Abstract
Keratolytic winter erythema (KWE) is a rare autosomal-dominant skin disorder characterized by recurrent episodes of palmoplantar erythema and epidermal peeling. KWE was previously mapped to 8p23.1–p22 (KWE critical region) in South African families. Using targeted resequencing of the KWE critical region in five South African families and SNP array and whole-genome sequencing in two Norwegian families, we identified two overlapping tandem duplications of 7.67 kb (South Africans) and 15.93 kb (Norwegians). The duplications segregated with the disease and were located upstream of CTSB, a gene encoding cathepsin B, a cysteine protease involved in keratinocyte homeostasis. Included in the 2.62 kb overlapping region of these duplications is an enhancer element that is active in epidermal keratinocytes. The activity of this enhancer correlated with CTSB expression in normal differentiating keratinocytes and other cell lines, but not with FDFT1 or NEIL2 expression. Gene expression (qPCR) analysis and immunohistochemistry of the palmar epidermis demonstrated significantly increased expression of CTSB, as well as stronger staining of cathepsin B in the stratum granulosum of affected individuals than in that of control individuals. Analysis of higher-order chromatin structure data and RNA polymerase II ChIA-PET data from MCF-7 cells did not suggest remote effects of the enhancer. In conclusion, KWE in South African and Norwegian families is caused by tandem duplications in a non-coding genomic region containing an active enhancer element for CTSB, resulting in upregulation of this gene in affected individuals.
Keywords: keratolytic winter erythema, KWE, CTSB, enhancer duplication, gene regulation, topological domains, keratoderma, autosomal-dominant trait, South Africa, Norway
Introduction
Keratolytic winter erythema (KWE [MIM: 148370]) is a rare autosomal-dominant epidermal skin disorder of unknown etiology. It was originally reported as “Oudtshoorn skin disease” in a large cohort living in South Africa.1, 2 KWE manifests during childhood with recurrent episodes of palmoplantar erythema and centrifugal epidermal peeling. Lateral and dorsal aspects of the hands and feet can be involved. A less common finding is a slowly migratory, annular erythema that is seen mostly on the extremities. Between flares, the skin can appear unremarkable. Itching can occur, and hyperhidrosis, associated with a pungent odor, is invariably present. Formation of vesicles or bullae is rare, whereas keratolysis that causes the formation of dry blisters is regularly seen. This is followed by peeling. Cold weather, moisture, febrile diseases, and physical and mental stress can trigger exacerbations. In severely affected individuals, skin manifestations persist unremittingly. Penetrance of the disease is high, but expressivity is variable, even within the same family.
All KWE-affected families in South Africa can be traced back to Captain Francois Renier Duminy (born in Lorient, France, in 1747), pointing to a founder effect as the cause of the high prevalence (1/7,200) of KWE in white Afrikaans speakers.3 KWE has also been described in Germany,4 Denmark,5 the US (in a family of Norwegian descent),6 and the UK (in a family linked to the South African pedigrees).7 Genome-wide linkage analysis in South African families and one German family mapped the disease locus to chromosomal region 8p23.1–22 from marker D8S1759 to D8S552.4, 8 The KWE critical region (∼1.2 Mb) contains 15 known protein-coding genes and is flanked by two olfactory repeat regions that are prone to non-allelic homologous recombination. This can result in chromosome rearrangements, including a common 4.7 Mb polymorphic 8p23 inversion.9 The South African families have a common haplotype within the KWE critical region, but this is not shared with the affected individuals from the large German family.4
Within the KWE critical region, the genes encoding cathepsin B (CTSB [MIM: 116810])10 and farnesyl-diphosphate farnesyltransferase (FDFT1 [MIM: 184420])11 are interesting candidate genes for this disease. However, sequencing of the coding regions in these genes,12 as well as studies of copy-number variation (CNV) in the KWE region,13 did not reveal pathogenic variants, and gene expression analyses were not conclusive.12
The aim of the present study was to identify the genetic cause of KWE by using targeted resequencing of the KWE critical region in five South African families and by using SNP array and whole-genome sequencing (WGS) in two Norwegian families. We identified two tandem duplications that segregate with the KWE phenotype in South African (7.67 kb) and Norwegian (15.93 kb) affected individuals. The duplications overlap at the site of a predicted enhancer region (2.62 kb) that is shown to be active in human epidermal keratinocytes (HEKs).
Subjects and Methods
Participants
South Africans
A total of 23 affected and 19 unaffected individuals from three KWE-affected families (A, B, and C; Figure 1) plus individuals III-2 and II-4 from families G and I, respectively (Figure S1), and seven ethnically matched control individuals (random white Afrikaans speakers) were included in the targeted sequencing of the KWE region. Individuals from families F–I (11 affected and 12 unaffected individuals; Figure S1), I-1 from family C (Figure 1), and 89 ethnically matched control individuals (random white Afrikaans speakers) were included in the validation process. Functional studies were performed from palmar skin biopsies of three affected individuals (a mother and her two sons with the validated 7.67 kb duplication, who were not part of the families described above) and three control individuals (two females and one male). All participants gave informed consent. The study was approved by the Human Research Ethics Committee (Medical) of the University of the Witwatersrand, South Africa (approval no. M140530).
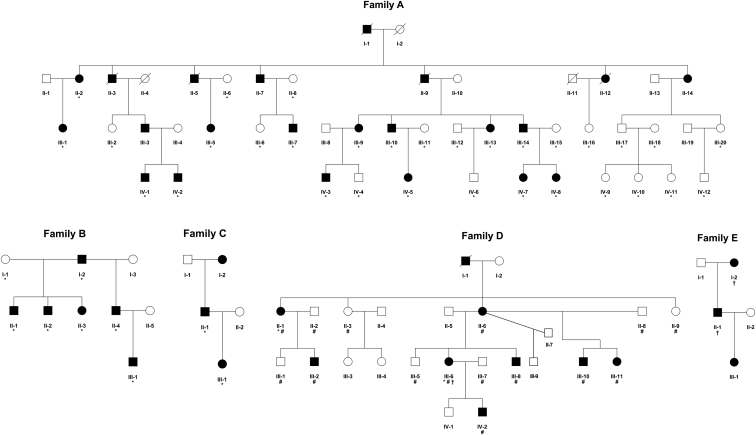
Figure 1.
Pedigrees of the South African and Norwegian KWE-Affected Families
South African (A–C) and Norwegian (D and E) families affected by KWE. The individuals whose samples were subjected to NGS sequencing (∗), the Affymetrix 6.0 SNP array (#), and the Affymetrix CytoScan HD Array (†) are marked as indicated here.
Norwegians
Linkage and CNV analyses were performed in family D, including eight affected and nine unaffected individuals (Figure 1), and CNV analysis was also performed in individuals I-2 and II-1 from family E. WGS was performed in individuals II-1 and III-6 from family D. Functional studies were performed on palmar skin biopsies of four affected individuals (III-6, II-6, II-1 from family D and I-2 from family E) and four female control individuals. All participants gave informed consent. The study was approved by the Regional Committee for Medical and Health Research Ethics in Western Norway (approval no. 2011/2453).
Linkage and CNV Analysis in Norwegians
Genomic DNA was extracted from peripheral blood with the QiaSymphony 101 instrument (QIAGEN). Genome-wide SNP genotyping analysis and CNV analysis were performed with the Affymetrix SNP Array 6.0 (1.8 million markers). Sample preparation and array hybridization were performed according to the supplier’s protocols, and arrays were scanned with the Affymetrix GeneChip Scanner 3000. Multi-point parametric linkage analysis was performed with Allegro v.2.6 software (Linux version) on a subset of 45,000 SNPs pruned for strong local linkage disequilibrium. A fully penetrant autosomal-dominant inheritance model and population disease allele frequency of 0.001 were used. Individuals III-6 (family D) and I-2 and II-1 (family E) were also analyzed with the Affymetrix CytoScan HD Array (2.6 million CNV markers). CNV data were generated with Affymetrix GeneChip Genotyping Console Software v.4.2 and analyzed by Affymetrix Chromosome Analysis Suite (ChAS) v.3.0.0.42.
