Abstract
Ca2+ is a ubiquitous intracellular messenger that regulates numerous physiological activities in humans, animals, plants, and bacteria. Cytosolic Ca2+ is kept at a low level, but subcellular organelles such as the endoplasmic reticulum (ER) and Golgi apparatus maintain high-concentration Ca2+ stores. Under resting conditions, store Ca2+ homeostasis is dynamically regulated to equilibrate between active Ca2+ uptake and passive Ca2+ leak processes. The evolutionarily conserved Transmembrane BAX Inhibitor-1 Motif-containing (TMBIM) proteins mediate Ca2+ homeostasis and cell death. This review focuses on recent advances in functional and structural analysis of TMBIM proteins in regulation of the two related functions. The roles of TMBIM proteins in pathogen infection and cancer are also discussed with prospects for treatment.
Keywords: Ca2+ homeostasis, Ca2+ signaling, Cell death, Apoptosis, Bax inhibitor-1, TMBIM, Membrane proteins, Cancer, Cellular stress, Ca2+ channel structure
1. Ca2+ homeostasis
Ca2+ is an essential signaling messenger in regulation of normal physiology and development. In solution, calcium exists as Ca2+ with two positive charges. This property allows it to interact with proteins, lipids and other biomolecules carrying negative charges. During the evolution of living organisms, from prokaryotes to plants and mammals, cytosolic Ca2+ concentration ([Ca2+]Cyto) has been restricted to and constantly maintained at a very low concentration of about 100 nM. In eukaryotes, cytosolic high Ca2+ concentration triggers cell death [1]. In order to prevent the accumulation of Ca2+ in cytosol, cells use a variety of Ca2+ toolkits such as pumps and channels to mobilize cytosolic Ca2+ to intracellular Ca2+ stores (Fig. 1). Endoplasmic reticulum (ER) and Golgi apparatus are two major Ca2+ stores [2–4] where [Ca2+] can be at the 0.3–0.5 mM range which is 3000–5000 fold higher than that in cytoplasm. At physiological conditions, [Ca2+] is balanced between cytosol and subcellular organelles so that Ca2+ stores are not overloaded and the cytosolic buffering capability is sufficient; and stress-induced Ca2+ release will trigger appropriate signaling events for cytoprotection [5–7]. However, under prolonged stress conditions, for example ER stress [7,8], Ca2+ could be released from ER, transferred into mitochondria, and causes Ca2+ -mediated cell death [1,9–11].
Fig.1.
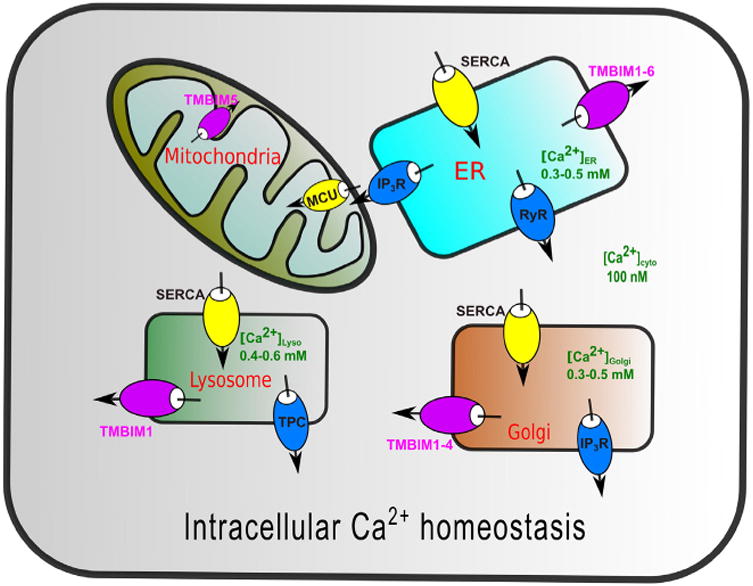
Intracellular Ca2+ homeostasis. Intracellular organelles are calcium stores where Ca2+ pumps and Ca2+ channels dynamically regulate intracellular Ca2+ homeostasis. Overloaded store Ca2+ causes Ca2+ transfer to mitochondria and induces Ca2+ -mediated cell death.
2. Apoptosis and its regulators
Apoptosis, or programmed cell death, controls the fate of cells in multi-cellular organisms during development and in response to cellular signals and stresses [12]. Appropriate apoptosis protects organisms from cellular damage and diseases. While uncontrolled apoptosis leads to malfunction and disrupted apoptosis leads to abnormity or carcinogenesis [13]. Two pathways regulate apoptosis: intrinsic and extrinsic [12,14]. In the intrinsic pathway, proapoptotic protein Bax or Bak is activated and translocates to the mitochondrial outer membrane. It subsequently undergoes a conformational change and oligomerizes to form a pore through which mitochondrial apoptotic factors are released [15, 16]. These mitochondrial apoptotic factors, including cytochrome c, apoptosis induce factor (AIF) and Smac/DIABLO, flux into the cytosol and trigger cell death through caspase activation [17]. In the extrinsic pathway, extracellular death signals activate cell surface receptor CD95 (also known as Fas) or tumor necrosis factor receptor (TNFR1) and induce an across-the-membrane signaling cascade leading to caspase activation and, ultimately, apoptosis.
Apoptosis is precisely regulated by at least two groups of regulators, e.g. proapoptotic regulators and antiapoptotic regulators related to B-cell lymphoma 2 (Bcl-2) family [18–20]. Proapoptotic regulators of the Bcl-2 family contain a BH3 domain (Fig. 2). Among these, Bim, Bad, Puma, Bid, Bik, Bmf, Hrk and Noxa have a BH3-only domain [21]; and Bax, Bak, and Bok have multiple domains (BH1–3) [22]. Antiapoptotic regulators of the Bcl-2 family include Bcl-2, Bcl-XL, Bcl-w, and Mcl-1 [23]. In addition to having the BH1–3 domains, antiapoptotic Bcl-2 regulators have an N-terminal BH4 domain that can bind directly or indirectly to the proapoptotic Bax to inactivate it [24,25]. Comparing to antiapoptotic regulators, proapoptotic regulators can function by either directly promoting the formation of the permeation transition pore (Bax or Bak) or competitively binding to antiapoptotic proteins to release Bax or Bak for initiation of apoptosis (BH3-only). Proapoptotic Bok is persistently active without apparent regulators except Mcl-1 [26]. It is proposed that Bok is regulated at expression levels through ER-associated degradation pathways [22,27].
Fig.2.
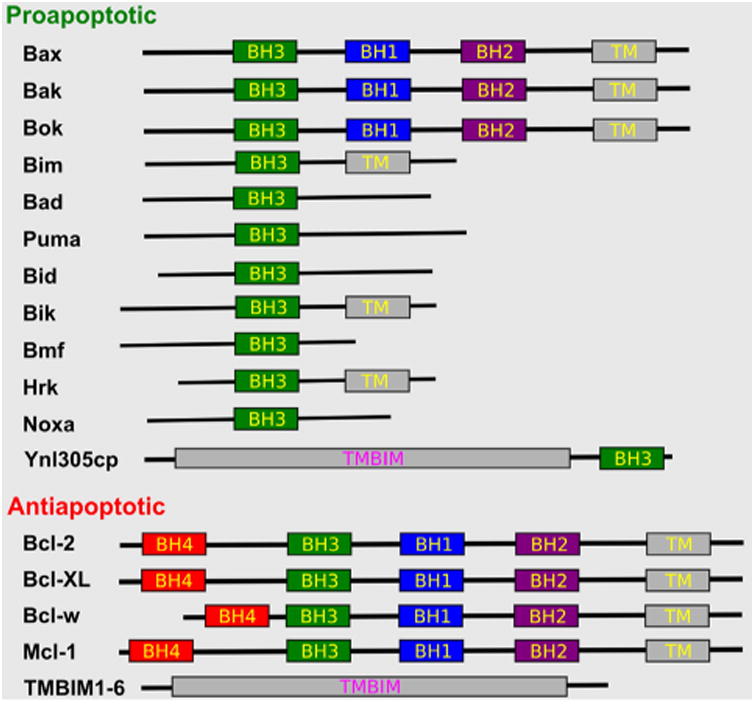
Apoptosis regulators of Bcl-2 and TMBIM families. Bcl-2 family regulators are either soluble or membrane anchored; while TMBIM family regulators are multi-pass membrane proteins. The BH domains, transmembrane domains (TMs), and multi-pass TMBIM domains (TMBIM) were colored differently.
