Abstract
Pituitary adenoma (PA) is one of the most common intracranial neoplasms. Several genetic predisposing factors for PA have been identified, but they account for a small portion of cases. In this study, we sought to identify the PA genetic risk factors by focusing on causative mutations for PAs. Among the 4 affected and 17 asymptomatic members from one family with familial PA, whole-exome sequencing identified cosegregation of the PA phenotype with the heterozygous missense mutation c.4136G>T (p.Arg1379Leu) in cadherin-related 23 (CDH23). This mutation causes an amino acid substitution in the calcium-binding motif of the extracellular cadherin (EC) domains of CDH23 and is predicted to impair cell-cell adhesion. Genomic screening in a total of 12 families with familial PA (20 individuals), 125 individuals with sporadic PA, and 260 control individuals showed that 33% of the families with familial PA (4/12) and 12% of individuals with sporadic PA (15/125) harbored functional CDH23 variants. In contrast, 0.8% of the healthy control individuals (2/260) carried functional CDH23 variants. Gene-based analysis also revealed a significant association between CDH23 genotype and PA (p = 5.54 × 10−7). Moreover, PA individuals who did not harbor functional CDH23 variants displayed tumors that were larger in size (p = 0.005) and more invasive (p < 0.001). Therefore, mutations in CDH23 are linked with familial and sporadic PA and could play important roles in the pathogenesis of PA.
Keywords: pituitary adenoma, familial pituitary adenoma, CDH23, mutation, whole-exome sequencing
Main Text
With a prevalence of 14%–22%, pituitary adenoma (PA [MIM: 102200]) is one of the most common neuroendocrine tumors and varies in different populations. PAs manifest with a variety of clinical features, including local invasion of surrounding structures and excessive hormone secretion.1 PAs are classified clinically as functional and non-functional (NF). According to the hormones produced by functional PAs, they are further classified into six subtypes: prolactin (PRL), growth hormone (GH), adrenocorticotropic hormone (ACTH), thyroid-stimulating hormone (TSH), gonadotropins hormone (GT), and multiple hormones (plurihormonal).2
Familial PAs are characterized by the development of PAs in multiple family members and are divided into (1) isolated groups, in which no other organs are involved in addition to the PA, and (2) syndromic groups. Each group can be caused by germline mutations in different genes predisposing to PA. For example, mutations in arylhydrocarbon-receptor-interacting protein (AIP [MIM: 605555]) and G-protein-coupled receptor 101 (GPR101 [MIM: 300393]) were discovered in familial isolated PAs, whereas mutations in multiple endocrine neoplastic type 1 (MEN1 [MIM: 613733]) and cyclin-dependent kinase inhibitor 1B (CDKN1B [MIM: 600778]) were found in syndromic PAs.3, 4 However, individuals with these mutations account for ∼20% of all cases of familial PA, and the genetic risk factors of most familial PAs remain unknown.1, 5, 6 A recent genome-wide association study by our group revealed that common variants at three loci are significantly associated with the occurrence of sporadic PAs.7 Genetic alterations of multiple tumor-suppressor genes, including RB1 (MIM: 614041) and MGMT (MIM: 156569), have also been identified in sporadic PAs.8 Moreover, some genetic risk factors for familial PA have been found in sporadic cases, e.g., mutations in AIP were detected in approximately 5% of individuals with sporadic PA.9 However, pathogenic variants in the majority of sporadic PAs have not yet been revealed.
In this study, we identified the germline genetic alterations in cadherin-related 23 (CDH23 [MIM: 605516]) by performing whole-exome sequencing (WES) and follow-up analyses of 12 families and 125 individuals with familial and sporadic PAs, respectively. Individuals with PAs were recruited at the Department of Neurosurgery at Huashan Hospital, an affiliate of the Fudan University Shanghai Medical College, from 2008 to 2015. This study was approved by the ethics committee at Huashan Hospital, and informed consent was obtained from each individual involved in the study.
