Abstract
We developed a variant-annotation method that combines sequence-based machine-learning classification with a context-dependent algorithm for selecting splice variants. Our approach is distinctive in that it compares the splice potential of a sequence bearing a variant with the splice potential of the reference sequence. After training, classification accurately identified 168 of 180 (93.3%) canonical splice sites of five genes. The combined method, CryptSplice, identified and correctly predicted the effect of 18 of 21 (86%) known splice-altering variants in CFTR, a well-studied gene whose loss-of-function variants cause cystic fibrosis (CF). Among 1,423 unannotated CFTR disease-associated variants, the method identified 32 potential exonic cryptic splice variants, two of which were experimentally evaluated and confirmed. After complete CFTR sequencing, the method found three cryptic intronic splice variants (one known and two experimentally verified) that completed the molecular diagnosis of CF in 6 of 14 individuals. CryptSplice interrogation of sequence data from six individuals with X-linked dyskeratosis congenita caused by an unknown disease-causing variant in DKC1 identified two splice-altering variants that were experimentally verified. To assess the extent to which disease-associated variants might activate cryptic splicing, we selected 458 pathogenic variants and 348 variants of uncertain significance (VUSs) classified as high confidence from ClinVar. Splice-site activation was predicted for 129 (28%) of the pathogenic variants and 75 (22%) of the VUSs. Our findings suggest that cryptic splice-site activation is more common than previously thought and should be routinely considered for all variants within the transcribed regions of genes.
Keywords: splicing, machine learning, splice variant, splice donor, splice acceptor, cryptic splicing, pseudoexon, minigene, cystic fibrosis
Introduction
Next-generation sequencing has enabled the detection of vast numbers of variants in the coding and non-coding regions of genes. Variants that have a deleterious effect on pre-mRNA splicing are thought to be limited primarily to the canonical splice sites. Exonic variants outside of the canonical splice sites are generally considered to affect RNA processing by disrupting splice enhancers and thereby causing a significant reduction in the spliceosomal recognition of the canonical splice sequence.1 However, exonic variants can also activate cryptic splice sites, leading to aberrant pre-mRNA splicing and loss of coding sequence.2 Such examples are sparse in the literature, and thus the frequency of exonic cryptic splicing is unknown. It has long been suspected, but not systematically shown, that variants that activate splice sites have been masquerading as exonic protein-altering variants or “benign” synonymous variants, leading to an under-appreciation of the frequency of this pathologic mechanism.3, 4, 5 Identifying whether exonic variation affects RNA rather than protein processing is vital as genetic medicine seeks to deploy personalized initiatives such as drug therapies aimed at addressing specific disease mechanisms.
By the same token, intronic variants outside of canonical splice sites can affect RNA processing by activating splice sites. Detection of intronic cryptic splice variants is challenging because the genomic space is much larger and there is far less evolutionary constraint, resulting in a higher degree of variation. Although diagnostic sequencing rarely interrogates complete introns, aberrant splicing due to deep intronic cryptic splice variants is often discovered in the analysis of RNA transcripts from individuals with unidentified disease-causing variants.6, 7, 8, 9 Deep intronic cryptic splice activation is considered a rare and exotic event, and the true frequency remains unknown. Given the higher number of variants in introns than in exons and the flexibility of splice sequences, it is possible that intronic variants could often activate cryptic splice sites. Indeed, case studies of deep intronic cryptic splice activation events have shown that a single-nucleotide substitution is sufficient to cause severe disease and that these variants do not always create novel GT or AG dinucleotides; usually, they occur near existing GT or AG dinucleotides.10, 11 These examples suggest that disease-associated variants in deep introns could be as culpable as those in exons and should be given similar scrutiny. Identifying the frequency of deep intronic cryptic splice events and the number of exonic variants that alter splicing is vital to the completion of Mendelian genotypes and our understanding of the molecular mechanisms of disease.
Material and Methods
The computational methods described here consist of two parts: the classification of splice sequences and the selection of splice variants. The foundation of classification is the application of predictive models trained on data from splice donors and acceptor sequences. Selection uses the predictions provided by the classifier models to compare the splice potentials of candidate splice variant sequences with the splice potentials of the reference sequences. Filtering by location during selection removes unlikely candidates and generates the final “high-confidence” candidate splice variants. Software and documentation for downloading, implementing, and modifying CryptSplice are publically available.
Classifying Splice Sequences
Training Data
Splice sequences from NN26912 and HS3D13 were utilized as training data. The NN269 and HS3D datasets contain “true” donor and acceptor sequences curated from GenBank genes annotated for canonical splice sites. Although a very small fraction of canonical donor sites utilize a GC consensus dinucleotide in lieu of GT,14, 15 the number was sufficiently small that considering GT-containing donors was determined to best enrich for good splice donor candidates. “False” sequences in the training data are sequences with GT or AG dinucleotides at least 60 bp away from canonical splice sites and shown to not be recognized by the spliceosome. Donor sequences extend from seven nucleotides upstream of GT (−7) to six nucleotides downstream of GT (+6). Acceptor sequences extend from 68 nucleotides upstream of AG (−68) to 20 nucleotides downstream of AG (+20). 1,116 true donors from NN269 and 2,796 true donors from HS3D were combined into a single training data set of true donors. 1,116 true acceptors from NN269 and 2,880 true acceptors from HS3D were combined into a single training data set of true acceptors. False sequences were randomly and proportionally selected from NN269 and HS3D to match the number of true sequences.
Features
Features were based upon previously published features16 and were chosen because of the comprehensiveness of the sequence information captured. Tables of statistical difference were not utilized for reducing bias and were substituted with binary coding of a feature present in the input. There were three types of features. In brief, the “component” feature separates splice sequences into left and right segments at the consensus dinucleotide and calculates the probability that a sequence substring will occur in the left and right segments. Probabilities for all possible sequence substrings up to a length of five nucleotides were calculated. Second, the “position” (pos) feature describes whether a given nucleotide exists at a given position across the length of the splice sequence. Third, the “adjacent position relationship” (apr) feature describes whether a given dinucleotide exists at a given position across the length of the splice sequence. Feature data were extracted by custom Python scripts.
Model Selection
Support vector machine (SVM) models were trained with custom scripts employing the Python scikit learn machine-learning library.17 Feature data were transformed with linear and radial basis function (RBF) kernels for determining whether reduction in data complexity was required for better model performance. Classifier models were trained with 10-fold cross-validation in which the training data were randomly split into ten equal pieces. Classifiers were trained on nine pieces and tested on the unseen tenth piece. This process was repeated ten times and averaged for determining model generalizability as measured by a distribution of accuracies. Classifiers were designed to provide probability estimates in addition to the typical binary classification of true or false. Probability estimates can be considered the splice potential of a sequence, where 1 reflects a perfect splice site and 0 indicates a sequence that does not resemble a splice site at all. Classifier models trained with RBF-transformed data had the best accuracies and were utilized in all further applications.
Creating Candidate Sequences
Candidate splice sequences were created from variants of interest and the surrounding NCBI RefSeq gene reference sequence with a custom Python script. For each gene, the first transcript listed by the Variant Effect Predictor was used.18 To standardize the numbering, we assigned the last nucleotide of the intron a 0 and the last nucleotide of the exon a 0. Variants of interest were able to generate candidate splice sequences if they fulfilled one of two criteria: (1) the variant occurred within −3 to +5 of an existing GT dinucleotide or within −22 to +1 of an existing AG dinucleotide, or (2) the variant created a GT or AG dinucleotide. If either or both criteria were satisfied, sequences were extracted within −7 to +6 of GT dinucleotides and/or within −68 to +20 of AG dinucleotides. In order to provide a contextual comparison for the change in a sequence’s splice potential when a variant was introduced, we subjected reference gene sequences to classification as well. Sequences were extracted within −7 to +6 of all GT dinucleotides and within −68 to +20 of all AG dinucleotides throughout the full reference sequences for all genes in this study.
Evaluating Candidate Sequences
All candidate splice sequences and reference sequences were subjected to classification utilizing the best-performing models as determined by the highest accuracies after training. Candidate sequences were given a binary classification of true or false and a probability estimate from 0 to 1.
Selecting Splice Variants
A selection algorithm to identify high-confidence candidate splice variants was developed to compare the splice potentials of variant sequences (P(var)) with those of the reference sequence (P(ref)) and the canonical splice site (P(canon)). A variant can cause aberrant splicing in three location scenarios, and two metrics were utilized for testing variants in these scenarios:
Δcanon is the splice potential of the canonical sequence P(canon) subtracted from the splice potential of the variant splice site P(var). Positive values indicate that the variant sequence has a greater splice potential than the canonical splice site. Negative values indicate that the variant sequence has a weaker splice potential than the canonical splice site. We tested multiple thresholds of Δcanon to see which captured all instances in a true set of known CFTR (MIM: 602421; GenBank: NM_000492.3) splice variants, leading us to set the default minimum threshold for this metric to |0.05|.
Δvariant is the splice potential of the reference sequence P(ref) subtracted from the splice potential of the variant sequence P(var). Positive values indicate an increase in the splice potential as a variant is introduced, and negative values indicate a weakening of the sequence’s splice potential. This metric can be calculated only for variants that do not create a GT or AG dinucleotide. (A variant that creates a GT or AG dinucleotide cannot be compared with the reference sequence because the GT or AG dinucleotide does not exist in the reference.) We tested multiple thresholds of Δvariant to see which captured all instances in a true set of known CFTR splice variants, leading us to set the default minimum threshold for this metric to |0.05|.