Proapoptotic and antiapoptotic proteins were originally thought to be only for soluble proteins or membrane anchored single-transmembrane proteins. Recently multi-pass transmembrane proteins of the TMBIM family were found to either suppress or promote cell death depending on whether there is a BH3-domain at the C-terminus [28,29]. Thus far, the majority of the TMBIM family is only comprised of antiapoptotic proteins, which inhibit apoptosis of both intrinsic and extrinsic pathways [30]. The only known proapoptotic TMBIM protein is the yeast homolog Ynl305cp in which a BH3 domain at the C-terminus promotes cell death [31] (Fig. 2).
3. TMBIM proteins in mediating cell death and Ca2+ homeostasis
In humans, there are six TMBIM homologs named as TMBIM1–6 based on the HUGO gene nomenclature. TMBIM proteins are defined by sequence conservations to Bax inhibitor-1 (BI-1, or TMBIM6), which was discovered as a human gene product capable of blocking Bax-induced cell death in yeast [32]. Human TMBIM proteins contain multiple predicted transmembrane helices varying mainly in their N-terminal extensions [29,30,33,34] (Fig. 3). The TMBIM family is essentially ubiquitous, present in humans, animals, plants, fungi and prokaryotes [29] (Fig. 3). Based on phylogenetic analysis, TMBIM1–3 are clustered together and TMBIM4–6 are separately clustered and remotely related [29,34].
Fig.3.

Sequence alignment of TMBIM proteins. The TMBIM family is highly conserved in humans, animals, worms, plants, fungi, bacteria, and certain viruses. The conserved residues were marked as “*”. The seven transmembrane helices based on the BsYetJ structure were drawn as cylinders and were colored differently.
TMBIM6 (BI-1 or TEGT) is an ER membrane protein with seven predicted transmembrane helices. TMBIM6 regulates apoptosis sensitive to ER-stress triggers such as etoposide and staurosporine, but not to extrinsic apoptosis ligand FasL (also known as CD95L), consistent with its sole ER localization [32,35]. Reduced [Ca2+]ER is cytoprotective [36, 37]; and TMBIM6 reduces resting [Ca2+]ER as well as [Ca2+]Cyto upon treatment with ER-stress inducers such as thapsigargin or tunicamycin [35,38]. TMBIM6 regulates Ca2+ flux in a pH-dependent manner [39–41].
TMBIM1 (RECS1, LFG3, PP1201, or PSEC0158) is localized to lysosome and endosome membranes in addition to ER and Golgi [42]. Relative to TMBIM6, TMBIM1 has a long N-terminal extension, presumably in which residing the localization information. TMBIM1 was identified from screening for genes responsive to centrifugal force and shear stress [43]. Deficiency of TMBIM1 in mice induced mice susceptibility to cystic medial degeneration and aortic dilation [44,45]. TMBIM1 suppresses Fas-mediated extrinsic apoptosis by interacting with Fas receptor [46] and mediates Ca2+ homeostasis by reducing [Ca2+]ER [42].
TMBIM2 (FAIM2, LFG, LFG2, NMP35, or KIAA0950) is localized to plasma membrane in addition to ER and Golgi [47]. TMBIM2 also suppresses Fas-induced apoptosis by interfering with caspase-8 activation [47]. Like TMBIM1, TMBIM2 has an N-terminal extension for localization. TMBIM2 is highly expressed in neurons and is rich at postsynaptic sites in dendrites [48]. The overexpression of TMBIM2 improved axon growth [49] and protected brain neurons from transient ischemia [50]. TMBIM2 is required for survival and maintenance of Purkinje and granule neuron cells [51]. TMBIM2 decreases releasable ER Ca2+ [42,52].
TMBIM3 (GRINA, LFG1, NMDAR1, or PM02) is localized to ER and Golgi apparatus and is mainly expressed in neuron cells [53,54]. TMBIM3 suppresses ER-stress induced cell death and reduces [Ca2+]ER. TMBIM3's function is correlated with TMBIM6 [54]. In fact, TMBIM3 interacts with TMBIM6 in ER membrane and synergizes its function in Ca2+ homeostasis and cell death. The binary interaction is also linked to unfolded protein response (UPR) and autophagy [54].
TMBIM4 (LFG4, S1R, Z-protein, or GAAP) is also localized to ER and Golgi where it mediates intracellular Ca2+ homeostasis and cell death [42,55,56]. Through modulation of Ca2+ homeostasis and Ca2+ signaling, TMBIM4 can mediate cell adhesion and migration [57,58]. TMBIM4 suppresses cell death of both intrinsic pathways induced by ceramide and H2O2, and extrinsic pathways induced by Fas ligand and TNF. TMBIM4 is essential for cell survival; and RNA interference induced spontaneous cell death [56].
TMBIM5 (GHITM, MICS1, or DERP2), also known as growth hormone-inducible transmembrane protein (GHITM), is localized to the mitochondrial inner membrane where it is required for maintenance of mitochondrial structure and release of apoptotic factor cytochrome c [59]. Different from other TMBIM proteins, TMIBM5 has eight predicted transmembrane helices, seven TMBIM-conserved plus an additional one at the N-terminus. The N-terminal helix is hypothesized to be involved in the translocation and positioning of the protein to mitochondrial inner membrane. Whether TMBIM5 can mediate mitochondrial Ca2+ homeostasis is unknown. However, premature cleavage of the protein may end up with a localization to ER where it indeed mediates Ca2+ homeostasis [42].
Interestingly, with an ER localization, all TMBIM proteins mediate ER Ca2+ homeostasis and suppress intrinsic apoptosis pathways by reducing [Ca2+]Cyto upon ER stress [42]. In addition to suppressing the intrinsic pathway, TMBIM1, 2 and 4 suppress extrinsic pathways originating from CD95 or TNF receptor which may accumulate at the Golgi prior to their traffic to plasma membrane for signaling [60,61]. In light that TMBIM1, 2 and 4 also have a Golgi localization [42,62,63], it is likely interactions between Golgi TMBIM proteins and CD95 or TNF receptor may block membrane trafficking and thus suppress extrinsic pathways [46]. TMBIM6 has only the ER localization [35], consistent with it being only sensitive to ER-stress induced intrinsic pathways. It is intriguing that TMBIM3 has both ER and Golgi localizations; while it is only sensitive to ER-stress induced intrinsic pathways [54]. All together, we can envision that the functions of TMBIM proteins in cell death and Ca2+ homeostasis are closely correlated with their subcellular localizations.
To explain how TMBIM proteins mediate Ca2+ homeostasis, two models were proposed based on TMBIM6. One is a Ca2+/H+ antiporter in which a proton gradient drives the Ca2+ flux from ER to cytoplasm. At the C-terminus of TMBIM6, there is a lysine-rich motif “EKDKKKEKK” which has certain similarity to a sodium channel and was thus proposed to form a pH-sensitive Ca2+ selectivity filter [32,39,40]. The other one is a channel in which the structure opens as a pore to allow the flow of Ca2+ along its gradient [64,65]. For both models, mediation of Ca2+ flux by TMBIM6 is pH dependent [41,66].
4. Ca2+-leak mechanism inspired by bacterial TMBIM structures
TMBIM proteins are conserved in almost the entire life kingdom (Fig. 2), suggesting that TMBIM-mediated cellular protection is an ancient trait, concomitant with the formation of single-cell organisms. The recent crystal structures of a bacterial TMBIM protein, YetJ from Bacillus subtilis (BsYetJ), provide substantial structural insights for mechanistic understanding the Ca2+ flux and homeostasis mechanisms [34].