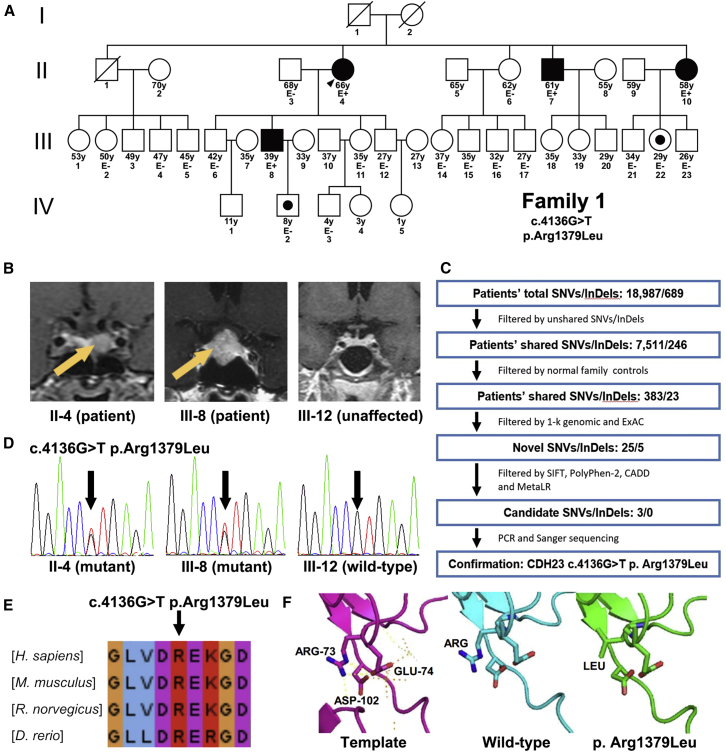
At first, we analyzed a PA family with four affected individuals (Figure 1A). Two of them (II-4 and III-8) were diagnosed with GH PAs, whereas the other two (II-7 and II-10) were diagnosed with NF PAs. An overview of the clinical findings is shown in Table S1 and Figure 1B. In brief, individual II-4 and III-8 suffered from high GH or IGF-1 levels and presented with acromegaly for at least 4 years. Neither of them received somatostatin analogs or any other medications before transsphenoidal surgery. Both clinical manifestations and hormone levels were significantly relieved 6 months after surgical resection. Individuals II-7 and II-10 were reported to experience occasional headaches for less than 1 year during the annual medical examinations and were further diagnosed with PAs according to the standard guidelines. No other symptoms, such as visual loss, acromegaly, amenorrhea, or menstrual abnormalities, were observed. Their hormone levels were within the normal ranges. Both individuals II-7 and II-10 declined to receive any surgical treatment, and no tumor enlargement was observed during the 9-month follow-up.
Figure 1.
CDH23 Mutations Identified in Familial PA
(A) Pedigree of family 1, who has familial PA. Two affected (II-4 and III-8) and two unaffected (II-3 and III-12) individuals were selected for exome sequencing. Black filled shapes represent affected siblings. White shapes represent asymptomatic subjects. White shapes with a black dot represent asymptomatic carriers of CDH23 mutations. Symbols crossed with a diagonal line indicate deceased relatives. The age of each member is indicated under each symbol. Family members with available clinical data and DNA samples are indicated with E− or E+ under their symbols. E− indicates that the person had a negative evaluation for PA, and E+ indicates that the individual has PA.
(B) MRI data of two individuals with PA (II-4 and III-8) and one unaffected person (III-12) who were included for exome sequencing. Yellow arrows mark tumors.
(C) Filters applied to variants and indels detected by exome sequencing in this family.
(D) Sequencing chromatograms of CDH23 in two individuals with PA (II-4 and III-8) and one unaffected person (III-12). All heterozygous variants were verified by Sanger sequencing (indicated by the black arrow). The mutation and amino acid substitution are shown on the top.
(E) Alignment of the CDH23 EC13 domain shows evolutionary conservation at the site of the identified nucleic acid changes (indicated by the black arrow).
(F) Protein pathogenicity prediction of wild-type and p.Arg1379Leu (c.4136G>T) CDH23 proteins. The pink model represents the homologous template for the EC domain (PDB: 2WHV). The blue model shows the structure of wild-type CDH23. The green model shows the structure of p.Arg1379Leu CDH23.