Targeted Sequencing of the KWE Critical Region in South Africans
DNA was extracted by the salting-out method.14 DNA libraries were prepared with the Illumina TruSeq DNA Sample Preparation Kit v.2 (FC-121-2001), and the KWE critical region (chr8: 11,477,641–12,742,458; UCSC Genome Browser hg19) was captured according to the NimbleGen SeqCap EZ SR protocol (6266304001). The libraries were sequenced on the Illumina HiSeq 2000 according to a 101 bp paired-end sequencing protocol. Reads were mapped to the reference human genome (hg19) with the Burrows-Wheeler Aligner. The average read depth was 795× across the critical region. Small variants were assessed with the Genome Analysis Toolkit,15, 16, 17 and large structural variants were analyzed with Pindel.18
WGS in Norwegians
WGS was performed with the Illumina X Ten platform according to a 150 bp paired-end sequencing protocol. Small variants were called with the Isaac read aligner and variant caller.19 Structural variants were called with Manta,20 and CNVs were called with CNV-seq.21 In two KWE samples, at least 97% of the genome was covered at least 15×. An average (two samples) of ∼3.4 million single-nucleotide variants (SNVs)—including ∼9,800 non-synonymous coding variants, ∼4,600 deletions, ∼1,600 insertions, ∼80 inversions, and ∼100 translocations—were called.
Variant Filtering
Variants were filtered with the use of structured query language (SQL) queries on a Vertica database (South African data) and RareVariantVis Bioconductor package (Norwegian data).22 Potential causal variants were considered if they were exclusive to affected individuals, were not present in public databases (such as dbSNP, the Database of Genomic Variants [DGV], or dbVar), and fit an autosomal-dominant model of inheritance. SNVs and small insertions and deletions (indels) were annotated with the Variant Effect Predictor (VEP)23 in the South African data and annotated with SnpEff24 in the Norwegian data. All coding, splice, and regulatory variants were prioritized, and when no variants segregating with the disease were identified, intronic and intergenic variants were considered.
Mapping Duplication Breakpoints
Genomic PCR and Sanger sequencing were performed for physical mapping of the exact breakpoints of the duplications. PCR primers (Table S1) were designed with either Primer325 (South Africans) or Oligo 6.3 (National Bioscience) (Norwegians). For South Africans, the details on validation of duplications are given in Table S1 and Figure S2.
Transcription Factors and Their Binding Sites
Transcription factor binding sites within the KWE duplication region in various cell lines were taken from the ENCODE Factorbook chromatin immunoprecipitation sequencing (ChIP-seq) data.26 With the use of RNA sequencing (RNA-seq) data from our previous experiment on differentiating human keratinocytes,27 transcription factors (within the KWE duplication region) were filtered for the subset that is expressed at a level of at least 10 FPKM (fragments per kilobase per million mapped reads) at any stage of human keratinocyte differentiation.
Enhancer Activity and Gene Expression in Normal Keratinocytes and Other Cells
Epigenomic Profiling Data of Differentiating HEKs
We previously performed ChIP-seq analyses of p63, H3K27ac, and RNA polymerase II (RNAPII) in HEKs at days 0, 2, 4, and 7 in culture.27 We quantitated the signals from H3K27ac and RNAPII ChIP-seq for the enhancer (chr8: 11,733,500–11,736,900) and three nearby potential target genes by calculating the reads per kilobase of transcript per million mapped reads (RPKM). This procedure normalizes the number of reads mapping to the enhancer or gene-body regions according to sequencing depth and gene and/or enhancer length.
ENCODE and Epigenome Roadmap Data
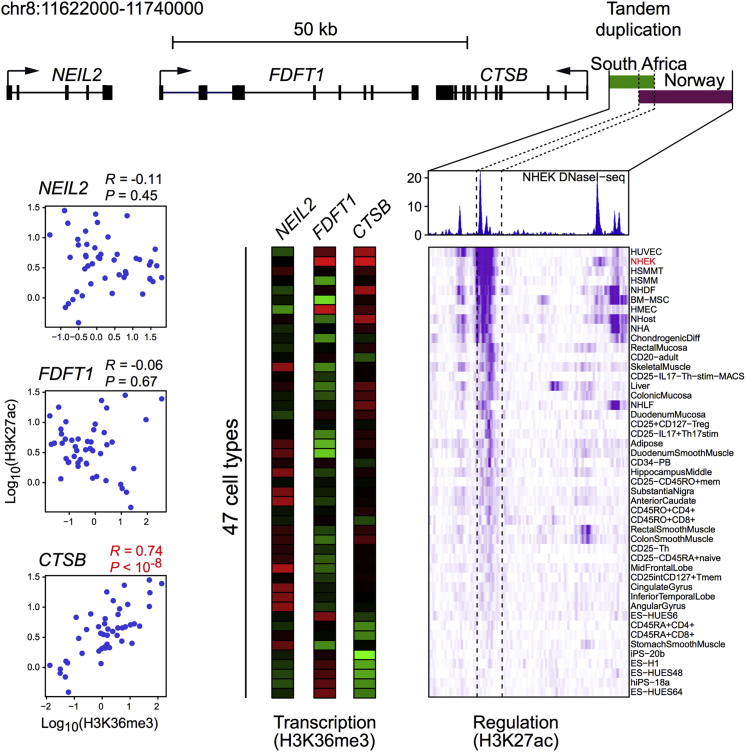
To identify potential regulatory elements within the KWE duplicated region, we used NHEK DNaseI sequencing data along with histone modification (H3K4me1 and H3K27ac) ChIP-seq data from ENCODE28 and Epigenome Roadmap29, 30 in 47 different human cell types. We calculated the average H3K27ac and H3K36me3 signals to predict overall enhancer activity and transcriptional activity of nearby genes, respectively. We performed Pearson’s correlation between the H3K27ac and H3K36me3 signals to predict the target genes of the active enhancer on the basis of the highest correlation.
Gene Expression and Protein Abundance in Palmar Biopsies from Affected and Healthy Control Individuals
Single 4 mm palmar skin punch biopsies were taken from the skin of the hypothenar (or thenar) regions of seven affected individuals who were not presenting with aggravated disease at the time of collection and seven healthy control individuals (three South Africans and four Norwegians in each group). The biopsies were cut in two perpendicularly to the skin surface. One half was used for examination of relative gene expression, and the other was processed for histology.
RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR
For RNA expression studies, we placed half of each biopsy in 1 mL dispase II protease (12 mg/mL) for 2–4 hr to separate the epidermis from the dermis. The separated epidermis was then placed in RNALater and stored at 4°C for 24 hr and then at −20°C until RNA extraction. Samples were homogenized with the TissueLyser II (QIAGEN), and total RNA was extracted with the RNeasy Mini kit (QIAGEN) and stored at −80°C. The quality and quantity of total RNA were determined with the Experion Automated Electrophoresis Station (Bio-Rad) and the NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies), respectively. All samples were stored at −80°C. One of the South African KWE samples was excluded from further analysis because of the low RNA quality and concentration. 250 ng total RNA from each sample was reverse transcribed to cDNA with the SuperScript VILO cDNA Synthesis Kit (ThermoFisher Scientific), and quantitative real-time PCR was conducted with the ABI Prism 7900HT Sequence Detection System (Applied Biosystems). The expression levels of CTSB, FDFT1, and nei-like DNA glycosylase 2 (NEIL2 [MIM: 608933]) were measured with TaqMan Gene Expression Assays (probe IDs: Hs00947433_m1, Hs00926054_m1, and Hs00979610h, respectively; ThermoFisher Scientific) and were normalized to that of the endogenous control RPLP0 (probe ID: Hs99999902m1).31 All samples were run in triplicate, and the relative gene expression levels were determined according to the comparative ΔCt method.32 Differences in gene expression levels between affected individuals (n = 6) and unaffected control individuals (n = 7) were determined, and p values were calculated with the Mann-Whitney U test.