Scenario 1: Weakening of a Canonical Splice Site
Candidate variant sequences qualified for this scenario if P(var) ≤ 0.85 and the variant occurred within −3 to +5 of a canonical splice donor or within −22 to +1 of a canonical splice acceptor. The Δcanon had to be ≤−0.05 to sufficiently weaken the canonical splice site in a biologically meaningful way, as determined by application to a true set of variants known to weaken CFTR canonical splice sites. Variants that altered the canonical AG or GT dinucleotides of the splice sequences were not analyzed because they are known to cause loss of splicing.19
Scenario 2: Cryptic Splice Activation That Outcompetes a Nearby Canonical Splice Site
Candidate variant sequences qualified for this scenario if P(var) for candidate donors was ≥0.7 or if P(var) for candidate acceptors was ≥0.6. These thresholds were set according to the minimum probability assigned to known canonical splice sites. The variant had to occur within 60 bp of a canonical splice site of the same predicted effect (i.e., cryptic donors near canonical donors) and, if applicable, have a Δvariant ≥ +0.05. To distinguish variants that activated splicing from those that cause loss of splicing, we excluded nucleotides at positions −3 to 0 in acceptors and −1 to +2 in donors from the analysis. If a Δvariant could not be calculated because a novel GT or AG dinucleotide had been created by the variant, Δcanon was calculated for the variant sequence. On the basis of a true set of variants known to activate cryptic sites that outcompete the canonical, in this scenario P(canon) was reduced by 0.1 to capture true examples. After this adjustment, the candidate variant sequence had to have a Δcanon ≥ +0.05.
Scenario 3: Deep Intronic Cryptic Splice Activation
Candidate variant sequences qualified for this scenario if P(var) for candidate donors was ≥0.7 or if P(var) for candidate acceptors was ≥0.6. These thresholds were set according to the minimum probability assigned to known canonical splice sites. The variant sequence had to occur at least 100 bp into the intron and, if applicable, have Δvariant ≥ +0.05. If a Δvariant could not be calculated because a novel GT or AG dinucleotide was created by the variant, the variant sequence was flagged if the first two criteria were satisfied (P(var) ≥ 0.7 [donors] or ≥ 0.6 [acceptors]).
Sequencing the Full CFTR Locus in Individuals with Cystic Fibrosis and One Known Disease-Causing Variant
Genomic DNA was extracted from whole blood according to a standard phenol-chloroform protocol. A custom-designed Agilent SureSelect capture was used to pull down the 215 kb region containing and surrounding CFTR in each sample. Samples were run on an Illumina HiSeq 2500. A custom-designed next-generation sequencing data-processing pipeline was used to align reads and call variants in all samples. This pipeline included alignment by the Burrows-Wheeler Aligner, duplicate removal by Picard, local realignment by the Genome Analysis Toolkit, and variant calling by four additional software programs. The intersection or merging of select variant callers was used for downstream analysis. Additionally, large indels and copy-number variants were called by three algorithms but primarily relied upon Conifer calls.
Introduction of Variants to Expression Minigenes
Expression minigenes were developed as described previously.20 Candidate splice variants were introduced into wild-type (WT) expression minigenes by site-directed mutagenesis (SDM) using oligonucleotides with the candidate splice variant and flanking 20 nt sequences identical to the region of interest. SDM reactions were performed in triplicate, pooled, and digested with Dpn1 for the removal of plasmid template. Purified SDM products were transformed with XL10 Gold supercompetent cells and clonally expanded overnight at 37°C with shaking in LB broth with ampicillin. DNA was extracted by MiniPrep, and mutagenesis was confirmed by Sanger sequencing.
Transient Transfection of HEK293 Cells with Expression Minigenes
4 μg of expression minigene plasmid was diluted in 250 μL of OptiMEM and combined with 7 μL of Lipofectamine 2000 (Invitrogen) diluted in 250 μL of OptiMEM. The Lipofectamine 2000 complexes were added to 6-well plates containing confluent HEK293 cells that had been grown in antibiotic-free media for 24 hr before transfection. 1 mL of antibiotic-free media was added to cultures 4 hr after transfection. Media were changed with antibiotic-free media 24 hr after transfection. Cells were lysed for analysis of protein and mRNA transcripts 48 hr after transfection.
Analysis of mRNA Transcripts and Protein from Transiently Transfected HEK293 Cells
Cells were washed twice with 1× PBS 48 hr after transfection. A standard RNA extraction procedure using TRIzol and chloroform was performed. RNA was prepared in a DNA-free bench with dedicated plasticware and pipets. RNA preparations were treated with DNase, and RT-PCR was immediately performed after RNA extraction. PCR of HEK-derived cDNAs was performed with primers lying in the exons, and aberrant splice products were visualized by gel electrophoresis. DNA fragments of interest were extracted and purified and verified by Sanger sequencing.
For protein analysis, cells were washed twice with 1× PBS 48 hr after transfection and lysed with 250 μL of RIPA buffer containing protease inhibitors and PMSF. Cell lysates were incubated on ice for 30 min and vortexed for 20 s every 10 min. Cell lysates were spun at 4°C for 15 min, and the supernatant was retained for western blotting. Loading samples were then prepared. For clear visualization of CFTR, the loading sample contained a minimum of 40 μg total protein. The amount of protein lysate required for 40 μg of total protein was calculated with the total protein concentrations previously determined by bicinchoninic acid assay. The volume of individual protein lysates, in combination with 1× PBS, accounted for three-quarters of each total sample volume. The remaining one-quarter of the total sample volume was a dye solution containing a 1:5 dilution of DTT to 4× Laemmli buffer. After being prepared, the loading samples were further denatured by incubation at 37°C for 15 min. Samples were loaded into a 7.5% Tris-HCL, 1.0 mm Criterion Precast Gel (Bio-Rad) and run with 1× running buffer prepared in house. The samples were bookended with All-Blue Precision Protein Standard Ladder (Bio-Rad) for protein-size comparison. Samples were run at 150 V for 2 hr. The gel was transferred to a polyvinylidene difluoride membrane with the Trans-Blot Turbo Transfer System at 2.5 A and 25 V for 10 min. The membrane was blocked for 1 hr in a 5% blocking solution of non-fat dry milk reconstituted in 1× PBS containing 0.1% Tween-20 (PBST). The membrane was washed in PBST and then incubated for 1 hr with primary anti-CFTR m570 or m596 antibody diluted at 1:5,000. The membrane was washed again with PBST for 30 min. The membrane was then incubated for 1 hr with an anti-mouse secondary antibody (GE Healthcare) diluted at 1:150,000. The secondary anti-mouse antibody was removed, and the membrane was washed thoroughly with PBST for 45 min before imaging.
Analysis of mRNA Transcripts from Individuals with Cystic Fibrosis
Human nasal epithelial (HNE) cells were collected with a nasal cytology brush, which brushed the inferior surface of the inferior nasal turbinate of each nostril from each individual as previously described.21, 22 HNE cells were collected under institutional review board (IRB) approval from Johns Hopkins University (IRB no. NA 00029159) and the University of Alabama at Birmingham (IRB no. F090916001). Written informed consent was obtained for all subjects. RNA was extracted from HNE cells with the QIAGEN RNeasyPlus Mini Kit according to the manufacturer’s protocol. RNA was eluted in 30 μL RNase-free water. The quantity and quality of RNA were determined by OD260 and OD260/OD280, respectively, with NanoDrop ND-1000.
RT-PCR was performed with the Bio-Rad iScript kit and 250 ng of RNA. cDNA was amplified with CFTR-specific primers—the forward primers were fluorescently labeled with 6-FAM (carboxyfluorescein) at the 5' end. The RT-PCR products (2 μL, 100-fold dilution) were mixed with 18 μL of Hi-Di Formamide (Applied Biosystems) and 0.25 μL of an internal size standard (GeneScan-500 Rox, Applied Biosystems). Products were separated by capillary electrophoreses on an ABI3100 Genetic Analyser with POP4 polymer (Applied Biosystems) and analyzed with GeneMapper Software v.3.7 (Applied Biosystems) at the Genetic Resource Core Facility of the Johns Hopkins University School of Medicine. Additionally, pyrosequencing was used for determining the percentage of the aberrantly spliced isoform as described previously. The RT-PCR products were obtained with biotin-labeled reverse primers. The products were sequenced according to the protocol of the PyroMark Q24 system (QIAGEN) with 0.4 μM of specific pyrosequencing primers, and pyrograms were analyzed with PyroMark Q24 v.2.0.6 (QIAGEN) at the Genetic Resource Core Facility of the Johns Hopkins University School of Medicine.
Analysis of mRNA Transcripts from Individuals with Dyskeratosis Congenita
Lymphocytes from whole blood were used to establish lymphoblastoid cell lines as previously described.23 RNA from lymphoblastoid cell lines was isolated with the QIAGEN RNeasy Mini Kit according to the manufacturer’s protocol. Reverse transcription was performed with the Invitrogen SuperScript First Strand Synthesis System, and the resulting cDNA was amplified with primers lying in distant exons. Products were separated, extracted, and purified by gel electrophoresis and cloned with the Thermo Fisher TOPO Cloning Kit. DNA isolated from clones was subjected to Sanger sequencing for verification of aberrantly spliced transcripts.
All genomic coordinates correspond to UCSC Genome Browser build hg19, and all CFTR variants and exons are identified by HGVS nomenclature and not with legacy names unless otherwise indicated.
Results
Training and Classification of Candidate Splice Donor and Acceptor Sequences
The foundation of our method was applying machine-learning models to agnostically evaluate the splicing potential of sequence surrounding any variant if the sequence contained a GT or AG dinucleotide. SVM donor and acceptor classifier models were trained with canonical “true” and “false” splice sequences from annotated gene sequences in GenBank and NCBI.12, 13 Classifier models were trained on feature data that reflected the essential distinguishing characteristics of a genuine splice sequence; therefore, feature data were entirely sequence based and included nucleotide content and substring frequency16 (see Material and Methods). Classifier models were internally validated by 10-fold cross-validation, and the highest-performing models were selected for classification of candidate splice sequences. Classifier models provide a binary classification (true or false) and a probability estimate from 0 to 1. Probability estimates can be considered approximations of a sequence’s splice potential, where a sequence with a score of 1 has a very high likelihood of being recognized by the spliceosome.