We performed WES in genomic DNAs from white blood cells of two affected (II-4 and III-8) and two asymptomatic (II-3 and III-12) family members to determine potential germline variants. In-solution exome capture was performed with the TruSeq Exome Enrichment Kit (Illumina) and the Agilent SureSelect Human All Exon V5+UTR Kit with 125-bp paired-end read sequences generated on a HiSeq 2500 (Illumina) and then analyzed by standard methods. UCSC Genome Browser build hg19 was used as the reference sequence. The mean sequencing depth was 84×, including >10× coverage for 88.2% of the target regions. Variants with allele frequencies greater than 0.1% in 1000 Genomes and the Exome Aggregation Consortium (ExAC) Browser were excluded from our analysis. A total of 383 nonsynonymous variants and 23 indels were shared by both PA individuals but not by the two unaffected family members. After filtration, we identified 25 variants and 5 heterozygous indels whose frequencies were less than 0.1% in 1000 Genomes and the ExAC Browser. Among them, three variants were predicted to affect the protein structure according to analyses by SIFT,10 PolyPhen-2,11 CADD,12 and MetaLR13 (Figure 1C). The threshold was determined according to the criteria reported before.14 Further Sanger sequencing of other family members ascertained that the heterozygous CDH23 c.4136G>T (p.Arg1379Leu) (GenBank: NM_022124.5) mutation segregated with PA disease (Figure 1D). This variant site was not identified in either 1000 Genomes or the ExAC Browser. Two phenotypically unaffected relatives (IV-2 and III-22) were found to carry CDH23 mutations. However, both of them are younger than 30 years old and could still develop PA in the future.
CDH23 is a member of the cadherin superfamily, which comprises calcium-dependent cell-cell adhesion glycoproteins.15 Germline mutations in CDH23 have been identified in people with Usher syndrome 1D (MIM: 601067) and nonsyndromic autosomal-recessive deafness (MIM: 601386), and Cdh23 mutant mice develop a quite similar disease.16, 17 Deregulated expression and activity of some cadherin proteins are frequently observed in human PAs and are inferred to contribute to the pathogenesis of PAs.18, 19, 20 In addition, CDH23 forms heterodimers and functions closely with protocadherin-related 15 (PCDH15), whose encoding gene is located near a genetic susceptibility locus for pituitary adenoma according to our previous work.7 These findings further suggest mutations in CDH23 as a potential genetic risk factor for this familial PA. The identified amino acid substitution is located in the second calcium-binding site of the extracellular cadherin 13 (EC13) domain, which is highly conserved among humans, mice, rats, and zebrafish (Figure 1E and Figure S1). Functions of cadherin members are typically mediated through their EC domains, especially through the highly conserved calcium-binding motifs.21 Thus, this substitution could lead to loss of calcium-binding ability and the function of the EC13 domain.
We performed further molecular modeling to evaluate the predicted functional effect of the discovered CDH23 mutation (Figure 1F). In the homologous template of the CDH23 domain (pink model [PDB: 2WHV]), amino acid position 1,379 (corresponding to Arg73) forms hydrogen bonds with Asp102 and Glu74 and thus greatly contributes to the calcium-binding ability and conformational stability of the EC domain. In comparison, the blue and green models represent the protein structures encoded by wild-type and mutant CDH23, respectively. The amino acid alteration caused by c.4136G>T (p.Arg1379Leu) was predicted to prevent the formation of hydrogen bonds and thus impair the calcium-binding ability and stability of the EC domain, suggesting that it is an inactivating mutation. We next evaluated whether this CDH23 mutation would affect expression and localization of CDH23. Immunohistochemistry staining showed that, regardless of the CDH23 genotype, CDH23 was present on the membrane of normal pituitary glands, as well as PAs, with similar expression levels, suggesting that the CDH23 mutation is not associated with reduced expression or mislocalization of the protein (Figure S2).
