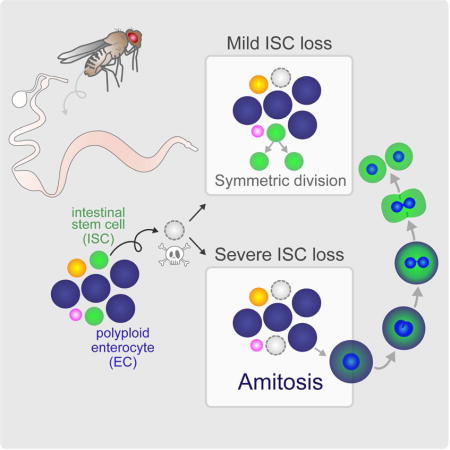
Summary
Organ fitness depends on appropriate maintenance of stem cell populations, and aberrations in functional stem cell numbers are associated with malignancies and aging. Symmetrical division is the best characterized mechanism of stem cell replacement, but other mechanisms could also be deployed, particularly in situations of high stress. Here, we show that after severe depletion, intestinal stem cells (ISCs) in the Drosophila midgut are replaced by spindle-independent ploidy reduction of cells in the enterocyte-lineage through a process known as amitosis. Amitosis is also induced by the functional loss of ISCs coupled with tissue demand and in aging flies, underscoring the generality of this mechanism. However, we also found that random homologous chromosome segregation during ploidy reduction can expose deleterious mutations through loss of heterozygosity. Together, our results highlight amitosis as an unappreciated mechanism for restoring stem cell homeostasis, but one with some associated risk in animals carrying mutations.
Keywords: Drosophila, midgut, intestinal stem cell, regeneration, dedifferentiation, depolyploidization, amitosis, starvation, injury, aging
Graphical abstract
Introduction
The intestinal epithelium is among the most actively self-renewing tissues and relies on resident stem cells for survival in the face of constant digestive, chemical and bacterial insults (Li and Jasper, 2016). As such, organ fitness is intimately linked to the number of functional stem cells. An abnormal increase of intestinal stem cell (ISC)-containing crypts is a hallmark of intestinal polyposis, a precancerous neoplasia (He et al., 2007), while a decline in functional ISCs severely hinders tissue homeostasis (Korinek et al., 1998) and is a hallmark of aging. Despite their pivotal role in preserving organ fitness, the mechanisms that replace ISCs under variable environmental challenges throughout the lifetime of the animal remain an enigma.
Expansions in stem cell number are thought to occur by symmetric stem cell divisions. Indeed, in the newly-eclosed fly, the balance of ISC mitotic events shifts from predominantly asymmetric to predominantly symmetric to expand the number of ISCs in the distal hairpin of the posterior midgut (PMG) (O’Brien et al., 2011). In addition, a decline in the number of ISC clones over time (de Navascues et al., 2012), in conjunction with the presence of symmetric divisions in adult flies (O’Brien et al., 2011), suggests that ISCs, themselves, are sporadically lost and replaced by this mechanism throughout the lifetime of the fly.
Mounting evidence suggests that alternate mechanisms also exist to replace stem cells. In the Drosophila ovary (Kai and Spradling, 2004) and testis (Brawley and Matunis, 2004), upon complete loss, germline stem cells (GSCs) are replaced by dedifferentiation of cystocytes and prospermatogonia, respectively. In the mouse airway epithelium (Tata et al., 2013), upon complete genetic ablation, stem cells are replenished by dedifferentiation of diploid secretory cells. Finally, in the mouse intestinal crypt, it has been suggested that upon genetic ablation, ISCs can be replaced by dedifferentiation of early enterocytes (Tetteh et al., 2016) or secretory (van Es et al., 2012) progenitors, underscoring a previously unappreciated plasticity in contending with stem cell loss.
While alternate mechanisms of stem cell replacement have been prompted by genetically ablating stem cells, the extent to which they are employed under environmental perturbations remain largely unexplored. Here, we developed a starvation assay, mimicking an environmental stressor, which induces a rapid and severe regional loss of ISCs in the Drosophila PMG. We demonstrate that, in areas nearly void of progenitor cells, ISCs are rapidly replaced upon re-feeding by a specialized dedifferentiation of polyploid cells in the EC-lineage, involving a depolyploidization event known as amitosis. Our data show that amitosis can be induced not only upon physical loss of ISCs during starvation, but also upon functional loss of ISCs during periods of proliferative demand, and during aging. Amitosis was first recognized as a specialized form of cell division in chicken red blood cells (Remak, 1841), and has been identified in a vast array of species, from primitive ciliates to mammals. Yet, the functional significance of this alternative cell division remains to be placed in somatic tissues of higher organisms. Together, our data highlight amitosis as a significant mechanism of stem cell renewal in the Drosophila intestinal epithelium that may extend to other tissues and organisms.
Results
ISC number remains at a steady-state in the PMG of young flies
Multipotent ISCs have been identified in both vertebrates (Barker et al., 2007) and invertebrates (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006) and give rise to all cell types in the epithelium. ISCs divide asymmetrically to give rise to an ISC and either a secretory enteroendocrine (ee) cell expressing the transcription factor Prospero (Pros) (Guo and Ohlstein, 2015; Zeng and Hou, 2015), or an enteroblast (EB) daughter cell that has high active Notch (N)-signaling (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2007) (Figure S1A). The EB subsequently enters the endocycle, and the resulting 4n polyploid pre-EC (Jiang et al., 2011; Zhou et al., 2015) further differentiates into an absorptive enterocyte (EC), ploidies of which typically range from 4n to 16n (Figure S1A).
To probe alternate mechanisms of ISC replacement, we first defined a time window during which ISC number is stable and perturbations can therefore be measured reliably. We used flies of the genotype NRE-lacZ; esg-GAL4, UAS-GFP (esg>GFP);, previously reported as a precise method of quantifying ISC and EB number (McLeod et al., 2010; O’Brien et al., 2011). In these flies, ISCs and EBs are marked by escargot (esg)-driven GFP expression and EBs are marked by strong lacZ expression under the control of a Notch response element (NRE) (Micchelli and Perrimon, 2006) (Figure S1A, B). Age-matched females were collected and maintained on a rich diet of standard cornmeal-molasses supplemented with yeast. Midguts were dissected at different time points, and the total number of ISCs per PMG was quantified by subtracting the number of diploid EBs (lacZ+ cells) from the total number of diploid ISCs and EBs (GFP+ cells) (Figure S1C). ISC number remained relatively constant in adults from 3 to 17 days PE (Figure S1D, steady-state phase), enabling us to precisely quantify and interpret any induced ISC loss and subsequent recovery during this time.
Starvation induces a rapid and severe regional loss of ISCs in the PMG
In a previous study, McLeod et al. observed a loss of ISCs in flies maintained on a restricted diet of 10% sucrose from 3 to 18 days PE. Recovery of ISC number was subsequently observed when animals were re-fed a protein-rich diet (McLeod et al., 2010), suggesting that ISC number changes in response to food availability. However, the interpretation of these results was confounded by the length of the restricted-diet assay, which extended into the period of the normal age-induced increase in ISC number reported in the PMG of well-fed animals (Biteau et al., 2008; Choi et al., 2008a; Choi et al., 2008b). In addition, the mechanism of ISC replacement was not explored.
Based on the study of McLeod et al., we supposed that complete starvation with only water may induce a more rapid loss of ISCs within the steady-state phase (Figure S1D). To this end, we developed an assay in which flies were maintained on a rich diet from 0–3 days PE and then transferred to vials containing only water for two days (Figure 1A).
Figure 1. Starvation Induces Loss of ISCs and EBs in the PMG.
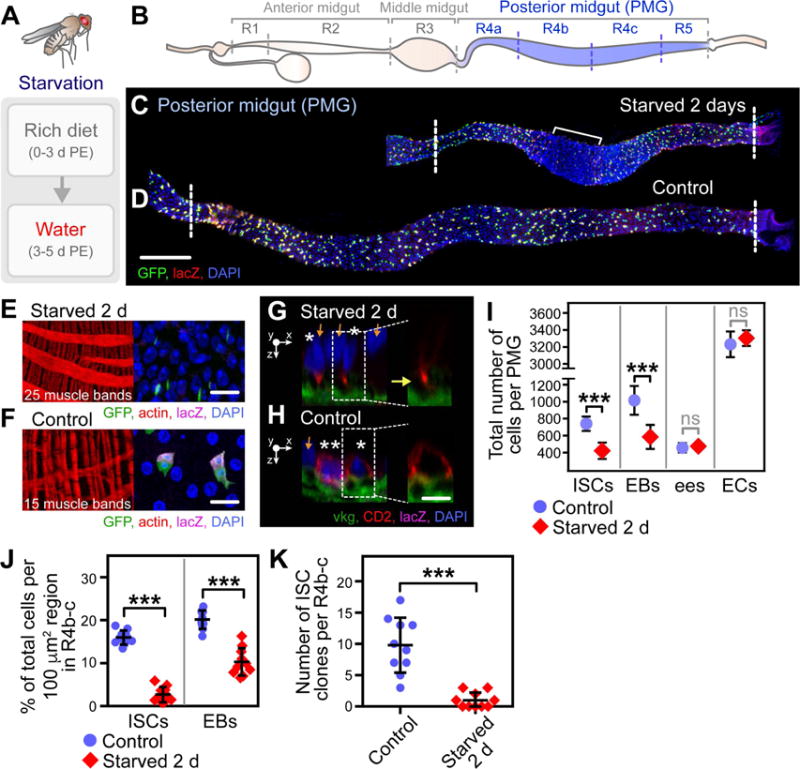
(A) Starvation assay used to induce ISC loss.
(B) Schematic representation of regions in the Drosophila midgut.
(C–D) Composite images of the PMG from starved (C) and control (D) flies. Bracket in (C) denotes an area within R4b-c that displays the most dramatic shortening. Scale bar = 100 μm. See also Figure S2A–B.
(E–F) Images displaying the visceral muscle (left panel) and cell density (right panel) in starved (E) and control (F) flies. The number of muscle bands per region is visualized by actin. Scale bar = 20 μm.
(G–H) Sagittal images displaying ISC location relative to the basement membrane, visualized by viking (vkg)-GFP, in starved (G) and control (H) flies (ISCs, asterisks; EB, double asterisks; ECs, orange arrows). Scale bar = 10 μm.
(I) Quantification of the number of each cell type in the PMG. n = 10. (***) denotes p < 0.001. See also Figure S1B–C.
(J) Quantification of the percent ISCs and EBs of total cells within a 100 μm × 100 μm region in R4b-c. n = 10. (***) denotes p < 0.001. See also Figure S2C for raw data.
(K) Quantification of the number of ISC MARCM clones in R4b-c. n = 10. (***) denotes p < 0.001.
(C–F, I, J) NRE-lacZ; esg>GFP;
(G–H) NRE-lacZ; esg>CD2, vkg-GFP;
(K) y, w, hsFLP tub>GFP; ; FRT82B tubGAL80/FRT82B ry506
Maintaining flies on water resulted in a dramatic shrinkage of the PMG (defined as regions R4a-R5) (Buchon et al., 2013) (Figure 1B) to approximately one-half the length (Figure 1C, Figure S2A–B) of age-matched controls (Figure 1D, Figure S2B). Tissue shrinkage resulted from regional muscle contraction (Figure 1C (bracket)), visualized by the condensed fibers of the visceral muscle in R4b-c, a subset of the PMG, of starved flies (Figure 1E, left panel, 25 muscle bands per region) relative to controls (Figure 1F, left panel, 15 muscle bands per region). Within the area of muscle contraction, cells in the epithelium were severely overcrowded (Figure 1E, right panel) compared to controls (Figure 1F, right panel). In addition, ISCs within R4b-c of starved flies delaminated from the basement membrane (Figure 1G, yellow arrow), likely by mechanical extrusion, while ISCs in control flies maintained direct contact with the basement membrane (Figure 1H).
