Abstract
Recent human-genetics studies have come to different conclusions regarding how and when modern humans spread out of Africa and into the rest of the world. I present here a simple parsimony-based analysis that suggests that East Asians and Melanesians are sister groups, and I discuss what implications this has for recent claims made about the demographic histories of non-African populations.
Introduction
Anatomically modern humans are thought to have evolved in sub-Saharan Africa 150,000–200,000 years ago1, 2 (150–200 Kya) and from there to have eventually colonized the rest of the world. It is unclear, though, how and when modern humans first left Africa and whether there was one major migration out of Africa or more than one. One theory, the “coastal migration model” (CMM), posits that the first modern humans to leave Africa departed from the Northeast through the Arabian peninsula, then along the coasts of India and Southeast Asia until they reached Australia roughly 50 Kya.3, 4 Descendants of this first wave of migration are hypothesized to include aboriginal Australians, Melanesians, and possibly so-called Negrito groups in South and Southeast Asia. Then, a second dispersal out of Africa led to the colonization of the rest of the world, including mainland Europe, Southeast Asia, and the Americas. The other major hypothesis posits that there was a single major migration out of Africa and that all extant non-African populations are descended from these first migrants.5, 6 (The focus on extant populations here excludes modern human groups that left no present-day descendants, cf. Fu et al., 2015 and Liu et al., 20157, 8). The evidence so far from early genetic studies and archeological studies has been mixed, and no clear scientific consensus has been reached.6, 9, 10
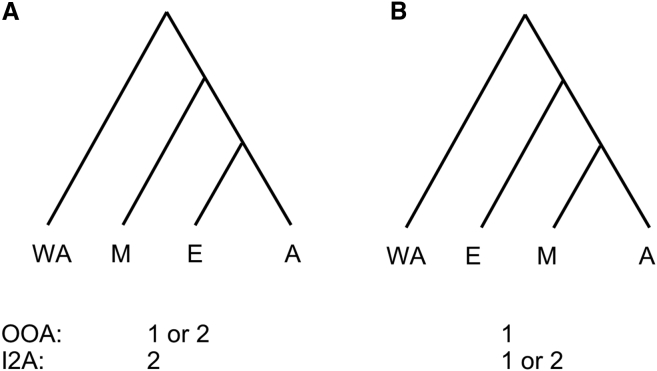
From a genetic perspective, it is not always clear what predictions different models make. Researchers have focused instead on questions that are more easily answerable. Figure 1 shows two possible branching orders for West Africans, Europeans, East Asians, and Melanesians, as well as interpretations in terms of waves of migration out of Africa (OOA) and into Asia (I2A). With the recent proliferation of whole-genome sequence datasets from diverse populations, it should be easier to distinguish between these competing demographic models. Rasmussen and colleagues11 published the genome of an aboriginal Australian individual and observed that this individual’s genome shows a greater divergence from African genomes than do genomes from mainland Eurasia. They interpreted this observation as evidence for the CMM and suggested that the greater divergence was due to an older separation time. However, this observation has an alternative explanation in light of the fact that aboriginal Australians (but not mainland Eurasians) have experienced a substantial amount of admixture from Denisovans.12, 13 In particular, the greater observed divergence could be due to the presence of introgressed Denisovan regions.
Figure 1.
Schematic of Two Different Potential Branching Orders for Major Non-African Populations
WA = West Africans (assumed to be the outgroup), E = Europeans, A = East Asians, and M = Melanesians. OOA = Number of major waves of migration out of Africa. I2A = Number of major waves of migration into East Asia.