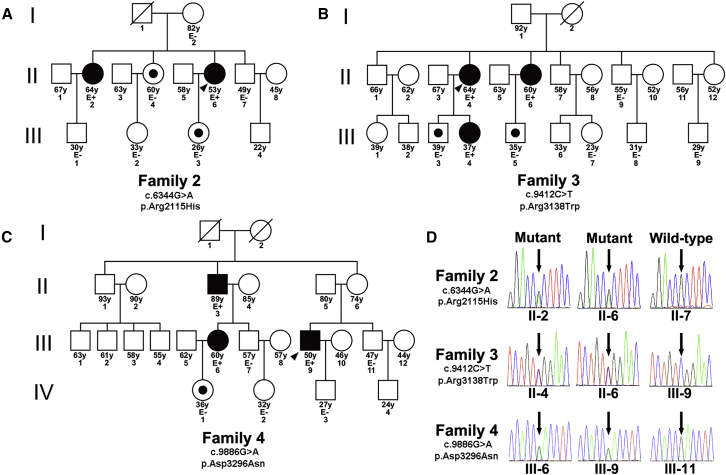
To further investigate whether CDH23 variants exist in other familial PAs, we recruited 11 additional families with familial PAs. Each family had two individuals with PA, except families 3 and 4 each had three affected individuals (Figures 2A–2C and Figure S3). WES was performed in one person per family. We found that three individuals with familial PA harbored CDH23 variants (c.6344G>A [p.Arg2115His], c.9412C>T [p.Arg3138Trp], and c.9886G>A [p.Asp3296Asn] in families 2, 3, and 4, respectively; GenBank: NM_022124.5; Figure 2D). All of these mutations were heterozygous, and the allele frequency of each variant in the ExAC Browser was less than 0.05% (Table S1). PolyPhen-2 scores of all three variants were greater than 0.99, indicating potential pathogenicity. Sanger sequencing of 8 affected and 17 unaffected relatives with available blood DNA samples verified the presence of the variants and showed that the CDH23 mutations co-segregated with clinical phenotypes (Figures 2A–2D and Figure S4). Notably, all three discovered alternative residues in CDH23 are highly conserved among humans, mice, rats, and zebrafish (Figure S5).
Figure 2.
Three Additional Families Affected by CDH23 Mutations
(A–C) Pedigrees of three families carrying CDH23 mutations. Black filled shapes represent affected siblings. White shapes represent asymptomatic subjects. White shapes with a black dot represent asymptomatic carriers of CDH23 mutations. Symbols crossed with a diagonal line indicate deceased relatives. The age of each member is indicated under each symbol. Family members with available clinical data and DNA samples are indicated with E− or E+ under their symbols. E− indicates that the person had a negative evaluation for PA, and E+ indicates that the individual has PA.
(D) Sanger sequencing showing CDH23 mutations in three affected individuals from different families. The black arrows indicate the mutated nucleotides. The mutations and amino acid substitutions are shown on the left.
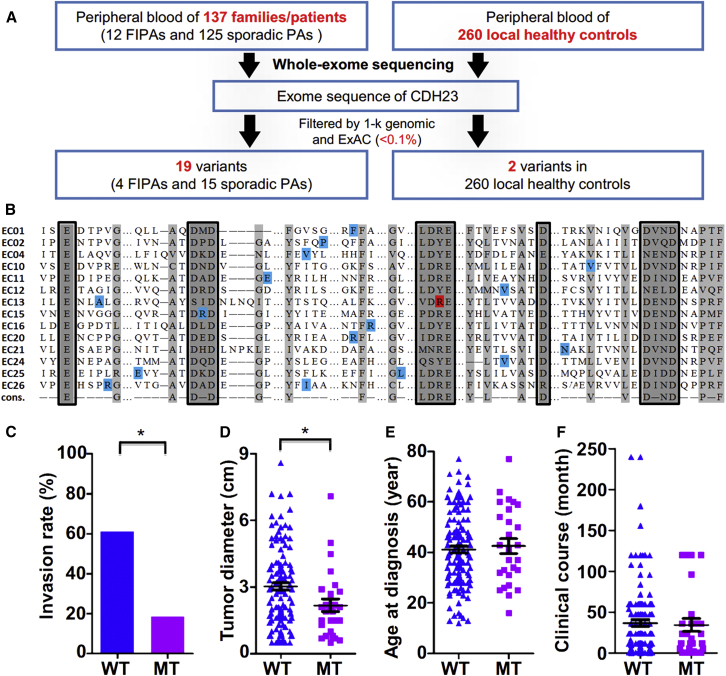
To identify whether CDH23 mutations are associated with sporadic PAs, we used WES to analyze genomic DNAs of white blood cells from 125 individuals with sporadic PAs. Subjects with any potential previous malignancies or family history of PAs were excluded. These PAs included 20 NF PAs, 20 PRL PAs, 20 GH PAs, 20 ACTH PAs, 20 GT PAs, 10 TSH PAs, and 15 plurihormonal PAs. The plurihormonal PAs in this study secreted GH and PRL (n = 11), GH and ACTH (n = 2), or GH and TSH (n = 2). We found that 15 (12.0%) individuals harbored CDH23 mutations, including one NF PA, three PRL PAs, two GH PAs, four ACTH PAs, one GT PA, three TSH PAs, and one plurihormonal (GH and TSH) PA (Figure 3A and Table S1). Most of these variants were heterozygous, but two were homozygous. Allele frequencies of all variants were less than 0.1% in 1000 Genomes and the ExAC Browser (Table S1). All 15 variants were predicted to be pathogenic by at least one in silico method (SIFT, PolyPhen-2, CADD, or MetaLR). Although the spectrum was diverse, all amino acid substitutions were located in the EC domains of CDH23 and were close to the conserved site of each domain (Figure 3B),21, 22 which could affect protein folding or calcium-binding affinity.