To determine the extent of ISC loss in starved flies, the total number of ISCs was quantified as described above (Figure S1C). After 2 days of starvation, the total number of ISCs per PMG decreased to an average of 420 ± 95, compared to an average of 740 ± 85 in controls (Figure 1I). EBs were also significantly depleted (Figure 1I). However, the number of differentiated ees (Pros+) and ECs (polyploid) was not affected (Figure 1I), giving rise to the severe cellular overcrowding in areas of muscle contraction.
As the number of ISCs and EBs appeared to be most severely depleted in R4b-c, we further quantified the percent of ISCs or EBs of total cells in a 100 μm × 100 μm region within R4b-c (Figure 1J, Figure S2C). ISCs and EBs constituted an average 16 ± 2 and 20 ± 3 percent of total cells per region in control flies, respectively (Figure 1J). In contrast, ISCs and EBs only constituted an average 3 ± 2 and 10 ± 4 percent of total cells per region in starved flies (Figure 1J), a severe decrease in the density of progenitor cells and particularly of ISCs.
To confirm ISC loss, we induced ISC clones using the Mosaic Analysis with a Repressible Cell Marker (MARCM) system (Lee and Luo, 2001) one day prior to starvation and quantified the number of ISC clones remaining in starved flies relative to controls. In accordance with our ISC number quantification, ISC clones were lost in starved flies, with the most dramatic loss in R4b-c (Figure 1K), corroborating that ISCs are lost in this region.
ISC number rapidly increases in re-fed animals
We next determined if ISCs are replaced by re-feeding flies a rich diet. Within 24 hours, the length of the PMG expanded to that of well-fed flies (Figure S2B). Following tissue expansion, a significant increase in ISC number was observed by 16 hours of re-feeding. ISC number expanded from an average of 420 ± 95 at t = 0 to 490 ± 62, 550 ± 70, 680 ± 90, and 760 ± 65 at t = 16, 24, 48, and 72 hours of re-feeding, respectively (Figure 2A), and was indistinguishable from that of age-matched controls by 72 hours of re-feeding.
Figure 2. While a Significant Increase in the Number of ISCs is Observed in Flies Re-fed for 16 Hours, Symmetric ISC Divisions and Mitoses are Negligible at this Time.
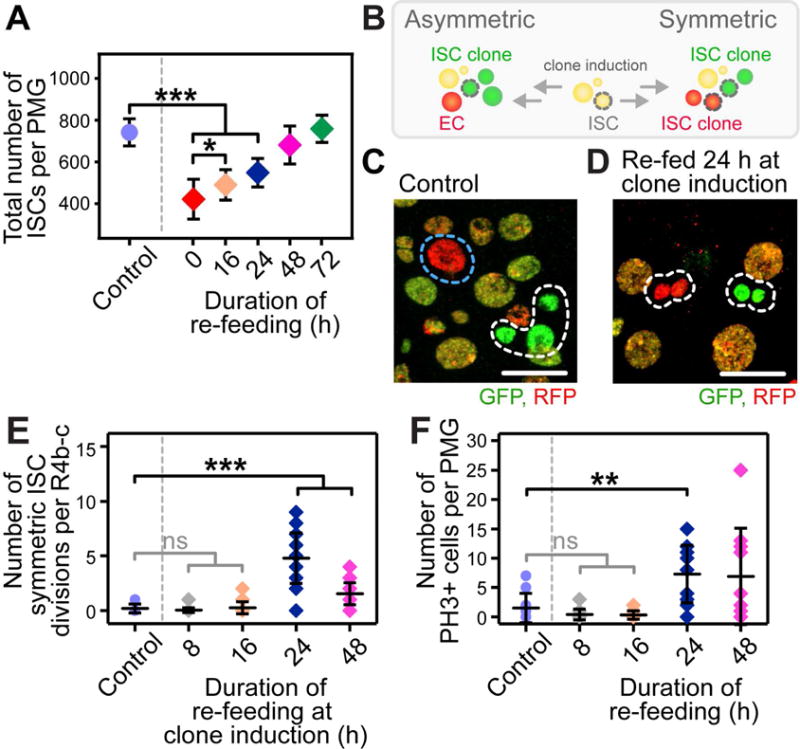
(A) Quantification of the number of ISCs in re-fed flies. n = 10. (*) and (***) denote p < 0.05 and p < 0.001, respectively.
(B) Schematic representation of the twin-spot lineage analysis used to quantify the number of symmetric ISC divisions.
(C–D) Images displaying an asymmetric (C, control) and symmetric (D, re-fed for 24 hours at clone induction) outcome of an ISC division (ISC clone, white dotted outline; transit clone, blue dotted outline). Scale bar = 20 μm.
(E) Quantification of the number of symmetric ISC divisions, defined as two adjacent ISC clones of different color (D), per R4b-c. n = 20. (***) denotes p < 0.001.
(F) Quantification of the number of PH3+ cells per PMG. n = 20. (**) denotes p < 0.01.
(A, F) NRE-lacZ; esg>GFP;
(B–E) y, w, hsFLP; ; FRT2A His-GFP/His2Av-mRFP, FRT2A
Symmetric ISC divisions and mitoses are negligible at 16 hours of re-feeding, when a significant increase in ISC number is observed
The ISC number increase in re-fed flies is reminiscent of that observed 0–3 days PE in the distal hairpin, which is linked to developmental tissue growth and governed by symmetric divisions of existing ISCs (O’Brien et al., 2011). However, R4b-c of starved flies was severely depleted of ISCs, as well as their immediate EB daughter cells (Figure 1I–K). As approximately 70 ISCs are replaced within the first 16 hours of re-feeding, we predicted that 70 symmetric ISC divisions would have to occur within this short time with few remaining ISCs in R4b-c.
To determine the contribution of symmetric ISC divisions to ISC replacement, we conducted twin-spot clonal analysis (Figure 2B) using flies of the genotype hsFLP; ; FRT2A His-GFP/His2AV-mRFP, FRT2A. In these flies, heat-shock (hs)-induced FLP/FRT-mediated recombination results in one GFP-marked and one RFP-marked daughter cell (Figure 2B). An asymmetric outcome is visualized by one multi-cellular ISC clone adjacent to one differentiated daughter cell of differing colors (Figure 2C). In contrast, a symmetric outcome is visualized by two adjacent multi-cellular ISC clones of differing colors (Figure 2D).
In control flies at 6 d PE, symmetric division of ISCs was negligible (Figure 2C, E). In re-fed flies, no significant increase in symmetric divisions was detectable at 8 or 16 hours of re-feeding (Figure 2E), despite an increase in ISC number from an average of 420 to 490 by 16 hours of re-feeding (Figure 2A). Symmetric divisions were only observed beginning at 24 hours of re-feeding (Figure 2D, E). Furthermore, by using phospho-histone-H3 (PH3) as a marker of mitosis, no increase in the number of mitoses were observed prior to 24 hours of re-feeding in the entire PMG (Figure 2F), suggesting that new ISCs did not arise from mitosis of remaining ISCs outside of R4b-c. Finally, while quantifying the number of PH3+ ISCs, we observed no evidence of ISCs migrating into this region. Together, these data demonstrate that symmetric ISC divisions cannot account for the initial increase in ISC number from 0–16 hours and point to an alternative initial source of new ISCs.
Polyploid cells undergoing amitosis are observed during the first 16 hours of re-feeding
The Snail-family of zinc-finger transcription factors are highly conserved across species, and are expressed in many types of stem cells in both Drosophila and mammals. Indeed, the Snail homolog escargot (esg) is expressed in ISCs and EBs of the fly midgut (Micchelli and Perrimon, 2006), and has been shown to maintain progenitor cells in an undifferentiated state (Korzelius et al., 2014; Loza-Coll et al., 2014). During the first 24 hours of re-feeding, we observed esg-GAL4-driven expression of GFP (Figure S3A) or the membrane-reporter CD2 (Figure 3A–E) in an average of 36 ± 25 4n polyploid cells in R4a-5 (Figure S3B), suggesting that ECs may adopt a progenitor-like state during this time.
Figure 3. Polyploid Cells Progressing Through Amitosis are Observed in Flies Starved and Re-fed for 8–16 hours.
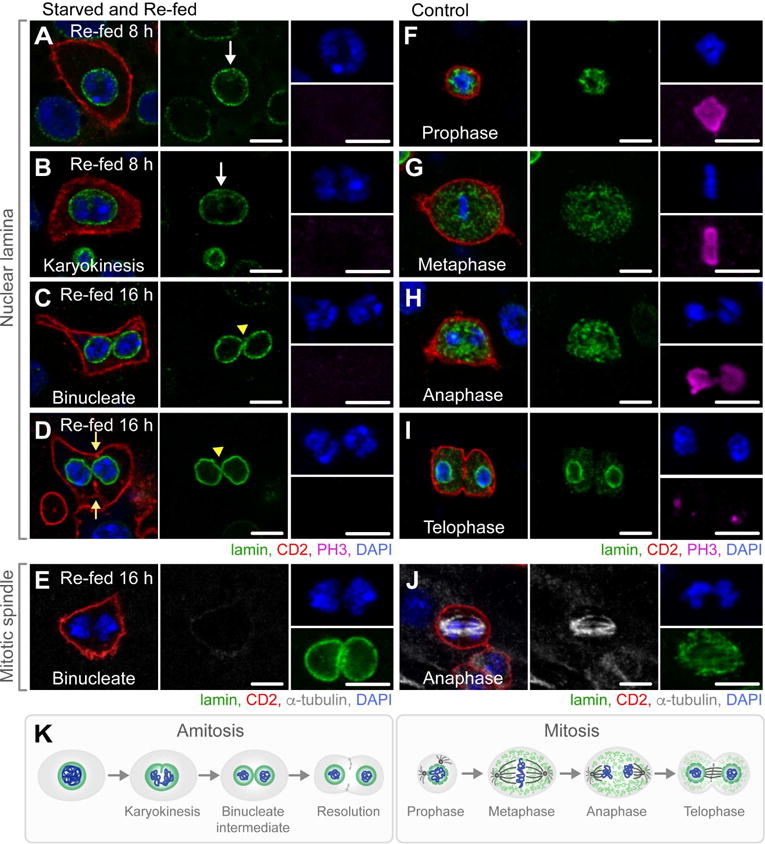
(A–D) Images of representative 4n polyploid cells undergoing amitosis in flies starved and re-fed for 8–16 hours. White arrows denote invagination of the intact nuclear lamina (A–B). Yellow arrow heads denote bridging of the nuclear lamina between two nuclei of a binucleate cell (C–D). Yellow arrows denote invagination of the cell membrane (D). Scale bar = 10 μm. See also Figure S3B–C for quantifications.
(E) A representative 4n polyploid binucleate cell lacking an α-tubulin-enriched spindle. Scale bar = 10 μm.
(F–I) Images of representative mitotic cells in control flies. Scale bar = 10 μm.
(J) A representative mitotic cell in anaphase with an α-tubulin-enriched spindle. Scale bar = 10 μm.
(K) Schematic representation of the progression through amitosis versus the progression through mitosis.
(A–J) NRE-lacZ; esg>CD2; See also Figure S3.
To characterize the ontogeny of these cells, midguts were dissected every 4 hours during the first 24 hours of re-feeding. Beginning at 8 hours of re-feeding, this subset of 4n polyploid CD2+ cells appeared to undergo a ploidy reduction (depolyploidization) (Figure 3A–E) exclusively in R4b-c where ISC depletion was most severe. Notably, these cells were negative for the mitotic marker PH3 (Figure 3A–D). The fate of these cells progressed over time, as follows:
At 8 hours of re-feeding, a subset of 4n polyploid CD2+ cells appeared to be undergoing karyokinesis and partitioning chromosomes within an intact nuclear lamina (Figure 3A–B), markedly differing from prophase and metaphase of a mitotic ISC division (Figure 3F–G), during which chromosomes condense and the nuclear lamina dissociates.