More recently, several large whole-genome sequencing studies14, 15, 16 have made conflicting claims regarding the historical branching order for the groups shown in Figure 1 (reviewed in Tucci and Akey, 201617). Specifically, Malaspinas et al.15 claimed that Melanesians are an outgroup in comparison to mainland Eurasians (i.e., Figure 1A), whereas Pagani et al.16 posited that Melanesians contain some ancestry from an older out-of-Africa migration (which we call “ghost” admixture) and implied (though did not state explicitly) that Figure 1A is the correct branching order (see, e.g., their Extended Data Figure 4A). In contrast Figures 1 and 3 in Mallick et al.14 suggested that Europeans are an outgroup in comparison to East Asians and Melanesians (i.e., Figure 1B). One difficulty in assessing the merits of these competing claims is that they are based on the output from complicated inference tools such as Fastsimcoal,18 MSMC19 or fineSTRUCTURE.20 Given the intricate nature of these methods, it is difficult to assess how sensitive they are to model assumptions and sampling biases or to determine whether they have been implemented correctly. In addition, both MSMC and fineSTRUCTURE require (haplotype) phased data, making them less reliable for analyses involving human populations not included in the HapMap21 or 1000 Genomes22 Projects.
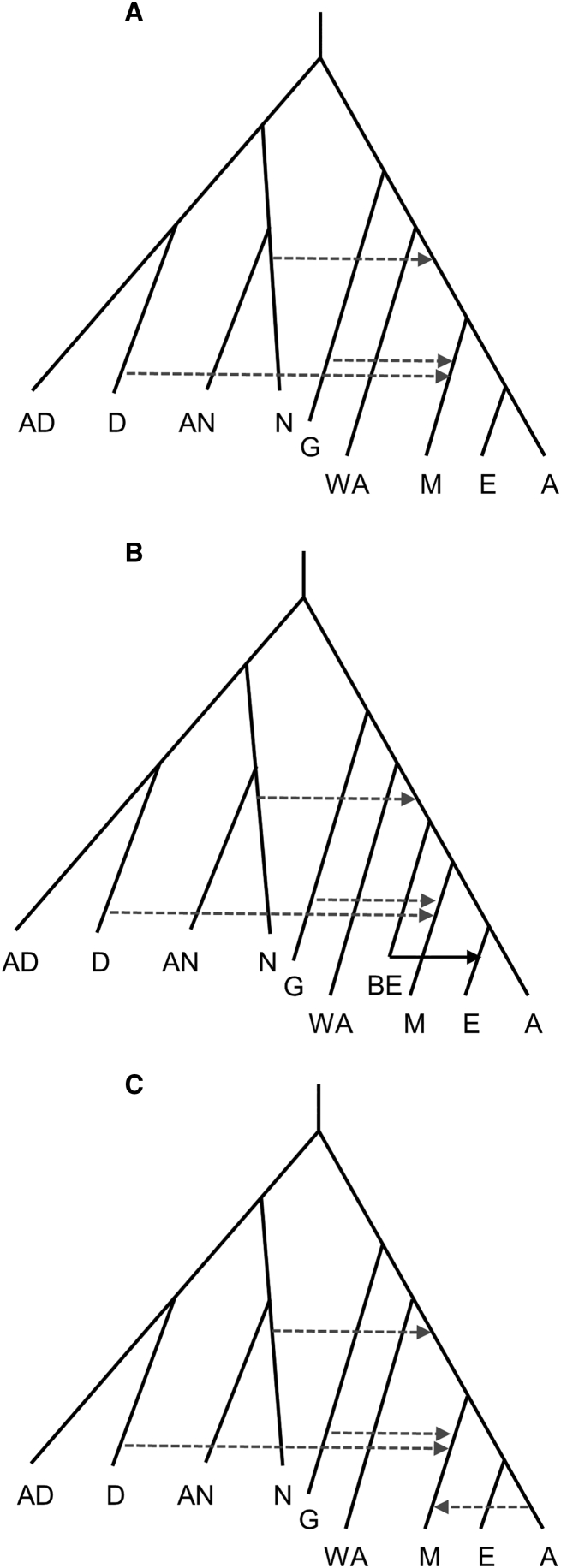
Figure 3.
Schematics of Population Models Used for Simulations
AD = Altai Denisovan population, D = admixing Denisovan population, AN = Altai Neanderthal population, N = admixing Neanderthal population, G = “ghost” population, WA = West African population, BE = Basal European population, M = Melanesian population, E = European population, and A = East Asian population. Dashed horizontal lines show admixture events. See text for all parameter values. Schematics are not to scale.