Figure 3.
CDH23 Mutations in a Large PA Cohort
(A) The frequency of CDH23 mutations in familial PAs, sporadic PAs, and local healthy control individuals.
(B) Mutant CDH23 variant sites in familial and sporadic PAs. All 14 EC domains with detected amino acid substitutions are shown. The ordinal of each EC domain is shown on the left. Corresponding EC domains are indicated to the left of the lineup. Light gray indicates conserved residues. Highly conserved calcium-binding elements are in gray and boxed. The residue in red indicates the mutant site discovered in family 1, and other variants are marked in blue.
(C) Percentages of individuals with invasions in both wild-type (WT) and mutant (MT) groups. The p values were calculated by Pearson’s χ2 test.
(D–F) Maximal tumor diameters of the WT and MT groups (D), ages at diagnosis of the WT and MT groups (E), and the clinical course of each person in the WT and MT groups (F). Each symbol represents an individual case. Error bars show the mean and standard errors of the data. The p values were calculated by the Mann-Whitney U test (∗p < 0.05 considered significant).
In contrast, among the 260 local healthy control individuals, two (0.8%) carried CDH23 mutations, one of which was predicted to be pathogenic (Table S2). The frequencies of CDH23 mutations were significantly different between people with sporadic PA and normal control individuals (p = 2 × 10−6, Pearson’s χ2 test). Sequence kernel association testing identified significant gene-based associations between PAs and CDH23 variants (p = 5.54 × 10−7), suggesting a significantly increased risk of PAs in subjects with CDH23 mutations.23 Very interestingly, none of the sporadic PAs harbored somatic CDH23 mutations on the basis of previous genomic studies.4 Similar to AIP variants, germline CDH23 variants are genetic risk factors for both familial and sporadic Pas.6, 24
On the basis of the fact that tumor growth and invasion are crucial for the treatment of PAs and that the cadherin superfamily is involved in cell-cell adhesion, we focused on the correlation between CDH23 genotypes and clinical characteristics, especially tumor diameter and invasiveness.25 Among the 145 cases considered (20 familial and 125 sporadic PAs), CDH23 mutations were identified in all subtypes. On the basis of the contrast-enhanced MRI and intra-operative findings, 72 of 118 (61.02%) CDH23 wild-type PAs extended into the parasellar spaces, including the cavernous sinus and the sphenoidal sinus. In contrast, the vast majority of CDH23 mutant PAs diffusely distributed within the sella: 5 of 27 cases (18.52%) displayed parasellar invasion (p < 0.001, Pearson’s χ2 test; Figure 3C and Table S3). This morphological characteristic is further supported by the fact that CDH23 mutant PAs are generally smaller than CDH23 wild-type PAs (1.90 versus 2.90 cm, p = 0.005, Mann-Whitney U test; Figure 3D and Table S3). To exclude the possibility that this finding was due to closer surveillance MRI scans detecting tumors in familial cases at earlier stages of their natural history, we analyzed only sporadic cases. As shown in Table S4, CDH23 mutations were significantly negatively related to both the invasiveness and tumor diameter in sporadic PAs (p = 0.003 and 0.046, respectively). CDH23 has been reported to play a role in tumor progression through regulating cell-cell adhesion. For example, CDH23 is related to the early stages of tumor metastasis and is upregulated in breast cancers (MIM: 114480).26 All of the identified CDH23 mutations seem to be inactivating mutations and could impair cell-cell adhesion because they are located close to or within the regions encoding the conserved calcium-binding motifs of CDH23, partially explaining why CDH23 mutations are related to the invasiveness of PA. Other clinical features, including age at diagnosis, clinical course, and radical resection rate, showed no significant differences between wild-type individuals and those with CDH23 mutations (Figures 3E and 3F and Tables S3 and S4).