At 16 hours of re-feeding, a subset of 4n polyploid CD2+ cells, approximately 1 ± 2 per R4b-c (Figure S3C), progressed through a binucleate intermediate containing two nuclei, each with a fully intact nuclear lamina, which were often bridged (Figure 3C–D, yellow arrow head), in contrast to anaphase or telophase of a mitotic division, during which the nuclear lamina remains completely or partially dissociated (Figure 3H–I). Binucleate cells lacked an organized α-tubulin-rich spindle (Figure 3E), in contrast to mitotic cells (Figure 3J), which rely on the spindle apparatus for proper separation of chromosomes. Finally, binucleate cells appeared to resolve by invagination of the cell membrane (Figure 3D, yellow arrows) in the absence of an anillin-rich contractile ring (Figure S3D), in contrast to a mitotic cell, in which anillin localizes to the cleavage furrow in anaphase (Figure S3E). Binucleate cells were fully laminated to the basement membrane (Figure S3F), and not an artifact of the membrane marker CD2, used as a reporter of esg-GAL4 expression, as these cells were also observed in NRE-lacZ; esg>GFP; flies (Figure S3G), in contrast to controls (Figure S3H).
The ontogeny of CD2+ polyploid cells suggested a process of depolyploidization unbeknownst to us. However, in searching the literature, we were able to identify each step of this process, documented in several different organisms. First, the nuclear invagination that we observed at 8 hours of re-feeding, more prominent on one side of the nucleus (Figure 3A–B), was reminiscent of that reported in the Tetrahymena macronucleus (Endo and Sugai, 2011), cricket egg follicle cells (Conklin, 1903), scorpion serosa (Johnson, 1892), rainbow trout erythrocytes (Wang et al., 2010), rat trophoblast cells (Zybina and Zybina, 2008), and human adrenal cells (Magalhaes et al., 1991) undergoing ploidy reduction. Second, the binucleate cell and characteristic bridge between the two nuclei that we observed at 16 hours of re-feeding (Figure 3C–D, yellow arrow heads) was like that reported in the Tetrahymena macronucleus (Endo and Sugai, 2011), scorpion serosa (Johnson, 1892), rainbow trout erythrocytes (Wang et al., 2010), mouse (Kuhn et al., 1991) and spotted skunk trophoblast cells (Isakova and Mead, 2004), and human fibroblasts (Walen, 2005) undergoing ploidy reduction. Finally, the lack of an α-tubulin-rich mitotic spindle was consistent with reports of cells in all above mentioned species and cell types. Together, the progression of depolyploidizing cells in re-fed flies matched the process of amitosis described in the literature across phyla and defined as a cell division in which nuclear invagination separates genetic material in the absence of a mitotic spindle, resulting in a binucleate cell that resolves into two daughter cells (Figure 3K). The localization of amitotic cells to R4b-c, and their appearance from 8 to 16 hours of re-feeding, the time at which we observed a significant increase in ISC number in the absence of symmetric ISC divisions or mitoses, pinpoints amitosis of 4n polyploid cells as the most likely source of new ISCs.
Cells in the EC-lineage serve as an alternate source of new ISCs
To confirm the contribution of cells in the EC-lineage to new ISCs, we conducted lineage analysis based on a flipout system (Theodosiou and Xu, 1998), in which a stop cassette flanked by FRT sites is excised upon GAL4-driven expression of UAS-flippase (UAS-FLP), resulting in Act5C-lacZ expression (Figure 4A). Two drivers, Myo1A-GAL4 (Morgan et al., 1995) and Mex-GAL4 (Phillips and Thomas, 2006), are commonly used to mark and genetically manipulate gene expression in ECs. However, we found Myo1A-GAL4 drove reporter expression in a sub-set of EBs after 5 d PE (Figure S4A) and in a sub-set of ISCs and EBs after 14 d PE (Figure S4B). In addition, Mex-GAL4-driven reporter expression was observed only in a subset of ECs in R4b-c and absent in Esg+ polyploid cells within this region in re-fed flies (Figure S4C). Therefore, to maintain stringency in our clonal analysis, we used 28E03-GAL4, identified in a screen of Janelia Farm lines (Jenett et al., 2012) as a driver expressed at high levels (Figure S4D–E) in a subset of EBs. Notably, 28E03-GAL4 is absent in all ees and ISCs within our analyses from 0–21 days PE. When coupled to the flipout system, the EB, pre-EC and fully differentiated ECs of increasing ploidy (EC-lineage) are marked (Figure 4B).
Figure 4. Cells in the EC-Lineage Give Rise to New ISCs in Re-fed Flies.
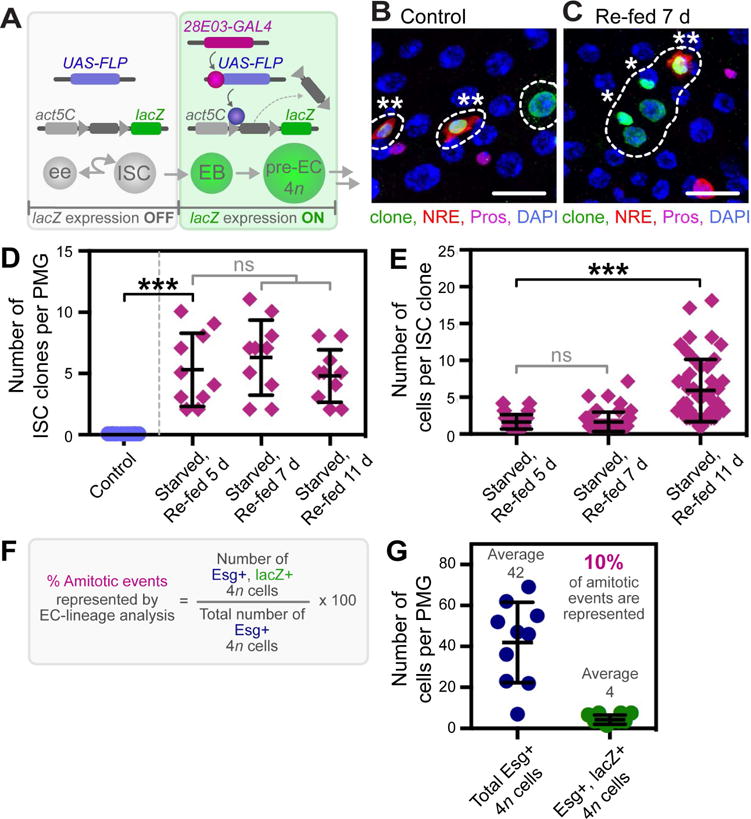
(A) Schematic representation of the flipout system used to determine the contribution of cells in the EC-lineage to ISC replacement. See also Figure S4D–G.
(B–C) Images from a representative control fly, in which only lineage-labeled EBs, pre-ECs and ECs are observed (B), and starved and re-fed fly, in which lineage-labeled ISC clones are observed (C) (lineage-labeled cells, dotted outline; lineage-labeled ISCs, asterisk; lineage-labeled EB, double asterisk). Scale bar = 30 μm.
(D) Quantification of the number of lineage-labeled ISC clones per PMG. n = 10. (***) denotes p < 0.001.
(E) Quantification of the number of cells per lineage-labeled ISC clone per PMG. n = 10. (***) denotes p < 0.001.
(F) Schematic representation of the calculation used to determine the percent of amitotic events represented by EC-lineage analysis.
(G) Quantification of the total number of Esg+ 4n cells and the number of Esg+, lacZ+ lineage-labeled 4n cells per PMG. n = 10.
(A–E) w ; pBID-UASC-FLP/Act5C FRT draf+ FRT lacZ; 28E03-GAL4/NRE-GFP
(G) w ; esg-GFP/Act5C FRT draf+ FRT lacZ; 28E03-GAL4/pBID-UASC-FLP
Flies of the genotype ; Act5C FRT draf+ FRT lacZ/pBID-UASC-FLP; 28E03-GAL4/NRE-GFP (EC-lineage flipout) (Figure 4A) were subjected to our starvation assay to determine the contribution of cells in the EC-lineage to new ISCs. No lineage-labeled ISCs were observed in control flies (Figure 4B, D). Likewise, no marked ISCs were observed immediately after re-feeding, ruling out the possibility that starvation induced the expression of 28E03-GAL4 in ISCs (Figure S4F–G″). In contrast, an average of 5 ± 3, 6 ± 3, and 5 ± 2 lineage-labeled ISCs were observed in flies that were starved and re-fed for 5, 7, and 11 days, respectively (Figure 4C–D), demonstrating that cells in the EC-lineage contribute to ISC replacement and that these newly formed ISCs are stable over time. In addition, lineage-labeled ISCs gave rise to multi-cell clones of increasing size over time, averaging 2 ± 1, 2 ± 1 and 6 ± 4 cells per clone at day 5, 7 and 11 of re-feeding, respectively (Figure 4E). Together, these data demonstrate that new, functional ISCs originate from cells in the EC-lineage, either by direct dedifferentiation of the EB or by amitosis of a 4n polyploid EC.
The frequency of flipout events is low in our EC-lineage analysis. Therefore, we sought to determine the percent of potential amitotic events represented by the number of ISC clones generated in our lineage analysis experiment. To this end, flies of the genotype ; esg-GFP/Act5C FRT draf+ FRT lacZ; 28E03-GAL4/pBID-UASC-FLP, incorporating a reporter of esg expression, were subjected to our starvation assay. We then quantified the total number of Esg+ 4n cells (42 ± 19) and the number of lineage-labeled Esg+ 4n cells (4 ± 2) in flies that were re-fed for 16 hours (Figure 4F–G). As 10% of the total Esg+ 4n cells were also lineage-labeled (Figure 4G), we estimate that the number of ISC clones originating from the EC-lineage represent 10% of the total potential amitotic events. Therefore, the 5 ± 3 ISC clones observed in flies re-fed for 5 days (Figure 4D) represent 20–80 ISCs originating from cells in the EC-lineage, consistent with an average of 70 ISCs gained within the first 16 hours of re-feeding (Figure 2A).
Amitosis gives rise to functional ISCs, but is also a source for loss of heterozygosity
During mitosis, spindle assembly ensures fidelity of chromosome segregation into each daughter cell. As such, heterozygous cells remain heterozygous. However, it has been shown that chromosomes can be randomly segregated into daughter cells during amitosis in ciliates (Prescott, 1994). In this case, a heterozygous polyploid cell could give rise to two daughter cells homozygous for one or more chromosomes. We took advantage of this property of amitosis in two ways. First, we hypothesized that random segregation of homologous chromosomes into each daughter cell could be used as lineage-analysis to directly determine if amitosis gives rise to ISCs. Second, we sought to probe the possible deleterious consequence amitosis may have in a fly carrying a heterozygous mutation.
We genetically probed for amitosis-induced random homologous chromosome segregation as follows. Flies of the genotype tub-GAL4, UAS-dsRed (tub>dsRed); ; +/TM6B, tubP-GAL80 were subjected to our starvation assay. In these flies, tub-GAL80 represses tub>dsRed expression. In the event of mitotic division or direct dedifferentiation of a diploid cell to an ISC, the resulting ISC should remain heterozygous on the third chromosome (+/TM6B, tubP-GAL80) (Figure 5A), and tub>dsRed expression will remain repressed by tub-GAL80. However, upon amitosis, a fraction of the resulting ISCs may inherit two copies of the wild-type third chromosome (+/+), restoring tub>dsRed expression and giving rise to a positively-labeled ISC clone (Figure 5A). Indeed, flies that were starved and re-fed contained an average of 6 ± 4 ISC clones with an average of 2 ± 2 cells per clone, per PMG (Figure 5C–D) (n = 10), supporting the idea that amitotic events give rise to new ISCs upon re-feeding. ISC clones contained all differentiated cell types (Figure 5C), demonstrating that ploidy reduction can result in fully functional, multipotent ISCs. In contrast, control flies contained none to rare positively-labeled clones (1 ± 1 ISC clones per PMG, with an average of 4 ± 2 cells per clone) (Figure 5B, D) (n = 10), presumably resulting from rare amitotic events occurring during other phases of ISC number expansion (Figure 6B).
Figure 5. Amitosis Gives Rise to New, Functional ISCs, but Can Also Lead to Loss of Heterozygosity.