In this study, I present a straightforward, easy-to-understand analysis of patterns of human genetic variation. Based on the phylogenetic concept of parsimony, this analysis can be thought of as an estimate of the average branch lengths, across the whole genome, of internal branches in the (unobserved) genealogies of sampled individuals, and it is similar to previously proposed approaches, such as D-statistics23 and the D4P statistic,11 for inferring branching orders and detecting admixture. The results reported here suggest that the true branching order of the major non-African populations is easy to recover, even with relatively unsophisticated analytical methods.
Material and Methods
Human Sequence Data
I started with the variant calls used in Pagani et al.16 (downloaded from the Estonian Biocenter website) and filtered them to consider only biallelic autosomal SNPs. I further considered only those variants where the reference allele was the ancestral allele and where the genotype was homozygous reference in 21 West and Central African (nine Yoruba, four Luhya, and eight Pygmy) genomes and in Neanderthal and Denisovan genomes. For each such variant, I then tabulated the frequency of the alternative allele across two test populations from Europe, East Asia, and Melanesia (i.e., Tuscan, Croat, Han, Japanese, Kosipe and Koinanbe).
Allele-Sharing Statistics
Suppose we have samples from a European, an East Asian, and a Melanesian population with sample sizes nE, nA, and nM, respectively. Then, for a set of S SNPs, we use ei, ai and mi to denote the number of copies of the derived allele at the i-th SNP in the European, East Asian, and Melanesian samples (0 ≤ ei ≤ 2nE, etc.). We then define KEA as follows:
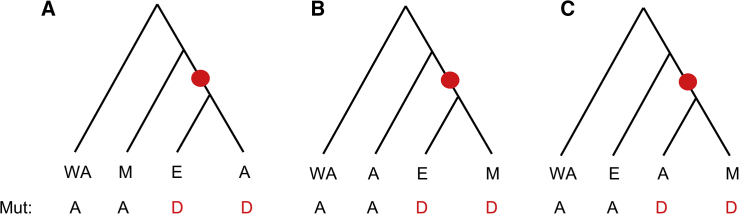
The term in the summation is the probability that randomly chosen single alleles from each of the three non-African populations will yield a derived allele shared between the European and East Asian samples and an ancestral allele in the Melanesian sample. KEM and KAM are defined analogously. Note that it is straightforward to modify the denominator to handle missing data in any of the samples. Finally, we define PEA, PEM, and PAM as KEA/(KEA+KEM+KAM), KEM/(KEA+KEM+KAM), and KAM/(KEA+KEM+KAM), respectively. Using a parsimony assumption, these three values reflect the proportions of phylogenetically informative SNPs that support each of the three possible tree topologies (Figure 2, assuming the West and Central African samples described above are an outgroup). Although the true topologies are expected to vary across the genome due to incomplete lineage sorting and additional demographic factors not considered here (e.g., migration), the relative number of variants supporting each topology is informative about the average genealogical history of the samples and thus the true branching order of the populations. This same approach was used in some of the work that showed that chimpanzees (and bonobos) are our closest living relatives.24, 25
Figure 2.
Possible Genealogies for Non-African Populations
This study assumes a single (haploid) sampled sequence from each of four populations, and it assumes that the African sequence is the outgroup. Each genealogy has a single, phylogenetically informative, internal branch (shown in red), leading to a unique pattern of ancestral (A) and derived (D) alleles. The proportion of phylogenetically informative sites supporting each topology is denoted by PEA, PEM, and PAM for Figures 2A, 2B, and 2C, respectively.
PEA, PEM, and PAM are similar to previously defined admixture-quantifying statistics, such as D-statistics,23 E-statistics,12 enhanced D-statistics,26 and the D4P statistic.11 All of these are counting statistics that condition on the presence or absence of derived alleles across individuals. For example, although enhanced D-statistics count sites that are homozygous ancestral in sub-Saharan Africans but derived in the Denisovan genome, the method introduced here counts variants that are homozygous ancestral in sub-Saharan Africans, Neanderthals, and Denisovans.
