Abstract
Interpretation of results from genome-wide association studies for T2D is challenging. Only very few loci have been replicated in African ancestry populations and the identification of the implicated functional genes remain largely undefined. We used genetic maps that capture detailed linkage disequilibrium information in European and African Americans and applied these to large T2D case-control samples in order to estimate locations for putative functional variants in both populations. Replicated T2D locations were tested for evidence of being regulatory hotspots using adipose expression. We validated a sample of our co-location intervals using next generation sequencing and functional annotation, including enhancers, transcription, and chromatin modifications. We identified 111 additional disease-susceptibility locations, 93 of which are cosmopolitan and 18 of which are European specific. We show that many previously known signals are also risk loci in African Americans. The majority of the disease locations appear to confer risk of T2D via the regulation of expression levels for a large number (266) of cis-regulated genes, the majority of which are not the nearest genes to the disease loci. Sequencing three cosmopolitan locations provided candidate functional variants that precisely co-locate with cell-specific chromatin domains and pancreatic islet enhancers. These variants have large effect sizes and are common across populations. Results show that disease-associated loci in different populations, gene expression, and cell-specific regulatory annotation can be effectively integrated by localizing these effects on high-resolution genetic maps. The cis-regulated genes provide insights into the complex molecular pathways involved and can be used as targets for sequencing and functional molecular studies.
Keywords: type 2 diabetes, gene mapping, linkage disequilibrium maps, expression quantitative trait loci, regulation
Introduction
No disease with a genetic predisposition has been more intensely investigated than type 2 diabetes (T2D [MIM: 125853]), the world’s most widespread and devastating metabolic disorder. Over the last 10 years, numerous consortia have undertaken to characterize the genetic causes of T2D through a very large number (>30) of genome-wide association studies (GWASs) and large-scale meta-analyses. Initially based on Europeans, the focus has now shifted to the replication of risk loci in additional ethnicities (trans-ethnic studies), motivated in part by the likely wider application of cosmopolitan variants for translational research, but also the desire for increasingly larger research sample sizes in order to try to boost study power.1 But since T2D is really a group of diseases,2 increased sample size should be met with skepticism unless accompanied by more detailed clinical phenotypes and strategies to minimize disease heterogeneity. Recent trans-ethnic meta-analysis of T2D for four populations (Europeans, East Asians, South Asians, and Mexican Americans) has identified 7 T2D loci,3 in addition to the previously published list of 69 loci.4 However, a large proportion of these 76 loci3 do not show evidence for nominal association for the same “lead” SNP. Since the lead SNP is unlikely to be the causal variant, this low replication rate is a general problem for trans-ethnic studies.5 The inability to account for genetic distance between neighboring SNPs and genetic heterogeneity (e.g., locus and allele heterogeneity and variation in LD between populations) are both potential thwarting factors in the endeavor to identify trans-ethnic disease loci.
On the other hand, the prevalence of T2D in African Americans (19%) at approximately twice that of European Americans (10%) and the existence of more genetic diversity in peoples of African ancestry, partly due to less extensive linkage disequilibrium (LD), also gives rise to a major opportunity for comparative fine-mapping studies.3, 6, 7 This possibility was missed by a recent major trans-ethnic meta-analyses that unfortunately excluded African-Americans.3 Here we seek to take advantage of this ancestry group by using a mapping approach to identify cosmopolitan T2D locations, which avoids the focus on lead SNPs. Instead we use high-resolution genetic maps to identify new cosmopolitan T2D susceptibility loci that are shared by both European and African American populations. Genetic distances from these maps accurately capture the genetic architecture of the relevant population and have been successfully used in gene-mapping studies for other common diseases.8, 9 We constructed genetic maps for each of these two populations and then used disease-associated location estimates on these maps as the basis for the precise co-localization and replication in both populations. We also analyzed all 76 previously known T2D loci3 to obtain refined location estimates on the same genetic maps. Since an estimated 90% of variants with a functional role in complex traits such as T2D are likely to be non-coding and regulatory,10 we assessed this scenario by exploiting publicly available subcutaneous adipose expression data. The hypothesis we tested is that T2D disease loci confer risk of disease by acting as expression quantitative trait loci (eQTL) that regulate the expression of neighboring (cis-) genes. To test this hypothesis (Figure 1), we used the same genetic maps to assess whether the location estimates for eQTL also precisely co-located with those mapped for T2D in this whole-genome analysis, thereby identifying potential cis-genes and pathways regulated by the disease loci. Finally, we performed fine mapping using targeted next generation sequencing (NGS) of the refined location estimates for one previously known locus (TCFL72) and two of the additional cosmopolitan loci from this study using independent case/control sample data, with the aim of identifying the candidate functional variants at the causal location estimates. These examples illustrate a way forward for the systematic identification of putative functional variants at these identified disease-associated eQTLs, coupled with the integration of functional annotation such as cell-specific chromatin domain modifications, enhancers, and transcription binding sites.
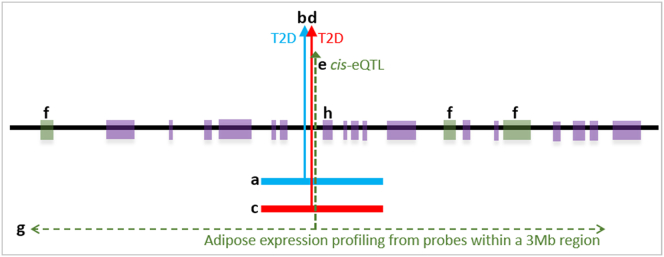
Figure 1.
A Schematic Presentation of the Functional Genomic Study Design
Shown are the linkage disequilibrium unit (LDU) window of the European (EUR) genetic map (a); location of the causal variant for T2D estimated on the EUR map using a EUR GWAS (b); the corresponding African American (AA) LDU genetic map for the same region (c); location of the causal variant for T2D estimated on the AA map using an AA GWAS (d); location of the cis-eQTL (e) for the three associated cis-genes (f) that are implicated using adipose expression data (g) for probes from genes within ±1.5 Mb distance either side of the T2D locations (b and d). In this example the nearest gene (h) is not the implicated regulated gene.
Subjects and Methods
Study Design
We analyzed two European (EUR) and one African American (AA) sample populations with a total of 5,800 T2D case subjects and 9,691 control subjects. The two independent EUR samples (SNP arrays for GWA and Metabochip) were obtained from the Wellcome Trust Case Control Consortium (WTCCC)11, 12 with a description of diagnostic criteria and sample matching provided in Supplemental Data. The AA GWA sample for a population of predominantly African ancestry was obtained from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK).13 Analyzing “one-SNP-at-a-time” ignores LD structure when testing for association with disease or gene expression. Here we use population-specific genetic maps, which provide (1) commensurability when making comparisons between different populations and SNP arrays; (2) the means to implement a multi-marker test of association;14 (3) genetic distances between loci when testing for association with disease or adipose expression; and (4) precise location estimates on the genetic map for potential functional variants, since these estimates are more efficient than using physical maps.15 We constructed two high-resolution genetic maps based upon HapMap data with genetic distances expressed in linkage disequilibrium units (LDU).16 The EUR LDU map was used for analyses of the two EUR T2D datasets and the AA LDU map was used for the analysis of the AA T2D dataset. The autosomal genome (sex chromosomes were not included in the analyses) was divided into 4,800 non-overlapping analytical windows, each with a minimum distance of 10 LDU. In addition to windows being the same minimum size on the genetic map, each window also had to include a minimum of 30 SNPs. These criteria yielded an average genetic length of 11 LDU. The identical boundaries in kilobases (kb) for all 4,800 analytical windows were used for the AA dataset, but with longer average genetic length (16 LDU), reflecting a population history of greater antiquity with additional historical recombination events. All SNPs in each analytical window were simultaneously used to test for association with disease using a multi-marker LDU model.14 The analysis returns one estimated location for a causal variant with the strongest signal, along with the association test p value for each window. Utilizing the genetic map in this way, the multi-marker test of association models the degree of regional LD when estimating the location of a putative causal variant on the genetic map. A schematic diagram of the functional genomic strategy used in the current study is provided in Figure 1. Strict criteria were used for the meta-analysis. Location estimates for genome-wide significant meta-analysis loci had to be nominally significant in both ancestry groups for the cosmopolitan loci and in both European samples for the European-specific loci. An interval criterion was used where location estimates from different datasets had to be within <100 kb of one another to qualify as a potential replication. Replicated loci had to pass a Bonferroni corrected meta-analysis p value threshold of 1 × 10−5, based on the total number of genomic tests performed (α = 0.05/4,800). We refer to the co-location interval (distance between location estimates) as the genomic region that most plausibly includes the functional variants that confer risk of T2D.
