Abstract
Pre-mRNA splicing is an important biological process that allows production of multiple proteins from a single gene in the genome, and mainly contributes to protein diversity in eukaryotic organisms. Alternative splicing is commonly governed by RNA binding proteins to meet the ever-changing demands of the cell. However, the mis-splicing may lead to human diseases. In the heart of human, mis-regulation of alternative splicing has been associated with heart failure. In this short review, we focus on alternative splicing of sarcomeric genes and review mis-splicing related heart failure with relatively well studied sarcomeric genes and splicing mechanisms with identified regulatory factors. The perspective of alternative splicing based therapeutic strategies in heart failure has also been discussed.
Keywords: alternative splicing, sarcomeric genes, splicing factor, heart failure
1. Introduction
More than 90% of human genes express multiple mRNAs via alternative splicing, and mRNA splicing has been proposed as a primary driver for generating transcriptional diversity and regulating gene expression in mammals [1, 2]. Considering that most of human genes are alternatively spliced and thus generating distinct protein isoforms with different functional and structural properties [1, 3], abnormal patterns of mRNA splicing are associated with heart diseases, and alternative splicing is broadly altered in human heart failure [4, 5]. Tissue-specific alternative splicing is usually regulated by tissue-specific splicing factors or a combination with ubiquitously expressed splicing factors to influence spliceosome assembly at splice sites [6]. The identification of tissue-specific splicing factors and their targets is essential to decode mechanisms of alternative splicing. Currently, only a few tissue-specific splicing factors and their potential targets have been reported. Here, we will focus on the discussion of alternative splicing events of sarcomeric genes with identified tissue-specific splicing factors. With specific gene examples, we discussed how sarcomeric gene alternative splicing is orchestrated by splicing factors and the impact of mis-splicing on heart function.
2. The core splicing mechanisms
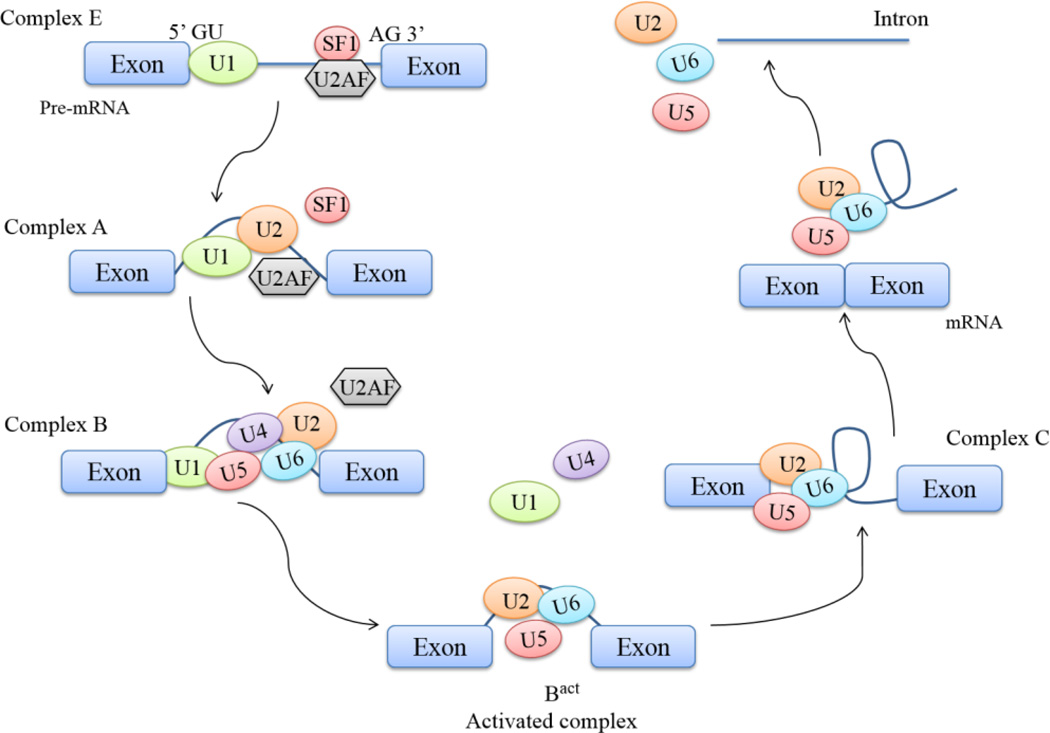
Biochemical studies have demonstrated that pre-mRNA splicing is carried out via a process that introns are removed and exons are linked together by the large ribonucleoprotein complex, called the spliceosome [7]. The spliceosome is a complex comprised of five small nuclear RNAs (snRNA) and more than 100 associated proteins [6]. The five snRNAs are U1, U2, U4, U5 and U6, and each of which can combine with associated proteins and make up an RNA-protein complex called snRNP [6–9]. The spliceosome complex functions in a dynamic assembly, reaction, and disassembly cycle, and a catalytically active spliceosome is formed upon recognizing an intron [7]. The core splicing signal for intron excision includes four consensus elements that are presenting in every intron for nearly all splicing: GU at 5' end of introns, AG at 3' end of introns, branch point sequence (BPS) located upstream of 3’ splice site and polypyrimidine tract located between BPS and 3’ splice site [9]. The initial step begins with: recognition of 5’ splice site of an intron by U1 snRNP; generation of complex E by coopperative binding of splicing factor 1 (SF1) and heterodimer U2AF65/U2AF35 to BPS region, polypyromidine tract and 3’ AG. Recruitment of U2 snRNP to BPS region to displace the SF1 is triggered by molecular interactions. Followed to complex A formation, U4–U6–U5 tri-snRNP joins the pre-spliceosome complex to form complex B [10]. At this stage, enzymatic activation occurs through a series of conformational change and compositional rearrangements to form the catalytically active complex B and complex C respectively [10, 11]. Within the assembled spliceosome, the 5’ splice site is cleaved with the lariat formation, and then the 3’ splice site is cleaved and ultimately the exons are ligated together. The spliceosome disassembles following exon ligation (Fig. 1).
Figure 1. Spliceosome assembly and disassembly in core splicing pathway.
The initial step begins with the recognition of 5’ SS by U1 snRNP and 3’ SS by SF1, U2AF (complex E). SF1 is replaced by U2 snRNP to branch point region (complex A). The formation of complex B is followed by recruitment of U4–U5–U6 tri-snRNP, displacing of U2AF protein. Complex B is remodeled via conformational changes and structure rearrangements by protein replacements. U1 and U4 snRNPs are released for recycling during remodeling, leading to the formation of a catalytically active complex (Bact). Further conformational rearrangements and protein displacements of the complex lead to the formation of a lariat intermediate (complex C). The intron lariat then is spliced at 5’ SS by an additional conformational change. Upon cleavage, intron is linearized for degradation and snRNPs recycled for a new assembly.