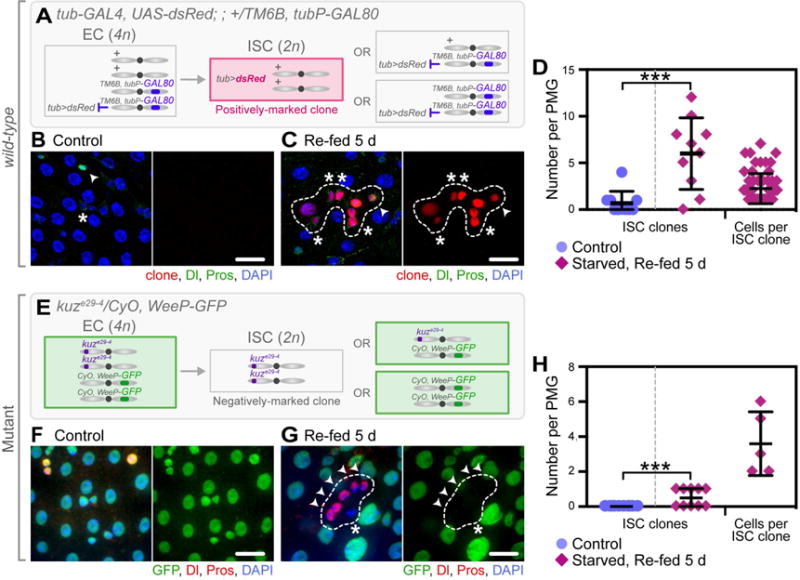
(A, E) Schematics of proposed amitosis-driven random homologous chromosome segregation per genotype, leading to either a positively-marked (A) or a negatively-marked (E) clone.
(B–C) Positively-marked ISC clones (third chromosome, +/+; loss of TM6B, tubP-GAL80) were observed in starved and re-fed flies (C, dashed outline), but rarely in controls (B) (ISCs, asterisk; ees, arrow head). Scale bar = 20 μm.
(D) Quantification of the number of wild-type positively-marked ISC clones and number of cells per clone, per PMG. n = 10. (***) denotes p < 0.001.
(F–G) Negatively-marked kuzbanian (kuz)-mutant ISC clones (second chromosome, kuze29-4/kuze29-4; loss of CyO, WeeP-GFP) were observed in starved and re-fed animals (G, dashed outline), but not in controls (F) (ISCs, asterisk; ees, arrow head). Scale bar = 20 μm.
(H) Quantification of the number of kuz negatively-marked mutant clones and number of cells per clone, per PMG. n = 10. (**) denotes p < 0.01.
(A–D) tub>dsRed; ; +/TM6B, tubP-GAL80, Tb
(E–H) ; kuze29-4/CyO, WeeP-GFP;
Figure 6. Amitosis Occurs in Response to ISC Dysfunction.
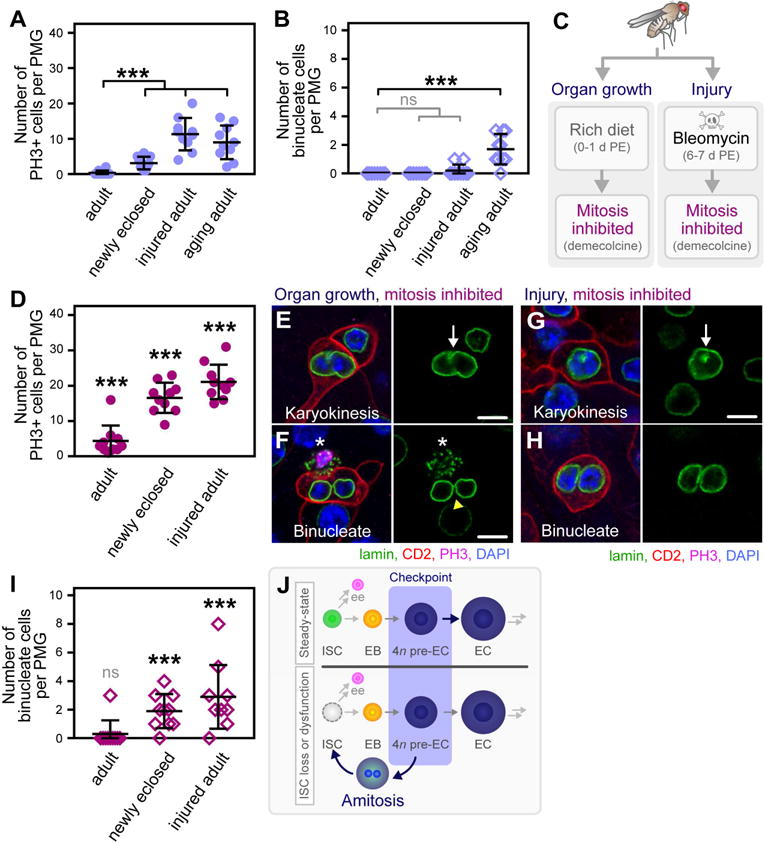
(A) Quantification of the number of PH3+ cells per PMG. n = 10. (***) denotes p < 0.001.
(B) Quantification of the number of binucleate amitotic cells per PMG. n = 10. (***) denotes p < 0.001.
(C) Assays used to inhibit mitosis, rendering ISCs dysfunctional during two phases of ISC proliferative demand: midgut growth in newly eclosed flies (left panel) and injury of the adult fly midgut (right panel).
(D) Quantification of the number of PH3+ cells per PMG in flies where mitosis is inhibited by demecolcine treatment. n = 10. (***) denotes p < 0.001.
(E–H) Images of representative amitotic cells in newly eclosed flies (E–F) and injured adult flies
(G–H) in which mitosis was inhibited. Scale bar = 10 μm. See also Figure S5A–B, D–E.
(I) Quantification of the number of binucleate amitotic cells per PMG. n = 10. (***) denotes p < 0.001.
(J) Model of amitosis as an alternate source of new ISCs.
(A–I) NRE-lacZ; esg>CD2;
As amitosis resulted in loss of heterozygosity in wild-type flies, we presumed that homozygous ISC mutant clones could be generated in starved and re-fed flies harboring a heterozygous mutation. To test this, we subjected flies heterozygous for a mutation in kuzbanian (kuze29-4/CyO, WeeP-GFP), an ADAM family metalloprotease involved in the cleavage of Notch (Lieber et al., 2002), to our starvation assay. Again, upon amitosis, a fraction of the resulting ISCs would inherit two copies of the kuze29-4 mutation on the second chromosome (kuze29-4/kuze29-4), resulting in a negatively-labeled ISC clone with a Notch-mutant phenotype (Figure 5E). Indeed, starved and re-fed flies contained an average of 1 ± 1 negatively-labeled mutant clones per PMG containing a single ISC and cluster of Pros+ ee cells (Figure G–H) (n = 10), a Notch-mutant phenotype previously reported in the Drosophila midgut (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006), and consistent with the loss of kuz-mediated S2 cleavage (Pan and Rubin, 1997). In control animals, negatively-labeled mutant clones were not observed (Figure 5F, H) (n = 10).
Amitosis occurs in response to ISC dysfunction during periods of proliferative demand
We hypothesized that, akin to physical loss, functional saturation or functional loss of the ISC during periods of increased demand may serve as an initiating factor of amitosis. Three additional periods of ISC number increases have been established. First, a developmental increase in ISC number, governed by symmetric ISC divisions, occurs in response to feeding and organ growth in newly eclosed flies (O’Brien et al., 2011). Second, a transient increase in ISC number is invoked by injury of the midgut and the ensuing proliferative demand necessary to repair the epithelium (Chatterjee and Ip, 2009; Choi et al., 2008a). Finally, an increase in ISC number has been reported in aging flies, due to the dysregulation of tissue homeostasis (Biteau et al., 2008; Choi et al., 2008a; Choi et al., 2008b). We sought to assess the generality by which amitosis is initiated throughout the lifetime of the fly in response to the conditional state of the ISC during these periods of ISC number increase.
We first quantified the number of PH3+ cells per PMG in newly eclosed (1 day PE), injured (7 days PE), and aging (26 days PE) flies relative to control adult flies (7 days PE). Consistent with previous reports, the rate of proliferation significantly increased during all three phases of ISC number expansion and proliferative demand (Figure 6A). We next quantified the number of binucleate cells, an indicator of amitosis, per PMG of flies in each condition. While not observed in control adult or newly eclosed flies, amitosis was observed at a low frequency in injured flies and significantly in aging flies (Figure 6B), suggesting that amitosis can be initiated by functional saturation or functional loss of the ISC, respectively.
To determine if amitosis initiates in response to ISC dyfunction, we sought to chemically inhibit mitosis by oral administration of demecolcine, a microtubule depolymerizing agent, in the following two assays. In the first assay, newly eclosed flies were fed a rich diet from 0–24 hours PE to enable organ growth and subsequent demand for an expansion of the ISC pool. Flies were then transferred to a rich diet containing 32 μg/mL demecolcine (Rebollo et al., 2004) (Figure 6C) for 16 hours. In the second assay, flies were fed a rich diet from 0–6 days PE and then transferred to a rich diet containing 25 μg/mL bleomycin, a damaging agent. After 24 hours of feeding, flies were removed from the damaging agent and allowed to recover for 16 hours on a rich diet containing demecolcine (Figure 6C).
Demecolcine effectively inhibited mitosis, as the number of PH3+ cells per PMG significantly increased in demecolcine-treated flies relative to non-treated cohorts (Figure 6D, treated, relative to Figure 6A, non-treated). Amitosis was not significantly observed in demecolcine-treated uninjured 7-day-old adults, in which an expansion of ISC number is not normally required (Figure 6I). Consistent with our hypothesis, blocking ISC mitoses in newly eclosed or injured flies led to the initiation of amitosis (Figure 6E–H). The number of binucleate cells increased from an average of zero (Figure 6B) to 2 ± 1 (Figure 6I) in newly eclosed flies and from an average of 0.2 ± 0.3 (Figure 6B) to 3 ± 2 (Figure 6I) in injured flies. Notably, amitosis only occurred in areas where all ISCs were blocked in mitosis (Figure S5A–B, asterisks), supporting that amitosis is initiated in response to ISC dysfunction during proliferative demand. By contrast, amitosis occurred in the presence of functional, PH3+, ISCs (Figure S5C, asterisk) in aging flies, pointing to dysregulation in the initiation of this process with age. Finally, rare tri-nucleate amitotic cells were observed when blocking ISC mitoses in newly eclosed or injured flies, suggesting that the process of amitosis can be error-prone (Figure S5D–E). Together, these data establish that amitosis is initiated in response to either the physical or functional loss of the ISC during periods of tissue demand and ISC number increases throughout the lifetime of the fly.
Discussion
Drosophila intestinal stem cell number is regulated by multiple mechanisms
Replenishment of differentiated cell types by stem cells has been an intensive focus of tissue homeostasis (Fuchs and Chen, 2013). However, homeostatic regulation of stem cell number, the very foundation of tissue homeostasis, remains poorly understood (Korinek et al., 1998). Increases in the number of stem cells can either be permanent, following stem cell loss, or transient, in response to increased tissue demand. Symmetric division of existing stem cells is viewed as the classical mode of stem cell expansion following loss, injury, or disease (Morrison and Kimble, 2006). However in the event of widespread stem cell loss or excessive demand for new cells, alternate mechanisms for generating new stem cells likely exist to satisfy short and long term tissue needs. Indeed, experiments involving genetic ablation of resident stem cells in a wide host of tissues and species, reveal that in the absence of stem cells, early stem cell daughter cells directly convert, by a process known as dedifferentiation, to new functional stem cells (Brawley and Matunis, 2004; Kai and Spradling, 2004; Nakagawa et al., 2007; Tata et al., 2013; Tetteh et al., 2016; van Es et al., 2012), underscoring the ability of tissues to employ alternative strategies to maintain stem cell number and tissue homeostasis.