We conducted in silico functional gene expression analyses to assess whether the same T2D loci are also eQTL that regulate the expression of neighboring cis-genes using data generated by the MuTHER consortium.17 Summary statistics for the probes and SNPs were available from the MuTHER website. Using adipose tissue mRNA expression probes as quantitative traits, we tested for cis-association at each disease locus by employing the same multi-locus LDU model, with potential regulated cis-genes defined to be within ±1.5 Mb distance either side of each replicated T2D causal location (Figure 1). We considered a disease locus to be a potential eQTL only if the estimated eQTL co-located to within 50 kb of the T2D location and passed Bonferroni correction for the total number of probes tested within ±1.5 Mb of each replicated disease locus. All LDU location estimates for both T2D and eQTL on the genetic map were converted back to kb B36 (NCBI36/hg18) for presentation purposes.
Finally, we conducted a NGS targeted re-sequencing experiment for three of the disease loci. Next generation sequencing was conducted using the Agilent SureSelectXT2 capture kit following manufacturer protocol guidelines for 100 ng of DNA. Blood DNA samples were sequenced for a total of 94 unrelated European individuals with T2D and 94 unaffected controls 1:1 matched for age, BMI, and sex. Case subjects with a family history of T2D (selection and diagnosis criteria described elsewhere)18 and control subjects were selected from families originally collected for an obesity (MIM: 601665) study without a history of T2D.19 Additional method details are provided in the Supplemental Data.
Results
Additional Loci for T2D
Tables 1 and 2 present the results for the 111 additional loci associated with T2D. Of the 111 loci, 93 provide evidence of being cosmopolitan (signals 1–93, Table 1), since these loci replicate for both EUR and AA samples, while 18 loci appear to be European specific (94–111, Table 2), with replication in European samples only. The distances between T2D location estimates for the majority of the 111 loci were narrow (<50 kb apart). Estimation of average pairwise D-prime (D′) for all HapMap SNPs found within all the identified 111 disease location intervals (ranging from 0 to <100 kb) is D′ = 0.86 in EUR and D′ = 0.78 in AA, which reflect the importance of using a genetic map in LDU distances for localization and the <100 kb interval as a criterion for replication. For the majority of the cosmopolitan loci (signals 6–80, Table 1), the Metabochip array was not informative due to the very low SNP coverage in many regions (symbol “–” in Table 1). Some signals for Metabochip passed the minimal number of SNPs (>30 per window) but did not provide significant evidence of association (“NS” in Table 1) due to the uneven genomic coverage of SNPs on the customized Metabochip design.12 For this reason, there were only 13 cosmopolitan loci (signals 81–93, Table 1) that provided replicated evidence for AA and Metabochip European samples.
Table 1.
Identified Cosmopolitan T2D Susceptibility Loci and Their Regulatory Role of Neighboring Gene Expression
| Signal | Chr. | Meta p Value | Distance between Locationsa | T2D Location GWAS-Eb | T2D Location GWAS-Ac | T2D Location metabo-Ed | Nearest Gene to T2D Locationse | No. of cis-Genesf | eQTL-Associated cis-Genesg | eQTL Distance from T2Dh |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 4p | 1.73 × 10−56 | 1 | 44,797 | 44,857 | 44,858 | – | 0 | – | – |
| 2 | 6q | 5.28 × 10−10 | 4 | 72,479 | 72,509 | 72,505 | – | 1 | FAM135A | 1 |
| 3 | 13q | 2.14 × 10−35 | 28 | 109,848 | 109,805 | 109,833 | COL4A2∗ | 1 | ANKRD10 | 20 |
| 4 | 17q | 2.80 × 10−12 | 7 | 65,769 | 65,762 | 65,769 | KCNJ2∗ | 1 | MAP2K6 | 37 |
| 5 | 20q | 1.83 × 10−6 | 77 | 44,104 | 44,181 | 44,104 | SLC12A5∗, CD40∗+ | 2 | CD40, CDH22 | 17 |
| 6 | 1p | 2.48 × 10−6 | 41 | 82,826 | 82,867 | – | – | 1 | LPHN2∗ | 8 |
| 7 | 1p | 5.20 × 10−7 | 73 | 84,451 | 84,524 | – | PRKACB∗+, SAMD13∗+ | 3 | PRKACB, SAMD13, C1orf52 | 6 |
| 8 | 1p | 1.79 × 10−11 | 94 | 106,441 | 106,535 | – | – | 1 | PRMT6 | 3 |
| 9 | 1q | 5.03 × 10−14 | 8 | 207,662 | 207,670 | – | MIR205HG∗ | 0 | – | – |
| 10 | 1q | 3.40 × 10−9 | 3 | 232,337 | 232,340 | – | SLC35F3∗ | 0 | – | – |
| 11 | 1q | 1.07 × 10−7 | 52 | 234,896 | 234,948 | – | ACTN2∗ | 3 | TBCE∗, B3GALNT2, ARID4B | 23 |
| 12 | 1q | 6.33 × 10−7 | 49 | 243,993 | 244,042 | – | SMYD3∗ | 0 | – | – |
| 13 | 2p | 4.57 × 10−7 | 95 | 12,139 | 12,044 | – | MIR3681HG∗ | 0 | – | – |
| 14 | 2p | 2.62 × 10−15 | 89 | 45,025 | 44,936 | – | SIX3∗, CAMKMT | 3 | PRKCE∗, DYNC2LI1, ABCG8 | 27 |
| 15 | 2q | 2.61 × 10−9 | 89 | 139,530 | 139,527 | – | – | 0 | – | – |
| 16 | 3p | 4.05 × 10−7 | 42 | 7,503 | 7,461 | – | GRM7∗ | 1 | RAD18 | 17 |
| 17 | 3p | 1.20 × 10−10 | 0 | 38,370 | 38,369 | NS | XYLB∗ | 0 | – | – |
| 18 | 3p | 5.10 × 10−11 | 95 | 41,078 | 41,173 | NS | CTNNB1 | 2 | CTNNB1∗, ZNF621 | 1 |
| 19 | 3p | 4.22 × 10−23 | 13 | 47,556 | 47,569 | NS | CSPG5 | 4 | IP6K2 (IHPK2)∗, KIF9, TESSP5, P4HTM | 3 |
| 20 | 3p | 8.39 × 10−6 | 20 | 54,986 | 54,965 | – | CACNA2D3∗ | 0 | – | – |
| 21 | 3p | 1.13 × 10−13 | 59 | 67,827 | 67,768 | – | SUCLG2 | 0 | – | – |
| 22 | 3p | 6.67 × 10−10 | 9 | 87,317 | 87,327 | NS | CHMP2B | 0 | – | – |
| 23 | 3q | 7.80 × 10−6 | 18 | 122,056 | 122,038 | – | BC032918 | 2 | CD86∗, ILDR1 | 4 |
| 24 | 3q | 5.39 × 10−8 | 49 | 184,743 | 184,694 | – | KLHL6∗ | 3 | AP2M1, ABCF3, MAGEF1 | 0 |
| 25 | 4q | 1.87 × 10−11 | 24 | 53,132 | 53,108 | NS | USP46∗+ | 1 | USP46 | 2 |
| 26 | 4q | 2.68 × 10−7 | 6 | 92,162 | 92,168 | – | CCSER1∗ | 2 | HSD17B13, PDHA2 | 0 |