Immunohistochemistry
For histology, half of each biopsy was fixed in buffered formalin for 4–8 hr. Paraffin sections (6 μm) were cut for histopathology (H&E staining) and immunohistochemistry with antisera against CTSB (R&D), FDFT1 (Sigma), and NEIL2 (Sigma). Antisera were raised in goat (CTSB), rabbit (FDFT1), and mouse (NEIL2). Deparaffinized and dehydrated sections were pretreated for 10 min in 10 mM sodium citrate buffer (pH 6.0) for antigen retrieval. Sections were blocked for endogenous peroxidase with 3% hydrogen peroxide. A blocking step using 5% normal serum from the animal in which the biotinylated conjugated serum had been raised was included for the reduction of background staining. Further staining and embedding were performed according to standard protocols with the avidine-biotine-peroxidase complex and amino-ethyl-carbazole as a peroxidase substrate.
CTSB staining was evaluated by two independent observers who were unaware of the identity of the samples. CTSB staining intensity was scored on a scale from 0 (no visible staining) to 3 (strong staining). Staining was scored separately for the stratum spinosum and the stratum granulosum. Scores for seven affected and seven control individuals were analyzed with a two-tailed Mann Whitney U test.
Prediction of Genomic Architecture
The genomic architecture of the region containing the duplication was evaluated with available keratinocyte Hi-C data.33 Putative CCTC-binding factor (CTCF)-mediated loops were predicted in normal HEKs (NHEKs) with NHEK-derived CTCF ChIP-seq data28 and an algorithm that exploits the relationship between CTCF motif orientation and interaction direction.34 Genomic architecture in the breast cancer epithelial cell line MCF-7 was determined with available data from CTCF Chromatin Interaction Analysis by Paired-End Tag Sequencing (ChIA-PET).28 RNAPII-targeted ChIA-PET data were used for identifying contacts between regulatory elements, such as enhancers and promoters, and determining possible interactions between the regulatory elements and their predicted target genes.35
Results
Clinical Description
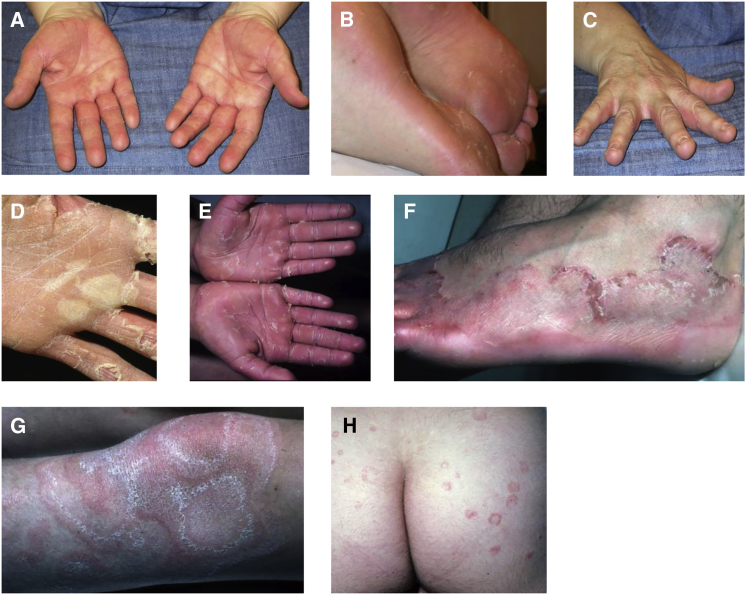
KWE is a highly penetrant disease (calculated at 92%)3 that has been well characterized in South African affected individuals.1, 2 The inclusion of two Norwegian families (Figure 1) in this study allowed interesting comparisons (Table 1). The age of onset is typically in early childhood, but it has been described at birth, and onset can be delayed to the early 20s. The South African affected individuals generally had more severe disease than the Norwegians, but palm and sole involvement was seen in all affected individuals of both nationalities (Figures 2A, 2B, 2D, and 2E). Web spaces, and even the dorsum of the feet in some cases, were also often affected (Figures 2C and 2F). Annular erythema was seen only on the extremities and/or buttocks (Figures 2G and 2H) and on the trunk or face (Table 1) of a few affected individuals. One female Norwegian affected individual (I-2 from family E) had unusual recurrent fixed indurated facial plaques. Severity and course of KWE varied considerably among those affected. Full remission was observed in a 45-year-old female (II-6 from family D; Figure 1), whereas a 19-year-old male (III-2 from family D; Figure 1) had continuous severe palmoplantar erythema and desquamation.
Table 1.
Clinical Data in Affected Individuals
| South African (n = 45) | Norwegian (n = 10) | |
|---|---|---|
| Age of Onset | <10 years in 84%3 | <2 years in 90% |
| Clinical Features in Palms and Soles | ||
| Keratolysis (formation of a dry “blister”) | yes | yes |
| Centrifugal peeling | yes | yes |
| Distinct erythematous margin | yes | yes |
| True blistering or bullae | no | exceptional |
| Web-space involvement | yes | yes |
| Transgrediens spread to dorsum of hands and feet | yes | yes |
| Hyperhidrosis with pungent odor of hands and feet | yes | yes |
| Annular Erythema | ||
| Extremities | rare | single individual |
| Trunk | rare | single individual |
| Face | single individual | no |
| Recurrent fixed indurated facial plaques | no | single individual |
| Factors Aggravating Symptoms | ||
| Cold weather | common | 2 of 10 individuals |
| Contact with water or moisture | yes | yes |
| General anesthesia | yes | yes |
| Infections or fever | yes | yes |
| Antibiotic use | yes | not noted |
| Topical steroids | yes | not noted |
| Mechanical pressure from shoes | not noted | yes |
| Factors Improving Symptoms | ||
| Pregnancy | yes, common | yes, common |
| Age | yes (in some individuals) | yes (in some individuals) |
Figure 2.
Affected Palmoplantar Regions and Extremities in Individuals with KWE
Palm and sole involvement was seen in all affected individuals of both nationalities. Exacerbation starts with an erythema that is often demarcated (A and B). It can be localized or affect the entire palm or sole and often spreads to the web spaces (C). This is followed by the appearance of opaque, dry “bullae,” which peel centrifugally, giving rise to a thick peel that can be easily gripped (D and E). In the palmar skin, the revealed base is red and glazed, and dermatoglyphics are retained (E). Transgrediens spread to the dorsum of the feet can be seen (F). Annular erythema was seen in only a few affected individuals on the extremities (G) and trunk and buttocks (H).
The characteristic seasonal worsening of symptoms during winter was observed in many South African families2 but was noted in only two of ten Norwegian affected individuals. Episodic peeling was mentioned by all Norwegians to be triggered by exposure to water and moist conditions. Exacerbations were also reported after infections, general anesthesia, the use of topical steroids, and in some cases, also the use of antibiotics. Premenstrual worsening was also noted, whereas pregnancy and increased age in some affected individuals seemed to improve the condition. Hair, nails, and mucous membranes were normal in all affected individuals. Impairment of social interaction during the teenage years, due to the effects of KWE, was reported by many Norwegian and South African affected individuals.
Identification of Overlapping Duplications in South African and Norwegian Affected Individuals
Extensive analyses of the protein-coding genes of the KWE critical region did not reveal any possible detrimental variants that segregated with the disease. Instead, a non-coding 7.67 kb tandem duplication was identified in the critical region on chr8: 11,729,286–11,736,955 in the South African affected individuals (Figures 1A–1C) with the use of targeted next-generation sequencing (NGS) data (Figure 3). The duplication was validated by PCR and Sanger sequencing (Table S1 and Figures S1 and S2) and segregated with KWE in all affected South African individuals tested (n = 38). It was not detected in any control samples (n = 127).
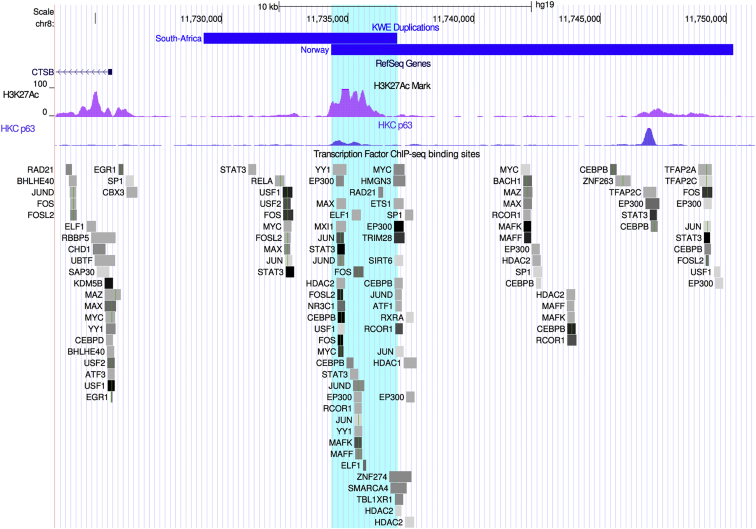
Figure 3.