Our data uncover an alternate mode of new stem cell production by a specialized process of depolyploidization known as amitosis. Following severe ISC loss, an increase in ISC number can be achieved in the absence of ISC mitoses. During this time, polyploid cells in the enterocyte lineage undergo amitosis to fully functional ISCs. Amitosis is also initiated in aging flies and in response to blocking the ISC’s ability to symmetrically divide in newly eclosed or injured adult flies. Together, these data underscore amitosis as a general alternative mode of stem cell replacement in the intestinal epithelium.
Means of achieving polyploidy and ploidy reduction
Two modes of ploidy reduction have been reported. Reductive mitotic division has been described in the mammalian liver (Duncan et al., 2009). Amitosis has been described in a host of cell types from primitive ciliates (Prescott, 1994) to plants (Miller, 1980), fish (Wang et al., 2010), birds (Patterson, 1908), skunk (Isakova and Mead, 2004), rodents (Kuhn et al., 1991; Zybina and Zybina, 2008), and humans (Magalhaes et al., 1991). In the absence of diploid progenitor cells, why might the Drosophila midgut replace ISCs through an amitotic division rather than a mitotic one? The answer may lie in how polyploidy arises in this tissue. Cells can achieve polyploidy by a variety of means including stalled mitoses, in which the mitotic program is active, or variations of endomitosis or endocycling, in which the mitotic program is partially or completely inactive. In the mammalian liver, hepatocytes are thought to achieve polyploidy through a stalled mitosis due to failed cytokinesis (Margall-Ducos et al., 2007), and subsequently undergo ploidy reduction through a reductive mitotic division (Duncan et al., 2010). In contrast, rodent trophoblast cells achieve polyploidy by exiting the mitotic program and entering the endocycle (MacAuley et al., 1998), subsequently undergoing ploidy reduction through amitosis. Along these lines, while polyploid cells in the Drosophila rectum can exit the endocycle and reenter the mitotic program, polyploid mitotic divisions are not reductive in this system (Fox et al., 2010). From work in these cell types, we would speculate that certain means of achieving polyploidy may direct the means of achieving depolyploidization. As is the case with mammalian trophoblasts, cells in the Drosophila enterocyte lineage become polyploid by entering the endocycle. Because the genetics underlying amitosis remain an enigma, nuances into how enterocytes achieve polyploidy may provide insight into signaling that also initiates the “amitotic cell cycle”.
If the initiation of amitosis is linked to the endocycle, can cells of any ploidy greater than 4n undergo amitosis in the midgut? We hypothesize that amitotic cells are at a specific stage within the lineage, just exiting the first endocycle, at which point they are still capable of expressing the stemness factor escargot. At this point, the 4n cell, presumably at a programmed checkpoint, can either progress to terminal differentiation or can immediately revert to diploid progenitors, depending on the needs of the tissue (Figure 6J). As such, we do not expect that terminally differentiated, highly polyploid (8n to 16n) cells would be capable of this process. Consistent with this hypothesis, it has been reported that ISC replacement is not observed upon genetic ablation of all cells expressing esg-GAL4 (Lu and Li, 2015), which includes progenitor cells, a sub-population of 4n ECs and the 4n amitotic cells observed herein. Finally, polyploid genomes are often not 2n replicates of the diploid genome, as certain genomic regions are underreplicated during endocycling (Gall et al., 1971). As underreplication often occurs in high ploidy cells with polytene chromosomes, such as the Drosophila salivary gland, or mammalian giant trophoblast cells (Schoenfelder and Fox, 2015), the first endocycle may give a cellular product with the most fidelity.
Amitosis as a conserved biological and pathological mechanism of cell propagation
Since Remak’s initial account of amitosis in 1841 (Remak, 1841), its occurrence has been found across phyla. Yet, the functional significance of this distinctive form of cell division remains largely unexplored. Our data demonstrate that amitosis is an integral component of stem cell homeostasis under physiological stress in the high-turnover tissue of the Drosophila midgut. Likewise, amitosis may also bear functional significance in maintaining homeostasis under physiological and pathological conditions in other tissues containing populations of polyploid cells, in which amitosis has not been previously explored. While polyploidy was once thought to be a rarity in vertebrates, polyploid cells have been identified in the uterus, placenta, myocardium, bone marrow, and perhaps most notably, the liver (Zielke et al., 2013). Although hepatocytes have been shown to undergo reductive mitotic divisions, it is worth noting that amitosis has been described in both the damaged rodent liver and human liver cancer tissue (Yiquan and Binkung, 1986), suggesting that amitosis may play a role in liver homeostasis.
In addition to developmentally-programmed polyploidy, polyploidy is often associated with tumorigenesis (Fox and Duronio, 2013). Cancer cells utilize endoreplication as a means to escape mitotic catastrophe, and subsequently undergo ploidy reduction to produce highly proliferative, aggressive cells. Following γ-irradiation, tetraploidization (Davoli and de Lange, 2011) has been reported in Barrett’s esophagus (Galipeau et al., 1996), prostate cancer (Montgomery et al., 1990) and colon adenomas (Park et al., 2011). Ploidy reversion from the tetraploid state results in two outcomes: aneuploidy (Davoli and de Lange, 2011) or depolyploidization, with all of the features of amitosis, to give rare but highly proliferative diploid cells that are morphologically indistinguishable from other diploid cells within a tumor (Erenpreisa et al., 2005a; Erenpreisa et al., 2005b; Erenpreisa et al., 2000).
As suggested in cancerous tissue, the process of amitosis bears ramifications. Our results demonstrate a beneficial outcome of amitosis – the generation of new, functional stem cells. However, our results also show a potential danger in the use of this mechanism, loss of heterozygosity and generation of cells homozygous for deleterious mutations. Mutation-driven endocycling leading to polyploid enterocytes has been identified in the human gastrointestinal tract (Clarke et al., 2007). However, whether these mutant enterocytes are capable of giving rise to new stem cells by amitosis under an array of physiological contexts has not been explored. Our findings raise the possibility that endocycling followed by amitosis may function as a cancer-initiating factor. Such errors, even at a low frequency, could have profound implications on tissue homeostasis and initiation of cancer.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Benjamin Ohlstein (bo2160@cumc.columbia.edu).
Experimental Model and Subject Details
Drosophila stocks and their use in this study
NRE-lacZ; esg-GAL4, UAS-GFP/CyO; – used to quantify the number of each cell type in the PMG (Figure 1, Figure S1, Figure S2, Figure 2A, F, and Figure 3A–B).
NRE-lacZ; esg-GAL4, UAS-CD2, vkg-GFP/CyO; – used to visualize the delamination of ISCs in starved flies (Figure 1).
y, w, hsFLP, tub-GAL4, UAS-nuc-GFP; ; FRT82B tubGAL80 (gift from G. Struhl) and NRE-lacZ;; FRT82B ry506/TM2 – used for MARCM clonal analysis of ISC loss following starvation (Figure 1).
w; ; His2Av-mRFP [w+], FRT 2A (BDSC: 34498) obtained from the Bloomington Drosophila Stock Center and y, w, hsFLP; ; FRT2A, His-GFP (Guo and Wang, 2009) – used for twin-spot clonal analysis (Figure 2B–E).
NRE-lacZ; esg-GAL4, UAS-CD2/CyO; – used to detect amitotic cells and quantify the number of binucleate cells (Figure 3, Figure S3C–F, Figure 6 and Figure S5).
UAS-anillin-GFP; Sp/CyO; P/TM3 and NRE-lacZ; esg-GAL4, UAS-CD2/CyO; – used to visualize the contractile ring (Figure S3D–E).
; Myo1A-GAL4, UAS-GFP/CyO; – used to test the specificity of Myo1A-GAL4 in the PMG (Figure S4A–B).
y w122; esg-GFP/CyO; 10× UAS-IVS-mCD8∷RFP/TM6B, Hu, Tb and ; Mex-GAL4; (gift from G. Thomas)– used to test the specificity of Mex-GAL4 in the PMG (Figure S4C).
NRE-lacZ; ; 28E03-GAL4, UAS-CD8∷GFP (made with: ; ; P{GMR28E03-GAL4}attP2 (BDSC: 45546) obtained from the Bloomington Drosophila Stock Center (Jenett et al., 2012)) – used to test the specificity of 28E03-GAL4 in the PMG (Figure S4D).
w; pBID-UASC-FLP/CyO; 28E03-GAL4/TM6B, Hu, Tb (made with: ; pBID-UASC-FLP attP16/SM6a; (transgenic line created and described in Method Details) and w; Act5C FRT draf+ FRT lacZ/CyO; NRE-GFP/TM2 (made with: ; Act5C FRT draf+ FRT lacZ/CyO; TM2/TM6B, Hu, Tb (gift from L. Johnston)) – used for EC-lineage clonal analysis (Figure 4 and Figure S4F–G).
w; Act5C FRT draf+ FRT lacZ/CyO; 28E03-GAL4/TM6B, Hu, Tb and w; esg-GFP/CyO; pBID-UASC-FLP/TM6B, Hu, Tb – used to determine the percent of amitotic events represented by EC-lineage clonal analysis (Figure 4F–G).
tub-GAL4, UAS-dsRed/FM7; ; MKRS/TM6B, tubP-GAL80, Tb (made with: w*; ; TM6B, P{w+mC=tubP-GAL80}OV3, Tb/TM3, Sb (BDSC: 9490) obtained from the Bloomington Drosophila Stock Center) and Oregon R. – used to test for random homologous chromosome segregation in wild-type flies (Figure 5A–D).
y w; kuze29-4/CyO, WeeP-GFP; (made with: ; kuze29-4/CyO; (BDSC: 5804) obtained from the Bloomington Drosophila Stock Center) – used to test for random homologous chromosome segregation in the presence of a heterozygous mutation (Figure 5E–H).
Drosophila husbandry
Female flies were used in all experiments. The age and rearing of flies used are noted within the text, figures, legends, and Method Details.
All fly lines were maintained on standard cornmeal-molasses medium (58.3 g cornmeal, 26.5 g yeast, 30 mL molasses, 6.0 g agar, 965 mL water, 1.38 g tegosept in 8.3 mL ethanol, and 5 mL propionic acid) at room temperature, unless otherwise indicated.
Method Details
Generation of pBID-UASC-FLP transgenic line
To eliminate non-specific UAS expression in our EC-lineage analysis experiments, a pBID-UASC-FLP transgenic line was generated by cloning flippase (FLP) (Pfeiffer et al., 2010) (amplified using the following primer sequences: AACTTAAAAAAAAAAATCAAAATGCCGCAGTTTGATATCC and CTATTAAATACGGCGATTGATGTAGGAGCTC) into a Gateway vector containing gypsy insulators flanking the insertion site (pBID-UASC-G, Addgene plasmid #35202) (Wang et al., 2012). Transgenic flies were generated by standard procedure (GenetiVision Corporation).
Assays used to induce amitosis
For all assays below, female and male flies were collected 0–2 hours post-eclosion at a 2:1 ratio and fed a standard cornmeal-molasses medium supplemented with live yeast (rich diet) for the time noted. Males were then discarded and well-fed female siblings were separated into control and experimental cohorts.
Starvation Assay
Female flies, maintained on a rich diet from 0–3 days PE, were separated into two cohorts. Cohort 1 was kept on a rich diet and served as controls. Cohort 2 was transferred to a vial containing a cellulose acetate plug (BuzzPlugs™, Fisher Scientific) soaked in tap water. Cohort 2 flies were kept in starvation conditions until midguts from dissected flies had shrunken to approximately half the length, in most cases 2 days. Midguts were dissected at this point for analysis of starvation phenotypes.
To assess recovery following starvation, starved flies were transferred to a vial of standard cornmeal-molasses supplemented with live yeast paste, and midguts were dissected over the time-course of re-feeding, as noted in the text.
Demecolcine feeding assay
Female flies were maintained on a rich diet for either 1 day or 7 days PE and then separated into two cohorts. Cohort 1 was transferred to a vial with yeast paste and a 3 × 3.5 cm piece of chromatography paper soaked with 5% sucrose in water, serving as controls. Cohort 2 was transferred to a vial with yeast paste containing 32 μg/mL demecolcine (diluted from a 25 mg/mL stock solution in ethanol) and a 3 × 3.5 cm piece of chromatography paper soaked with 5% sucrose and 32 μg/mL demecolcine in water.