Simulations Using the Malaspinas et al. Model
I obtained the exact simulation parameters used for generating data under the model shown in Malaspinas et al.’s Figure S07.315 from Vitor Sousa. I then used these parameter values and the program ms0ancient216 to simulate sequence data. I assumed (diploid) sample sizes of 21, 3, 3, and 3 for the West African, European, East Asian, and Australian populations. ms0ancient2 is a simple modification of Hudson’s ms;27 it changes branch lengths to allow for the past sampling of two archaic genomes. To sample non-African sequences roughly 42 Kya, I shifted the scaled times of events by 0.032324 (I used the same time scaling as in Malaspinas et al.15). Both sets of simulations included a total of 2.05 Gb of simulated sequence to roughly approximate the total length of the genome that was available after filtering.
Simulations Modeled after Pagani et al.
I constructed five simple branching models (without migration) based on the split times described in Pagani et al.16 All models used a scaling of 2N generations = 1 million years and, when possible, assumed that the median genetic split times as estimated from MSMC19 are actual population split times. Models 1–3 are simple branching models (Figure 3A) with a split time between Europeans and East Asians of 30 Kya, a split time between Eurasians and Papuans of 40 Kya, and a split time between Eurasians and West Africans of 75 Kya. Papuans also derived 0%–5% of their ancestry from an unsampled “ghost” population that was completely isolated from 38–120 Kya. Models 1, 2, and 3 have 0%, 2%, and 5% “ghost” admixture, respectively. These models also assumed 4% admixture from Denisovans into Papuans 35 Kya, 2% Neanderthal admixture into non-Africans 65 Kya, an Altai Neanderthal sampling time of 130 Kya, a Denisovan sampling time of 100 Kya, an intra-Neanderthal split time of 150 Kya, an intra-Denisovan split time of 350 Kya, a Neanderthal-Denisovan split time of 425 Kya and an archaic-modern split time of 650 Kya. These parameter values, while somewhat arbitrary, were taken from the literature28 whenever possible.
I also tried to construct a model that followed a similar branching order as in Extended Data Figure from Pagani et al.16 (but without the West European hunter-gatherer population). Specifically, I assumed that the ancestral European population split 10 Kya into two groups (i.e., ANE and Basal European). One of these (ANE) merged with the ancestral East Asian population 30 Kya, whereas the other (Basal European) merged with the ancestral Melanesian–East-Asian population 45 Kya (Figure 3B). All other model parameters were assumed to be the same as above. Models 4 and 5 assumed a 20%–80% or an 80%–20% split of European ancestry into the ANE and Basal European groups, respectively.
Additional Simulations
I tested additional demographic models to better understand how specific parameter values affect the relative values of PEA, PEM, and PAM. These models were similar to the Pagani models 1–3 described above. One set of simulations took Pagani model 2 and added a single pulse of migration from the East Asian population into the Melanesian population 10–20 Kya (Figure 3C). This pulse accounted for 20%–90% of the subsequent Melanesian gene pool. The other set of simulations used the same population branching order as in Figure 1B, but with all other model parameters (e.g., Neanderthal and Denisovan admixture, “ghost” admixture, and African–non-African divergence time) being the same as in Pagani models 1–3. These simulations had East-Asian–Melanesian split times from 40–45 Kya and European–East-Asian split times of 50–60 Kya. For each parameter combination, we simulated 2.05 Gb of sequence and estimated PEM, PEA, and PAM. We also calculated these parameters on sites that conditioned on the homozygous reference allele in only the 21 sub-Saharan Africans and the Neanderthal, to see what effect the Denisovan conditioning had on the proportions.
Results
I focused on variants that were not present in sub-Saharan Africans or archaic humans and that are thus likely to have arisen after the dispersal of modern humans out of Africa. I also assumed that West Africans are an outgroup with respect to all non-Africans (although this assumption can be relaxed). Given a single representative sequence from each of three non-African groups, there are three possible tree topologies, each containing a single internal branch and a unique phylogenetically informative variant (PIV) pattern (Figure 2). Specifically, if exactly two out of three of the sequences share a derived allele, then the genealogy at this site has (under the parsimony assumption) the two populations with a derived allele as sister groups.