3. Alternative splicing regulatory elements and factors
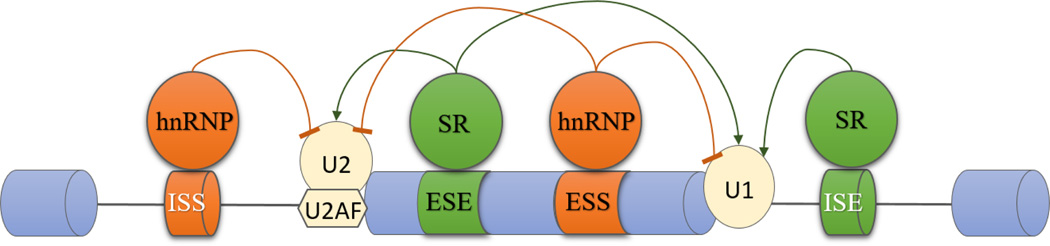
The core splicing sequences usually contain too little information to unambiguously identify splice site. Recognition and regulation of alternative splicing are modulated by additional sequences which are referred to as auxiliary cis-regulatory sequences. The auxiliary cis-regulatory sequences are located in exons and neighbouring introns. These sequence elements are known as exonic or intronic splicing enhancer (ESE or ISE), or exonic or intronic splicing silencer (ESS or ISS) [8, 9]. These auxiliary cis-regulatory sequence elements can recruit trans-regulatory factors to orchestrate alternative splicing of the whole transcript networks [12]. Trans-regulatory factors include Ser/Arg-rich (SR) proteins and heterogeneous nuclear ribonucleoprotein (hnRNP) families. In general, the ESE/ISE elements are recognized by SR proteins [13], which the interaction between ESE/ISE and SR proteins facilitates recruitment of the spliceosome and exon inclusions in the mature mRNA, while the ESS/ISS binds the hnRNP family to interfere with spliceosome assembly resulting in exclusion of an exon or exon skipping [8, 9, 12] (Fig. 2). SR proteins are frequently phosphorylated and phosphorylation of the SR proteins plays an important role in the initiation of the assembly of the spliceosome [11]. SR proteins bind to an ESE in mRNAs and recruit snRNPs simultaneously, which allow the interaction of U1 with 5’ splice site in an intron [14] and induce exon inclusion. As for the exon exclusion, the molecular mechanism is less well understood. Recently, it has been shown that hnRNP can prevent exchange of U1 for U6 by changing the way of U1 binding on the exon, which impedes splicing catalytic process and inhibits exon inclusion eventually [15].
Figure 2. Alternative splicing regulation with splicing elements and factors.
Splicing is orchestrated by cis-regulatory sequences and trans-regulatory factors in pre-mRNA. Exonic splicing enhancers (ESEs) and exonic splicing silencers (ESSs) locate in exon, and intronic splicing enhancers (ISEs) while intronic splicing silencers (ISSs) locate in intron. SR proteins and hnRNPs are two major families of alternative splicing regulatory proteins, which are recruited by splicing enhancers and silencers. Generally, SR proteins bind to splicing enhancers, facilitating spliceosome assembly, whereas hnRNP proteins are recruited by splicing silencers, interfering with the spliceosome assembly.
In addition, several factors have been identified as muscle tissue-specific regulators (Table 1). These different factors can work independently to manage exclusive splicing event of genes, or they can act as cooperative or antagonistic pairs in deciding single gene splicing [17]. Furthermore, the same trans-regulatory factor can act as a splicing activator or a repressor depending on its position of binding sites and context [18]. For an instance, RBM20 is a SR protein, but it acts as a splicing repressor [19]. RBM20 recognizes splicing silencers at the intronic positions and interacts with the spliceosome complex A but not the complex B resulting in splicing repression [20].
Table 1.
Muscle specific splicing factors and regulated sarcomeric genes
| Splicing factor | Major target sarcomeric genes |
Splicing events | Pathological consequence | Citation |
|---|---|---|---|---|
| RBM20 |
TTN, LDB3, TPM1 Required for 31 genes alternative splicing in heart; |
Titin N2B:N2BA ratio; LDB3 exon 4 inclusion and exon 5–6 exclusion; Regulates exon exclusion |
When N2B:N2BA decreases: - passive tension decreases - systolic dysfunction - dilated cardiomyopathy (DCM) Exon 4 downregulation and exon 5–6 upregulation in DCM through impaired binding of PGM1. |
Guo et al. [45] Maatz et al. [20] |
| RBM24 |
Tnnt2, Tpm1, Tpm2, Actb |
Essential for embryonic cardiac development and required for 68 splicing events; Regulates exon inclusion |
Hypertrophic and dilated cardiomyopathies |
Yang et al. [16] |
| MNBL proteins: MNBL1 |
LDB3, TNNT2 | LDB3 exon 11 exclusion; cTnT exon 5 exclusion |
Myotonic dystrophy type 1 (DM1) | Charlet-B et al. [80] Warf et al. [79] |
| CELF family: CUGBP1 CUGBP2 |
LDB3, TNNT2 | LDB3 exon 11 inclusion; cTnT exon 5 inclusion Antagonize MNBL function on exon choice control |
Myotonic dystrophy type 1 (DM1) cardiomyopathy Fetal cTnT 1 is replaced by adult cTnT 3 through exon 5 exclusion; Re-expression of embryonic cTnT in failing heart; embryonic cTnT produce higher Ca2+ sensitivity associates with cardiac arrhythmias; Exon 11 inclusion of LDB3 leads to low binding affinity to PKC |
Yamashita et al. [61] |
| RBFox family: RBFox1 |
NRAP, MYBPC3, LDB3 |
LDB3 exon 4 inclusion increases in RBFOX1−/− |
Cardiac hypertrophy and heart failure with “fetal-like” mRNA changes |
Pedrotti et al. [63] |
4. Sarcomeric gene alternative splicing and heart failure
4.1 Sarcomeric structure and protein function in striated muscle
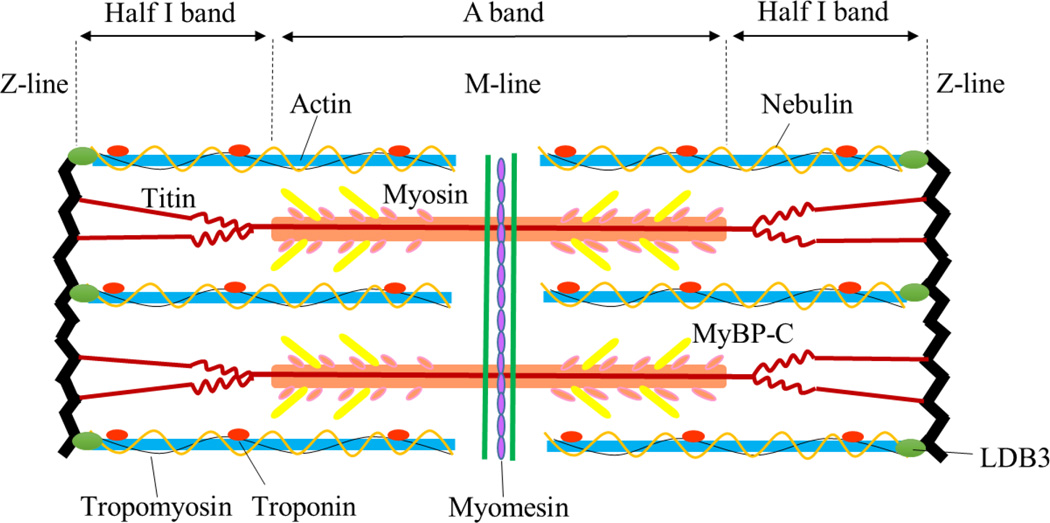
A sarcomere is basic unit of myofibrils in striated muscle. Repeating sections of individual sarcomeres compose myofibrils, which form muscle cells. Sarcomere backbone is composed of three major filament systems: myosin-based thick filament, associated with ctyoskeletal protein myosin-binding protein-C (MyBP-C); actin-based thin filament, accompanied with regulatory proteins troponin complex and tropomyosin, and third filament system titin [21]. Parallel arrays of actin-containing thin filaments span the I-band and overlap with myosin-containing thick filaments in A-band, and titin spans across each half of the sarcomere from Z-line to M-line, or center of the sarcomere [22, 23]. The sarcomere contains more than 20 major myofibrillar proteins. Figure 3 shows sarcomere structure with most abundant sarmoceric proteins and their specific positions.
Figure 3. The striated sarcomere structure.
Location of the major and most abundant sarcomeric proteins in the sarcomere: thick filament contains myosin and MyBP-C and locates in the A band. Thin filament is composed of actin, tropomyosin and troponin complex, spans I band and overlaps thick filament in A band. Titin spans half sarcomere from Z-line to M-line, with its N-terminus binding to actin at Z-line and C-terminus attaching to myosin. Nebulin is the fourth filament system in skeletal muscle which spans alone with actin. Myomesin is the M-line sarcomeric protein which crosslinks myosin filaments to M-line. LDB3 locates in Z-line and maintains Z-line integrity during muscle contraction.
