA), which inhibited C3 mRNA expression by 85–90% 60–72 hours post-transduction (Figure S5C) (in four hiPSC-RPE lines from three donors: AMD 2, AMD 2’, AMD 4 and Control 1 in Table S1). Three weeks post-transduction, we observed inhibition of C3 protein secretion in supernatants from hiPSC-RPE treated with C3shRNA (55.4%±21.04% reduction), which was comparable to that achieved by NAM treatment (49.7%±23.6% reduction) (Figure 5C). To ascertain whether C3 knockdown can inhibit other AMD biomarkers, we assessed the expression of the first panel of AMD/drusen associated protein transcripts by qPCR (Figure 5D). C3shRNA significantly reduced the expression of most of this panel. However, unlike NAM treatment, C3shRNA did not inhibit VEGF-A and APOJ secretion in all the hiPSC-RPE lines tested (Figure 5E). We then assessed whether C3shRNA can reduce other complement and inflammatory protein transcripts. While NAM inhibited most of the complement components analyzed, C3shRNA only inhibited the downstream C5 transcript, but not CFI or CFH (Figure 5F). Further, C3shRNA treatment did not reduce the inflammatory transcripts (MCP-1 and ANG), which were effectively reduced by NAM (Figure 5F).
A notable difference between C3shRNA and NAM treatment was the effect on overall RPE cell viability. LDH levels in the culture supernatants were similar in C3shRNA and vehicle-treated conditions, but were significantly decreased in 10mM NAM (Figure 5G). Further, C3shRNA showed a trend towards inhibition of RPE markers, with BEST1 being significantly reduced, while NAM maintained RPE markers expression similar to vehicle (Figure 5H). While both NAM and C3shRNA inhibited the expression of TP53, a pro-apoptotic gene in the PI3K-Akt pathway (Khoo et al., 2014), the levels were significantly different for C3shRNA (6.41%±1.8% inhibition) and NAM (31.22%±7.2% inhibition) (p<0.05; Figure 5H), which might contribute to the observed difference in their effect on RPE cell survival.
Dampening of the complement pathway, for example using C3 inhibitors, is being actively explored as a therapeutic avenue for AMD, with several drugs undergoing clinical trials (Holz et al., 2014). Our results suggest that while the inhibition of C3 might reduce downstream complement activation and some drusen-protein expression, it fails to dampen production of inflammatory factors and it does not improve RPE health and survival, which NAM treatment accomplishes. Hence, C3 inhibition does not mimic the multiple beneficial effects of NAM in the hiPSC-RPE AMD model.
NAM modulates aging associated pathways in RPE cells
To further dissect the effect of NAM on hiPSC-RPE, we focused separately on upregulated and downregulated genes. GO enrichment (FDR p≤0.1 and two-fold change) for exclusively downregulated genes overlapped with those shown in Figure 3B, indicating that a major effect of NAM was gene repression. Interestingly, GO enrichment for only upregulated genes showed that NAM primarily regulates genes involved in DNA packaging and nucleosome assembly (Figure 6A). These findings, and the significantly increased expression of histone coding genes and the DNA methyltransferase DNMT1 with concurrent inhibition of the DNA demethylase gene TET1 (Figure 6B–C), indicate that NAM could be involved in maintaining chromatin and DNA methylation states.
Figure 6. NAM acts on aging associated pathways and increases expression of SIRT1 protein and NAD biosynthetic pathway in hiPSC-RPE.
(A) GO functional enrichment for cellular components of exclusively upregulated genes in NAM treated hiPSC-RPE compared to vehicle from RNA-seq analysis, using the STRING database (n=7 donors; 4 AMD and 3 control; 7 lines).
(B) Altered expression of DNA methylation associated genes detected by RNA-seq analysis of NAM treatment (veh: vehicle) (n=7 donors; 4 AMD and 3 control; 7 lines).
(C) Altered expression of functionally related genes detected by RNA-seq analysis of NAM treatment (veh: vehicle) (n=7 donors; 4 AMD and 3 control; 7 lines).
(D) SIRT1 protein and total NAD (NAD+NADH) levels measured 40–48 hours and 12–18 hours, respectively after the last medium change from 10mM NAM treated hiPSC-RPE relative to vehicle (baseline defined as 100%). Data are expressed as mean± SEM (n=5 donors; 2 AMD and 3 Control; 6 lines). Paired Student’s t-test (two-tailed) was used for statistical analysis (*= p<0.05).
In-depth analysis of the genes showing significant changes (FDR p≤0.1) revealed that NAM induced similar expression changes (induction or repression) in several functionally related gene clusters (Figure 6C). NAM increased the expression of ribosomal synthesis genes (including mitochondrial) and DNA/RNA polymerase and histones. Aging is associated with decreased histone expression, reduced global DNA methylation and reduced biogenesis of mitochondria (Lopez-Otin et al., 2013; Pal and Tyler, 2016), and we show that NAM could potentially reverse these changes by maintaining histone, mitochondrial ribosomal and electron transport chain (ETC) subunits, and increasing global DNA methylation. NAM inhibited TGF-β, collagen and insulin growth factor associated genes, all of which are implicated in aging (Ewald et al., 2015; Shaw et al., 2007).
Consistent with a potential anti-aging effect, we found that NAM treatment significantly increased Sirtuin 1 (SIRT1) protein levels in hiPSC-RPE lysates (Figure 6D) (six hiPSC-RPE lines from five donors; AMD 2, AMD 2’, AMD 4, Control 1, Control 2 and Control 3 in Table S1). SIRT1 is reduced with aging, which can result in dysregulated inflammation, possibly by hypomethylation of promoters of certain inflammatory genes (Liu and McCall, 2013). Further, NAM showed marginally increased levels (P=0.055) of the central metabolite NADt (total NAD+NADH) in hiPSC-RPE lysates (Figure 6D). Similar, long-term Nicotinamide Mononucleotide treatment in mice causes NAD upregulation and prevents age-associated gene expression changes (Mills et al., 2016). NAD/NADH are key regulators of metabolism implicated in aging, oxidative stress, apoptosis, inflammation and calcium homeostasis (Ying, 2008), which are also major pathways implicated in AMD.
Discussion
AMD is a complex disorder with multiple genetic and environmental components. A number of rodent models have been used for AMD in vivo studies, but none recapitulates the complete repertoire of human disease features, likely due to differences in species and longevity as well as the lack of an anatomical macula in rodents. Non-human primate models that do have a macula can show signs of AMD, but the slow disease time course and the nature of primate models make these difficult to use experimentally (Pennesi et al., 2012), underscoring the need to develop additional human AMD-related disease models. RPE cells can be obtained from human iPSCs, which facilitates modeling AMD in culture to dissect environmental and genetic effects. Homozygosity for the ARMS2 risk allele and the linked HTRA1 risk allele, the frequency of which is approximately 8% globally and 2–5% in European and US populations (1000 genomes project), have been shown to increase the risk of advanced AMD by 7–8 fold (Scholl et al., 2007). Notably, ARMS2 is an evolutionary recent primate-specific gene (Francis et al., 2008), which adds to the relevance of human iPSC-RPE-based AMD models.