| 27 | 4q | 5.24 × 10−13 | 56 | 96,319 | 96,376 | – | UNC5C∗ | 1 | PDHA2 | 2 |
| 28 | 4q | 4.02 × 10−13 | 3 | 102,095 | 102,098 | – | PPP3CA | 0 | – | – |
| 29 | 4q | 1.50 × 10−6 | 74 | 148,449 | 148,375 | >100 kb | – | 1 | EDNRA | 1 |
| 30 | 5q | 8.84 × 10−6 | 44 | 52,127 | 52,083 | – | PELO∗ | 1 | ITGA1 | 20 |
| 31 | 5q | 1.56 × 10−8 | 14 | 59,263 | 59,277 | – | PDE4D∗+ | 1 | PDE4D∗ | 38 |
| 32 | 5q | 2.81 × 10−6 | 77 | 97,282 | 97,359 | – | – | 0 | – | – |
| 33 | 6p | 1.00 × 10−7 | 86 | 40,514 | 40,429 | – | LRFN2∗, BC132805∗ | 2 | KCNK5, UNC5CL∗ | 0 |
| 34 | 6p | 1.30 × 10−15 | 72 | 44,790 | 44,718 | – | BX647715 | 1 | PTK7 | 4 |
| 35 | 6q | 3.04 × 10−7 | 5 | 168,842 | 168,837 | – | SMOC2 | 3 | MLLT4∗, CCR6, C6orf122 | 1 |
| 36 | 7p | 6.93 × 10−8 | 85 | 24,390 | 24,305 | – | NPY | 2 | CCDC126, OSBPL3 | 35 |
| 37 | 7p | 1.49 × 10−8 | 4 | 35,510 | 35,514 | – | HERPUD2 | 1 | AAA1 | 34 |
| 38 | 7p | 5.97 × 10−6 | 75 | 37,471 | 37,396 | – | ELMO1∗+ | 2 | ELMO1, GPR141 | 30 |
| 39 | 7q | 4.00 × 10−6 | 77 | 82,225 | 82,149 | – | PCLO∗ | 1 | HGF∗ | 26 |
| 40 | 7q | 4.78 × 10−12 | 62 | 132,516 | 132,454 | – | CHCHD3 | 0 | – | – |
| 41 | 7q | 1.30 × 10−9 | 73 | 133,716 | 133,789 | – | SLC35B4, AKR1B1∗ | 0 | – | – |
| 42 | 7q | 4.44 × 10−10 | 14 | 134,336 | 134,322 | – | AGBL3∗ | 1 | TMEM140∗ | 0 |
| 43 | 7q | 6.98 × 10−14 | 75 | 141,873 | 141,798 | – | TCRBV20S1∗, TCRB∗ | 4 | FAM131B∗, ZYX, OR2A25, OR2F1 | 0 |
| 44 | 8p | 4.81 × 10−8 | 93 | 14,339 | 14,246 | – | SGCZ∗ | 2 | C8orf79, TUSC3 | 11 |
| 45 | 8q | 7.00 × 10−8 | 87 | 70,416 | 70,503 | – | SULF1 | 1 | PRDM14 | 46 |
| 46 | 9q | 6.46 × 10−8 | 13 | 110,613 | 110,626 | NS | ACTL7B | 2 | EPB41L4B∗, KLF4 | 29 |
| 47 | 9q | 8.64 × 10−7 | 19 | 132,799 | 132,781 | – | FIBCD1∗ | 4 | ABL1∗, AIF1L, RAPGEF1∗PRDM12 | 23 |
| 48 | 10q | 5.13 × 10−10 | 11 | 44,467 | 44,456 | >100 kb | – | 1 | ZNF22∗ | 32 |
| 49 | 10q | 1.28 × 10−6 | 8 | 57,261 | 57,268 | – | – | 1 | CDC2 | 0 |
| 50 | 10q | 9.29 × 10−9 | 0 | 65,408 | 65,408 | NS | CR622643 | 0 | – | – |
| 51 | 10q | 8.37 × 10−11 | 66 | 77,817 | 77,883 | – | C10orf11∗+ | 2 | C10orf11, DUPD1 | 1 |
| 52 | 11p | 4.29 × 10−9 | 5 | 22,257 | 22,262 | – | ANO5∗ | 0 | – | – |
| 53 | 11p | 5.53 × 10−13 | 88 | 32,453 | 32,364 | – | WT1∗+ | 4 | PRRG4∗, FBXO3∗, ELP4, WT1 | 14 |
| 54 | 11q | 4.12 × 10−8 | 0 | 57,558 | 57,558 | NS | OR9Q1∗ | 1 | MPEG1 | 33 |
| 55 | 11q | 1.11 × 10−9 | 21 | 59,295 | 59,315 | – | STX3∗ | 0 | – | – |
| 56 | 12p | 1.40 × 10−7 | 31 | 25,404 | 25,373 | – | KRAS | 1 | SSPN | 0 |
| 57 | 12p | 8.75 × 10−7 | 6 | 29,568 | 29,574 | – | TMTC1∗ | 0 | – | – |
| 58 | 12q | 3.45 × 10−6 | 91 | 57,035 | 57,127 | NS | – | 2 | SLC26A10, MBD6 | 1 |
| 59 | 12q | 8.78 × 10−6 | 34 | 61,602 | 61,636 | – | PPM1H∗ | 0 | – | – |
| 60 | 12q | 2.45 × 10−6 | 62 | 98,425 | 98,363 | – | ANKS1B∗ | 0 | – | – |
| 61 | 12q | 1.45 × 10−6 | 22 | 104,931 | 104,909 | – | NUAK1 | 1 | MTERFD3 | 0 |
| 62 | 13q | 2.09 × 10−7 | 85 | 26,479 | 26,564 | – | USP12∗ | 0 | – | – |
| 63 | 13q | 8.19 × 10−6 | 40 | 47,133 | 47,173 | – | – | 0 | – | – |
| 64 | 13q | 7.16 × 10−14 | 13 | 65,393 | 65,380 | – | MIR548X2 | 0 | – | – |
| 65 | 13q | 1.10 × 10−14 | 82 | 66,885 | 66,803 | – | – | 0 | – | – |
| 66 | 13q | 5.74 × 10−7 | 44 | 101,207 | 101,251 | – | FGF14∗+ | 3 | TMTC4, PCCA, FGF14 | 0 |
| 67 | 15q | 1.13 × 10−6 | 16 | 45,938 | 45,954 | – | LINC01494 | 0 | – | – |
| 68 | 16p | 2.90 × 10−10 | 80 | 1,700 | 1,781 | – | MAPK8IP3, IGFALS | 7 | HAGH∗, PGP∗, TPSAB1∗, FAHD1, GFER, GNPTG, PRR25 | 0 |
| 69 | 16p | 2.24 × 10−6 | 32 | 12,605 | 12,636 | – | SNX29 | 3 | CLEC16A∗, SOCS1, LITAF | 10 |
| 70 | 16q | 1.75 × 10−26 | 4 | 70,999 | 70,995 | >100 kb | AK055364 | 1 | HYDIN | 2 |
| 71 | 16q | 1.03 × 10−11 | 20 | 77,036 | 77,016 | – | WWOX∗ | 0 | – | – |
| 72 | 16q | 5.52 × 10−19 | 29 | 78,435 | 78,406 | – | LOC101928248 | 2 | MAF∗, WWOX | 11 |
| 73 | 17q | 4.15 × 10−7 | 47 | 34,640 | 34,593 | >100 kb | STAC2, CACNB1∗ | 4 | PLXDC1, CRKRS, ZPBP2, KRT222 | 6 |
| 74 | 18q | 9.58 × 10−10 | 0 | 36,270 | 36,271 | – | – | 0 | – | – |
| 75 | 18q | 6.84 × 10−6 | 72 | 74,880 | 74,809 | – | SALL3 | 0 | – | – |
| 76 | 21q | 3.09 × 10−9 | 30 | 20,521 | 20,490 | – | DYRK1A | 0 | – | – |
| 77 | 21q | 9.56 × 10−8 | 61 | 27,738 | 27,677 | – | MIR5009 | 0 | – | – |
| 78 | 21q | 1.35 × 10−10 | 39 | 34,148 | 34,109 | – | ITSN∗ | 2 | C21orf66, TCP10L | 0 |
| 79 | 21q | 1.53 × 10−6 | 1 | 37,635 | 37,633 | – | DYRK1A | 1 | KCNJ6 | 0 |
| 80 | 22q | 7.29 × 10−9 | 0 | 31,376 | 31,376 | – | SYN3∗ | 1 | PISD | 12 |
| 81 | 2p | 6.46 × 10−6 | 7 | NS | 21,294 | 21,301 | – | 1 | SDC1 | 33 |
| 82 | 2p | 5.02 × 10−9 | 0 | >100 kb | 26,774 | 26,774 | KCNK3 | 6 | OTOF∗, GAREM2, CCDC121, ASXL2, DNMT3A, CENPA | 3 |
| 83 | 2q | 1.02 × 10−11 | 1 | >100 kb | 135,313 | 135,313 | ACMSD∗ | 0 | – | – |
| 84 | 2q | 2.25 × 10−11 | 0 | NS | 203,732 | 203,732 | NBEAL1 | 0 | – | – |
| 85 | 4q | 2.77 × 10−9 | 8 | >100 kb | 103,404 | 103,395 | SLC39A8∗+ | 1 | SLC39A8 | 0 |
| 86 | 6p | 2.09 × 10−13 | 60 | NS | 28,525 | 28,586 | ZSCAN23, PX6 | 8 | ZKSCAN3, PGBD1∗, MAS1L, OR2W1, OR11A1, HLA-F, IFITM4P, ZNF184 | 6 |
| 87 | 8p | 1.54 × 10−18 | 44 | >100 kb | 18,194 | 18,238 | NAT1, NAT2 | 3 | PDGFRL∗, CSGALNACT1, PCM1 | 1 |
| 88 | 11p | 1.86 × 10−20 | 86 | >100 kb | 8,508 | 8,594 | STK33∗, TRIM66∗ | 0 | – | – |
| 89 | 12q | 4.10 × 10−16 | 44 | >100 kb | 48,574 | 48,618 | FAIM2∗+ | 1 | FAIM2 | 10 |
| 90 | 15q | 1.93 × 10−7 | 77 | NS | 61,133 | 61,210 | TPM1∗+, LACTB∗+ | 9 | TPM1, RPS27L∗, DAPK2∗, SNX1, LACTB, APH1B, HERC1, FAM96A, VPS13C | 1 |
| 91 | 17q | 2.02 × 10−6 | 86 | >100 kb | 44,633 | 44,548 | GNGT2∗+, B4GALNT2 | 3 | GNGT2, SAMD14∗, HOXB3∗ | 19 |
| 92 | 17q | 3.95 × 10−8 | 85 | >100 kb | 60,951 | 60,866 | AXIN2∗+ | 1 | AXIN2 | 38 |