During cardiac development and under disease conditions, most of sarcomeric proteins show isoform switching to adapt to the ever changing physiological conditions. Some protein isoforms are generated from distinct genes, such as myosin, actin, troponin I and C, MyBP-C and tropomyosin; others arise from a single gene by alternative splicing, like titin, troponin T, tropomyosin (some isoforms produced by a signle gene) and LDB3. Muscle-specific alternative splicing was one of the first observed tissue-specific events [24]. Muscle tissue mis-splicing of key sarcomeric genes has been found in heart failure [25]. Unfortunately, only a few muscle tissue-specific splicing factors have been identified so far, and very little is known about their molecular mechanisms that mediate muscle-specific splicing (Table 1). Here, we focus on the four sarcomeric genes whose alternative splicing events have been associated with heart failure.
4.2 Sarcomeric gene mis-splicing in heart failure
4.2.1 Titin
Titin is a protein initially described as connectin [26] and later named titin, because of its apparently gigantic size determined on polyacrylamide gels [27, 28]. Titin is the largest known polypeptide, and is made up of 363 exons and 38,138 amino acid residues in humans, equating to a potential total size of approximately 4.2 MDa [29]. Most of the sarcomeric proteins are orchestrated by interactions with titin, which acts as the giant blueprint protein of the sarcomere [30]. Titin is responsible for maintaining passive tension in striated muscle, which explains its flexibility and extensible abilities [31].
Titin is expressed in potentially millions of different isoforms generated by alternative splicing from the transcript of a single gene TTN [23]. Across mammalian species, titin molecules in striated muscle are classified based on their N2-unique domain expression [32]. The isoforms contain the N2B sequence only expressed in cardiac muscle and the isoforms with a different unique region called N2A are found in both cardiac and skeletal muscles [23]. The isoforms with N2A unique sequence in cardiac muscle called N2BA which also contains N2B unique domain, while in skeletal muscle the isoforms are called N2A which solely has N2A unique domain. Therefore, there are two major classes of titin isoforms expressed in cardiac muscle: the smaller and stiffer N2B (approximately 3.0 MDa) and the larger and more complaint N2BAs (between ~3.2 and 3.8 MDa) [33]. These two classes of titin isoforms are co-expressed in cardiac muscle with the altered ratios depending on species and developmental stage. During prenatal or perinatal development, the large fetal isoform is replaced by the smaller size isoform that means the N2BA isoforms decrease in proportion, and the N2B isoform predominates in mature animal [34, 35]. The titin isoforms express at a ratio of ~35% N2BA to ~65% N2B in normal adult human left ventricle [36]. The adult wild-type rat hearts express less than 10% N2BA and more than 90% N2B isoform [27]. There is an intimate relationship between the size of the I-band region of titin and titin-based passive tension, that is, a larger elastic I-band regions corresponding to a lower passive tension, so titin-based passive tension is largely determined by titin isoform expression ratio in the heart. Overall, a decreased ratio of N2BA induces elevated stiffness, whereas an increased ratio indicates reduced myofibrillar stiffness. Altered splicing of TTN has been found in a series of heart diseases, such as heart failure, hypertrophic cardiomyopathy, and ischemic heart disease [37]. Analyses have already shown that the left ventricle biopsies from patients with diastolic heart failure and hypertension rats had a reduced N2BA/N2B titin which is associated with increased myocardial stiffness [38–40]. In conditions with eccentric remodeling and volume overload, the left ventricle tissues from the hypertrophy mice with both systolic and diastolic dysfunction showed a marked reduction in N2BA/N2B ratio [41]. In human patients with non-tachycardia-induced dilated cardiomyopathy (DCM), an increase in the N2BA/N2B ratio was observed in DCM patients [42–44]. Therefore, in order to develop a therapeutic strategy for heart failure originated by titin isoform switching, it is critical important to understand the mechanisms controlling titin splicing.
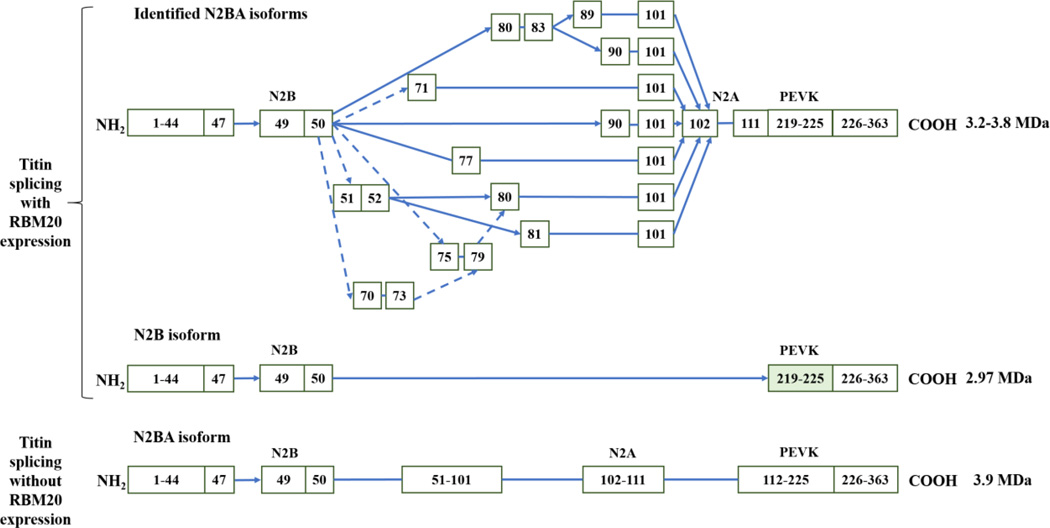
Recently, RBM20 has been identified as a major regulator of titin splicing [45] (Fig. 4). In an engineered mouse model, an in-frame deletion of the RNA recognition motif (RRM) of Rbm20 further verified the role of Rbm20 in the regulation of titin splicing [46]. Mutations with loss of function in RBM20 have been associated with development of heart failure [47–53]. Mutations in exon 9 of RBM20 were found in 3% of all DCM cases, and in over 13% of those with a history of sudden death [53]. In addition, Maatz et al. [20] suggested that the difference in endogenous RBM20 expression significantly affects splicing of orthologous exons for genes like TTN and LDB3. This difference was based on the comparison of the splicing patterns of RBM20-modulated exons in heart tissues from end-stage heart failure patients, implying the endogenous RBM20 expression levels are pivotal for the regulation of cardiac function. Presently, core sequence of UCUU in titin gene has been recommended as the cis-acting RNA elements recognized by RBM20 [20], and the ratio of other factors such as SFRS1 (SF2/ASF), U2AF65 and hnRNP L has been predicted to cooperatively regulate titin splicing with RBM20 [19]. However, blasting UCUU against titin gene indicates that the core sequence appears multiple times in the most of introns and exons of titin, therefore, it is unkonwn which UCUU core-containing sequence can be bound by RBM20, and future studies need be warranted to identify the specific UCUU-core containing sequence for RBM20 binding. Also cooperative regulation with other regulatory factors hasn’t been validated yet. Although RBM20 has been known as the major regulator of titin splicing, the detailed mechanisms are still unclear.
Figure 4. RBM20-regulated titin alternative splicing patterns (modified from [22]).
In the heart with RBM20 expression, cardiac titin mRNA undergoes extensive exon usages in regions corresponding to middle Ig and PEVK domain, while these exon splicing events seem inhibited with RBM20 deficiency. Arrows indicate the exons spliced, solid line connections denote consecutive exons.