This study shows that RPE cells derived from hiPSCs can synthesize several major AMD-associated factors, including drusen proteins, and in the future we will investigate whether these accumulate together in sub-RPE deposits. Our observations suggest a clear involvement of genetic susceptibility towards AMD, notably in the inflammatory and complement factors associated with AMD pathogenesis. The tissue of origin (cornea or RPE) was not the same for all hiPSC-RPE lines which could result in some epigenetic differences; however, in a recent study, we found that such different originating tissues did not affect hiPSC-RPE phenotype or functionality (Miyagishima et al., 2016), and we found no significant differences related to tissue of origin or to cell passage. Some AMD-associated factors did not show significant differences between AMD and control lines, but these could emerge if oxidative stressors (Rabin et al., 2013) or complement components such as C1q (Johnson et al., 2011) or molecules that accelerate aging such as progerin (Miller et al., 2013) were added. Nevertheless, we show that this model, even without added perturbagens, demonstrates patient-associated differences and can be used to identify potential drugs that inhibit production of AMD-associated proteins.
While evidence supports the involvement of blood-derived immune cells in wet AMD, dry AMD is believed to originate from dysregulation of the complex immune and inflammatory activities of the RPE itself (Ambati et al., 2013). As we show here, pure populations of hiPSC-RPE, even in the absence of immune cells or microglia, can be a major source of most of the inflammatory cytokines known to be involved in the progression to advanced AMD.
Complement dysregulation is regarded as a critical factor in AMD progression (Bradley et al., 2011), and so it is noteworthy that we recapitulate these phenotypes in the hiPSC-RPE based AMD model. C1S has been previously documented in AMD (Newman et al., 2012) and we show that in addition to C1S, C1R is also expressed more by AMD hiPSC-RPE, which suggests the involvement of the classical complement cascade in addition to the alternative cascade. The classical pathway requires a complex of C1q, C1s and C1r. We found that C1Q is not expressed by RPE, but C1Q is expressed in retina (Luo et al., 2011) and increases in the CNS during aging (Stephan et al., 2013). We hypothesize that C1q from retinal sources could complex with C1r and C1s produced by RPE cells to activate the classical cascade increasingly with age, an effect potentially exacerbated given the higher expression of C1R and C1S in the RPE of AMD patients. Due to a 100% linkage disequilibrium between ARMS2 and HTRA1 risk alleles, it has not been possible to establish the causal variant for AMD between these two genes; however, it has been suggested that HTRA1 might be the causal variant, as it encodes for a serine peptidase that can cleave APOJ and vitronectin, and thereby possibly activate complement cascades (An et al., 2010; Bradley et al., 2011). Our finding that the ARMS2/HTRA1 risk locus appears to strongly affect the complement system also supports this hypothesis. Since the AMD lines with the homozygous ARMS2/HTRA1 risk allele were also heterozygous for the CFH risk allele, as were all the other AMD lines and one control line (expected as the frequency of the CFH heterozygous genotype is approximately 45% in the US), our findings regarding enhanced complement expression in the ARMS2/HTRA1 homozygous lines could be ascribed to synergistic interaction between the ARMS2/HTRA1 and CFH genotypes. In the future, a larger collection of hiPSC lines with different AMD genotypes would enable separation of the impact of each locus.
Here for the first time we report that NAM can effectively repress the complement pathway, including C3, which plays a pivotal role in both the classical and the alternative cascades, and is sufficient to chronically activate the pathway even in the absence of other complement components (Bradley et al., 2011). We found that C3 inhibition alone did not inhibit secretion of some key factors associated with AMD progression, including VEGF-A and APOJ, nor benefit RPE cell survival, unlike NAM. This suggests that complement inhibition might not be able to prevent AMD progression nor rescue the RPE atrophy in AMD as well as NAM, which likely targets multiple independent pathways; for example, NAM affects the PI3K-AKT pathway (Figure 3B), which is suggested to modulate angiogenesis by increasing VEGF-A secretion (Karar and Maity, 2011).
The effect of NAM in lowering VEGF-A is particularly important in potentially preventing progression to wet AMD. Further, we noted that ICAM1 was strongly upregulated in AMD ARMS2/HTRA1 hiPSC-RPE and markedly inhibited by NAM. The ARMS2/HTRA1 genotype increases the risk of both the dry and wet form of AMD, and ICAM1 has been linked to development of the wet form of AMD (Jonas et al., 2012). Current therapies for wet AMD are focused on anti-VEGF-A, although some patients eventually resist the treatment and neovascularization recommences or dry AMD progresses, which is a growing concern (Ehlken et al., 2014). Augmentation of anti-VEGF with anti-PDGF therapies are being developed as a more effective treatment for wet AMD (Holz et al., 2014). Our data suggest that inhibition of ICAM1 should be explored in a dual approach with VEGF-A targeted therapies, especially in patients with the ARMS2/HTRA1 homozygous genotype.
To understand more about the breadth of NAM’s actions, we provide an unbiased, RNA-seq on a single, pure population of human RPE cells, which has enabled us to identify the major genes and pathways impacted. It is notable that the major effect of NAM on gene expression is repressive, while the genes that are upregulated are mostly involved in DNA and chromatin pathways. In addition to an up-regulation of SIRT1, a chromatin modifying enzyme already known to be regulated by NAM (Jang et al., 2012), we found an increase in other genes involved in nucleosome production and modification, and changes in DNMT1 and TET1 consistent with maintenance of DNA methylation. Hence, it will be worthwhile to explore more comprehensively the impact of NAM on the epigenome in the RPE, and on epigenomic regulators in addition to SIRT1.
In conclusion, based on these findings, we propose that NAM may be an effective agent in developing therapies for AMD, as it represses AMD-associated protein production, complement and inflammatory pathways and VEGF-A production, and improves cell survival and maintenance of RPE genes. Our findings suggest that further studies of action of NAM, especially on chromatin stability, could illuminate potential therapeutic approaches for preventing the progression of AMD and preserving vision in the elderly.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests of reagents can be directed to and fulfilled by the corresponding author, Sally Temple (sallytemple@neuralsci.org).
MTAs required.