Several mutations in CDH23 are associated with inherited hearing loss and blindness.16 However, none of the variants found in this study were linked to any symptoms of deafness or blindness. This could be explained by the observation that no shared variants were identified in either hearing loss or PA. Another possible explanation is that deafness-related CDH23 mutations are mostly homozygous or compound heterozygous,16, 27, 28 whereas most CDH23 mutations in this study were heterozygous. It is conceivable that carriers of CDH23 mutations are at a high risk of developing hearing loss. Because most individuals included in this study are younger than 65 years old, age-related hearing loss (MIM: 612448) cannot be excluded.29 Extensive clinical investigation should be undertaken to determine whether pituitary MRI scans should be adopted in the screening of CDH23-related diseases, including Usher syndrome 1D and age-related hearing loss.
It is common that familial and sporadic human diseases, including PA, share the same susceptibility genes. For example, APC (MIM: 611731) mutations are found in both familial and sporadic human colorectal cancers (MIM: 114500).30 CDH23 is another case, further supporting the notion that sporadic and familial PAs can be attributed to similar genetic predisposing factors.
In summary, we have identified CDH23 mutations as a genetic risk factor for both familial and sporadic PAs. Further functional studies of this gene are necessary for gaining insights into the pathogenic mechanisms involved in PAs.
Acknowledgments
We thank and acknowledge all of the participants in this study. We gratefully thank Mrs. Qiuwei Song and Mrs. Yun Zhang for sample collection. We thank Prof. Xingdang Liu, Prof. Yun Lu, Mrs. Jue Ji, Ms. Jie Zheng, and Mrs. Xiaolan Qin for technique support. We appreciate Dr. Daizhan Zhou for analysis consultation. We also thank Beijing Pangenomics Technology Co., Ltd. (Genetron Health) for help with the genomic data analysis. This work was supported by the China Pituitary Adenoma Specialist Council, the Chang Jiang Scholars Program (to Y. Zhao), the National Program for Support of Top-Notch Young Professionals (to Y. Zhao and Y. Shi), the Program for New-Century Excellent Talents in University (NCET-10-0356 to Y. Zhao), the Shanghai Rising-Star Tracking Program (12QH1400400 to Y. Zhao), the National High Technology Research and Development Program of China (2014AA020611 to Y.Z.), the National Key Basic Research Program of China (973 Program; 2015CB559100 to Y. Shi), the National Natural Science Foundation of China (31325014 and 81421061 to Y. Shi), the Program of Shanghai Academic Research Leader (15XD1502200 to Y. Shi), the Shanghai Key Laboratory of Psychotic Disorders (13dz2260500 to Y.S.), the 1000 Youth Elite Program and Shanghai Pujiang Scholarship (15PJ1407400 to C.H.), the Natural Science Foundation and Major Basic Research Program of Shanghai (16JC1420100 to Y.M.), and the National Natural Science Foundation of China (81602191 to M.S.).
Published: April 13, 2017
Footnotes
Supplemental Data include five figures and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.03.011.
Contributor Information
Chuanxin Huang, Email: huangcx@shsmu.edu.cn.
Yao Zhao, Email: zhaoyaohs@vip.sina.com.