In analyzing the whole-genome sequence data described in Pagani et al.,16 I considered a panel of 21 West and Central African samples (nine Yoruba, four Luhya, and eight pygmies), two representative European populations (Tuscans and Croats), two representative East Asian populations (Han and Japanese) and two representative Melanesian groups (Kosipe and Koinanbe). I tabulated the proportion of PIVs supporting each topology (PEA, PEM, and PAM, cf. Figure 2) across each of the eight combinations of one European, one East Asian, and one Melanesian population; these proportions were averaged over all possible choices of a single haploid sequence from each population. The results were highly consistent across the different population combinations; PAM ranged from 0.418–0.430, and PEA and PEM were always <0.3 (Table 1). So, although the “true” demographic history of non-African populations was certainly very complex, on average Melanesian and East Asian sequences are more genetically similar (and share a more recent common ancestor) than do European and East Asian or European and Melanesian sequences, and Figure 1B is likely to be the true population branching order. Note that the same conclusion was reached in the neighbor-joining tree shown in Figure 1a of Mallick et al.14 and in the unrooted tree in Extended Data Figure 1 of Malaspinas et al.15 If we assume that sub-Saharan Africans are ancestral, then this branching order was proposed by Cavalli-Sforza and colleagues more than 50 years ago.29
Table 1.
Proportion of Sites Supporting Each Topology for Both Real and Simulated Sequence Data
| Populations | PEA(%) | PEM(%) | PAM(%) |
|---|---|---|---|
| Tuscan, Han, Kosipe | 29.1 | 28.0 | 42.9 |
| Tuscan, Han, Koinanbe | 29.3 | 28.2 | 42.5 |
| Tuscan, Japanese, Kosipe | 28.8 | 28.2 | 43.0 |
| Tuscan, Japanese, Koinanbe | 29.0 | 28.4 | 42.6 |
| Croat, Han, Kosipe | 29.4 | 28.4 | 42.2 |
| Croat, Han, Koinanbe | 29.6 | 28.5 | 41.9 |
| Croat, Japanese, Kosipe | 29.3 | 28.5 | 42.1 |
| Croat, Japanese, Koinanbe | 29.5 | 28.7 | 41.8 |
| Malaspinas model | 28.1 | 26.7 | 45.2 |
| Pagani model 1 | 38.0 | 30.7 | 31.3 |
| Pagani model 2 | 38.6 | 30.8 | 30.6 |
| Pagani model 3 | 39.2 | 30.4 | 30.3 |
| Pagani model 4 | 37.1 | 31.2 | 31.7 |
| Pagani model 5 | 34.2 | 31.8 | 34.1 |
See also Figure 2.
Next, I compared the expectations for PEA, PEM, and PAM under the demographic model proposed by Malaspinas and colleagues (cf. Figure S07.315). My a priori expectation was that PEA would be slightly larger than PEM and PAM because their model, like Figure 1A, has Melanesians as an outgroup with respect to Europeans and East Asians. Surprisingly, I found instead that simulations under their model produce large PAM values, as expected under the branching order shown in Figure 1B (with Europeans as an outgroup with respect to East Asians and Melanesians) and as observed in the actual data (Table 1). A closer look at the parameters in their model (Tables S07.3 and S07.5) explains this apparent discrepancy. The estimated split times for Aboriginal Australians (58 Kya) and ancestral Eurasians (57 Kya) are almost identical, and more importantly, these two ancestral populations are connected by high scaled migration rates (4Nm > 10, cf. Table S07.5 in Masaspinas et al.15) after the split. One consequence of the high migration rates is that the ancestral Australians do not start diverging genetically from ancestral Europeans and ancestral East Asians until after the European–East-Asian split 42 Kya (when the migration rates decrease). I verified that this is the case by simulating sequences that were sampled from each non-African population right after the European–East-Asian split under the Malaspinas model. I found that the expected FST30 between ancestral Australians and ancestral Eurasians sampled at this time was <0.001 (i.e., no genetic differentiation). Finally, the higher modeled migration rates between aboriginal Australians and East Asians in the past 42 Kyr leads to the observed greater genetic similarity between these two populations. I conclude that, in practice, the Malaspinas model behaves just like the simple branching model shown in Figure 1B. Furthermore, any model, including models with all possible branching orders for non-African populations, that has the European, East Asian, and Australian populations splitting roughly simultaneously 42 Kya (and with the same migration rates since then) should have roughly the same likelihood as the model presented in the Malaspinas paper.