| 93 | 20p | 1.90 × 10−19 | 31 | >100 kb | 25,718 | 25,687 | FAM182B∗ | 0 | – | – |
T2D-associated intervals in kb (<100) that harbor T2D susceptibility loci in both populations; the minimum distance is provided for signals 1–5
Location estimates for the European (E) GWASS
Location estimates for the African American (A) GWASS
Location estimates for the Metaboship European (E) samples; signals with low SNP coverage (indicated by “–”) were not meta-analyzed
Genes with asterisk (∗) denote the intragenic localization and genes with plus sign (+) indicate self-regulatory
Number of cis-genes regulated by the eQTL
List of cis-genes associated with eQTLs that co-located within <50 kb of the T2D locations; cis-genes with asterisk (∗) have previously shown evidence of association between body mass index in morbidly obese and adipose/liver expression profiles20
Distance in kb (<50) between eQTL and T2D locations; the minimum is given when more than one cis-gene is implicated
Table 2.
Identified European-Specific T2D Susceptibility Loci and Their Regulatory Role of Neighboring Gene Expression
| Signal | Chr. | Meta p Value | Distance of T2D Locationsa | T2D Location GWAS-Eb | T2D Location GWAS-Ac | T2D Location metabo-Ed | Nearest Gene to T2D Locationse | No. of cis-Genesf | eQTL Associated cis-Genesg | eQTL Distance from T2Dh |
|---|---|---|---|---|---|---|---|---|---|---|
| 94 | 1p | 2.02 × 10−7 | 3 | 25,879 | NS | 25,876 | MAN1C1∗ | 2 | DHDDS, C1orf172 | 0 |
| 95 | 1q | 6.94 × 10−12 | 21 | 228,322 | NS | 228,344 | GALNT2∗ | 0 | – | – |
| 96 | 2p | 4.11 × 10−34 | 34 | 605 | NS | 639 | TMEM18 | 0 | – | – |
| 97 | 2p | 1.18 × 10−7 | 46 | 40,320 | >100 kb | 40,274 | SLC8A1-AS1∗ | 4 | THUMPD2, TMEM178, MORN2, DHX57 | 0 |
| 98 | 3p | 1.03 × 10−9 | 5 | 53,518 | NS | 53,513 | CACNA1D∗ | 0 | – | – |
| 99 | 3q | 9.35 × 10−7 | 7 | 133,919 | NS | 133,912 | ACAD11∗+ | 8 | ACAD11, SLC02A1, RYK, NPHP3, ACKR4, SRPRB, CDV3, RAB6B | 0 |
| 100 | 4q | 1.86 × 10−22 | 66 | 104,224 | >100 kb | 104,157 | BDH2, NHEDC1 | 0 | – | – |
| 101 | 5q | 2.62 × 10−9 | 12 | 76,449 | NS | 76,461 | ZBED3 | 1 | PDE8B∗ | 15 |
| 102 | 6p | 2.64 × 10−24 | 13 | 29,662 | NS | 29,674 | GABBR1 | 7 | GNL1∗, TRIM10, TRIM27, DDR1, TRIM40, TRIM15, OR10C1 | 9 |
| 103 | 6p | 1.46 × 10−28 | 6 | 31,709 | >100 kb | 31,704 | BAT2, AIF1 | 14 | AIF1∗, TRIM39∗, AGER, BAT4, CCHCR1, DOM3Z, EHMT2, HLA-DMB, HLA-DPA1, HSPA1B, LST1, TRIM10, NOTCH4, HLA-DRA | 0 |
| 104 | 6q | 6.29 × 10−56 | 0 | 118,801 | >100 kb | 118,801 | SLC35F1 | 0 | – | – |
| 105 | 6q | 4.23 × 10−17 | 37 | 127,581 | NS | 127,544 | RSPO3∗+ | 2 | RSPO3, TRMT11 | 1 |
| 106 | 10q | 3.39 × 10−24 | 40 | 104,790 | >100 kb | 104,830 | CNNM2∗ | 0 | – | – |
| 107 | 11p | 2.50 × 10−6 | 1 | 43,836 | NS | 43,836 | HSD17B12 | 1 | HSD17B12 | 1 |
| 108 | 11q | 6.39 × 10−6 | 21 | 65,337 | >100 kb | 65,357 | OVOL1, SNX32∗+ | 4 | SNX32, MAJIN∗, SNX15, EFEMP2 | 9 |
| 109 | 12q | 6.16 × 10−6 | 4 | 54,905 | >100 kb | 54,909 | OBFC2B∗+ | 4 | OBFC2B, RGL1∗, RPS26, STAT2 | 0 |
| 110 | 12q | 4.66 × 10−8 | 3 | 111,476 | >100 kb | 111,473 | PTPN11 | 4 | ERP29, TMEM116, MAPKAPK5, NAP1 | 20 |
| 111 | 17q | 3.05 × 10−9 | 51 | 25,003 | >100 kb | 25,054 | SSH2∗ | 1 | CORO6 | 9 |
T2D associated intervals in kb (<100) that harbor T2D susceptibility loci in two European populations
T2D location estimates for the European (E) GWAS
The African American (A) GWAS yielded either significant but distant locations from the European T2D location (>100 kb) or not significant (NS) estimates
Location estimates for the Metaboship European (E) samples that were within <100 kb of the GWAS-E location
Genes with asterisk (∗) denote the intragenic localization and genes with plus sign (+) indicate self-regulatory genes
Number of cis-genes regulated by the eQTLs
List of cis-genes associated with eQTLs that co-located within <50 kb of the T2D locus, cis-genes with asterisk (∗) have previously shown evidence of association between body mass index for morbidly obese and adipose/liver expression profiles20
Distance in kb (<50) between eQTL and T2D locations, the minimum is given when more than one cis-gene is implicated
For the 111 additional T2D loci, half of the location estimates are intragenic and half are intergenic. For the latter, we follow the convention of labeling the disease loci using the nearest gene symbol (within 100 kb from the T2D location). The in silico expression analyses, however, indicate which cis-genes are regulated and therefore functionally implicated by the identified T2D loci. Two-thirds (71/111) of the disease loci also show strong evidence of being eQTLs using our stringent criteria but the remaining one-third may well reflect that these replicated loci could be eQTLs for a T2D-relevant tissue other than subcutaneous adipose. The 71 eQTL signals regulate the expression of a conservatively estimated total of 183 cis-genes (Tables 1 and 2), the majority of which are not genes that are the nearest to the disease locus. Indeed, further investigation of the 183 cis-genes substantiates quantitatively what has previously been suspected: namely, that the physical kb distance of the eQTL to the nearest gene (Figure 1) is entirely unrelated (p > 0.05) to the distance between the same eQTL and the actual (cis-associated) functional gene (see Figure S1). This result demonstrates that the assumption that the nearest gene is also the most likely candidate functional gene is not justified. Further analysis of the 183 cis-genes also showed that the distance between the eQTL and the T2D location estimates is not biased by the distance between the T2D sample location estimates within the <100 kb interval (see Figure S2).