Duplications in Affected Individuals Overlap at an Enhancer Region
Schematic overview of the KWE critical region in chr8: 11,723,410–11,751,135 (a scale is shown at the top). The South African (7.67 kb) and Norwegian (15.93 kb) tandem duplications are displayed as blue horizontal bars located upstream of CTSB. The 2.62 kb overlap (chr8: 11,734,333–11,736,955) between the two duplications (turquoise shading) is clearly positioned at the site of an H3K27Ac histone mark (pink peaks) in the ENCODE NHEK cell line. Such histone marks are often found at the site of active regulatory elements and enhancers. We showed that the keratinocyte master regulator p63 binds to the enhancer region of the histone mark H3K27Ac in differentiating HEKs (HKC p63, blue peaks). Several other transcription factors are also known to bind to this region in different cell types (ChIP-seq data from ENCODE, 90 cell lines). Only transcription factors that are expressed in differentiating keratinocytes (cut off at FPKM = 10) are shown. The different transcription factors are displayed as boxes, and the binding strength is represented by a color scale from light gray (weaker) to black (stronger).
Independently, significant linkage (maximum LOD score of 3.3) between KWE and an approximately 6.6 Mb region (chr8: 9,231,148–15,837,977) encompassing the KWE critical region was found in Norwegian family D (Figure 1). Subsequently, WGS and CNV analyses identified a 15.93 kb duplication overlapping the South African duplication in all affected individuals from the two Norwegian families (Figure 3). Sanger sequencing (Table S1) revealed that it was a tandem duplication at chr8: 11,734,333–11,750,263 and that a 95 bp triplication (chr8: 11,744,352–11,744,446) was located between the tandemly duplicated regions. Such triplications are not uncommon in tandem duplications.36
The South African and Norwegian duplications are located upstream of CTSB and overlap at a 2.62 kb region (Figure 3). No similar duplications or a duplication of the same size as the 2.62 kb overlap have been reported in the DGV. However, several duplications that encompass larger regions, including the 2.62 kb overlap region, CTSB, and even FTFD1 (Figure S3), have been described. We also found one such large duplication when analyzing the SNP array data in a Dutch cohort of 1,416 healthy students (Figure S3).37
An Active Enhancer in the Region of Overlap between the Duplications
Next, we examined the genomic region of the 2.62 kb overlap between the South African and Norwegian duplications (chr8: 11,734,333–11,736,955). A strong H3K27ac signal probably representing an active enhancer was found within this region in epidermal keratinocytes (Figure 3).27, 28, 38 Several transcription factors were shown to bind to this regulatory region.26 By analyzing our previously published RNA-seq data,27 we found that most of these transcription factors seem to be expressed in differentiating keratinocytes (Figure 3).27, 38 These include p63 (the master regulator of keratinocyte proliferation and differentiation),39 STAT3,40, 41 and CEBPB.42
Correlation between Enhancer Activity and CTSB Transcription
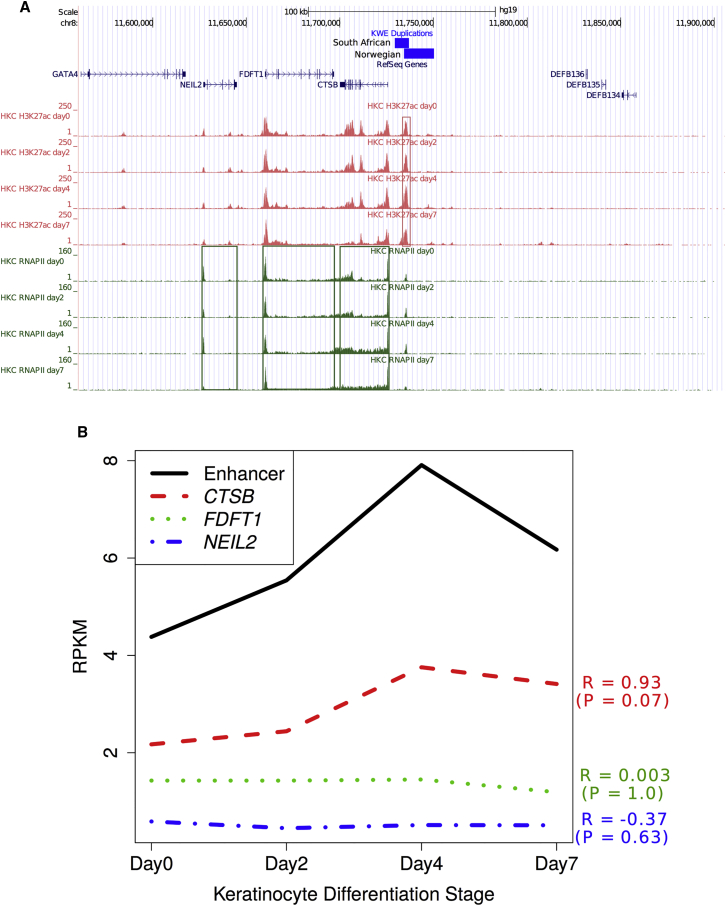
To investigate the target gene(s) of the enhancer in the KWE region (chr8: 11,734,333–11,736,955), we first re-analyzed our ChIP-seq dataset from differentiating keratinocytes.27 The closest genes to the enhancer included the strong candidates CTSB and FDFT112 and NEIL2. We used H3K27ac and RNAPII ChIP-seq data27 to investigate enhancer activity and transcription, respectively, of these three genes. Both enhancer activity and CTSB expression increased during early differentiation (days 0–4) and then decreased in subsequent differentiation stages (days 4–7) (Figure 4A).27 FDFT1 expression was maintained at a high level during differentiation, whereas NEIL2 seemed to be expressed at a low level, possibly because of the poised RNAPII at the promoter (Figure 4A). We found a high correlation, although not statistically significant given the small number of data points, between the enhancer’s activity and CTSB expression during keratinocyte differentiation (p = 0.07, R = 0.93), but no correlation with FDFT1 (p = 1.0, R = 0.003) or NEIL2 (p = 0.63, R = −0.37) expression (Figure 4B), suggesting that this enhancer regulates CTSB during normal keratinocyte differentiation. ENCODE ChIP-seq datasets with histone marks H3K27ac and H3K36me3 also showed a strong correlation between the enhancer’s activity and CTSB expression (p < 10−9 and R = 0.74) across 47 different cell types, whereas no correlation was observed with FDFT1 (p = 0.67 and R = −0.06) or NEIL2 (p = 0.45 and R = 0.11) expression (Figure 5).
Figure 4.
Correlation of Enhancer Activity and Transcription of Nearby Genes during Keratinocyte Differentiation
(A) Schematic overview of the KWE critical region in chr8: 11,560,500–11,905,700 (a scale is shown at the top). The South African (7.67 kb) and Norwegian (15.93 kb) tandem duplications are displayed as blue horizontal bars. ChIP-seq analyses of H3K27ac and RNAPII during human primary keratinocyte (HKC) differentiation were assessed. The H3K27ac histone modification signal represents enhancer activity (red panels), and RNAPII binding levels represent transcription rate (green panels).
(B) There was a strong correlation between enhancer activity and CTSB expression (R = 0.93, p = 0.07) during keratinocyte differentiation, but not between enhancer activity and FDFT1 or NEIL2 expression.27, 38
Figure 5.