To visualize feeding, blue food coloring was added to all media for both controls and experiments at a final concentration of 1%. Flies that had not fed within 2 hours of transfer (clear abdomen) were removed from the vials.
Demecolcine was administered for 16 hours, upon which midguts were dissected and analyzed as noted in the text.
Bleomycin feeding assay
Female flies were maintained on a rich diet for 6 days PE and then separated into two cohorts. Cohort 1 was transferred to a vial with yeast paste and a 3 × 3.5 cm piece of chromatography paper soaked with 5% sucrose in water, serving as controls. Cohort 2 was transferred to a vial with yeast paste containing 25 μg/mL bleomycin sulfate from Streptomyces verticillus (diluted from a 10 mg/mL stock solution in DMSO) and a 3 × 3.5 cm piece of chromatography paper soaked with 5% sucrose and 25 μg/mL bleomycin in water.
To visualize feeding, blue food coloring was added to all media for both controls and experiments at a final concentration of 1%. Flies that had not fed within 2 hours of transfer (clear abdomen) were removed from the vials.
Bleomycin was administered for 24 hours. Flies were then allowed to recover by transferring to either a vial of standard cornmeal-molasses supplemented with yeast paste for 16 hours or a vial with yeast paste containing 32 μg/mL demecolcine and a 3 × 3.5 cm piece of chromatography paper soaked with 5% sucrose and 32 μg/mL demecolcine in water for 16 hours. Midguts were then dissected and analyzed as noted in the text.
Lineage analysis
MARCM clonal analysis
To corroborate loss of ISCs in flies subjected to our starvation assay, we generated ISC clones using the Mosaic Analysis with a Repressible Cell Marker (MARCM) system (Lee and Luo, 2001) as follows. Flies of the genotype y, w, hsFLP, tub-GAL4, UAS-nuc-GFP; ; FRT82B tubGAL80 and NRE-lacZ; ; FRT82B ry506/TM2 were crossed. Females of the genotype y, w, hsFLP, tub-GAL4, UAS-nuc-GFP/NRE-lacZ; ; FRT82B tubGAL80/FRT82B ry506 were collected upon eclosion and maintained on a rich diet from 0–3 days PE.
Clones were induced in 3-day-old flies, 8 hours prior to starvation, by heat-shock at 37 °C for 45 minutes. Flies were then separated into control and experimental cohorts and subjected to the starvation assay outlined above. The number of clones per R4b-c was quantified after 2 days of starvation (2 days after clone induction).
Twin-spot lineage analysis
To determine the contribution of symmetric ISC divisions to ISC replacement, we used the following twin-spot lineage analysis. Flies of the genotype w; ; His2Av-mRFP [w+], FRT 2A and y, w, hsFLP; ; FRT2A, His-GFP were crossed. Females of the genotype y, w, hsFLP/w; ; His2Av-mRFP [w+], FRT2A/FRT2A, His-GFP were collected upon eclosion, maintained on a rich diet from 0–3 days PE and separated into control and experimental cohorts subjected to the starvation assay outlined above.
ISC clones were generated by FLP/FRT-mediated mitotic recombination upon heat-shock at different points of re-feeding and in age-matched controls, as noted in the text. All heat-shocks were conducted at 37 °C for 45 minutes. An asymmetric outcome is defined as an ISC clone and a single differentiated daughter cell of different colors. In contrast, a symmetric outcome is defined as two adjacent ISC clones of different colors. The number of asymmetric and symmetric clones were quantified.
EC-lineage analysis
To determine the contribution of cells in the EC-lineage to ISC replacement, we used lineage analysis based on a flipout system (Theodosiou and Xu, 1998), in which a stop cassette flanked by FRT sites is excised upon expression of flippase (FLP) using the GAL4/UAS system. Using the GAL4 driver 28E03-GAL4, we analyzed the fate of cells in the EC-lineage as follows. Flies of the genotype w; pBID-UASC-FLP/CyO; 28E03-GAL4/TM6B, Hu, Tb and w; Act5C FRT draf+ FRT lacZ/CyO; NRE-GFP/TM2 were crossed. Females of the genotype w; pBID-UASC-FLP/Act5C FRT draf+ FRT lacZ; 28E03-GAL4/NRE-GFP were collected upon eclosion, maintained on a rich diet from 0–3 days PE, and separated into control and experimental cohorts subjected to the starvation assay outlined above.
Flies were dissected and analyzed after starvation and after 5, 7, and 11 days of re-feeding. Lineage-labeled ISCs (ISC clones) were defined as Act5C-lacZ positive, NRE-GFP negative, Pros negative cells. The number of ISC clones per PMG, as well as the number of cells per ISC clone were quantified.
Amitotic events represented by EC-lineage analysis
To eliminate non-specific UAS expression, we used our insulated pBID-UASC-FLP transgenic line for EC-lineage analysis. While use of this flippase ensures stringent lineage analysis, it is with the tradeoff of low frequency in the labeling of cells. Therefore, to determine the percent of potential amitotic events represented by our lineage analysis, we used the following approach.
Flies of the genotype w; Act5C FRT draf+ FRT lacZ/CyO; 28E03-GAL4/TM6B, Hu, Tb and w; esg-GFP/CyO; pBID-UASC-FLP/TM6B, Hu, Tb were crossed. Females of the genotype w; Act5C FRT draf+ FRT lacZ/esg-GFP; 28E03-GAL4/pBID-UASC-FLP were collected upon eclosion, maintained on a rich diet from 0–3 days PE, and separated into control and experimental cohorts subjected to the starvation assay outlined above.
Flies were dissected and analyzed after 16 hours of re-feeding. The total number of Esg+ 4n cells as well as the number of lineage-labeled Esg+ 4n cells were quantified. The percent lineage-labeled of total Esg+ 4n cells was then calculated and this percentage was used as a scaling factor to estimate the number of amitotic events represented by the number of lineage-labeled ISCs originating from the EC-lineage.
Random segregation lineage analysis
We genetically probed for amitosis-induced random homologous chromosome segregation as follows. Flies of the genotype tub-GAL4, UAS-dsRed/FM7; ; MKRS/TM6B, tubP-GAL80, Tb and Oregon R. were crossed. Females of the genotype +/tub-GAL4, UAS-dsRed; ; +/TM6B, tubP-GAL80, Tb were collected upon eclosion, maintained on a rich diet from 0–3 days PE, and separated into control and experimental cohorts subjected to the starvation assay outlined above.
To determine if loss of heterozygosity can be deleterious in flies harboring a mutation, we repeated the above experiment by subjecting females of the genotype kuze29-4/CyO, WeeP-GFP (Clyne et al., 2003) to our starvation assay.
Flies in both the wild-type and mutant experiments were dissected and analyzed after 5 days of re-feeding. The number of ISC clones per PMG was quantified.
It is important to note that when genetically testing for amitosis-induced random homologous chromosome segregation, we used tubP-GAL80 or WeeP-GFP within a balancer chromosome (TM6B or CyO, respectively) to rule out the possibility of somatic recombination.
Dissection and immunohistochemistry
Dissection
Prior to all dissections, flies were anesthetized with CO2. Midguts were dissected in 2× gut buffer (200 mM glutamic acid, 50 mM KCl, 40 mM MgSO4, 4 mM NaH2PO4, 4 mM Na2HPO4 dibasic, and 2 mM MgCl2) (Ohlstein and Spradling, 2006) and immediately transferred to fixation solution (4% formaldehyde in 1× gut buffer) for 2 hours at room temperature.
Immunohistochemistry
All washes were conducted using 1× PBT (1× PBS and 0.5% Triton X-100) containing 0.5% BSA. Antisera were diluted in 1× PBT containing 0.5% BSA and midguts were incubated in antisera at overnight at 4 °C. Secondary antibodies were diluted in 1× PBT containing 0.5% BSA and incubated for 1 hour at room temperature. Midguts were counterstained with 1 μg/mL 4,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich) in 1× PBT and mounted in Vectashield mounting medium (Vector Laboratories). Samples were counterstained with DAPI in 1× PBT containing 0.5% BSA for 5 minutes. Rhodamine phalloidin (1:4,000) was added with secondary antibodies to visualize actin. All midguts were mounted in Vectashield mounting medium.
Anti-sera were used at the following final concentrations:
| Chicken polyclonal anti-GFP | 1:2,000 |
| Rabbit polyclonal anti-β-galactosidase | 1:10,000 |
| Mouse monoclonal anti-rat CD2 | 1:2,000 |
| Mouse monoclonal anti-Prospero | 1:200 |
| Rabbit polyclonal anti-RFP | 1:10,000 |
| Mouse monoclonal anti-lamin DmO | 1:200 |
| Rat monoclonal anti-alpha-tubulin | 1:500 |
| Rat monoclonal anti-cadherin, DE-, extracellular domain | 1:20 |
| Rat monoclonal anti-Delta, extracellular domain | 1:200 |
| Rabbit polyclonal anti-phospho-Histone H3 (Ser10) | 1:2,000 |
| Rabbit polyclonal anti-GFP | 1:2,000 |
All secondary antibodies were used at a final concentration of 1:2000.
Image acquisition and processing
Image acquisition
Images were obtained using a either a DSU spinning disc confocal microscope (Olympus) equipped with UPLFLN ×20, ×40 oil immersion and ×60 oil-immersion objectives and 512×512 EM-CCD camera (ImagEM Enhanced C9100-13, Hamamatsu Photonics) or a Leica TCS SP5 II confocal microscope (Leica Microsystems) equipped with an ×40 oil-immersion and ×63 oil-immersion objective, 405 nm diode, 458, 476, 488, 496 and 514 nm Ar, 543 nm HeNe and 633 nm HeNe lasers and digital zoom. Images were acquired using SlideBook (version 4.2, Intelligent Imaging Innovations) or Leica Application Suite (Leica Microsystems) software.
For all images, Z-stacks through the midgut were acquired every 0.2–1 μm through the tissue.
Image processing
Images acquired in SlideBook and Leica Application Suite were processed using Fiji software. Composite images of Z-stacks in each channel were generated by summing slices. Composites of each channel were then false-colored according to fluorophore using Photoshop CS3 or CC (Adobe) and channels were overlayed as individual layers. For whole midgut images, each Z-composite at different X-Y positions along the midgut were manually stitched together in Photoshop. All figures were then compiled in Illustrator CS3 or CC (Adobe) using final merged images from Photoshop.
Quantification and Statistical Analysis
For all quantifications, n represents the number of midguts analyzed, error bars represent standard deviation, and statistical significance was determined using the Welch’s T-test with equal sample size and unequal variance, unless otherwise noted, and expressed as P-values. (*) denotes p < 0.05, (**) denotes p < 0.01, (***) denotes p < 0.001 and (ns) denotes values whose difference was not significant.
With the exception of total cell number counts per PMG (Figure 1I and Figure 2A), all graphs are scatter plots of raw data to present the full distribution of values observed, with averages reported as a black line.