Experimental Model and Subject details
Culture of RPESC derived RPE, hiPSC derived RPE, Corneal fibroblasts and Mouse embryonic feeders
Adult human cadaver donor eyes were obtained from eye banks. The donor demographic information is provided in Table S1 and Table S2. A protocol for the use of human cadaver eye tissue to generate cell lines was submitted to the Albany Medical College IRB. The IRB determined that the protocol was exempt category 4. Two types of ocular tissues were used: RPE for the generation of adult RPESC-RPE and cornea, and both these cells were used for iPSC generation.
RPE dissection and culturing to generate RPESC-RPE was performed as described previously (Blenkinsop et al., 2015; Blenkinsop et al., 2013; Salero et al., 2012).
Cells from the cornea of cadaver eyes were isolated by digesting the tissue with collagenase (Cat. # 17104-019; Thermo Fisher) and then cultured in standard fibroblast medium consisting of DMEM (Cat. # 11960-044; Thermo Fisher) with 10% FBS (Cat. # 10082-147; Thermo Fisher). Fibroblast medium was supplemented with 1% Penicillin/Streptomycin (Cat. # 15140-122; Thermo Fisher), 2mM l-Glutamine (Cat. # 25030-081; Thermo Fisher) and 1mM Sodium Pyruvate (Cat. # 11360-070; Thermo Fisher). Irradiated mouse embryonic feeders (Cat. # GSE-6301G; GlobalStem) used for growing hiPSC were also maintained in the same standard fibroblast media.
hiPSCs were generated and characterized as described in Method Details. The resulting iPSC lines were authenticated by genotyping, karyotyping, and in vitro tri-lineage differentiation was used to establish the pluripotency in the hiPSC lines.
RPESC derived RPE and hiPSC-RPE were cultured and maintained in a medium consisting of DMEM/F12 (Cat. # 10-092CM; Corning) and MEM alpha modification (Cat. # M4526; Sigma) in equal proportion with 2% FBS (Cat. # 10082-147; Thermo Fisher). RPE medium was supplemented with 1% Penicillin/Streptomycin (Cat. # 15140-122; Thermo Fisher), 1% MEM Non-essential amino acids (Cat. # 11140-050; Thermo Fisher), 1mM l-Glutamine (Cat. # 25030-081; Thermo Fisher), 1mM Sodium Pyruvate (Cat. # 11360-070; Thermo Fisher), 0.5% N1 (Cat. # N6530; Sigma), 0.25 mg/mL taurine (Cat. # T8691; Sigma), 0.02 µg/mL hydrocortisone (Cat. # H0888; Sigma), and 0.013 ng/mL tri-iodothyronine (Cat. # T5516; Sigma). The RPE medium was further supplemented with 1mM or 10mM Nicotinamide (Cat. # N0636; Sigma) or vehicle, for experiments described in the manuscript.
Culture of 293 FT cells
293 FT cells (Cat. # R70007; Thermo Fisher) were cultured according to the vendor’s instructions in the following medium: DMEM (Cat. # 11960-044; Thermo Fisher) supplemented with 10% FBS (Cat. # 10082-147; Thermo Fisher), 1% MEM Non-essential amino acids (Cat. # 11140-050; Thermo Fisher), 6mM l-Glutamine (Cat. # 25030-081; Thermo Fisher), 1mM Sodium Pyruvate (Cat. # 11360-070; Thermo Fisher), 1% Penicillin/Streptomycin (Cat. # 15140-122; Thermo Fisher) and 500µg/ml Geneticin (Cat. # 11811-031; Thermo Fisher).
Method Details
hiPSC generation and characterization
Cadaver donor globes from patients with AMD as well as age-matched unaffected individuals were received from the National Disease Research Interchange (Philadelphia, PA, USA) and the Eye-Bank for Sight Restoration, Inc. (New York, NY, USA), and the ocular tissues used to establish either cornea-associated fibroblasts or RPE cells for production of hiPSCs. The RPE cells were isolated and grown to establish a functional RPE monolayer (Blenkinsop et al., 2015; Blenkinsop et al., 2013). To derive hiPSC from these cells, either Sendai virus-based (CytoTune -iPS Sendai Reprogramming kit; Cat. # A13780-01; Thermo Fisher) or mRNA-based (mRNA Reprogramming kit; Cat. # 00-0071; Stemgent) was used (i.e. non-integrating reprogramming methods) according to manufacturer’s instructions. Approximately three weeks after the transduction with Sendai virus or mRNA transfection, colonies with the typical iPSC morphology appeared. Cells were live-stained with directly conjugated AlexaFluor 488-anti TRA-1-60 (Cat. # 560173; BD), positive colonies were then hand-picked and sub-cloned in order to be expanded and characterized. In general, three clones were generated and characterized per hiPSC line.
Analysis of Pluripotency
Cells were stained with pluripotency markers for immunostaining and flow cytometry. Briefly, hiPSC were fixed for 20 min in 4% Paraformaldehyde in PBS (Cat. # sc-281692; Santa Cruz), washed twice with PBS, permeabilized with 0.2% Triton X-100 (Cat. # T9284; Sigma) for 10 min at room temperature, then rinsed twice with PBS+1% BSA (Cat. # sc-2323; Santa Cruz). Blocking for non-specific antibody binding was performed using Protein block serum-free (Cat. # X0909; Dako) for 30 mins, cells were then incubated for 2 hours with PBS + 1% BSA containing the respective primary antibody. All incubations were performed at room temperature. For surface marker staining, after fixation, cells were incubated for 2 hours in PBS+1% BSA containing the respective conjugated antibody. The following antibodies were used: KLF4 (Cat. # sc-20691; Santa Cruz), c-MYC (Cat. # sc-764; Santa Cruz), OCT4 (Cat. # 2840S; Cell Signaling), SOX2 (Cat. # sc-17319; Santa Cruz), NANOG (Cat. # AF1997; R&D systems), AlexaFluor 488-anti TRA-1-60 (Cat. # 560173; BD). Cells were washed twice in PBS + 1% BSA, followed by staining with the secondary antibodies (KLF4, OCT4 and c-MYC: Donkey Anti-Rabbit IgG (H+L); Cat. # 711-545-152; Jackson ImmunoResearch; SOX2 and NANOG: Donkey Anti-Goat IgG (H+L); Cat. # 705-485-147; Jackson ImmunoResearch). For FACS analysis, hiPSC were dissociated with Accutase (Cat. # A11105-01; Thermo Fisher) for 5 mins at room temperature, washed with DMEM/F12 media with 10% KSR (Cat. # 10828-028; Thermo Fisher) and incubated with AlexaFluor 647-anti TRA-1-60 (Cat. # 560122; BD) and AlexaFluor 488-anti SSEA4 (Cat. # 560308; BD) for 30 min at room temperature. Stained cells were washed twice in PBS+1% BSA. Cells were analyzed on a FACS Aria 2 Cell Sorter (BD) with 10,000 events acquired for each sample. Data were analyzed with FlowJo software.