Correlation of Enhancer Activity and Transcription of Nearby Genes in Cell Lines Reported by the ENCODE Consortium
The region of the South African (7.67 kb, green bar) and Norwegian (15.93 kb, purple bar) tandem duplications is displayed (a scale is shown at the top). The enhancer within the overlapping region was identified on the basis of a DNaseI hypersensitivity assay performed in NHEKs (blue peaks).28 Beneath this panel, the histone modification H3K27ac levels (ChIP-seq data) across the enhancer region in 47 human tissue and cell types are shown as a regulation heatmap (purple). Darker shades of purple indicate higher signal of the histone marker (H3K27ac), correlated with enhancer activity in the different tissues. Transcription of three nearby genes (CTSB, FDFT1, and NEIL2) was predicted by H3K36me3 levels (ChIP-seq data) across the genes in the same 47 different cell types. The transcription data are displayed as three parallel vertical bars in which red denotes high transcription activity and green denotes low activity. The correlations between H3K27ac and H3K36me3 levels are shown in the plots to the left. There was a highly significant positive correlation between the enhancer’s activity and CTSB expression (R = 0.74, p < 10−8).
Individuals with KWE Have Increased Relative Expression of CTSB in the Epidermis
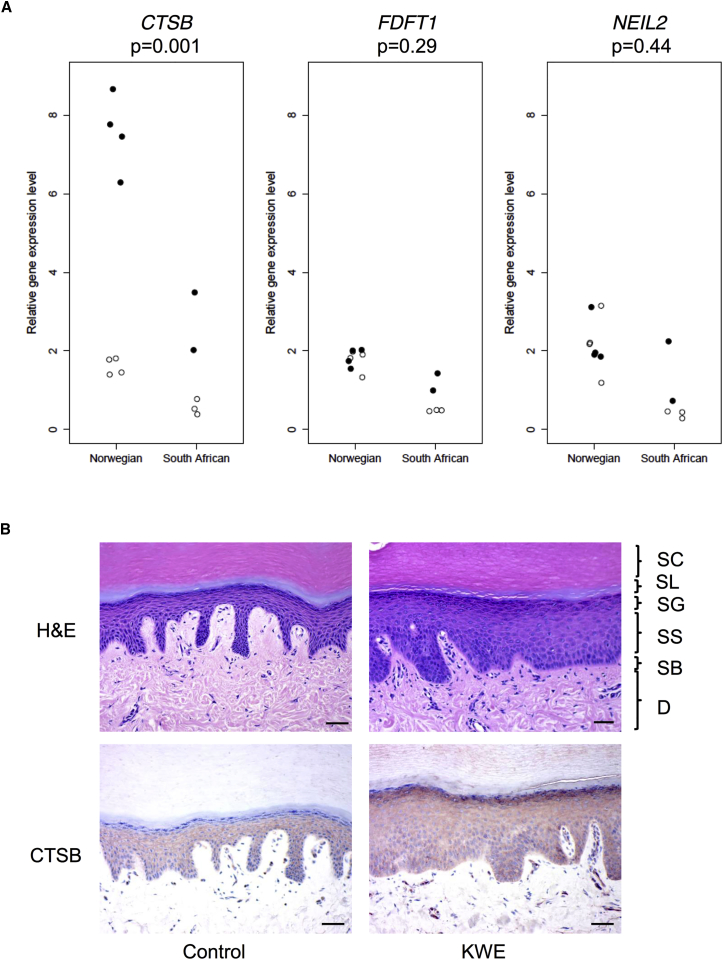
Using quantitative real-time PCR, we compared the relative expression levels of CTSB, FDFT1, and NEIL2 in the palmar epidermis between affected individuals (two South Africans and four Norwegians) and unaffected individuals (three South Africans and four Norwegians) (Figure 6A). Given the small sample size, we conducted a combined analysis (i.e., South African and Norwegian samples analyzed together) and found that the relative expression level of CTSB was significantly higher in the samples from the affected individuals than in control individuals (p = 0.001) (Figure 6A). There was no statistically significant difference in the expression of FDFT1 (p = 0.29) or NEIL2 (p = 0.44) between the two groups. Fold changes for each gene are shown in Tables S2–S4.
Figure 6.
Comparison of the Expression of CTSB, FDFT1, and NEIL2 and Abundance of CTSB in Skin from KWE-Affected Individuals and Ethnically Matched Control Individuals
(A) Quantitative real-time PCR was used for determining the expression of CTSB, FDFT1, and NEIL2 in RNA samples isolated from epidermal tissue of the skin biopsies obtained from eight Norwegian and five South African individuals. Affected and control individuals are shown as closed and open circles, respectively. For each gene, the individual expression levels are given as a fold change relative to the mean value for all healthy control individuals (both Norwegian and South African). The p values were calculated with the Mann-Whitney U test, including all healthy control individuals (n = 7) and all KWE-affected individuals (n = 6).
(B) Immunohistochemistry showing H&E staining (upper panels) of palmar skin biopsies of a healthy control individual (left) and KWE-affected individual (right). Abbreviations are as follows: SC, stratum corneum; SL, stratum lucidum; SG, stratum granulosum; SS, stratum spinosum; SB, stratum basale; and D, dermis. The scale bar represents 50 μm. Note the increased epidermal thickness of the KWE skin. Lower panels show CTSB staining of serial sections of the same individuals. CTSB is present in a granular pattern in the stratum spinosum of control and KWE skin. Note that CTSB staining is absent in the stratum granulosum of control skin, whereas it is abundant in the stratum granulosum of KWE-affected skin.
Immunohistochemistry of CTSB, FDFT1, and NEIL2
The abundance and localization of CTSB, FDFT1, and NEIL2 were examined by histopathology and immunohistochemistry on biopsies of palmar skin from affected and control individuals. As shown in Figure 6B, H&E staining of the samples from affected individuals showed a thicker epidermis and stratum corneum than did staining of control palmar skin. Morphology was largely normal, such that all epidermal layers and an orthokeratotic stratum corneum were present. Because the biopsies were taken from non-lesioned skin, no overt blistering or stratum corneum splitting was visible and no parakeratosis was evident.1 CTSB was found to be present in the stratum spinosum of normal skin in a granular pattern throughout the epidermis, consistent with a lysosomal localization, whereas the protein was absent (five of seven individuals) or weak (two of seven individuals) in the stratum granulosum (Table S2 and Figure 6B). In biopsies from affected individuals, CTSB was found in the stratum spinosum in a pattern similar to that in normal control individuals and was always present in the stratum granulosum, although it varied from weak (three of seven individuals) to strong (four of seven individuals) (Table S2 and Figure 6B). Semiquantitative scores for CTSB stratum granulosum staining were 0.3 ± 0.5 (mean ± SD) for normal skin and 1.7 ± 1.0 for KWE-affected skin (p = 0.006, Mann-Whitney U test). We found very faint staining of FDFT1 in the epidermal layers but strong staining of the sweat glands, and there was no apparent difference between affected and control individuals (data not shown). We failed to obtain a reliable staining for NEIL2.
Chromatin Architecture of the Genomic Region Encompassing the Enhancer
Enhancers can also affect the expression of remotely located genes, and we therefore investigated the chromatin architecture of this genomic region. Low-resolution Hi-C data generated in epidermal keratinocytes33 indicated that the enhancer lies within a chromatin subdomain that includes CTSB and FDFT1 (Figure S4A). Domain demarcations are defined by CTCF binding and interactions, and CTCF chromosomal interaction loops (e.g., investigated by ChIA-PET) can be used for identifying topological domains.43 There are no available CTCF ChIA-PET data in epidermal keratinocytes, but CTCF binding sites in keratinocytes and breast epithelial MCF-7 cells are highly conserved in this region (Figure S4B). We therefore investigated CTCF loops detected in a CTCF ChIA-PET study on MCF-7 cells.28 These data on CTCF chromosomal interaction identified a domain that included CTSB, FDFT1, and NEIL2, as well as the enhancer, and a smaller subdomain containing only CTSB and the enhancer within this region (Figure S4D). This hierarchical CTCF-mediated domain architecture was predicted to also occur in keratinocytes on the basis of an algorithm that exploits the orientation of the CTCF motif in the binding sites to predict CTCF-mediated interactions (Figure S4C).34 Next, we used RNAPII ChIA-PET data for the MCF-7 cell line to detect interaction loops between transcriptionally active promoters and enhancers.28 We identified RNAPII interaction loops between the identified enhancer and the CTSB promoter and extensive loops within CTSB in both of the two RNAPII ChIA-PET replicates (Figure S4E), consistent with our other data showing that the enhancer regulates CTSB. Notably, we did not detect any RNAPII interaction loops between the enhancer and FDFT1 or NEIL2 (Figure S4E), or any other more remotely located genes.
