Abstract
Epigenetic dysregulation has emerged as a recurring mechanism in the etiology of neurodevelopmental disorders. Two such disorders, CHARGE and Kabuki syndromes, result from loss of function mutations in chromodomain helicase DNA-binding protein 7 (CHD7LOF) and lysine (K) methyltransferase 2D (KMT2DLOF), respectively. Although these two syndromes are clinically distinct, there is significant phenotypic overlap. We therefore expected that epigenetically driven developmental pathways regulated by CHD7 and KMT2D would overlap and that DNA methylation (DNAm) alterations downstream of the mutations in these genes would identify common target genes, elucidating a mechanistic link between these two conditions, as well as specific target genes for each disorder. Genome-wide DNAm profiles in individuals with CHARGE and Kabuki syndromes with CHD7LOF or KMT2DLOF identified distinct sets of DNAm differences in each of the disorders, which were used to generate two unique, highly specific and sensitive DNAm signatures. These DNAm signatures were able to differentiate pathogenic mutations in these two genes from controls and from each other. Analysis of the DNAm targets in each gene-specific signature identified both common gene targets, including homeobox A5 (HOXA5), which could account for some of the clinical overlap in CHARGE and Kabuki syndromes, as well as distinct gene targets. Our findings demonstrate how characterization of the epigenome can contribute to our understanding of disease pathophysiology for epigenetic disorders, paving the way for explorations of novel therapeutics.
Keywords: CHARGE syndrome, Kabuki syndrome, chromodomain helicase DNA-binding protein 7 (CHD7), lysine (K) methyltransferase 2D (KMT2D), DNA methylation, Epigenetics
Introduction
Genes that function in epigenetic regulation (epigenes), including those involved in chromatin remodeling and histone modifications, are increasingly being identified in the etiology of a variety of neurodevelopmental disorders. Two such disorders include CHARGE syndrome [MIM: 214800], caused by heterozygous mutations in chromodomain helicase DNA-binding protein 7 (CHD7 [MIM: 608892]), and Kabuki syndrome [MIM: 147920] caused by heterozygous mutations in lysine (K)-specific methyltransferase 2D (KMT2D [MIM: 602113]).1, 2 CHARGE syndrome is characterized by Coloboma, Heart defects, Atresia of the choanae, Retardation of growth and development, Genital hypoplasia, and Ear abnormalities including deafness and vestibular disorders. Kabuki syndrome is characterized by a typical facial gestalt, postnatal growth deficiency, congenital heart defects, hearing loss and intellectual disability as well as skeletal, dermatoglyphic, genitourinary, and ophthalmologic anomalies (including coloboma). There is extensive clinical overlap between these two syndromes and clinical distinction can be particularly challenging in early life as the characteristic facial features of Kabuki syndrome often become apparent with age.3, 4 A molecular link between CHD7 and KMT2D function has been proposed via their known interaction with members of the WAR complex (WDR5 [WD-repeat protein 5], ASH2L [absent, small, homeotic discs-2-like], and also RBBP5 [retinoblastoma-binding protein-5]), which has been shown to be involved in histone methylation.5, 6 It has been suggested that CHD7 and KMT2D might regulate a common subset of genes via their interaction with the WAR complex, which might explain the overlapping features in CHARGE and Kabuki syndromes.7
We and others have previously demonstrated that loss of function (LOF) mutations in epigenes can be associated with specific patterns of DNA methylation (DNAm) alterations that constitute unique signatures.8, 9, 10 Specifically, unique DNAm signatures are observed in individuals harboring mutations in lysine-specific demethylase 5C (KDM5C [MIM: 314690]), which encodes an H3K4 demethylase and causes non-syndromic intellectual disability [MIM: 300534], DNA methyltransferase 1 (DNMT1 [MIM: 126375]), which cause adult-onset autosomal dominant cerebellar ataxia with deafness and narcolepsy (ADCA-DN [MIM: 604121]), and nuclear receptor binding SET domain protein 1(NSD1 [MIM: 606681]) which encodes a histone methyltransferase and cause Sotos syndrome [MIM:117550].8, 9, 10 Moreover, for NSD1 we have shown that genes encoding proteins in growth and neurodevelopmental pathways are highly represented in the DNAm signature reflecting the pathophysiology of the disorder.8 We hypothesized that comparison of genome-wide DNAm alterations in individuals with heterozygous LOF mutations in CHD7 and KMT2D, respectively, would identify two disease-specific DNAm signatures that would include common target genes and biological pathways, reflecting the clinical overlap of these two conditions, as well as distinct target genes reflecting divergent clinical features.
Here we analyze DNAm using whole blood from individuals with a clinical diagnosis of either CHARGE or Kabuki syndrome with LOF mutations in CHD7 (CHD7LOF) or KMT2D (KMT2DLOF), respectively. When compared to controls, CHD7LOF and KMT2DLOF demonstrate specific sets of differentially methylated CpGs, which we defined as two unique DNAm signatures. We show that the CHD7LOF and KMT2DLOF DNAm signatures both demonstrate high sensitivity and specificity, not only differentiating individuals with LOF mutations from controls but also differentiating individuals with CHD7LOF and KMT2DLOF from each other. As such, these DNAm signatures can offer a molecular means of distinguishing between CHARGE and Kabuki syndromes when clinical distinction is challenging. We also demonstrate that the specific DNAm signatures can be used to differentiate pathogenic mutations in CHD7 and KMT2D from benign sequence variants. Finally, analysis of the DNAm gene targets identified in the CHD7LOF and KMT2DLOF-specific DNAm signatures show gain of DNAm at homeobox A5 (HOXA5 [MIM: 142592]) in both signatures, which may account for some of the clinical overlap observed in CHARGE and Kabuki syndromes. There are also distinct DNAm alterations in each of the signatures that likely drive molecular pathways contributing to the distinct clinical features in these two syndromes.
Material and Methods
Research Participants
Discovery Cohort
Individuals with a clinical diagnosis of CHARGE syndrome and CHD7 LOF mutations (nonsense, frameshift mutations resulting in a premature stop, exonic deletions, and splice site mutations) were recruited through the Division of Clinical and Metabolic Genetics at the Hospital for Sick Children in Toronto, Ontario, Service de Génétique, Centre de Référence Anomalies du Développement de l’Ouest, CHU Poitiers, France and Our Lady’s Hospital for Sick Children in Dublin, Ireland (n = 19). Individuals with a clinical diagnosis of Kabuki syndrome and KMT2D LOF mutations (nonsense and frameshift mutations resulting in a premature stop) were recruited at the Hospital for Sick Children and the Center for Human Genetics, Inc. (n = 11). A detailed list of the specific CHD7 (GenBank: NM_017780.3) and KMT2D (GenBank: NM_003482.3) LOF mutations, designated CHD7LOF and KMT2DLOF can be found in Tables S1 and S2. Phenotypic information on individuals with CHD7LOF and KMT2DLOF is provided in Tables S3 and S4. The majority of the individuals in our Discovery and control cohorts are of European descent: CHD7LOF (16/19); CHD7LOF cohort controls (28/29); KMT2DLOF (10/11); KMT2DLOF cohort controls (9/11). These cohorts were used for the derivation of the DNAm signatures. Informed consent was obtained from all research participants according to the protocol approved by the Research Ethics Board of the Hospital for Sick Children (REB#0019980189).
Validation Cohort
Anonymized DNA samples from individuals with CHD7 or KMT2D sequence variants (n = 56) including pathogenic, likely pathogenic, and variants of uncertain significance (VUS) were obtained from PreventionGenetics, USA (Table S5). There was also one sample from an individual with a pathogenic mutation in lysine demethylase 6A (KDM6A c.2668_2669dupTA (p.Pro891Thrfs∗8); GenBank: NM_021140.2; [MIM: 300128]; Kabuki syndrome 2 [MIM: 300867]) in the anonymized cohort. All variants which differed from the reference sequences were interpreted using American College of Medical Genetics and Genomics (ACMG) Guidelines11 with slight modifications, which include previously unpublished de novo LOF variants being reported as likely pathogenic, instead of pathogenic as is in the ACMG guidelines.
Cohort with CHD7 and KMT2D Sequence Variants, Excluding LOF
Individuals with CHD7 (n = 13) or KMT2D (n = 10) VUS including missense and splice site variants were recruited from the same institutions as the CHD7 and KMT2D Discovery Cohorts (see above). Independent in silico prediction algorithms, namely PolyPhen-2,12 SIFT,13 Mutation Taster,14 and ESE finder15, 16 were used to evaluate the pathogenicity of the mutations in each case (Tables 1 and 2). Phenotypic information available for the individuals is provided in Tables S3 and S4. Individuals with CHD7 sequence variants underwent clinical classification utilizing the criteria established by Verloes17 and Hale18 (Table 1).
Table 1.
Classification of CHD7 Sequence Variants Utilizing DNAm Signatures, Clinical Criteria and In Silico Prediction Algorithms
| Sample ID | Mutation | Protein Change | Inheritance | Signature (positive/negative) | Verloes criteria (2005) | Hale proposed criteria (2015) | SIFT (score) | Mutation Taster (p value) | PolyPhen-2 prediction effect (score) |
|---|---|---|---|---|---|---|---|---|---|
| CHD7-20 | c.6322G>T | p.Gly2108Trp | de novo | positive | N | N∗ | Deleterious (0) | Disease Causing (1) | Probably Damaging (1) |
| CHD7-21 | c.3746G>A | p.Arg1249Gln | de novo | positive | N | I | Deleterious (0) | Disease Causing (1) | Probably Damaging (1) |
| CHD7-22 | c2751G>A | p. Thr917 = | inherited | negative | P | Y | ND | ND | ND |
| CHD7-23 | c.-15G>A | NDa | inherited | positive | A | Y | ND | ND | ND |
| CHD7-24 | c.4225G>A | p.Val1409Met | not maternal | positive | P | Y | Deleterious (0) | Disease Causing (1) | Probably Damaging (0.997) |
| CHD7-25 | c.5436C>G | p.Asp1812Glu | de novo | positive | I | I | Deleterious (0) | Disease Causing (1) | Probably Damaging (1) |
| CHD7-26 | c.5633A>G | p.Asp1878Gly | inherited | negative | I | I | Deleterious (0.01) | Disease Causing (1) | Benign (0.394) |
| CHD7-27 | c.5848G>A | p.Ala1950Thr | inherited | negative | I | I | Deleterious (0) | Disease Causing (1) | Benign (0.057) |
| CHD7-28 | c.6304G>T | p.Val2102Phe | inherited | negative | I | I | Deleterious (0.05) | Disease Causing (0.969) | Benign (0.389) |
| CHD7-29 | c.3566G>A | p.Arg1189His | inherited | negative | I | I | Deleterious (0) | Disease Causing (1) | Probably Damaging (1) |
| CHD7-30 | c2238+1del | NDb | de novo | positive | N | N∗ | ND | ND | ND |
| CHD7-31 | c.2049_2050insAAAGCA | p.Ala685_Lys686dup | inherited | negative | I | I | ND | ND | ND |
| CHD7-32 | c.6377A>T | p.Asp2126Val | inherited | negative | N | N | Tolerated (0.13) | Disease Causing (1) | Benign (0.014) |
CHARGE Criteria Legend: N, does not meet criteria; p, partial CHARGE; A, atypical CHARGE; Y, meets criteria;
I, insufficient information to classify; N∗, meets criteria if variant considered pathogenic; ND, not determined.