Web Resources
1000 Genomes, http://www.1000genomes.org/
ExAC Browser, http://exac.broadinstitute.org/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
OMIM, http://www.omim.org
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
References
- 1.Lecoq A.L., Kamenický P., Guiochon-Mantel A., Chanson P. Genetic mutations in sporadic pituitary adenomas--what to screen for? Nat. Rev. Endocrinol. 2015;11:43–54. doi: 10.1038/nrendo.2014.181. [DOI] [PubMed] [Google Scholar]
- 2.Saeger W., Lüdecke D.K., Buchfelder M., Fahlbusch R., Quabbe H.J., Petersenn S. Pathohistological classification of pituitary tumors: 10 years of experience with the German Pituitary Tumor Registry. Eur. J. Endocrinol. 2007;156:203–216. doi: 10.1530/eje.1.02326. [DOI] [PubMed] [Google Scholar]
- 3.Elston M.S., McDonald K.L., Clifton-Bligh R.J., Robinson B.G. Familial pituitary tumor syndromes. Nat. Rev. Endocrinol. 2009;5:453–461. doi: 10.1038/nrendo.2009.126. [DOI] [PubMed] [Google Scholar]
- 4.Caimari F., Korbonits M. Novel Genetic Causes of Pituitary Adenomas. Clin. Cancer Res. 2016;22:5030–5042. doi: 10.1158/1078-0432.CCR-16-0452. [DOI] [PubMed] [Google Scholar]
- 5.Toledo R.A., Lourenço D.M., Jr., Toledo S.P. Familial isolated pituitary adenoma: evidence for genetic heterogeneity. Front. Horm. Res. 2010;38:77–86. doi: 10.1159/000318497. [DOI] [PubMed] [Google Scholar]
- 6.Preda V., Korbonits M., Cudlip S., Karavitaki N., Grossman A.B. Low rate of germline AIP mutations in patients with apparently sporadic pituitary adenomas before the age of 40: a single-centre adult cohort. Eur. J. Endocrinol. 2014;171:659–666. doi: 10.1530/EJE-14-0426. [DOI] [PubMed] [Google Scholar]
- 7.Ye Z., Li Z., Wang Y., Mao Y., Shen M., Zhang Q., Li S., Zhou L., Shou X., Chen J. Common variants at 10p12.31, 10q21.1 and 13q12.13 are associated with sporadic pituitary adenoma. Nat. Genet. 2015;47:793–797. doi: 10.1038/ng.3322. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Y., Zhang X., Klibanski A. Genetic and epigenetic mutations of tumor suppressive genes in sporadic pituitary adenoma. Mol. Cell. Endocrinol. 2014;386:16–33. doi: 10.1016/j.mce.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai F., Zhang Y.D., Zhao X., Yang Y.K., Ma S.H., Dai C.X., Liu X.H., Yao Y., Feng M., Wei J.J. Screening for AIP gene mutations in a Han Chinese pituitary adenoma cohort followed by LOH analysis. Eur. J. Endocrinol. 2013;169:867–884. doi: 10.1530/EJE-13-0442. [DOI] [PubMed] [Google Scholar]
- 10.Ng P.C., Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jagadeesh K.A., Wenger A.M., Berger M.J., Guturu H., Stenson P.D., Cooper D.N., Bernstein J.A., Bejerano G. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 2016;48:1581–1586. doi: 10.1038/ng.3703. [DOI] [PubMed] [Google Scholar]
- 15.Sotomayor M., Gaudet R., Corey D.P. Sorting out a promiscuous superfamily: towards cadherin connectomics. Trends Cell Biol. 2014;24:524–536. doi: 10.1016/j.tcb.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz J.M., Bhatti R., Madeo A.C., Turriff A., Muskett J.A., Zalewski C.K., King K.A., Ahmed Z.M., Riazuddin S., Ahmad N. Allelic hierarchy of CDH23 mutations causing non-syndromic deafness DFNB12 or Usher syndrome USH1D in compound heterozygotes. J. Med. Genet. 2011;48:767–775. doi: 10.1136/jmedgenet-2011-100262. [DOI] [PubMed] [Google Scholar]
- 17.Manji S.S., Miller K.A., Williams L.H., Andreasen L., Siboe M., Rose E., Bahlo M., Kuiper M., Dahl H.H. An ENU-induced mutation of Cdh23 causes congenital hearing loss, but no vestibular dysfunction, in mice. Am. J. Pathol. 2011;179:903–914. doi: 10.1016/j.ajpath.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fougner S.L., Lekva T., Borota O.C., Hald J.K., Bollerslev J., Berg J.P. The expression of E-cadherin in somatotroph pituitary adenomas is related to tumor size, invasiveness, and somatostatin analog response. J. Clin. Endocrinol. Metab. 2010;95:2334–2342. doi: 10.1210/jc.2009-2197. [DOI] [PubMed] [Google Scholar]