4.2.2. Cypher/LDB3
LIM domain binding 3 (LDB3), also known as Z-band alternatively spliced PDZ-motif (ZASP) in human or Cypher in mice, belongs to PDZ-LIM domain family [54]. It complexes with other Z-line associated proteins, like α-actinin, and plays a critical role in muscle ultrastructure and functions as maintaining Z-line integrity during muscle contraction [55]. The global cypher-null mice develop congenital dilated cardiomyopathy, and are found with severe defects in striated muscle which are postnatal lethal [56]. Cardiac specific deletion of cypher also leads to DCM and premature adult lethality [57]. Cypher contains a total of 17 exons and has at least six alternatively spliced isoforms. Cardiac and skeletal muscle each contains two long isoforms and one short isoform, and the expression of these isoforms is strictly regulated during muscle development [58].
Cypher long (Cypher 1 and 3) and short (Cypher 2) isoforms are defined by the presence and absence of three C-terminal LIM domains, respectively. The long isoform has been suggested to be more important than the short isoform physiologically. Depletion of a selective cypher long isoform has been associated to lead to late-onset cardiomyopathy [59]. Exon 10 is last exon of Cypher short isoform. Exon 11 is differentially spliced to generate Cypher 1 (included) or Cypher 3 (excluded) isoforms. Previous report showed that the exon 11 inclusion isoform is decreased during mouse heart development, which is only regulated by MBNL1 [60]. Later, another study suggested that the splicing of exon 11 is regulated by splicing factors MBNL1 and CUG triplet repeat RNA-binding protein (CUGBP1, a member of CELF family) in myotonic dystrophy type 1 (DM1) skeletal muscle [61]. In DM1 skeletal muscle, CTG repeats expand in the 3’-untranslated region (UTR) of the DMPK gene. The CUG repeat RNA causes sequestration of MBNL1, however, upregulation of CUGBP1 leads to inclusion of LDB3 exon 11. Inclusion of exon 11 alters binding affinity of LDB3 to protein kinase C (PKC) and contributes to inappropriate activation of PKC, leading to CUGBP1 phosphorylation and its overexpression in the pathogenesis of DM1 [61].
Interestingly, the Ldb3 gene harbors two classes of splicing variants that are skeletal and cardiac muscle-specific. The cardiac specific region is only defined by exon 4, whereas the skeletal muscle specific region is defined by three exons, exons 5–7 [58]. RBM20 has been identified as a splicing factor that regulates LDB3 exon 4 and exons 5–7 splicing [45]. Inclusion of exon 4 or exons 5–7 is mutually exclusive which is regulated by RBM20. In the Rbm20-deficient rats or human subject with the RBM20 missense mutation, expression of exon 4 is downregulated in the left ventricular tissue, whereas expression of exons 5–7 is upregulated. Therefore, RBM20 mediates the heart specific splicing of LDB3 rather than the alternation between the “long isoform” and “short isoform” of LDB3 described above. Similarly, the cardiac isoform of cypher is switched to the skeletal isoform in ASF/SF2 (a prototypical SR protein) deficient mice [62]. Interestingly, it has been reported that exon 4 shows an increase of inclusion in RBFox1−/− muscle [63]. Together, RBM20, ASF/SF2, and RBFox1 may co-regulate the exon 4 or exons 5–7 specific splicing of LDB3 in the heart muscle. Mutations in exon 4 of LDB3 have been shown to be associated with dilated cardiomyopathy through impaired binding of the glycolytic enzyme phosphoglucomutase 1 (PGM1) [64], which is a key enzyme involving in glucose consumption and energy management. These results demonstrate that exon 4 inclusion isoforms may play a pivotal role in the heart muscle development.
4.2.3. Troponin T
Troponin plays a crucial role in the regulation of contraction and relaxation of striated muscles. Troponin is a complex consisted of three regulatory proteins which are calcium binding protein troponin C (TnC), inhibitory protein troponin I (TnI), and tropomyosin-binding protein troponin T (TnT) [65]. Each isoform of TnC and TnI is encoded from individual distinct genes. So far, no splicing events as well as involved splicing factors have been reported for TnC and TnI during heart development and/or pathological conditions [66]. Nonetheless, TnT undergoes alternative splicing to generate multiple isoforms [67].
Mammalian cTnT gene contains 17 exons of which exon 4, exon5 and exon 13 are alternatively spliced [68]. In terms of the exon 4 and 5 alternative splicing, four human cardiac isoforms are generated, cTnT1 (all exon present), cTnT2 (missing exon 4), cTnT3 (missing exon 5), and cTnT4 (missing both exon 4 and 5). In the heart, alternative splice forms of cardiac TnT (cTnT) are under developmental change. The expressed cTnT isoforms are regulated during embryonic and postnatal heart development, and altered splicing patterns are found in failing human heart, diabetic rat heart, and familial hypertrophic cardiomyopathy human heart [69–72]. Splicing of cTnT gene is strictly governed, and the abnormal splicing will cause cardiac dysfunction. A healthy adult mouse heart solely expresses cTnT3, while coexistence of two or three cTnT variants in adult mouse hearts will cause decreased myocardial contractility and ventricular pumping efficiency [73]. Moreover, studies using reconstituted myofilaments showed embryonic cTnT produced high Ca2+ sensitivity in comparison to adult cTnT [74], which implies that re-expression of embryonic cTnT in adult heart may increase susceptibility to arrhythmia that is associated with the increased myofilamental Ca2+ sensitivity [75,76].
It is well documented that alternative splicing of cTnT exons 4 and 5 is developmentally regulated. Exon inclusion is predominantly in embryonic and neonatal heart but is gradually altered to exon exclusion during postnatal heart development [77]. Xu et al. [62] found that ASF/SF2 is involved in the regulation of tissue-specific alternative splicing of cTnT. Ablation of ASF/SF2 in heart results in cTnT exon 4 efficiently included in 4-week-old mouse hearts [62], suggesting that ASF/SF2 plays a role in coordination with developmental isoform transition of cTnT. With regard to the cTnT role in muscle contraction, the elevated exon 4 inclusion leads to less relaxation of muscle fibers in the absence of Ca2+ [78], which may contribute to the hypercontraction phenotype in ASF/SF2 knockout mice [62]. In the case of exon 5, Warf et al. [79] demonstrated that the protein factors MBNL and U2AF65 regulate the cTnT exon 5 splicing. MBNL1 directly binds to cTnT pre-mRNA and controls the exclusion of exon 5 by competing with the essential splicing factor U2AF65 for binding at the 3’ end of intron 4. When U2AF65 is sequestered from binding to the pre-mRNA, the U2 snRNP can no longer be recruited for spliceosome assembly and the exon 5 is excluded. On the other hands Charlet-B et al. [80] indicated that inclusion of cTnT exon 5 in cTnT1 isoform in embryonic heart requires binding of splicing factor ETR-3 (also known as CUGBP2, a member of CELF family). Binding and activation by ETR-3, however, are directly antagonized by the splicing factor polypyrimidine tract binding protein (PTB). Furthermore, Goo and Cooper [81] showed that the activation of cTnT exon 5 inclusion by ETR-3 is promoted by direct interaction with components of the activated U2 snRNP and enhanced binding of U2 snRNP and complex A assembly. Taken together, development of cTnT isoform switching is regulated by the splicing factors of ASF/SF2, MBNL1, ETR-3 and PTB. Balance between these competing splicing factors determines the isoform expressions of cTnT and thus affects the development of associated heart diseases. Strikingly, like exon 5, the ASF/SF2 governed exon 4 splicing is tightly bound with a developmental program, which indicates that ASF/SF2 cannot act alone in the regulation. Interestingly, cTnT has been identified as one of the RBM20 regulated genes through HITS-CLIP [20], however, another study showed that cTnT is not a target of RBM20 [82]. These conflicted results suggest more studies are needed for cTnT splicing.