ESEfinder Splicing Prediction for CHD7-23 identifies loss of a SF2/ASF site.
ESEfinder Splicing Prediction for CHD7-30 identifies a gain of a SC35 site.
Table 2.
Classification of KMT2D Sequence Variants Utilizing DNAm Signatures and In Silico Prediction Algorithms
| Sample ID | Mutation | Protein change | Inheritance | Signature (positive/negative) | SIFT (score) | Mutation Taster(p value) | PolyPhen-2 prediction effect (score) |
|---|---|---|---|---|---|---|---|
| KMT2D-12 | c.15143G>A | p.Arg5048His | de novo | positive | Deleterious (0) | Disease Causing (1) | Probably Damaging (1) |
| KMT2D-13 | c.12028T>C | p.Ser4010Pro | inherited | negative | Tolerated (0.28) | Polymorphism (1) | Benign (0.001) |
| KMT2D-14 | c.16522-5_16522-4delTT | NDa | de novo | positive | ND | ND | ND |
| KMT2D-15 | c.15910A>G | p.Ile5304Val | inherited | negative | Tolerated (0.06) | Disease Causing (1) | Probably Damaging (0.997) |
| KMT2D-16 | c.15659G>A | p.Arg5220His | inherited | negative | Deleterious (0.01) | Disease Causing (1) | Probably Damaging (1) |
| KMT2D-17 | c.10256A>G | p.Asp3419Gly | inherited | negative | Tolerated (0.2) | Disease Causing (1) | Probably Damaging (1) |
| KMT2D-18 | c.8974G>A | p.Glu2992Lys | inherited | negative | Tolerated (0.08) | Disease Causing (1) | Probably Damaging (0.0996) |
| KMT2D-19 | c.8831A>G | p.Asn2944Ser | inherited | negative | Tolerated (0.27) | Polymorphism (1) | Benign (0.013) |
| KMT2D-20 | c.832G>A | p.Ala278Thr | inherited | negative | Tolerated (0.53) | Polymorphism (0.98) | Benign (0) |
| KMT2D-21 | c.682C>G | p.Arg228Gly | inherited | negative | Deleterious (0.02) | Polymorphism (0.883) | Benign (0.36) |
ND = not determined
ESEfinder Splicing Prediction for KMT2D-14 identifies loss of a SF2/ASF site.
Control DNA Samples
Age- and sex- matched controls for our discovery and validation cohorts were obtained from three sources (Table S6). These included the Simons Simplex Collection (SSC; Simons Foundation Autism Research Initiative). 19 The Hospital for Sick Children, and The University of Michigan (Dr. Gregory Hanna).20 The controls were screened using various neurodevelopmental assessments.19, 20
DNAm Data from Public Databases
Publically available Illumina HumanMethylation450 microarray data for an additional 162 blood controls, which were not expected to contain pathogenic mutations in either CHD7 or KMT2D, were downloaded from the Gene Expression Omnibus (GEO) data repository, chosen from individuals younger than 50 years of age in five GEO series (GEO: GSE32148, GEO: GSE40279, GEO: GSE41169, GEO: GSE46648, GEO: GSE53128; see Table S7).
DNAm Data Processing
DNA samples were converted using sodium bisulfite (EpiTect PLUS Bisulfite Kit, QIAGEN). The sodium bisulfite converted DNA was then hybridized to the Illumina Infinium HumanMethylation450 BeadChip Array to interrogate over 480,000 CpG sites in the human genome. For both the Discovery and Validation cohorts, cases and controls were randomized on the arrays (modified from21). Illumina Genome Studio software was used to perform control probe normalization and background subtraction and to extract DNAm values (β values) for each CpG, which represent the percentage of methylated cytosines. These β values ranged between 0 (no methylation) and 1 (full methylation). We excluded probes located on sex chromosomes, autosomal probes that cross-react with sex chromosome probes, non-specific probes, and probes targeting CpG sites within 5bp of a SNP that has a minor allele frequency above 1%.22, 23 Subsequent analyses were performed on the remaining 363,979 probes.
CHD7LOF and KMT2DLOF DNAm Signatures
Differential DNAm between individuals with LOF mutations and controls at individual CpGs was identified using three criteria. First, we applied regression modeling implemented in the limma Bioconductor package to detect statistically significant differences in DNAm (false discovery rate [FDR] corrected p value < 0.01) attributed to either CHD7LOF or KMT2DLOF versus controls, while accounting for sex and age (Figure S1) as confounding factors in the model design matrix.24 The limma models were applied to DNAm data logit-transformed into M-values.25 Second, to account for possible effects related to non-normal distribution of DNAm values at individual CpG sites, we used the non-parametric Mann-Whitney U test at each probe to detect statistically significant DNAm differences (FDR corrected p values < 0.01) between the respective Discovery Cohorts (CHD7LOF or KMT2DLOF) and the control groups. Third, to ensure robust results, statistically significant probes were additionally filtered for effect size. Delta beta (Δβ) was defined for each probe as the difference between average DNAm in each Discovery Cohort and its control group, respectively. We retained only those significant probes for which the DNAm difference (Δβ) between CHD7LOF or KMT2DLOF and their controls was greater than 0.10 (10% DNAm difference).
The choice of the significance level p < 0.01 and the 10% effect size threshold for both CHD7LOF and KMT2DLOF was guided by the volcano plots of the limma regression and the Mann-Whitney U test (Figure S2). The CpG sites that satisfied all three criteria defined the CHD7LOF and KMT2DLOF DNAm signatures, respectively (Tables S8 and S9).
Building Classification Models with the CHD7LOF and KMT2DLOF DNAm Signatures
Using the two DNAm signatures, we developed predictive models for scoring individuals with CHD7 or KMT2D sequence variants, based on their DNAm profiles. As the DNAm signatures for CHD7LOF and KMT2DLOF often appeared in groups corresponding to the same gene or region, the CpG sites were filtered for redundancy in order to be used as predictive data features for the respective models. We applied R caret software package to identify highly correlated CpGs and removed redundant data features (using the default threshold of 0.90 correlation in the findCorrelations function of caret). The remaining CpG sites were used to build the CHD7LOF and KMT2DLOF classification models.
We then built a support vector machine (SVM) model with linear kernel, using the non-redundant DNAm signatures as data features to predict the putative pathogenicity of each sequence variant. Model training was performed by the R kernlab software package via caret. The training set comprised the CHD7LOF or the KMT2DLOF and control samples from the Discovery Cohort. The model was set to return a quantitative predictive score between 0 and 1. The SVM scores were derived in kernlab using internally randomized cross-validation and thus exhibit slight variability. Therefore for each individual we determined the average score over 100 scoring trials. We then applied these models to samples from the Validation Cohort, another cohort with CHD7 or KMT2D sequence variants (excluding LOF mutations) and GEO controls. High scores indicate putative pathogenicity of sequence variants in CHD7 or KMT2D, and low scores suggest that the sequence variants are benign.
Assessment of Blood Cell Type Composition Effect
To ensure that the CHD7LOF and KMT2DLOF predictive models were not affected by variation in blood cell type composition, we examined 60 DNAm data samples from Reinius et al., 201226 (GEO: GSE35069). These data represent six healthy control whole-blood samples that were sorted into each of the following cell types: peripheral blood mononuclear cells (PBMC), granulocytes, neutrophils, eosinophils, as well as isolated cell populations (CD4+ T cells, CD8+ T cells, CD56+ NK cells, CD19+ B cells, CD14+ monocytes). The CHD7LOF and KMT2DLOF DNAm signature classification results were plotted for the cell-type samples from Reinius et al., 2012,26 together with the CHD7LOF or KMT2DLOF samples and the matching control samples from our Discovery Cohort (Figure S3). Our goal was to verify that none of the blood-cell subtypes were given high scores by either CHD7LOF or KMT2DLOF predictive models, confirming the lack of a confounding effect between blood-cell type composition and the DNAm signature-based predictions.
Differentially Methylated Regions in the CHD7 and KMT2D Discovery Cohorts
To find genomic regions with DNAm differences in CHD7LOF or KMT2DLOF, we used the bump hunting method,27 which strengthens the detection of regional differences by combining differential-methylation patterns across neighboring CpG sites.28 The bump hunting design matrix accounted for the potential confounding effects of the sex and age factors. The analysis initially considered CpGs with Δβ > 5% by magnitude between cases and controls as candidates for the differentially methylation regions (DMRs), with gaps no more than 500 bp between neighboring CpGs. Statistical significance was established using 1,000 randomized bootstrap iterations, as is recommended in the Bioconductor bumphunter package documentation when accounting for confounders. The resulting DMRs were post-filtered to retain only those with p value < 0.01 and average methylation difference Δβ > 10% by magnitude across the DMR. To further improve robustness, we also required these DMRs to comprise at least three neighboring CpGs, of which at least one has been already been included in the DNAm signature set for either CHD7LOF or KMT2DLOF, as described above (Tables S10 and S11). DMRs are presented using visualization methods adapted from DMRcate software (Figure S4).29
Functional Enrichment Analysis
To identify prominent functional enrichment patterns, we analyzed the list of differentially methylated CpGs in the context of wider genomic regions using GREAT.30 We retained only the Gene Ontology (GO) Biological Process31 functional categories for which at least three genes (including their genomic neighborhoods) were targeted by the CHD7LOF or KMT2DLOF specific DNAm signatures (Tables S12 and S13). The background set of probes to which the comparison was made was defined as the 363,979 autosomal CpGs used as the initial input to our analysis pipeline. All p values were FDR corrected.
DNAm Validation by Sodium Bisulfite Pyrosequencing
Differential DNAm between the CHD7LOF and KMT2DLOF individuals in the Discovery Cohorts and matching controls was validated for selected genomic loci using pyrosequencing assays. These assays were designed using QIAGEN Assay Design Software v1.0.6 to target-specific CpGs identified by the microarray experiment, as well as adjacent sites (Table S14). DNAm was assessed for the following CpG sites: cg01370449, cg04863892 and cg19759481 HOXA5, cg16787483; cg24626752 and cg09823859 (SLITRK5; SLIT and NTRK like family member 5 [MIM: 609680]); cg18546840 and cg18871253 (FOXP2; Forkhead box P2 [MIM: 605317]),) and cg15254671 (MYO1F; myosin IF [MIM: 601480]). Pyrosequencing was done using the PyroMark Q24 system and Pyrosequencing Gold Reagents (QIAGEN). Testing for a statistical difference between all groups was performed using a Kruskal-Wallis test (p < 0.0001).