The sequence alignment of BsYetJ with human TMBIM proteins are quite well in particular for the C-terminal region (TM6-7) where two aspartic acid residues (Asp171 on TM6 and Asp195 on TM7 in BsYetJ) are highly conserved (Fig. 3). BsYetJ functionally uptakes Ca2+ in E. coli and mediates a pH-dependent Ca2+ flux in proteoliposomes [34]. The BsYetJ structure was solved at three difference pH conditions, each corresponding to a different conformational state: closed at pH 8, open at pH 6 and in equilibrium at pH 7 (Fig. 4A–F). In these structures, BsYetJ is a monomer and contains seven transmembrane helices as predicted. The structure has a novel fold consisting of two inversely assembled triple-helix-sandwich repeats that wraps the TM7 in the middle. At pH 7, both the pH 8 and pH 6 structures coexist in equilibrium (Fig. 4E–F), with TM2 underwent conformational changes by as large as 14 Å. Consisting of only 14 residues in the closed-state structure, TM2 is short, with relatively long connecting loops to TM1 and TM3, consistent with it being flexible for conformational changes.
Fig.4.
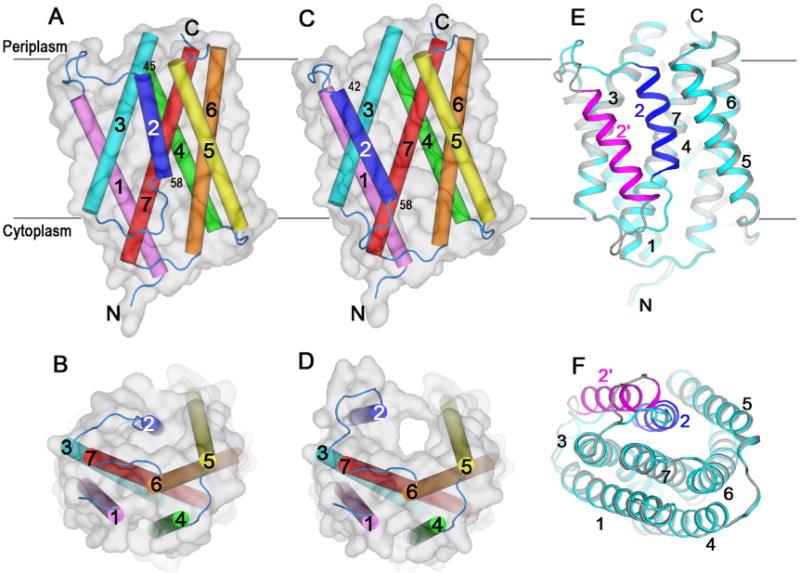
Structures of BsYetJ in multiple conformational states. (A–B) Closed conformation structure at pH 8 in two orientations. (C-D) Open conformation structure at pH 6 in two orientations. (E-F) Equilibrated conformations of closed and open structures at pH 7 in two orientations. (A), (C) and (E) are side views and (B), (D), and (F) are top views from the periplasm. The seven transmembrane helices were color-coded. The grey transparent surfaces in (A-D) show the overall shapes of these structures. In (E) and (F), the closed conformation was colored in blue for TM2 and cyan for the rest; and the open conformation was colored in magenta for TM2 and grey for the rest.
Structurally, TM2 can regulate the Ca2+ flux. In the structure, the C-terminal residues Asp171 and Asp195 form an H-bond (Fig. 5A). At pH 8 the carboxylate group of Asp171 is negatively charged and forms a hydrogen-bonded salt bridge with Arg60 from TM2 and latches TM2 in the closed conformation. At pH 6, a predicted protonation of Asp171 disrupts its interaction with Arg60 and thus opens the structure (Fig. 4D). Asp171 and Asp195 are TMBIM-family conserved (Fig. 3) and may form a pH sensor that can sense environmental pH and regulate the pH-dependent Ca2+ flux.
Fig.5.
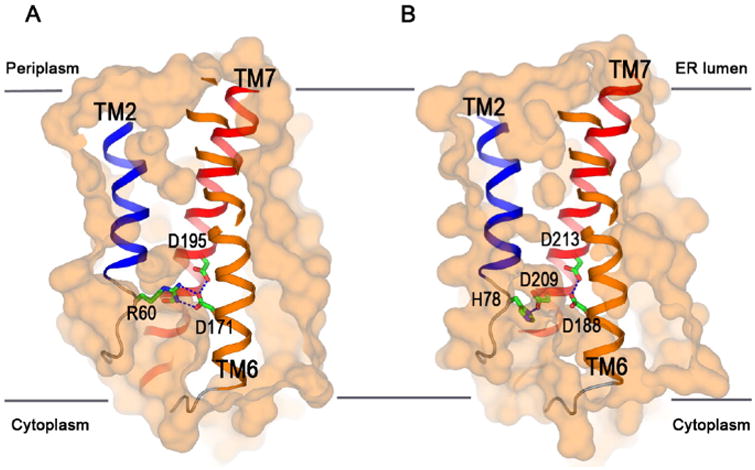
Molecular basis for Ca2+ leak. (A) BsYetJ structure in the closed conformation with the Asp171-Asp195 pH sensor. The sensor forms two H-bonds with Arg60 from TM2 and locks the structure in a closed conformation. The protonation of Asp171 at a neutral pH would disrupt TM2 and allow a Ca2+ leak. (B) Homology model of human TMBIM6 in the closed conformation with an equivalent pH sensor comprising of Asp188, Asp209 and Asp213. It is proposed that the protonation states of these aspartate residues will regulate Ca2+ leak in a pH-dependent manner.
Based on the BsYetJ structure and the sequence alignment of the TMBIM family (Figs. 2 and 3), insights into the molecular mechanisms of human TMBIM-mediated Ca2+ homeostasis may be proposed. First, all six human TMBIM proteins have the two conserved aspartic acid residues; and they should necessarily form an H-bond and allow the aspartic acid residue on TM6 to latch/release a positively charged residue from TM2 for regulation of Ca2+ leak. Sequence alignment shows most homologs have an arginine at the latch position, suggesting a conserved pH-dependent Ca2+ leak mechanism. However, TMBIM5 has an aspartic acid and TMBIM6 has a histidine, suggesting existence of alternative latch mechanisms and sensitivity to environmental pHs. For example, in the TMBIM homology model, deprotonated Asp209 (Ser191 in BsYetJ) on TM6 favors an H-bond with His78 from TM2 (Fig. 5B). Asp209 is unique to TMBIM6 and is in close proximity to the D188–D213 pH sensor (Asp171–Asp195 in BsYetJ). Asp 209 and Asp213 in TMBIM6 have been demonstrated critically important in TMBIM6-mediated Ca2+ homeostasis and cell death [64]. It is likely that the three aspartic residues will sense pH and regulate the Ca2+ flux with a bell-shaped pH-dependence [34,67]. At pH 8, TMBIM6 would be closed due to deprotonation of Asp209 and formation of an H-bond with His78 (Fig. 5B). At a more neutral pH of 7, the pKa of Asp209 would increase; and Asp209 will be partially protonated which disrupts its interaction with His78 and allows for formation of a negatively charged pore for Ca2+ passage. At pH 5–6, the pore is open; while the complete protonation of Asp209, Asp188 and Asp213 makes the pore hydrophobic and unfavorable for Ca2+ flux. In addition, lipid hydrocarbon tails might also fill up the hydrophobic pore which further seals the pore to prevent Ca2+ to pass through. This model fits with the experimental data obtained for TMBIM6-mediated Ca2+ release [41].
Different from other known Ca2+ channels in which there is a defined filter selectively for Ca2+ [68,69], TMBIM proteins are unique in that there is no clear Ca2+ filter inside, consistent with it being functioning as a monomer and with TM2 opening and closing laterally. Although a higher TMBIM oligomer does exist, it is not required for regulation of either Ca2+ homeostasis or cell death [70]. The unique structures of TMBIM proteins make them suitable as intrinsic Ca2+ leak channels to prevent the overload of intracellular Ca2+ stores. Necessarily, the leaky capacity of TMBIM proteins must be regulated to reach an equilibrium with Ca2+ loading capacity by pumps as depleted Ca2+ stores also cause cell death.
5. Functional conservation of TMBIM proteins in plant and bacteria
TMIBM proteins, originally identified in humans and mammals, were also identified to be present ubiquitously in plants and bacteria [34,71–74].
In plants, the first TMBIM protein was identified by functional screening [72,75] in a way similar to TMBIM6. Plant TMBIM proteins mediate Ca2+ homeostasis [76] and have resistance to various biotic and abiotic stresses including Bax protein, heat and cold shock, fungal toxin, tunicamycin, fungal pathogen, sucrose starvation, and H2O2 [77]. The growing plant TMBIM functions include suppression of Bax-induced cell death [73,77], regulation of calmodulin binding [76], enhanced sphingolipid synthesis upon cold stress [78], and enhanced drought tolerance [79].