Interestingly, approximately 40% of the eQTL signals observed in this study have at least one cis-gene whose expression (in adipose or liver) is also strongly associated with BMI in morbidly obese individuals.20 We also show that a number of the identified T2D loci also regulate the expression levels of nuclear-encoded mitochondrial genes. For example, many cis-genes implicate T2D through the regulation of the molecular functions of fatty acid metabolism (PCCA [MIM: 232000], ACAD11 [MIM: 614288], signals 66 and 99, respectively), glycerophospholipid metabolism (PISD [MIM: 612770], GPAM [MIM: 602395], signals 80 and 117), pyruvate metabolism (PDHA2 [MIM: 179061], HAGH [MIM: 138760], signals 26/27 and 68), mitochondrial transcription and translation (MTERFD3 [MIM: 616929], LACTB [MIM: 608440], TRMT11 [MIM: 609752], signals 61, 90, 105), and mitochondrial protein transport (GFER [MIM: 600924], signal 68). The cis-genes PDHA2 and PCCA (signals 26/27 and 66, respectively) directly implicate the genetic dysregulation of Krebs cycle function as a risk factor for T2D.
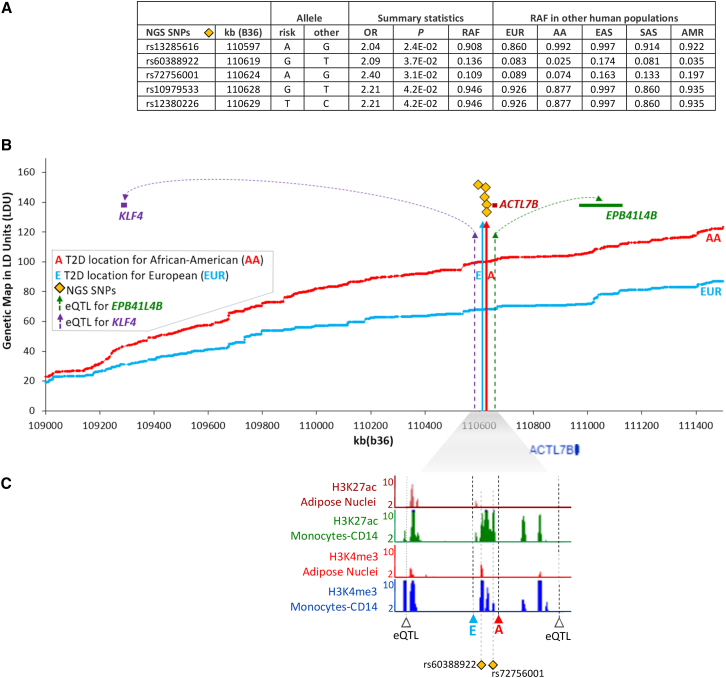
To further characterize the observed T2D and associated eQTL location estimates in relation to potential functional variants, three of the cosmopolitan loci were targeted for sequencing using an independent sample of Europeans. Figure 2 presents an example of a regulatory intergenic hotspot near ACTL7B (MIM: 604304) on chromosome 9q31.3 (signal 46, Table 1). The two genetic maps (y axis in LDU) for AA and EUR are plotted against the physical genomic region (x axis in kb), with each data point representing a HapMap SNP from which the genetic LDU maps were inferred (Figure 2B). Cumulative LDU plots the non-linear relationship between physical distance and the underlying LD, which is typically a “Block-Step” structure. Blocks of LD (SNPs with the same LDU location) represent areas of conserved LD and low haplotype diversity, and steps (increasing LDU distances) define LD breakdown, primarily caused by recombination, since crossover profiles agree precisely with the corresponding LDU steps.16 The maps show numerous short blocks across the entire region with total LDU length being greater for AA (greater LD breakdown) than the EUR population. The functional location estimates A and E (13 kb apart) indicated by the vertical solid line arrows are associated with T2D in AA and EUR samples, respectively. The dotted lines, in close proximity to the T2D locations (<30 kb), represent the location of eQTLs that regulate the expression of KLF4 (MIM: 602253) and EPB41L4B (MIM: 610340) in subcutaneous adipose (nine genes reside between the two illustrated, but for clarity only the cis-genes regulated by the T2D-associated eQTL are plotted). KLF4 and EPB41L4B are 1.3 Mb downstream and 350 kb upstream, respectively. Targeted next-generation re-sequencing of the 39 kb region centered on these A and E locations shows evidence of association between T2D and variants, which coincide with the estimates for the T2D-associated eQTL (summary statistics of the NGS SNPs are provided in Figure 2A). Although only nominally significant (p < 0.05) due to small sample size (94 case subjects and 94 control subjects), these common variants both confirm the expected location estimate and account for a relatively large risk of disease (odds ratio [OR] of 2.0–2.4), with the risk allele frequencies (RAF) being similar in a number of human populations from the 1000 Genomes Project. Examination of the epigenetic chromatin marks from trimethylation of histone H3 at lysine 4 (H3K4me3) and acetylation of histone H3 at lysine 27 (H3K27ac), which highlight regulatory elements such as active promoters and enhancers,21 has previously been shown to overlap with T2D loci,22 but such marks are often cell type specific.23 Figure 2C plots the −log10 p values of the chromatin profiles, demonstrating that T2D causal locations also co-localize with chromatin domains for CD14+ monocytes and adipose nuclei. The most intense chromatin peaks were observed in CD14+ monocytes at the precise eQTL location for KLF4 and at rs60388922 and rs72756001 SNPs, which reside within the E and A T2D interval. Hence both of these SNPs are good causal candidate variants.
Figure 2.
Candidate Causal Variants at T2D Locations in the ACTL7B Region and Their Regulatory Role
(A) Identified T2D-associated SNPs using targeted next generation sequencing (NGS) of the functional location estimates on the LDU genetic maps and their risk allele frequency (RAF) in Europeans (EUR), African Americans (AA), East Asian (EAS), South Asian (SAS), and Mexican Americans (AMR).
(B) The two LDU genetic maps (y axis in LDU) for AA and EUR are plotted against the physical genomic region (x axis in kb). Vertical solid arrows represent the functional location estimates A and E that are associated with T2D in AA and EUR samples, respectively. The dotted line arrows are the locations of eQTLs.
(C) −log10 p values of cell type-specific chromatin profiles are plotted against the kb map. T2D, eQTL, and NGS SNP locations overlap with the co-ordinates of the chromatin peaks.