Results
DNA Methylation Signatures for CHD7LOF and KDM2DLOF
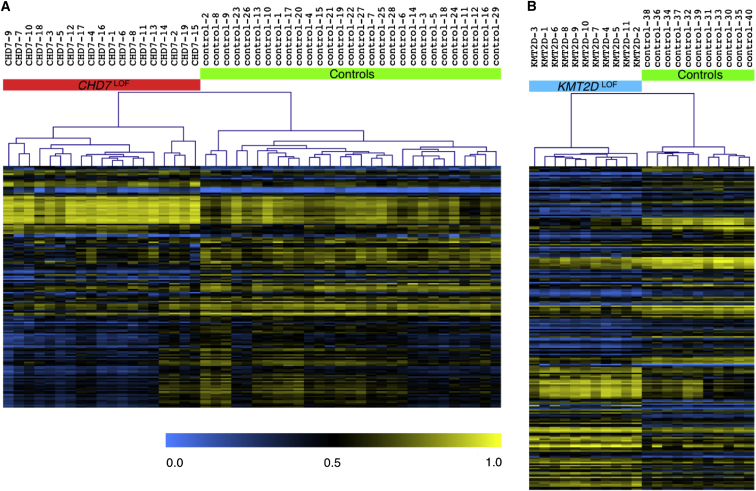
To determine whether LOF mutations in CHD7 (CHD7LOF) generate a specific pattern of DNAm alterations, we compared genome-wide DNAm patterns in 19 CHD7LOF individuals (CHD7LOF Discovery Cohort) and 29 matched controls. Volcano plots of the results of statistical criteria, specifically limma regression modeling, and non-parametric Mann-Whitney U tests, were used to determine a significance level p < 0.01 and an effect size of a 10% Δβ threshold (Figures S2A and S2B). We identified a set of 163 significant differentially methylated CpG sites for the CHD7LOF Discovery Cohort, defined as the CHD7LOF DNAm signature (Table S8). Unsupervised hierarchical clustering of the DNAm of each of the samples for these 163 CpGs sites clearly distinguished the CHD7LOF individuals from controls (Figure 1A).
Figure 1.
Hierarchical Clustering of the Discovery Cohorts Using the CHD7LOF and KMT2DLOF DNAm Signatures
The heatmap shows the unsupervised hierarchical clustering of (A) 19 CHD7LOF individuals and 29 matching controls samples, using only 163 differentially methylated CpG sites specific to CHD7LOF. The color gradient of the heatmap indicates the methylation level, from low (blue) to high (yellow). DNAm profiles fall into two separate clusters corresponding to CHD7LOF mutations (red) and controls (green). Euclidean distance metric is used in the clustering.
(B) 11 KMT2DLOF individuals and 11 matching controls samples, using only 221 differentially methylated CpG sites specific to KMT2DLOF. DNAm profiles fall into two separate clusters corresponding to KMT2DLOF mutations (blue) and controls (green).
Similarly, analysis of genome-wide DNAm patterns for 11 KMT2DLOF individuals (KMT2DLOF Discovery Cohort) and 11 matched controls identified a set of 221 significant differentially methylated CpGs for KMT2DLOF, defined as the KMT2DLOF DNAm signature (Table S9). Volcano plots of the limma regression and the Mann-Whitney U test for the KMT2DLOF Discovery Cohort were used to determine the significance level p < 0.01 and the 10% effect size (Figures S2C and S2D) to identify this set of CpGs. Unsupervised hierarchical clustering using the KMT2DLOF DNAm signature clearly distinguished the KMT2DLOF individuals from controls (Figure 1B).
Predictive Modeling Using DNAm Signatures for CHD7LOF and KMT2DLOF
We used the DNAm signatures for the CHD7 and KMT2D Discovery Cohorts to derive the classification signatures, by removing redundant data features. Removing redundancies from the initial collections of CpG sites (see Methods) resulted in a CHD7LOF DNAm classification signature comprising 75 CpGs and a KMT2DLOF DNAm classification signature comprised of 112 CpGs (Tables S8 and S9). Two SVM classification models were then built using the CHD7 and KMT2D Discovery Cohort samples as training sets and the corresponding non-redundant DNAm signatures as data features.
CHD7LOF and KMT2DLOF DNAm Signatures Are Independent of Blood Cell Type Composition
Both CHARGE and Kabuki syndromes can be associated with immune dysfunction although the sub-populations of T cells that are potentially altered represent a small fraction of the total lymphocytes.32, 33 To ensure that the predictive models were not affected by the variation in blood cell types, the two predictive models were applied to DNAm data from normal whole blood, peripheral blood mononuclear cells (PBMC), granulocytes, and isolated cell populations (CD4+ T cells, CD8+ T cells, CD56+ NK cells, CD19+ B cells, CD14+ monocytes).26 Prediction results demonstrated that all individual cell types received low predictive scores, placing them near other controls in our data (Figure S3). This suggests that the prediction of either CHD7LOF or KMT2DLOF status using the respective DNAm signatures is not influenced by a particular blood cell type within a sample. Clinical blood counts and differentials were available for 7 individuals with CHD7 mutations (3 LOF; 4 sequence variants) and for 2 individuals with KMT2D mutations (1 LOF and 1 sequence variant), and all were within the normal range.
Specificity of CHD7LOF and KMT2DLOF DNAm Classification Signatures
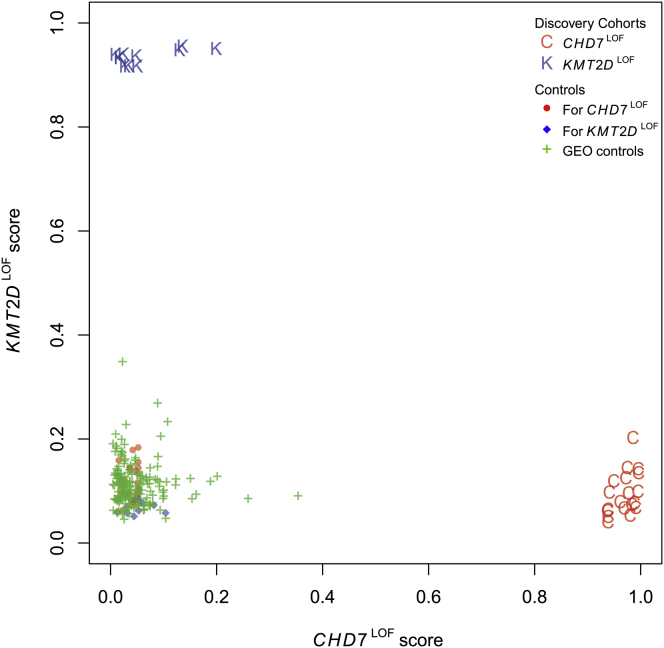
To quantify the specificity of the DNAm classification signatures and to confirm that the two models do not generate overlapping predictions, we applied the CHD7LOF predictive model to the KMT2DLOF cohort, and conversely the KMT2DLOF predictive model to the CHD7LOF cohort. None of the KMT2DLOF individuals or controls scored as putative CHD7LOF using the CHD7LOF classification signature, confirming that the CHD7LOF predictive model was specific to CHD7LOF (Figure 2). Similarly, none of the CHD7LOF individuals or controls scored as KMT2DLOF, confirming the specificity of the second model (Figure 2). Next we assessed the specificity of the predictive analysis on the collection of control blood DNAm data extracted from the GEO repository. All 162 GEO samples had low prediction scores for both CHD7LOF and KMT2DLOF, i.e., they were predicted not to have pathogenic mutations in CHD7 or KMT2D by the corresponding models, demonstrating 100% specificity of the DNAm signatures (Figure 2).
Figure 2.
Specificity of the CHD7LOF and KMT2DLOF DNAm Classification Signatures
The plot shows the predictions for all samples from the original Discovery Cohorts, as well as for 162 normal blood samples extracted from the GEO repository. The x axis shows the predictive scores generated from the CHD7LOF-specific predictive model derived using the CHD7LOF individuals and matching controls. The y axis shows the predictive score of the KMT2DLOF-specific predictive model derived using the KMT2DLOF individuals and matching controls. Importantly, using the KMT2DLOF-specific model all 19 CHD7LOF (red C) received low scores, along with all CHD7LOF matching controls (red circles) and all GEO samples (green crosses). Similarly, using the CHD7LOF-specific model all 11 KMT2DLOF (blue K) received low scores, along with all KMT2DLOF matching controls (blue diamonds) and all GEO samples (green crosses).
Validation of CHD7LOF and KMT2DLOF DNAm Classification Signatures Using a Blinded Cohort
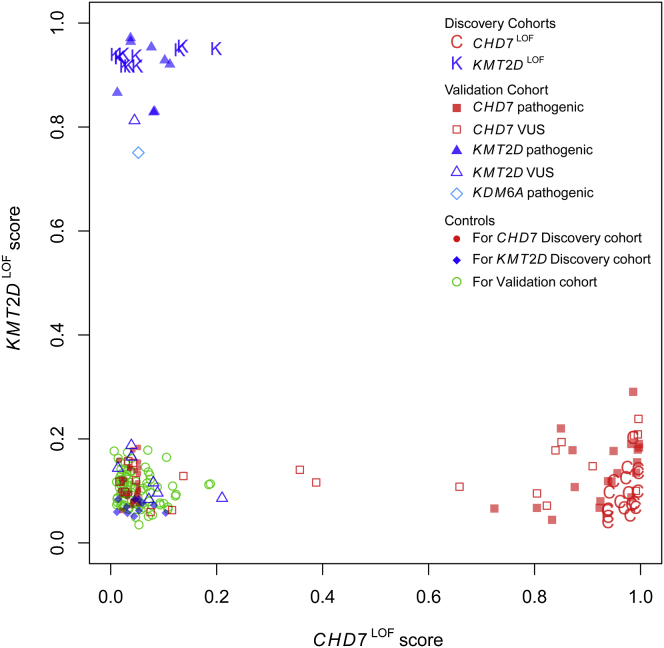
Next we applied the predictive models for CHD7LOF and KMT2DLOF DNAm classification signatures in a blinded fashion to a cohort of individuals with either CHD7 or KMT2D mutations classified as pathogenic, likely pathogenic or variants of unknown significance (VUS; Validation Cohort). After the prediction scores were generated, the mutation classification was unblinded for comparison to the reported variant classification (Figure 3). The CHD7LOF DNAm signature predictive model generated high scores for all of the CHD7 mutation samples (n = 20) that were classified as pathogenic or likely pathogenic. The same was true of the KMT2DLOF model (n = 8). Thus, both predictive models demonstrated the ability to correctly classify pathogenic mutations in their respective genes, CHD7 or KMT2D (100% sensitivity), while giving low scores to samples with mutations in the other gene and to all controls in the Validation Cohort (100% specificity).
Figure 3.