4.2.4. Tropomyosin
In muscle, tropomyosin (TPM) exists as a rod-shaped coiled-coil dimer that forms a head-to-tail polymer along the length of an actin filament [83, 84]. TPM is responsible for the regulation of many properties of actin, including stabilizing actin filaments [85], and one of major role of TPM is mediating muscle contraction via regulation of actin-myosin interaction. In the relaxed state, TPM and troponin complex blocks myosin binding sites on the actin filament, thus preventing cross-bridge formation and, ultimately, muscle contraction. Upon releasing of intracellular calcium by neuronal stimulation, troponin binds to calcium, which leads to lateral movement of TPM. Movement of TPM exposes myosin-binding site of actin, thus allowing formation of cross-bridge and muscle contraction [86].
In humans, four genes, TPM1, TPM2, TPM3 and TPM4, have been characterized and give rise to multiple isoforms via alternative splicing, use of different promoters, and different poly (A) addition sites [87]. In human striated muscle, mainly expressed isoforms are TPM1α and TPM1κ alternatively spliced from TPM1 gene, TPM2β from TPM2 gene, and TPM3γ from TPM3 gene [88]. TPM1κ and TPM1α have an identical exon composition except for exon 2 where TPM1κ contains smooth muscle exon 2a instead of striated muscle exon 2b as in TPM1α. Expression of TPM1κ isoforms is differentially regulated in heart development (3–5% of total TPM isoform population), upregulation of TPM1κ isoform (approximately doubled) was found in patients with DCM and heart failure [89, 90]. Physiological changes with increased expression of TPM1κ isoform include decreased contractility of left ventricle, systolic and diastolic dysfunctions, reduced myofilament calcium sensitivity of tension development, and weaker interaction between thin filaments [89]. Previous studies demonstrate that expressions of TPM1α and TPM2β in the heart are governed by developmental changes, both isoforms are expressed during embryogenesis and fetal development, however, TPM2β isoform reduces dramatically after birth [91]. In transgenic mice with a level of 55% TPM2β causes an increase in myofilament Ca2+ sensitivity, a decrease in the maximum rate of relaxation, and an increase in the time to one-half relaxation of the hearts [92]. In a mouse model with overexpression of TPM3γ isoform the heart shows increased heart rate and decreased Ca2+ sensitivity [92]. Mechanisms of TPM alternative splicing in the heart have not been well-recognized, however, TPM1 has recently been identified as a target of RBM20 [45], but it is still unknown which specific splicing event is regulated by RBM20. Thus it would be exciting to know whether RBM20 controls the expression of TPM1κ and TPM1α isoforms and whether TPM1κ upregulation is associated with RBM20.
Although splicing events in the heart are unclear, several splicing factors have been recognized in mediating TPM splicing in other cell types. For example, hnRNP H, a member of the hnRNP family, enhances the activity of an ESS and participates in exclusion of exon 7 of TPM2β in nonmuscle cells [93, 94]. Ubiquitously expressed hnRNP H binds to ESS sequences at the 5’ end of exon 7. Binding of hnRNP H to ESS correlates with silencer activity resulting in exon 7 skipping [93]. Inclusion of exons 2 and 3 of TPM1α is mutually exclusive. Exon2 is selected in smooth muscle cells, whereas exon 3 is selected in skeletal muscle, heart and brain [95, 96]. 9G8, a member of SR protein family, was found to function as a splicing activator of exon 2. However, hnRNP H and F antagonize with 9G8 by competing for and binding to the same element sequences on pre-mRNA [97]. On the other hand, repression of exon 3 in smooth muscle is mediated by PTB (also known as hnRNP I) and MBNL1. MBNL1 binds to negative regulatory elements consisting of UGC clusters or CUG motifs, which acts as repressors of exon 3. Binding of MBNL1 to pre-mRNA promotes its interaction with another factor PTB. Therefore, skipping of exon 3 involves the cooperation between PTB and MNBL1 [98]. Since exon 3 is favored in striated muscle, an interesting next step would be to explore if MBNL1 and PTB govern the splicing of TPM1α in the heart and control the inclusion of exon 3 in cardiac muscle for maintaining splicing mode in a cardiac-specific manner.
5. Future perspectives
Alternative splicing is highly regulated and frequently occurred in eukaryotic organisms, and diseases can be caused by abnormal alternative splicing. This makes alternative splicing a therapeutic target for treatment of mis-splicing associated diseases. Currently, alternative splicing-based therapy is attracting researchers’ attention as a potential therapeutic strategy for treatment of heart diseases since mis-splicing-caused cardiac disease is rising. Over last two decades, antisense oligonucleotides (AONs) have been studied to interfere with the core splicing or the regulatory factors binding sites. AONs have been applied to the Duchene muscular dystrophy models. Skipping of certain mutated exons by using AONs was promoted to avoid premature truncation of the protein [99]. In a myotonic dystrophy type 1 mouse model, splicing factor MNBL1 is sequestered by CUG expansion in DMPK mRNAs. By using AONs, CUG repeats are blocked, allowing MBNL1 to reach its natural targets [100]. Additional splicing misregulation events have been found in skeletal muscles of DM1 patients. For instance, re-expression of an embryonic dystrophin isoform due to aberrant splicing mediation of DMD exon 78 has been strongly related to the severity of DM1. Recently, it has been reported that DMD exon 78 splicing is also regulated by MBNL1 during skeletal muscle development, and mis-exclusion of exon 78 is a direct consequence of MBNL1 loss-of-function caused by CUG repeats [101]. Up to date, two antisense drugs have been approved by U.S. Food and Drug Administration (FDA) as a treatment of cytomegalovirus retinitis [102, 103] and homozygous familial hypercholesterolemia [104]. However, AONs have still not been extensively used due to lack of understanding of detailed splicing mechanisms. Upregulation of cardioprotective isoforms may also serve as an attractive therapeutic approach. As discussed above, an increase of N2BA/N2B titin isoform ratio has been found in patients with dilated cardiomyopathy. Studies have demonstrated that hormones, like insulin and thyroid hormone, can increase the expression of N2B/N2BA titin isoform ratio via the PI3K/AKT signaling pathway [105–107]. Therefore, further understanding of how splicing signaling pathway of sarcomeric genes is regulated may eventually enable us to shift isoforms expression to reduce pathological effects. On the other hand, regulation of activity of splicing factors may also be a promising therapeutic option for manipulating specific gene splicing in pathological conditions. Since RBM20 is the major regulator of titin isoform transition, N2B titin isoform increases in the presence of RBM20, while N2BA titin isoform increases in the absence of RBM20 [45]. Therefore, by mediating the expression level or activity of RBM20, the ratio between N2B and N2BA titin isoforms can be elaborately manipulated. This is just an example for the emerging application by correcting the abnormal protein isoform ratios resulting from mRNA mis-splicing. Taken together, in the future, correcting the abnormal isoform ratios for treatment of heart failure will be a promising strategy.