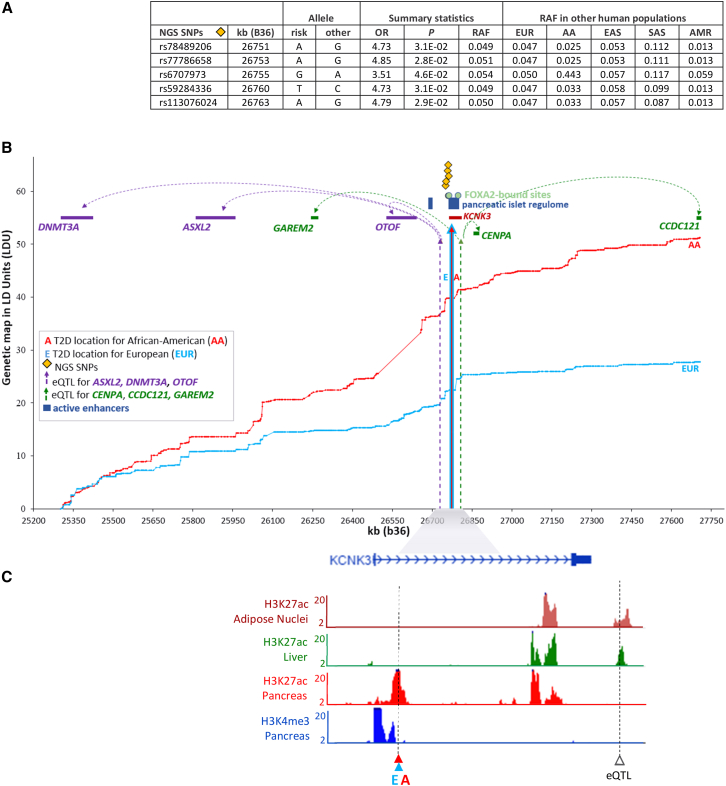
Figure 3 presents the regulatory potassium channel, subfamily k, member 3 (KCNK3 [MIM: 603220]) locus (signal 82, Table 1) on chromosome 2p23.3 with identical location estimates for both AA and EUR samples (Figure 3B). Despite the disease locus residing within KCNK3, expression analyses indicate that this locus may not be functionally related to this gene, but instead is a cis-eQTL that regulates the distant genes DNMT3A (MIM: 602769), ASXL2 (MIM: 612991), GAREM2, OTOF (MIM: 603681), CENPA (MIM: 117139), and CCDC121, with DNMT3A and CCDC121 being 1.5 Mb and 896 kb away from the eQTL, respectively. This is a gene-rich region (57 genes in total), but for clarity only the six identified cis-genes have been plotted. Figure 3A presents summary statistics for the variants associated with T2D for a 42 kb targeted re-sequence region. These associated variants (p value < 0.05) coincide with the promoter region of KCNK3, 11 kb upstream of the T2D location estimate and account for a high risk of disease (OR = 3.5–4.8). The RAF are approximately 0.05 and the results show that these variants are indeed cosmopolitan, since they are common not only in EUR and AA but also in other human populations (e.g., East and South Asians and Mexican Americans). Using information from the human pancreatic islet regulome,22 where sequences targeted by islet transcription factors highlight active enhancers, we observed that the identified T2D location resides within a cluster of such active enhancers. The transcription factor FOXA2-bound sites are also plotted within the kb boundaries of the active enhancer cluster. Chromatin peaks also overlap with the regulatory T2D and eQTL locations for pancreas and liver cell types (Figure 3C), suggesting evidence for more than one functional mutation within this region. The full interval for the T2D and eQTL locations were not entirely covered by NGS data, but nevertheless, the associated NGS SNPs reside between two active islet enhancers (Figure 3B). In contrast to imputation methods that use high-resolution “out-of-sample” marker panels to infer missing SNPs to subsequently test one at a time, LDU analysis uses marker panels to infer high-resolution genetic maps. Subsequently, multi-marker tests of association use genetic distances for SNP arrays placed on those maps to infer the location of disease-associated functional variants. Fine mapping is therefore achieved by inferring fine-scale genetic maps, not by imputing SNPs. It is worth noting in relation to this that using the KCNK3 locus as an example, the NGS SNPs genotyped for the case control data that were associated with T2D status (Figure 3A) could not be imputed based on the 1000G data as the reference panel.
Figure 3.
Candidate Causal Variants at T2D Locations in the KCNK3 Region and Their Regulatory Role
(A) Identified T2D-associated SNPs using targeted next generation sequencing (NGS) of the functional location estimates on the LDU genetic maps and their risk allele frequency (RAF) in Europeans (EUR), African Americans (AA), East Asian (EAS), South Asian (SAS), and Mexican Americans (AMR).
(B) The two LDU genetic maps (y axis in LDU) for AA and EUR are plotted against the physical genomic region (x axis in kb). Vertical solid arrows represent the functional location estimates A and E that are associated with T2D in AA and EUR samples, respectively. The dotted arrows are the locations of eQTLs.
(C) −log10 p values of cell type-specific chromatin profiles are plotted against the kb map. T2D and eQTL locations overlap with the co-ordinates of the chromatin peaks.
Findings at Previously Known T2D Loci
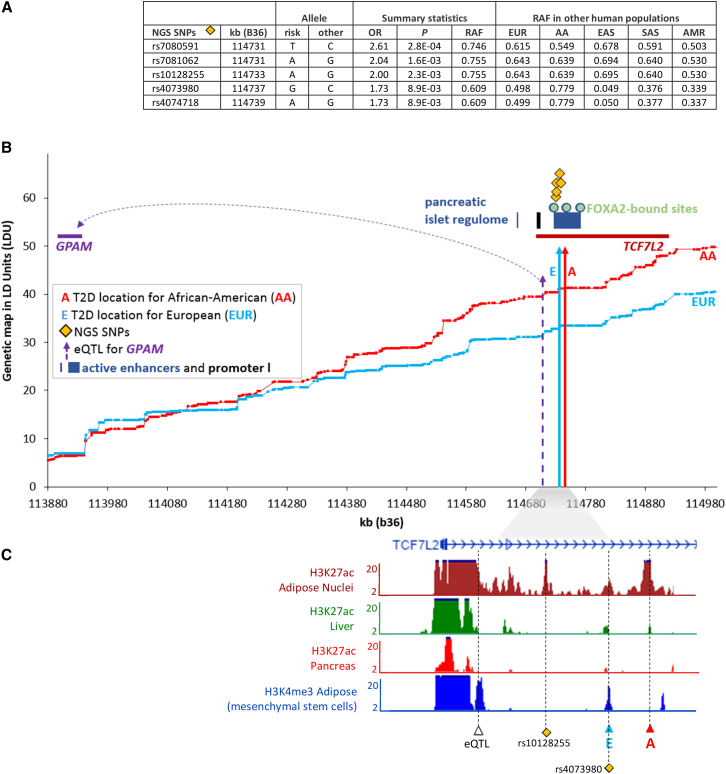
Using the same data and analytical methods, we have confirmed disease location estimates for 62 out of 76 previously known loci3 (Table S1). Of these, about half (33/62) show evidence of being eQTL with the majority regulating cis-genes more than 1 Mb away. In addition, more than one-third (22/62) of the loci replicate for AA samples, which were previously excluded from trans-ethnic meta-analysis.3 We investigated the Transcription factor 7-like 2 (TCF7L2 [MIM: 602228]) locus (signal 117, Table S1). Figure 4B plots the T2D locations for EUR and AA and shows that this signal harbors a cis-eQTL for the distant GPAM. The T2D re-sequence variants we identified, which co-locate to the <30 kb interval between the T2D and eQTL locations, account for a large risk of disease (OR = 1.7–2.6). The summary statistics in Figure 4A show that these risk variants are the major allele in all other human populations. The T2D locations for both populations reside within an active enhancer cluster that is targeted by transcription factor FOXA2 (kb locations of the regulatory elements are plotted on the x axis). Inferring the likely transcriptional activity by observing the chromatin state, we show that the eQTL, T2D co-locations, and NGS SNPs all map precisely to highly significant H3K4me3 and H3K27ac peaks, in particular in adipose cells (Figure 4C). This illustrates the importance of co-locating within an interval on the genetic map, since it allows for potential allelic heterogeneity.
Figure 4.
Candidate Causal Variants at T2D Locations in the TCF7L2 Region and Their Regulatory Role
(A) Identified T2D-associated SNPs using targeted next generation sequencing (NGS) of the functional location estimates on the LDU genetic maps and their risk allele frequency (RAF) in Europeans (EUR), African Americans (AA), East Asian (EAS), South Asian (SAS), and Mexican Americans (AMR).
(B) The two LDU genetic maps (y axis in LDU) for AA and EUR are plotted against the physical genomic region (x axis in kb). Vertical solid arrows represent the functional location estimates A and E that are associated with T2D in AA and EUR samples, respectively. The dotted arrow is the location of eQTL.
(C) −log10 p values of cell type-specific chromatin profiles are plotted against the kb map. T2D, eQTL, and NGS SNP locations overlap with the co-ordinates of the chromatin peaks.
Discussion
This study provides a comprehensive genomic catalog of susceptibility loci for T2D in European and African ancestry populations and evidence that the majority of the additional 111 and 62 previously known disease loci are eQTLs for 183 and 83 cis-genes, respectively. This implies that these disease loci confer risk of T2D via the cis-regulation of the expression levels in tissue relevant to T2D for a large number (266 in total) of neighboring genes. This study identifies a large number of disease loci at regulatory hotspots and replicates them in both European and African American populations, with 84% (93/111) of the additional loci being cosmopolitan. This replication was made possible by analyses that make use of, rather than being confounded by, the fine-scale differences in LD between these two populations, where causal locations and also eQTLs are estimated on an LDU map, avoiding the ambivalence of interpreting individual GWAS SNPs. Interestingly, recent results in the literature support our conclusion that cosmopolitan loci are more widespread than previously thought. For example, it was not until recently that the most established bona fide TCF7L2 locus for T2D in Europeans was also confirmed in African ancestry groups in an extensive study that included 17 African American GWA samples.7 This is a locus that was identified in the present study, which included only one African American sample.