- 19.Qian Z.R., Sano T., Yoshimoto K., Asa S.L., Yamada S., Mizusawa N., Kudo E. Tumor-specific downregulation and methylation of the CDH13 (H-cadherin) and CDH1 (E-cadherin) genes correlate with aggressiveness of human pituitary adenomas. Mod. Pathol. 2007;20:1269–1277. doi: 10.1038/modpathol.3800965. [DOI] [PubMed] [Google Scholar]
- 20.Chauvet N., Romanò N., Meunier A.C., Galibert E., Fontanaud P., Mathieu M.N., Osterstock G., Osterstock P., Baccino E., Rigau V. Combining Cadherin Expression with Molecular Markers Discriminates Invasiveness in Growth Hormone and Prolactin Pituitary Adenomas. J. Neuroendocrinol. 2016;28:12352. doi: 10.1111/jne.12352. [DOI] [PubMed] [Google Scholar]
- 21.de Brouwer A.P., Pennings R.J., Roeters M., Van Hauwe P., Astuto L.M., Hoefsloot L.H., Huygen P.L., van den Helm B., Deutman A.F., Bork J.M. Mutations in the calcium-binding motifs of CDH23 and the 35delG mutation in GJB2 cause hearing loss in one family. Hum. Genet. 2003;112:156–163. doi: 10.1007/s00439-002-0833-0. [DOI] [PubMed] [Google Scholar]
- 22.Astuto L.M., Bork J.M., Weston M.D., Askew J.W., Fields R.R., Orten D.J., Ohliger S.J., Riazuddin S., Morell R.J., Khan S. CDH23 mutation and phenotype heterogeneity: a profile of 107 diverse families with Usher syndrome and nonsyndromic deafness. Am. J. Hum. Genet. 2002;71:262–275. doi: 10.1086/341558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beckers A., Aaltonen L.A., Daly A.F., Karhu A. Familial isolated pituitary adenomas (FIPA) and the pituitary adenoma predisposition due to mutations in the aryl hydrocarbon receptor interacting protein (AIP) gene. Endocr. Rev. 2013;34:239–277. doi: 10.1210/er.2012-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colao A., Grasso L.F., Pivonello R., Lombardi G. Therapy of aggressive pituitary tumors. Expert Opin. Pharmacother. 2011;12:1561–1570. doi: 10.1517/14656566.2011.568478. [DOI] [PubMed] [Google Scholar]
- 26.Apostolopoulou M., Ligon L. Cadherin-23 mediates heterotypic cell-cell adhesion between breast cancer epithelial cells and fibroblasts. PLoS ONE. 2012;7:e33289. doi: 10.1371/journal.pone.0033289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schultz J.M., Yang Y., Caride A.J., Filoteo A.G., Penheiter A.R., Lagziel A., Morell R.J., Mohiddin S.A., Fananapazir L., Madeo A.C. Modification of human hearing loss by plasma-membrane calcium pump PMCA2. N. Engl. J. Med. 2005;352:1557–1564. doi: 10.1056/NEJMoa043899. [DOI] [PubMed] [Google Scholar]
- 28.Miyagawa M., Nishio S.Y., Usami S. Prevalence and clinical features of hearing loss patients with CDH23 mutations: a large cohort study. PLoS ONE. 2012;7:e40366. doi: 10.1371/journal.pone.0040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang J.H., Liu K.S., Wu C.C., Liu T.C. Association of cadherin23 single nucleotide polymorphism with age-related hearing impairment in Han Chinese. Otolaryngol. Head Neck Surg. 2012;147:531–534. doi: 10.1177/0194599812446904. [DOI] [PubMed] [Google Scholar]
- 30.Hadjisavvas A., Papasavva T., Loizidou M., Malas S., Potamitis G., Christodoulou C., Pavlides G., Papamichael D., Klonis C., Nasioulas G. Novel germline mutations in the APC gene of Cypriot patients with familial and sporadic adenomatous polyposis. Clin. Genet. 2006;69:404–409. doi: 10.1111/j.1399-0004.2006.00617.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.