Discussion
Twenty years ago, KWE was mapped to the short arm of chromosome 8 (8p23.1–p22),4 but the causal mutation remained elusive. Here, we report two tandem non-coding DNA duplications that are located in the KWE critical region and segregate with the disease in eight South African (7.67 kb) and two Norwegian (15.93 kb) families. The duplications have a 2.62 kb overlap at the site of an active enhancer upstream of CTSB (Figure 3), a region also known to bind several transcription factors, including the keratinocyte master regulator p63 (Figure 3). No similar small duplications in non-coding DNA, including this enhancer region, have previously been reported (Figure S3), whereas large duplications overlapping the enhancer region and at least CTSB or both CTSB and FDFT1 (and several genes upstream) have been described in many unaffected individuals (Figure S3). This is in line with previous reports on absent or minor phenotypic effects associated with large duplications encompassing both a regulatory region and its target genes44 and stronger effects caused by smaller duplications.45 Interestingly, tandem duplications of regulatory regions have thus far primarily been described as causative for limb- and skull-malformation disorders and sex-reversal phenotypes and not for disorders with recurrent symptoms, such as KWE.45, 46, 47, 48, 49 Our data point to the duplicated enhancer as the genetic cause of the KWE phenotype and hence to an unusual and intriguing disease mechanism.
Duplication of an enhancer region can have complex long-range cis-regulatory effects and/or affect the topological chromatin domain structure, causing unexpected dysregulation of even remotely located genes.49 However, the most common effect of such a duplication is increased expression of the proximal target gene(s),47 and our data point to the latter mechanism in KWE. There was a significant increase in relative CTSB expression in the epidermis, as well as stronger CTSB staining in the stratum granulosum of non-lesioned palmar skin from KWE-affected individuals than in that of healthy control individuals (Figures 6A and 6B). Additional data also point to CTSB as the normal target for the non-duplicated enhancer. First, the activity of the enhancer correlated with the expression of CTSB, but not FDFT1 or NEIL2, during keratinocyte differentiation (Figure 4), and this correlation was strongly supported by ENCODE histone marker data in several cell lines (Figure 5). Second, the enhancer was involved only in RNAPII chromatin interaction loops with the promoter of CTSB and not with the FDFT1 or NEIL2 promoters (Figure S4E).
Our analysis of published chromatin data on NHEK and MCF-7 cells did not support interactions between the enhancer and remote targets (Figure S4). These data displayed different topological subdomains in the KWE critical region: one encompassing only the enhancer and CTSB and a larger one also including FDFT1 and possibly NEIL2 (Figures S4A–S4D). The interdomain interactions in the KWE critical region (in MCF-7 cells) were contained almost exclusively within the largest of these predicted subdomains, including NEIL2 (Figure S4E), making it less likely that the enhancer targets genes outside this domain.
CTSB encodes CTSB, a lysosomal protease that is localized throughout the epidermal layers and secreted into pericellular spaces.10 Keratinocyte cornification and desquamation are controlled by a finely tuned balance between different proteases (including several cathepsins) and their inhibitors.50, 51 Interestingly, loss-of-function mutations in the gene encoding cystatin A (CSTA [MIM: 184600]), an important inhibitor of CTSB, causes acral peeling skin syndrome (MIM: 607936)52 and exfoliative ichthyosis (MIM: 607936),53 which have skin desquamation phenotypes very similar to KWE. Deficiency of cystatin M/E, a cysteine protease inhibitor that also targets CTSB, causes an ichthyosis phenotype in mice.54 Loss-of-function mutations in other cathepsins have also been shown to cause skin phenotypes in both humans and animals;10, 55, 56, 57, 58, 59, 60, 61 a pertinent example is Papillon Lefèvre syndrome (MIM: 245000), which is characterized by palmoplantar hyperkeratosis, periodontal disease, and mutations in cathepsin C (CTSC [MIM: 602365]).61
The puzzling intermittent occurrence of the epidermal peeling, the predilection of lesions to palmoplantar skin, and the variable inter- and intrafamilial manifestation of KWE point to a labile state caused by the dysregulated expression of CTSB in the epidermis. This situation could imply a risk of shifting the normal balance between CTSB activity and its inhibitors, such as CSTA, which is also present in the stratum spinosum and granulosum62 and secreted in normal sweat.63 It is likely that triggering environmental factors could disturb this balance, for example, by inducing changes in transcription factor levels and/or altering the chromatin topology of the region, causing sudden fluctuations in the expression of CTSB. Interestingly, more CTSB was confined to the stratum granulosum in KWE-affected individuals than in control individuals (Figure 6B). This layer is subjected to apoptosis and temporary degradation during KWE exacerbation,13 possibly because the secretion of CTSB rises above a threshold and triggers the lysosomal apoptosis pathway.64 Further studies are clearly warranted for the investigation of the pathophysiological effects of upregulated CTSB, as well as the therapeutic potential of CSTA and other protease inhibitors in KWE and related diseases.
In conclusion, we have shown that KWE in South African and Norwegian families is caused by two different tandem duplications in a non-coding genomic region upstream of CTSB. The duplications, which overlap at the site of an active enhancer element in keratinocytes, are associated with increased epidermal accumulation of CTSB, a crucial cysteine protease for keratinocyte differentiation and desquamation.
Conflicts of Interest
R.B. is the owner of Congenomics LLC and provides consulting services to Novartis. E.J.O., F.Y., J.Z., M.S., and T.M.M. are employed by Novartis Pharma AG, and J.S. is employed by Bristol-Myers Squibb.
Acknowledgments
The authors wish to thank all the participants of this joint study. Technical assistance from Rita Holdhus, Jorunn Bringsli, Hilde Eldevik Rusaas, Atle Brendehaug, Anita Fernandez, Virginie Petitjean, Marc Altorfer, and Moritz Frei and contributions from Jayne Hehir-Kwa, Christian Gilissen, Alexander Hoischen, Kjell Petersen, Shaun Aron, Karen Koch, and Laeeka Moosa were highly appreciated. The South African study was supported by the South African Medical Research Council, the National Research Foundation, the University of the Witwatersrand, and the National Health Laboratory Service. The South African next-generation sequencing (NGS) study was funded by the Novartis Next Generation Scientist Program, and T.N. was supported by this program and the Novartis Visiting Scholars Program. M.R. is a South African Research Chair in Genomics and Bioinformatics of African populations, which is hosted by the University of the Witwatersrand, funded by the Department of Science and Technology, and administered by the National Research Foundation of South Africa. The Norwegian NGS study was performed by the Genomics Core Facility at the University of Bergen and supported by a grant from the Bergen Research Foundation (807964), including funding of the postdoctoral scholarship of T.S.
Published: April 27, 2017
Footnotes
Supplemental Data include four figures and four tables and can be found with this article at http://dx.doi.org/10.1016/j.ajhg.2017.03.012.
Contributor Information
Torunn Fiskerstrand, Email: torunn.fiskerstrand@helse-bergen.no.
Michele Ramsay, Email: michele.ramsay@wits.ac.za.