In bacteria, TMBIM proteins are classified as BI-1 family in the uniprot database (www.uniprot.org). However, so far, the physiological functions of these bacterial TMBIM proteins are not clear. In Escherichia coli (strain K12), there are three TMBIM proteins YccA, YbhL and YbhM. Among these, YccA was identified to be an inhibitor of FtsH AAA+ protease that can otherwise degrade translocon subunit SecY under stress conditions [80]. As BsYetJ can uptake Ca2+ when overexpressed in E. coli [34], likely these bacterial TMBIM proteins have similar Ca2+ uptake activities. The consequence of Ca2+ uptake in bacteria remains to be defined in particular under physiological and stress conditions. It is noted that ybhL and ybhM genes are physically clustered in the E. coli genome, suggesting that their expression might be regulated by a common promotor and transcription factor(s). In analogy to their eukaryotic relatives in cytoprotection, it may be hypothesized that under stress conditions such as heat and cold shock, bacterial TMBIM proteins might be involved in cytoprotection through an as-yet-unknown mechanism.
6. TMBIM proteins hijacked for invasion
In eukaryotic cells, most TMBIM proteins are located in subcellular membranes rather than plasma membranes. Due to their critical roles in playing with the life and death, TMBIM proteins have been hijacked by certain pathogens for infection. Below are a few examples of demonstrating how the TMBIM pathway is being jeopardized. Likely new ways of TMBIM-mediated pathogen attacks are to be identified.
Camelpox virus (CMLV), cowpox virus (CPXV) and certain vaccinia viruses (VACV) encode virulent membrane proteins of TMBIM4 like [56,57]. These viral TMBIM4 proteins share ∼70% sequence identity with human TMBIM4 and can mimic TMBIM4 to prevent host cell death during their infection processes. During the virus infection, viral TMBIM4 proteins are dumped into host cells together with other virulence factors. The viral TMBIM4 proteins then translocate to ER and Golgi to suppress the cell death so that viruses can replicate in host cells without killing them. These viral TMBIM proteins regulate host cell Ca2+ homeostasis and suppression of cell death indistinguishable to that of human TMBIM4 [55,70].
In addition to the TMBIM4 pathway, TMBIM6 is also the target for attack by some pathogen bacteria enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic Escherichia coli (EHEC) through an effector protein NleH [81]. NleH, after injection into host cells through the bacterial type II secretion system, can recognize and bind to the N-terminus of TMBIM6 to gain antiapoptosis function. It is intriguing that the binding of NleH to TMBIM6 increased [Ca2+]Cyto, while TMBIM6 alone reduced [Ca2+]Cyto. Except TMBIM5, which is mainly in the mitochondrial inner membrane and less likely to be attacked by cytosolic pathogenic effectors, TMBIM1–4 could also be possible targets to be hijacked by bacterial pathogens for assisting their infection and survival. Future studies on pathogen-TMBIM1–4 interactions may be expected.
Based on a BLAST search against human TMBIM proteins, there are ten human cytomegalovirus (HCMV) viral proteins of the US12 family (US12–21) with appreciable homology to TMBIM proteins [82–84]. Like TMBIM proteins, these US12 proteins all have a predicted 7TM topology. These proteins are essential for virulence through yet a not clear pathway. Nevertheless, based on their similarities to TMBIM proteins, it can be hypothesized that these remote TMBIM homologs may localize to subcellular organelles and contribute to the modulation of Ca2+ homeostasis and cell death pathways for their virulence.
7. TMBIM proteins as regulators of Ca2+ signaling
In addition to functioning as standalone proteins in mediation of Ca2+ homeostasis, TMBIM proteins interact with other Ca2+ signaling proteins for regulation of cell life and death.
TMBIM6 inhibits Bax-induced cell death through the intrinsic pathway. It is puzzling that TMBIM6 does not interact with Bax. Rather, it interacts with antiapoptotic proteins Bcl-2 and Bcl-XL involving the BH4 domain [32]. Whether TMBIM6 interacts with other BH4-domaining containing antiapoptotic proteins, e.g. Bcl-w and Mcl-1 (Fig. 2) is waiting for experiments. Recently TMBIM2 and TMBIM4 were also shown to interact with Bcl-2 and Bcl-XL, thus broadening the generality of the Ca2+ signaling mediated by the two families of apoptosis regulators [52]. It is noted that the BH4 domain is amphiphilic; and the TM6 and TM7 in TMBIM proteins are also amphiphilic and contribute to formation of a transmembrane pore. Hence the interactions between Bcl-2 and TMBIM proteins might have the amphiphilic BH4 domain helix in the TMBIM pore [34]. Because Bcl-2 and Bcl-XL mediate Ca2+ homeostasis upstream of TMBIM6 [85], such interactions should allow a Ca2+ leak to reduce [Ca2+]ER and to suppress cell death [39,85].
TMBIM proteins, atleast TMBIM3, TMBIM4 and TMBIM6, have interactions with IP3 activated Ca2+ channels (IP3Rs) for regulation of ER stress and autophagy [86–89]. Bcl-2 family members have been longtime thought to mediate Ca2+ signaling pathways [90,91]. For example, Bcl-2 and Bcl-XL can directly interact with IP3R channels to regulate their Ca2+-flux properties [92–99]. It is possible that TMBIM proteins function as docking proteins for Bcl-2- and Bcl-XL-mediated IP3R function. However, a challenging concept for this is that the BH4 domain of Bcl-2 implicated in interaction with TMBIM6 is also responsible for binding IP3Rs. Therefore, it is also possible that TMBIM6 negatively regulates the interaction of Bcl-2 with IP3Rs and vice versa. Future in vivo and in vitro studies of interplays between TMBIM, Bcl-2/Bcl-XL and IP3R proteins would be pivotal to understand the associated Ca2+ signaling and cell death.
8. TMBIM proteins in cancer
Apoptosis plays a crucial role in carcinogenesis as well as in resistance to chemotherapy and radiotherapy [100]. Despite differences in genetic background and lineage among cancer cells, they all share characteristic hallmarks such as evasion from apoptosis surveillance [13, 101]. TMBIM6 expression is upregulated in certain cancers including prostate, breast, glioma, uterine, ovarian and lung [86,102–104]. Knockdown of TMBIM6 expression by RNA interference induced spontaneous cell death of cancer cells in prostate and breast [102,103]. Clearly these cancers have escaped the surveillance of apoptosis by overexpression of antiapoptotic proteins including TMBIM6.
In addition to TMBIM6, TMBIM3 expression is up regulated in breast cancer, colorectal cancer, gastric cancer, head and neck cancer; and TMBIM4 expression is upregulated in colorectal cancer and leukemia [28]. Conversely, TMBIM1 is significantly down regulated in breast cancer, leukemia and sarcoma; and TMBIM2 is down regulated in brain and CNS cancers [105]. TMBIM5, although with an inner mitochondrial membrane localization, has an expression that is up regulated in head and neck cancer and down regulated in leukemia [28]. Because TMBIM proteins regulate Ca2+ homeostasis, abnormal TMBIM expression (up or down) in cancer cells may have changed the Ca2+ homeostasis toward favoring cancer cell survival and progression. Altered Ca2+ homeostasis mediates many hallmarks of cancer [13,106,107]. Therefore, remodeling of the Ca2+ homeostasis in cancer mediated by TMBIM proteins might provide a novel strategy for treatment.
9. Conclusion and prospects
TMBIM proteins are evolutionary conserved membrane proteins regulating both Ca2+ homeostasis and cell death. TMBIM proteins have novel 7TM-fold structures that allow the leak of Ca2+ for regulating Ca2+ homeostasis. Their pH-dependence and interactions within TMBIM proteins and with Bcl-2 and IP3R family proteins add another level of regulation of Ca2+ signaling and cell death. For the future, it is needed to study the detailed mechanisms of TMBIM proteins in organelle-specific cell death mechanisms, pathogen-host interactions, plant and bacteria stress resistance, pH-dependent regulation of Ca2+ signaling, and remodelling of Ca2+ signaling in cancer. The work will pave the way to harness the highly conserved and ubiquitously expressed TMBIM proteins for biological and biomedical applications.