Validation of CHD7LOF and KMT2DLOF DNAm Classification Signatures on a Blinded Cohort
We derived the scores for each sample using the two predictive models built for the CHD7LOF and KMT2DLOF DNAm classification signatures (x axis and y axis, respectively; see Figure 2), for a validation set of DNAm samples. This set included both pathogenic mutations and VUS in CHD7 (red squares), KMT2D (blue triangles) and KDM6A (turquoise diamond). The mutation and their pathogenicity were initially blinded and were revealed only after the prediction scores were determined. Importantly, all CHD7 mutations received low scores by the KMT2D-specific predictive model, and vice versa. Pathogenic mutations in CHD7 (filled red squares) received high scores from the CHD7LOF model, and pathogenic mutations in KMT2D (filled blue triangles) received very high scores from the KMT2DLOF model. Interestingly, a pathogenic mutation in the Kabuki-associated gene KDM6A also received a very high score from the KMT2DLOF model, indicating a potential methylation-signature overlap between these two genes.
The Validation Cohort included one individual with a mutation in KDM6A, a second gene in which mutations are associated with Kabuki syndrome.34 Interestingly, the KDM6A mutation received a high score from the KMT2DLOF DNAm classification signature model, indicating that it is more similar to KMT2DLOF than controls or CHD7LOF.
Analysis of Sequence Variants in CHD7 and KMT2D Using Gene-Specific DNAm Classification Signatures
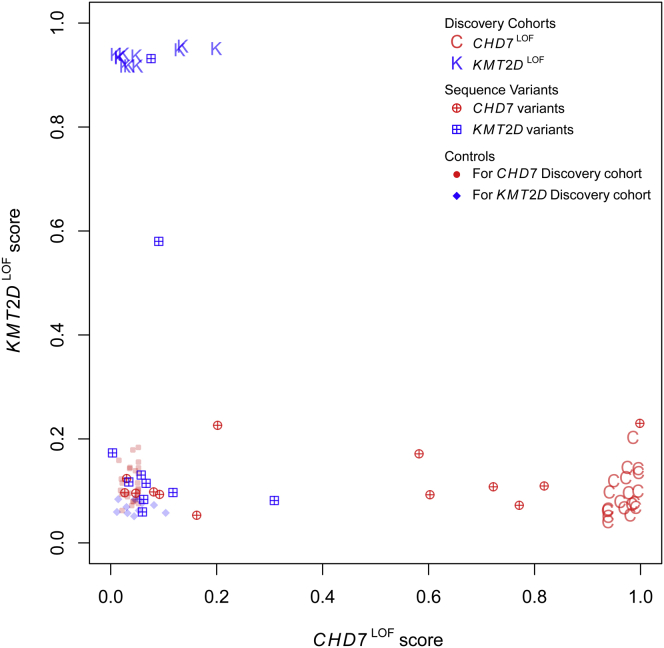
The predictive models were next applied to cohorts of individuals with CHD7 and KMT2D sequence variants of uncertain significance (VUS) that included missense and splice site mutations (Figure 4; Tables S1 and S2). Phenotypic information for these individuals is summarized in Tables S3 and S4. Of 13 individuals with VUS in CHD7, 6 clustered with the CHD7LOF Discovery Cohort, suggesting that these variants (CHD7-20, 21, 23, 24, 25, and 30) are pathogenic, and 7 clustered with the control samples suggesting that these variants (CHD7-22, 26, 27, 28, 29, 31, and 32) are benign (Figure 4). The 6 variants predicted to be pathogenic included 2 splice site mutations (one inherited and one de novo) and 4 missense mutations (3 de novo, one inheritance unknown). The inherited splice site mutation was identified in a parent with no overt clinical manifestations of CHARGE syndrome. The parent had a normal ophthalmology exam and did not have a history of hearing loss, but did not undergo imaging of the middle/inner ear. All of the 7 variants predicted to be benign are inherited missense mutations. Of the 10 individuals with VUS in KMT2D, 1 clustered with the KMT2DLOF Discovery Cohort (KMT2D-12), suggesting that this variant is pathogenic and 8 clustered with the control samples suggesting that these variants (KMT2D-13, 15, 16, 17, 18, 19, 20, and 21) are benign. The KMT2D-14 variant had an intermediate score between the KMT2DLOF Discovery Cohort and the control samples. The KMT2D-12 variant predicted to be pathogenic is a de novo missense mutation. The KMT2D-14 variant with the intermediate score is a de novo splice site mutation. The 8 variants predicted to be benign are all inherited missense mutations.
Figure 4.
Sequence Variants in CHD7 and KMT2D Sorted Using the CHD7LOF and KMT2DLOF DNAm Classification Signatures
We derived the scores for each individual using the two models generated for CHD7LOF and KMT2DLOF DNAm classification signatures (x axis and y axis, respectively; see Figure 2), for a set of 13 mutation variants in CHD7 (red crossed circles) and 10 mutation variants in KMT2D (blue crossed squares). The details of the sample classification are shown in Tables 1 and 2.
Diagnostic Classification by DNAm Signature versus Clinical Criteria
A comparison of the CHD7LOF DNAm classification signature prediction (pathogenic or benign) to the Verloes17 diagnostic criteria for CHARGE syndrome and that recently proposed by Hale18 revealed discordant results for four individuals within our CHD7 variant cohort (Table 1). Specifically, three individuals who did not meet criteria for a diagnosis of CHARGE using either the Verloes or Hale criteria (CHD7-20, CHD7-21, and CHD7-30) were determined to have pathogenic CHD7 mutations when classified utilizing the CHD7LOF DNAm classification signature. Of note, individual CHD7-21 did not have sufficient clinical information to be classified using the Hale criteria. All three of these mutations were de novo. The fourth individual (CHD7-22) was classified as “partial” CHARGE using the Verloes criteria and met a clinical diagnosis for CHARGE using the Hale criteria but the CHD7LOF DNAm classification signature predicted this to be a benign CHD7 variant. This variant was an inherited synonymous mutation.
Comparison of the CHD7LOF DNAm classification signature predictions to independent in silico prediction algorithms, specifically PolyPhen-212, SIFT,13 Mutation Taster,14 and ESE finder15, 16 also revealed discordant results for 5 individuals with missense mutations (CHD7-26, CHD7-27, CHD7-28, CHD7-29, and CHD7-32). Specifically, 5 results were discordant with Mutation Taster, 4 results were discordant with SIFT and 1 result was discordant with PolyPhen-2. However, none of these predictive algorithms were in complete agreement with each other.
Consensus clinical diagnostic criteria for Kabuki syndrome have not been established. Individuals with this condition have characteristic facial features, in addition to a variety of congenital anomalies (intellectual disability, short stature, persistent fingertip pads, and skeletal anomalies), which suggest the diagnosis. Individual KMT2D-12 who clustered with the KMT2DLOF Discovery Cohort when the KMT2DLOF DNAm classification signature was applied had a clinical diagnosis of Kabuki syndrome (established by O.C.; Table 2). Individual KMT2D-14 who had an intermediate score between the KMT2DLOF Discovery Cohort and the control samples also had a clinical diagnosis of Kabuki syndrome (established by R.M.-L.). Notably, both individuals had typical facial features in addition to other characteristic clinical features.
Genes with Differential DNAm in the CHD7LOF and KMT2DLOF DNAm Signatures
The majority of CpG sites differentially methylated for CHD7LOF and KMT2DLOF were specific to each DNAm signature. The CHD7LOF CpG sites were located within the bodies or promoter regions (up to 1500 bp upstream of the transcription start site) of 86 known RefSeq genes (Table S8).35 Several genes demonstrated differential DNAm at multiple probes, including FOXP2, HOXA transcript antisense RNA, myeloid-specific 1 (HOTAIRM1), homeobox A1 (HOXA1 [MIM: 142955]), homeobox A6 (HOXA6 [MIM: 142951]), HOXA5 and SLITRK5. Analysis of differentially methylated regions, using bump hunting,27 detected consistent patterns of DNAm gain or loss in the vicinity of these genes (Table S10). Enrichment analysis of the probes within the CHD7LOF DNAm signature confirmed a statistically significant over-representation in GO Biological Process31 categories related to growth and embryonic development of the brain, ear, digestive, endocrine, and neural systems, as well as additional functional categories that are highly relevant to the phenotypic features associated with CHARGE syndrome (Table S12).
The KMT2DLOF CpG sites were located within the bodies or promoter regions of 105 known genes (Table S9). Among these, multiple CpG sites mapped to HOTAIRM1, homeobox A4 (HOXA4 [MIM: 142953]), HOXA5, and SLITRK5. These genes were also identified as corresponding to KMT2DLOF associated differentially methylated regions detected by bump hunting (Table S11). Enrichment analysis of the probes within the KMT2DLOF DNAm signature confirmed a statistically significant over-representation in GO biological processes categories related to skeletal, lung and digestive system development; as well there were functional categories similar to those observed for the CHD7LOF signature including pattern specification and embryonic morphogenesis (Table S13).
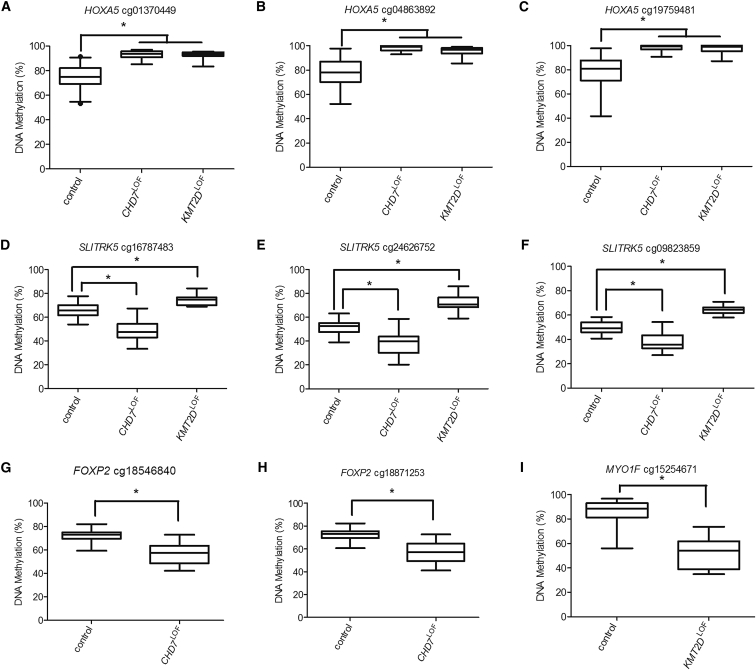
Due to the presence of both overlapping and clinically distinct features seen in CHARGE and Kabuki syndromes, we examined the CHD7LOF and KMT2DLOF DNAm signatures for both shared and distinct CpG targets. There were 14 CpG sites shared by the CHD7LOF and KMT2DLOF DNAm signatures: 11 corresponding to HOXA5 and 3 to SLITRK5. The HOXA5 CpGs demonstrated a gain of DNAm in both the CHD7LOF and KMT2DLOF signatures. DNAm was validated using sodium bisulfite pyrosequencing for 3 CpG sites in the HOXA5 promoter (cg01370449, cg04863892, and cg19759481; Figures 5A–5C). The average gain of DNAm for the three sites is CHD7LOF was 18%, 20%, and 20% respectively, compared to 18%, 18%, and 19% for KMT2DLOF. Furthermore, the analysis of DMRs for CHD7LOF and KMT2DLOF using bump hunting also confirmed gain of DNAm at overlapping DMRs in the vicinity of HOXA5 (Figure S4A). The shared CpG sites near SLITRK5 demonstrated DNAm changes in opposite directions in the two signatures, with a loss of DNAm in CHD7LOF but gain of DNAm in KMT2DLOF. For SLITRK5, 3 CpG sites (cg16787483, cg24626752, and cg09823859; Figures 5D–5F) were validated using pyrosequencing. An average loss of DNAm of 20%, 14%, and 12% in the CHD7LOF samples was confirmed, whereas an average gain of DNAm of 21%, 24%, and 24% was confirmed in KMT2DLOF samples. This finding was confirmed by bumphunting in the overlapping DMRs (Figure S4B).