I also tried to explore the claims made by Pagani and colleagues.16 Although they do not propose a specific demographic model, they do assume in the text that median genetic-split times estimated from MSMC analyses can be thought of as genetic divergence times between populations. I constructed five simple population models by using MSMC-estimated split times and assuming that Papuans have 0%–5% of their ancestry from an unsampled “ghost” population, which branched off from other modern human groups 120 Kya (Figure 3). Three of these models used the topology shown in Figure 1A because Pagani et al. estimated a Papuan-Eurasian MSMC split time of ∼40 Kya and a European–East-Asian MSMC split time of ∼30 Kya,16 whereas two models (based on Pagani et al.’s extended Data Figure 10) incorporate heterogeneity in the true population topology across the genome (see Material and Methods for details).
As expected, simulations under these models produce larger PEA values and smaller PAM values than what was found in the actual data (Table 1). These results are insensitive to assumed “ghost” admixture proportions (i.e., there is little difference between the results of models 1–3) but rather are a simple consequence of the models’ underlying population topology.
To test whether other, similar demographic models might produce results more in-line with observations, I also ran simulations under a model with the topology shown in Figure 1A, but with recent (one-way) migration from East Asians into Melanesians (Figure 3C), and under a model with the same topology as in Figure 1B. These simulations show that one-way migration increases the relative value of PAM (Table S1 in the Supplemental Data available with this article online). Qualitatively, this is because those regions of the genome affected by recent migration have a true topology with Europeans as an outgroup, and the overall values of PEA, PEM, and PAM reflect a weighted average of the different true genealogical trees across the genome. However, the magnitude of this effect does not seem to be enough to produce PAM values as large as those that are observed, even if Melanesians derive 80%–90% of their genome from East Asian migrants as recently as 10 Kya. However, such an extreme migration model would be inconsistent with the roughly constant levels of Denisovan ancestry observed across Melanesian and aboriginal Australian populations.15
My simulations with Europeans as an outgroup with respect to other non-African groups showed several things. First, as expected, increasing the difference between the European and East Asian and between the East Asian and Melanesian divergence times increases the relative value of PAM because it increases the average internal branch length in Figure 1B. Second, there are parameter combinations that produce PEA, PEM, and PAM values similar to those that are observed, but these require Europeans to be a clear outgroup relative to East Asians and Melanesians (Table S1). Third, both sets of simulations show that conditioning on the absence of alleles in the Denisovan genome has almost no effect on PEA, PEM, and PAM. Finally, the presence of low (e.g., ≤5%) levels of “ghost” admixture has only a minor effect on allele-sharing statistics and is completely unnecessary for explaining the observed values of PEA, PEM, and PAM.
Discussion
In summary, we have shown that East Asian and Melanesian samples have an excess of shared derived variants, reflecting their genetic similarity to each other. (I reached the same qualitative conclusions with the Malaspinas et al.15 dataset, but the genomes are not fully publicly available.) These results are incompatible with simple versions of the CMM because an older divergence of Melanesian populations would lead to larger PEA values. My focus on shared variants is crucial because it concentrates on the internal branches present during the times when non-African populations diverged from each other. If instead I had just counted the number of derived variants that were present in non-Africans and homozygous ancestral in 21 sub-Saharan Africans, the Neanderthal genome, and the Denisovan genome, then Melanesians would have more of these sites (an average of 62,546) compared with East Asians (an average of 57,574) or Europeans (an average of 47,054). These “non-African alleles” are less informative about the true population branching order because they reflect a combination of other demographic factors, such as ancient admixture between Melanesians and Denisovans, as well as more recent admixture between Southern Europeans and sub-Saharan Africans.