Acknowledgments
The author thanks reviewers for their insightful suggestions and apologizes for not being able to cite all high-quality references relevant to the topic. This work was supported in part by the National Institutes of Health grant GM107462 and Brookhaven National Laboratory LDRD 15-034.
Footnotes
This article is part of a Special Issue entitled: ECS Meeting edited by Claus Heizmann, Joachim Krebs and Jacques Haiech.
Transparency document: The Transparency document associated with this article can be found, in online version.
Conflict of interest: The author declares no conflict of interest.
References
- 1.Pinton P, Giorgi C, Siviero R, Zecchini E, Rizzuto R. Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis. Oncogene. 2008;27:6407–6418. doi: 10.1038/onc.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinton P, Pozzan T, Rizzuto R. The Golgi apparatus is an inositol 1, 4, 5-trisphosphate-sensitive Ca2+ store, with functional properties distinct from those of the endoplasmic reticulum. EMBO J. 1998;17:5298–5308. doi: 10.1093/emboj/17.18.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sammels E, Parys JB, Missiaen L, De Smedt H, Bultynck G. Intracellular Ca2+ storage in health and disease: a dynamic equilibrium. Cell Calcium. 2010;47:297–314. doi: 10.1016/j.ceca.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Camello C, Lomax R, Petersen O, Tepikin A. Calcium leak from intracellular stores—the enigma of calcium signalling. Cell Calcium. 2002;32:355–361. doi: 10.1016/s0143416002001926. [DOI] [PubMed] [Google Scholar]
- 5.Farfariello V, Iamshanova O, Germain E, Fliniaux I, Prevarskaya N. Calcium homeostasis in cancer: a focus on senescence. Biochim Biophys Acta. 2015;1853:1974–1979. doi: 10.1016/j.bbamcr.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Arruda AP, Hotamisligil GS. Calcium homeostasis and organelle function in the pathogenesis of obesity and diabetes. Cell Metab. 2015;22:381–397. doi: 10.1016/j.cmet.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krebs J, Agellon LB, Michalak M. Ca2+ homeostasis and endoplasmic reticulum (ER) stress: an integrated view of calcium signaling. Biochem Biophys Res Commun. 2015;460:114–121. doi: 10.1016/j.bbrc.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta, Mol Cell Res. 2013;1833:3460–3470. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchi S, Patergnani S, Pinton P. The endoplasmic reticulum–mitochondria connection: one touch, multiple functions. Biochim Biophys Acta Bioenerg. 2014;1837:461–469. doi: 10.1016/j.bbabio.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 10.Orrenius S, Zhivotovsky B, Nicotera P. Regulation of cell death: the calcium-apoptosis link. Nat Rev Mol Cell Biol. 2003;4:552–565. doi: 10.1038/nrm1150. [DOI] [PubMed] [Google Scholar]
- 11.Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115:2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Rudin CM, Thompson CB. Apoptosis and disease: regulation and clinical relevance of programmed cell death. Annu Rev Med. 1997;48:267–281. doi: 10.1146/annurev.med.48.1.267. [DOI] [PubMed] [Google Scholar]
- 15.Czabotar PE, Westphal D, Dewson G, Ma S, Hockings C, Fairlie WD, Lee EF, Yao SG, Robin AY, Smith BJ, Huang DCS, Kluck RM, Adams JM, Colman PM. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell. 2013;152:519–531. doi: 10.1016/j.cell.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 16.Bleicken S, Jeschke G, Stegmueller C, Salvador-Gallego R, Garcia-Saez AJ, Bordignon E. Structural model of active Bax at the membrane. Mol Cell. 2014;56:496–505. doi: 10.1016/j.molcel.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Favaloro B, Allocati N, Graziano V, Di Ilio C, De Laurenzi V. Role of apoptosis in disease. Aging. 2012;4:330–349. doi: 10.18632/aging.100459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardwick JM, Soane L. Multiple functions of BCL-2 family proteins. Cold Spring Harb Perspect Biol. 2013;5:a008722. doi: 10.1101/cshperspect.a008722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szegezdi E, Macdonald DC, Ni Chonghaile T, Gupta S, Samali A. Bcl-2 family on guard at the ER. Am J Phys Cell Phys. 2009;296:C941–C953. doi: 10.1152/ajpcell.00612.2008. [DOI] [PubMed] [Google Scholar]
- 20.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 21.Kelly PN, Strasser A. The role of Bcl-2 and its pro-survival relatives in tumourigenesis and cancer therapy. Cell Death Differ. 2011;18:1414–1424. doi: 10.1038/cdd.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Einsele-Scholz S, Malmsheimer S, Bertram K, Stehle D, Johanning J, Manz M, Daniel PT, Gillissen BF, Schulze-Osthoff K, Essmann F. Bok is a genuine multi-BH-domain protein that triggers apoptosis in the absence of Bax and Bak. J Cell Sci. 2016;129:2213–2223. doi: 10.1242/jcs.181727. [DOI] [PubMed] [Google Scholar]
- 23.Akl H, Vervloessem T, Kiviluoto S, Bittremieux M, Parys JB, De Smedt H, Bultynck G. A dual role for the anti-apoptotic Bcl-2 protein in cancer: mitochondria versus endoplasmic reticulum. Biochim Biophys Acta, Mol Cell Res. 2014;1843:2240–2252. doi: 10.1016/j.bbamcr.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 24.Barclay LA, Wales TE, Garner TP, Wachter F, Lee S, Guerra RM, Stewart ML, Braun CR, Bird GH, Gavathiotis E, Engen JR, Walensky LD. Inhibition of pro-apoptotic BAX by a noncanonical interaction mechanism. Mol Cell. 2015;57:873–886. doi: 10.1016/j.molcel.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vervloessem T, La Rovere R, Bultynck G. Antagonizing Bcl-2's BH4 domain in cancer. Aging. 2015;7:748–749. doi: 10.18632/aging.100828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu SY, Kaipia A, McGee E, Lomeli M, Hsueh AJW. Bok is a pro-apoptotic Bcl-2 protein with restricted expression in reproductive tissues and heterodimerizes with selective anti-apoptotic Bcl-2 family members. Proc Natl Acad Sci U S A. 1997;94:12401–12406. doi: 10.1073/pnas.94.23.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Llambi F, Wang YM, Victor B, Yang M, Schneider DM, Gingras S, Parsons MJ, Zheng JH, Brown SA, Pelletier S, Moldoveanu T, Chen TS, Green DR. BOK is a non-canonical BCL-2 family effector of apoptosis regulated by ER-associated degradation. Cell. 2016;165:421–433. doi: 10.1016/j.cell.2016.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rojas-Rivera D, Hetz C. TMBIM protein family: ancestral regulators of cell death. Oncogene. 2015;34:269–280. doi: 10.1038/onc.2014.6. [DOI] [PubMed] [Google Scholar]
- 29.Henke N, Lisak DA, Schneider L, Habicht J, Pergande M, Methner A. The ancient cell death suppressor BAX inhibitor-1. Cell Calcium. 2011;50:251–260. doi: 10.1016/j.ceca.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Hu L, Smith TF, Goldberger G. LFG: a candidate apoptosis regulatory gene family. Apoptosis. 2009;14:1255–1265. doi: 10.1007/s10495-009-0402-2. [DOI] [PubMed] [Google Scholar]
- 31.Buttner S, Ruli D, Vogtle FN, Galluzzi L, Moitzi B, Eisenberg T, Kepp O, Habernig L, Carmona-Gutierrez D, Rockenfeller P, Laun P, Breitenbach M, Khoury C, Frohlich KU, Rechberger G, Meisinger C, Kroemer G, Madeo F. A yeast BH3-only protein mediates the mitochondrial pathway of apoptosis. EMBO J. 2011;30:2779–2792. doi: 10.1038/emboj.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Q, Reed JC. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol Cell. 1998;1:337–346. doi: 10.1016/s1097-2765(00)80034-9. [DOI] [PubMed] [Google Scholar]