Figure 5.
Targeted Sodium Bisulfite Pyrosequencing Validation of DNAm Alterations in CHD7 and KMT2D Discovery Cohorts
(A–C) DNAm was assessed for three CpG sites in the promoter of HOXA5 (cg01370449, cg04863892, and cg19759481). The gain of DNAm for the three sites in CHD7LOF: 18%, 20%, and 20%. For KMT2DLOF there was also a gain of DNAm: 18%, 18%, and 19%, respectively. Both the CHD7LOF and KMT2DLOFgroup are statistically different from the controls for all three probes, but not from each other.
(D–F) DNAm was assessed for three CpG sites in the gene body of SLITRK5 (cg16787483, cg24626752, and cg09823859). A loss of DNAm of 20%, 14%, and 12% in the CHD7LOF samples and a gain of DNAm of 21%, 24%, and 24% in KMT2DLOF samples are shown. Both the CHD7LOF and KMT2DLOFgroup are statistically different from the controls for all three probes, and from each other.
(G and H) DNAm was analyzed for FOXP2 (cg18546840 and cg18871253) in CHD7LOF, which had a 15% loss of DNAm compared to controls.
(I) DNAm was analyzed for MYO1F (cg15254671) in KMT2DLOF, which had a loss of DNAm of 33% compared to controls. Testing for a statistical difference between all groups was performed using a Kruskal-Wallis test; ∗p < 0.0001.
DNAm was also validated for CpG sites in genes specific to each DNAm signature. For the CHD7LOF DNAm signature two CpG sites in FOXP2 (cg18546840 and cg18871253) were selected because of the critical role of FOXP2 in brain and craniofacial development.36, 37 Both of these CpG sites exhibit an average loss of DNAm of 15% (Figures 5G and 5H). For the KMT2DLOF DNAm signature a CpG site in MYO1F (cg15254671) with an average 33% loss of DNAm (Figure 5I) was validated. This CpG site, along with four others in the same CpG island (spanning exon 23 to 24), carry chromatin marks classified as a promoter or enhancer in different cell types.38, 39
Discussion
We have identified two unique DNAm signatures associated with loss of function mutations in CHD7 (CHD7LOF) and KMT2D (KMT2DLOF), further enhancing our understanding of the critical role of epigenetic dysregulation in neurodevelopmental disorders. These two gene-specific signatures demonstrate 100% specificity and 100% sensitivity, enabling differentiation between pathogenic and benign mutations in CHD7 and KMT2D, respectively. That is, these DNAm signatures can function as tools to classify variants of unknown significance (VUS) in these genes. Interestingly, comparisons of the differentially methylated sites within the CHD7LOF and KMT2DLOF DNAm signatures provide evidence that CHD7 and KMT2D regulate common biological pathways likely reflecting the clinical overlap between CHARGE and Kabuki syndromes.
Functional Roles of CHD7 and KMT2D
Mutations in CHD7 were initially identified as the etiology of CHARGE syndrome in 2004.1 Since that time, much has been learned regarding the biological function of this gene. CHD7, an ATP-dependent chromodomain helicase chromatin remodeling protein is involved in the formation of several large protein complexes that regulate the movement of nucleosomes along DNA, and as such affects the activity of numerous signaling pathways during embryonic development.40 These CHD7-containing protein complexes bind to DNA at specific sites, the majority of which overlap with regulatory elements such as gene promoters or enhancers.41, 42 The epigenetic effects of CHD7 on chromatin and gene regulation appear to vary both temporally and spatially, depending largely upon the function of the protein complex with which it interacts (for review see 43). CHD7 is expressed in embryonic stem cells; its expression becomes restricted to specific tissues, including the brain, eye, heart, and ear, during differentiation.44 Gene-expression studies in mouse embryos carrying a homozygous deletion of Chd7 demonstrate significant expression differences in many genes important for brain development.45
Mutations in KMT2D were initially identified as the major cause of Kabuki syndrome in 2010.2 KMT2D, a lysine methyltransferase, adds a tri-methyl mark to histone 3 lysine 4 (H3K4), which promotes gene expression by facilitating open chromatin conformation.46 KMT2D belongs to the family of mixed-lineage leukemia (MLL) genes, which are involved in controlling other genes essential for embryogenesis, including the HOX genes.47, 48 In a mouse model of Kabuki syndrome, heterozygous disruption of Kmt2d is associated with a genome-wide reduction of H3K4 tri-methylation.49 Histone methylation patterns in cardiac tissue from embryos with homozygous deletions of Kmt2d show a global decrease in H3K4 mono- and di-methylation, when compared to controls, suggesting that KMT2D functions at both enhancers and gene promoters to regulate gene expression.47
Utility of CHD7LOF and KMT2DLOF DNAm Signatures
Diagnosis of CHARGE and Kabuki syndromes in the clinical setting can be challenging. CHARGE syndrome in particular has been shown to have extensive intra- and interfamilial clinical variability.50, 51 Since the first description of CHARGE syndrome in 1981, diagnostic criteria for this condition have undergone several iterations, reflecting a broadening of the phenotype.17, 18, 52, 53 The challenges in establishing a clinical diagnosis of CHARGE syndrome using existing diagnostic criteria (Table 1) are highlighted by data for four individuals in the current study, that demonstrate incongruities between the criteria-based clinical classifications of CHARGE syndrome and our CHD7LOF DNAm classification signature predictions. Specifically, three individuals who did not meet clinical criteria for a diagnosis of CHARGE syndrome were identified to have the DNAm signature (predicting pathogenicity) and one individual who did meet clinical criteria for a diagnosis of CHARGE syndrome did not have the DNAm signature (predicting a benign variant). Existing in silico prediction tools often provide different and contradictory results as seen for several of the individuals in our CHD7 variant cohort.54 We propose that our CHD7LOF DNAm classification signature could be used as a functional molecular test to aid in the interpretation of the pathogenicity of CHD7 sequence variants, providing a valuable tool to facilitate in the diagnosis of CHARGE syndrome.
Similarly, the KMT2DLOF DNAm classification signature could be used to assess the pathogenicity of KMT2D sequence variants (Table 2). For example, in the case of the two individuals with KMT2D VUS, the DNAm classification signature provided functional molecular validation for a suspected clinical diagnosis when sequence analysis did not provide a definitive answer.
For individuals reported to have putative pathogenic variants in CHD7, 17% do not fulfill diagnostic criteria for CHARGE syndrome, further demonstrating the phenotypic variability and the imperfect alignment between clinical classification and molecular test data.55, 56 Further, although pathogenic mutations in CHD7 are identified in 90% of individuals fulfilling Blake’s criteria, only 65%–70% of individuals with typical or suspected CHARGE syndrome are identified to have a pathogenic CHD7 mutation.55, 57 Similarly, only ∼70% of individuals with a clinical diagnosis of Kabuki syndrome are identified to have pathogenic mutations in KMT2D or KDM6A58. It is not clear whether the missing 30% of mutations occur in promoters or enhancers not identified by current sequencing techniques or if there might be locus heterogeneity. The CHD7LOF and KMT2DLOF DNAm signatures could provide a means of enhancing the molecular diagnostic rates for these syndromes, because they could detect loss of function mutations that might not be detected by current sequence-based testing. In future, further validation of the clinical utility of these signatures could be derived from RNA sequence-based functional assays.
The utility of DNAm signatures for the epigenes CHD7 and KMT2D constitute a generalization of our previous work with epigenes, in which we demonstrated the utility of a DNAm signature in classifying VUS in NSD1 associated with Sotos syndrome.8 The NSD1-specific DNAm signature also enabled molecular distinction between Sotos syndrome and the clinically overlapping overgrowth condition Weaver syndrome [MIM: 277590] resulting from mutations in enhancer of zeste, Drosophila, homolog 2 (EZH2 [MIM: 601573]), another epigene encoding a histone methyltransferase.59, 60
Overlapping Molecular Mechanisms for CHARGE and Kabuki Syndromes
Clinical overlap between CHARGE and Kabuki syndromes was initially reported by Ming et al. (2003) over 10 years ago.3 Since then, several reports have demonstrated the difficulty in distinguishing between these two conditions, especially in infancy when the typical facial gestalt of Kabuki syndrome might not yet be apparent.4, 61, 62 Overlapping clinical features include postnatal growth retardation, cleft lip/palate, hearing loss, congenital heart defects, urogenital malformations, developmental delay, and intellectual disability. As well, ocular coloboma, which is a major diagnostic criterion for CHARGE syndrome, has occasionally been reported in individuals with Kabuki syndrome.4, 61, 62 The genes associated with CHARGE (CHD7) and Kabuki (KMT2D and KDM6A) syndromes all play a role in chromatin remodeling. Evidence supporting a functional connection between CHD7 and KMT2D comes from studies showing that both these protein interact with members of the WAR complex (WDR5, RBBP5, and ASH2L).6, 7, 63 On the basis of these findings, it was suggested that CHD7 and KMT2D regulate a common subset of genes.7 Our finding that the unique CHD7LOF and KMT2DLOF DNAm signatures have common CpG targets, specifically within HOXA5 and SLITRK5, provides additional evidence for molecular mechanistic connections between CHARGE and Kabuki syndromes and also provides important functional data to explain the pathophysiologic basis of the overlapping features in these two conditions.
Our previous work on NSD1 demonstrated the relevance of the DNAm targets in blood to our understanding of the pathophysiology of the disease. In the case of NSD1 loss of function mutations, DNAm gene targets were enriched for neural and cellular development pathways, reflecting the cardinal features of Sotos syndrome (overgrowth and developmental delay).8 Our finding that the CHD7 and KMT2D DNAm targets appear to relate to genes involved in the embryonic development of cell types and tissues demonstrating malformations in CHARGE and Kabuki syndromes further supports the functional significance of these DNAm signatures and provides valuable data relevant to the pathophysiology of these conditions.