To predict the impact of a variant on the splice potential of a given sequence, we introduced variants into NCBI reference gene sequences to create candidate splice sequences for classification. Variants created candidate splice sequences if they satisfied these criteria: (1) the variant created a novel GT or AG or occurred within −3 to +5 of an existing GT (in relation to the last nucleotide of the exon, which was assigned a 0) and/or within −22 to +1 of an existing AG (in relation to the last nucleotide of the intron, which was assigned a 0), and (2) the variant did not occur outside the exons and introns of the gene of interest (i.e., in the UTRs) (Figure 1A). If these criteria were met, sequence windows were extracted within −7 to +6 of the GT dinucleotide and/or within −68 to +20 of the AG dinucleotide. These larger window sizes were chosen to maximize the inclusion of variants likely to affect splicing according to numerous splice-site studies.24 Variants often occurred in sufficient proximity to multiple GT or AG dinucleotides to result in multiple overlapping sequences. Thus, multiple candidate splice sequences were assessed for some variants.
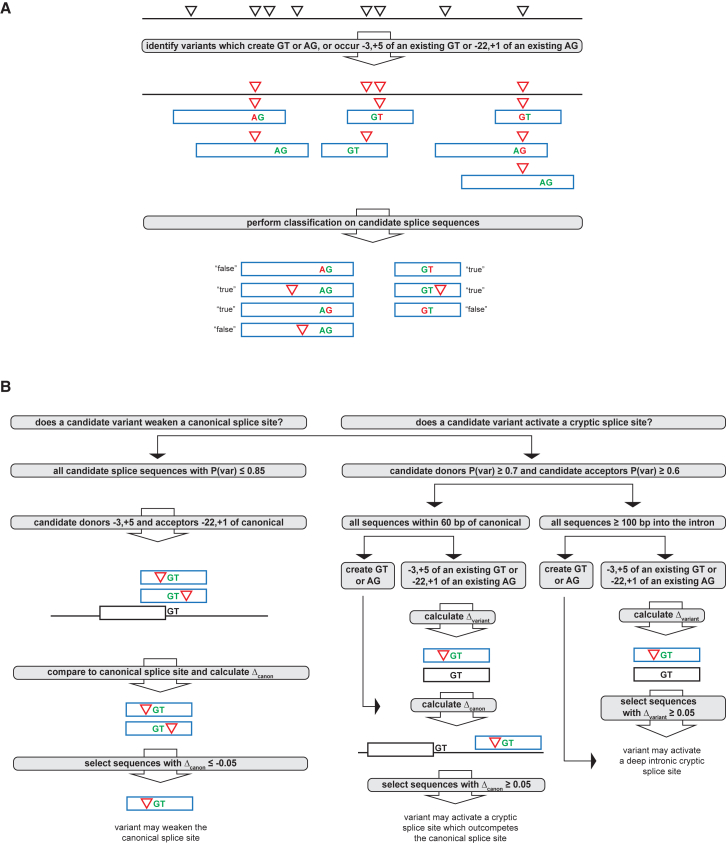
Figure 1.
Overview of Splice Sequence Classification and Selection of High-Confidence Candidate Splice Variants
(A) Variants (black triangles) were chosen for splice-sequence classification if they either create a GT or AG dinucleotide or occur within the specified ranges of an existing GT or AG dinucleotide. Candidate splice sequences containing chosen variants are indicated by the blue rectangles, and candidate variants are represented by red triangles or, in the event that the variant creates a GT or AG dinucleotide, red text. Candidate splice sequences were subjected to classification and were given binary classifications (true or false) and probability estimates of splice potential (P(var) for variant sequences).
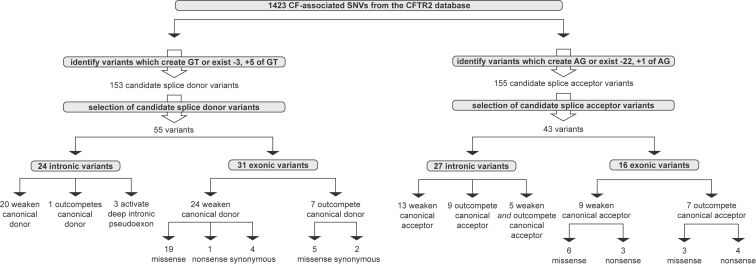
(B) To determine which variants were likely to weaken the canonical splice site (left), activate a cryptic splice site that could outcompete the canonical (center), or activate a cryptic splice site in the deep intron (right), we employed a custom algorithm that utilizes variant distance from canonical splice sites (black rectangles) and changes in splice potential (Δvariant and Δcanon) to select high-confidence candidate splice sequences (blue rectangles). See Material and Methods for more information.
A unique feature of our method is the context-dependent determination of how a variant alters the splice potential of its native context. The genome is littered with many sequences with high splice potential even in the absence of variation, in some cases because of the inactivation of ancient exons over evolutionary time.25, 26, 27 These cryptic splice sites are especially vulnerable to activation and could have deleterious consequences if they occur within sufficient proximity to canonical splice sites or other latent deep intronic splice sites (to allow the formation of a pseudoexon). The accurate identification of cryptic splice sites therefore requires comparison of the splice potential of the variant splice sequence and the splice potential of the reference sequence. To assess the splice potential of native reference sequences, we also subjected full reference gene sequences to classification.
Using Canonical Splice Sites to Validate Splice Sequence Classifier Models
To evaluate the sensitivity of the classifier models, we classified the entire gene sequences for five genes with well-established exon-intron junctions and splicing patterns: BRCA2 (GenBank: NM_000059.3; MIM: 600185), CFTR (GenBank: NM_000492.3; MIM: 602421), DKC1 (GenBank: NM__001142463.1; MIM: 300126), HEXB (GenBank: NM_000521.3; MIM: 606873), and LMNA (GenBank: NM_170707.3; MIM: 150330). 88 of 90 (97.8% sensitivity) canonical donors across these five genes were accurately given a “true” classification, and 80 of 90 (88.9% sensitivity) canonical acceptors were accurately given a “true” classification. The combined sensitivity of the donor and acceptor classifier models was 93.3%. An evaluation of the specificity was not possible given the difficulty in ascertaining “true negative” splice variants. We used the probability estimates of these 90 canonical splice sites to set the minimum thresholds for the P(var) of selected candidate variant sequences as described above (i.e., canonical donors had probabilities over 0.7, and canonical acceptors had probabilities over 0.6; see Material and Methods).
Selection of High-Confidence Splice Variants
The premise of this work was that variants could activate or inactivate splice sites; therefore, any method that accurately detects cryptic splice activation should also be able to detect the disruption of canonical splicing. We designed a selection algorithm to assess each variant for its possible effect on splicing by comparing the splice probabilities of candidate splice sequences bearing variants of interest (P(var)) with the splice probabilities of reference sequences (P(ref)) and/or nearby canonical splice sites (P(canon)) (Figure 1B). The Δcanon metric is defined as the splice potential of the variant sequence (P(var)) minus the splice potential of the canonical site (P(canon)). The Δvariant metric is defined as the splice potential of the variant sequence (P(var)) minus the splice potential of the reference sequence (P(ref)). (Δvariant cannot be calculated for variants that create GT or AG dinucleotides given that the corresponding reference sequences lack the necessary GT or AG dinucleotide.) The thresholds for Δcanon and Δvariant were established with known standards in variant annotation and calling (i.e., true sets) of canonical splice sequences and known splice variants (see Material and Methods). The Δcanon and Δvariant metrics enabled the selection of high-confidence splice variants on the basis of three possible scenarios of aberrant splicing: (1) weakening of the canonical splice site (Figure 1B, left), (2) activation of a cryptic splice site that outcompetes the canonical splice site (Figure 1B, center), and (3) activation of a deep intronic cryptic splice site and subsequent pseudoexon inclusion (Figure 1B, right).
Validation of the Context-Specific Selection Algorithm on Known CFTR Splice-Altering Variants
The sensitivity of the splice-variant selection algorithm was tested with 21 CFTR splice-altering variants manually curated from the literature, none of which alter a GT or AG dinucleotide10, 20, 28, 29, 30, 31 (also see the CFTR2 database in the Web Resources; the list is available upon request). 8 of the 21 CFTR splice-altering variants cause cryptic splice-site activation either in the deep intron or near enough to a canonical splice site to outcompete it. The remaining 13 known CFTR splice-altering variants weaken the canonical splice sites in which they occur. All but 3 of the 21 known splice variants were correctly predicted to alter splicing by their previously reported mechanisms with default thresholds set to Δvariant = 0.05 and Δcanon = 0.05. Two variants were classified by the predictive model to allow splicing because they generated high probability estimates: c.2988G>A (rs121908797, P(var) = 0.889) and c.1766+3A>G (rs397508298, P(var) = 0.940); however, in vitro experimental studies indicate that these variants disrupt splicing.20, 31 Upon further inspection, it became apparent that the P(var) values for the candidate splice sequences with either c.2988G>A or c.1766+3A>G were statistical outliers (≥3 SD above the mean [0.263 ± 0.313]) in the distribution of P(var) for variants reported to abolish CFTR canonical splice sites. The third variant, c.2816A>G (rs397508440), was excluded during splice-variant selection because of the specification that cryptic splice sites that outcompete the canonical splice site lie within 60 bp of the canonical splice site. This 60 bp threshold was established because exonic cryptic splice sites are unlikely to be utilized if the remaining exon is of insufficient length.32 c.2816A>G activates a cryptic donor that outcompetes the canonical splice site (see experimental data below) but is 97 bp upstream of the canonical donor. The selection algorithm had a sensitivity of 85.7% in this validation exercise. Most importantly, the 18 selected known splice variants were assigned the correct mechanism by which they altered splicing according to previous reports (i.e., c.3700A>G activates an exonic cryptic site2).