- 33.Ishikawa T, Watanabe N, Nagano M, Kawai-Yamada M, Lam E. Bax inhibitor-1: a highly conserved endoplasmic reticulum-resident cell death suppressor. Cell Death Differ. 2011;18:1271–1278. doi: 10.1038/cdd.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang Y, Bruni R, Kloss B, Assur Z, Kloppmann E, Rost B, Hendrickson WA, Liu Q. Structural basis for a pH-sensitive calcium leak across membranes. Science. 2014;344:1131–1135. doi: 10.1126/science.1252043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chae HJ, Kim HR, Xu CY, Bailly-Maitre B, Krajewska M, Krajewski S, Banares S, Cui J, Digicaylioglu M, Ke N, Kitada S, Monosov E, Thomas M, Kress CL, Babendure JR, Tsien RY, Lipton SA, Reed JC. BI-1 regulates an apoptosis pathway linked to endoplasmic reticulum stress. Mol Cell. 2004;15:355–366. doi: 10.1016/j.molcel.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 36.Pozzan T, Rizzuto R. High tide of calcium in mitochondria. Nat Cell Biol. 2000;2:E25–E27. doi: 10.1038/35000095. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura K, Bossy-Wetzel E, Burns K, Fadel MP, Lozyk M, Goping IS, Opas M, Bleackley RC, Green DR, Michalak M. Changes in endoplasmic reticulum luminal environment affect cell sensitivity to apoptosis. J Cell Biol. 2000;150:731–740. doi: 10.1083/jcb.150.4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westphalen BC, Wessig J, Leypoldt F, Arnold S, Methner A. BI-1 protects cells from oxygen glucose deprivation by reducing the calcium content of the endoplasmic reticulum. Cell Death Differ. 2005;12:304–306. doi: 10.1038/sj.cdd.4401547. [DOI] [PubMed] [Google Scholar]
- 39.Ahn T, Yun CH, Kim HR, Chae HJ. Cardiolipin, phosphatidylserine, and BH4 domain of Bcl-2 family regulate Ca2+/H+ antiporter activity of human Bax inhibitor-1. Cell Calcium. 2010;47:387–396. doi: 10.1016/j.ceca.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 40.Ahn T, Yun CH, Chae HZ, Kim HR, Chae HJ. Ca2+/H+ antiporter-like activity of human recombinant Bax inhibitor-1 reconstituted into liposomes. FEBS J. 2009;276:2285–2291. doi: 10.1111/j.1742-4658.2009.06956.x. [DOI] [PubMed] [Google Scholar]
- 41.Kiviluoto S, Luyten T, Schneider L, Lisak D, Rojas-Rivera D, Welkenhuyzen K, Missaen L, De Smedt H, Parys JB, Hetz C, Methner A, Bultynck G. Bax inhibitor-1-mediated Ca2+ leak is decreased by cytosolic acidosis. Cell Calcium. 2013;54:186–192. doi: 10.1016/j.ceca.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 42.Lisak DA, Schacht T, Enders V, Habicht J, Kiviluoto S, Schneider J, Henke N, Bultynck G, Methner A. The transmembrane Bax inhibitor motif (TMBIM) containing protein family: tissue expression, intracellular localization and effects on the ER CA2+-filling state. Biochim Biophys Acta, Mol Cell Res. 2015;1853:2104–2114. doi: 10.1016/j.bbamcr.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 43.Yoshisue H, Suzuki K, Kawabata A, Ohya T, Zhao H, Sakurada K, Taba Y, Sasaguri T, Sakai N, Yamashita S, Matsuzawa Y, Nojima H. Large scale isolation of non-uniform shear stress-responsive genes from cultured human endothelial cells through the preparation of a subtracted cDNA library. Atherosclerosis. 2002;162:323–334. doi: 10.1016/s0021-9150(01)00735-3. [DOI] [PubMed] [Google Scholar]
- 44.Zhao H, Ito A, Kimura SH, Yabuta N, Sakai N, Ikawa M, Okabe M, Matsuzawa Y, Yamashita S, Nojima H. RECS1 deficiency in mice induces susceptibility to cystic medial degeneration. Genes Genet Syst. 2006;81:41–50. doi: 10.1266/ggs.81.41. [DOI] [PubMed] [Google Scholar]
- 45.Zhao HJ, Ito A, Sakai N, Matsuzawa Y, Yamashita S, Nojima H. RECS1 is a negative regulator of matrix metalloproteinase-9 production and aged RECS1 knockout mice are prone to aortic dilation. Circ J. 2006;70:615–624. doi: 10.1253/circj.70.615. [DOI] [PubMed] [Google Scholar]
- 46.Shukla S, Fujita KI, Xiao Q, Liao ZY, Garfield S, Srinivasula SM. A shear stress responsive gene product PP1201 protects against Fas-mediated apoptosis by reducing Fas expression on the cell surface. Apoptosis. 2011;16:162–173. doi: 10.1007/s10495-010-0556-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandez M, Segura MF, Sole C, Colino A, Comella JX, Cena V. Lifeguard/neuro-nal membrane protein 35 regulates Fas ligand-mediated apoptosis in neurons via microdomain recruitment. J Neurochem. 2007;103:190–203. doi: 10.1111/j.1471-4159.2007.04767.x. [DOI] [PubMed] [Google Scholar]
- 48.Schweitzer B, Suter U, Taylor V. Neural membrane protein 35/Lifeguard is localized at postsynaptic sites and in dendrites. Mol Brain Res. 2002;107:47–56. doi: 10.1016/s0169-328x(02)00445-x. [DOI] [PubMed] [Google Scholar]
- 49.Merianda TT, Vuppalanchi D, Yoo S, Blesch A, Twiss JL. Axonal transport of neural membrane protein 35 mRNA increases axon growth. J Cell Sci. 2013;126:90–102. doi: 10.1242/jcs.107268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reich A, Spering C, Gertz K, Harms C, Gerhardt E, Kronenberg G, Nave KA, Schwab M, Tauber SC, Drinkut A, Harms K, Beier CP, Voigt A, Gobbels S, Endres M, Schulz JB. Fas/CD95 regulatory protein Faim2 is neuroprotective after transient brain ischemia. J Neurosci. 2011;31:225–233. doi: 10.1523/JNEUROSCI.2188-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Mendoza TH, Perez-Garcia CG, Kroll TT, Hoong NH, O'Leary DDM, Verma IM. Antiapoptotic protein lifeguard is required for survival and maintenance of Purkinje and granular cells. Proc Natl Acad Sci U S A. 2011;108:17189–17194. doi: 10.1073/pnas.1114226108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Urresti J, Ruiz-Meana M, Coccia E, Arevalo JC, Castellano J, Fernandez-Sanz C, Galenkamp KMO, Planells-Ferrer L, Moubarak RS, Llecha-Cano N, Reix S, Garcia-Dorado D, Barneda-Zahonero B, Comella JX. Lifeguard inhibits Fas ligand-mediated endoplasmic reticulum-calcium release mandatory for apoptosis in type II apoptotic cells. J Biol Chem. 2016;291:1221–1234. doi: 10.1074/jbc.M115.677682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nielsen JA, Chambers MA, Romm E, Lee LY, Berndt JA, Hudson LD. Mouse transmembrane BAX inhibitor motif 3 (Tmbim3) encodes a 38 kDa transmembrane protein expressed in the central nervous system. Mol Cell Biochem. 2011;357:73–81. doi: 10.1007/s11010-011-0877-3. [DOI] [PubMed] [Google Scholar]