The T2D-associated common genetic variation in TCF7L2 is well established, but the mechanism by which risk of disease is conferred remains elusive. Our refined localization of this locus reveals that this is a regulatory site for the distant nuclear-encoded mitochondrial gene, GPAM, and identifies additional candidate causal variants. GPAM is interesting as it is a key rate-limiting enzyme in lipogenesis and highly expressed in liver and adipose tissue. Nuclear-encoded mitochondrial genes are of particular interest in relation to T2D, since mitochondrial function has been demonstrated to impact upon a myriad of molecular and cellular functional processes implicated in T2D.24, 25, 26 However, to date human genetic association studies have identified few, if any, nuclear-encoded mitochondrial genes that directly confer risk of T2D in its common form. In this study we have identified at least 11 further nuclear-encoded mitochondria genes, which are regulated by eQTLs that also appear to confer risk of T2D. This indicates that one of the molecular mechanisms that contribute to inherited risk of T2D is mitochondrial dysfunction in relation to energy metabolism. These findings are also supported by an independent study design that utilizes transcriptome and proteome data to reconstruct metabolic pathways in myocytes and identify the same mitochondrial pathways implicated in our study for adipose tissue (namely, fatty acid beta-oxidation, Krebs cycle, pyruvate metabolism, and branched-chain amino acid [valine, leucine, and isoleucine] metabolism).27
Recent large-scale whole-genome and -exome association studies28 have empirically questioned any major role for coding variants in the etiology of T2D and therefore make regulatory loci such as the ones highlighted in this study all the more important to investigate. We believe that the targeted re-sequencing of informative refined regions using case-control data has the power to dissect the genetic epidemiology of T2D. While imputation methods can successfully infer missing genotypes for most genomic regions using population data as the reference data, ironically it appears that imputation methods are likely to fail to impute or to make correct inferences where it matters most (e.g., at disease loci with allelic spectra that differ from the general population). This important point requires further investigation.
Chromatin analyses and targeted re-sequencing of these refined regions can be used to identify potential causal variant locations. The two identified signals (ACTL7B and KCNK3) that we have investigated lends support to this approach. A cosmopolitan disease location interval that includes KCNK3 was observed to be a hotspot for the regulation of six neighboring genes, with OTOF of particular interest because of its known association with hearing loss (autosomal-recessive forms of deafness29). According to the NIDDK, the prevalence of low- or mid-frequency hearing impairment among diabetics is three times that of non-diabetics (28% compared with 9%)30 with the potential mechanism operating via microvascular and neural damage due to long-term hyperglycemia.31 Interestingly, adipose gene expression for OTOF has also been previously shown to be associated with BMI in the morbidly obese,29 providing further evidence of a functional role through a regulatory mechanism. Targeted re-sequencing within the KCNK3 location interval identified candidate causal variants with large effect sizes. As with the TCF7L2 locus, the T2D and eQTL locations were found to reside in pancreatic islet enhancer and FOXA2 transcription factor-binding sites. Studies have shown that dysregulation of islet enhancers is relevant to the underlying mechanisms of T2D22 and a recent examination of some of the previously found T2D loci have been found to overlap with FOXA2-bound sites.32
The cosmopolitan T2D location interval near ACTL7B overlapped with chromatin peaks for CD14+ monocytes and included an eQTL for KLF4 and EPB41L4B. KLF4 is highly expressed in CD14+ monocytes and belongs to the Krüppel-like factor (KLF) family that consists of transcription factors that can activate or repress different genes involved in processes such as differentiation, development, and cell cycle progression, with several of these proteins implicated in glucose homeostasis.33 Klf4 is also used experimentally to induce pluripotent cells that can differentiate into insulin-producing cells.34 EPB41L4B codes for an erythrocyte membrane protein (EMP) with greatly increased EMP glycosylation observed in T2D-affected subjects35 with likely clinical implications.36 It is recognized clinically that both obesity and T2D are associated with a state of abnormal inflammatory response. Here we show that the T2D variants and eQTL locations for KLF4 and EPB41L4B reside in regions with chromatin modifications mainly observed in CD14+ monocytes. Monocytes play a pivotal role in innate immunity and are involved in metabolic regulation.37 It has been shown that unbalanced proinflammatory/anti-inflammatory markers of CD14+ cells is associated with metabolic disorder in obese T2D-affected individuals.38 KLF4 is a critical regulator of monocyte differentiation39 and EPB41L4B expression in subcutaneous and omental adipose is strongly associated with BMI in morbidly obese individuals.20 Therefore, these T2D intragenic variants and the regulated cis-genes (KLF4 and EPB41L4B) are likely to be involved in an inflammatory pathway for obesity and T2D.
The complex causal chain between a gene and its effect on susceptibility cannot be unravelled until we have a full understanding of the regulatory genetic architecture that underpins T2D and until the causal changes have been localized in the DNA sequence.40 Our results show that disease-associated loci in different populations, gene expression, and cell-specific regulatory annotation can be effectively integrated by localizing these effects on high-resolution linkage disequilibrium maps. By exploiting these maps to refine causal location estimates, we have identified a genomic catalog of cosmopolitan and European disease loci with correspondingly important clinical implications that provides important molecular insights and opportunities to understand the molecular basis of this devastating common disease.
Acknowledgments
We would like to thank the WTCCC, UK, for making the WTCCC T2D genomic data available. A full list of the investigators who contributed to the generation of the data is available from http://www.wtccc.org.uk. We are grateful to the NIDDK for making the AA T2D phenotype and genomic data available to us. The NIDDK whole-genome association search for T2D genes in African Americans was conducted by Donald Bowden, Center for Human Genomics, Center for Diabetes Research, Wake Forest University School of Medicine, with support from the NIDDK. The datasets used were obtained from the database of Genotypes and Phenotypes (dbGaP) at accession number phs000140. This manuscript was not prepared in collaboration with the labs of any of the investigators responsible for generating the data, and does not necessarily reflect the views or opinions of these investigators. T.A. would like to acknowledge the Medical Research Council UK (Investigator Award 91993) for supporting his work. All authors are grateful to Professor Dallas Swallow (UCL) for her valuable comments on the manuscript, Professor Philippe Froguel (Imperial College, CNRS 8199, EGID 59045) for generously supplying us with European DNA samples for the NGS pilot work, and Aminah Ali (UCL) for her valuable contributions to processing the NGS data. N.M. would like to acknowledge Newton E. Morton (University of Southampton) for the previous body of work on LDU maps.