Highlights.
mis-splicing of sarcomeric genes is asscociated with heart failure
fundamental mechanisms of pre-mRNA splicing are critical for heart failure treatment
muscle specific splicing factors play a cardinal role in heart failure development
manipulation of sarcomeric mRNA splicing is a therapeutic strategy for heart failure
Acknowledgments
Funding: This work was supported by National Institute of General Medical Sciences (1003088F), American Heart Association (AHA) (1003018) and Agricultural Experiment Station’s competitive Grant from University of Wyoming (1003093K).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson JM, Castle J, Garrett-Engele P, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez JM, Maietta P, Ezkurdia I, et al. APPRIS: Annotation of principal and alternative splice isoforms. Nucleic Acids Res. 2013;41:D110–D117. doi: 10.1093/nar/gks1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis FJ, Gupta M, Pogwizd SM, et al. Increased expression of alternatively spliced dominant-negative isoform of SRF in human failing hearts. Am J Physiol Heart Circ Physiol. 2002;282:H1521–H1533. doi: 10.1152/ajpheart.00844.2001. [DOI] [PubMed] [Google Scholar]
- 5.Kong SW, Hu YW, Ho JW, et al. Heart failure-associated changes in RNA splicing of sarcomere genes. Circ Cardiovasc Genet. 2010;3:138–146. doi: 10.1161/CIRCGENETICS.109.904698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black DL. Mechanisms of Alternative Pre-Messenger RNA Splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y, Rio DC. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu Rev Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.House AE, Lynch KW. Regulation of alternative splicing: More than just the ABCs. J Biol Chem. 2008;283:1217–1221. doi: 10.1074/jbc.R700031200. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Burge CB. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daguenet E, Dujardin G, Valcárcel J. The pathogenicity of splicing defects: mechanistic insights into pre-mRNA processing inform novel therapeutic approaches. EMBO Rep. 2015;16:1640–1655. doi: 10.15252/embr.201541116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Lara-Pezzi E, Gómez-Salinero J, Gatto A, et al. The Alternative Heart: Impact of Alternative Splicing in Heart Disease. J of Cardiovasc Trans Res. 2013;6:945–955. doi: 10.1007/s12265-013-9482-z. [DOI] [PubMed] [Google Scholar]
- 13.Blencowe BJ. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 14.Cho S, Hoang A, Sinha R, et al. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc Natl Acad Sci. 2011;108:8233–8238. doi: 10.1073/pnas.1017700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiou NT, Shankarling G, Lynch KW. hnRNP L and hnRNPA1 induce extended U1 snRNA interactions with an exon to repress spliceosome assembly. Mol Cell. 2013;49:972–982. doi: 10.1016/j.molcel.2012.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Hung LH, Licht T, et al. RBM24 is a major regulator of muscle-specific alternative splicing. Dev Cell. 2014;31:87–99. doi: 10.1016/j.devcel.2014.08.025. [DOI] [PubMed] [Google Scholar]
- 17.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu XD, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S, Guo W, Dewey CN, et al. Rbm20 regulates titin alternative splicing as a splicing repressor. Nucleic Acids Res. 2013;41:2659–2672. doi: 10.1093/nar/gks1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maatz H, Jens M, Liss M, et al. RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing. J Clin Invest. 2014;124:3419–3430. doi: 10.1172/JCI74523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krüger M, Kötter S. Titin, a Central Mediator for Hypertrophic Signaling, Exercise-Induced Mechanosignaling and Skeletal Muscle Remodeling. Front Physiol. 2016;7:1–8. doi: 10.3389/fphys.2016.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo W, Bharmal SJ, Esbona K, et al. Titin diversity--alternative splicing gone wild. J Biomed Biotechnol. 2010:753675. doi: 10.1155/2010/753675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labeit S, Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- 24.Llorian M, Smith CW. Decoding muscle alternative splicing. Curr Opin Genet Dev. 2011;21:380–387. doi: 10.1016/j.gde.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Kong SW, Hu YW, Ho JW, et al. Heart failure-associated changes in RNA splicing of sarcomere genes. Circ Cardiovasc Genet. 2010;3:138–146. doi: 10.1161/CIRCGENETICS.109.904698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maruyama K, Kimura S, Kuroda M, et al. Connectin, an elastic protein of muscle. Its abundance in cardiac myofibrils. J Biochem. 1977;82:347–350. [PubMed] [Google Scholar]
- 27.Neagoe C, Opitz CA, Makarenko I, et al. Gigantic variety: expression patterns of titin isoforms in striated muscles and consequences for myofibrillar passive stiffness. J Muscle Res Cell Motil. 2003;24:175–189. doi: 10.1023/a:1026053530766. [DOI] [PubMed] [Google Scholar]
- 28.Wang K, McClure J, Tu A. Titin: major myofibrillar components of striated muscle. Proc Natl Acad Sci USA. 1979;76:3698–3702. doi: 10.1073/pnas.76.8.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bang ML, Centner T, Fornoff F, et al. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- 30.Tskhovrebova L, Trinick J. Titin: properties and family relationships. Nat Rev Mol Cell Biol. 2003;4:679–689. doi: 10.1038/nrm1198. [DOI] [PubMed] [Google Scholar]
- 31.Fukuda N, Terui T, Ohtsuki I, et al. Titin and troponin: Central players in the Frank-Starling mechanism of the heart. Curr Cardiol Rev. 2009;5:119–124. doi: 10.2174/157340309788166714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vikhlyantsev IM, Podlubnaya ZA. New titin (connectin) isoforms and their functional role in striated muscles of mammals: facts and suppositions. Biochemistry (Moscow) 2012;77:1515–1535. doi: 10.1134/S0006297912130093. [DOI] [PubMed] [Google Scholar]
- 33.Cazorla O, Freiburg A, Helmes M, et al. Differential expression of cardiac titin isoforms and modulation of cellular stiffness. Circ Res. 2000;86:59–67. doi: 10.1161/01.res.86.1.59. [DOI] [PubMed] [Google Scholar]
- 34.Warren CM, Krzesinski PR, Campbell KS, et al. Titin isoform changes in rat myocardium during development. Mech Dev. 2004;121:1301–1312. doi: 10.1016/j.mod.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Opitz CA, Leake MC, Makarenko I, et al. Developmentally regulated switching of titin size alters myofibrillar stiffness in the perinatal heart. Circ Res. 2004;94:967–975. doi: 10.1161/01.RES.0000124301.48193.E1. [DOI] [PubMed] [Google Scholar]
- 36.Neagoe C, Kulke M, del Monte F, et al. Titin isoform switch in ischemic human heart disease. Circulation. 2002;106:1333–1341. doi: 10.1161/01.cir.0000029803.93022.93. [DOI] [PubMed] [Google Scholar]
- 37.Chauveau C, Rowell J, Ferreiro A. A rising titan: TTN review and mutation update. Hum Mutat. 2014;35:1046–1059. doi: 10.1002/humu.22611. [DOI] [PubMed] [Google Scholar]
- 38.van Heerebeek L, Borbély A, Niessen HW, et al. Myocardial structure and function differ in systolic and diastolic heart failure. Circulation. 2006;113:1966–1973. doi: 10.1161/CIRCULATIONAHA.105.587519. [DOI] [PubMed] [Google Scholar]
- 39.Williams L, Howell N, Pagano D, et al. Titin isoform expression in aortic stenosis. Clin Sci (Lond) 2009;117:237–242. doi: 10.1042/CS20080248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warren CM, Jordan MC, Roos KP, et al. Titin isoform expression in normal and hypertensive myocardium. Cardiovasc Res. 2003;59:86–94. doi: 10.1016/s0008-6363(03)00328-6. [DOI] [PubMed] [Google Scholar]