- 54.Rojas-Rivera D, Armisen R, Colombo A, Martinez G, Eguiguren AL, Diaz A, Kiviluoto S, Rodriguez D, Patron M, Rizzuto R, Bultynck G, Concha ML, Sierralta J, Stutzin A, Hetz C. TMBIM3/GRINA is a novel unfolded protein response (UPR) target gene that controls apoptosis through the modulation of ER calcium homeostasis. Cell Death Differ. 2012;19:1013–1026. doi: 10.1038/cdd.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Mattia F, Gubser C, van Dommelen MM, Visch HJ, Distelmaier F, Postigo A, Luyten T, Parys JB, de Smedt H, Smith GL, Willems PH, van Kuppeveld FJ. Human Golgi antiapoptotic protein modulates intracellular calcium fluxes. Mol Biol Cell. 2009;20:3638–3645. doi: 10.1091/mbc.E09-05-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gubser C, Bergamaschi D, Hollinshead M, Lu X, van Kuppeveld FJ, Smith GL. A new inhibitor of apoptosis from vaccinia virus and eukaryotes. PLoS Pathog. 2007;3:e17. doi: 10.1371/journal.ppat.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carrara G, Saraiva N, Parsons M, Byrne B, Prole DL, Taylor CW, Smith GL. Golgi anti-apoptotic proteins are highly conserved ion channels that affect apoptosis and cell migration. J Biol Chem. 2015;290:11785–11801. doi: 10.1074/jbc.M115.637306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saraiva N, Prole DL, Carrara G, Johnson BF, Taylor CW, Parsons M, Smith GL. hGAAP promotes cell adhesion and migration via the stimulation of store-operated Ca2+ entry and calpain 2. J Cell Biol. 2013;202:699–713. doi: 10.1083/jcb.201301016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oka T, Sayano T, Tamai S, Yokota S, Kato H, Fujii G, Mihara K. Identification of a novel protein MICS1 that is involved in maintenance of mitochondrial morphology and apoptotic release of cytochrome c. Mol Biol Cell. 2008;19:2597–2608. doi: 10.1091/mbc.E07-12-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones SJ, Ledgerwood EC, Prins JB, Galbraith J, Johnson DR, Pober JS, Bradley JR. TNF recruits TRADDto the plasma membrane but not the trans-Golgi network, the principal subcellular location of TNF-R1. J Immunol. 1999;162:1042–1048. [PubMed] [Google Scholar]
- 61.Bennett M, Macdonald K, Chan SW, Luzio JP, Simari R, Weissberg P. Cell surface trafficking of Fas: a rapid mechanism of p53-mediated apoptosis. Science. 1998;282:290–293. doi: 10.1126/science.282.5387.290. [DOI] [PubMed] [Google Scholar]
- 62.Yamaji T, Nishikawa K, Hanada K. Transmembrane BAX inhibitor motif containing (TMBIM) family proteins perturbs a trans-Golgi network enzyme, Gb3 synthase, and reduces Gb3 biosynthesis. J Biol Chem. 2010;285:35505–35518. doi: 10.1074/jbc.M110.154229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Somia NV, Schmitt MJ, Vetter DE, Van Antwerp D, Heinemann SF, Verma IM. LFG: an anti-apoptotic gene that provides protection from Fas-mediated cell death. Proc Natl Acad Sci U S A. 1999;96:12667–12672. doi: 10.1073/pnas.96.22.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bultynck G, Kiviluoto S, Henke N, Ivanova H, Schneider L, Rybalchenko V, Luyten T, Nuyts K, De Borggraeve W, Bezprozvanny I. The C terminus of Bax inhibitor-1 forms a Ca2+-permeable channel pore. J Biol Chem. 2012;287:2544–2557. doi: 10.1074/jbc.M111.275354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim HR, Lee GH, Ha KC, Ahn T, Moon JY, Lee BJ, Cho SG, Kim S, Seo YR, Shin YJ. Bax inhibitor-1 Is a pH-dependent regulator of Ca2+ channel activity in the endoplasmic reticulum. J Biol Chem. 2008;283:15946–15955. doi: 10.1074/jbc.M800075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee GH, Hwang JD, Choi JY, Park HJ, Cho JY, Kim KW, Chae HJ, Kim HR. An acidic pH environment increases cell death and pro-inflammatory cytokine release in osteoblasts: the involvement of BAX inhibitor-1. Int J Biochem Cell Biol. 2011;43:1305–1317. doi: 10.1016/j.biocel.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 67.Bultynck G, Kiviluoto S, Methner A. Bax inhibitor-1 is likely a pH-sensitive calcium leak channel, not a H+/Ca2+ exchanger. Sci Signal. 2014;7:pe22. doi: 10.1126/scisignal.2005764. [DOI] [PubMed] [Google Scholar]
- 68.Tang L, El-Din TMG, Payandeh J, Martinez GQ, Heard TM, Scheuer T, Zheng N, Catterall WA. Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature. 2014;505:56–61. doi: 10.1038/nature12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu J, Yan Z, Li Z, Qian X, Lu S, Dong M, Zhou Q, Yan N. Structure of the voltage-gated calcium channel Cav1. 1 at 3.6 Å resolution. Nature. 2016 doi: 10.1038/nature19321. [DOI] [PubMed] [Google Scholar]
- 70.Saraiva N, Prole DL, Carrara G, Maluquer de Motes C, Johnson BF, Byrne B, Taylor CW, Smith GL. Human and viral Golgi anti-apoptotic protein (GAAP) oligomerize via different mechanisms and monomeric GAAP inhibits apoptosis and modulates calcium. J Biol Chem. 2013;288:13057–13067. doi: 10.1074/jbc.M112.414367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chae HJ, Ke N, Kim HR, Chen SR, Godzik A, Dickman M, Reed JC. Evolutionarily conserved cytoprotection provided by Bax inhibitor-1 homologs from animals, plants, and yeast. Gene. 2003;323:101–113. doi: 10.1016/j.gene.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 72.Kawai M, Pan L, Reed JC, Uchimiya H. Evolutionally conserved plant homologue of the Bax inhibitor-1 (BI-1) gene capable of suppressing Bax-induced cell death in yeast(1) FEBS Lett. 1999;464:143–147. doi: 10.1016/s0014-5793(99)01695-6. [DOI] [PubMed] [Google Scholar]
- 73.Watanabe N, Lam E. Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. Plant J. 2006;45:884–894. doi: 10.1111/j.1365-313X.2006.02654.x. [DOI] [PubMed] [Google Scholar]
- 74.Huckelhoven R. BAX inhibitor-1, an ancient cell death suppressor in animals and plants with prokaryotic relatives. Apoptosis. 2004;9:299–307. doi: 10.1023/b:appt.0000025806.71000.1c. [DOI] [PubMed] [Google Scholar]
- 75.Sanchez P, Zabala MD, Grant M. AtBI-1, a plant homologue of Bax inhibitor-1, suppresses Bax-induced cell death in yeast and is rapidly upregulated during wounding and pathogen challenge. Plant J. 2000;21:393–399. doi: 10.1046/j.1365-313x.2000.00690.x. [DOI] [PubMed] [Google Scholar]
- 76.Ihara-Ohori Y, Nagano M, Muto S, Uchimiya H, Kawai-Yamada M. Cell death suppressor Arabidopsis Bax inhibitor-1 is associated with calmodulin binding and ion homeostasis. Plant Physiol. 2007;143:650–660. doi: 10.1104/pp.106.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watanabe N, Lam E. Bax inhibitor-1, a conserved cell death suppressor, is a key molecular switch downstream from a variety of biotic and abiotic stress signals in plants. Int J Mol Sci. 2009;10:3149–3167. doi: 10.3390/ijms10073149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nagano M, Ishikawa T, Ogawa Y, Iwabuchi M, Nakasone A, Shimamoto K, Uchimiya H, Kawai-Yamada M. Arabidopsis Bax inhibitor-1 promotes sphingolipid synthesis during cold stress by interacting with ceramide-modifying enzymes. Planta. 2014;240:77–89. doi: 10.1007/s00425-014-2065-7. [DOI] [PubMed] [Google Scholar]