Published: May 4, 2017
Footnotes
Supplemental Data include two figures, two tables, and Supplemental Methods and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.04.007.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
International HapMap Project, ftp://ftp.ncbi.nlm.nih.gov/hapmap/
Islet Regulome Browser, http://gattaca.imppc.org/isletregulome/home
OMIM, http://www.omim.org/
Roadmap, http://www.roadmapepigenomics.org/
Supplemental Data
References
- 1.Altshuler D., Daly M. Guilt beyond a reasonable doubt. Nat. Genet. 2007;39:813–815. doi: 10.1038/ng0707-813. [DOI] [PubMed] [Google Scholar]
- 2.Lebovitz H.E. Type 2 diabetes: an overview. Clin. Chem. 1999;45:1339–1345. [PubMed] [Google Scholar]
- 3.Mahajan A., Go M.J., Zhang W., Below J.E., Gaulton K.J., Ferreira T., Horikoshi M., Johnson A.D., Ng M.C., Prokopenko I., DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium. South Asian Type 2 Diabetes (SAT2D) Consortium. Mexican American Type 2 Diabetes (MAT2D) Consortium. Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segrè A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A., Wellcome Trust Case Control Consortium. Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators. Genetic Investigation of ANthropometric Traits (GIANT) Consortium. Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium. South Asian Type 2 Diabetes (SAT2D) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y.R., Keating B.J. Trans-ethnic genome-wide association studies: advantages and challenges of mapping in diverse populations. Genome Med. 2014;6:91. doi: 10.1186/s13073-014-0091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asimit J.L., Hatzikotoulas K., McCarthy M., Morris A.P., Zeggini E. Trans-ethnic study design approaches for fine-mapping. Eur. J. Hum. Genet. 2016;24:1330–1336. doi: 10.1038/ejhg.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng M.C., Shriner D., Chen B.H., Li J., Chen W.M., Guo X., Liu J., Bielinski S.J., Yanek L.R., Nalls M.A., FIND Consortium. eMERGE Consortium. DIAGRAM Consortium. MuTHER Consortium. MEta-analysis of type 2 DIabetes in African Americans Consortium Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLoS Genet. 2014;10:e1004517. doi: 10.1371/journal.pgen.1004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Direk K., Lau W., Small K.S., Maniatis N., Andrew T. ABCC5 transporter is a novel type 2 diabetes susceptibility gene in European and African American populations. Ann. Hum. Genet. 2014;78:333–344. doi: 10.1111/ahg.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elding H., Lau W., Swallow D.M., Maniatis N. Refinement in localization and identification of gene regions associated with Crohn disease. Am. J. Hum. Genet. 2013;92:107–113. doi: 10.1016/j.ajhg.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X., Bailey S.D., Lupien M. Laying a solid foundation for Manhattan--‘setting the functional basis for the post-GWAS era’. Trends Genet. 2014;30:140–149. doi: 10.1016/j.tig.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voight B.F., Kang H.M., Ding J., Palmer C.D., Sidore C., Chines P.S., Burtt N.P., Fuchsberger C., Li Y., Erdmann J. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer N.D., McDonough C.W., Hicks P.J., Roh B.H., Wing M.R., An S.S., Hester J.M., Cooke J.N., Bostrom M.A., Rudock M.E., DIAGRAM Consortium. MAGIC Investigators A genome-wide association search for type 2 diabetes genes in African Americans. PLoS ONE. 2012;7:e29202. doi: 10.1371/journal.pone.0029202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maniatis N., Collins A., Morton N.E. Effects of single SNPs, haplotypes, and whole-genome LD maps on accuracy of association mapping. Genet. Epidemiol. 2007;31:179–188. doi: 10.1002/gepi.20199. [DOI] [PubMed] [Google Scholar]
- 15.Maniatis N., Collins A., Gibson J., Zhang W., Tapper W., Morton N.E. Positional cloning by linkage disequilibrium. Am. J. Hum. Genet. 2004;74:846–855. doi: 10.1086/383589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maniatis N., Collins A., Xu C.F., McCarthy L.C., Hewett D.R., Tapper W., Ennis S., Ke X., Morton N.E. The first linkage disequilibrium (LD) maps: delineation of hot and cold blocks by diplotype analysis. Proc. Natl. Acad. Sci. USA. 2002;99:2228–2233. doi: 10.1073/pnas.042680999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grundberg E., Small K.S., Hedman A.K., Nica A.C., Buil A., Keildson S., Bell J.T., Yang T.P., Meduri E., Barrett A., Multiple Tissue Human Expression Resource (MuTHER) Consortium Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vionnet N., Hani E.H., Dupont S., Gallina S., Francke S., Dotte S., De Matos F., Durand E., Leprêtre F., Lecoeur C. Genomewide search for type 2 diabetes-susceptibility genes in French whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2-diabetes locus on chromosome 1q21-q24. Am. J. Hum. Genet. 2000;67:1470–1480. doi: 10.1086/316887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyre D., Lecoeur C., Delplanque J., Francke S., Vatin V., Durand E., Weill J., Dina C., Froguel P. A genome-wide scan for childhood obesity-associated traits in French families shows significant linkage on chromosome 6q22.31-q23.2. Diabetes. 2004;53:803–811. doi: 10.2337/diabetes.53.3.803. [DOI] [PubMed] [Google Scholar]
- 20.Greenawalt D.M., Dobrin R., Chudin E., Hatoum I.J., Suver C., Beaulaurier J., Zhang B., Castro V., Zhu J., Sieberts S.K. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 2011;21:1008–1016. doi: 10.1101/gr.112821.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Consortium E.P., ENCODE Project Consortium A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasquali L., Gaulton K.J., Rodríguez-Seguí S.A., Mularoni L., Miguel-Escalada I., Akerman I., Tena J.J., Morán I., Gómez-Marín C., van de Bunt M. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet. 2014;46:136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trynka G., Sandor C., Han B., Xu H., Stranger B.E., Liu X.S., Raychaudhuri S. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 2013;45:124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowell B.B., Shulman G.I. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- 25.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 26.Patti M.E., Corvera S. The role of mitochondria in the pathogenesis of type 2 diabetes. Endocr. Rev. 2010;31:364–395. doi: 10.1210/er.2009-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Väremo L., Scheele C., Broholm C., Mardinoglu A., Kampf C., Asplund A., Nookaew I., Uhlén M., Pedersen B.K., Nielsen J. Proteome- and transcriptome-driven reconstruction of the human myocyte metabolic network and its use for identification of markers for diabetes. Cell Rep. 2015;11:921–933. doi: 10.1016/j.celrep.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 28.Fuchsberger C., Flannick J., Teslovich T.M., Mahajan A., Agarwala V., Gaulton K.J., Ma C., Fontanillas P., Moutsianas L., McCarthy D.J. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duman D., Tekin M. Autosomal recessive nonsyndromic deafness genes: a review. Front. Biosci. (Landmark Ed.) 2012;17:2213–2236. doi: 10.2741/4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bainbridge K.E., Hoffman H.J., Cowie C.C. Diabetes and hearing impairment in the United States: audiometric evidence from the National Health and Nutrition Examination Survey, 1999 to 2004. Ann. Intern. Med. 2008;149:1–10. doi: 10.7326/0003-4819-149-1-200807010-00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bainbridge K.E., Cheng Y.J., Cowie C.C. Potential mediators of diabetes-related hearing impairment in the U.S. population: National Health and Nutrition Examination Survey 1999-2004. Diabetes Care. 2010;33:811–816. doi: 10.2337/dc09-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaulton K.J., Ferreira T., Lee Y., Raimondo A., Mägi R., Reschen M.E., Mahajan A., Locke A., Rayner N.W., Robertson N., DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat. Genet. 2015;47:1415–1425. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray S., Feinberg M.W., Hull S., Kuo C.T., Watanabe M., Sen-Banerjee S., DePina A., Haspel R., Jain M.K. The Krüppel-like factor KLF15 regulates the insulin-sensitive glucose transporter GLUT4. J. Biol. Chem. 2002;277:34322–34328. doi: 10.1074/jbc.M201304200. [DOI] [PubMed] [Google Scholar]
- 34.Noguchi H. Recent advances in stem cell research for the treatment of diabetes. World J. Stem Cells. 2009;1:36–42. doi: 10.4252/wjsc.v1.i1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamaguchi M., Nakamura N., Nakano K., Kitagawa Y., Shigeta H., Hasegawa G., Ienaga K., Nakamura K., Nakazawa Y., Fukui I. Immunochemical quantification of crossline as a fluorescent advanced glycation endproduct in erythrocyte membrane proteins from diabetic patients with or without retinopathy. Diabet. Med. 1998;15:458–462. doi: 10.1002/(SICI)1096-9136(199806)15:6<458::AID-DIA601>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 36.Adewoye E.O., Akinlade K.S., Olorunsogo O.O. Erythrocyte membrane protein alteration in diabetics. East Afr. Med. J. 2001;78:438–440. doi: 10.4314/eamj.v78i8.8998. [DOI] [PubMed] [Google Scholar]
- 37.Fernández-Real J.M., Pickup J.C. Innate immunity, insulin resistance and type 2 diabetes. Trends Endocrinol. Metab. 2008;19:10–16. doi: 10.1016/j.tem.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Satoh N., Shimatsu A., Himeno A., Sasaki Y., Yamakage H., Yamada K., Suganami T., Ogawa Y. Unbalanced M1/M2 phenotype of peripheral blood monocytes in obese diabetic patients: effect of pioglitazone. Diabetes Care. 2010;33:e7. doi: 10.2337/dc09-1315. [DOI] [PubMed] [Google Scholar]
- 39.Feinberg M.W., Wara A.K., Cao Z., Lebedeva M.A., Rosenbauer F., Iwasaki H., Hirai H., Katz J.P., Haspel R.L., Gray S. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 2007;26:4138–4148. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morton N.E. Linkage disequilibrium maps and association mapping. J. Clin. Invest. 2005;115:1425–1430. doi: 10.1172/JCI25032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.