In vitro tri-lineage differentiation, karyotyping and quantification of RPE gene expression in hiPSCs
For in vitro tri-lineage differentiation, FGF2 was removed from the culture medium to allow the hiPSCs to spontaneously differentiate for approximately three weeks. Cells were then fixed and stained for 2 hours at room temperature using the following primary antibodies: AFP (Cat. # RB-365-A1; Thermo Fisher) and Sox17-NL557 (Cat. # NL1924R; R&D systems) for endoderm; β-tubulin III (Cat. # T8660; Sigma) for ectoderm and α-SMA (Cat. # MS-113-P0; Thermo Fisher) for mesoderm. Cells were then washed twice in PBS+1% BSA, and incubated with the secondary antibodies (AFP: Goat Anti-Rabbit IgG (H+L); Cat. # 111-585-003; Jackson ImmunoResearch; β-tubulin III: Goat Anti-Mouse IgG, Fcγ Subclass 2b Specific; Cat. # 115-485-207; Jackson ImmunoResearch; α-SMA: Goat anti-Mouse IgG2a Secondary Antibody; Cat. # A21241; Thermo Fisher) for 1 hour at room temperature. After washing twice with PBS+1% BSA, cells were incubated with DAPI for 20 mins at room temperature for nuclear staining. G-band karyotyping was performed by the Cytogenetics laboratory of the Wadsworth Center at the David Axelrod Institute (Albany, NY), and by Cell Guidance Systems Genetics Service (St. Louis, MO). To confirm that no residual RPE donor cells contaminated the hiPSC cultures, total RNA was isolated (Cat. # 74034; Qiagen) from each hiPSC line, cDNA was reverse transcribed (Cat. # 4387406; Thermo Fisher) and qPCR performed using primers specific for the RPE gene RPE65 (Table S4).
Differentiation of hiPSC into RPE
hiPSC lines were grown on irradiated mouse embryonic feeders (Cat. # GSE-6301G; GlobalStem) in serum-free medium supplemented with 4ng/ml FGF2 (Cat. # 233-FB; R&D systems). Prior to differentiation, passage 6–26 hiPSCs were feeder-depleted by passaging them onto Matrigel-coated plates (Corning) in the presence of mTeSR (Cat. # 05850; Stem Cell Technologies) medium. A modification of a recently published method (Ferrer et al., 2014) was used to differentiate hiPSC into RPE. Briefly, 100nM LDN193189 (Cat. # 04-0074; Stemgent) and 10uM SB431542 (Cat. # 1614; Tocris) were added to serum-free differentiation medium every day for 3 days to drive neural induction from hiPSCs. At day 5 (D5), to specify RPE differentiation, 10mM Nicotinamide (Cat. # N0636; Sigma) and 150ng/ml Activin A (Cat. # 338-AC; R&D System) were added. Colonies with typical RPE morphology and pigmentation appeared around D30-D40, then medium was changed to RPE-medium (Blenkinsop et al., 2013). RPE were then manually picked and plated onto a 24-well plate (Corning, Primaria) at 1–2×105 cells/well. When confluent, RPE cells were passaged by splitting the monolayer with 0.25% Trypsin-EDTA (Cat. # 25200-172; Thermo Fisher) supplemented with 12ng/ml DNase (Cat. # DN-25; Sigma). A pigmented monolayer of RPE was obtained approximately 1 month after splitting, with some variation in time depending on the success of enrichment at each passage.
For the experiments described, pure hiPCS-RPE were replated onto transwell inserts (Corning) for 12-well plates or onto 24 or 48 well cell culture plates (Cat. # 3527 and Cat. # 3548; Corning) at 1×105 cells/well for further analysis.
Genotyping
TaqMan genotyping assays (Thermo Fisher) were used for genotyping SNP rs10490924 in the ARMS2 gene (Assay ID: C__29934973_20; Cat. # 4351379), SNP rs11200638 in the HTRA1 gene (Assay ID: C__31018186_10; Cat. # 4351379), SNP rs1061170 in the CFH gene (Custom SNP genotyping assay; Assay ID: AHI1TPW; Cat. # 4331349) and SNP rs2230199 in the C3 gene (Assay ID: C__26330755_10; Cat. # 4351379). For further validation, the ARMS2 gene was sequenced in all the samples.
hiPSC-RPE culture, Nicotinamide treatment and collection of samples for analysis
After plating, hiPSC-RPE and human RPESC-RPE cells were cultured in Nicotinamide-containing RPE culture medium with 10mM Nicotinamide (Cat. # N0636; Sigma) for 3–4 weeks, until they formed cobblestone monolayers. Cells were fed on alternate days.
For comparing the AMD/drusen and complement/inflammatory protein transcript expression in AMD and control hiPSC-RPE by qPCR, cells were collected in RNA protect 3–4 weeks after plating and mRNA was extracted for qPCR (described later). For protein expression in AMD and control hiPSC-RPE, supernatant was collected approximately 5 weeks after plating and ELISA was carried out (described later).
For assessing the effect of Nicotinamide, hiPSC-RPE lines and human RPESC derived RPE lines that were confluent after 3–4 weeks of growth were treated with 1mM Nicotinamide, 10mM Nicotinamide or vehicle (cell culture grade water) at 1:100 dilution for an additional 3 or 12 weeks (depending on experiment).
To analyze the expression of AMD associated protein transcripts in Nicotinamide and vehicle treated hiPSC-RPE by qPCR in the 3–5 weeks long experiments, cells were collected in RNA protect approximately 3 weeks after beginning the treatment with Nicotinamide versus vehicle.
To analyze protein expression in Nicotinamide and vehicle treated hiPSC-RPE by ELISA, supernatant was collected 2–3 weeks after beginning the treatment with Nicotinamide or vehicle.
To analyze protein expression in Nicotinamide and vehicle treated hiPSC-RPE by western blotting (described later), lysates were collected 4–5 weeks after beginning the treatment with Nicotinamide or vehicle.