Quantification of total cell numbers per PMG
Z-stack images through one-half the width of the PMG were acquired at X-Y positions along the length of the PMG from R4a to R5. Maximum projections of each Z-stack were obtained using the Stack Arithmetic drop-in of MetaMorph software. Total number of each cell type was counted using the Manually Count Objects drop-in of MetaMorph and exported to Microscoft Excel and Igor Pro for further statistical analysis. All numbers reported are two times the number counted per one-half of the PMG imaged in the Z-direction.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken polyclonal anti-GFP | Abcam | Cat# ab13970; RRID:AB_300798 |
| Rabbit polyclonal anti-β-galactosidase | MP Biomedicals | Cat# 0855978; RRID:AB_2334737 |
| Mouse monoclonal anti-rat CD2 | Bio-Rad (Formerly AbD Serotec) | Cat# MCA154GA; RRID: AB_566608 |
| Mouse monoclonal anti-Prospero | DSHB | Cat# MR1A; RRID: AB_528440 |
| Rabbit polyclonal anti-RFP | MBL International | Cat# PM005; RRID:AB_591279 |
| Mouse monoclonal anti-lamin DmO | DSHB | Cat# ADL84.12; RRID: AB_528338 |
| Rat monoclonal anti-alpha-tubulin | Bio-Rad (Formerly AbD Serotec) | Cat# MCA78G; RRID: AB_325005 |
| Rat monoclonal anti-cadherin, DE- (extracellular domain) | DSHB | Cat# DCAD2; RRID: AB_528120 |
| Mouse monoclonal anti-Delta, extracellular domain | DSHB | Cat# C594.9B; RRID:AB_528194 |
| Rabbit polyclonal anti-phospho-Histone H3 (Ser10) | Millipore | Cat# 06-570; RRID: AB_310177 |
| Rabbit polyclonal anti-GFP | Torrey Pines Biolabs | Cat# TP401 071519; RRID: AB_10013661 |
| Goat anti-chicken IgY (H+L) secondary antibody, Alexa Fluor® 488 | Abcam | Cat# ab7114; RRID: AB_955348 |
| Goat anti-chicken IgY (H+L) secondary antibody, Alexa Fluor® 488 | ThermoFisher Scientific | Cat# A11039; RRID: AB_2534096 |
| Donkey anti-rabbit IgG (H+L) secondary antibody, Alexa Fluor® 488 | ThermoFisher Scientific | Cat# A21206; RRID:AB_2535792 |
| Donkey anti-rabbit IgG (H+L) secondary antibody, Alexa Fluor® 546 | ThermoFisher Scientific | Cat# A10040; RRID: AB_2534016 |
| Goat anti-rabbit IgG (H+L) secondary antibody, Alexa Fluor® 633 | ThermoFisher Scientific | Cat# A21070; RRID: AB_2535731 |
| Donkey anti-mouse IgG (H+L) secondary antibody, Alexa Fluor® 555 | ThermoFisher Scientific | Cat# A31570; RRID: AB_2536180 |
| Goat anti-mouse IgG (H+L) secondary antibody, Alexa Fluor® 633 | ThermoFisher Scientific | Cat# A21050; RRID: AB_2535718 |
| Goat anti-mouse IgG2a secondary antibody, Alexa Fluor® 555 | ThermoFisher Scientific | Cat# A21137; RRID: AB_2535776 |
| Goat anti-mouse IgG1 secondary antibody, Alexa Fluor® 488 | ThermoFisher Scientific | Cat# A21121; RRID: AB_2535764 |
| Goat anti-mouse IgG1 secondary antibody, Alexa Fluor® 647 | ThermoFisher Scientific | Cat# A21240; RRID: AB_2535809 |
| Goat anti-rat IgG (H+L) secondary antibody, Alexa Fluor® 633 | ThermoFisher Scientific | Cat# A21094; RRID: AB_2535749 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Demecolcine ≥98 % (HPLC) | Sigma | Cat# D7385; CAS: 477-30-5 |
| Bleomycin (sulfate) ≥95% | Cayman Chemical Company | Cat# 13877; CAS: 9041-93-4 |
| Sucrose, ultra-pure grade | LabScientific | Cat# 0335; CAS: 57-50-1 |
| Deep blue food coloring | Esco Foods | Cat# 34-400 |
| Pierce™ 16% Formaldehyde (w/v), methanol-free | ThermoFisher Scientific | Cat# 28908; CAS: 50-00-0 |
| Molecular Probes™ Rhodamine Phalloidin | Invitrogen | Cat# R415 |
| 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI) | Sigma | Cat# D9542; CAS: 28718-90-3 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: NRE-lacZ; ;TM2/TM6B, Hu, Tb | Sarah Bray; Furriols et al., 2001 | N/A |
| D. melanogaster: ; esg-GAL4, UAS-GFP; | Craig Micchelli; Micchelli et al., 2006 | N/A |
| D. melanogaster: NRE-lacZ; esg-GAL4, UAS-GFP/CyO; | This paper | N/A |
| D. melanogaster: w[*]; P{w[+mC]=UAS-CD2}5; | Bloomington Drosophila Stock Center | BDSC: 1284; FlyBase: FBst0001284 |
| D. melanogaster: ; vkg-GFP/CyO; | Jin Jiang; Tian et al., 2014 | N/A |
| D. melanogaster: NRE-lacZ; esg-GAL4, UAS-CD2, vkg-GFP/CyO; | This paper | N/A |
| D. melanogaster: y, w, hsFLP, tub-GAL4, UAS-nuc-GFP; ; FRT82B tubGAL80 | Gary Struhl | N/A |
| D. melanogaster: ; ; P{ry[+t7.2]=neoFRT}82B ry[506] | Bloomington Drosophila Stock Center | BDSC: 2035; FlyBase: FBst0002035 |
| D. melanogaster: NRE-lacZ; ; FRT82B ry[506]/TM2 | This paper | N/A |
| D. melanogaster: wcall net [1118]; ; P{w[+mC]=His2Av-mRFP1}III.1 P{w[+mW.hs]=FRT(w[hs])}2A | Bloomington Drosophila Stock Center | BDSC: 34498; FlyBase: FBst0034498 |
| D. melanogaster: y, w, hsFLP; ; FRT2A, His-GFP | Zheng Guo; Guo et al., 2009 | N/A |
| D. melanogaster: NRE-lacZ; esg-GAL4, UAS-CD2/CyO; | This paper | N/A |
| D. melanogaster: UAS-anillin-GFP; Sp/CyO; P/TM3 | Don Fox | N/A |
| D. melanogaster: ; Myo1A-GAL4, UAS-GFP/CyO; | Bruce Edgar; Jiang et al., 2009 | N/A |
| D. melanogaster: ; esg-GFP[P01986]; | Alan Spradling; Buszczak et al., 2007 | |
| D. melanogaster: w[*]; ; P{y[+t7.7] w[+mC]=10×UAS-IVS-mCD8∷RFP}attP2 | Bloomington Drosophila Stock Center | BDSC: 32218; FlyBase: FBst0032218 |
| D. melanogaster: y, w[122]; esg-GFP/CyO; 10× UAS-IVS-mCD8∷RFP/TM6B, Hu, Tb | This paper | N/A |
| D. melanogaster: P{w[+mC] GAL4∷Hsp70B [mex.P5′2.1]=mexGAL4} | Philips et al., 2006 | N/A |
| D. melanogaster: ; ; P{GMR28E03-GAL4}attP2 | Bloomington Drosophila Stock Center | BDSC: 45546; FlyBase: FBsf0000163290 |
| D. melanogaster: y[1] w[*]; Pin[Yt]/CyO; P{w[+mC]=UAS-mCD8∷GFP.L}LL6 | Bloomington Drosophila Stock Center | BDSC: 5130; FlyBase: FBst0005130 |
| D. melanogaster: NRE-lacZ; ; 28E03-GAL4, UAS-mCD8∷GFP | This paper | N/A |
| D. melanogaster: ; pBID-UASC-FLP attP16/SM6a; | This paper | N/A |
| D. melanogaster: w; pBID-UASC-FLP/CyO; 28E03-GAL4/TM6B, Hu, Tb | This paper | N/A |
| D. melanogaster: ; Act5C FRT draf+ FRT lacZ/CyO; TM2/TM6B, Hu, Tb | Laura Johnston | N/A |
| D. melanogaster: w[1118]; P{w[+m*]=NRE-EGFP.S}1 | Bloomington Drosophila Stock Center | BDSC: 30728; FlyBase: FBst0030728 |
| D. melanogaster: w; Act5C FRT draf+ FRT lacZ/CyO; NRE-EGFP/TM2 | This paper | N/A |
| D. melanogaster: w; Act5C FRT draf+ FRT lacZ/CyO; 28E03-GAL4/TM6B, Hu, Tb | This paper | N/A |
| D. melanogaster: w; esg-GFP/CyO; pBID-UASCFLP/TM6B, Hu, Tb | This paper | N/A |
| D. melanogaster: w[*]; ; TM6B, P{w[+mC]=tubPGAL80} OV3, Tb[1]/TM3, Sb[1] | Bloomington Drosophila Stock Center | BDSC: 9490; FlyBase: FBst0009490 |
| D. melanogaster: tub-GAL4, UAS-dsRed/FM7; ; MKRS/TM6B, tubP-GAL80, Tb | This paper | N/A |
| D. melanogaster: Oregon R. | Bloomington Drosophila Stock Center | BDSC: 2376; FlyBase: FBst0002376 |
| D. melanogaster: ; kuz[e29-4]/CyO; | Bloomington Drosophila Stock Center | BDSC: 5804; FlyBase: FBst0005804 |
| D. melanogaster: ; ln2LR, Gla, Bc/CyO, WeeP-GFP; | Brian McCabe | N/A |
| D. melanogaster: y w; kuz[e29-4]/CyO, WeeP-GFP | This paper | N/A |
| Oligonucleotides | ||
| Primer for cloning of flippase (FLP), forward: AACTTAAAAAAAAAAATCAAAATGCCGCAGTTTGATATCC | eurofins, Fisher Scientific | www.operon.com/fihersci |
| Primer for cloning of flippase (FLP), reverse: CTATTAAATACGGCGATTGATGTAGGAGCTC | eurofins, Fisher Scientific | www.operon.com/fishersci |
| Recombinant DNA | ||
| Plasmid: pBID-UASC-G | Wang et al., 2012 | Addgene plasmid #35202 |
| Software and Algorithms | ||
| Fiji | Schindelin et al., 2012 | https://fiji.sc/ |
| MetaMorph | Molecular Devices | https://www.moleculardevices.com/ |
| Igor Pro | WaveMetrics | https://www.wavemetrics.com/ |
| Prism 6 | GraphPad | http://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| BuzzPlugs™ vial closures | Fisher Scientific | Cat# AS273 |
| Whatman™ 3MM chromatography paper | Fisher Scientific | Cat# 05-714-5 |
Supplementary Material
Acknowledgments
We would like to thank the Developmental Studies Hybridoma Bank, the Bloomington Drosophila Stock Center, Gary Struhl, and Laura Johnston for kindly providing reagents, Gary Struhl for assistance with confocal microscopy, and members of the Ohlstein and Spradling laboratories for assistance with the screen of Janelia Farm GAL4 lines. We would like to thank Neus Rafel, Zheng Guo, and Andrew Tomlinson for critical comments and discussions on this manuscript, and in particular, Brian McCabe for discussions on random homologous chromosome segregation. The work in this manuscript was funded by the National Institutes of Health Grant R01 DK107702-01, the American Cancer Society Grant 124994-RSG-13-382-01-CSM, the Hirschl Research Scientist Award and the HHMI Faculty Scholar award (to B.O.) and the Columbia University Medical Training Grant 5-T32-HD055165-01 (to E.M.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, B.O. and E.M.L.; Methodology, B.O. and E.M.L.; Investigation, E.M.L.; Resources, B.O.; Writing – Original Draft, E.M.L.; Writing – Review & Editing, B.O. and E.M.L.; Visualization, E.M.L.; Funding Acquisition, B.O. and E.M.L.