- 79.Ramiro DA, Melotto-Passarin DM, Barbosa Mde A, Santos FD, Gomez SG, Massola Junior NS, Lam E, Carrer H. Expression of Arabidopsis Bax inhibitor-1 in transgenic sugarcane confers drought tolerance. Plant Biotechnol J. 2016;14:1826–1837. doi: 10.1111/pbi.12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Stelten J, Silva F, Belin D, Silhavy TJ. Effects of antibiotics and a proto-onco-gene homolog on destruction of protein translocator SecY. Science. 2009;325:753–756. doi: 10.1126/science.1172221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hemrajani C, Berger CN, Robinson KS, Marches O, Mousnier A, Frankel G. NleH effectors interact with Bax inhibitor-1 to block apoptosis during enteropathogenic Escherichia coli infection. Proc Natl Acad Sci U S A. 2010;107:3129–3134. doi: 10.1073/pnas.0911609106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lesniewski M, Das S, Skomorovska-Prokvolit Y, Wang FZ, Pellett PE. Primate cytomegalovirus US12 gene family: a distinct and diverse clade of seven-transmembrane proteins. Virology. 2006;354:286–298. doi: 10.1016/j.virol.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 83.Cavaletto N, Luganini A, Gribaudo G. Inactivation of the human cytomegalovirus US20 gene hampers productive viral replication in endothelial cells. J Virol. 2015;89:11092–11106. doi: 10.1128/JVI.01141-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bronzini M, Luganini A, Dell'Oste V, De Andrea M, Landolfo S, Gribaudo G. The US16 gene of human cytomegalovirus is required for efficient viral infection of endothelial and epithelial cells. J Virol. 2012;86:6875–6888. doi: 10.1128/JVI.06310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu C, Xu W, Palmer AE, Reed JC. BI-1 regulates endoplasmic reticulum Ca2+ homeostasis downstream of Bcl-2 family proteins. J Biol Chem. 2008;283:11477–11484. doi: 10.1074/jbc.M708385200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sano R, Hou YCC, Hedvat M, Correa RG, Shu CW, Krajewska M, Diaz PW, Tamble CM, Quarato G, Gottlieb RA, Yamaguchi M, Nizet V, Dahl R, Thomas DD, Tait SW, Green DR, Fisher PB, Matsuzawa SI, Reed JC. Endoplasmic reticulum protein BI-1 regulates Ca2+-mediated bioenergetics to promote autophagy. Genes Dev. 2012;26:1041–1054. doi: 10.1101/gad.184325.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ivanova H, Vervliet T, Missiaen L, Parys JB, De Smedt H, Bultynck G. Inositol 1,4,5-trisphosphate receptor-isoform diversity in cell death and survival. Biochim Biophys Acta, Mol Cell Res. 2014;1843:2164–2183. doi: 10.1016/j.bbamcr.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 88.Kiviluoto S, Vervliet T, Ivanova H, Decuypere JP, De Smedt H, Missiaen L, Bultynck G, Parys JB. Regulation of inositol 1,4,5-trisphosphate receptors during endoplasmic reticulum stress. Biochim Biophys Acta. 2013;1833:1612–1624. doi: 10.1016/j.bbamcr.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 89.Akl H, Bultynck G. Altered Ca2+ signaling in cancer cells: proto-oncogenes and tumor suppressors targeting IP3 receptors. Biochim Biophys Acta Rev Cancer. 2013;1835:180–193. doi: 10.1016/j.bbcan.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 90.Vervliet T, Parys JB, Bultynck G. Bcl-2 proteins and calcium signaling: complexity beneath the surface. Oncogene. 2016;35:5079–5092. doi: 10.1038/onc.2016.31. [DOI] [PubMed] [Google Scholar]
- 91.Rong Y, Distelhorst CW. Bcl-2 protein family members: versatile regulators of calcium signaling in cell survival and apoptosis. Annu Rev Physiol. 2008;70:73–91. doi: 10.1146/annurev.physiol.70.021507.105852. [DOI] [PubMed] [Google Scholar]
- 92.Chen R, Valencia I, Zhong F, McColl KS, Roderick HL, Bootman MD, Berridge MJ, Conway SJ, Holmes AB, Mignery GA, Velez P, Distelhorst CW. Bcl-2 functionally interacts with inositol 1,4,5-trisphosphate receptors to regulate calcium release from the ER in response to inositol 1,4,5-trisphosphate. J Cell Biol. 2004;166:193–203. doi: 10.1083/jcb.200309146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rong YP, Aromolaran AS, Bultynck G, Zhong F, Li X, McColl K, Matsuyama S, Herlitze S, Roderick HL, Bootman MD, Mignery GA, Parys JB, De Smedt H, Distelhorst CW. Targeting Bcl-2-IP3 receptor interaction to reverse Bcl-2's inhibition of apoptotic calcium signals. Mol Cell. 2008;31:255–265. doi: 10.1016/j.molcel.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rong YP, Bultynck G, Aromolaran AS, Zhong F, Parys JB, De Smedt H, Mignery GA, Roderick HL, Bootman MD, Distelhorst CW. The BH4 domain of Bcl-2 inhibits ER calcium release and apoptosis by binding the regulatory and coupling domain of the IP3 receptor. Proc Natl Acad Sci U S A. 2009;106:14397–14402. doi: 10.1073/pnas.0907555106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Monaco G, Decrock E, Akl H, Ponsaerts R, Vervliet T, Luyten T, De Maeyer M, Missiaen L, Distelhorst CW, De Smedt H, Parys JB, Leybaert L, Bultynck G. Selective regulation of IP3-receptor-mediated Ca2+ signaling and apoptosis by the BH4 domain of Bcl-2 versus Bcl-Xl. Cell Death Differ. 2012;19:295–309. doi: 10.1038/cdd.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ivanova H, Ritaine A, Wagner L, Luyten T, Shapovalov G, Welkenhuyzen K, Seitaj B, Monaco G, De Smedt H, Prevarskaya N, Yule DI, Parys JB, Bultynck G. The transmembrane domain of Bcl-2alpha, but not its hydrophobic cleft, is a critical determinant for efficient IP3 receptor inhibition. Oncotarget. 2016:55704–55720. doi: 10.18632/oncotarget.11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.White C, Li C, Yang J, Petrenko NB, Madesh M, Thompson CB, Foskett JK. The endoplasmic reticulum gateway to apoptosis by Bcl-X(L) modulation of the InsP3R. Nat Cell Biol. 2005;7:1021–1028. doi: 10.1038/ncb1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li C, Wang X, Vais H, Thompson CB, Foskett JK, White C. Apoptosis regulation by Bcl-x(L) modulation of mammalian inositol 1,4,5-trisphosphate receptor channel isoform gating. Proc Natl Acad Sci U S A. 2007;104:12565–12570. doi: 10.1073/pnas.0702489104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang J, Vais H, Gu W, Foskett JK. Biphasic regulation of InsP3 receptor gating by dual Ca2+ release channel BH3-like domains mediates Bcl-xL control of cell viability. Proc Natl Acad Sci U S A. 2016;113:E1953–E1962. doi: 10.1073/pnas.1517935113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT, Liu B, Bao JK. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45:487–498. doi: 10.1111/j.1365-2184.2012.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hainaut P, Plymoth A. Targeting the hallmarks of cancer: towards a rational approach to next-generation cancer therapy. Curr Opin Oncol. 2013;25:50–51. doi: 10.1097/CCO.0b013e32835b651e. [DOI] [PubMed] [Google Scholar]
- 102.Grzmil M, Kaulfuss S, Thelen P, Hemmerlein B, Schweyer S, Obenauer S, Kang TW, Burfeind P. Expression and functional analysis of Bax inhibitor-1 in human breast cancer cells. J Pathol. 2006;208:340–349. doi: 10.1002/path.1902. [DOI] [PubMed] [Google Scholar]
- 103.Grzmil M, Thelen P, Hemmerlein B, Schweyer S, Voigt S, Mury D, Burfeind P. Bax inhibitor-1 is overexpressed in prostate cancer and its specific down-regulation by RNA interference leads to cell death in human prostate carcinoma cells. Am J Pathol. 2003;163:543–552. doi: 10.1016/S0002-9440(10)63682-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tanaka R, Ishiyama T, Uchihara T, Inadome Y, Iijima T, Morishita Y, Kano J, Goya T, Noguchi M. Expression of the Bax inhibitor-1 gene in pulmonary adenocarcinoma. Cancer. 2006;106:648–653. doi: 10.1002/cncr.21639. [DOI] [PubMed] [Google Scholar]
- 105.Planells-Ferrer L, Urresti J, Soriano A, Reix S, Murphy DM, Ferreres JC, Borras F, Gallego S, Stallings RL, Moubarak RS, Segura MF, Comella JX. MYCN repression of lifeguard/FAIM2 enhances neuroblastoma aggressiveness. Cell Death Dis. 2014;5 doi: 10.1038/cddis.2014.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stewart TA, Yapa KTDS, Monteith GR. Altered calcium signaling in cancer cells. Biochim Biophys Acta Biomembr. 2015;1848:2502–2511. doi: 10.1016/j.bbamem.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 107.Marchi S, Pinton P. Alterations of calcium homeostasis in cancer cells. Curr Opin Pharmacol. 2016;29:1–6. doi: 10.1016/j.coph.2016.03.002. [DOI] [PubMed] [Google Scholar]