C3shRNA, Lentivirus preparation and transduction
shRNA plasmids were generated for C3 (Coding sequence: GGAGAATTGCTTCATACAA) and Scrambled control (Sequence: TTCTCCGAACGTGTCACGT), using FUGW-H1 lentiviral construct as previously described (Phoenix and Temple, 2010). Plasmid constructs were co-transfected with pCMV-VSVG and pCMV-dvpr into 293FT cells (Cat. # R70007; Thermo Fisher) for packaging lentivirus. Supernatant was harvested 2 and 3 days after transfection and virus particles were concentrated by ultra-centrifugation. Lentiviruses were used at 2 MOI for cell transduction with polybrene transfection reagent (Cat. # TR-1003-G; Millipore). After passaging, hiPSC-RPE lines were cultured in Nicotinamide-containing RPE culture medium for 3–4 weeks, until they formed cobblestone monolayers. hiPSC-RPE lines were transduced with C3shRNA lentivirus or Scrambled virus in vehicle (cell culture grade water) containing media. In parallel, hiPSC-RPE lines in 10mM Nicotinamide were transduced with Scrambled virus (positive control). C3 knockdown efficiency in the hiPSC-RPE lines was quantified by qPCR 60–72 hours post transduction. Similar to Nicotinamide treatment regimen, 3 weeks post transduction with C3shRNA lentivirus, mRNA was extracted from hiPSC-RPE lines and cDNA was synthesized for qPCR (described later), and medium was collected for ELISA 60–72 hours after the last medium change (described later). ELISA was performed for C3 and VEGF-A, and the LDH assay was used for quantifying cytotoxicity (described later).
Transepithelial electrical resistance (TER)
hiPSC-RPE cells were plated on human placental extracellular matrix (10ug/ml, Cat. # 354237; Corning) coated trans-well inserts (Cat. # 3460; Corning) at a density of 1×105 cells per well. Cells were cultured for approximately 4 weeks until they formed a cobblestone monolayer and TER was measured using the EVOM2 Epithelial Volt-ohmmeter (World precision instruments).
Immunocytochemistry for RPE markers
hiPSC-RPE were fixed on transwell inserts (Cat. # 3460; Corning) and in 24 or 48 well cell culture plates (Cat. # 3527 and Cat. # 3548; Corning) using 4% Paraformaldehyde (Cat. # sc-281692; Santa Cruz) in PBS for 10 minutes, followed by rinsing three times with PBS. Fixed cells were blocked and permeabilized for an hour in a solution consisting of 0.01% Saponin (Cat. # S7900; Sigma) and 5% normal goat serum (Cat. # 005-000-121; Jackson ImmunoResearch) in 1% BSA (Cat. # sc-2323; Santa Cruz). Fixed cells were then incubated overnight at 4°C with primary antibodies for OTX2 (Cat. # Ab92515; Abcam), CRALBP (Cat. # Ab15051; Abcam), MCT-1 (Cat. # HPA003324; Sigma), MITF (Cat. # Ab3201; Abcam) and Phalloidin (Cat. # A12379; Thermo Fisher). Cells were rinsed three times with PBS and incubated with the secondary antibodies (OTX2 and MCT1: Goat anti-Rabbit IgG (H+L) Secondary Antibody: Cat. # A11035 and A21245; Thermo Fisher; CRALBP: Goat anti-Mouse IgG (H+L) Secondary Antibody; Cat. # A-11003; Thermo Fisher; MITF: Goat anti-Mouse IgG (H+L) Secondary Antibody; Cat. # A-21235; Thermo Fisher) at room temperature for 1 hour.
RNA extraction and synthesis of cDNA
To extract RNA, hiPSC-RPE cells were scraped from the culture wells and collected in RNA protect (Cat. # 76526; Qiagen). RNA was purified using the RNeasy plus micro kit (Cat. # 74034; Qiagen). RNA concentration and quality was determined by Nanodrop. Total RNA was converted to cDNA using the High capacity RNA to cDNA kit (Cat. # 4387406; Thermo Fisher).
Quantitative RT PCR
Quantitative RT-PCR was carried out with SYBR Green PCR master mix (Cat. # 4312704; Thermo Fisher) on QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher) in 20µl reaction volumes. All reactions were carried out in either duplicate or triplicate and Ct values were obtained. Relative difference in the gene expression was normalized to expression levels of a housekeeping gene, ribosomal protein S18 (RPS18). Primer sequences are shown in Table S4.
RNA-seq analysis
Total RNA was extracted as described previously and Agilent 2100 was used to estimate the RNA integrity numbers, which were 8.4–10. 1X75 Single-read high output RNA-seq was carried out by the Center for Functional Genomics, University at Albany, using Clontech SMARTer Stranded Total RNA Seq Kit Pico Input mammalian. 10ng of Total RNA per sample was fragmented (94°C for 4 minutes), then synthesized to cDNA. Illumina adaptors and indexes were added to cDNA followed by Aline PCR Clean DX magnetic bead purification. Ribosomal cDNA was depleted with ZapR and R-Probes and the remaining library was amplified using 12-cycles of PCR. Final library was purified using Aline PCR Clean DX magnetic beads and validated using the Agilent HS DNA Kit and Qubit HS dsDNA Quant Kit. Each library was diluted to 4nM and then equal volumes of each 4nM library were combined to form the Library Pool. 5ul of the Library Pool was denatured with 0.1N NaOH for 5 minutes then diluted with HT1 buffer. To this 1.6pM library, 10% PhiX control was added, and the mixture was loaded onto the Illumina NextSeq 500 flowcell (1X75 Single-read). The quality of the reads was verified by FASTQC. Raw reads were aligned to the human RefSeq mRNA database using the STAR aligner (Dobin et al., 2013). The bedtools and samtools utilities were used to remove sequences that mapped to ribosomal regions. Uniquely mapped reads were used for further analysis. Differential expression of the mapped reads was analyzed using the DESeq/DESeq2 R package. Normalized reads for each gene were generated and FDR p≤0.1 was used to identify differentially expressed genes.
Enzyme-Linked Immunosorbent Assay (ELISA) in culture supernatants
Total supernatant was collected from hiPSC-RPE culture wells 60–72 hours after the last medium change (600µl for 24well and 300µl for 48well plates). ELISA kits (Key Resources Table) were used to measure the secretion of Aβ-40 (dilution of supernatant 1:2 to 1:4), Aβ-42 (dilution of supernatant 1:2), VEGF-A (dilution of supernatant 1:50), APOJ (dilution of supernatant 1:32) and Complement component 3 (dilution of supernatant 1:50) in the culture supernatants. Note that dilutions were made to ensure protein of interest was within the concentration range for detection and quantification. All the samples were tested in duplicates following the manufacturer’s instructions. Immediately after adding the stop solution, optical densities/absorbance readings were obtained at 450nm using a plate reader (1420 multi-label counter; Perkin Elmer). The average of the duplicate readings was used to estimate the concentration using the standard curve.