Web Resources
1000 Genomes, http://browser.1000genomes.org
Ensembl VEP, http://grch37.ensembl.org/Homo_sapiens/Tools/VEP
OMIM, http://www.omim.org/
Primer3, http://bioinfo.ut.ee/primer3-04.0/
RareVariantVis, https://www.bioconductor.org/packages/release/bioc/html/RareVariantVis.html
SnpEff, http://snpeff.sourceforge.net/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Findlay G.H., Morrison J.G. Erythrokeratolysis hiemalis--keratolytic winter erythema or ‘Oudtshoorn Skin’. A new epidermal genodermatosis with its histological features. Br. J. Dermatol. 1978;98:491–495. doi: 10.1111/j.1365-2133.1978.tb01934.x. [DOI] [PubMed] [Google Scholar]
- 2.Findlay G.H., Nurse G.T., Heyl T., Hull P.R., Jenkins T., Klevansky H., Morrison J.G., Sher J., Schulz E.J., Swart E. Keratolytic winter erythema or ‘oudtshoorn skin’: a newly recognized inherited dermatosis prevalent in South Africa. S. Afr. Med. J. 1977;52:871–874. [PubMed] [Google Scholar]
- 3.Hull P. University of the Witwatersrand; 1986. Keratolytic Winter Erythema (Oudtshoorn disease): clinical, genetic and ultraspectural aspects. PhD. [Google Scholar]
- 4.Starfield M., Hennies H.C., Jung M., Jenkins T., Wienker T., Hull P., Spurdle A., Küster W., Ramsay M., Reis A. Localization of the gene causing keratolytic winter erythema to chromosome 8p22-p23, and evidence for a founder effect in South African Afrikaans-speakers. Am. J. Hum. Genet. 1997;61:370–378. doi: 10.1086/514848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danielsen A.G., Weismann K., Thomsen H.K. Erythrokeratolysis hiemalis (keratolytic winter erythema): a case report from Denmark. J. Eur. Acad. Dermatol. Venereol. 2001;15:255–256. doi: 10.1046/j.1468-3083.2001.00258.x. [DOI] [PubMed] [Google Scholar]
- 6.Huntington M.K., Jassim A.D. Genetic heterogeneity in keratolytic winter erythema (Oudtshoorn skin disease) Arch. Dermatol. 2006;142:1073–1074. doi: 10.1001/archderm.142.8.1073. [DOI] [PubMed] [Google Scholar]
- 7.Amin A.N., DeGiovanni C.V., Farrant P.B., Hull P.R., Woollons A. Photodynamic therapy for the treatment of keratolytic winter erythema. Clin. Exp. Dermatol. 2011;36:668–669. doi: 10.1111/j.1365-2230.2010.03927.x. [DOI] [PubMed] [Google Scholar]
- 8.Appel S., Filter M., Reis A., Hennies H.C., Bergheim A., Ogilvie E., Arndt S., Simmons A., Lovett M., Hide W. Physical and transcriptional map of the critical region for keratolytic winter erythema (KWE) on chromosome 8p22-p23 between D8S550 and D8S1759. Eur. J. Hum. Genet. 2002;10:17–25. doi: 10.1038/sj.ejhg.5200750. [DOI] [PubMed] [Google Scholar]
- 9.Giglio S., Calvari V., Gregato G., Gimelli G., Camanini S., Giorda R., Ragusa A., Guerneri S., Selicorni A., Stumm M. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am. J. Hum. Genet. 2002;71:276–285. doi: 10.1086/341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Büth H., Wolters B., Hartwig B., Meier-Bornheim R., Veith H., Hansen M., Sommerhoff C.P., Schaschke N., Machleidt W., Fusenig N.E. HaCaT keratinocytes secrete lysosomal cysteine proteinases during migration. Eur. J. Cell Biol. 2004;83:781–795. doi: 10.1078/0171-9335-00428. [DOI] [PubMed] [Google Scholar]
- 11.Iwai I., Han H., den Hollander L., Svensson S., Ofverstedt L.G., Anwar J., Brewer J., Bloksgaard M., Laloeuf A., Nosek D. The human skin barrier is organized as stacked bilayers of fully extended ceramides with cholesterol molecules associated with the ceramide sphingoid moiety. J. Invest. Dermatol. 2012;132:2215–2225. doi: 10.1038/jid.2012.43. [DOI] [PubMed] [Google Scholar]
- 12.Hobbs A., Aron S., Hartshorne S., Hull P.R., Ramsay M. Exclusion of CTSB and FDFT1 as positional and functional candidate genes for keratolytic winter erythema (KWE) J. Dermatol. Sci. 2012;65:58–62. doi: 10.1016/j.jdermsci.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Hull P.R., Hobbs A., Aron S., Ramsay M. The elusive gene for keratolytic winter erythema. S. Afr. Med. J. 2013;103(12, Suppl 1):961–965. doi: 10.7196/samj.7253. [DOI] [PubMed] [Google Scholar]
- 14.Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:1–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye K., Schulz M.H., Long Q., Apweiler R., Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raczy C., Petrovski R., Saunders C.T., Chorny I., Kruglyak S., Margulies E.H., Chuang H.Y., Källberg M., Kumar S.A., Liao A. Isaac: ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics. 2013;29:2041–2043. doi: 10.1093/bioinformatics/btt314. [DOI] [PubMed] [Google Scholar]
- 20.Chen X., Schulz-Trieglaff O., Shaw R., Barnes B., Schlesinger F., Källberg M., Cox A.J., Kruglyak S., Saunders C.T. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 21.Xie C., Tammi M.T. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics. 2009;10:80. doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stokowy T., Garbulowski M., Fiskerstrand T., Holdhus R., Labun K., Sztromwasser P., Gilissen C., Hoischen A., Houge G., Petersen K. RareVariantVis: new tool for visualization of causative variants in rare monogenic disorders using whole genome sequencing data. Bioinformatics. 2016;32:3018–3020. doi: 10.1093/bioinformatics/btw359. [DOI] [PubMed] [Google Scholar]
- 23.McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., Zhuang J., Iyer S., Lin X.Y., Greven M.C., Kim B.H., Moore J., Pierce B.G., Dong X., Virgil D. Factorbook.org: a Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Res. 2013;41:D171–D176. doi: 10.1093/nar/gks1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kouwenhoven E.N., Oti M., Niehues H., van Heeringen S.J., Schalkwijk J., Stunnenberg H.G., van Bokhoven H., Zhou H. Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation. EMBO Rep. 2015;16:863–878. doi: 10.15252/embr.201439941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu J., Adli M., Zou J.Y., Verstappen G., Coyne M., Zhang X., Durham T., Miri M., Deshpande V., De Jager P.L. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minner F., Poumay Y. Candidate housekeeping genes require evaluation before their selection for studies of human epidermal keratinocytes. J. Invest. Dermatol. 2009;129:770–773. doi: 10.1038/jid.2008.247. [DOI] [PubMed] [Google Scholar]
- 32.Winer J., Jung C.K., Shackel I., Williams P.M. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 33.Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oti M., Falck J., Huynen M.A., Zhou H. CTCF-mediated chromatin loops enclose inducible gene regulatory domains. BMC Genomics. 2016;17:252. doi: 10.1186/s12864-016-2516-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li G., Ruan X., Auerbach R.K., Sandhu K.S., Zheng M., Wang P., Poh H.M., Goh Y., Lim J., Zhang J. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carvalho C.M., Lupski J.R. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 2016;17:224–238. doi: 10.1038/nrg.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bralten J., Arias-Vásquez A., Makkinje R., Veltman J.A., Brunner H.G., Fernández G., Rijpkema M., Franke B. Association of the Alzheimer’s gene SORL1 with hippocampal volume in young, healthy adults. Am. J. Psychiatry. 2011;168:1083–1089. doi: 10.1176/appi.ajp.2011.10101509. [DOI] [PubMed] [Google Scholar]