Evaluation of CFTR Variants Associated with Cystic Fibrosis for Their Effect on Pre-mRNA Splicing
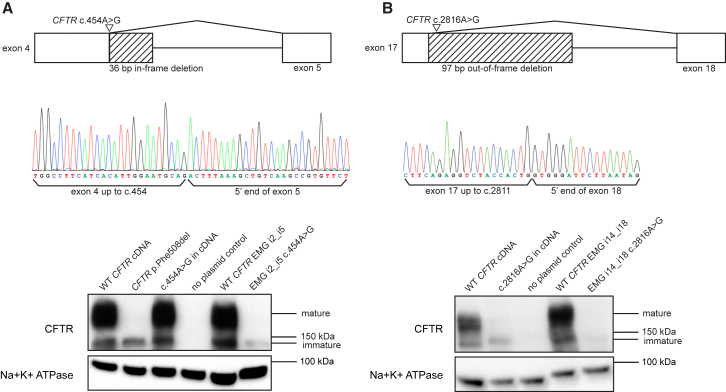
To test the ability of CryptSplice to identify splice variants, we evaluated CFTR variants that had been detected in individuals with cystic fibrosis (CF [MIM: 219700]). 1,477 variants were obtained from the CFTR2 database, an extensive catalog of CFTR variants.33 After the removal of variants with nonstandard HGVS cDNA nomenclature, variants in the promoter and UTRs, and variants with indeterminate breakpoints, 1,423 variants from the CFTR2 database remained for evaluation. 308 of the 1,423 variants either created a GT or AG dinucleotide or occurred within sufficient proximity of an existing GT or AG dinucleotide to create 3,315 potential donor and acceptor sequences. 98 of the 308 (31.8%) donor and acceptor variants were selected as candidates for altering native splice patterns (Figure 2 and Table S1; see Material and Methods for criteria). All candidate variants were assessed for the manner in which they were predicted to disrupt RNA processing, as well as the predicted protein impact. 32 (32.7%) of the 98 selected variants were predicted to activate cryptic splice sites that would either outcompete a nearby canonical splice site (29 selected variants) or lead to pseudoexon inclusion (three selected variants). 5 of the 32 cryptic splice variants were predicted to weaken the canonical acceptor while simultaneously activating a cryptic acceptor. Two of these five variants (c.1585−8G>A [rs193922503] and c.1585−9T>A [rs397508234]) have been previously reported to activate cryptic acceptors that outcompete the canonical acceptor.20 Notably, 33 (70%) of the 47 selected exonic variants were predicted to be “missense” variants, suggesting that a significant portion of variants with a high likelihood of causing aberrant splicing could be mistaken as protein altering.
Figure 2.
Selection of High-Confidence Candidate Splice Variants from the CFTR2 Database
Out of 1,477 variants in the CFTR2 database, 1,423 had standard HGVS cDNA nomenclature, were not in the promoter or UTRs, and did not have indeterminate breakpoints and were thus subjected to splice-sequence classification and splice-variant selection as described in Figure 1. Of 153 candidate splice donor variants (left), 55 were selected as high-confidence candidates. Of 155 candidate splice acceptor variants (right), 43 were selected as high-confidence candidates.
Experimental Evaluation of Predicted Exonic Cryptic Splice Variants in CFTR
Two of the missense exonic variants predicted to activate cryptic splice sites were selected for in vitro investigation with expression minigenes. Expression minigenes containing multiple entire or abridged introns and the entire coding region of CFTR enable simultaneous analysis of the consequence of variants upon pre-mRNA splicing and protein translation in a near-native context.1, 20 c.454A>G (p.Met152Val) (rs397508721) was predicted to create a novel GT dinucleotide and thus activate an exon 4 cryptic donor (P(var) = 0.996) that could outcompete the canonical donor (Δcanon = +0.162) and result in a 36 bp in-frame deletion (p.Met152_Lys163del) (Figure 3A, top). To test this prediction, we introduced c.454A>G into exon 4 of a CFTR expression minigene with sequences from flanking introns (2–5). RNA extracted from HEK293 cells transiently transfected with the resulting expression minigene was reverse transcribed and amplified by PCR (RT-PCR). Sanger sequencing of this cDNA confirmed the predicted 36 bp deletion (Figure 3A, middle). Mature protein was generated by the WT CFTR expression minigene i2_i5 (Figure 3A, bottom, lane 5) but not in the presence of c.454A>G (lane 6). A faint signal corresponding to the molecular mass of immature CFTR was observed, consistent with the presence of incompletely glycosylated protein misfolded because of an in-frame deletion of 12 amino acids. To demonstrate the importance of studying variants in a near native context including relevant introns, we introduced c.454A>G into a plasmid bearing CFTR cDNA and no introns. Western blotting showed mature, properly folded protein similar to that seen in WT controls (Figure 3A, bottom, lane 3). The observation of stable and normally processed CFTR is consistent with the in silico prediction that c.454A>G leads to a benign amino acid substitution (Met to Val) by PolyPhen and SIFT.
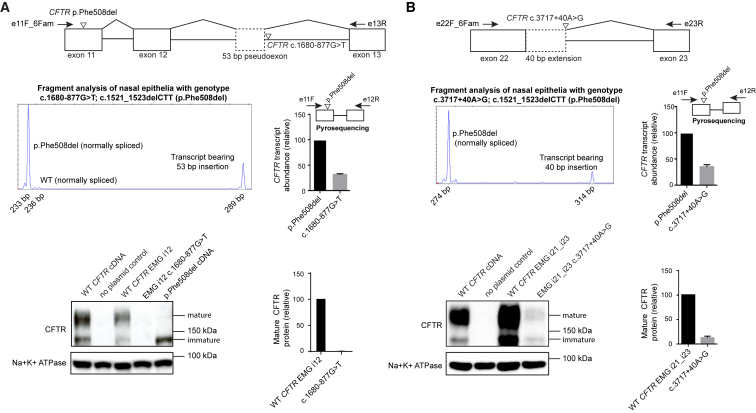
Figure 3.
Experimental Validation of Exonic CFTR Cryptic Splice Variants
(A) Top: c.454A>G (predicted missense: p.Met152Val, inverted triangle) was predicted to activate a cryptic donor upstream of the exon 4 canonical donor and result in an in-frame 36 bp deletion leading to a 12 amino acid deletion (diagonally hashed rectangle). Middle: Sequence analysis of RNA transcripts from HEK293 cells transfected with an expression minigene bearing c.454A>G and flanking introns showed deletion of the last 36 exonic nucleotides from the final processed transcript as a result of utilization of the cryptic donor at c.454 over the canonical exon 4 donor. Bottom: western blotting of an expression minigene with c.454 A>G (EMG i2_i5 c.454A>G) showed a drastic loss of normal CFTR and a very faint amount of immature protein. By contrast, c.454A>G in a plasmid bearing CFTR cDNA with no intronic sequences showed CFTR identical to that seen in WT transfections. CFTR bearing the p.Phe508del variant that causes severe misfolding that precluded glycosylation is included to show the location of immature CFTR.
(B) Top: c.2816A>G (predicted missense: p.His939Arg, inverted triangle) was predicted to activate a cryptic donor upstream of the exon 17 canonical donor and result in a 97 bp deletion (diagonally hashed rectangle). Middle: sequencing of transcripts from HEK293 cells transfected with expression minigenes bearing c.2816A>G and flanking introns showed deletion of the last 97 exonic nucleotides as a result of utilization of the cryptic donor at c.2811. Bottom: western blotting of expression minigenes with c.2816A>G (EMG i14_i18 c.2816A>G) showed a near complete loss of normal protein and a very faint amount of immature protein, consistent with translation of a small amount of normally spliced transcript bearing a deleterious missense mutation causing misfolded protein, as illustrated by the immature protein generated by CFTR cDNA with c.2816A>G.
The second missense variant studied, c.2816A>G (p.His939Arg) (rs397508440), occurs in the +3 position downstream of an existing GT in the middle of exon 17. c.2816A>G was predicted to activate a cryptic donor (Δvariant = +0.783) 97 bp upstream of the canonical donor and result in a frameshift starting at codon position 939 and the introduction of a premature termination codon (PTC) ten amino acids downstream (p.His939Glyfs∗10) (Figure 3B, top). To test this prediction, we introduced c.2816A>G into exon 17 of an expression minigene with sequences from CFTR introns 14–18. Sequencing of RT-PCR products from transiently transfected HEK293 cells confirmed the predicted 97 bp deletion (Figure 3B, middle). Mature protein was produced by the normally spliced WT expression minigene (Figure 3B, bottom, lane 4) but not by the expression minigene bearing c.2816A>G (lane 5), consistent with the expectation of nonsense-mediated RNA decay (NMD) of aberrantly spliced transcript bearing a PTC. Very low amounts of immature protein were observed for c.2816A>G, most likely as a result of the generation of some normally spliced transcript bearing the predicted His-to-Arg substitution. In support of this conjecture, CFTR cDNA bearing c.2816A>G and no introns was also found to generate immature protein of the same molecular mass (Figure 3B, bottom, lane 2). These data show that c.2816A>G should be annotated as disease causing because missplicing of pre-mRNA RNA leads to pathogenic decrease in RNA transcript. Together, these results verify that c.454A>G and c.2816A>G, although annotated as missense variants, activate exonic cryptic splice sites and lead to the loss of mature CFTR. The molecular consequence of these variants is consistent with the severe CF phenotype observed in individuals carrying c.454A>G (n = 3) or c.2816A>G (n = 5) in trans with a known severe disease-causing variant (data not shown). These experiments also emphasize the importance of studying exonic variants in a near-native context incorporating relevant introns to accurately determine the underlying molecular mechanism of disease.