- 41.Hutchinson KR, Saripalli C, Chung CS, et al. Increased myocardial stiffness due to cardiac titin isoform switching in a mouse model of volume overload limits eccentric remodeling. J Mol Cell Cardiol. 2015;79:104–114. doi: 10.1016/j.yjmcc.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makarenko I, Opitz CA, Leake MC, et al. Passive stiffness changes caused by upregulation of compliant titin isoforms in human dilated cardiomyopathy hearts. Circ Res. 2004;95:708–716. doi: 10.1161/01.RES.0000143901.37063.2f. [DOI] [PubMed] [Google Scholar]
- 43.Nagueh SF, Shah G, Wu Y, et al. Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation. 2004;110:155–162. doi: 10.1161/01.CIR.0000135591.37759.AF. [DOI] [PubMed] [Google Scholar]
- 44.Borbély A, van der Velden J, Papp Z, et al. Cardiomyocyte stiffness in diastolic heart failure. Circulation. 2005;111:774–781. doi: 10.1161/01.CIR.0000155257.33485.6D. [DOI] [PubMed] [Google Scholar]
- 45.Guo W, Schafer S, Greaser ML, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18:766–773. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Methawasin M, Hutchinson KR, Lee EJ, et al. Experimentally Increasing Titin Compliance in a Novel Mouse Model Attenuates the Frank-Starling Mechanism but has a Beneficial Effect on Diastole. Circulation. 2014;129:1924–1936. doi: 10.1161/CIRCULATIONAHA.113.005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Refaat MM, Lubitz SA, Makino S, et al. Genetic variation in the alternative splicing regulator RBM20 is associated with dilated cardiomyopathy. Heart Rhythm. 2012;9:390–396. doi: 10.1016/j.hrthm.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beraldi R, Li X, Martinez Fernandez A, et al. Rbm20-deficient cardiogenesis reveals early disruption of RNA processing and sarcomere remodeling establishing a developmental etiology for dilated cardiomyopathy. Hum Mol Genet. 2014;23:3779–3791. doi: 10.1093/hmg/ddu091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li D, Morales A, Gonzalez-Quintana J, et al. Identification of novel mutations in RBM20 in patients with dilated cardiomyopathy. Clin Transl Sci. 2010;3:90–97. doi: 10.1111/j.1752-8062.2010.00198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rampersaud E, Siegfried JD, Norton N, et al. Rare variant mutations identified in pediatric patients with dilated cardiomyopathy. Prog Pediatr Cardiol. 2011;31:39–47. doi: 10.1016/j.ppedcard.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao Y, Feng Y, Zhang YM, et al. Targeted next-generation sequencing of candidate genes reveals novel mutations in patients with dilated cardiomyopathy. Int J Mol Med. 2015;36:1479–1486. doi: 10.3892/ijmm.2015.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Millat G, Bouvagnet P, Chevalier P, et al. Clinical and mutational spectrum in a cohort of 105 unrelated patients with dilated cardiomyopathy. Eur J Med Genet. 2011;54:e570–e575. doi: 10.1016/j.ejmg.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 53.Brauch KM, Karst ML, Herron KJ, et al. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol. 2009;54:930–941. doi: 10.1016/j.jacc.2009.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou Q, Ruiz-Lozano P, Martone ME, et al. Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to alpha-actinin-2 and protein kinase C. J Biol Chem. 1999;274:19807–19813. doi: 10.1074/jbc.274.28.19807. [DOI] [PubMed] [Google Scholar]
- 55.Zheng M, Cheng H, Banerjee I, et al. ALP/Enigma PDZ–LIM domain proteins in the heart. J Mol Cell Biol. 2010;2:96–102. doi: 10.1093/jmcb/mjp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Q, Chu PH, Huang C, et al. Ablation of Cypher, a PDZ– LIM domain Z-line protein, causes a severe form of congenital myopathy. J Cell Biol. 2001;155:605–612. doi: 10.1083/jcb.200107092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng M, Cheng H, Li X, et al. Cardiac-specific ablation of Cypher leads to a severe form of dilated cardiomyopathy with premature death. Hum Mol Genet. 2009;18:701–713. doi: 10.1093/hmg/ddn400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang CQ, Zhou Q, Liang PH, et al. Characterization and in vivo functional analysis of splice variants of cypher. J Biol Chem. 2003;278:7360–7365. doi: 10.1074/jbc.M211875200. [DOI] [PubMed] [Google Scholar]
- 59.Cheng H, Zheng M, Peter AK, et al. Selective deletion of long but not short Cypher isoforms leads to late-onset dilated cardiomyopathy. Hum Mol Genet. 2011;20:1751–1762. doi: 10.1093/hmg/ddr050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalsotra A, Xiao X, Ward A, et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamashita Y, Matsuura T, Kurosaki T, et al. LDB3 splicing abnormalities are specific to skeletal muscles of patients with myotonic dystrophy type 1 and alter its PKC binding affinity. Neurobiol Dis. 2014;69:200–205. doi: 10.1016/j.nbd.2014.05.026. [DOI] [PubMed] [Google Scholar]
- 62.Xu X, Yang D, Ding JH, et al. ASF/SF2-Regulated CaMKIIδ Alternative Splicing Temporally Reprograms Excitation-Contraction Coupling in Cardiac Muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 63.Pedrotti S, Giudice J, Dagnino-Acosta A, et al. The RNA-binding protein Rbfox1 regulates splicing required for skeletal muscle structure and function. Hum Mol Genet. 2015;24:2360–2374. doi: 10.1093/hmg/ddv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arimura T, Inagaki N, Hayashi T, et al. Impaired binding of ZASP/Cypher with phosphoglucomutase 1 is associated with dilated cardiomyopathy. Cardiovasc Res. 2009;83:80–88. doi: 10.1093/cvr/cvp119. [DOI] [PubMed] [Google Scholar]
- 65.Greaser ML, Gergely J. Purification and properties of the components from troponin. J Biol Chem. 1973;248:2125–2133. [PubMed] [Google Scholar]
- 66.Sheng JJ, Jin JP. Gene regulation, alternative splicing, and posttranslational modification of troponin subunits in cardiac development and adaptation: a focused review. Front Physiol. 2014;5:165. doi: 10.3389/fphys.2014.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jin JP, Huang QQ, Yeh HI, et al. Complete nucleotide sequence and structural organization of rat cardiac troponin T gene. A single gene generates embryonic and adult isoforms via developmentally regulated alternative splicing. J Mol Biol. 1992;227:1269–1276. doi: 10.1016/0022-2836(92)90540-z. [DOI] [PubMed] [Google Scholar]
- 68.Jin JP, Wang J, Zhang J. Expression of cDNAs encoding mouse cardiac troponin T isoforms: characterization of a large sample of independent clones. Gene. 1996;168:217–221. doi: 10.1016/0378-1119(95)00803-9. [DOI] [PubMed] [Google Scholar]
- 69.Ricchiuti V, Apple FS. RNA expression of cardiac troponin T isoforms in diseased human skeletal muscle. Clin Chem. 1999;45:2129–2135. [PubMed] [Google Scholar]
- 70.Anderson PA, Malouf NN, Oakeley AE, et al. Troponin T isoform expression in humans. A comparison among normal and failing adult heart, fetal heart, and adult and fetal skeletal muscle. Circ Res. 1991;69:1226–1233. doi: 10.1161/01.res.69.5.1226. [DOI] [PubMed] [Google Scholar]
- 71.Akella AB, Ding XL, Cheng R, et al. Diminished Ca2+ sensitivity of skinned cardiac muscle contractility coincident with troponin T-band shifts in the diabetic rat. Circ Res. 1995;76:600–606. doi: 10.1161/01.res.76.4.600. [DOI] [PubMed] [Google Scholar]
- 72.Thierfelder L, Watkins H, MacRae C, et al. Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell. 1994;77:701–712. doi: 10.1016/0092-8674(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 73.Feng HZ, Jin JP. Coexistence of cardiac troponin T variants reduces heart efficiency. Am J Physiol Heart Circ Physiol. 2010;299:H97–H105. doi: 10.1152/ajpheart.01105.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gomes AV, Venkatraman G, Davis JP, et al. Cardiac troponin T isoforms affect the Ca(2+) sensitivity of force development in the presence of slow skeletal troponin I: insights into the role of troponin T isoforms in the fetal heart. J Biol Chem. 2004;279:49579–49587. doi: 10.1074/jbc.M407340200. [DOI] [PubMed] [Google Scholar]