ELISA in lysates
Quantification of NAD/NADH (Cat. # ab65348; Abcam) was measured by ELISA in the cell lysates. The total protein concentration in the lysates was calculated using the Pierce BCA Protein Assay (Cat. # 23225; Thermo Fisher) to normalize the Nicotinamide and vehicle treated samples. All the samples were tested in duplicates, following the manufacturer’s instructions. Multiple optical densities/absorbance readings were obtained during 1–4 hours at 450nm, using a plate reader (1420 multi-label counter; Perkin Elmer) and the maximum signal was observed at 3 hours after the addition of the developer solution. The average of the duplicate readings was used to estimate the concentration using the standard curve.
The amount of SIRT1 (Cat. # ab171573; Abcam) was measured by ELISA in the cell lysates. The total protein concentration in the lysates was calculated using the Pierce BCA Protein Assay (Cat. # 23225; Thermo Fisher) to normalize the Nicotinamide and vehicle treated samples. All the samples were tested in duplicates, following the manufacturer’s instructions. Optical densities/absorbance readings were obtained immediately after adding the stop solution at 450nm, using a plate reader (1420 multi-label counter; Perkin Elmer). The average of the duplicate readings was used to estimate the concentration using the standard curve.
Western blotting
hiPSC-RPE cells were lysed in RIPA Lysis and Extraction buffer (Cat. # 89901; Thermo Fisher) with Halt Protease Inhibitor Cocktail (Cat. # 78429; Thermo Fisher). The total protein concentration in Nicotinamide and vehicle treated hiPSC-RPE samples was normalized using the Pierce BCA Protein Assay (Cat. # 23225; Thermo Fisher). Proteins at equal concentration along with the molecular weight western blotting standards (Cat. # 161-0376; Bio-Rad) underwent electrophoresis in 10% Bis-Tris pre-cast polyacrylamide gels (Cat. # NP0316BOX; Thermo Fisher). Proteins were transferred from gel to membrane using iBlot gel transfer stacks (Cat. # IB4010-02; Thermo Fisher) and the iBlot Gel Transfer Device (Cat. # IB1001; Thermo Fisher). The membranes were blocked with 5% BSA in TBS-T (Tris-buffered saline with 0.1% Tween 20) and incubated with primary antibodies for APOE (Cat. # AB947; Millipore) at 1:500 dilution and β-tubulin (Cat. # E7; DSHB) at 1:2500 dilution, overnight at 4°C. Membranes were washed three times with TBS-T to remove the primary antibodies and incubated with HRP conjugated secondary antibodies against goat antibody (Cat. # 705-035-003; Jackson ImmunoResearch) and mouse antibody (Cat. # sc-2005; Santa Cruz). The western blot membrane was developed using the Pico Chemiluminescent substrate (Cat. # 34080; Thermo Fisher) and Femto maximum sensitivity substrate (Cat. #34095; Thermo Fisher), and images were acquired on the ImageQuant LAS 4000 gel imager (GE Healthcare). The pixel density of the protein bands was analyzed using ImageJ. Pixel density values for the background were subtracted from the pixel density values for APOE and β-tubulin. The expression of APOE was normalized to the loading control β-tubulin.
Cytokine analysis
Cytokines were analyzed in the culture supernatants of hiPSC-RPE lines collected 60–72 hours after the last medium change. The human XL cytokine array (Cat. # ARY022; R&D systems) was used to quantify the secretion of various cytokines in the vehicle and Nicotinamide treated hiPSC-RPE lines, following the manufacturer’s instructions. 500µl (maximum suggested volume) of supernatant diluted to a final volume of 1.5ml with array buffer 6 (included in the kit) was used for each hiPSC-RPE line. Capture and control antibodies were spotted in duplicate on the nitrocellulose membranes in the XL cytokine array and the signal produced at each spot corresponded to the amount of protein bound. The pixel densities in the array images were quantified using the ‘Microarray profile’ plug-in of ImageJ. Pixel density values for the background were subtracted from the pixel density values for each protein. After quantifying the pixel intensity of the cytokine array, those cytokines/proteins showing substantial variation between the paired technical replicates in one or more lines were discounted from further analysis, narrowing down to 29 cytokines. The mean of the duplicate spots for every protein/cytokine was calculated.
Lactate Dehydrogenase (LDH) assay
Supernatant was collected from hiPSC-RPE culture wells 60–72 hours after the last medium change. Quantification of LDH was done using the Roche Cytotoxicity Detection kit plus (Cat. # 04744926001), following the manufacturer’s instructions. Absorbance for the samples was measured at 490nm, using a plate reader (1420 multi-label counter; Perkin Elmer). Absorbance measured in the medium from a well with completely lysed cells of the corresponding line was used to define 100% cytotoxicity. Absorbance was measured for the blank medium (without cells) and also for Nicotinamide and vehicle. Absorbance for blank medium was subtracted from absorbance for the corresponding experimental wells and completely lysed cells. The percentage cytotoxicity was calculated by dividing the absorbance for the experimental wells by absorbance for 100% cytotoxicity.
Bioinformatic analysis
Functional networks and gene connectivity data were extracted using the STRING database. STRING provided gene connectivity data based on several evidence types (direct interaction, co-localization, gene-regulation and co-citation), grouping together genes, which are closely related to each other with the highest confidence (0.9 interaction score). Connectivity data from STRING was then analyzed using the iGraph package with the R statistical language. Network analysis of the extracted connectivity data, employing edge-betweenness and random walk methods, was used to highlight subnetworks or neighborhoods. The resulting neighborhoods were tested for enrichment of relevant biological/pathway terms using the STRING database. Gene set enrichment analysis for disease terms was conducted using the ENRICHR gene enrichment tool (Chen et al., 2013).
Quantification and Statistical Analysis
GraphPad Prism version 5.0 was used for all statistical analysis. Statistical parameters, including the type of tests, number of samples (n), descriptive statistics and significance are reported in the figures and figure legends. A one-tailed Student’s t test was used when the assumption/hypothesis was stated in advance regarding the direction of expected outcome.
Data and Software Availability
Data Resources
RNA-seq data was submitted to the NCBI Gene Expression Omnibus repository (GEO: GSE90889; http://www.ncbi.nlm.nih.gov/geo/).
Supplementary Material
Highlights.