Both CHD7 and KMT2D have been previously linked to expression of various homeobox-containing genes. The homeobox (HOX) genes encode highly conserved transcription factors that are expressed in a spatially and temporally regulated manner during development.64 HOX expression during development is tightly regulated in part by chromatin structure and epigenetic modifications, including DNAm.65 In the mouse, the developing neural tube of Chd7−/− embryos also showed altered expression of other homeobox genes, such as orthodenticle homeobox 2 (Otx2) and gastrulation brain homeobox 2 (Gbx2).66 Chromatin immunoprecipitation assays have shown that CHD7 binds to other chromatin-associated proteins at genomic sites within the HOXA1, HOXA5, and HOXA6.41 In fibroblast cells from individuals with loss of function KMT2D mutations, targeted expression analysis of homeobox C6 (HOXC6 [MIM: 142972]) showed a decreased transcript level compared to controls.67 KMT2D has also been shown to bind to DNA in the HoxA cluster in mouse embryonic-stem-cell-derived cardiomyocytes.47
The finding that HOXA5 is regulated by both CHD7 and KMT2D provides us with further clues to the pathophysiology of CHARGE and Kabuki syndromes. In both the CHD7LOFand KMT2DLOF DNAm signatures, a gain in DNAm was observed at the HOXA5 promoter. Functional studies in a wide range of vertebrate species have established the conserved roles of HOX genes as transcription factors that regulate axial patterning of the developing embryo.68 A study of the mouse promoter HoxA5 showed that it is unmethylated in specific embryonic tissues, including liver, intestine, and spleen.69 In those same tissues postnatally, the HoxA5 promoter becomes completely (spleen), or partially (intestine) unmethylated, or remains unmethylated (liver). Therefore, DNAm appears to be a crucial element in the developmental regulation of Hox activity postnatally. Our findings of a 20% increase in DNAm is likely to be functionally relevant given data from a mouse model wherein a high-fat diet is associated with a 25% increase in DNA methylation of the Hoxa5promoter in adipose tissue resulting in a significant reduction in mRNA and protein expression of this gene.70
Some of the clinical features shared by CHARGE and Kabuki syndromes could be mediated by reduced expression of HOXA5. Based on functional studies of HOXA5 in mice, we propose that these could include growth deficiency, skeletal and limb anomalies, renal dysgenesis, and neural development.71, 72, 73, 74, 75 In one mouse model with heterozygote Hoxa5 truncating mutations, the mutant mice were phenotypically indistinguishable from their wild-type littermates but on further evaluation of skeletal morphology, an increased rate of rib anomalies/vertebral defect were identified.76 Of interest, individuals with CHARGE and Kabuki syndromes demonstrate vertebral anomalies and individuals with CHARGE syndrome can have missing ribs.77 Additional studies focused on the brain and behavior of these mice could demonstrate important changes in brain function and/or behavior given a recent report characterizing the expression profile and the neuroanatomical localization of HOXA5 in the fetal, postnatal, and adult brain.75 They identified Hoxa5 transcripts in the medulla oblongata and the pons from fetal to adult stages, and in the thalamus and the cortex from postnatal stages through adulthood. They also demonstrated that Hoxa5 is transcribed in the adult cerebellum and that the HOXA5 protein is present in all the Hoxa5-expressing hindbrain nuclei in adulthood. This suggests that HOXA5 in these nuclei might be required for processes beyond the early developmental patterning and neuronal migration phases, including axonal growth and synapse formation during circuit establishment, refinement of neural circuits during early postnatal life in response to environmental cues, or adult synaptic plasticity.75 Interestingly, in each of the DNAm signatures there are multiple HOXA genes that have differential DNAm, including HOTAIRM, HOXA1, and HOXA6 in CHD7LOF and HOXA4 in KMT2DLOF.
HOTAIRM is a long non-coding RNA (lncRNA), which has been found to interact with different chromatin-modifying complexes.78 Recent work by Wang and Dostie (2016) found that HOTAIRM1 contributes to three-dimensional changes in chromatin organization required for the temporal collinear activation of HOXA genes.79 Their findings also demonstrate that lncRNAs derived from the HOTAIRM1 gene can activate and/or repress HOXA gene expression in different cell types. Our finding that CHD7LOF is associated with differential DNAm at HOTAIRM suggests multiple layers of epigenetic dysregulation impacting cell-type-specific HOXA gene expression in CHARGE syndrome. HOTAIRM1 is also known to modulate β-integrin signaling, which has been shown to be critical for the expansion of neural stem cells in the development of the cerebral cortex of model organisms.80, 81 These data suggest specific pathophysiologic mechanisms that could account for neurodevelopmental anomalies in CHARGE syndrome.
A second gene, SLITRK5, also had shared differentially methylated CpG sites in the gene body and surrounding shores in each of the DNAm signatures with a loss of DNAm in CHD7LOF and a gain of DNAm in KMT2DLOF. The SLITRK family encodes transmembrane proteins which function at synapses82, 83. SLITRK5 has been shown to function in synaptic adhesion and in tropomyosin receptor kinase B (TRKB) signaling upon brain-derived neurotrophic factor (BDNF) stimulation.84 In Chd7−/− mouse whole embryos, expression of SLITRK5 is decreased.45 Mice with homozygous deletion of Slitrk5 develop an over-grooming phenotype and deficiencies in corticostriatal transmission as well as a reduction in glutamate receptor subunits.85 We expect that dysregulation of SLITRK5, either gain or loss, could impact neuronal development due to its role in many tightly regulated processes at the synapse.
Epigenetic Targets Specific to CHARGE and Kabuki Syndromes
One of the genes specific to the CHD7LOF DNAm signature is FOXP2. In mice, FOXP2 has been shown to be expressed in the motor related circuitry in the brain, which controls craniofacial development including muscles of the face and striatal brain development.36, 37 In mouse embryonic stem cells and neural progenitors, chromatin immunoprecipitation assays show that CHD7 binds to the Foxp2 promoter.41 As mutations in FOXP2 have been identified in individuals with speech and language deficits,86 it is possible that altered FOXP2 expression might contribute to the speech and language difficulties in individuals with CHARGE syndrome.
Specific to the KMT2DLOF DNAm signature are CpG sites within the gene body of MYO1F. These sites are located in a CpG island carrying chromatin marks which classify the region as regulatory, specifically as a promoter or enhancer in different cell types.38, 39 This unconventional myosin is expressed in the inner ear and heterozygous missense mutations in this gene have been identified in individuals with hereditary hearing loss.87
Overlapping Epigenetic Targets for KMT2D and KDM6A in Kabuki Syndrome
Our observation that the single sample from with a KDM6A pathogenic mutation in the Validation cohort clustered with the pathogenic KMT2D samples suggests that these two genes regulate overlapping sets of genes. This is in keeping with evidence from earlier studies that showed these two proteins share HOX targets.88 KDM6A, a histone demethylase, has been shown to interact with KMT2D as part of a multi-protein complex.89 More recently, the majority of KDM6A target genes have been shown to be co-regulated by KMT2D.90 Knockdown of the zebrafish orthologs of KMT2D and KDM6A confirmed the role of these proteins in craniofacial, heart, and brain development, providing direct evidence of the overlapping, functional roles of these genes in the development of tissues and organs affected in Kabuki syndrome.91 DNAm analysis of additional samples with KDM6A pathogenic mutations will be required to assess for potentially overlapping yet distinct DNAm signatures. Such a study would also allow for a comparison of the target genes regulated by KMT2D and KDM6A furthering our understanding of the overlapping and distinct functions of these two genes in the etiology of Kabuki syndrome.
Potential for Therapeutic Interventions Based on Epigenetic Targets
As our knowledge of epigenetic regulation in neurodevelopmental disorders increases, so does the opportunity for therapeutic interventions. The potential positive impact of such interventions is heightened by compelling evidence for the potential to reverse postnatally neurological phenotypes in mouse models of Rett syndrome (RTT [MIM: 312750]) caused by mutations in the epigene methyl-CpG binding protein 2 (MECP2 [MIM: 300005]).92 Recently, Bjornsson et al. (2014) showed that memory deficits in a mouse model of Kabuki syndrome (Kmt2d +/ βGeo) can be prevented or even reversed through systemic delivery of a histone deacetylase (HDAC) inhibitor, which promotes open chromatin states.49 Their findings provide support for the hypothesis that neurodevelopmental deficits in Kabuki syndrome are maintained by an impairment of adult neurogenesis because of an imbalance between open and closed chromatin states for critical target genes. We propose that HOXA5 (regulated by both CHD7 and KMT2D), which has recently been found to be transcribed in the adult brain might be one of these critical target genes.93 A more general potential therapeutic role for HDAC inhibitors in neurodevelopmental syndromes caused by certain epigenes is supported by the fact that these inhibitors have also been shown to reverse the long term memory deficit in mouse models of Rubinstein-Taybi syndrome (RSTS1 [MIM: 180849]) caused by haploinsufficient mutations in another epigene, histone acetyltransferase CREBBP (cAMP-responsive element binding protein binding protein [MIM: 600140]).93
Conclusion
In this article, we present DNAm signatures for CHD7 and KMT2D in human blood cells which hold promise for translational clinical use. These signatures, which have a high degree of sensitivity and specificity, identify specific target genes in human blood cells regulated by CHD7 and KMT2D. Our findings provide evidence that CHARGE and Kabuki syndromes result from dysregulatrion of key genes involved embryonal development that are expressed in a tissue-specific manner. Future studies in developmental model systems such as human induced pluripotent stem cells will help to enhance our understanding of epigenetic regulation in diverse cell-types, including neuronal cells. The field of epigenomics offers a host of opportunities to positively impact precision medicine including a robust means of classifying pathogenicity of VUS and understanding disease pathophysiology in the context of genome-wide targets of epigenes. The identification of a multitude of gene-specific targets across the genome provides tangible opportunities to explore novel therapeutics to reverse neurodevelopmental deficits caused by epigenetic dysregulation.
Acknowledgments
We would like to thank all of the patients and families for their participation in our research study, and the physicians, genetic counselors, and clinical staff for their assistance with recruitment. We would also like to thank Youliang Lou, Chunhua Zhao, and Khadine Wiltshire for their invaluable contributions to this work. Thank you as well to Dr. Greg Hanna (University of Michigan) for contributing blood DNA samples from neurotypical control individuals who had undergone cognitive/behavioral assessments. This research was funded by the Canadian Institutes of Health Research (R.W., D.T.B., M.T.S., and S.C.), the Ontario Brain Institute (R.W., D.T.B., and M.T.S.) and the Rare Diseases Foundation (R.W. and D.T.B.). This work was also supported by the Canadian Centre for Computational Genomics (C3G), part of the Genome Innovation Network (GIN), funded by Genome Canada through Genome Quebec and Ontario Genomics (M.B. and A.L.T.), and Genome Canada through Ontario Genomics (A.L.T., S.C., M.B., and R.W.).
Published: May 4, 2017
Footnotes
Supplemental Information includes four figures and fourteen tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.04.004.
Accession Numbers
The accession number for the DNA methylation data reported in this paper is GEO: GSE97362.