References
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawley C, Matunis E. Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science. 2004;304:1331–1334. doi: 10.1126/science.1097676. [DOI] [PubMed] [Google Scholar]
- Buchon N, Osman D, David FP, Fang HY, Boquete JP, Deplancke B, Lemaitre B. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 2013;3:1725–1738. doi: 10.1016/j.celrep.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Ip YT. Pathogenic stimulation of intestinal stem cell response in Drosophila. J Cell Physiol. 2009;220:664–671. doi: 10.1002/jcp.21808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi NH, Kim JG, Yang DJ, Kim YS, Yoo MA. Age-related changes in Drosophila midgut are associated with PVF2, a PDGF/VEGF-like growth factor. Aging Cell. 2008a;7:318–334. doi: 10.1111/j.1474-9726.2008.00380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Hwang MS, Park JS, Bae SK, Kim YS, Yoo MA. Age-related upregulation of Drosophila caudal gene via NF-kappaB in the adult posterior midgut. Biochimica et biophysica acta. 2008b;1780:1093–1100. doi: 10.1016/j.bbagen.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Clarke AR, Jones N, Pryde F, Adachi Y, Sansom OJ. 53BP1 deficiency in intestinal enterocytes does not alter the immediate response to ionizing radiation, but leads to increased nuclear area consistent with polyploidy. Oncogene. 2007;26:6349–6355. doi: 10.1038/sj.onc.1210457. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Brotman JS, Sweeney ST, Davis G. Green fluorescent protein tagging Drosophila proteins at their native genomic loci with small P elements. Genetics. 2003;165:1433–1441. doi: 10.1093/genetics/165.3.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin EG. Amitosis in-the egg follicle cells of the cricket. Am Nat. 1903;37:667–675. [Google Scholar]
- Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. In: Schekman R, Goldstein L, Lehmann R, editors. Annual Review of Cell and Developmental Biology. Vol. 27. 2011. pp. 585–610. [DOI] [PubMed] [Google Scholar]
- de Navascues J, Perdigoto CN, Bian Y, Schneider MH, Bardin AJ, Martinez-Arias A, Simons BD. Drosophila midgut homeostasis involves neutral competition between symmetrically dividing intestinal stem cells. The EMBO J. 2012;31:2473–2485. doi: 10.1038/emboj.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Hickey RD, Paulk NK, Culberson AJ, Olson SB, Finegold MJ, Grompe M. Ploidy reductions in murine fusion-derived hepatocytes. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Taylor MH, Hickey RD, Newell AEH, Lenzi ML, Olson SB, Finegold MJ, Grompe M. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Sugai T. Amitotic division of the macronucleus in Tetrahymena thermophila: DNA distribution by genomic unit. Zool Sci. 2011;28:482–490. doi: 10.2108/zsj.28.482. [DOI] [PubMed] [Google Scholar]
- Erenpreisa J, Kalejs M, Cragg MS. Mitotic catastrophe and endomitosis in tumour cells: An evolutionary key to a molecular solution. Cell Biol Int. 2005a;29:1012–1018. doi: 10.1016/j.cellbi.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Erenpreisa J, Kalejs M, Ianzini F, Kosmacek EA, Mackey MA, Emzinsh D, Cragg MS, Ivanov A, Illidge TM. Segregation of genomes in polyploid tumour cells following mitotic catastrophe. Cell Biol Int. 2005b;29:1005–1011. doi: 10.1016/j.cellbi.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Erenpreisa JA, Cragg MS, Fringes B, Sharakhov I, Illidge TM. Release of mitotic descendants by giant cells from irradiated Burkitt’s lymphoma cell lines. Cell Biol Int. 2000;24:635–648. doi: 10.1006/cbir.2000.0558. [DOI] [PubMed] [Google Scholar]
- Fox DT, Duronio RJ. Endoreplication and polyploidy: insights into development and disease. Development. 2013;140:3–12. doi: 10.1242/dev.080531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Gall JG, Spradling AC. Error-prone polyploid mitosis during normal Drosophila development. Genes Dev. 2010;24:2294–2302. doi: 10.1101/gad.1952710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Chen T. A matter of life and death: self-renewal in stem cells. EMBO Rep. 2013;14:39–48. doi: 10.1038/embor.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galipeau PC, Cowan DS, Sanchez CA, Barrett MT, Emond MJ, Levine DS, Rabinovitch PS, Reid BJ. 17p (p53) allelic losses, 4N (G(2)/tetraploid) populations, and progression to aneuploidy in Barrett’s esophagus. PNAS. 1996;93:7081–7084. doi: 10.1073/pnas.93.14.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Cohen EH, Polan ML. Repetitive DNA sequences in Drosophila. Chromosoma. 1971;33:319–&. doi: 10.1007/BF00284948. [DOI] [PubMed] [Google Scholar]
- Guo Z, Ohlstein B. Bidirectional Notch signaling regulates Drosophila intestinal stem cell multipotency. Science. 2015;350 doi: 10.1126/science.aab0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XC, Yin T, Grindley JC, Tian Q, Sato T, Tao WA, Dirisina R, Porter-Westpfahl KS, Hembree M, Johnson T, et al. PTEN-deficient intestinal stem cells initiate intestinal polyposis. Nature Genet. 2007;39:189–198. doi: 10.1038/ng1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakova GK, Mead RA. Occurrence of amitotic division of trophoblast cell nuclei in blastocysts of the western spotted skunk (Spilogale putorius latifrons) Hereditas. 2004;140:177–184. doi: 10.1111/j.1601-5223.2004.01749.x. [DOI] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TTB, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HQ, Grenley MO, Bravo MJ, Blumhagen RZ, Edgar BA. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell. 2011;8:84–95. doi: 10.1016/j.stem.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson HP. Amitosis in the embryonal envelopes of the scorpion. Bull Mus Comp Zool. 1892;22:127–161. [Google Scholar]
- Kai T, Spradling A. Differentiating germ cells can revert into functional stem cells in Drosophila melanogaster ovaries. Nature. 2004;428:564–569. doi: 10.1038/nature02436. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nature Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- Korzelius J, Naumann SK, Loza-Coll MA, Chan JSK, Dutta D, Oberheim J, Glaesser C, Southall TD, Brand AH, Jones DL, et al. escargot maintains stemness and suppresses differentiation in Drosophila intestinal stem cells. EMBO J. 2014;33:2967–2982. doi: 10.15252/embj.201489072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn EM, Therman E, Susman B. Amitosis and endocycles in early cultured mouse trophoblast. Placenta. 1991;12:251–261. doi: 10.1016/0143-4004(91)90006-2. [DOI] [PubMed] [Google Scholar]
- Lee TM, Luo LQ. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Li H, Jasper H. Gastrointestinal stem cells in health and disease: from flies to humans. Dis Model Mech. 2016;9:487–499. doi: 10.1242/dmm.024232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber T, Kidd S, Young MW. Kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 2002;16:209–221. doi: 10.1101/gad.942302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loza-Coll MA, Southall TD, Sandall SL, Brand AH, Jones DL. Regulation of Drosophila intestinal stem cell maintenance and differentiation by the transcription factor escargot. EMBO J. 2014;33:2983–2996. doi: 10.15252/embj.201489050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YF, Li ZH. No intestinal stem cell regeneration after complete progenitor ablation in Drosophila adult midgut. J Genet Genomics. 2015;42:83–86. doi: 10.1016/j.jgg.2014.10.002. [DOI] [PubMed] [Google Scholar]
- MacAuley A, Cross JC, Werb Z. Reprogramming the cell cycle for endoreduplication in rodent trophoblast cells. Mol Biol Cell. 1998;9:795–807. doi: 10.1091/mbc.9.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes MC, Pignatelli D, Magalhaes MM. Amitosis in human adrenal cells. Histol Histopathol. 1991;6:251–256. [PubMed] [Google Scholar]
- Margall-Ducos G, Celton-Morizur S, Couton D, Bregerie O, Desdouets C. Liver tetraploidization is controlled by a new process of incomplete cytokinesis. J Cell Sci. 2007;120:3633–3639. doi: 10.1242/jcs.016907. [DOI] [PubMed] [Google Scholar]
- McLeod CJ, Wang L, Wong C, Jones DL. Stem cell dynamics in response to nutrient availability. Current Biol. 2010;20:2100–2105. doi: 10.1016/j.cub.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Miller RH. Amitosis and endocytogenesis in the fruit of Malus-sylvestris. Ann Bot-London. 1980;46:567–575. [Google Scholar]
- Montgomery BT, Nativ O, Blute ML, Farrow GM, Myers RP, Zincke H, Therneau TM, Lieber MM. Stage-B prostate adenocarcinoma-Flow cytometric nuclear-DNA analysis. Arch Surg. 1990;125:327–331. doi: 10.1001/archsurg.1990.01410150049010. [DOI] [PubMed] [Google Scholar]
- Morgan NS, Heintzelman MB, Mooseker MS. Characterization of Myosin-IA and Myosin-IB, 2 unconventional myosins associated with the Drosophila brush-border cytoskeleton. Dev Biol. 1995;172:51–71. doi: 10.1006/dbio.1995.0005. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Nabeshima YI, Yoshida S. Functional identification of the actual and potential stem cell compartments in mouse spermatogenesis. Dev Cell. 2007;12:195–206. doi: 10.1016/j.devcel.2007.01.002. [DOI] [PubMed] [Google Scholar]
- O’Brien LE, Soliman SS, Li X, Bilder D. Altered modes of stem cell division drive adaptive intestinal growth. Cell. 2011;147:603–614. doi: 10.1016/j.cell.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential Notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- Pan DJ, Rubin GM. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Mol Biol Cell. 1997;8:34–34. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- Park SU, Choi ES, Jang YS, Hong SH, Kim IH, Chang DK. Effects of chromosomal polyploidy on survival of colon cancer cells. Korean J Gastroenterol. 2011;57:150–157. doi: 10.4166/2011.57.3.150. [DOI] [PubMed] [Google Scholar]
- Patterson JT. Amitosis in the Pigeon’s egg. Anatomischer Anzeiger. 1908;32:117–125. [Google Scholar]
- Phillips MD, Thomas GH. Brush border spectrin is required for early endosome recycling in Drosophila. J Cell Sci. 2006;119:1361–1370. doi: 10.1242/jcs.02839. [DOI] [PubMed] [Google Scholar]
- Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo E, Llamazares S, Reina J, Gonzalez C. Contribution of noncentrosomal microtubules to spindle assembly in Drosophila spermatocytes. PLoS Biol. 2004;2:E8. doi: 10.1371/journal.pbio.0020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remak R. Ueber die entstehung der blutkörperchen. Medizinische Zeitung. 1841;10:127. [Google Scholar]
- Schoenfelder KP, Fox DT. The expanding implications of polyploidy. J Cell Biol. 2015;209:485–491. doi: 10.1083/jcb.201502016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata PR, Mou H, Pardo-Saganta A, Zhao R, Prabhu M, Law BM, Vinarsky V, Cho JL, Breton S, Sahay A, et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–+. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetteh PW, Basak O, Farin HF, Wiebrands K, Kretzschmar K, Begthel H, van den Born M, Korving J, de Sauvage F, van Es JH, et al. Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell. 2016;18:203–213. doi: 10.1016/j.stem.2016.01.001. [DOI] [PubMed] [Google Scholar]
- Theodosiou NA, Xu T. Use of FLP/FRT system to study Drosophila development. MCME. 1998;14:355–365. doi: 10.1006/meth.1998.0591. [DOI] [PubMed] [Google Scholar]
- van Es JH, Sato T, van de Wetering M, Lyubimova A, Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nature Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walen KH. Budded karyoplasts from multinucleated fibroblast cells contain centrosomes and change their morphology to mitotic cells. Cell Biol Int. 2005;29:1057–1065. doi: 10.1016/j.cellbi.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Wang B, Liu Y, Chen X, Fan Z. Amitosis-like nuclear division in erythrocytes of triploid rainbow trout Oncorhynchus mykiss. J Fish Biol. 2010;76:1205–1211. doi: 10.1111/j.1095-8649.2010.02556.x. [DOI] [PubMed] [Google Scholar]
- Yiquan C, Binkung W. A study on amitosis of the nucleus of the mammalian cell I. A study under the light and transmission electron microscope. Acta Anat. 1986;127:69–76. [PubMed] [Google Scholar]
- Zeng X, Hou SX. Enteroendocrine cells are generated from stem cells through a distinct progenitor in the adult Drosophila posterior midgut. Development. 2015;142:644–653. doi: 10.1242/dev.113357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Florescu S, Boettcher AL, Luo L, Dutta D, Kerr G, Cai Y, Edgar BA, Boutros M. Dpp/Gbb signaling is required for normal intestinal regeneration during infection. Dev Biol. 2015;399:189–203. doi: 10.1016/j.ydbio.2014.12.017. [DOI] [PubMed] [Google Scholar]
- Zielke N, Edgar BA, DePamphilis ML. Endoreplication. CSH Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybina EV, Zybina TG. Modifications of nuclear envelope during differentiation and depolyploidization of rat trophoblast cells. Micron. 2008;39:593–606. doi: 10.1016/j.micron.2007.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