Complement and inflammatory factors are upregulated in AMD hiPSC derived RPE
Nicotinamide inhibits AMD biomarkers including drusen components and VEGFA
Nicotinamide suppresses production of complement and inflammatory factors
Nicotinamide increases RPE cell survival while targeting aging associated pathways
Acknowledgments
We thank Melissa Campbell, Patty Lederman, Carol Charniga and Susan Borden for technical assistance. We are grateful to the eye bank donors and their families. This work was supported by NIH/NIA grant 1RF1AG042932-01, NIH/NEI grant R01EY022079, the Regenerative Research Foundation and by the Empire State Stem Cell Fund through New York State Department of Health Contract # C028504. J.D.M. was supported by NIH/NEI grant F32EY025931. Opinions expressed here are solely those of the authors and do not necessarily reflect those of the Empire State Stem Cell Board, the New York State Department of Health, or the State of New York.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, J.S.S., B.C., T.A.B., J.H.S., and S.T.; Methodology, J.S.S., B.C., J.D.M., Q.W., T.A.B., and S.T.; Software, T.R.K.; Validation, J.S.S.; Formal Analysis, J.S.S., N.C.B., and T.R.K.; Investigation, J.S.S., B.C., and J.D.M.; Data Curation, J.S.S., and T.R.K.; Writing-Original Manuscript, J.S.S., B.C., and S.T.; Writing-Review and Editing, J.S.S., B.C., J.D.M., and S.T.; Visualization, J.S.S.; Supervision, Project Administration and Funding Acquisition, S.T.
References
- Alcazar O, Hawkridge AM, Collier TS, Cousins SW, Bhattacharya SK, Muddiman DC, Marin-Castano ME. Proteomics characterization of cell membrane blebs in human retinal pigment epithelium cells. Molecular & cellular proteomics : MCP. 2009;8:2201–2211. doi: 10.1074/mcp.M900203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambati J, Atkinson JP, Gelfand BD. Immunology of age-related macular degeneration. Nature reviews Immunology. 2013;13:438–451. doi: 10.1038/nri3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An E, Lu X, Flippin J, Devaney JM, Halligan B, Hoffman EP, Strunnikova N, Csaky K, Hathout Y. Secreted proteome profiling in human RPE cell cultures derived from donors with age related macular degeneration and age matched healthy donors. Journal of proteome research. 2006;5:2599–2610. doi: 10.1021/pr060121j. [DOI] [PubMed] [Google Scholar]
- An E, Sen S, Park SK, Gordish-Dressman H, Hathout Y. Identification of novel substrates for the serine protease HTRA1 in the human RPE secretome. Investigative ophthalmology & visual science. 2010;51:3379–3386. doi: 10.1167/iovs.09-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkinsop TA, Saini JS, Maminishkis A, Bharti K, Wan Q, Banzon T, Lotfi M, Davis J, Singh D, Rizzolo LJ, et al. Human Adult Retinal Pigment Epithelial Stem Cell-Derived RPE Monolayers Exhibit Key Physiological Characteristics of Native Tissue. Investigative ophthalmology & visual science. 2015;56:7085–7099. doi: 10.1167/iovs.14-16246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkinsop TA, Salero E, Stern JH, Temple S. The culture and maintenance of functional retinal pigment epithelial monolayers from adult human eye. Methods Mol Biol. 2013;945:45–65. doi: 10.1007/978-1-62703-125-7_4. [DOI] [PubMed] [Google Scholar]
- Bradley DT, Zipfel PF, Hughes AE. Complement in age-related macular degeneration: a focus on function. Eye (Lond) 2011;25:683–693. doi: 10.1038/eye.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb JW. The proteomics of drusen. Cold Spring Harbor perspectives in medicine. 2014;4:a017194. doi: 10.1101/cshperspect.a017194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Rabin DM, Salero E, Lederman PL, Temple S, Stern JH. Human retinal pigment epithelium cell changes and expression of alphaB-crystallin: a biomarker for retinal pigment epithelium cell change in age-related macular degeneration. Archives of ophthalmology. 2007;125:641–645. doi: 10.1001/archopht.125.5.641. [DOI] [PubMed] [Google Scholar]
- Deangelis MM, Silveira AC, Carr EA, Kim IK. Genetics of age-related macular degeneration: current concepts, future directions. Seminars in ophthalmology. 2011;26:77–93. doi: 10.3109/08820538.2011.577129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlken C, Jungmann S, Bohringer D, Agostini HT, Junker B, Pielen A. Switch of anti-VEGF agents is an option for nonresponders in the treatment of AMD. Eye (Lond) 2014;28:538–545. doi: 10.1038/eye.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald CY, Landis JN, Porter Abate J, Murphy CT, Blackwell TK. Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature. 2015;519:97–101. doi: 10.1038/nature14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer M, Corneo B, Davis J, Wan Q, Miyagishima KJ, King R, Maminishkis A, Marugan J, Sharma R, Shure M, et al. A multiplex high-throughput gene expression assay to simultaneously detect disease and functional markers in induced pluripotent stem cell-derived retinal pigment epithelium. Stem cells translational medicine. 2014;3:911–922. doi: 10.5966/sctm.2013-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis PJ, Appukuttan B, Simmons E, Landauer N, Stoddard J, Hamon S, Ott J, Ferguson B, Klein M, Stout JT, et al. Rhesus monkeys and humans share common susceptibility genes for age-related macular disease. Human molecular genetics. 2008;17:2673–2680. doi: 10.1093/hmg/ddn167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Chen W, Schu M, Yaspan BL, Yu Y, Thorleifsson G, Zack DJ, Arakawa S, Cipriani V, Ripke S, et al. Seven new loci associated with age-related macular degeneration. Nature genetics. 2013;45:433–439. doi: 10.1038/ng.2578. 439e431-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Fariss RN, Stambolian D, Abecasis GR, Curcio CA, Swaroop A. Age-related macular degeneration: genetics and biology coming together. Annual review of genomics and human genetics. 2014;15:151–171. doi: 10.1146/annurev-genom-090413-025610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KN, Steffan JS, Martinez-Coria H, Sun X, Schreiber SS, Thompson LM, LaFerla FM. Nicotinamide restores cognition in Alzheimer’s disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phosphotau. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:11500–11510. doi: 10.1523/JNEUROSCI.3203-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annual review of immunology. 2014;32:659–702. doi: 10.1146/annurev-immunol-032713-120145. [DOI] [PubMed] [Google Scholar]
- He S, Incardona F, Jin M, Ryan SJ, Hinton DR. Thrombospondin-1 expression in RPE and choroidal neovascular membranes. Yan ke xue bao = Eye science / “Yan ke xue bao” bian ji bu. 2006;22:265–274. [PubMed] [Google Scholar]