Although I cannot formally rule out Pagani et al.’s claim of “ghost” admixture into the ancestors of Melanesians,16 there are several factors that make this highly unlikely. My simulations suggest that the large observed PAM values are only possible with models having Europeans as an outgroup with respect to East Asians and Melanesians (Table 1 and Table S1). However, this branching order is inconsistent with the observed MSMC split times,16 and these same split times were used prominently in Pagani et al.’s justification for their claim of “ghost” admixture. Simulations suggest that haplotype-based methods such as MSMC are extremely sensitive to phasing errors,31 and this problem is most likely exacerbated by the small number of Melanesians included in the Pagani et al. study (just three Kosipe and three Koinanbe).
Finally, the old Papuan–West-African split time estimated by Pagani and colleagues,16 and used as motivation for their claim of “ghost” admixture, is probably a simple consequence of Denisovan admixture into Melanesians. It is important to emphasize that the large genetic distance between the Altai Denisovan genome and the genomes of the Denisovans that interbred with the Melanesian lineage12, 26 make it impossible to successfully “mask” out all Denisovan ancestry tracts in contemporary human genomes. This is evident from the fact that PEA > PEM even in simulations without “ghost” admixture and from the small effect that masking putative Denisovan SNPs has on PIV proportions (Table S1).
In addition, PEA and PEM correspond to groups 2 and 1, respectively, in the D4P test,11 so Rasmussen et al.’s evidence for the CMM (i.e., that group 2 is larger than group 1, or equivalently that PEA > PEM) can be directly ascribed to the effects of Denisovan admixture. Furthermore, my analyses suggest that effective methods for distinguishing between the models listed in Rasmussen et al.’s Figure 1A would include sites ancestral in Europeans but derived in East Asians and Melanesians or aboriginal Australians (i.e., PAM).
The analyses presented here have been primarily qualitative. Although PEA, PEM, and PAM could in principle be used as summary statistics in a likelihood-based parameter estimation framework, they are more useful as heuristic descriptors of the data. As such, they can serve as a “sanity-check” on the conclusions of more sophisticated (but also more opaque) analytical tools. The challenge then remains for proponents of models featuring multiple major waves of migration of modern humans out of Africa to produce an explicit, testable model of human demography that at a minimum can produce the same qualitative patterns of genetic variation as those described above.
Published: May 4, 2017
Footnotes
The Supplemental Data include one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.04.002.
Web Resources
Estonian Biocenter Human Genome Diversity Panel (EGDP), http://www.ebc.ee/free_data
Supplemental Data
References
- 1.White T.D., Asfaw B., DeGusta D., Gilbert H., Richards G.D., Suwa G., Howell F.C. Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature. 2003;423:742–747. doi: 10.1038/nature01669. [DOI] [PubMed] [Google Scholar]
- 2.Clark J.D., Beyene Y., WoldeGabriel G., Hart W.K., Renne P.R., Gilbert H., Defleur A., Suwa G., Katoh S., Ludwig K.R. Stratigraphic, chronological and behavioural contexts of Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature. 2003;423:747–752. doi: 10.1038/nature01670. [DOI] [PubMed] [Google Scholar]
- 3.Lahr M.M., Foley R.A. Multiple dispersals and modern human origins. Evol. Anthropol. 1994;3:48–60. [Google Scholar]
- 4.Cavalli-Sforza L.L., Menozzi P., Piazza A. Princeton University Press; Princeton, NJ: 1994. The History and Geography of Human Genes. [Google Scholar]
- 5.Vigilant L., Stoneking M., Harpending H., Hawkes K., Wilson A.C. African populations and the evolution of human mitochondrial DNA. Science. 1991;253:1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- 6.Ramachandran S., Deshpande O., Roseman C.C., Rosenberg N.A., Feldman M.W., Cavalli-Sforza L.L. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl. Acad. Sci. USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Q., Hajdinjak M., Moldovan O.T., Constantin S., Mallick S., Skoglund P., Patterson N., Rohland N., Lazaridis I., Nickel B. An early modern human from Romania with a recent Neanderthal ancestor. Nature. 2015;524:216–219. doi: 10.1038/nature14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W., Martinón-Torres M., Cai Y.J., Xing S., Tong H.W., Pei S.W., Sier M.J., Wu X.H., Edwards R.L., Cheng H. The earliest unequivocally modern humans in southern China. Nature. 2015;526:696–699. doi: 10.1038/nature15696. [DOI] [PubMed] [Google Scholar]