- 38.Kouwenhoven E.N., van Heeringen S.J., Tena J.J., Oti M., Dutilh B.E., Alonso M.E., de la Calle-Mustienes E., Smeenk L., Rinne T., Parsaulian L. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 2010;6:e1001065. doi: 10.1371/journal.pgen.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sethi I., Sinha S., Buck M.J. Role of chromatin and transcriptional co-regulators in mediating p63-genome interactions in keratinocytes. BMC Genomics. 2014;15:1042. doi: 10.1186/1471-2164-15-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orecchia V., Regis G., Tassone B., Valenti C., Avalle L., Saoncella S., Calautti E., Poli V. Constitutive STAT3 activation in epidermal keratinocytes enhances cell clonogenicity and favours spontaneous immortalization by opposing differentiation and senescence checkpoints. Exp. Dermatol. 2015;24:29–34. doi: 10.1111/exd.12585. [DOI] [PubMed] [Google Scholar]
- 41.Sano S., Chan K.S., DiGiovanni J. Impact of Stat3 activation upon skin biology: a dichotomy of its role between homeostasis and diseases. J. Dermatol. Sci. 2008;50:1–14. doi: 10.1016/j.jdermsci.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 42.Rozenberg J.M., Bhattacharya P., Chatterjee R., Glass K., Vinson C. Combinatorial recruitment of CREB, C/EBPβ and c-Jun determines activation of promoters upon keratinocyte differentiation. PLoS ONE. 2013;8:e78179. doi: 10.1371/journal.pone.0078179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips-Cremins J.E., Sauria M.E., Sanyal A., Gerasimova T.I., Lajoie B.R., Bell J.S., Ong C.T., Hookway T.A., Guo C., Sun Y. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montavon T., Thevenet L., Duboule D. Impact of copy number variations (CNVs) on long-range gene regulation at the HoxD locus. Proc. Natl. Acad. Sci. USA. 2012;109:20204–20211. doi: 10.1073/pnas.1217659109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lohan S., Spielmann M., Doelken S.C., Flöttmann R., Muhammad F., Baig S.M., Wajid M., Hülsemann W., Habenicht R., Kjaer K.W. Microduplications encompassing the Sonic hedgehog limb enhancer ZRS are associated with Haas-type polysyndactyly and Laurin-Sandrow syndrome. Clin. Genet. 2014;86:318–325. doi: 10.1111/cge.12352. [DOI] [PubMed] [Google Scholar]
- 46.Klopocki E., Lohan S., Brancati F., Koll R., Brehm A., Seemann P., Dathe K., Stricker S., Hecht J., Bosse K. Copy-number variations involving the IHH locus are associated with syndactyly and craniosynostosis. Am. J. Hum. Genet. 2011;88:70–75. doi: 10.1016/j.ajhg.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dathe K., Kjaer K.W., Brehm A., Meinecke P., Nürnberg P., Neto J.C., Brunoni D., Tommerup N., Ott C.E., Klopocki E. Duplications involving a conserved regulatory element downstream of BMP2 are associated with brachydactyly type A2. Am. J. Hum. Genet. 2009;84:483–492. doi: 10.1016/j.ajhg.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurth I., Klopocki E., Stricker S., van Oosterwijk J., Vanek S., Altmann J., Santos H.G., van Harssel J.J., de Ravel T., Wilkie A.O. Duplications of noncoding elements 5′ of SOX9 are associated with brachydactyly-anonychia. Nat. Genet. 2009;41:862–863. doi: 10.1038/ng0809-862. [DOI] [PubMed] [Google Scholar]
- 49.Zhang F., Lupski J.R. Non-coding genetic variants in human disease. Hum. Mol. Genet. 2015;24(R1):R102–R110. doi: 10.1093/hmg/ddv259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brocklehurst K., Philpott M.P. Cysteine proteases: mode of action and role in epidermal differentiation. Cell Tissue Res. 2013;351:237–244. doi: 10.1007/s00441-013-1557-2. [DOI] [PubMed] [Google Scholar]
- 51.Zeeuwen P.L. Epidermal differentiation: the role of proteases and their inhibitors. Eur. J. Cell Biol. 2004;83:761–773. doi: 10.1078/0171-9335-00388. [DOI] [PubMed] [Google Scholar]
- 52.Krunic A.L., Stone K.L., Simpson M.A., McGrath J.A. Acral peeling skin syndrome resulting from a homozygous nonsense mutation in the CSTA gene encoding cystatin A. Pediatr. Dermatol. 2013;30:e87–e88. doi: 10.1111/pde.12092. [DOI] [PubMed] [Google Scholar]
- 53.Blaydon D.C., Nitoiu D., Eckl K.M., Cabral R.M., Bland P., Hausser I., van Heel D.A., Rajpopat S., Fischer J., Oji V. Mutations in CSTA, encoding Cystatin A, underlie exfoliative ichthyosis and reveal a role for this protease inhibitor in cell-cell adhesion. Am. J. Hum. Genet. 2011;89:564–571. doi: 10.1016/j.ajhg.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeeuwen P.L., van Vlijmen-Willems I.M., Hendriks W., Merkx G.F., Schalkwijk J. A null mutation in the cystatin M/E gene of ichq mice causes juvenile lethality and defects in epidermal cornification. Hum. Mol. Genet. 2002;11:2867–2875. doi: 10.1093/hmg/11.23.2867. [DOI] [PubMed] [Google Scholar]
- 55.Egberts F., Heinrich M., Jensen J.M., Winoto-Morbach S., Pfeiffer S., Wickel M., Schunck M., Steude J., Saftig P., Proksch E., Schütze S. Cathepsin D is involved in the regulation of transglutaminase 1 and epidermal differentiation. J. Cell Sci. 2004;117:2295–2307. doi: 10.1242/jcs.01075. [DOI] [PubMed] [Google Scholar]
- 56.Horikoshi T., Arany I., Rajaraman S., Chen S.H., Brysk H., Lei G., Tyring S.K., Brysk M.M. Isoforms of cathepsin D and human epidermal differentiation. Biochimie. 1998;80:605–612. doi: 10.1016/s0300-9084(98)80013-8. [DOI] [PubMed] [Google Scholar]
- 57.Nishimura F., Naruishi H., Naruishi K., Yamada T., Sasaki J., Peters C., Uchiyama Y., Murayama Y. Cathepsin-L, a key molecule in the pathogenesis of drug-induced and I-cell disease-mediated gingival overgrowth: a study with cathepsin-L-deficient mice. Am. J. Pathol. 2002;161:2047–2052. doi: 10.1016/S0002-9440(10)64483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reinheckel T., Hagemann S., Dollwet-Mack S., Martinez E., Lohmüller T., Zlatkovic G., Tobin D.J., Maas-Szabowski N., Peters C. The lysosomal cysteine protease cathepsin L regulates keratinocyte proliferation by control of growth factor recycling. J. Cell Sci. 2005;118:3387–3395. doi: 10.1242/jcs.02469. [DOI] [PubMed] [Google Scholar]
- 59.Roth W., Deussing J., Botchkarev V.A., Pauly-Evers M., Saftig P., Hafner A., Schmidt P., Schmahl W., Scherer J., Anton-Lamprecht I. Cathepsin L deficiency as molecular defect of furless: hyperproliferation of keratinocytes and pertubation of hair follicle cycling. FASEB J. 2000;14:2075–2086. doi: 10.1096/fj.99-0970com. [DOI] [PubMed] [Google Scholar]
- 60.Schwarz G., Boehncke W.H., Braun M., Schröter C.J., Burster T., Flad T., Dressel D., Weber E., Schmid H., Kalbacher H. Cathepsin S activity is detectable in human keratinocytes and is selectively upregulated upon stimulation with interferon-gamma. J. Invest. Dermatol. 2002;119:44–49. doi: 10.1046/j.1523-1747.2002.01800.x. [DOI] [PubMed] [Google Scholar]
- 61.Toomes C., James J., Wood A.J., Wu C.L., McCormick D., Lench N., Hewitt C., Moynihan L., Roberts E., Woods C.G. Loss-of-function mutations in the cathepsin C gene result in periodontal disease and palmoplantar keratosis. Nat. Genet. 1999;23:421–424. doi: 10.1038/70525. [DOI] [PubMed] [Google Scholar]
- 62.Palungwachira P., Kakuta M., Yamazaki M., Yaguchi H., Tsuboi R., Takamori K., Ogawa H. Immunohistochemical localization of cathepsin L and cystatin A in normal skin and skin tumors. J. Dermatol. 2002;29:573–579. doi: 10.1111/j.1346-8138.2002.tb00182.x. [DOI] [PubMed] [Google Scholar]
- 63.Kato T., Takai T., Mitsuishi K., Okumura K., Ogawa H. Cystatin A inhibits IL-8 production by keratinocytes stimulated with Der p 1 and Der f 1: biochemical skin barrier against mite cysteine proteases. J. Allergy Clin. Immunol. 2005;116:169–176. doi: 10.1016/j.jaci.2005.03.044. [DOI] [PubMed] [Google Scholar]
- 64.de Castro M.A., Bunt G., Wouters F.S. Cathepsin B launches an apoptotic exit effort upon cell death-associated disruption of lysosomes. Cell Death Dis. 2016;2:16012. doi: 10.1038/cddiscovery.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.