Web Resources
Supplemental Data
References
- 1.Vissers L.E., van Ravenswaaij C.M., Admiraal R., Hurst J.A., de Vries B.B., Janssen I.M., van der Vliet W.A., Huys E.H., de Jong P.J., Hamel B.C. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat. Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 2.Ng S.B., Bigham A.W., Buckingham K.J., Hannibal M.C., McMillin M.J., Gildersleeve H.I., Beck A.E., Tabor H.K., Cooper G.M., Mefford H.C. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ming J.E., Russell K.L., Bason L., McDonald-McGinn D.M., Zackai E.H. Coloboma and other ophthalmologic anomalies in Kabuki syndrome: distinction from charge association. Am. J. Med. Genet. A. 2003;123A:249–252. doi: 10.1002/ajmg.a.20277. [DOI] [PubMed] [Google Scholar]
- 4.Patel N., Alkuraya F.S. Overlap between CHARGE and Kabuki syndromes: more than an interesting clinical observation? Am. J. Med. Genet. A. 2015;167A:259–260. doi: 10.1002/ajmg.a.36804. [DOI] [PubMed] [Google Scholar]
- 5.Dou Y., Milne T.A., Ruthenburg A.J., Lee S., Lee J.W., Verdine G.L., Allis C.D., Roeder R.G. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 6.Zhang P., Lee H., Brunzelle J.S., Couture J.F. The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases. Nucleic Acids Res. 2012;40:4237–4246. doi: 10.1093/nar/gkr1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulz Y., Freese L., Mänz J., Zoll B., Völter C., Brockmann K., Bögershausen N., Becker J., Wollnik B., Pauli S. CHARGE and Kabuki syndromes: a phenotypic and molecular link. Hum. Mol. Genet. 2014;23:4396–4405. doi: 10.1093/hmg/ddu156. [DOI] [PubMed] [Google Scholar]
- 8.Choufani S., Cytrynbaum C., Chung B.H., Turinsky A.L., Grafodatskaya D., Chen Y.A., Cohen A.S., Dupuis L., Butcher D.T., Siu M.T. NSD1 mutations generate a genome-wide DNA methylation signature. Nat. Commun. 2015;6:10207. doi: 10.1038/ncomms10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grafodatskaya D., Chung B.H., Butcher D.T., Turinsky A.L., Goodman S.J., Choufani S., Chen Y.A., Lou Y., Zhao C., Rajendram R. Multilocus loss of DNA methylation in individuals with mutations in the histone H3 lysine 4 demethylase KDM5C. BMC Med. Genomics. 2013;6:1. doi: 10.1186/1755-8794-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kernohan K.D., Cigana Schenkel L., Huang L., Smith A., Pare G., Ainsworth P., Boycott K.M., Warman-Chardon J., Sadikovic B., Care4Rare Canada Consortium Identification of a methylation profile for DNMT1-associated autosomal dominant cerebellar ataxia, deafness, and narcolepsy. Clin. Epigenetics. 2016;8:91. doi: 10.1186/s13148-016-0254-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sim N.L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 15.Smith P.J., Zhang C., Wang J., Chew S.L., Zhang M.Q., Krainer A.R. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 16.Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verloes A. Updated diagnostic criteria for CHARGE syndrome: a proposal. Am. J. Med. Genet. A. 2005;133A:306–308. doi: 10.1002/ajmg.a.30559. [DOI] [PubMed] [Google Scholar]
- 18.Hale C.L., Niederriter A.N., Green G.E., Martin D.M. Atypical phenotypes associated with pathogenic CHD7 variants and a proposal for broadening CHARGE syndrome clinical diagnostic criteria. Am. J. Med. Genet. A. 2016;170A:344–354. doi: 10.1002/ajmg.a.37435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischbach G.D., Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Hanna G.L., Liu Y., Isaacs Y.E., Ayoub A.M., Torres J.J., O’Hara N.B., Gehring W.J. Withdrawn/Depressed Behaviors and Error-Related Brain Activity in Youth With Obsessive-Compulsive Disorder. J Am Acad Child Adolesc Psychiatry. 2016;55:906–913 e902. doi: 10.1016/j.jaac.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buhule O.D., Minster R.L., Hawley N.L., Medvedovic M., Sun G., Viali S., Deka R., McGarvey S.T., Weeks D.E. Stratified randomization controls better for batch effects in 450K methylation analysis: a cautionary tale. Front. Genet. 2014;5:354. doi: 10.3389/fgene.2014.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y.A., Choufani S., Grafodatskaya D., Butcher D.T., Ferreira J.C., Weksberg R. Cross-reactive DNA microarray probes lead to false discovery of autosomal sex-associated DNA methylation. Am. J. Hum. Genet. 2012;91:762–764. doi: 10.1016/j.ajhg.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y.A., Lemire M., Choufani S., Butcher D.T., Grafodatskaya D., Zanke B.W., Gallinger S., Hudson T.J., Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du P., Zhang X., Huang C.C., Jafari N., Kibbe W.A., Hou L., Lin S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reinius L.E., Acevedo N., Joerink M., Pershagen G., Dahlén S.E., Greco D., Söderhäll C., Scheynius A., Kere J. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaffe A.E., Murakami P., Lee H., Leek J.T., Fallin M.D., Feinberg A.P., Irizarry R.A. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int. J. Epidemiol. 2012;41:200–209. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D., Xie Z., Pape M.L., Dye T. An evaluation of statistical methods for DNA methylation microarray data analysis. BMC Bioinformatics. 2015;16:217. doi: 10.1186/s12859-015-0641-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters T.J., Buckley M.J., Statham A.L., Pidsley R., Samaras K., V Lord R., Clark S.J., Molloy P.L. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin. 2015;8:6. doi: 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Consortium T.G.O., Gene Ontology Consortium Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin J.L., Lee W.I., Huang J.L., Chen P.K., Chan K.C., Lo L.J., You Y.J., Shih Y.F., Tseng T.Y., Wu M.C. Immunologic assessment and KMT2D mutation detection in Kabuki syndrome. Clin. Genet. 2015;88:255–260. doi: 10.1111/cge.12484. [DOI] [PubMed] [Google Scholar]
- 33.Wong M.T., Lambeck A.J., van der Burg M., la Bastide-van Gemert S., Hogendorf L.A., van Ravenswaaij-Arts C.M., Schölvinck E.H. Immune Dysfunction in Children with CHARGE Syndrome: A Cross-Sectional Study. PLoS ONE. 2015;10:e0142350. doi: 10.1371/journal.pone.0142350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banka S., Lederer D., Benoit V., Jenkins E., Howard E., Bunstone S., Kerr B., McKee S., Lloyd I.C., Shears D. Novel KDM6A (UTX) mutations and a clinical and molecular review of the X-linked Kabuki syndrome (KS2) Clin. Genet. 2015;87:252–258. doi: 10.1111/cge.12363. [DOI] [PubMed] [Google Scholar]
- 35.Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cesario J.M., Almaidhan A.A., Jeong J. Expression of forkhead box transcription factor genes Foxp1 and Foxp2 during jaw development. Gene Expr. Patterns. 2016;20:111–119. doi: 10.1016/j.gep.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Rhijn J.R., Vernes S.C. Retinoic Acid Signaling: A New Piece in the Spoken Language Puzzle. Front. Psychol. 2015;6:1816. doi: 10.3389/fpsyg.2015.01816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouazoune K., Kingston R.E. Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders. Proc. Natl. Acad. Sci. USA. 2012;109:19238–19243. doi: 10.1073/pnas.1213825109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnetz M.P., Bartels C.F., Shastri K., Balasubramanian D., Zentner G.E., Balaji R., Zhang X., Song L., Wang Z., Laframboise T. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19:590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zentner G.E., Tesar P.J., Scacheri P.C. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin D.M. Epigenetic Developmental Disorders: CHARGE syndrome, a case study. Curr. Genet. Med. Rep. 2015;3:1–7. doi: 10.1007/s40142-014-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin D.M. Chromatin remodeling in development and disease: focus on CHD7. PLoS Genet. 2010;6:e1001010. doi: 10.1371/journal.pgen.1001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schulz Y., Wehner P., Opitz L., Salinas-Riester G., Bongers E.M., van Ravenswaaij-Arts C.M., Wincent J., Schoumans J., Kohlhase J., Borchers A., Pauli S. CHD7, the gene mutated in CHARGE syndrome, regulates genes involved in neural crest cell guidance. Hum. Genet. 2014;133:997–1009. doi: 10.1007/s00439-014-1444-2. [DOI] [PubMed] [Google Scholar]
- 46.Vallianatos C.N., Iwase S. Disrupted intricacy of histone H3K4 methylation in neurodevelopmental disorders. Epigenomics. 2015;7:503–519. doi: 10.2217/epi.15.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ang S.Y., Uebersohn A., Spencer C.I., Huang Y., Lee J.E., Ge K., Bruneau B.G. KMT2D regulates specific programs in heart development via histone H3 lysine 4 di-methylation. Development. 2016;143:810–821. doi: 10.1242/dev.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eissenberg J.C., Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 2010;339:240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bjornsson H.T., Benjamin J.S., Zhang L., Weissman J., Gerber E.E., Chen Y.C., Vaurio R.G., Potter M.C., Hansen K.D., Dietz H.C. Histone deacetylase inhibition rescues structural and functional brain deficits in a mouse model of Kabuki syndrome. Sci. Transl. Med. 2014;6:256ra135. doi: 10.1126/scitranslmed.3009278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delahaye A., Sznajer Y., Lyonnet S., Elmaleh-Bergès M., Delpierre I., Audollent S., Wiener-Vacher S., Mansbach A.L., Amiel J., Baumann C. Familial CHARGE syndrome because of CHD7 mutation: clinical intra- and interfamilial variability. Clin. Genet. 2007;72:112–121. doi: 10.1111/j.1399-0004.2007.00821.x. [DOI] [PubMed] [Google Scholar]
- 51.Jongmans M.C., Hoefsloot L.H., van der Donk K.P., Admiraal R.J., Magee A., van de Laar I., Hendriks Y., Verheij J.B., Walpole I., Brunner H.G., van Ravenswaaij C.M. Familial CHARGE syndrome and the CHD7 gene: a recurrent missense mutation, intrafamilial recurrence and variability. Am. J. Med. Genet. A. 2008;146A:43–50. doi: 10.1002/ajmg.a.31921. [DOI] [PubMed] [Google Scholar]
- 52.Blake K.D., Davenport S.L., Hall B.D., Hefner M.A., Pagon R.A., Williams M.S., Lin A.E., Graham J.M., Jr. CHARGE association: an update and review for the primary pediatrician. Clin. Pediatr. (Phila.) 1998;37:159–173. doi: 10.1177/000992289803700302. [DOI] [PubMed] [Google Scholar]