Identification of Intronic Cryptic Splice Variants in CFTR
The entire CFTR was sequenced in 14 individuals with diagnostically elevated sweat chloride concentrations and phenotypic features of CF, but only one disease-causing variant was identified after analysis of all exons and flanking introns. Because these individuals had compelling clinical evidence of CF, it was likely that they carried a second deleterious variant outside of the regions analyzed, such as an intronic variant that disrupted normal CFTR pre-mRNA splicing.6 Sequencing identified 41 candidate intronic CFTR variants in these 14 subjects after the exclusion of 388 intronic variants that were found in cis with CF-causing variants in 31 CF individuals with two known CF-causing alleles. The 41 intronic variants created 27 candidate donor sequences and 48 candidate acceptor sequences. Given the small number of candidate sequences generated and the assumption of deep intronic cryptic splice activation, all candidate sequences with a “true” classification were evaluated manually. Five different intronic variants created sequences that were classified as true splice sequences. One variant, c.3140−26A>G (rs76151804), identified in three individuals, had been previously characterized as CF causing.30 This variant creates an AG dinucleotide and activates a splice acceptor (P(var) = 0.896). The remaining four variants were predicted to activate deep intronic cryptic donors: c.1680−877G>T (two subjects; rs397508261, Δvariant = +0.228), c.3717+40A>G (rs397508595, Δvariant = +0.071), c.1584+689G>A (chr7: 117,200,398; Δvariant = +0.361), and c.2491−1190C>T (chr7: 117,233,794; P(var) = 0.994).
Experimental Evaluation of Predicted Deep Intronic Cryptic Splice Variants in CFTR
c.1680−877G>T (legacy 1811+1643G>T or c.1679+1643G>T) was identified in 2 of the 14 subjects. Both were of Hispanic ancestry, and each carried CFTR p.Phe508del (c.1521_1523delCTT, legacy F508del) as their single identified CF-causing mutation. The variant is in the +4 position downstream of an existing GT dinucleotide and was predicted to create a deep intronic donor (Δvariant = +0.228). An intronic acceptor (chr7: 117,229,470; P(ref) = 0.870) was predicted 60 bp upstream of the variant. Spliceosomal recognition of the intronic acceptor at chr7: 117,229,470 and the cryptic donor activated by c.1680−877G>T would result in the inclusion of a 53 bp pseudoexon between exons 12 and 13 in the final processed mRNA (p.Ala561Serfs∗15) (Figure 4A, top). Notably, the variant is 9 bp downstream of a different, previously identified CF-causing deep intronic cryptic donor created by c.1680−886A>G (CFTR legacy name 1811+1.6kbA>G, rs39750826610). c.1680−886A>G activates a cryptic exon by using the same acceptor that was predicted for c.1680−877G>T. RNA was isolated from primary nasal epithelial cells in an individual carrying c.1680−877G>T with p.Phe508del in trans. RT-PCR was performed, and RNA transcripts were inspected by fragment analysis and pyrosequencing. The primers were selected such that both p.Phe508del and c.1680−877G>T transcripts could be amplified for determining the relative abundance of each transcript. A transcript bearing the predicted 53 bp cryptic pseudoexon between exons 12 and 13 was detected and found to be in lower abundance (31.38% ± 1.18%, n = 3) than the p.Phe508del transcript, most likely because of NMD (Figure 4A, middle). Insertion of the expected 53 bp pseudoexon sequence was confirmed by Sanger sequencing (data not shown). To determine the effect of the variant upon protein production, we collected cell lysates from HEK293 cells transiently transfected with expression minigenes with c.1680−877G>T and sequence from intron 12. As expected, the expression minigene bearing c.1680−877G>T showed a complete loss of normal mature or immature CFTR (n = 4) (Figure 4A, bottom, lane 4). The finding of complete loss of normal splicing due to activation of a cryptic donor by c.1680−877G>T is consistent with the severe CF phenotype observed in individuals carrying this variant in homozygosity (n = 4) and in individuals carrying at least one copy of c.1680−877G>T in trans with a known severe loss-of-function variant (n=19 CF individuals; CFTR2 database).
Figure 4.
Experimental Validation of Intronic CFTR Cryptic Splice Variants
(A) Top: c.1680−877G>T (inverted triangle) was predicted to activate a deep intronic cryptic donor (dashed-edge rectangle) and a latent acceptor 55 bp upstream that could enable pseudogene inclusion. Middle: RT-PCR of RNA isolated from primary nasal epithelial cells of an individual with genotype c.1680−877G>T;c.1521_1523delCTT (p.Phe508del) was subjected to fragment analysis and pyrosequencing. The identification of a 289 bp peak by fragment analysis and subsequent sequencing confirmed incorporation of the 53 bp cryptic pseudoexon, as indicated by the dotted-edge rectangle in the upper panel. Pyrosequencing confirmed the lower abundance of transcript bearing c.1680−877G>T (n = 3). Bottom: western blotting revealed loss of normal CFTR from the expression minigene bearing c.1680−877G>T (EMG i12 c.1680−877G>T), consistent with the prediction of a premature termination codon, NMD, and protein instability (n = 3 in plot). Error bars indicate standard error of the mean (SEM).
(B) Top: c.3717+40A>G (inverted open triangle) was predicted to activate a cryptic donor 40 nucleotides downstream of the exon 22 canonical donor and result in exon extension (dashed-edge rectangle), frameshift, and a premature termination. Middle: RT-PCR was performed on primary nasal epithelial cells of an individual with genotype c.3717+40A>G;c.1521_1523delCTT (p.Phe508del). Fragment analysis using primers from exons 22 and 23 confirmed retention of the first 40 nucleotides of intron 22 (314 bp peak). Pyrosequencing revealed lower abundance of the c.3717+40A>G transcript than of the p.Phe508del transcript (n = 3). Bottom: western blotting showed residual amounts of both mature fully glycosylated and immature core-glycosylated forms of CFTR generated by expression minigene bearing c.3717+40A>G (EMG i21_i23 c.3717+40A>G) as a result of leaky splicing (n = 3 in plot). Error bars indicate SEM.
c.3717+40A>G was identified in an adult individual carrying p.Phe508del and exhibiting features consistent with a mild CF phenotype (late diagnosis at age 14 years, mean sweat chloride concentration of 83 mEq/L, and exocrine pancreatic sufficiency). c.3717+40A>G was predicted to activate a splice donor sequence (Δvariant = +0.071) 40 bp from a canonical splice donor. Furthermore, the splice donor sequence activated by c.3717+40A>G was predicted to outcompete the nearby canonical donor (Δcanon = +0.261) (Figure 4B, top). RT-PCR was performed on RNA isolated from nasal epithelial biopsies obtained from this individual. Fragment analysis revealed transcript retaining the first 40 nucleotides of intron 22, consistent with cryptic splice activation at c.3717+40A>G (Figure 4B, middle). Retained intron 22 sequence was confirmed by Sanger sequencing (data not shown). As previously done, we performed pyrosequencing with primers to select transcripts that included p.Phe508del so we could measure the relative abundance of the transcripts. The c.3717+40A>G transcript was substantially less abundant (34.69% ± 2.62%, n = 3) than the p.Phe508del transcript. In vitro analysis using expression minigenes bearing c.3717+40A>G and sequence from introns 21–23 further corroborated the findings from primary nasal cells (data not shown). The inclusion of 40 intronic nucleotides was predicted to cause a frameshift at amino acid position 1,241 and the introduction of a PTC nine amino acids downstream (p.Gly1241Argfs∗9). Western blotting of cell lysates from expression minigene transfections showed very low amounts of protein (12.55% ± 1.99%; n = 3) migrating at a size consistent with mature wild-type protein (Figure 4B, bottom). These results indicate that c.3717+40A>G does not completely ablate normal splicing of this region and results in the translation of a small quantity of WT CFTR, consistent with the mild CF phenotype observed in this individual.
c.1584+689G>A was predicted to activate a donor (Δvariant = +0.361) that would create a deep intronic cryptic pseudoexon and thus lead to transcript loss due to NMD. This variant was identified in an individual whose known CF-causing variant was c.489+1G>T (legacy 621+1G>T). Previous studies of RNA from this individual’s nasal epithelia did not show inclusion of a pseudoexon; however, RNA transcription from the chromosome not bearing c.489+1G>T was significantly diminished, consistent with the prediction of NMD.34 The last candidate variant, c.2491−1190C>T, was predicted to create a novel GT (P(var) = 0.994) that would activate a deep intronic cryptic donor. However, it was not considered a strong candidate to activate cryptic exon splicing given the absence of a strong upstream acceptor. To test this prediction, we analyzed CFTR RNA transcripts from the nasal epithelial cells of a pair of dizygotic CF twins carrying this variant in trans with p.Phe508del. No aberrant splicing of transcript from the non-p.Phe508del allele was observed in either subject (data not shown).