- 9.Macaulay V., Hill C., Achilli A., Rengo C., Clarke D., Meehan W., Blackburn J., Semino O., Scozzari R., Cruciani F. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science. 2005;308:1034–1036. doi: 10.1126/science.1109792. [DOI] [PubMed] [Google Scholar]
- 10.Lahr M.M., Foley R.A. Towards a theory of modern human origins: Geography, demography, and diversity in recent human evolution. Am. J. Phys. Anthropol. 1998;(Suppl 27):137–176. doi: 10.1002/(sici)1096-8644(1998)107:27+<137::aid-ajpa6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 11.Rasmussen M., Guo X., Wang Y., Lohmueller K.E., Rasmussen S., Albrechtsen A., Skotte L., Lindgreen S., Metspalu M., Jombart T. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334:94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reich D., Green R.E., Kircher M., Krause J., Patterson N., Durand E.Y., Viola B., Briggs A.W., Stenzel U., Johnson P.L. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reich D., Patterson N., Kircher M., Delfin F., Nandineni M.R., Pugach I., Ko A.M., Ko Y.C., Jinam T.A., Phipps M.E. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am. J. Hum. Genet. 2011;89:516–528. doi: 10.1016/j.ajhg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallick S., Li H., Lipson M., Mathieson I., Gymrek M., Racimo F., Zhao M., Chennagiri N., Nordenfelt S., Tandon A. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016;538:201–206. doi: 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malaspinas A.S., Westaway M.C., Muller C., Sousa V.C., Lao O., Alves I., Bergström A., Athanasiadis G., Cheng J.Y., Crawford J.E. A genomic history of Aboriginal Australia. Nature. 2016;538:207–214. doi: 10.1038/nature18299. [DOI] [PubMed] [Google Scholar]
- 16.Pagani L., Lawson D.J., Jagoda E., Mörseburg A., Eriksson A., Mitt M., Clemente F., Hudjashov G., DeGiorgio M., Saag L. Genomic analyses inform on migration events during the peopling of Eurasia. Nature. 2016;538:238–242. doi: 10.1038/nature19792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tucci S., Akey J.M. Population genetics: A map of human wanderlust. Nature. 2016;538:179–180. doi: 10.1038/nature19472. [DOI] [PubMed] [Google Scholar]
- 18.Excoffier L., Dupanloup I., Huerta-Sánchez E., Sousa V.C., Foll M. Robust demographic inference from genomic and SNP data. PLoS Genet. 2013;9:e1003905. doi: 10.1371/journal.pgen.1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schiffels S., Durbin R. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 2014;46:919–925. doi: 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawson D.J., Hellenthal G., Myers S., Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8:e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.International HapMap 3 Consortium. Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green R.E., Krause J., Briggs A.W., Maricic T., Stenzel U., Kircher M., Patterson N., Li H., Zhai W., Fritz M.H. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satta Y., Klein J., Takahata N. DNA archives and our nearest relative: the trichotomy problem revisited. Mol. Phylogenet. Evol. 2000;14:259–275. doi: 10.1006/mpev.2000.0704. [DOI] [PubMed] [Google Scholar]
- 25.Chen F.C., Li W.H. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer M., Kircher M., Gansauge M.T., Li H., Racimo F., Mallick S., Schraiber J.G., Jay F., Prüfer K., de Filippo C. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hudson R.R. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 28.Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P.H., de Filippo C. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavalli-Sforza L.L., Barrai I., Edwards A.W.F. Analysis of human evolution under random genetic drift. Cold Spring Harb. Symp. Quant. Biol. 1964;29:9–20. doi: 10.1101/sqb.1964.029.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Hudson R.R., Slatkin M., Maddison W.P. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terhorst J., Kamm J.A., Song Y.S. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 2017;49:303–309. doi: 10.1038/ng.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.