- 53.Blake K.D., Prasad C. CHARGE syndrome. Orphanet J. Rare Dis. 2006;1:34. doi: 10.1186/1750-1172-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vihinen M. Majority vote and other problems when using computational tools. Hum. Mutat. 2014;35:912–914. doi: 10.1002/humu.22600. [DOI] [PubMed] [Google Scholar]
- 55.Jongmans M.C., Admiraal R.J., van der Donk K.P., Vissers L.E., Baas A.F., Kapusta L., van Hagen J.M., Donnai D., de Ravel T.J., Veltman J.A. CHARGE syndrome: the phenotypic spectrum of mutations in the CHD7 gene. J. Med. Genet. 2006;43:306–314. doi: 10.1136/jmg.2005.036061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bergman J.E., Janssen N., Hoefsloot L.H., Jongmans M.C., Hofstra R.M., van Ravenswaaij-Arts C.M. CHD7 mutations and CHARGE syndrome: the clinical implications of an expanding phenotype. J. Med. Genet. 2011;48:334–342. doi: 10.1136/jmg.2010.087106. [DOI] [PubMed] [Google Scholar]
- 57.Bergman J.E., Janssen N., van der Sloot A.M., de Walle H.E., Schoots J., Rendtorff N.D., Tranebjaerg L., Hoefsloot L.H., van Ravenswaaij-Arts C.M., Hofstra R.M. A novel classification system to predict the pathogenic effects of CHD7 missense variants in CHARGE syndrome. Hum. Mutat. 2012;33:1251–1260. doi: 10.1002/humu.22106. [DOI] [PubMed] [Google Scholar]
- 58.Bögershausen N., Wollnik B. Unmasking Kabuki syndrome. Clin. Genet. 2013;83:201–211. doi: 10.1111/cge.12051. [DOI] [PubMed] [Google Scholar]
- 59.Gibson W.T., Hood R.L., Zhan S.H., Bulman D.E., Fejes A.P., Moore R., Mungall A.J., Eydoux P., Babul-Hirji R., An J., FORGE Canada Consortium Mutations in EZH2 cause Weaver syndrome. Am. J. Hum. Genet. 2012;90:110–118. doi: 10.1016/j.ajhg.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tatton-Brown K., Hanks S., Ruark E., Zachariou A., Duarte Sdel.V., Ramsay E., Snape K., Murray A., Perdeaux E.R., Seal S., Childhood Overgrowth Collaboration Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget. 2011;2:1127–1133. doi: 10.18632/oncotarget.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Geneviève D., Amiel J., Viot G., Le Merrer M., Sanlaville D., Urtizberea A., Gérard M., Munnich A., Cormier-Daire V., Lyonnet S. Atypical findings in Kabuki syndrome: report of 8 patients in a series of 20 and review of the literature. Am. J. Med. Genet. A. 2004;129A:64–68. doi: 10.1002/ajmg.a.30144. [DOI] [PubMed] [Google Scholar]
- 62.Verhagen J.M., Oostdijk W., Terwisscha van Scheltinga C.E., Schalij-Delfos N.E., van Bever Y. An unusual presentation of Kabuki syndrome: clinical overlap with CHARGE syndrome. Eur. J. Med. Genet. 2014;57:510–512. doi: 10.1016/j.ejmg.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 63.Li D.D., Wang Z.H., Chen W.L., Xie Y.Y., You Q.D., Guo X.K. Structure-based design of ester compounds to inhibit MLL complex catalytic activity by targeting mixed lineage leukemia 1 (MLL1)-WDR5 interaction. Bioorg. Med. Chem. 2016;24:6109–6118. doi: 10.1016/j.bmc.2016.09.073. [DOI] [PubMed] [Google Scholar]
- 64.Montavon T., Soshnikova N. Hox gene regulation and timing in embryogenesis. Semin. Cell Dev. Biol. 2014;34:76–84. doi: 10.1016/j.semcdb.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 65.Barber B.A., Rastegar M. Epigenetic control of Hox genes during neurogenesis, development, and disease. Ann. Anat. 2010;192:261–274. doi: 10.1016/j.aanat.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 66.Basson M.A. Epistatic interactions between Chd7 and Fgf8 during cerebellar development: Implications for CHARGE syndrome. Rare Dis. 2014;2:e28688. doi: 10.4161/rdis.28688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Micale L., Augello B., Maffeo C., Selicorni A., Zucchetti F., Fusco C., De Nittis P., Pellico M.T., Mandriani B., Fischetto R. Molecular analysis, pathogenic mechanisms, and readthrough therapy on a large cohort of Kabuki syndrome patients. Hum. Mutat. 2014;35:841–850. doi: 10.1002/humu.22547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mallo M., Wellik D.M., Deschamps J. Hox genes and regional patterning of the vertebrate body plan. Dev. Biol. 2010;344:7–15. doi: 10.1016/j.ydbio.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hershko A.Y., Kafri T., Fainsod A., Razin A. Methylation of HoxA5 and HoxB5 and its relevance to expression during mouse development. Gene. 2003;302:65–72. doi: 10.1016/s0378111902010910. [DOI] [PubMed] [Google Scholar]
- 70.Parrillo L., Costa V., Raciti G.A., Longo M., Spinelli R., Esposito R., Nigro C., Vastolo V., Desiderio A., Zatterale F. Hoxa5 undergoes dynamic DNA methylation and transcriptional repression in the adipose tissue of mice exposed to high-fat diet. Int. J. Obes. 2016;40:929–937. doi: 10.1038/ijo.2016.36. [DOI] [PubMed] [Google Scholar]
- 71.Svingen T., Tonissen K.F. Hox transcription factors and their elusive mammalian gene targets. Heredity (Edinb) 2006;97:88–96. doi: 10.1038/sj.hdy.6800847. [DOI] [PubMed] [Google Scholar]
- 72.Joksimovic M., Jeannotte L., Tuggle C.K. Dynamic expression of murine HOXA5 protein in the central nervous system. Gene Expr. Patterns. 2005;5:792–800. doi: 10.1016/j.modgep.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 73.Larochelle C., Tremblay M., Bernier D., Aubin J., Jeannotte L. Multiple cis-acting regulatory regions are required for restricted spatio-temporal Hoxa5 gene expression. Dev. Dyn. 1999;214:127–140. doi: 10.1002/(SICI)1097-0177(199902)214:2<127::AID-AJA3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 74.Hutlet B., Theys N., Coste C., Ahn M.T., Doshishti-Agolli K., Lizen B., Gofflot F. Systematic expression analysis of Hox genes at adulthood reveals novel patterns in the central nervous system. Brain Struct. Funct. 2016;221:1223–1243. doi: 10.1007/s00429-014-0965-8. [DOI] [PubMed] [Google Scholar]
- 75.Lizen B., Hutlet B., Bissen D., Sauvegarde D., Hermant M., Ahn M.T., Gofflot F. HOXA5 localization in postnatal and adult mouse brain is suggestive of regulatory roles in postmitotic neurons. J. Comp. Neurol. 2017;525:1155–1175. doi: 10.1002/cne.24123. [DOI] [PubMed] [Google Scholar]
- 76.Jeannotte L., Lemieux M., Charron J., Poirier F., Robertson E.J. Specification of axial identity in the mouse: role of the Hoxa-5 (Hox1.3) gene. Genes Dev. 1993;7:2085–2096. doi: 10.1101/gad.7.11.2085. [DOI] [PubMed] [Google Scholar]
- 77.Lalani, S.R., Hefner, M.A., Belmont, J.W., and Davenport, S.L.H. (1993). CHARGE Syndrome. In GeneReviews(R), R.A. Pagon, M.P. Adam, H.H. Ardinger, S.E. Wallace, A. Amemiya, L.J.H. Bean, T.D. Bird, N. Ledbetter, H.C. Mefford, R.J.H. Smith, et al., eds. (Seattle, WA).
- 78.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang X.Q., Dostie J. Reciprocal regulation of chromatin state and architecture by HOTAIRM1 contributes to temporal collinear HOXA gene activation. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw966. Published online October 26, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Long K., Moss L., Laursen L., Boulter L., ffrench-Constant C. Integrin signalling regulates the expansion of neuroepithelial progenitors and neurogenesis via Wnt7a and Decorin. Nat. Commun. 2016;7:10354. doi: 10.1038/ncomms10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Loulier K., Lathia J.D., Marthiens V., Relucio J., Mughal M.R., Tang S.C., Coksaygan T., Hall P.E., Chigurupati S., Patton B. beta1 integrin maintains integrity of the embryonic neocortical stem cell niche. PLoS Biol. 2009;7:e1000176. doi: 10.1371/journal.pbio.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takahashi H., Katayama K., Sohya K., Miyamoto H., Prasad T., Matsumoto Y., Ota M., Yasuda H., Tsumoto T., Aruga J. Selective control of inhibitory synapse development by Slitrk3-PTPdelta trans-synaptic interaction. Nat Neurosci. 2012;15:389–398. doi: 10.1038/nn.3040. S381-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aruga J., Mikoshiba K. Identification and characterization of Slitrk, a novel neuronal transmembrane protein family controlling neurite outgrowth. Mol. Cell. Neurosci. 2003;24:117–129. doi: 10.1016/s1044-7431(03)00129-5. [DOI] [PubMed] [Google Scholar]
- 84.Song M., Giza J., Proenca C.C., Jing D., Elliott M., Dincheva I., Shmelkov S.V., Kim J., Schreiner R., Huang S.H. Slitrk5 Mediates BDNF-Dependent TrkB Receptor Trafficking and Signaling. Dev. Cell. 2015;33:690–702. doi: 10.1016/j.devcel.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shmelkov S.V., Hormigo A., Jing D., Proenca C.C., Bath K.G., Milde T., Shmelkov E., Kushner J.S., Baljevic M., Dincheva I. Slitrk5 deficiency impairs corticostriatal circuitry and leads to obsessive-compulsive-like behaviors in mice. Nat Med. 2010;16:598–602. doi: 10.1038/nm.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vargha-Khadem F., Gadian D.G., Copp A., Mishkin M. FOXP2 and the neuroanatomy of speech and language. Nat Rev Neurosci. 1993;6:131–138. doi: 10.1038/nrn1605. [DOI] [PubMed] [Google Scholar]
- 87.Zadro C., Alemanno M.S., Bellacchio E., Ficarella R., Donaudy F., Melchionda S., Zelante L., Rabionet R., Hilgert N., Estivill X. Are MYO1C and MYO1F associated with hearing loss? Biochim. Biophys. Acta. 2009;1792:27–32. doi: 10.1016/j.bbadis.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 88.Lee M.G., Villa R., Trojer P., Norman J., Yan K.P., Reinberg D., Di Croce L., Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 89.Issaeva I., Zonis Y., Rozovskaia T., Orlovsky K., Croce C.M., Nakamura T., Mazo A., Eisenbach L., Canaani E. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell. Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim J.H., Sharma A., Dhar S.S., Lee S.H., Gu B., Chan C.H., Lin H.K., Lee M.G. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014;74:1705–1717. doi: 10.1158/0008-5472.CAN-13-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Van Laarhoven P.M., Neitzel L.R., Quintana A.M., Geiger E.A., Zackai E.H., Clouthier D.E., Artinger K.B., Ming J.E., Shaikh T.H. Kabuki syndrome genes KMT2D and KDM6A: functional analyses demonstrate critical roles in craniofacial, heart and brain development. Hum. Mol. Genet. 2015;24:4443–4453. doi: 10.1093/hmg/ddv180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guy J., Gan J., Selfridge J., Cobb S., Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Alarcón J.M., Malleret G., Touzani K., Vronskaya S., Ishii S., Kandel E.R., Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/- mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.