Identification and Characterization of DKC1 Intronic Splice Variants in Individuals with Dyskeratosis Congenita
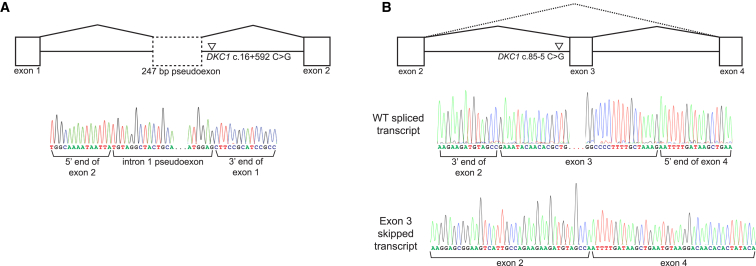
To verify that CryptSplice is broadly applicable, we analyzed variants identified in individuals with dyskeratosis congenita (DC [MIM: 305000]). DC is a genetically heterogeneous multiorgan disease of shortened telomeres and is defined by mucocutaneous abnormalities with bone marrow failure and pulmonary fibrosis as the primary causes of mortality.35 An X-linked recessively inherited form of DC is most common, and 50%–70% of cases have been attributed to mutations in DKC1 (MIM: 300126),23, 36 leaving a significant number of X-linked DC-affected individuals with no identified disease-causing DKC variant.37 A variant call format file extracted from a 3 Mb region surrounding the 15 kb DKC1 locus from exome and genome sequencing of five DC individuals and a single variant identified by Sanger sequencing in the exon 3 canonical splice acceptor of a sixth DC individual were subjected to classification. The 4,685 unique variants present in the six individuals resulted in five candidate donor sequences and 12 candidate acceptor sequences. After splice variants were selected, two variants were predicted to affect native splice patterns. The first variant, DKC1 c.16+592C>G, was adjacent to a GT dinucleotide and was predicted to activate a splice donor (chrX: 153,991,848; Δvariant = +0.709). This variant was found in an individual presenting with mild DC and had been identified in an additional DC individual.23, 38 An acceptor of moderate splice potential (chrX: 153,991,611; P(ref) = 0.579) was identified 235 bp upstream of c.16+592. Sequencing of RNA transcripts from the DC individual confirmed activation of the predicted cryptic donor and inclusion of a pseudoexon (Figure 5A). Interestingly, the deep intronic acceptor used by this pseudoexon was not the one identified by the classifier model but rather a sequence 14 bp upstream that had a low splice potential (P(ref) = 0.134). Normally spliced DKC1 transcripts were also detected in relatively equal proportion, which corroborates previous findings in which half of the normal dyskerin protein was detected by western blotting of cells derived from the affected individual.23 The second variant, DKC1 c.85−5C>G, was predicted to moderately decrease the splice potential of the exon 3 canonical acceptor (chrX: 153,993,717; Δcanon = −0.152). Given the higher P(var) of this candidate splice sequence than of validation sequences bearing variants known experimentally to completely abolish normal splicing, two transcripts were predicted: normally spliced WT transcript and transcript with exon 3 skipped (Figure 5B, top). RT-PCR and subsequent Sanger sequencing showed three products: the first two predicted transcripts (Figure 5B, bottom) plus one transcript with intron 2 retained (data not shown). The transcript retaining intron 2, which is 477 bp long, had very low expression in relation to the WT and exon-3-skipped transcripts. Retention of intron 2 (p.Glu29Valfs∗38) and skipping of exon 3 (p.Glu29_Lys57del) are both consistent with the prediction that c.85−5C>G would disrupt the normal splicing of exon 3. An acceptor of moderate splice potential (P(ref) = 0.684) was identified 82 bp upstream of the exon 3 canonical acceptor, yielding the possibility of a fourth transcript with an extension of exon 3, causing a frameshift, premature termination, and NMD; however, this product was not detected.39
Figure 5.
Experimental Validation of DKC1 Splice Variants
(A) DKC1 c.16+592C>G (inverted triangle) was predicted to activate a deep intronic cryptic donor and result in the inclusion of a pseudoexon in the final processed mRNA (dashed-edge rectangle). Sanger sequencing of transcripts from lymphoblastoid cell lines derived from a DC individual confirmed the inclusion of a cryptic pseudoexon (a reverse-complement DNA sequence is shown).
(B) DKC1 c.85−5C>G (inverted triangle) was predicted to moderately weaken the canonical exon 3 acceptor and lead to two predicted transcripts: skipping of exon 3 (dotted lines joining) and normally spliced transcript (solid lines joining exons). Amplification and Sanger sequencing of RNA transcripts from lymphoblastoid cell lines derived from the affected individual confirmed the two predicted transcripts (WT and exon 3 skipped).
Identification of Candidate Splice Variants among ClinVar SNVs
The identification of splice variants from amongst CF-associated CFTR variants demonstrated that splice variants could be under-ascertained in databases of disease variants. To test this hypothesis, we applied CryptSplice to 24,787 SNVs labeled “pathogenic” or “probably pathogenic” in the October 2015 ClinVar FTP download. Over 15,000 unique SNVs generated nearly 29,000 candidate splice sequences by creating a new GT or AG dinucleotide or existing in sufficient proximity to an existing GT or AG dinucleotide (Figure S1). 459 candidate splice variants were selected as having a high likelihood of altering native splice patterns (Table S2) and were evaluated for their effect on RNA processing. 129 (28.1%) of the 459 high-confidence splice variants were predicted to activate cryptic splice sites. Interestingly, although most of the selected variants involving splice donors were predicted to weaken nearby canonical donors (315 of 379 [83%]), most of the selected variants involving splice acceptors were predicted to activate cryptic acceptors (65 of 80 [81.3%]). 293 (63.8%) of the 459 selected splice variants were exonic, and 205 (44.7%) of all selected variants were annotated as missense mutations in ClinVar.
To explore whether variants designated in ClinVar as variants of uncertain significance (VUSs) might affect splicing, we applied CryptSplice to 28,147 VUSs in the October 2015 ClinVar FTP download. Over 18,500 unique VUSs created GT or AG dinucleotides or occurred within sufficient proximity to existing GT or AG dinucleotides to generate over 35,000 candidate donor and acceptor sequences (Figure S2). 348 candidate splice variants were selected as having a high likelihood of altering native splice patterns (Table S3), and the majority (273 [78.4%]) of selected VUSs were predicted to weaken nearby canonical donors or acceptors. 75 (21.6%) of 348 selected VUSs were predicted to result in cryptic splice site activation either near canonical donors or in the deep intron. 137 of the selected VUSs were exonic, and 113 (32.5%) of all selected VUSs were annotated as missense mutations in ClinVar. Overall, 28.1% of ClinVar pathogenic variants and 21.6% of ClinVar VUSs selected with high confidence to affect splicing were predicted to activate cryptic splice sites (Figures S1 and S2), a rate similar to that observed with variants in the CFTR2 database (32.7%).
Discussion
The activation of cryptic splice sites by SNVs is a well understood pathologic mechanism. Indeed, one of the earliest β+ thalassemia (MIM: 613985)-associated variants identified activated a cryptic acceptor 19 bp upstream of the canonical exon 2 acceptor of the β-globin gene (HBB [MIM: 141900]).40 Using a systematic method for the in silico identification of variants that activate cryptic splice sites, we discovered that this mechanism is not rare. Consequently, we propose that cryptic splice activation should be considered in the evaluation of the pathogenicity of exonic and intronic variants. Although we present Mendelian autosomal and X-linked recessive examples whereby cryptic splice activation causes substantial reductions in normally spliced transcripts, it is reasonable to posit that the same mechanism could be the cause of diseases with dominant and complex inheritance patterns. Variants that activate cryptic splice sites and lead to the loss of protein or aberrant protein could underlie dominant disorders caused by haploinsufficient, dominant-negative, or gain-of-function mechanisms. Variability in splicing efficiency shown here was associated with mild phenotypes, suggesting that some variants could have subtle effects on protein expression and that these subtle effects could be predicted computationally. Thus, intronic variants associated with complex traits should be considered for splice-site activation.
Exonic SNVs are often assumed to primarily affect protein, especially if the variant is non-synonymous. For example, there are efforts to determine the impact of every exonic variant on protein processing and function.41, 42 However, these massively parallel mutagenesis methods presume that exonic variants will not affect mRNA processing and thus interrogate variants in cDNA in the absence of introns. The importance of considering whether variants affect splicing is illustrated by the dramatic differences in the effect of CFTR c.454A>G upon protein synthesis when cDNA (without introns) or expression minigenes (with introns) were employed (see Figure 3A). Correct assessment of variant effect upon gene function is essential to inform treatment.33, 43, 44, 45 Of particular note, a recent clinical trial evaluated the efficacy of the small molecule ivacaftor in CF-affected individuals carrying variants that permitted residual protein function,46 as determined by in vitro chloride-conductance measurements of mutant CFTR.47 At the completion of the clinical trial, all individuals except for those carrying p.Gly970Arg had positive clinical responses to ivacaftor. The variant that causes p.Gly970Arg (c.2908G>C) alters the last nucleotide of the exon and was predicted by the method presented here to cause a complete loss of normal splicing at the canonical donor (Table S1). Thus, the lack of clinical response to ivacaftor in CF-affected individuals carrying c.2908G>C is explained if the underlying molecular defect affects mRNA processing instead of protein function. The importance of elucidating molecular mechanisms will become increasingly relevant to disease-research communities that seek to deploy small-molecule therapies.48
Deep intronic variants are largely dismissed as disease causing given the high degree of intronic variation, the distance of these variants from coding or other highly conserved regions, and the practical issues inherent to obtaining and working with RNA from clinical specimens.49 Experimental detection of aberrantly spliced transcripts with premature termination codons can be difficult, especially in primary tissues, as a result of RNA degradation caused by NMD. For example, the deep intronic variant CFTR c.1584+689G>A was predicted to result in activation of a cryptic pseudoexon. The individual carrying this variant had been shown in previous studies34 to have severely decreased RNA transcription from the chromosome bearing c.1584+689G>A. Amplification of cDNA derived from this individual’s nasal epithelial RNA did not show pseudoexon inclusion. However, it is reasonable to posit that transcripts containing this predicted pseudoexon were sufficiently degraded by NMD to preclude detection by PCR. Of note, we were able to find intronic variants that caused aberrant splicing in a substantial fraction of CF (7 of 14) and DC (2 of 6) individuals with undiscovered disease-causing variants. These findings suggest that deleterious splice variants are likely to be present in the introns of other genes associated with loss-of-function Mendelian disorders; therefore, the introns of the suspected disease-associated genes should be inspected in cases where exome sequencing is unable to assign a disease-causing variant in an individual with unambiguous Mendelian disease.