- Holz FG, Schmitz-Valckenberg S, Fleckenstein M. Recent developments in the treatment of age-related macular degeneration. The Journal of clinical investigation. 2014;124:1430–1438. doi: 10.1172/JCI71029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager RD, Mieler WF, Miller JW. Age-related macular degeneration. The New England journal of medicine. 2008;358:2606–2617. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- Jang SY, Kang HT, Hwang ES. Nicotinamide-induced mitophagy: event mediated by high NAD+/NADH ratio and SIRT1 protein activation. The Journal of biological chemistry. 2012;287:19304–19314. doi: 10.1074/jbc.M112.363747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LV, Forest DL, Banna CD, Radeke CM, Maloney MA, Hu J, Spencer CN, Walker AM, Tsie MS, Bok D, et al. Cell culture model that mimics drusen formation and triggers complement activation associated with age-related macular degeneration. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18277–18282. doi: 10.1073/pnas.1109703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas JB, Tao Y, Neumaier M, Findeisen P. Cytokine concentration in aqueous humour of eyes with exudative age-related macular degeneration. Acta ophthalmologica. 2012;90:e381–e388. doi: 10.1111/j.1755-3768.2012.02414.x. [DOI] [PubMed] [Google Scholar]
- Karar J, Maity A. PI3K/AKT/mTOR Pathway in Angiogenesis. Frontiers in molecular neuroscience. 2011;4:51. doi: 10.3389/fnmol.2011.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo KH, Verma CS, Lane DP. Drugging the p53 pathway: understanding the route to clinical efficacy. Nature reviews Drug discovery. 2014;13:217–236. doi: 10.1038/nrd4236. [DOI] [PubMed] [Google Scholar]
- Liu TF, McCall CE. Deacetylation by SIRT1 Reprograms Inflammation and Cancer. Genes & cancer. 2013;4:135–147. doi: 10.1177/1947601913476948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Chen M, Xu H. Complement gene expression and regulation in mouse retina and retinal pigment epithelium/choroid. Molecular vision. 2011;17:1588–1597. [PMC free article] [PubMed] [Google Scholar]
- Maiese K, Chong ZZ, Hou J, Shang YC. The vitamin nicotinamide: translating nutrition into clinical care. Molecules. 2009;14:3446–3485. doi: 10.3390/molecules14093446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, Tu EY, Mandal PK, Vera E, Shim JW, Kriks S, et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell stem cell. 2013;13:691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills KF, Yoshida S, Stein LR, Grozio A, Kubota S, Sasaki Y, Redpath P, Migaud ME, Apte RS, Uchida K, et al. Long-Term Administration of Nicotinamide Mononucleotide Mitigates Age-Associated Physiological Decline in Mice. Cell Metabolism. 2016;24:795–806. doi: 10.1016/j.cmet.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima KJ, Wan Q, Corneo B, Sharma R, Lotfi MR, Boles NC, Hua F, Maminishkis A, Zhang C, Blenkinsop T, et al. In Pursuit of Authenticity: Induced Pluripotent Stem Cell-Derived Retinal Pigment Epithelium for Clinical Applications. Stem cells translational medicine. 2016 doi: 10.5966/sctm.2016-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, Gallo NB, Hancox LS, Miller NJ, Radeke CM, Maloney MA, Cooper JB, Hageman GS, Anderson DH, Johnson LV, et al. Systems-level analysis of age-related macular degeneration reveals global biomarkers and phenotype-specific functional networks. Genome medicine. 2012;4:16. doi: 10.1186/gm315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno-Matsui K. Parallel findings in age-related macular degeneration and Alzheimer’s disease. Progress in retinal and eye research. 2011;30:217–238. doi: 10.1016/j.preteyeres.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Pal S, Tyler JK. Epigenetics and aging. Science advances. 2016;2:e1600584. doi: 10.1126/sciadv.1600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennesi ME, Neuringer M, Courtney RJ. Animal models of age related macular degeneration. Molecular aspects of medicine. 2012;33:487–509. doi: 10.1016/j.mam.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoenix TN, Temple S. Spred1, a negative regulator of Ras-MAPK-ERK, is enriched in CNS germinal zones, dampens NSC proliferation, and maintains ventricular zone structure. Genes & development. 2010;24:45–56. doi: 10.1101/gad.1839510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin DM, Rabin RL, Blenkinsop TA, Temple S, Stern JH. Chronic oxidative stress upregulates Drusen-related protein expression in adult human RPE stem cell-derived RPE cells: a novel culture model for dry AMD. Aging. 2013;5:51–66. doi: 10.18632/aging.100516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein DB, Wittenborn JS, Zhang X, Honeycutt AA, Lesesne SB, Saaddine J. Forecasting age-related macular degeneration through the year 2050: the potential impact of new treatments. Archives of ophthalmology. 2009;127:533–540. doi: 10.1001/archophthalmol.2009.58. [DOI] [PubMed] [Google Scholar]
- Saini JS, Temple S, Stern JH. Human Retinal Pigment Epithelium Stem Cell (RPESC) Advances in experimental medicine and biology. 2016;854:557–562. doi: 10.1007/978-3-319-17121-0_74. [DOI] [PubMed] [Google Scholar]
- Salero E, Blenkinsop TA, Corneo B, Harris A, Rabin D, Stern JH, Temple S. Adult human RPE can be activated into a multipotent stem cell that produces mesenchymal derivatives. Cell stem cell. 2012;10:88–95. doi: 10.1016/j.stem.2011.11.018. [DOI] [PubMed] [Google Scholar]
- Scholl HP, Fleckenstein M, Charbel Issa P, Keilhauer C, Holz FG, Weber BH. An update on the genetics of age-related macular degeneration. Molecular vision. 2007;13:196–205. [PMC free article] [PubMed] [Google Scholar]
- Shaw WM, Luo S, Landis J, Ashraf J, Murphy CT. The C. elegans TGF-beta Dauer pathway regulates longevity via insulin signaling. Current biology : CB. 2007;17:1635–1645. doi: 10.1016/j.cub.2007.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan AH, Madison DV, Mateos JM, Fraser DA, Lovelett EA, Coutellier L, Kim L, Tsai HH, Huang EJ, Rowitch DH, et al. A dramatic increase of C1q protein in the CNS during normal aging. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:13460–13474. doi: 10.1523/JNEUROSCI.1333-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss O. The retinal pigment epithelium in visual function. Physiological reviews. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- Wang L, Clark ME, Crossman DK, Kojima K, Messinger JD, Mobley JA, Curcio CA. Abundant lipid and protein components of drusen. PloS one. 2010;5:e10329. doi: 10.1371/journal.pone.0010329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Li Y, Chan L, Tsai YT, Wu WH, Nguyen HV, Hsu CW, Li X, Brown LM, Egli D, et al. Validation of genome-wide association study (GWAS)-identified disease risk alleles with patient-specific stem cell lines. Human molecular genetics. 2014;23:3445–3455. doi: 10.1093/hmg/ddu053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxidants & redox signaling. 2008;10:179–206. doi: 10.1089/ars.2007.1672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.