- 75.Baudenbacher F, Schober T, Pinto JR, et al. Myofilament Ca2+ sensitization causes susceptibility to cardiac arrhythmia in mice. J Clin Invest. 2008;118:3893–3903. doi: 10.1172/JCI36642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wei B, Jin JP. Troponin T isoforms and posttranscriptional modifications: Evolution, regulation and function. Arch Biochem Biophys. 2011;505:144–154. doi: 10.1016/j.abb.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jin JP. Alternative RNA splicing-generated cardiac troponin T isoform switching: a non-heart-restricted genetic programming synchronized in developing cardiac and skeletal muscles. Biochem Biophys Res Commun. 1996;225:883–889. doi: 10.1006/bbrc.1996.1267. [DOI] [PubMed] [Google Scholar]
- 78.Gomes AV, Guzman G, Zhao J, et al. Cardiac troponin T isoforms affect the Ca2+ sensitivity and inhibition of force development. Insights into the role of troponin T isoforms in the heart. J Biol Chem. 2002;277:35341–35349. doi: 10.1074/jbc.M204118200. [DOI] [PubMed] [Google Scholar]
- 79.Warf MB, Diegel JV, von Hippel PH, et al. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. PNAS. 2009;106:9203–9208. doi: 10.1073/pnas.0900342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Charlet-B N, Logan P, Singh G, et al. Dynamic Antagonism between ETR-3 and PTB Regulates Cell Type-Specific Alternative Splicing. Mol Cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 81.Goo YH, Cooper TA. CUGBP2 directly interacts with U2 17S snRNP components and promotes U2 snRNA binding to cardiac troponin T pre-mRNA. Nucleic Acids Research. 2009;13:4275–4286. doi: 10.1093/nar/gkp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Greaser ML, Warren CM, Esbona K, et al. Mutation that dramatically alters rat titin isoform expression and cardiomyocyte passive tension. Journal of Molecular and Cellular Cardiology. 2008;44:983–991. doi: 10.1016/j.yjmcc.2008.02.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin JJ, Warren KS, Wamboldt DD, et al. Tropomyosin isoforms in nonmuscle cells. Int Rev Cytol. 1997;170:1–38. doi: 10.1016/s0074-7696(08)61619-8. [DOI] [PubMed] [Google Scholar]
- 84.Phillips GN, Lattman EE, Cummins P, et al. Crystal structure and molecular interactions of tropomyosin. Nature. 1979;278:413–417. doi: 10.1038/278413a0. [DOI] [PubMed] [Google Scholar]
- 85.Cooper JA. Actin dynamics: tropomyosin provides stability. Curr Biol. 2002;12:R523–R525. doi: 10.1016/s0960-9822(02)01028-x. [DOI] [PubMed] [Google Scholar]
- 86.Yin Z, Ren J, Guo W. Sarcomeric protein isoform transitions in cardiac muscle: a journey to heart failure. Biochim Biophys Acta. 2015;1852:47–52. doi: 10.1016/j.bbadis.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perry SV. Vertebrate tropomyosin: distribution, properties and function. J Muscle Res Cell Motil. 2001;22:5–49. doi: 10.1023/a:1010303732441. [DOI] [PubMed] [Google Scholar]
- 88.Denz CR, Narshi A, Zajdel RW, et al. Expression of a novel cardiac-specific tropomyosin isoform in humans. Biochem Biophys Res Commun. 2004;320:1291–1297. doi: 10.1016/j.bbrc.2004.06.084. [DOI] [PubMed] [Google Scholar]
- 89.Rajan S, Jagatheesan G, Karam CN, et al. Molecular and functional characterization of a novel cardiac-specific human tropomyosin isoform. Circulation. 2010;121:410–418. doi: 10.1161/CIRCULATIONAHA.109.889725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Karam CN, Warren CM, Rajan S. Expression of tropomyosin-κ induces dilated cardiomyopathy and depresses cardiac myofilament tension by mechanisms involving cross-bridge dependent activation and altered tropomyosin phosphorylation. J Muscle Res Cell Motil. 2011;31:315–322. doi: 10.1007/s10974-010-9237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Muthuchamy M, Pajak L, Howles P, et al. Developmental analysis of tropomyosin gene expression in embryonic stem cells and mouse embryos. Mol Cell Biol. 1993;13:3311–3323. doi: 10.1128/mcb.13.6.3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jagatheesan G, Rajan S, Wieczorek DF. Investigations into tropomyosin function using mouse models. J Mol Cell Cardiol. 2010;48:893–898. doi: 10.1016/j.yjmcc.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen CD, Kobayashi R, Helfman DM. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat β-tropomyosin gene. Genes Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guo W, Mulligan GJ, Wormsley S, et al. Alternative splicing of beta-tropomyosin pre-mRNA: cis-acting elements and cellular factors that block the use of a skeletal muscle exon in nonmuscle cells. Genes Dev. 1991;5:2096–2107. doi: 10.1101/gad.5.11.2096. [DOI] [PubMed] [Google Scholar]
- 95.Smith CW, Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989;56:749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- 96.Ellis PD, Smith CW, Kemp P. Regulated tissue-specific alternative splicing of enhanced green fluorescent protein transgenes conferred by alpha-tropomyosin regulatory elements in transgenic mice. J Biol Chem. 2004;279:36660–36669. doi: 10.1074/jbc.M405380200. [DOI] [PubMed] [Google Scholar]
- 97.Crawford JB, Patton JG. Activation of α-Tropomyosin Exon 2 Is Regulated by the SR Protein 9G8 and Heterogeneous Nuclear Ribonucleoproteins H and F. Mol Cell Biol. 2006;26:8791–8802. doi: 10.1128/MCB.01677-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gooding C, Edge C, Lorenz M, et al. MBNL1 and PTB cooperate to repress splicing of Tpm1 exon 3. Nucleic Acids Res. 2013;41:4765–4782. doi: 10.1093/nar/gkt168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lu QL, Yokota T, Takeda S, et al. The status of exon skipping as a therapeutic approach to Duchenne muscular dystrophy. Mol Ther. 2011;19:9–15. doi: 10.1038/mt.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wheeler TM, Sobczak K, Lueck JD, et al. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rau F, Lainé J, Ramanoudjame L, et al. Abnormal splicing switch of DMD's penultimate exon compromises muscle fibre maintenance in myotonic dystrophy. Nat Commun. 2015;6:7205. doi: 10.1038/ncomms8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Azad RF, Driver VB, Tanaka K, et al. Antiviral activity of a phosphorothioate oligonucleotide complementary to RNA of the human cytomegalovirus major immediate-early region. Antimicrob Agents Chemother. 1993;37:1945–1954. doi: 10.1128/aac.37.9.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Anderson KP, Fox MC, Brown-Driver V, et al. Inhibition of human cytomegalovirus immediate-early gene expression by an antisense oligonucleotide complementary to immediate-early RNA. Antimicrob Agents Chemother. 1996;40:2004–2011. doi: 10.1128/aac.40.9.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Merki E, Graham MJ, Mullick AE, et al. Antisense oligonucleotide directed to human apolipoprotein B-100 reduces lipoprotein(a) levels and oxidized phospholipids on human apolipoprotein B-100 particles in lipoprotein(a) transgenic mice. Circulation. 2008;118:743–753. doi: 10.1161/CIRCULATIONAHA.108.786822. [DOI] [PubMed] [Google Scholar]
- 105.Kruger M, Sachse C, Zimmermann WH, et al. Thyroid hormone regulates developmental titin isoform transitions via the phosphatidylinositol-3-kinase/AKT pathway. Circ Res. 2008;102:439–447. doi: 10.1161/CIRCRESAHA.107.162719. [DOI] [PubMed] [Google Scholar]
- 106.Krüger M, Babicz K, von Frieling-Salewsky M, et al. Insulin signaling regulates cardiac titin properties in heart development and diabetic cardiomyopathy. J Mol Cell Cardiol. 2010;48:910–916. doi: 10.1016/j.yjmcc.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 107.Zhu C, Yin Z, Ren J, et al. RBM20 is an essential factor for thyroid hormone-regulated titin isoform transition. J Mol Cell Biol. 2015;7:88–90. doi: 10.1093/jmcb/mjv002. [DOI] [PMC free article] [PubMed] [Google Scholar]