Assessing the effect of variants occurring outside of highly conserved positions in acceptor sites is known to be difficult. We observed a lower detection rate for canonical acceptors (88.9% sensitivity) than for canonical donors (97.8% sensitivity) in our validation using the canonical sites of five genes. Further, we were unable to computationally predict the acceptor that paired with the donor cryptically activated by DKC1 c.16+592C>G. Multiple factors most likely contribute to the lower ascertainment of acceptors than of donors by the method presented here. The longer length of acceptors and the flexibility of the polypyrimidine tract allow for a greater diversity of nucleotide combinations. Further, the splice-variant scoring algorithm is designed to find variants with a high likelihood of altering native splice patterns through comparisons of splice potentials (Δvariant and Δcanon, where applicable). Δvariant and Δcanon metrics rely on the probability estimates provided by the splice sequence classifiers and therefore do not consider non-sequence contributors to splice-site recognition. The role of splice enhancers and inhibitors in the maintenance of normal splicing patterns is well established,5 and the incorporation of predictive tools that model the functions of splice enhancers and inhibitors50, 51 into the method described here could possibly lead to better ascertainment of cryptic splice sites, particularly acceptors. Indeed, in our validation using known CFTR splice variants, all five acceptor variants were correctly identified. Although using such stringent criteria could increase the number of false negatives, it also decreases the number of false positives, making each selected candidate splice variant a high-confidence candidate. Furthermore, the parameters used here were chosen to optimize the detection of cryptic splice sites rather than the loss of splicing at canonical splice sites. Modification of P(var), Δvariant, and Δcanon thresholds will allow for variability in the permissiveness of the selection algorithm, enabling the user to accept a more or less stringent sensitivity when searching for variants that lead to loss of splicing as opposed to cryptic splicing.
Our findings in both CF and DC individuals reveal that the deep introns are an untapped reservoir of cryptic splice variants sufficient to cause severe, life-limiting disease. These variants can create GT or AG dinucleotides that could immediately bring splicing mechanisms to mind; however, the majority of these variants lie adjacent to existing GT or AG dinucleotides and can thus be assessed more easily and accurately with the aid of artificial intelligence tools such as the classification-based method described here.52 Although existing in silico splice prediction tools have demonstrated success, other classifiers are limited by simple human-delineated rules that do not capture the complexity of sequence information required for spliceosome recognition. NNSplice, for example, uses neural networks based on the frequencies of dinucleotides within a sequence;12 in the training of the splice sequence classifiers described here, dinucleotide frequency is but one of three sequence features used. Other commonly used splice prediction tools utilize thermodynamics and conservation data, both of which are descriptive of spliceosome recognition but not deterministic (as is nucleotide sequence), to assess splice potential. Further, the utilization of the scoring algorithm refines the classifier predictions from statistically significant to biologically meaningful findings according to the unique genomic context of each candidate variant. The comparison of a variant sequence’s splice potential with the splice potential of the reference provides a powerful filter with demonstrable accuracy. Context dependency is still relatively new to variant annotation tools but should enable more precise annotation of potential disease-causing variants. Finally, our findings suggest that all variants should be considered as possible splice variants, even if they appear to cause amino acid substitutions predicted to have deleterious consequences on the resulting protein.
Acknowledgments
The authors would like to acknowledge Alice Sanchez and Sean Griffith for developing and testing CryptSplice software; Zhiyong Liu and Heather Hathorne at the Gregory Fleming James Cystic Fibrosis Research Center at the University of Alabama at Birmingham for assisting with the collection of RNA from an individual with cystic fibrosis (CF); Deepika Polineni and Lyndsay Marriott at the University of Kansas Medical Center for coordinating enrollment and specimen collection; Douglas Walker and Pamela Zeitlin at the Johns Hopkins Cystic Fibrosis Center for assisting with specimen collection; Aravinda Chakravarti, Michael Beer, and Michael Parsons for their thoughtful suggestions on this work and this manuscript; and CF- and dyskeratosis-congenita-affected individuals who graciously participated in our research studies. This work was supported by the NIH (P30DK072482 to G.M.S., R01CA160433 and R01HL119476 to M.A., and R01DK44003 to G.R.C.), the Cystic Fibrosis Foundation (CUTTIN13A2 to G.R.C. and Sorscher15YO to G.M.S.), Cystic Fibrosis Foundation Therapeutics (CUTTIN15XX1 to G.R.C.), the Commonwealth Foundation Personalized Medicine Initiative at Johns Hopkins (to M.A.), and Gilead Sciences Inc. (125828 to N.S.).
Published: May 4, 2017
Footnotes
Supplemental Data include two figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.04.001.
Web Resources
CFTR2, http://www.cftr2.org
ClinVar, http://www.ncbi.nlm.nih.gov/clinvar/
CryptSplice, https://bitbucket.org/jhucidr/cryptsplice
OMIM, http://www.omim.org
Variant Effect Predictor, https://genomebiology.biomedcentral.com/articles/10.1186/s13059-016-0974-4
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
Predicted high-confidence splice variants from CFTR2 with predicted splicing consequence. Some variants are predicted to alter in splicing in multiple ways (i.e., weaken canonical splice site and create a novel splice site) and therefore have multiple predictions.
Predicted high-confidence splice variants with hg19 coordinates, RefSeq transcript accession number, ClinVar cDNA and protein names, and predicted splicing consequence. Some variants are predicted to alter in splicing in multiple ways (i.e., weaken canonical splice site and create a novel splice site) and therefore have multiple predictions.
Predicted high-confidence splice variants with hg19 coordinates, RefSeq transcript accession number, ClinVar cDNA and protein names, and predicted splicing consequence. Some variants are predicted to alter in splicing in multiple ways (i.e., weaken canonical splice site and create a novel splice site) and therefore have multiple predictions.
References
- 1.Soukarieh O., Gaildrat P., Hamieh M., Drouet A., Baert-Desurmont S., Frébourg T., Tosi M., Martins A. Exonic Splicing Mutations Are More Prevalent than Currently Estimated and Can Be Predicted by Using In Silico Tools. PLoS Genet. 2016;12:e1005756. doi: 10.1371/journal.pgen.1005756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molinski S.V., Gonska T., Huan L.J., Baskin B., Janahi I.A., Ray P.N., Bear C.E. Genetic, cell biological, and clinical interrogation of the CFTR mutation c.3700 A>G (p.Ile1234Val) informs strategies for future medical intervention. Genet. Med. 2014;16:625–632. doi: 10.1038/gim.2014.4. [DOI] [PubMed] [Google Scholar]
- 3.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 4.Wang G.S., Cooper T.A. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 5.Singh R.K., Cooper T.A. Pre-mRNA splicing in disease and therapeutics. Trends Mol. Med. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonini J., Varilh J., Raynal C., Thèze C., Beyne E., Audrezet M.P., Ferec C., Bienvenu T., Girodon E., Tuffery-Giraud S. Small-scale high-throughput sequencing-based identification of new therapeutic tools in cystic fibrosis. Genet. Med. 2015;17:796–806. doi: 10.1038/gim.2014.194. [DOI] [PubMed] [Google Scholar]
- 7.Bonifert T., Karle K.N., Tonagel F., Batra M., Wilhelm C., Theurer Y., Schoenfeld C., Kluba T., Kamenisch Y., Carelli V. Pure and syndromic optic atrophy explained by deep intronic OPA1 mutations and an intralocus modifier. Brain. 2014;137:2164–2177. doi: 10.1093/brain/awu165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pezeshkpoor B., Zimmer N., Marquardt N., Nanda I., Haaf T., Budde U., Oldenburg J., El-Maarri O. Deep intronic ‘mutations’ cause hemophilia A: application of next generation sequencing in patients without detectable mutation in F8 cDNA. J. Thromb. Haemost. 2013;11:1679–1687. doi: 10.1111/jth.12339. [DOI] [PubMed] [Google Scholar]
- 9.den Hollander A.I., Koenekoop R.K., Yzer S., Lopez I., Arends M.L., Voesenek K.E., Zonneveld M.N., Strom T.M., Meitinger T., Brunner H.G. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chillón M., Dörk T., Casals T., Giménez J., Fonknechten N., Will K., Ramos D., Nunes V., Estivill X. A novel donor splice site in intron 11 of the CFTR gene, created by mutation 1811+1.6kbA-->G, produces a new exon: high frequency in Spanish cystic fibrosis chromosomes and association with severe phenotype. Am. J. Hum. Genet. 1995;56:623–629. [PMC free article] [PubMed] [Google Scholar]
- 11.Reboul M.P., Bieth E., Fayon M., Biteau N., Barbier R., Dromer C., Desgeorges M., Claustres M., Bremont F., Lacombe D., Iron A. Splice mutation 1811+1.6kbA>G causes severe cystic fibrosis with pancreatic insufficiency: report of 11 compound heterozygous and two homozygous patients. J. Med. Genet. 2002;39:e73. doi: 10.1136/jmg.39.11.e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reese M.G., Eeckman F.H., Kulp D., Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 13.Pollastro P., Rampone S. HS3D, A dataset of Homo Sapiens splice regions, and its extraction procedure from a major public database. Int. J. Mod. Phys. C. 2002;13:1105–1117. [Google Scholar]
- 14.Burset M., Seledtsov I.A., Solovyev V.V. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 2000;28:4364–4375. doi: 10.1093/nar/28.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson I.J. A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res. 1991;19:3795–3798. doi: 10.1093/nar/19.14.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J.L., Wang L.F., Wang H.Y., Bai L.Y., Yuan Z.M. High-accuracy splice site prediction based on sequence component and position features. Genet. Mol. Res. 2012;11:3432–3451. doi: 10.4238/2012.September.25.12. [DOI] [PubMed] [Google Scholar]
- 17.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 18.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma N., Sosnay P.R., Ramalho A.S., Douville C., Franca A., Gottschalk L.B., Park J., Lee M., Vecchio-Pagan B., Raraigh K.S. Experimental assessment of splicing variants using expression minigenes and comparison with in silico predictions. Hum. Mutat. 2014;35:1249–1259. doi: 10.1002/humu.22624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reynolds S.D., Rios C., Wesolowska-Andersen A., Zhuang Y., Pinter M., Happoldt C., Hill C.L., Lallier S.W., Cosgrove G.P., Solomon G.M. Airway Progenitor Clone Formation Is Enhanced by Y-27632-Dependent Changes in the Transcriptome. Am. J. Respir. Cell Mol. Biol. 2016;55:323–336. doi: 10.1165/rcmb.2015-0274MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poole A., Urbanek C., Eng C., Schageman J., Jacobson S., O’Connor B.P., Galanter J.M., Gignoux C.R., Roth L.A., Kumar R. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J. Allergy Clin. Immunol. 2014;133 doi: 10.1016/j.jaci.2013.11.025. 670–8.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parry E.M., Alder J.K., Lee S.S., Phillips J.A., 3rd, Loyd J.E., Duggal P., Armanios M. Decreased dyskerin levels as a mechanism of telomere shortening in X-linked dyskeratosis congenita. J. Med. Genet. 2011;48:327–333. doi: 10.1136/jmg.2010.085100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pagani F., Baralle F.E. Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 25.Xing Y., Lee C. Alternative splicing and RNA selection pressure--evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- 26.Alekseyenko A.V., Kim N., Lee C.J. Global analysis of exon creation versus loss and the role of alternative splicing in 17 vertebrate genomes. RNA. 2007;13:661–670. doi: 10.1261/rna.325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J., Lu Z.X., Tokheim C.J., Miller S.E., Xing Y. Species-specific exon loss in human transcriptomes. Mol. Biol. Evol. 2015;32:481–494. doi: 10.1093/molbev/msu317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Highsmith W.E., Burch L.H., Zhou Z., Olsen J.C., Boat T.E., Spock A., Gorvoy J.D., Quittel L., Friedman K.J., Silverman L.M. A novel mutation in the cystic fibrosis gene in patients with pulmonary disease but normal sweat chloride concentrations. N. Engl. J. Med. 1994;331:974–980. doi: 10.1056/NEJM199410133311503. [DOI] [PubMed] [Google Scholar]
- 29.Tzetis M., Efthymiadou A., Doudounakis S., Kanavakis E. Qualitative and quantitative analysis of mRNA associated with four putative splicing mutations (621+3A-->G, 2751+2T-->A, 296+1G-->C, 1717-9T-->C-D565G) and one nonsense mutation (E822X) in the CFTR gene. Hum. Genet. 2001;109:592–601. doi: 10.1007/s00439-001-0631-0. [DOI] [PubMed] [Google Scholar]
- 30.Amaral M.D., Pacheco P., Beck S., Farinha C.M., Penque D., Nogueira P., Barreto C., Lopes B., Casals T., Dapena J. Cystic fibrosis patients with the 3272-26A>G splicing mutation have milder disease than F508del homozygotes: a large European study. J. Med. Genet. 2001;38:777–783. doi: 10.1136/jmg.38.11.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dujardin G., Commandeur D., Le Jossic-Corcos C., Ferec C., Corcos L. Splicing defects in the CFTR gene: minigene analysis of two mutations, 1811+1G>C and 1898+3A>G. J. Cyst. Fibros. 2011;10:212–216. doi: 10.1016/j.jcf.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 32.Yu J., Yang Z., Kibukawa M., Paddock M., Passey D.A., Wong G.K. Minimal introns are not “junk”. Genome Res. 2002;12:1185–1189. doi: 10.1101/gr.224602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sosnay P.R., Siklosi K.R., Van Goor F., Kaniecki K., Yu H., Sharma N., Ramalho A.S., Amaral M.D., Dorfman R., Zielenski J. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat. Genet. 2013;45:1160–1167. doi: 10.1038/ng.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheridan M.B., Hefferon T.W., Wang N., Merlo C., Milla C., Borowitz D., Green E.D., Mogayzel P.J., Jr., Cutting G.R. CFTR transcription defects in pancreatic sufficient cystic fibrosis patients with only one mutation in the coding region of CFTR. J. Med. Genet. 2011;48:235–241. doi: 10.1136/jmg.2010.083287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dokal I. Dyskeratosis congenita in all its forms. Br. J. Haematol. 2000;110:768–779. doi: 10.1046/j.1365-2141.2000.02109.x. [DOI] [PubMed] [Google Scholar]
- 36.Heiss N.S., Knight S.W., Vulliamy T.J., Klauck S.M., Wiemann S., Mason P.J., Poustka A., Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 37.Walter J.E., Armanios M., Shah U., Friedmann A.M., Spitzer T., Sharatz S.M., Hagen C. CASE RECORDS of the MASSACHUSETTS GENERAL HOSPITAL. Case 41-2015. A 14-Year-Old Boy with Immune and Liver Abnormalities. N. Engl. J. Med. 2015;373:2664–2676. doi: 10.1056/NEJMcpc1408595. [DOI] [PubMed] [Google Scholar]
- 38.Knight S.W., Vulliamy T.J., Morgan B., Devriendt K., Mason P.J., Dokal I. Identification of novel DKC1 mutations in patients with dyskeratosis congenita: implications for pathophysiology and diagnosis. Hum. Genet. 2001;108:299–303. doi: 10.1007/s004390100494. [DOI] [PubMed] [Google Scholar]
- 39.Knight S.W., Heiss N.S., Vulliamy T.J., Greschner S., Stavrides G., Pai G.S., Lestringant G., Varma N., Mason P.J., Dokal I., Poustka A. X-linked dyskeratosis congenita is predominantly caused by missense mutations in the DKC1 gene. Am. J. Hum. Genet. 1999;65:50–58. doi: 10.1086/302446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Busslinger M., Moschonas N., Flavell R.A. Beta + thalassemia: aberrant splicing results from a single point mutation in an intron. Cell. 1981;27:289–298. doi: 10.1016/0092-8674(81)90412-8. [DOI] [PubMed] [Google Scholar]
- 41.Kitzman J.O., Starita L.M., Lo R.S., Fields S., Shendure J. Massively parallel single-amino-acid mutagenesis. Nat. Methods. 2015;12:203–206. doi: 10.1038/nmeth.3223. 4, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Starita L.M., Young D.L., Islam M., Kitzman J.O., Gullingsrud J., Hause R.J., Fowler D.M., Parvin J.D., Shendure J., Fields S. Massively Parallel Functional Analysis of BRCA1 RING Domain Variants. Genetics. 2015;200:413–422. doi: 10.1534/genetics.115.175802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lynch T.J., Bell D.W., Sordella R., Gurubhagavatula S., Okimoto R.A., Brannigan B.W., Harris P.L., Haserlat S.M., Supko J.G., Haluska F.G. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 44.Dietz H.C. New therapeutic approaches to mendelian disorders. N. Engl. J. Med. 2010;363:852–863. doi: 10.1056/NEJMra0907180. [DOI] [PubMed] [Google Scholar]
- 45.Finkel R.S., Flanigan K.M., Wong B., Bönnemann C., Sampson J., Sweeney H.L., Reha A., Northcutt V.J., Elfring G., Barth J., Peltz S.W. Phase 2a study of ataluren-mediated dystrophin production in patients with nonsense mutation Duchenne muscular dystrophy. PLoS ONE. 2013;8:e81302. doi: 10.1371/journal.pone.0081302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Committee, F.P.-A.D.A. (2014). Ivacaftor for the Treatment of Cystic Fibrosis in Patients Age 6 Years and Older with an R117H-CFTR Mutation in the CFTR Gene.
- 47.Yu H., Burton B., Huang C.J., Worley J., Cao D., Johnson J.P., Jr., Urrutia A., Joubran J., Seepersaud S., Sussky K. Ivacaftor potentiation of multiple CFTR channels with gating mutations. J. Cyst. Fibros. 2012;11:237–245. doi: 10.1016/j.jcf.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 48.Scotti M.M., Swanson M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baralle D., Baralle M. Splicing in action: assessing disease causing sequence changes. J. Med. Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fairbrother W.G., Yeh R.F., Sharp P.A., Burge C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 51.Smith P.J., Zhang C., Wang J., Chew S.L., Zhang M.Q., Krainer A.R. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 52.Libbrecht M.W., Noble W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015;16:321–332. doi: 10.1038/nrg3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Predicted high-confidence splice variants from CFTR2 with predicted splicing consequence. Some variants are predicted to alter in splicing in multiple ways (i.e., weaken canonical splice site and create a novel splice site) and therefore have multiple predictions.
Predicted high-confidence splice variants with hg19 coordinates, RefSeq transcript accession number, ClinVar cDNA and protein names, and predicted splicing consequence. Some variants are predicted to alter in splicing in multiple ways (i.e., weaken canonical splice site and create a novel splice site) and therefore have multiple predictions.
Predicted high-confidence splice variants with hg19 coordinates, RefSeq transcript accession number, ClinVar cDNA and protein names, and predicted splicing consequence. Some variants are predicted to alter in splicing in multiple ways (i.e., weaken canonical splice site and create a novel splice site) and therefore have multiple predictions.